Abstract
The contribution of low-frequency and damaging genetic variants associated with platelet function to ischemic stroke (IS) susceptibility remains unknown. We employed a deep re-sequencing approach in Polish patients in order to investigate the contribution of rare variants (minor allele frequency, MAF < 1%) to the IS genetic susceptibility in this population. The genes selected for re-sequencing consisted of 26 genes coding for proteins associated with the surface membrane of platelets. Targeted pooled re-sequencing (Illumina HiSeq 2500) was performed on genomic DNA of 500 cases (patients with history of clinically proven diagnosis of large-vessel IS) and 500 controls. After quality control and prioritization based on allele frequency and damaging probability, follow-up individual genotyping of deleterious rare variants was performed in patients from the original cohort. Gene-based analyses identified an association between IS and 6 rare functional and damaging variants in the purinergic genes (P2RY1 and P2RY12 locus). The predicted properties of the most damaging rare variants in P2RY1 and P2RY12 were confirmed by using mouse fibroblast cell cultures transfected with plasmid constructs containing cDNA of mutated variants (FLIPR on FlexStation3). This study identified a putative role for rare variants in P2RY1 and P2RY12 genes involved in platelet reactivity on large-vessel IS susceptibility in a Polish population.
Keywords: DNA sequencing, platelets, genetic polymorphism, cerebrovascular stroke, purinergic receptors, large-vessel ischemic stroke, Polish population
1. Introduction
The genetics of complex diseases, including ischemic stroke (IS), has been previously investigated in genome wide association studies (GWAS), which identified large numbers of common single nucleotide variants associated with disease susceptibility [1]. While relevant disease pathways have been identified by several GWAS, and one targeted re-sequencing study focused on common genetic variants, IS-associated common variants only explain <10% of variance in disease onset [2]. Therefore, research looking into the missing heritability in IS has been focused on the evaluation of the contribution of low frequency and rare variants [3]. Sequencing studies have revealed that low frequency (i.e., minor allele frequency or MAF between 1% and 5%), and in particular, rare (MAF < 1%) genetic variants, are more likely to have a deleterious effect on health compared to common variants [4,5]. Few re-sequencing studies investigating IS in European populations have been performed [6,7]. These studies showed that low frequency and rare protein coding variants in several genes are associated with stroke (p < 1 × 10−6) [7].
The pathogenesis of IS is strongly influenced by the activation of platelets and subsequent release of the bioactive materials they transport. Platelets can also exert a far-reaching influence, when activated, by releasing microparticles containing lipids, receptors, proteins, and genetic material into circulation. Apart from their well-established role in hemostasis, platelets have been identified as key players in inflammation, angiogenesis, and central nervous system repair [8]. For that reason, we have selected genes associated with platelet plasma membrane receptors as the target for this re-sequencing study. Only one previous re-sequencing study, in association with IS, was performed in the Polish population, and focused on platelets’ common genetic variants [9].
Thus, we aimed to further investigate the contribution of rare, presumably large effect genetic variants within a selection of previously described 26 genes encoding platelet surface receptors, to IS susceptibility in the Polish population [10].
2. Results
The study design and flowchart is presented in Figure 1. Pooled targeted enrichment, with the custom Agilent SureSelect capturing kit, resulted in coverage of 99.6%. Sequencing of 10 pools (five each for control and stroke groups) was performed on the Illumina HiSeq2500 sequencer, and generated an average of 36.1 (22.7–45.9 range) million pair-end 101 bp reads, and 5.3 (3–7 range) Gbp per pooled sample consisting of 100 subjects. It corresponds to mean coverage per pool of 12000×, associated with a mean of 120× per individual sample (range 21–369).
Figure 1.
Study-flow diagram. TOAST; Trial of Org 10172 in Acute Stroke Treatment, hx; history.
The frequency of the investigated damaging allele was presented as combined MAF (cMAF) which encompassed all rare damaging variants in the sequenced gene or region. In total, 477 unique single nucleotide variants (SNVs) with sufficient quality coverage were detected after subsequent stringent quality control. Sixty nine percent (69%) of SNVs were known in the single nucleotide polymorphism (SNP) database (dbSNP) version dbSNP138 (see complete list of known variants in Tables S1 and S2). In all, 248 of the 477 variants (51.9%) were coding variants within target exons of sequenced genes, and the remainder was located in untranslated (introns) and intergenic regions. The final 38 rare and damaging variants were selected out of 129 non-synonymous coding variants based on MAF < 1%, and predicted deleteriousness of single variants, from significant gene-based tests using Combined Annotation Dependent Depletion (CADD), with a minimum scaled CADD score of 10 (corresponding to the top 10% deleterious variants in the genome, as indicated by authors) as a threshold for predicted deleteriousness or damaging properties [11]. The selected variants consisted of 28 known (by dbSNP149 November 2016) and 10 novel (previously not listed) variants.
The rare SNPs (n = 38, Table 1) with the most damaging properties were submitted for verification with individual genotyping, of which 31 passed the design of iPLEX Design suite. The individual genotyping was performed in all patients (n = 1000) from the original cohort of patients used for pooled targeted re-sequencing. The presence of all 31 variants was verified in at least one carrier from investigated cohorts. In addition, we repeated the calculations in the remaining 605 patients, after exclusion of subjects (from both groups) with a known medical history which could interfere with the burden analysis (including coronary artery disease—CAD, congestive heart failure—CHF, and diabetes mellitus—DM), because the prevalence of these conditions differ between control and stroke groups (Table S3).
Table 1.
List of all rare (predicted MAF < 0.1%) non-synonymous and deleterious single nucleotide variants observed in the investigated Polish patients (n = 1000) after pooled resequencing of exons in 26 genes.
| Chr | Gene | Position | Ref | Alt | dbSNP149 | cDNA | Protein AA | CADD | MAF Ctrl | MAF Stroke |
|---|---|---|---|---|---|---|---|---|---|---|
| chr3 | ITPR1 | 4714920 | A | G | rs35789999 | c.A2260G | p.M754V | 15.92 | 0.0014 | |
| chr3 | ITPR1 | 4716885 | C | T | rs201519806 | c.C2687T | p.A896V | 12.80 | 0.0011 | |
| chr3 | ITPR1 | 4774887 | G | T | c.G5147T | p.G1716V | 15.77 | 0.0014 | ||
| chr3 | ITPR1 | 4821291 | G | T | rs373973399 | c.G6160T | p.A2054S | 20.2 | 0.0013 | |
| chr3 | ITPR1 | 4842276 | G | A | rs201144431 | c.G6910A | p.A2304T | 16.91 | 0.0015 | 0.001 |
| chr17 | GP1BA | 4837662 | T | C | rs201408072 | c.T1763C | p.V588A | 15.12 | 0.0013 | |
| chr3 | RAF1 | 12641707 | C | T | rs555034652 | c.G934A | p.V312M | 12.39 | 0.0017 | |
| chr1 | PTAFR | 28477192 | T | C | rs138629813 | c.A341G | p.N114S | 20.8 | 0.0018 | |
| chr1 | PTAFR | 28477408 | C | A | c.G125T | p.R42L | 14.45 | 0.0015 | ||
| chr17 | ITGA2B | 42453084 | C | T | rs74988902 | c.G2602A | p.V868M | 13.50 | 0.0014 | |
| chr17 | ITGA2B | 42455791 | G | A | rs200481952 | c.C2033T | p.A678V | 20.4 | 0.0015 | |
| chr17 | ITGA2B | 42457474 | G | A | rs548977341 | c.C1648T | p.R550W | 22.3 | 0.0018 | |
| chr17 | ITGA2B | 42463054 | G | C | rs76066357 | c.C439G | p.L147V | 11.09 | 0.0025 | |
| chr17 | ITGB3 | 45363765 | A | G | rs56173532 | c.A754G | p.I252V | 13.67 | 0.0012 | |
| chr17 | ITGB3 | 45376796 | G | A | rs144884023 | c.G1813A | p.G605S | 35 | 0.0013 | |
| chr19 | PTGIR | 47126849 | G | A | rs4987262 | c.C634T | p.R212C | 22.3 | 0.0034 | |
| chr5 | ITGA2 | 52344487 | A | G | rs55973669 | c.A517G | p.I173V | 12.10 | 0.0022 | |
| chr19 | GP6 | 55543660 | G | A | rs199588110 | c.C172T | p.R58C | 18.28 | 0.0025 | 0.0016 |
| chr19 | GP6 | 55543660 | G | A | rs199588110 | c.C172T | p.R58C | 18.28 | 0.0023 | |
| chr19 | GP6 | 55543692 | C | T | rs750889036 | c.G140A | p.R47Q | 10.75 | 0.0019 | |
| chr11 | FERMT3 | 63974970 | T | G | rs759179590 | c.T134G | p.V45G | 33 | 0.0035 | 0.0042 |
| chr11 | FERMT3 | 63974995 | C | G | rs142815441 | c.C159G | p.I53M | 12.17 | 0.0013 | |
| chr11 | FERMT3 | 63978538 | G | A | rs762181713 | c.G409A | p.E137K | 29.0 | 0.0014 | |
| chr11 | P2RY2 | 72945434 | T | C | rs148391446 | c.T230C | p.V77A | 14.97 | 0.0017 | |
| chr11 | P2RY2 | 72945799 | A | G | rs141776297 | c.A595G | p.S199G | 15.02 | 0.0021 | |
| chr11 | P2RY2 | 72945799 | A | G | rs141776297 | c.A595G | p.S199G | 15.02 | 0.0019 | 0.0026 |
| chr11 | P2RY2 | 72946279 | T | C | rs74472890 | c.T1075C | p.S359P | 12.54 | 0.0024 | |
| chr3 | GP9 | 128781048 | G | A | rs3796130 | c.G466A | p.A156T | 12.21 | 0.0015 | |
| chr3 | P2RY12 * | 151055962 | C | A | c.G672T | p.R224S | 15 | 0.0021 | ||
| chr3 | P2RY12 * | 151056084 | G | T | c.C550A | p.L184I | 15.13 | 0.0013 | ||
| chr3 | P2RY1 * | 152554155 | G | A | c.G584A | p.R195H | 18.35 | 0.0012 | 0.0034 | |
| chr3 | P2RY1 * | 152554326 | C | A | c.C755A | p.S252Y | 22.4 | 0.0015 | ||
| chr3 | P2RY1 * | 152554395 | C | A | c.C824A | p.P275H | 22.8 | 0.0013 | ||
| chr3 | P2RY1 * | 152554482 | C | T | rs868057570 | c.C911T | p.A304V | 12.84 | 0.0026 | |
| chr1 | PEAR1 | 156878116 | C | T | c.C1099T | p.R367W | 18.01 | 0.0012 | ||
| chr1 | SELP | 169576246 | G | A | c.C1460T | p.A487V | 12.76 | 0.0017 | ||
| chr1 | SELP | 169581608 | G | A | rs139249907 | c.C808T | p.R270X | 13.86 | 0.0016 | |
| chr3 | GP5 | 194117640 | C | A | c.G1372T | p.A458S | 10.81 | 0.0012 |
* Functional and damaging variants selected for FlexStation3 analyzes.
The initial statistical analysis was performed using Pearson’s chi-squared test for the comparison of the total (cumulative) frequencies of all cMAF for rare deleterious variants in all sequenced genes between controls and IS cohort, as confirmed by individual genotyping. The obtained data are shown in Table 2. There was a highly statistically significant (p = 0.0005) increase in cMAF for all damaging variants in the IS group when compared with controls. Subsequent calculation of cMAF in the study patients remaining after removal of results of subjects with known CAD, CHF, and DM (in both groups) provided a similar difference in the cMAF for all deleterious variants between control and IS cohorts.
Table 2.
List of cumulative minor allele frequencies (cMAF) for damaging non-synonymous variants in the individually genotyped subjects from the control (ctrl) and study (stroke) groups in all patients used for pooled sequencing (left panel) and remaining patients after subtracting cardiac conditions (right panel).
| Gene | All Individuals (n = 1000). Number of Variant Carriers for Each Locus and Cohort in Brackets |
Subjects without Cardiac Disease (n = 605). Number of Variants Carriers for Each Locus and Cohort in Brackets |
||||
|---|---|---|---|---|---|---|
| Region | cMAF ctrl |
cMAF stroke |
CMAT P/Fisher | cMAF ctrl |
cMAF stroke |
CMAT P/Fisher |
| GP6 | 0.006 (3) |
0.002 (1) |
0.3700 | 0.000 (0) |
0.000 (0) |
NA |
| ITGA2 | 0.000 (0) |
0.004 (2) |
0.4900 | 0.0000 (0) |
0.002 (2) |
0.51 |
| ITGA2B/ITGB3 | 0.004 (2) |
0.014 (7) |
0.1200 | 0.005 (2) |
0.006 (5) |
0.62 |
| ITPR1 | 0.004 (2) |
0.006 (3) |
0.4900 | 0.0025 (1) |
0.002 (2) |
0.90 |
| P2RY1/P2RY12 | 0.002 (1) |
0.02 (10) |
0.0007 ** | 0.0025 (1) |
0.123 (10) |
0.002 ** |
| P2RY2 | 0.004 (2) |
0.006 (3) |
0.2100 | 0.0006 (1) |
0.004 (3) |
0.62 |
| PEAR1 | 0.0000 (0) |
0.002 (1) |
1.0000 | 0.000 (0) |
0.000 (0) |
NA |
| PTAFR | 0.0000 (0) |
0.002 (1) |
1.0000 | 0.0000 (0) |
0.001 (1) |
0.99 |
| SELP | 0.0000 (0) |
0.004 (2) |
0.1200 | 0.0000 (0) |
0.002 (2) |
0.14 |
| PTGIR | 0.0000 (0) |
0.004 (2) |
0.2500 | 0.0000 (0) |
0.002 (2) |
0.26 |
| Total # | 0.02 (10) |
0.064 (32) |
0.0005 * | 0.0125 (5) |
0.033 (27) |
0.03 * |
| OR | 3.4 (7.6–6.9) | 2.8 (1.1–7.3) | ||||
** Statistical significance (p value) calculated using burden CMAT test (for data with cMAF for variants in present both control and stroke groups) or Fisher exact test (for variants with cMAF only in one studies group and not observed in another group). * Statistical analysis performed using Pearson’s chi-squared test; #—combined cMAF for all observed damaging variants in one of the study group. OR—odds ratio.
The pooled analysis of multiple variants within unique regions or genes, which was based on pooled association test (CMAT), demonstrated a statistically significant difference (p = 0.0007) between control and IS cohorts for the P2RY1–P2RY12 location on chromosome 3 (Table 2). It contained five novel and one known rare and deleterious (CADD score range 12.8–22.8) variant. The region-based, Bonferroni-corrected significance threshold was p = 0.0021 (0.05/24 analyzed regions). Similar results were obtained after repeating CMAT analysis for control and IS cohorts remaining after removal of patients with CAD and DM (p = 0.03).
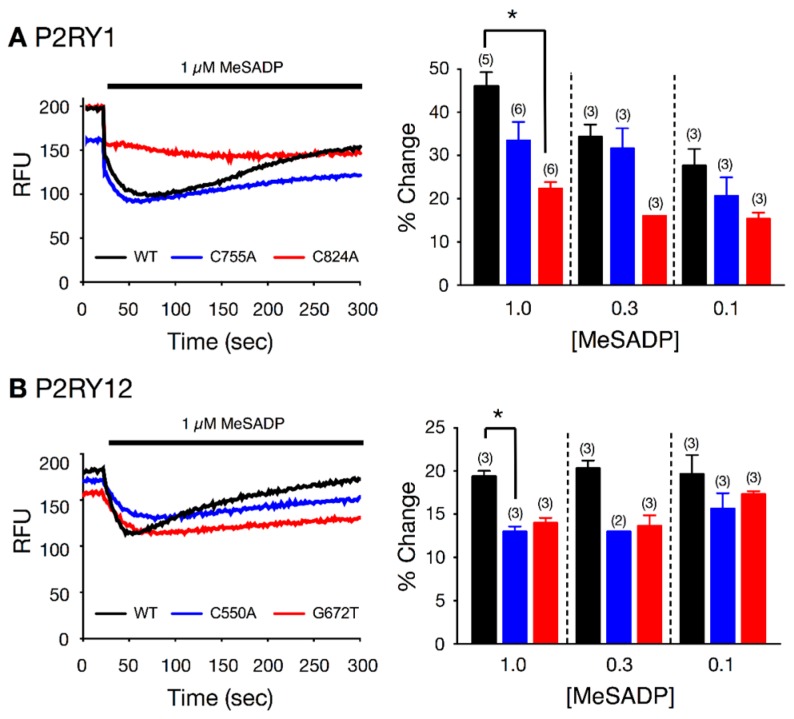
To determine whether the SNPs exert a deleterious effect on P2RY1 and P2RY12 function, we chose the two presumably most damaging variants (with highest CADD) from each gene, and examined coupling between the heterologously expressed mutant receptors and G protein inwardly-rectifying K+ (GIRK) channels in mouse fibroblast (L cells). Figure 2 shows the fluorescence signals of 3 individual wells with L cells expressing wild type P2RY1 (black trace), P2RY1 C755A (blue trace), and P2RY1 C824A (red trace) before and following 2-methylthioadenosine diphosphate (MeSADP) (1 μM) exposure. Following MeSADP application, the fluorescence decreased rapidly, indicative of cell hyperpolarization. It can be seen that the decrease in fluorescence for the C824A variant is approximately half of that obtained with the wild type-expressing L cells. Similarly, the decreased magnitude in fluorescence of C755A P2RY1-expressing cells was not as high as that observed in control. The summary plot illustrates that MeSADP produced a dose-dependent increase in hyperpolarization for wild type P2RY1-expressing cells, while that obtained with C824A was significantly (p < 0.05) attenuated with application of 1 μM MeSADP. On the other hand, the MeSADP-mediated hyperpolarization in C755A-expressing mutants was lower when compared to wild type P2RY1, but did not reach significance (p = 0.10). The fluorescence tracings shown in Figure 2B depict the effect of MeSADP (1 µM) in L cells expressing wild type P2RY12 (black trace), P2RY12 C550A (blue trace), and P2RY12 G672T (red trace) variants. The summary plot shows that MeSADP (1 µM) exposure produced a significantly (p < 0.05) lower change in P2RY12 C550A-expressing cells than the wild type receptor. The P2RY12 G672T-expressing cells also exhibited a diminished change (p = 0.08) following receptor activation.
Figure 2.
Effect of MeSADP-stimulated fluorescence changes in L cells heterologously expressing P2RY1 (A) and P2RY12 (B) wild type and variant receptors. The left panel (A) shows sample fluorescence signals (raw fluorescence units, RFU) from L cells transfected with wild type (black trace), C755A (blue), and C824A (red) P2RY1 cDNA constructs, before and during MeSADP (1 µM solid line) application. Left panel (B) depicts the fluorescence signals (RFU) in cells expressing wild type (black trace), C550A (blue trace) and G672T P2RY12 receptors. Panels on the right (A,B) are summary plots showing the mean (+SEM) changes of fluorescence signals following MeSADP application. * p < 0.05 employing ANOVA.
3. Discussion
In the present sequencing study of a relatively large cohort of 1000 Polish subjects, we investigated the contribution of rare non-synonymous variants in genes coding for platelet plasma membrane surface proteins, to the genetic susceptibility of large-vessel IS. The comparison of MAF for all rare and damaging non-synonymous variants across all sequenced genes and regions demonstrated that there was a statistically significant increase in the cumulative frequency of these variants in the IS group when compared with controls. These results allowed us to examine the contribution of rare coding variants to population variation in IS. By prioritizing rare variants using gene- and region-based tests, we identified novel associations, not previously detected by GWAS. We found association of 6 rare non-synonymous variants in P2RY1–P2RY12 coding region with large-vessel IS. Grouping rare variants by gene units allowed us to observe associations we were underpowered to detect when only examining single variants, as shown in previous studies of other traits [9]. By doing so, we had not only been gaining insight into the biology underlying platelet membrane proteins, but show that grouping variants by functional annotations could be an effective future strategy. It is important to note that, with exception of one variant, all of the other five investigated rare non-synonymous variants in the P2RY1–P2RY12 were novel, and never reported before (based on review of >300 rare non-synonymous variants in dbSNP149, and 1000 Genome database listings in March 2017). This might raise the possibility that the observed variants might be specific for the Polish cohort. It is important to mention that Polish patients have been poorly, or not at all, represented in 1000 Genomes and dbSNP. In addition, the recent work of Visschedijk et al. indicates that, at least in case of rare damaging variants associated with ulcerative colitis, the associated variants in the Dutch population could not be replicated in a German replication cohort [12]. Moreover, the first trans-ancestry association study in inflammatory bowel disease performed in several thousands of European individuals and individuals from East Asia, India, or Iran show that the majority of the loci based on MAF > 5% were shared between different ancestry groups [13]. However, this study also found genetic heterogeneity between different ancestry groups for less frequent alleles. Rare variants are even more likely to be specific to a particular population, as was demonstrated by a recent sequencing study in Korean population [14]. It was also demonstrated that the rare variants differ strongly among populations, even between closely related UK populations [15]. As far as the results of our study are concerned, it is interesting that we were able to confirm the presence only one (out of several hundreds) of previously listed rare non-synonymous variants in the P2RY1–P2RY12 region, which either indicated population-specific distribution of these variants, or might also indicate the limited power of this study. Further studies in both Polish and other populations are obviously needed to confirm that the rare deleterious variants in the purinergic receptor genes region on chromosome 3 are associated with large-vessel stroke, but also with other types of IS (small-vessel and embolic).
Different coding and non-coding SNPs (mostly common type) in P2RY1 and P2RY12 were previously evaluated in various IS population, however, the reported results have been conflicting so far [16,17,18,19]. It is worth mentioning that the possible associations of P2RY12 genetic variants, in a “gain-of-function” haplotype H2 of P2Y12, with thromboembolic events (myocardial infarction (MI), IS, or deep venous thromboembolism/pulmonary embolism (DVT/PE)), were not confirmed in a prospective analysis of 14,916 initially healthy American men [17]. The reported conflicting findings could be partly attributable to allelic heterogeneity, case-control selection criteria, phenotype/trait definition, and different population backgrounds.
The hyperpolarization response observed in the cell culture model served as a measure of adenine diphosphate (ADP)-induced purinergic activation, and was significantly attenuated by the investigated variants. Therefore, it can be assumed that the mutations, when expressed in platelets, could cause subsequent alterations in aggregation and clot formation. The reverse situation (more loss-of-function mutations in the IS group), as observed in this study, indicates that other mechanisms could be responsible for pro-stroke activity associated with P2RY1/P2RY12 receptors. It was previously demonstrated that the ADP-initiated activation of purinergic receptors causes release of endothelium-derived relaxing factor and nitric oxide, with subsequent activation of GIRK channel, hyperpolarization, and vasodilatation of small arteries [20,21]. It can be therefore hypothesized that the attenuation of this mechanism (presence of loss-of-function mutations in P2RY1/P2RY12 receptors) could produce a greater tendency to vasospasm and hypoperfusion associated with IS. It is interesting to note, in this regard, that a protective role for SNVs within coding regions of P2RY1 in MI was postulated by Ignatovica et al. in the Latvian population [22].
No previous studies addressed the impact or association of rare damaging polymorphisms in P2RY genes and IS. On the other hand, different SNPs (mostly common type) in P2RY1 and P2RY12 were previously evaluated in various IS cohorts, however, the reported results have been conflicting so far [16,17,18,19,20]. Out of all P2RY receptors, the greatest focus has been put on P2RY12, which is expressed in megakaryocyte/platelet lineage, and contributes to progression of thrombosis and hemostasis to cerebrovascular events [23]. To date, most of the studies focused on association between common SNPs within P2RY12 gene and antiplatelet drug response, mainly clopidogrel [24]. However, only few studies aimed to evaluate the relation between genetic polymorphism within P2RY receptor genes’ family, and risk of stroke and data are conflicting. Ziegler et al. found increased risk of ischemic cerebrovascular events in patients with peripheral artery disease treated with clopidogrel, that was associated with of rs6785930 (which is not part of the “H2” haplotype), but not rs6809699 (which is part of the “H2” haplotype) [16]. In the latest study performed in order to investigate the relationship between genetic polymorphisms and poor clinical outcomes in IS patients who underwent stenting for extracranial or intracranial arterial stenosis, four common SNPs within P2RY12 gene were included (i.e., rs6787801, rs6798347, rs2046934, and rs6801273). Only A-allele carriers of rs2046934 of P2RY12 had a significant association with an increased risk of clinical outcome events (transient ischemic attack (TIA), IS, MI, and death). In another study, the C allele in P2RY12 (rs2046934) was predicted to be a protective factor for clopidogrel resistance in IS patients [25]. The latest study aimed to evaluate the association of 2 common SNPs in P2RY12 (i.e., rs16863323, and rs9859538), 3 common SNPs in P2RY1 (i.e., rs701265, rs1439010, and rs1371097), and 2 SNPs in GPIIIa gene, as well as their interactions with antiplatelet drug responsiveness, and adverse clinical events after minor IS. The high-risk interactive genotypes (i.e., rs16863323TT in P2RY12) were independently associated with poor antiplatelet drug responsiveness and increased risk of primary outcomes, defined as composite adverse events of recurrent IS, MI, and death within 90 days after treatment [26]. However, in a prospective analysis of 14,916 American men, there was no evidence for an association of any of the variants or the haplotype H2 (composed of dbSNP rs10935838, rs2046934, rs5853517, and rs6809699) tested with risk of MI, or IS [17].
It has been previously reported that genetic variability of P2RY2 receptor had substantial influence on occurrence of IS. In 237 Japanese patients with a history of IS, five single SNPs within P2RY2 gene (rs4944831, rs1783596, rs4944832, rs4382936, rs10898909) were genotyped, and using a dominant model for rs4944832 phenotype, it was found that the GG genotype of this SNP is a genetic marker for IS, particularly in women. Moreover, the overall distribution of the haplotype defined by rs1783596–rs4382936–rs10898909 was significantly different between IS and the control groups [27]. The reported conflicting findings could be partly attributable to allelic heterogeneity, case-control selection criteria, phenotype/trait definition, and different population backgrounds.
Our confirmatory in vitro results also indicate that the MeSADP-mediated hyperpolarization of L cells expressing P2RY1 and P2RY12 variants was attenuated when compared to cells expressing the wild type receptors. These findings suggest that the signaling is likely altered in platelets expressing the mutant P2RY receptors.
4. Materials and Methods
4.1. Patients
The local ethics committee of the Institute of Psychiatry and Neurology, Warsaw, Poland, approved both the study protocol (including DNA sample collection and genotyping) and the informed consent form (identification code KB IPiN 06/2011). The study was conducted in accordance with the current version of the Declaration of Helsinki at the time when the study was designed, and informed written consent was obtained from all enrolled patients. The study population consisted of 500 patients with the diagnosis of the acute IS based on clinical features according to the World Health Organization definition and always supported by brain imaging (CT or MRI) that was selected for the existing dataset, as previously described [9,28,29,30]. Briefly, it included information about patients’ demographics, comorbidities, laboratory findings, and the course of stroke. Based on the Trial of Org 10172 in Acute Stroke Treatment (TOAST) classification, we included (i) all patients classified as having IS due to large-vessel atherosclerosis, and (ii) a subset of patients classified as having IS of unknown etiology, provided they had at least 50% stenosis of the carotid artery ipsilateral to the infarct side, and no evidence or no history of atrial fibrillation. The controls consisted of 500 age- and gender-matched patients, free of stroke, with multiple risk factors for cardiovascular disease or present CAD. Blood sampling and DNA extraction was performed as described before [9].
4.2. Genotyping
The target for genotyping consisted of 26 genes (Table 3) coding for platelet plasma membrane functions and containing total of 241 exons, as well as 10 flanking bases beyond each exon on both sides, which were selected using the human (Homo sapiens, hg19, GRCh37, February 2009) database. Pooled targeted enrichment of DNA, from 500 Polish large-vessel IS patients (100 individuals per pool × 5 pools) and 500 age-, gender-matched control patients (without any type of stroke history) (100 individuals per pool × 5 pools), was performed using a custom-made kit (SureSelect from Agilent Santa Clara, CA, USA) in accordance with the manufacturer’s instructions. The customized library was created by the SureDesign platform from Agilent Technologies, with average coverage of 99.6% of the selected exons. A detailed description of sequencing and data analysis is given in the Materials and Methods Supplementary Materials.
Table 3.
List of sequenced genes involved with the platelet membrane functions.
| Gene | Protein Product | Chr Location |
|---|---|---|
| P2RY2 | purinergic receptor P2Y, G-protein coupled, 2 | 11q13.5-q14.1 |
| P2RY12 | purinergic receptor P2Y, G-protein coupled, 12 | 3q25.1 |
| P2RY1 | Purinergic receptor P2Y, G-protein coupled, 1 | 3q25.2 |
| ITGB3 | integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) | 17q21.32 |
| ITGA2B | integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) | 17q21.32 |
| GP5 | glycoprotein V | 3q29 |
| GP9 | glycoprotein IX | 3q21.3 |
| GP6 | glycoprotein VI | 19q13.42 |
| GP1Bα | glycoprotein 1bα | 17p13.2 |
| GP1BB | glycoprotein Ib (platelet), beta polypeptide, antigen CD42c | 22q11.21 |
| ITGA2 (GPIa) | integrin, alpha 2 (alpha 2 subunit of VLA-2 receptor, antigen CD49B) | 5q11.2 |
| ADRA2A | adrenoceptor alpha 2A | 10q25.2 |
| TBXA2R | thromboxane A2 receptor | 19p13.3 |
| HTR2A | 5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled | 13q14-q21 |
| F2R (PAR-1) | proteinase-activated receptor 1 (PAR1), coagulation factor II (thrombin) receptor | 5q13 |
| F2RL3 (PAR-4) | protease activated receptor 4 (PAR-4), coagulation factor II (thrombin) receptor-like 3 | 19p12 |
| PEAR1 | platelet endothelial aggregation receptor-1 | 1q23.1 |
| GNB3 | guanine nucleotide binding protein (G protein), beta polypeptide 3 | 12p13 |
| CD148 | receptor-type protein tyrosine phosphatase | 11p11.1 |
| ITPR1 | inositol 1,4,5-trisphosphate receptor, type 1 | 3p26.1 |
| CD36 | thrombospondin receptor, antigen CD36 | 7q11.2 |
| CD40 | TNF receptor superfamily member 5, antigen CD40 | 20q12-q13.2 |
| EPR1 | effector cell peptidase receptor 1 | 17q25 |
| PECAM-1 | platelet/endothelial cell adhesion molecule 1 | 17q23.3 |
| FERMT3 | ferritin family member 3 | 11q13.1 |
| PTAFR | platelet-activating factor receptor | 1p35-p34.3 |
| PTGIR | prostaglandin I2 (prostacyclin) receptor (IP) | 19q13. |
4.2.1. Statistical Tests and Calculations
The initial analysis was performed by Pearson’s chi-squared test to ascertain statistical significance for the difference in the total number of alleles in re-sequenced genes containing damaging, rare, non-synonymous variants between controls and patients with large-vessel ischemia. The frequency of the investigated damaging allele was presented as cMAF, which encompassed all rare damaging variants in the sequenced gene or region. Since all observed genes were selected for MAF < 0.05, gene- or region-based burden analysis with the cumulative minor-allele test (CMAT) test (10,000× permutations) was performed in all targeted regions separately, to estimate the statistical significance of the observed differences in the accumulation of non-synonymous SNVs in large-vessel stroke in the investigated cohort when compared to controls. Statistical significance threshold was adjusted to number of target regions re-sequenced in the study (Bonferroni correction).
4.2.2. Sample Size and Power Considerations
In power calculations, instead of using individual rare variants, we decided to use predicted cMAF for all deleterious rare variants in single genes or regions. In this study, we have followed a self-sufficient, closed-form, maximum-likelihood estimator for allele frequencies that account for errors associated with sequencing, and a likelihood-ratio test statistic that provides a simple means for evaluating the null hypothesis of monomorphism [31,32]. Unbiased estimates of allele frequencies 10/n (where n is the number of individuals sampled) appear to be achievable, and near-certain identification of a SNP requires a cMAF (which is 10 variants for all pooled samples in the given cohort cMAF ~ 0.01). In addition, because the power to detect significant allele-frequency differences between two populations is limited, we set both the number of sampled individuals (500 in the cohort) and depth of sequencing coverage in excess of 100. The level of significance (p = 0.001) was assumed for a Fisher’s exact test for frequency differences in cMAF between populations.
4.3. Fluorescence-Based Functional Assay for P2RY1 and P2RY12 Receptor Activation in L Cells
Mouse fibroblast (L cells) cultures were transiently transfected by electroporation with cDNA constructs of the identified purinergic receptor genes within the pcDNA3.1 plasmid vector. GIRK channels, effectors for purinergic receptors, are not natively expressed in L cells. Thus, L cells were co-transfected with the GIRK4 S143T cDNA construct. The heterologously expressed purinergic receptors were stimulated with the P2Y1 and P2Y12 selective agonist, MeSADP. Purinergic receptor stimulation in this system cause specific G proteins to activate GIRK channels, resulting in membrane hyperpolarization (details provided in the Materials and Methods Supplementary Materials).
5. Limitations
The main limitation of the study is the lack of verification of observed variants and its association with IS in the independent verification cohort. It should be, however, noted that several previous studies demonstrated that the rare variant associations, because of their private character, are often limited to very limited populations, and are very hard to reproduce, unless the verification groups are very large (in this case, several tens of thousands patients). The other limitation of the study is that only a fraction of all known relevant genes related to platelet reactivity were sequenced in this study. Since the aim of our study was to analyze the impact of rare variants within genes related to platelet reactivity on IS risk, we did not measure platelet reactivity and its association with genetic polymorphism. Also, important limitation of the study was that we matched controls based on gender and age; however, comorbid clinical factors such as DM, CAD, or heart failure were observed more often in the control group. Based on the results of the multivariate logistic regression, after correcting for these clinical factors, one can conclude that the impact of observed polymorphism is unrelated to well-known risk factors for atherothrombotic disease, which further increases the strength of the results. Finally, the iPLEX-Sequenom method used with individual genotyping has a very high level of specificity (~100%), and its sensitivity depends very much on the primer design and stringency, which is usually optimized for detection of frequent SNPs, but not so much for singletons (e.g., only one mutated allele among hundreds of wild type alleles). We did our own spot check to confirm the heterozygous calls, and they were made with high confidence. However, because neither original Sequenom’s calling algorithm nor our post-processing scripts have been optimized for detection of very rare variants, we may have missed other samples with heterozygous or alternate allele homozygous genotypes.
6. Conclusions
In summary, the results of our study indicate that the distributions of investigated rare deleterious variants in the coding regions of selected platelet membrane genes (and in particular, located in the region of purinergic receptor P2RY1–P2RY12), in the cohort of Polish patients, could be associated with the large-vessel IS. The mechanisms by which these variants interact with the purinergic transmission and IS remains unclear, and will require further investigations. It is also not certain if our findings could be translated directly to other populations, as the variants driving the observed associations are rare, and appear to be limited to the Polish population. Further studies in larger, and also different populations, would be needed to clarify this question.
Acknowledgments
Research subject was implemented with CEPT infrastructure financed by the European Union—the European Regional Development Fund within the Operational Program “Innovative economy” for 2007–2013. The study was supported financially as part of the research grant from the Ministry of Science and Higher Education “Iuventus Plus” research grant (grant number IP2014 038473). The authors would like to thank Michal Karlinski, Agnieszka Cudna, Lukasz Milanowski, Marta Solarska, and Pawel Wylezol for preparing samples and database for further analysis.
Abbreviations
| IS | Ischemic stroke |
| GWAS | Genome wide association studies |
| MAF | Minor allele frequency |
| SNVs | Single nucleotide variants |
| SNPs | Single nucleotide polymorphism |
| CADD | Combined Annotation Dependent Depletion |
| TOAST | Trial of Org 10172 in Acute Stroke Treatment |
| CAD | Coronary artery disease |
| CHF | Congestive heart failure |
| DM | Diabetes mellitus |
| cMAF | Combined MAF |
| CMAT | Cumulative minor-allele test |
| SNP | Single nucleotide polymorphism |
| GIRK | G protein inwardly-rectifying K+ |
| MeSAD | 2-Methylthioadenosine diphosphate |
| TIA | Transient ischemic attack |
Supplementary Materials
Supplementary materials can be found at www.mdpi.com/1422-0067/18/12/2678/s1.
Author Contributions
Piotr K. Janicki substantial contribution to concept and design, analysis and interpretation of data, critical writing or revising the intellectual content, and final approval of the version to be published. Ceren Eyileten analysis and interpretation of data, critical writing or revising the intellectual content, final approval of the version to be published. Victor Ruiz-Velasco substantial contribution to concept and design, analysis and interpretation of data, critical writing or revising the intellectual content, and final approval of the version to be published. Khaled Anwar Sedeek analysis and interpretation of data, final approval of the version to be published. Justyna Pordzik analysis and interpretation of data, final approval of the version to be published. Anna Czlonkowska substantial contribution to concept and design, final approval of the version to be published. Iwona Kurkowska-Jastrzebska substantial contribution to concept and design, final approval of the version to be published. Shigekazu Sugino analysis and interpretation of data, final approval of the version to be published. Yuka Imamura-Kawasawa substantial contribution to concept and design, analysis of data, final approval of the version to be published. Dagmara Mirowska-Guzel substantial contribution to concept and design, final approval of the version to be published. Marek Postula substantial contribution to concept and design, analysis and interpretation of data, critical writing or revising the intellectual content, and final approval of the version to be published.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holliday E.G., Maguire J.M., Evans T.J., Koblar S.A., Jannes J., Sturm J.W., Hankey G.J., Baker R., Golledge J., Parsons M.W., et al. Common variants at 6p21.1 are associated with large artery atherosclerotic stroke. Nat. Genet. 2012;44:1147–1151. doi: 10.1038/ng.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bevan S., Traylor M., Adib-Samii P., Malik R., Paul N.L., Jackson C., Farrall M., Rothwell P.M., Sudlow C., Dichgans M., et al. Genetic heritability of ischemic stroke and the contribution of previously reported candidate gene and genomewide associations. Stroke. 2012;43:3161–3167. doi: 10.1161/STROKEAHA.112.665760. [DOI] [PubMed] [Google Scholar]
- 4.Gibson G. Rare and common variants: Twenty arguments. Nat. Rev. Genet. 2011;13:135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Francioli L.C., Menelaou A., Pulit S.L., van Dijk F., Palamara P.F., Elbers C.C., Neerincx P.B., Ye K., Guryev V., Kloosterman W.P., et al. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet. 2014;46:818–825. doi: 10.1038/ng.3021. [DOI] [PubMed] [Google Scholar]
- 6.Cheng Y.C., Cole J.W., Kittner S.J., Mitchell B.D. Genetics of ischemic stroke in young adults. Circ. Cardiovasc. Genet. 2014;7:383–392. doi: 10.1161/CIRCGENETICS.113.000390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Auer P.L., Nalls M., Meschia J.F., Worrall B.B., Longstreth W.T., Jr., Seshadri S., Kooperberg C., Burger K.M., Carlson C.S., Carty C.L., et al. Rare and Coding Region Genetic Variants Associated With Risk of Ischemic Stroke: The NHLBI Exome Sequence Project. JAMA Neurol. 2015;72:781–788. doi: 10.1001/jamaneurol.2015.0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anyanwu C., Hahn M., Nath M., Li J., Barone F.C., Rosenbaum D.M., Zhou J. Platelets Pleiotropic Roles in Ischemic Stroke. Austin J. Cerebrovasc. Dis. Stroke. 2016;3:1048. [Google Scholar]
- 9.Postula M., Janicki P.K., Milanowski L., Pordzik J., Eyileten C., Karlinski M., Wylezol P., Solarska M., Czlonkowka A., Kurkowska-Jastrzebka I., et al. Association of frequent genetic variants in platelet activation pathway genes with large-vessel ischemic stroke in Polish population. Platelets. 2017;28:66–73. doi: 10.1080/09537104.2016.1203404. [DOI] [PubMed] [Google Scholar]
- 10.Milanowski L., Pordzik J., Janicki P.K., Postula M. Common genetic variants in platelet surface receptors and its association with ischemic stroke. Pharmacogenomics. 2016;17:953–971. doi: 10.2217/pgs.16.21. [DOI] [PubMed] [Google Scholar]
- 11.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Visschedijk M.C., Alberts R., Mucha S., Deelen P., de Jong D.J., Pierik M., Spekhorst L.M., Imhann F., van der Meulen-de A.E., van der Woude C.J., et al. Pooled Resequencing of 122 Ulcerative Colitis Genes in a Large Dutch Cohort Suggests Population-Specific Associations of Rare Variants in MUC2. PLoS ONE. 2016;11:e0159609. doi: 10.1371/journal.pone.0159609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J.Z., van Sommeren S., Huang H., Ng S.C., Alberts R., Takahashi A., Ripke S., Lee J.C., Jostins L., Shah T., et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong K.W., Shin M.S., Ahn Y.B., Lee H.J., Kim H.D. Genomewide association study on chronic periodontitis in Korean population: Results from the Yangpyeong health cohort. J. Clin. Periodontol. 2015;42:703–710. doi: 10.1111/jcpe.12437. [DOI] [PubMed] [Google Scholar]
- 15.Prescott N.J., Lehne B., Stone K., Lee J.C., Taylor K., Knight J., Papouli E., Mirza M.M., Simpson M.A., Spain S.L., et al. Pooled Sequencing of 531 Genes in Inflammatory Bowel Disease Identifies an Associated Rare Variant in BTNL2 and Implicates Other Immune Related Genes. PLoS Genet. 2015;11:e1004955. doi: 10.1371/journal.pgen.1004955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ziegler S., Schillinger M., Funk M., Felber K., Exner M., Mlekusch W., Sabeti S., Amighi J., Minar E., Brunner M., et al. Association of a functional polymorphism in the clopidogrel target receptor gene, P2Y12, and the risk for ischemic cerebrovascular events in patients with peripheral artery disease. Stroke. 2005;36:1394–1399. doi: 10.1161/01.STR.0000169922.79281.a5. [DOI] [PubMed] [Google Scholar]
- 17.Zee R.Y., Michaud S.E., Diehl K.A., Chasman D.I., Emmerich J., Gaussem P., Aiach M., Ridker P.M. Purinergic receptor P2Y, G-protein coupled, 12 gene variants and risk of incident ischemic stroke, myocardial infarction, and venous thromboembolism. Atherosclerosis. 2008;197:694–699. doi: 10.1016/j.atherosclerosis.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Wang Z., Nakayama T., Sato N., Yamaguchi M., Izumi Y., Kasamaki Y., Ohta M., Soma M., Aoi N., Ozawa Y., et al. Purinergic receptor P2Y, G-protein coupled, 2 (P2RY2) gene is associated with cerebral infarction in Japanese subjects. Hypertens. Res. 2009;32:989–996. doi: 10.1038/hr.2009.136. [DOI] [PubMed] [Google Scholar]
- 19.Yi X., Zhou Q., Wang C., Lin J., Liu P., Fu C. Platelet receptor Gene (P2Y12, P2Y1) and platelet glycoprotein Gene (GPIIIa) polymorphisms are associated with antiplatelet drug responsiveness and clinical outcomes after acute minor ischemic stroke. Eur. J. Clin. Pharmacol. 2017;73:437–443. doi: 10.1007/s00228-017-2198-2. [DOI] [PubMed] [Google Scholar]
- 20.You J., Johnson T.D., Marrelli S.P., Mombouli J.V., Bryan R.M., Jr. P2u receptor-mediated release of endothelium-derived relaxing factor/nitric oxide and endothelium-derived hyperpolarizing factor from cerebrovascular endothelium in rats. Stroke. 1999;30:1125–1133. doi: 10.1161/01.STR.30.5.1125. [DOI] [PubMed] [Google Scholar]
- 21.Marrelli S.P. Altered endothelial Ca2+ regulation after ischemia/reperfusion produces potentiated endothelium-derived hyperpolarizing factor-mediated dilations. Stroke. 2002;33:2285–2291. doi: 10.1161/01.STR.0000027439.61501.39. [DOI] [PubMed] [Google Scholar]
- 22.Ignatovica V., Latkovskis G., Peculis R., Megnis K., Schioth H.B., Vaivade I., Fridmanis D., Pirags V., Erglis A., Klovins J. Single nucleotide polymorphisms of the purinergic 1 receptor are not associated with myocardial infarction in a Latvian population. Mol. Biol. Rep. 2012;39:1917–1925. doi: 10.1007/s11033-011-0938-4. [DOI] [PubMed] [Google Scholar]
- 23.Abbracchio M.P., Burnstock G., Boeynaems J.-M., Barnard E.A., Boyer J.L., Kennedy C., Knight G.E., Fumagalli M., Gachet C., Jacobson K.A., et al. International Union of Pharmacology LVIII: Update on the P2Y G Protein-Coupled Nucleotide Receptors: From Molecular Mechanisms and Pathophysiology to Therapy. Pharmacol. Rev. 2006;58:281–341. doi: 10.1124/pr.58.3.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui G., Zhang S., Zou J., Chen Y., Chen H. P2Y12 receptor gene polymorphism and the risk of resistance to clopidogrel: A meta-analysis and review of the literature. Adv. Clin. Exp. Med. 2017;26:343–349. doi: 10.17219/acem/63745. [DOI] [PubMed] [Google Scholar]
- 25.Liu R., Zhou Z., Chen Y., Li J.L., Jin J., Huang M., Zhao M., Yu W.B., Chen X.M., Cai Y.F., et al. Associations of CYP3A4, NR1I2, CYP2C19 and P2RY12 polymorphisms with clopidogrel resistance in Chinese patients with ischemic stroke. Acta Pharmacol. Sin. 2016;37:882–888. doi: 10.1038/aps.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yi X., Lin J., Wang Y., Zhou J., Zhou Q. Interaction among CYP2C8, GPIIIa and P2Y12 variants increase susceptibility to ischemic stroke in Chinese population. Oncotarget. 2017;8:70811–70820. doi: 10.18632/oncotarget.19991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z., Nakayama T., Sato N., Izumi Y., Kasamaki Y., Ohta M., Soma M., Aoi N., Ozawa Y., Ma Y. The purinergic receptor P2Y, G-protein coupled, 2 (P2RY2) gene associated with essential hypertension in Japanese men. J. Hum. Hypertens. 2010;24:327–335. doi: 10.1038/jhh.2009.67. [DOI] [PubMed] [Google Scholar]
- 28.WHO Task Force on Stroke and Other Cerebrovascular Diseases Recommendations on stroke prevention, diagnosis, and therapy. Report of the WHO task force on stroke and other cerebrovascular disorders. Stroke. 1989;20:1407–1431. doi: 10.1161/01.str.20.10.1407. [DOI] [PubMed] [Google Scholar]
- 29.Foulkes M., Wolf P., Price T., Mohr J., Hier D. The Stroke Data Bank: Design, methods, and baseline characteristics. Stroke. 1988;19:547–554. doi: 10.1161/01.STR.19.5.547. [DOI] [PubMed] [Google Scholar]
- 30.Grabska K., Gromadzka G., Członkowska A. Infections and ischemic stroke outcome. Neurol. Res. Int. 2011;2011:691348. doi: 10.1155/2011/691348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S., Abecasis G.R., Boehnke M., Lin X. Rare-variant association analysis: Study designs and statistical tests. Am. J. Hum. Genet. 2014;95:5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lynch M., Bost D., Wilson S., Maruki T., Harrison S. Population-genetic inference from pooled-sequencing data. Genome Biol. Evol. 2014;6:1210–1218. doi: 10.1093/gbe/evu085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.