Abstract
Leaf senescence, the final stage of leaf development, is a complex and highly regulated process that involves a series of coordinated actions at the cellular, tissue, organ, and organism levels under the control of a highly regulated genetic program. In the last decade, the use of mutants with different levels of leaf senescence phenotypes has led to the cloning and functional characterizations of a few genes, which has greatly improved the understanding of genetic mechanisms underlying leaf senescence. In this review, we summarize the recent achievements in the genetic mechanisms in rice leaf senescence.
Keywords: rice (Oryza sativa L.), leaf senescence, mutants, senescence-associated genes
1. Introduction
Leaf senescence is the final stage of leaf development. The green leaves gradually turn to yellow, orange, red, and eventually brown and die. This process is accompanied by a series of changes at the cellular, tissue, organ, and organism levels [1]. As a form of programmed cell death (PCD), leaf senescence is primarily an age-dependent process; however, it can also be triggered prematurely by internal and external factors [2]. By integrating environmental and endogenous factors, leaf senescence provides the optimal fitness for plant development [1].
Leaf senescence is an active rather than passive process to death, and the main functions of leaf senescence are to (a) recycle and re-use the nutrients from senescing leaves into newly developing organs or offspring and (b) enhance the chance of plant survival to adapt to biotic/abiotic stresses [3,4,5]. For grain crops, leaf senescence affects grain yield and quality such as nutrient loss and incomplete filling, etc. [6]. Thus, studying the molecular mechanisms of leaf senescence will not only facilitate the understanding of this fundamental biological process, but may also provide a way to regulate leaf senescence for improving the agricultural traits of crop plants [1].
To date, many excellent reviews have described the molecular processes involved in leaf senescence in plants [5,6,7,8]. The molecular and genetic understanding of leaf senescence has been mainly gained through the use of the model plant Arabidoposis, which is a Dicot species. Genes controlling leaf senescence are termed as senescence-associated genes (SAGs), and many senescence-associated genes (SAGs) have been identified in plants [9,10]. Our knowledge on the molecular mechanisms underlying leaf senescence in monocots including the major cereals crops such as rice, maize, wheat, barley, and sorghum is still limited. However, with the development of molecular biology and genomics, much inspirational progress has been made in elucidating the molecular mechanisms of leaf senescence in rice. The leaf senescence database currently contains more than 130 SAGs experimentally identified in rice [11]. The objective of this review is to briefly summarize recent progress in this field.
2. Chloroplast Degradation Involved in Leaf Senescence
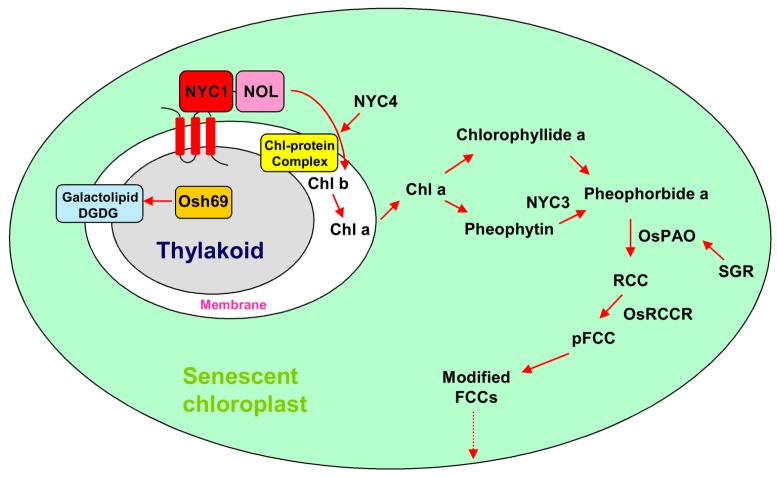
During leaf senescence, chloroplasts are the first organelles to be dismantled, which can induce the production of reactive oxygen species (ROS) such as hydrogen peroxide (H2O2), superoxide anion radicals (O2−), hydroxyl radicals (OH·), and singlet oxygen (1O2). As signaling triggers, ROS influence the expression of nuclear genes, thereby causing oxidative stress and damage to the cell [12,13,14,15]. The chlorophyll (Chl) degradation pathways involved in leaf senescence have been well established in recent years [16]. Based on the current literature, several SAGs are reported to relate to chlorophyll degradation in rice by using mutations that exhibit a stay-green phenotype in the process of leaf senescence (Table 1). During the degradation of chlorophyll, the first step is the conversion from Chl b to Chl a. NON-YELLOW COLORING 1 (NYC1), a chlorophyll b reductase for catalyzing the degradation chlorophyll b, encodes a chloroplast-localized short-chain dehydrogenase/reductase (SDR) and plays an important role in the degradation of the light-harvesting complex II (LHC II) and the thylakoid membrane (Figure 1) [17,18]. NYC1-LIKE (NOL), a thylakoid membrane location protein, is functionally similar to NYC1. NOL and NYC1 may form a complex to function as a chlorophyll b reductase in rice (Figure 1) [18]. Next, Chl a degradation may start with the de-chelation of Mg2+ by a magnesium-chelating substance which then removes phytol by pheophytinase (PPH) [19]. In rice, NON-YELLOW COLORING 3 (NYC3), which encodes a plastid-localizing α/β hydrolase-fold family protein with an esterase/lipase motif, may function in removing phytol residues from pheophytin a [20]. The STAY GREEN RICE (SGR) gene encodes a senescence-inducible chloroplast stay-green protein 1. The SGR mutant showed chlorophyll retention, stable chlorophyll-protein complexes, and thylakoid membrane structures, but lost its photosynthetic competence during leaf senescence. Further research showed that SGR may be involved in regulating or participating in the activity of pheophorbide a oxygenase (PAO), thereby influencing the degradation of chlorophyll and pigment-protein complexes (Figure 1) [21]. NYC4 (ortholog of Arabidopsis THF1) is also involved in the degradation of chlorophyll–protein complexes during leaf senescence, but its function is distinct from SGR. NYC4 is mainly involved in the degradation of chlorophyll-protein complexes, rather than in the regulation of chlorophyll breakdown [22]. As the downstream of SGR, PAO, and red chlorophyll catabolite reductase (RCCR) are the keys in catalyzing chlorophyll degradation. Knockdown of OsPAO and OsRCCR1 increased the production of ROS, resulting in leaf death and lesion mimic spots (Figure 1) [16].
Table 1.
Leaf senescence related genes in rice.
| Gene | Accession Number | Functional Annotation | Mutant Phenotype | Overexpression Phenotype | Regulatory Role Δ | Ref. |
|---|---|---|---|---|---|---|
| OsPAO | LOC_Os03g05310 | Pheophorbide a oxygenase | early | unknown | − | [16] |
| OsRCCR1 | LOC_Os10g25030 | Red chlorophyll catabolite reductase | early | unknown | − | [16] |
| NYC1 | LOC_Os01g12710 | Short-chain dehydrogenase/reductase | delayed | unknown | + | [17] |
| NOL | LOC_Os03g45194 | Short-chain dehydrogenase/reductase | delayed | unknown | + | [18] |
| NYC3 | LOC_Os06g24730 | α/β hydrolase-fold family protein | delayed | unknown | + | [20] |
| SGR | LOC_Os09g36200 | Senescence-inducible chloroplast stay-green protein 1 | delayed | early | + | [21] |
| NYC4 | LOC_Os07g37250 | THYLAKOID FORMATION 1 | delayed | unknown | + | [22] |
| RLS1 | LOC_Os02g10900 | NB-ARC domain containing protein | early | early | − | [23] |
| Osh69 | LOC_Os08g38710 | Alkaline α-galactosidase | unknown | unknown | + | [24] |
| OsMYC2 | LOC_Os10g42430 | JA-inducible basic helix-loop-helix transcription factor | unknown | early | + | [26] |
| OsCOI1b | LOC_Os05g37690 | F-box domain and LRR containing protein | delayed | unknown | + | [27] |
| OsPME1 | LOC_Os01g57854 | Pectinesterase | delayed | early | + | [28] |
| OsTSD2 | LOC_Os02g51860 | Pectin methyltransferase | delayed | unknown | + | [28] |
| OsFBK12 | LOC_Os03g07530 | F-box protein containing a kelch repeat motif | early | delayed | − | [29] |
| OsPLS1 | LOC_Os06g45120 | Vacuolar H+-ATPase subunit A1 | early | unknown | − | [30] |
| OsGATA12 | LOC_Os03g61570 | GATA-like zinc finger transcription factor | unknown | delayed | − | [31] |
| OsWRKY42 | LOC_Os02g26430 | Nuclear transcriptional repressor | unknown | early | + | [32] |
| OsHox33 | LOC_Os12g41860 | Class III homeodomain-leucine zipper gene family | early | unknown | − | [33] |
| ONAC106 | LOC_Os01g66120 | NAC domain transcription factor | delayed | unknown | + | [34] |
| OsNAP/PS1 | LOC_Os03g21060 | No apical meristem | delayed | early | + | [35] |
| SUB1A * | No | Submergence tolerance regulator | unknown | delayed | − | [36] |
| OsDOS | LOC_Os01g09620 | Nuclear-localized CCCH-type zinc finger protein | early | delayed | − | [37] |
| OsTZF1 | LOC_Os05g10670 | CCCH-tandem zinc finger protein | early | delayed | − | [38] |
| LTS1/OsNaPRT1 | LOC_Os03g62110 | Nicotinate phosphoribosyltransferase | early | unknown | − | [39] |
| OsSRT1 | LOC_Os04g20270 | NAD+-dependent histone deacetylases | early | delayed | − | [40] |
| OsFd-GOGAT | LOC_Os07g46460 | Ferredoxin-dependent glutamate synthase | early | unknown | − | [41] |
| Osl2 | LOC_Os04g52450 | γ-aminobutyric acid (GABA):pyruvate transaminase | unknown | unknown | + | [42] |
| DEL1 | LOC_Os10g31910 | Pectate lyase precursor | early | unknown | − | [43] |
| SPL29 | LOC_Os08g10600 | UDP-N-acetylglucosamine pyrophosphorylase 1 | early | unknown | − | [44] |
| ES1/TUTOU1 | LOC_Os01g11040 | SCAR-like protein 2 | early | unknown | − | [45] |
| OsSWEET5 | LOC_Os05g51090 | Sugar transporter family | unknown | early | + | [46] |
| OsGDCH | LOC_Os10g37180 | Glycine decarboxylase complex H subunit | early | Unknown | − | [47] |
Δ + positive regulation; − negative regulation; * absent in Nipponbare and therefore without an LOC number. NB-ARC: nucleotide-binding, Apaf-1, R proteins, and Ced-4; JA: jasmonate; LRR: Leucine-rich repeat; GATA: GATA motif; NAC: NAM/ATAF1/ATAFC2; CCCH: C-x8-C-x5-C-x3-H; UDP: uridine diphosphate; SCAR: suppressor of cAMP receptor.
Figure 1.
Chlorophyll degradation pathway involved in rice during leaf senescence. RCC: red chlorophyll catabolite; FCC: fluorescent chlorophyll catabolite.
The chloroplast degradation mutants above-mentioned all showed a stay-green phenotype; however, Jiao et al. [23] identified a mutant rapid leaf senescence 1 (rls1), which displayed a rapid leaf senescence during chloroplast degradation. RLS1 encodes an NB-containing protein with an ARM domain at the carboxyl terminus. The NB domain consists of three motifs and is found in many plant disease resistance proteins [23].
Galactolipids digalactosyl diacylglycerol (DGDG) and monogalactosyl diacylglycerol (MGDG) are the most abundant lipids of thylakoid membranes [24]. At the early stage of leaf senescence, the thylakoid membrane gradually breaks down, and the photosynthetic apparatus disassembles [25]. Osh69, a family of glycosyl hydrolases, encodes alkaline α-galactosidase. The Osh69 protein can cleave the terminal α-galactosidic bond of the galactolipid DGDG [24]. In addition, Osh69 upregulation can be induced by many factors including darkness, hormones, and stress [24].
3. Phytohormones and Transcription Factors Involved in Rice Leaf Senescence
Phytohormones play vital roles in plant development including leaf senescence (Table 1) [1].
In rice, the plant hormone methyl jasmonate (MeJA) and its precursor jasmonate (JA) were the first identified senescence promoting substances [26]. CORONATINE INSENSITIVE 1b (OsCOI1b) encodes a homolog of the Arabidopsis jasmonate (JA) receptor COI1. The mutation of OsCOI1b showed methyl jasmonate (MeJA) insensitivity and delayed leaf senescence [27]. By using a metabolite-based genome-wide association study (mGWAS), Fang et al. [28] identified two major quantitative genes OsPME1 (encoding pectin esterase) and OsTSD2 (encoding pectin methyltransferase) that affected the content of JA. Pectin methyl esterfication is the major source of MeOH. Subsequent investigations using mutants and transgenic lines revealed an MeOH–jasmonates cascade and its epigenetic that regulates leaf senescence [28]. F-box proteins are components of E3 ubiquitin ligase with functions in a wide variety of biological processes [48]. OsFBK12, encoding an F-box protein containing a kelch repeat motif, was involved in 26S proteasome-mediated degradation by interacting with Oryza sativa S-PHASEKINASE-ASSOCIATED PROTEIN1-LIKE PROTEIN (OSK) and targeted the substrate S-ADENOSYL-L-METHIONINE SYNTHETASE1 (SAMS1), triggering changes in ethylene (ETH) levels for the regulation of leaf senescence [29]. ORYZA SATIVA PREMATURE LEAF SENESCENCE (OsPLS1) encoding a vacuolar H+-ATPase subunit A1, plays a negative regulatory role in the onset of rice leaf senescence. The ospls1 mutant showed higher salicylic acid (SA) levels, increased ROS accumulation, and upregulation of WRKY genes [30]. In addition, Yamada et al. [49] found that strigolactone (SL)-deficient mutants in rice, such as d10, d17, and d27, showed accelerated dark-induced leaf senescence, implying that SL is involved in leaf senescence.
Several senescence-related transcription factors (TFs) are important for regulating leaf senescence (Table 1), for example, the zinc finger transcription factor OsGATA12, whose overexpression causes delayed leaf senescence, the reduction of leaf and tiller number, and improved rice yield. Further study showed that OsGATA12 may be involved in decreased chlorophyll degradation [31]. Overexpression of OsWRKY42 showed an accumulation of ROS and promoted leaf senescence by repressing OsMT1d expression via binding its W-box promoter in rice [32]. The class III homeodomain-leucine zipper (HD-Zip III) gene family plays important roles in plant growth and development [50]. Knockdown of an HD-Zip III member, OsHox33, accelerates leaf senescence in rice [33]. ONAC106, a senescence-associated NACs (NAM/ATAF1/ATAF2/CUC2) transcription factor, negatively regulates leaf senescence [34].
Many studies have clearly shown that transcription factors and phytohormones interactively regulate the leaf senescence process (Table 1). NACs are plant-specific transcription factors and some NACs have been confirmed to play important roles in regulating leaf senescence [51,52,53,54]. In rice, OsNAP/PS1 encodes a plant-specific NAC transcriptional activator and is induced specifically by abscisic acid (ABA). Overexpression of OsNAP/PS1 significantly promoted premature leaf senescence, whereas knockdown of OsNAP/PS1 produced an obvious delay of leaf senescence [35]. The transcription factor SUBMERGENCE1A (SUB1A), a key regulator of submergence in rice, significantly delays dark-induced senescence by the restriction of MeJA responsiveness and ETH production [36]. A nuclear-localized zinc finger/CCCH transcription factor protein OsDOS (delay of the onset of senescence) was found to take parts of the JA pathway. Overexpression of OsDOS showed delayed leaf senescence, whereas knockdown caused accelerated age-dependent leaf senescence, indicating it was a negative regulator for leaf senescence [37]. In contrast, the rice OsTZF1, which encodes a zinc finger CCCH type family protein, is induced by many factors including ABA, JA, SA, drought, high-salt, and H2O2. Overexpression of OsTZF1 showed delayed seed germination, growth retardation, delayed leaf senescence, improved tolerance to high-salt and drought stresses, and caused opposite phenotypes [38]. OsMYC2, a JA-inducible basic helix-loop-helix transcriptional factor, is a positive regulator of leaf senescence by the direct regulation of some SAGs in rice. Overexpression of OsMYC2 significantly promoted leaf senescence and a reduction in chlorophyll content, and was negatively regulated by OsJAZ8 (a JA ZIM-domain protein), involved in the JA signaling pathway in rice [26]. In addition, a recent study showed that miR319-controlled TCP transcription factors were involved in regulating JA content and leaf senescence [55].
Aside from the transcription factors mentioned above, based on microarray data, Liu et al. (2016) concluded that the W-box and G-box cis-elements may function as positive regulators affecting rice leaf senescence (Table 1) [56].
4. Energy Metabolism Pathway Regulated Rice Leaf Senescence
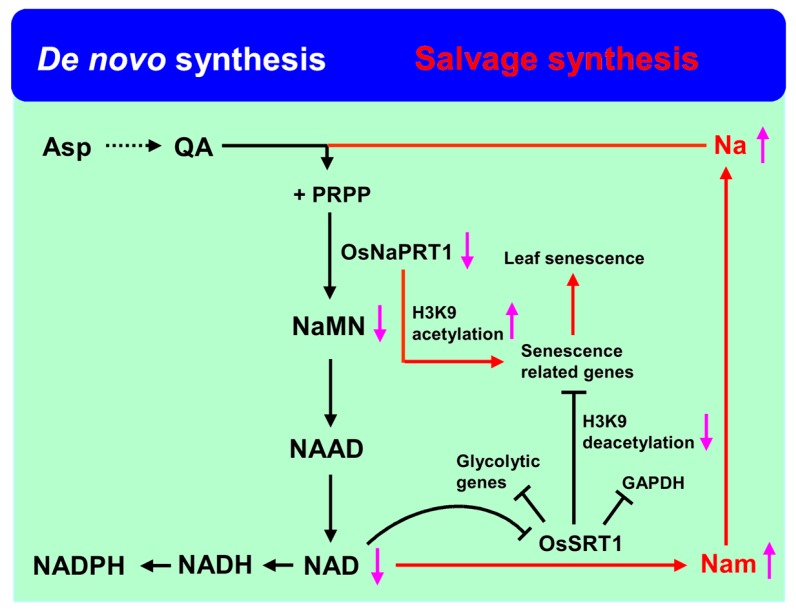
Nicotinamide adenine dinucleotide (NAD) and its derivative nicotinamide adenine dinucleo-tide phosphate (NADP) are important energy metabolite pathways involved in redox reactions in living organisms [57,58]. It was shown that NAD depletion could prevent cell death in vivo to maintain the balance of the internal environment [59]. In Arabidopsis, there are two NADP biosynthetic pathways: de novo and the salvage pathway [60,61]. In the salvage pathway, SIR2, an NAD+-dependent histone deacetylase, plays a crucial role in converting NAD to nicotinamide (Nam) [39,40]. In rice, there are two SIR2 homologous genes, OsSRT1 (OsSIRT701) and OsSRT2 (OsSIRT702) [62]. RNA interference of OsSRT1 results in an increase of histone H3K9 acetylation and a decrease of H3K9 dimethylation, H2O2 accumulation, DNA fragmentation, programmed cell death, and mimicking plant lesions, and its overexpression enhances the tolerance of redox [40]. Recent research indicated that OsSRT1 could regulate carbon metabolic flux through the repression of glycolysis by the deacetylation of both histone and glycolytic glyceraldehyde-3-phosphatedehydrogenase (GAPDH) (Figure 2) [63].
Figure 2.
NAD synthesis and catabolic pathways involved in rice leaf senescence. Asp: aspartate, QA: quinolinic acid, Na: nicotinic acid, Nam: nicotinamide, PRPP: 5-phosphoribosyl-1-pyrophosphate, NaMN: nicotinate mononucleotide, NAAD: nicotinic acid adenine dinucleotide, NAD: nicotinamide adenine dinucleotide.
Downstream of NAD, Nam from nicotinate mononucleotide (NaMN) was catalyzed by two enzymes: nicotinamidase and nicotinate phosphoribosyltransferase (NaPRTase) [59,61]. In rice, a mutation of NaPRTase, LTS1, revealed increased concentrations of nicotinate and nicotinamide as well as decreased NAD content. Further research indicated that the decreased NAD repressed the expression of OsSRTs and would result in a lower deacetylation ability of OsSRTs, hence activating senescence-related genes by increasing the acetylation of histone H3K9, leading to leaf senescence in rice (Figure 2) [39].
5. Nitrogen Remobilization Involved in Rice Leaf Senescence
Nitrogen remobilization increases nitrogen use efficiency and plays an important role in sustainable agriculture. Nitrogen molecules have a major presence in proteins and nucleic acids, and are transported in the form of amino acids (particularly glutamine and asparagine) from the senescence leaves to new parts [64]. The metabolism of glutamate and γ-aminobutyric acid (GABA) plays an important role in nitrogen circulation [65]. During glutamate metabolism, glutamine synthetase catalyzes ammonia and 2-oxoglutarate into glutamine, whereas glutamate synthase (or glutamine 2-oxoglutarate aminotransferase, GOGAT) catalyzes the reversible conversion of glutamine into glutamate [65,66]. In higher plants, GOGAT has two isoforms: Fd-GOGAT and NADH-GOGAT. Fd-GOGAT is predominantly located in the chloroplasts of photosynthetic tissues, and NADH-GOGAT is present in non-photosynthesizing cells [66]. In rice, the gogat1 mutant exhibited chlorosis under natural conditions and less extent premature leaf senescence under low light conditions. Meanwhile, the gogat1 mutant showed a reduced seed setting rate and increased grain protein and amino acid content. This result showed that OsFd-GOGAT plays an important role in nitrogen remobilization during leaf senescence [41].
The transferring glutamate to succinate via GABA is called the GABA metabolism or GABA shunt [65]. As a temporary storage of nitrogen, enhanced GABA can inhibit the synthesis of glutamine during senescence [67]. GABA:pyruvate-transaminase catalyzes GABA into succinic semialdehyde (SSA). SSA is then catalyzed into succinate by succinic semialdehyde dehydrogenase (SSADH) and goes into a tricarboxylic acid (TCA) cycle [65]. In rice, Osl2, encoding γ-aminobutyric acid (GABA):pyruvate transaminase, is upregulated and plays a key role in nitrogen metabolism during leaf senescence [42,65].
6. Other Genes Involved in Leaf Senescence
Recent research has shown that cell-wall-related genes are involved in the regulation of leaf senescence. The DWARF AND EARLY-SENESCENCE 1 (DEL1) gene encodes a pectate lyase precursor. Loss of function of DEL1 decreased total pectate lyase (PEL) activity, increased the degree of methylesterified homogalacturonan (HG), and perturbed cell wall composition and structure, resulting in triggering ROS activity, thereby leading to leaf senescence [43].
UDP-N-acetylglucosamine pyrophosphorylase (UAP) is widely distributed in living organisms [44]. Wang et al. [44] cloned the SPOTTED LEAF 29 (SPL29) gene, which encodes UAP1 in rice. The spl29 mutant displayed many changes involved in chloroplast degradation, chlorophyll loss and photosystem II decline, enhanced resistance to bacterial blight inoculation, increased malondialdehyde content and ROS, upregulated SAGs and defence response genes, downregulated photosynthesis-related genes, etc. [44].
Actin filament plays an important role in many endomembrane processes such as vacuole formation, endocytosis of plasma membrane (PL), and vesicle transport from the Golgi complex, etc. [68,69,70,71,72]. The ARP2/3 complex as a key regulator of actin filament nucleation can be inactive by itself and active by the SCAR/wave complex in plants [73]. The SCAR/WAVE complex is highly conserved, and deficiency in the SCAR/WAVE complex in plants often leads to morphological changes [45]. In rice, EARLY SENESCENCE 1 (ES1) encodes a SCAR-LIKE PROTEIN2, which plays an important role in leaf senescence. The es1 mutant shows a short and irregular arrangement of actin filaments. The changes to the actin filaments increase the water loss of leaves, thereby leading to leaf senescence [45].
The SWEET family plays important roles in plant growth and development. In rice, overexpression of OsSWEET5, a novel sugar transporter family, caused growth retardation and precocious senescence at the seedling stage [46].
Glycine decarboxylase complex (GDC) is a multi-protein complex, which plays a major role in the photorespiration of plants [74]. Under ambient CO2, knockdown of OsGDCH caused leaf senescence due to chlorophyll loss, protein degradation, chloroplast breakdown, and autophagy, as well as ROS accumulation [47].
7. Perspectives
Leaf senescence is a very complex phenomenon. It is an evolutionarily acquired developmental strategy to adapt to internal and external factors [2]. Leaves are the main locations of plant photosynthesis, most of the carbon in mature rice grains originates from leaf photosynthesis [75]. Timely leaf senescence can make plants accumulate enough nutrients for assimilation, while excessive leaf senescence can lead to decreased plant photosynthetic capacity and assimilation capacity, thereby reducing crop yield and quality. In addition, adequate remobilization of nutrients increases the usage efficiency of crops, thereby reducing the use of fertilizers [8].
Rice (Oryza sativa L.) is an important staple food that feeds more than half of the world’s population, mainly in Asia [76]. Achieving increases in rice grain-yield is a permanent topic of concern for over-increasing populations [77]. Delaying leaf senescence, particularly of the flag leaf, would help to increase grain yield [3]. The stay-green traits have been used in breeding to enhance stress resistance and increase grain yield, although the relationships between leaf senescence and crop yield and quality have not yet been well characterized [78]. Systematically elucidating the molecular mechanisms of leaf senescence will provide breeders with new tools/options for further improving many important agronomic traits in future.
As summarized in this review, significant progress has been made in the cloning and functional characterization of leaf SAGs in rice in the last few decades. However, the discussion in this review only focuses on the role of single genes in the onset of senescence. The main reason for this is based on the study progress of leaf senescence in rice. Therefore, a deeper understanding of leaf senescence will provide more insights into the improvement of crop productivity.
Acknowledgments
We thank Long Chen (China National Rice Research Institute) for assistance on document arrangement. This research was supported by the National Natural Science Foundation of China (Grant No. 3161143006, 91735303, 91435105) and the Science and Technology Innovation Project of the Chinese Academy of Agriculture Sciences.
Abbreviations
| ABA | abscisic acid |
| Chl | chlorophyll |
| ETH | ethylene |
| DGDG | digalactosyl diacylglycerol |
| GAPDH | glyceraldehyde-3-phosphatedehydrogenase |
| HG | homogalacturonan |
| H2O2 | hydrogen peroxide |
| JA | jasmonate |
| LHCⅡ | light-havesting complexⅡ |
| MeJA | methyl jasmonate |
| MGDG | monogalactosyl diacylglycerol |
| mGWAS | metabolite-based genome-wide association study |
| NAD | nicotinamide adenine dinucleotide |
| NADP | nicotinamide adenine dinucleotide phosphate |
| Nam | nicotinamide |
| NaMN | nicotinate mononucleotide |
| 1O2 | singlet oxygen |
| O2− | superoxide anion radical |
| OH | hydroxyl radical |
| PCD | programmed cell death |
| PEL | pectate lyase |
| PL | plasma membrane |
| PPH | pheophytinase |
| ROS | reactive oxygen species |
| SA | salicylic acid |
| SAGs | senescence-associated genes |
| SL | strigolactone |
| SSA | succinic semialdehyde |
| SSADH | succinic semialdehyde dehydrogenase |
| TCA | tricarboxylic acid |
| TFs | transcription factors |
Author Contributions
Yujia Leng and Dali Zeng wrote the manuscript. Guoyou Ye and Dali Zeng revised the manuscript. All authors approved the final version of the manuscript.
Conflicts of interest
The authors declare no conflict of interest.
References
- 1.Lim P.O., Kim H.J., Nam H.G. Leaf senescence. Annu. Rev. Plant Biol. 2007;58:115–136. doi: 10.1146/annurev.arplant.57.032905.105316. [DOI] [PubMed] [Google Scholar]
- 2.Quirino B.F., Noh Y.S., Himelblau E., Amasino R.M. Molecular Aspect of Leaf Senescence. Trends Plant Sci. 2000;5:278–282. doi: 10.1016/S1360-1385(00)01655-1. [DOI] [PubMed] [Google Scholar]
- 3.Buchanan-Wollaston V., Earl S., Harrison E., Mathas E., Navabpour S., Page T., Pink D. The molecular analysis of leaf senescence—A genomics approach. Plant Biotechnol. J. 2003;1:3–22. doi: 10.1046/j.1467-7652.2003.00004.x. [DOI] [PubMed] [Google Scholar]
- 4.Dani K.G.S., Fineschi S., Michelozzi M., Loreto F. Do cytokinins, volatile isoprenoids and carotenoids synergically delay leaf senescence? Plant Cell Environ. 2016;39:1103–1111. doi: 10.1111/pce.12705. [DOI] [PubMed] [Google Scholar]
- 5.Kim J., Woo R.H., Nam H.G. Toward systems understanding of leaf senescence: An integrated multi-omics perspective on leaf senescence research. Mol. Plant. 2016;9:813–825. doi: 10.1016/j.molp.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 6.Schippers J.H.M., Schmidt R., Wagstaff C., Jing H.C. Living to die and dying to live: The survival strategy behind leaf senescence. Plant Physiol. 2015;169:914–930. doi: 10.1104/pp.15.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J., Chang C., Tucker M.L. To grow old: Regulatory role of ethylene and jasmonic acid in senescence. Front. Plant Sci. 2015;6:1–7. doi: 10.3389/fpls.2015.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schippers J.H.M. Transcriptional networks in leaf senescence. Curr. Opin. Plant Biol. 2015;27:77–83. doi: 10.1016/j.pbi.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 9.Buchanan-wollaston V. The molecular biology of leaf senescence. J. Exp. Bot. 1997;48:181–199. doi: 10.1093/jxb/48.2.181. [DOI] [Google Scholar]
- 10.Liu L., Zhou Y., Zhou G., Ye Z.J., Zhao L.N., Li X.H., Lin Y.J. Identification of early senescence-associated genes in rice flag leaves. Plant Mol. Biol. 2008;67:37–55. doi: 10.1007/s11103-008-9300-1. [DOI] [PubMed] [Google Scholar]
- 11.Liu X.C., Li Z.H., Jiang Z.Q., Zhao Y., Peng J.Y., Jin J.P., Guo H.W., Luo J.C. LSD: A leaf senescence database. Nucleic Acids Res. 2011;39:D1103–D1107. doi: 10.1093/nar/gkq1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dodge J.D. Changes in chloroplast fine structure during the autumnal senescence of Betula leaves. Ann. Bot. 1970;34:817–824. doi: 10.1093/oxfordjournals.aob.a084412. [DOI] [Google Scholar]
- 13.Apel K., Hirt H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 14.Gechev T.S., van Breusegem F., Stone J.M., Denev I., Laloi C. Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. BioEssays. 2006;28:1091–1101. doi: 10.1002/bies.20493. [DOI] [PubMed] [Google Scholar]
- 15.Balazadeh S., Jaspert N., Arif M., Mueller-Roeber B., Maurina V.G. Expression of ROS-responsive genes and transcription factors after metabolic formation of H2O2 in chloroplasts. Front. Plant Sci. 2012;3:234. doi: 10.3389/fpls.2012.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Y.Y., Li M.R., Chen Y.P., Wu P.Z., Wu G.J., Jiang H.W. Knockdown of OsPAO and OsRCCR1 cause different plant death phenotypes in rice. J. Plant Physiol. 2011;168:1952–1959. doi: 10.1016/j.jplph.2011.05.026. [DOI] [PubMed] [Google Scholar]
- 17.Kusaba M., Ito H., Morita R., Lida S., Sato Y., Fujimoto M., Kawasaki S., Tanaka R., Hirochik H., Nishimura M., et al. Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell. 2007;19:1362–1375. doi: 10.1105/tpc.106.042911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sato Y., Morita R., Katsuma S., Nishimura M., Tanaka A., Kusaba M. Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 2009;57:120–131. doi: 10.1111/j.1365-313X.2008.03670.x. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki T., Shioi Y. Re-examination of Mg-dechelation reaction in the degradation of chlorophylls using chlorophyllin α as a substrate. Photosynth. Res. 2002;74:217–223. doi: 10.1023/A:1020915812770. [DOI] [PubMed] [Google Scholar]
- 20.Morita R., Sato Y., Masuda Y., Nishimura M., Kusaba M. Defect in non-yellow coloring 3, an α/β hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J. 2009;59:940–952. doi: 10.1111/j.1365-313X.2009.03919.x. [DOI] [PubMed] [Google Scholar]
- 21.Jiang H.W., Li M.R., Liang N.T., Han H.B., Wei Y.B., Xu X.L., Liu J., Xu Z.F., Chen F., Wu G.J. Molecular cloning and function analysis of the stay green gene in rice. Plant J. 2007;52:197–209. doi: 10.1111/j.1365-313X.2007.03221.x. [DOI] [PubMed] [Google Scholar]
- 22.Yamatani H., Sato Y., Masuda Y., Kato T., Morita R., Fujunaga K., Nagamura Y., Nishimura M., Sakamoto W., Tanaka A., et al. NYC4, the rice ortholog of Arabidopsis THF1, is involved in the degradation of chlorophyll-protein complexes during leaf senescence. Plant J. 2013;74:652–662. doi: 10.1111/tpj.12154. [DOI] [PubMed] [Google Scholar]
- 23.Jiao B.B., Wang J.J., Zhu X.D., Zeng L.J., Li Q., He Z.H. A novel protein RLS1 with NB-ARM domains is involved in chloroplast degradation during leaf senescence in rice. Mol. Plant. 2012;5:205–217. doi: 10.1093/mp/ssr081. [DOI] [PubMed] [Google Scholar]
- 24.Lee R.H., Lin M.C., Chen S.C.G. A novel alkaline α-galactosidase gene is involved in rice leaf senescence. Plant Mol. Biol. 2004;55:281–295. doi: 10.1007/s11103-004-0641-0. [DOI] [PubMed] [Google Scholar]
- 25.Lee R.H., Chen S.C.G. Programmed cell death during rice leaf senescence is non-apoptotic. New Phytol. 2002;55:25–32. doi: 10.1046/j.1469-8137.2002.00443.x. [DOI] [PubMed] [Google Scholar]
- 26.Uji Y., Akimitsu K., Gomi K. Identification of OsMYC2-regulated senescence-associated genes in rice. Planta. 2017;245:1–6. doi: 10.1007/s00425-017-2697-5. [DOI] [PubMed] [Google Scholar]
- 27.Lee S.H., Sakuraba Y., Lee T., Kim K.W., An G., Lee H.Y., Paek N.C. Mutation of Oryza sativa CORONATINE INSENSITIVE 1b (OsCOI1b) delays leaf senescence. J. Integr. Plant Biol. 2015;57:562–576. doi: 10.1111/jipb.12276. [DOI] [PubMed] [Google Scholar]
- 28.Fang C.Y., Hua Z., Jian W., Wu Y.Y., Li K., Jin C., Chen W., Wang S.C., Wang W.S., Zhang H.W., et al. Control of leaf senescence by an MeOH-Jasmonates cascade that is epigenetically regulated by OsSRT1 in rice. Mol. Plant. 2016;9:1366–1378. doi: 10.1016/j.molp.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y., Xu Y.Y., Luo W., Li W.X., Chen N., Zhang D.J., Chong K. The F-box protein OsFBK12 targets OsSAMS1 for degradation and affects pleiotropic phenotypes, including leaf senescence, in Rice. Plant Physiol. 2013;163:1673–1685. doi: 10.1104/pp.113.224527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang X., Pan G., Li K.Y., Huang F.D., Cheng F.M., Pan G. A single cytosine deletion in the OsPLS1 gene encoding vacuolar-type H+-ATPase subunit A1 leads to premature leaf senescence and seed dormancy in rice. J. Exp. Bot. 2016;67:2761–2776. doi: 10.1093/jxb/erw109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu G.W., Casaretto J.A., Ying S., Mahmood K., Liu F., Bi Y.M., Rothstein S.J. Overexpression of OsGATA12, regulates chlorophyll content, delays plant senescence and improves rice yield under high density planting. Plant Mol. Biol. 2017;94:1–13. doi: 10.1007/s11103-017-0604-x. [DOI] [PubMed] [Google Scholar]
- 32.Han M., Kim C.Y., Lee J., Lee S.K., Jeon J.S. OsWRKY42 represses OsMT1d and induces rective oxygen species and leaf senescence in rice. Mol. Cells. 2014;37:532–539. doi: 10.14348/molcells.2014.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luan W.J., Shen A., Jin Z.P., Song S.S., Long L.Z., Sha A.H. Knockdown of OsHox33, a member of the class III homeodomain-leucine zipper gene family, accelerates leaf senescence in rice. Sci. China Life Sci. 2013;56:1113–1123. doi: 10.1007/s11427-013-4565-2. [DOI] [PubMed] [Google Scholar]
- 34.Sakuraba S., Piao W.L., Lim J.H., Han S.H., Kim Y.S., An G., Paek N.C. Rice ONAC106 inhibits leaf senescence and increases salt tolerance and tiller angle. Plant Cell Physiol. 2015;56:2325–2339. doi: 10.1093/pcp/pcv144. [DOI] [PubMed] [Google Scholar]
- 35.Liang C.Z., Wang Y.Q., Zhu Y.N., Tang J.Y., Hu B., Liu L.C., Ou S.J., Wu H.K., Sun X.H., Chu J.F., et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. USA. 2014;111:10013–10018. doi: 10.1073/pnas.1321568111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukao T., Yeung E., Bailey-Serres J. The submergence tolerance gene SUB1A delays leaf senescence under prolonged darkness through hormonal regulation in rice. Plant Physiol. 2012;160:1795–1807. doi: 10.1104/pp.112.207738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kong Z.S., Li M.N., Yang W.Q., Xu W.Y., Xue Y.B. A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol. 2006;141:1376–1388. doi: 10.1104/pp.106.082941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jan A., Maruyama K., Todaka D., Kidokoro S., Abo M., Yoshimura E., Shinozaki K., Nakashima K., Yamaguchi-Shinozaki K. OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol. 2013;161:1202–1216. doi: 10.1104/pp.112.205385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu L.W., Ren D.Y., Hu S.K., Li G.M., Dong G.J., Jiang L., Hu X.M., Ye W.J., Cui Y.T., Zhu L., et al. Down-Regulation of a Nicotinate Phosphoribosyltransferase Gene, OsNaPRT1, Leads to Withered Leaf Tips. Plant Physiol. 2016;171:1085–1098. doi: 10.1104/pp.15.01898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang L.M., Sun Q.W., Qin F.J., Li C., Zhao Y., Zhou D.X. Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol. 2007;144:1508–1519. doi: 10.1104/pp.107.099473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng D.D., Qin R., Li M., Alamin M., Jin X.L., Liu Y., Shi C.H. The ferredoxin-dependent glutamate synthase (OsFd-GOGAT) participates in leaf senescence and the nitrogen remobilization in rice. Mol. Genet. Genom. 2016;292:1–11. doi: 10.1007/s00438-016-1275-z. [DOI] [PubMed] [Google Scholar]
- 42.Ansari M.I., Lee R.H., Chen S.C.G. A novel senescence-associated gene encoding γ-aminobutyric acid (GABA):Pyruvate transaminase is upregulated during rice leaf senescence. Physiol. Plant. 2005;123:1–8. doi: 10.1111/j.1399-3054.2004.00430.x. [DOI] [Google Scholar]
- 43.Leng Y.J., Yang Y.L., Ren D.Y., Huang L.C., Dai L.P., Wang Y.Q., Chen L., Tu Z.J., Gao Y.H., Li X.Y., et al. A rice PECTATE LYASE-LIKE gene is required for plant growth and leaf senescence. Plant Physiol. 2017;174:1151–1166. doi: 10.1104/pp.16.01625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z.H., Wang Y., Hong X., Hu D.H., Liu C.X., Yang J., Li Y., Huang Y.Q., Feng Y.Q., Gong G.Y., et al. Functional inactivation of UDP-N-acetylglucosamine pyrophosphorylase 1 (UAP1) induces early leaf senescence and defence responses in rice. J. Exp. Bot. 2015;66:973–987. doi: 10.1093/jxb/eru456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rao Y.C., Yang Y.L., Xu J., Li X.J., Leng Y.J., Dai L.P., Huang L.C., Shao G.S., Ren D.Y., Hu J., et al. EARLY SENESCENCE1 encodes a SCAR-LIKE PROTEIN2 that affects water loss in rice. Plant Physiol. 2015;169:1225–1239. doi: 10.1104/pp.15.00991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Y., Liu L., Huang W.F., Yuan M., Zhou F., Li X.H., Lin Y.J. Overexpression of OsSWEET5 in rice causes growth retardation and precocious senescence. PLoS ONE. 2014;9:e94210. doi: 10.1371/journal.pone.0094210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Q.Y., Yu Q., Wang Z.Q., Pan Y.F., Lv W.T., Zhu L.L., Chen R.Z., He G.C. Knockdown of GDCH gene reveals reactive oxygen species-induced leaf senescence in rice. Plant Cell Environ. 2013;36:1476–1489. doi: 10.1111/pce.12078. [DOI] [PubMed] [Google Scholar]
- 48.Schulman B.A., Carrano A.C., Jeffrey P.D., Bowen Z., Kinnucan E.R., Finnin M.S., Elledge S.J., Harper J.W., Pagano M., Pavletich N.P. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature. 2000;408:381–386. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- 49.Yamada Y., Furusawa S., Nagasaka S., Shimomura K., Yamaguchi S., Umehara M. Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency. Planta. 2014;240:399–408. doi: 10.1007/s00425-014-2096-0. [DOI] [PubMed] [Google Scholar]
- 50.Ariel F.D., Manavella P.A., Dezar C.A., Chan R. The true story of the HD-Zip family. Trends Plant Sci. 2007;12:419–426. doi: 10.1016/j.tplants.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 51.Oh S.A., Park J.H., Lee G.I., Paek K.H., Park S.K., Nam H.G. Identification of three genetic loci controlling leaf senescence in Arabidopsis thaliana. Plant J. 1997;12:527–535. doi: 10.1046/j.1365-313X.1997.00527.x. [DOI] [PubMed] [Google Scholar]
- 52.Guo Y., Cai Z., Gan S. Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 2004;27:521–549. doi: 10.1111/j.1365-3040.2003.01158.x. [DOI] [Google Scholar]
- 53.Uauy C., Distelfeld A., Fahima T., Blechl A., Dubcovsky J. A NAC gene regulating senescence improves grain protein, Zinc, and iron content in wheat. Science. 2006;314:1298–1301. doi: 10.1126/science.1133649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou Y., Huang W., Liu L., Chen T.Y., Zhou F., Lin Y.J. Identification and functional characterization of a rice NAC gene involved in the regulation of leaf senescence. BMC Plant Biol. 2013;13:132. doi: 10.1186/1471-2229-13-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schommer C., Palatnik J.F., Aggarwal P., Chetelat A., Cubas P., Farmer E.E., Nath U., Weigel D. Control of Jasmonate Biosynthesis and Senescence by miR319 Targets. PLoS Biol. 2008;6:e230. doi: 10.1371/journal.pbio.0060230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu L., Xu W., Hu X.S., Liu H.J., Lin Y.J. W-box and G-box elements play important roles in early senescence of rice flag leaf. Sci. Rep. 2016;6:20881. doi: 10.1038/srep20881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berger F., Ramirez-Hernandez M.H., Ziegler M. The new life of a centenarian: Signalling functions of NAD(P) Trends Biochem. Sci. 2004;29:111–118. doi: 10.1016/j.tibs.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 58.Noctor G., Queval G., Gakiere B. NAD(P) synthesis and pyridine nucleotide cycling in plants and their potential importance in stress conditions. J. Exp. Bot. 2006;57:1603–1620. doi: 10.1093/jxb/erj202. [DOI] [PubMed] [Google Scholar]
- 59.Hashida S.N., Takahashi H., Uchimiya H. The role of NAD biosynthesis in plant development and stress responses. Ann. Bot. 2009;103:819–824. doi: 10.1093/aob/mcp019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katoh A., Hashimoto T. Molecular biology of pyridine nucleotide and nicotine biosynthesis. Front. Biosci. 2004;9:1577–1586. doi: 10.2741/1350. [DOI] [PubMed] [Google Scholar]
- 61.Wang G., Pichersky E. Nicotinamidase participates in the salvage pathway of NAD biosynthesis in Arabidopsis. Plant J. 2007;49:1020–1029. doi: 10.1111/j.1365-313X.2006.03013.x. [DOI] [PubMed] [Google Scholar]
- 62.Pandey R., Muller A., Napoli C.A., Selinger D.A., Pikaard C.S., Richards E.J., Bender J., Mount D.W., Jorgensen R.A. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002;30:5036–5055. doi: 10.1093/nar/gkf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang H., Zhao Y., Zhou D.X. Rice NAD+-dependent histone deacetylase OsSRT1 represses glycolysis and regulates the moonlighting function of GAPDH as a transcriptional activator of glycolytic genes. Nucleic Acids Res. 2017;45 doi: 10.1093/nar/gkx825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feller U., Fischer A. Nitrogen metabolism in senescing leaves. Crit. Rev. Plant Sci. 1994;13:241–273. doi: 10.1080/07352689409701916. [DOI] [Google Scholar]
- 65.Ansari M.I., Chen S.C.G. Biochemical characterization of gamma-aminobutyric acid (GABA):Pyruvate transaminase during rice leaf senescence. Int. J. Integr. Biol. 2009;6:27–32. [Google Scholar]
- 66.Ishizaki T., Ohsumi C., Totsuka K., Igarashi D. Analysis of glutamate homeostasis by overexpression of Fd-GOGAT gene in Arabidopsis thaliana. Amino Acids. 2010;38:943–950. doi: 10.1007/s00726-009-0303-2. [DOI] [PubMed] [Google Scholar]
- 67.Bown A.W., Shelp B.J. The metabolism and function of gamma-aminobutyric acid. Plant Physiol. 1997;115:1–5. doi: 10.1104/pp.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Svitkina T.M., Borisy G.G. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eitzen G., Wang L., Thorngren N., Wickner W. Remodeling of organelle-bound actin is required for yeast vacuole fusion. J. Cell Biol. 2002;158:669–679. doi: 10.1083/jcb.200204089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaksonen M., Sun Y., Drubin D.G. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/S0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- 71.Mathur J., Mathur N., Kernebeck B., Hulskamp M. Mutations in actin-related proteins 2 and 3 affect cell shape development in Arabidopsis. Plant Cell. 2003;15:1632–1645. doi: 10.1105/tpc.011676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen J.L., Lacomis L., Erdjument-Bromage H., Tempst P., Stamnes M. Cytosol-derived proteins are sufficient for Arp2/3 recruitment and ARF/coatomer-dependent actin polymerization on Golgi membranes. FEBS Lett. 2004;566:281–286. doi: 10.1016/j.febslet.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 73.Zhang C., Mallery E.L., Schlueter J., Huang S., Fan Y., Brankle S., Staiger C.J., Szymanski D.B. Arabidopsis SCARs function interchangeably to meet actin-related protein 2/3 activation thresholds during morphogenesis. Plant Cell. 2008;20:995–1011. doi: 10.1105/tpc.107.055350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Engel N., van den Daele K., Kolukisaoglu U., Morgenthal K., Weckwerth W., Parnik T., Keerberg O., Bauwe H. Deletion of glycine decarboxylase in Arabidopsis is lethal under nonphotorespiratory conditions. Plant Physiol. 2007;144:1328–1335. doi: 10.1104/pp.107.099317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang A.H., Lu Q.T., Yan Y., Ding S.H., Wen X.G., Lu C.M. Comparative proteomic analysis provides new insights into the regulation of carbon metabolism during leaf senescence of rice grown under field conditions. J. Plant Physiol. 2010;167:1380–1389. doi: 10.1016/j.jplph.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 76.Zeng D.L., Tian Z.X., Rao Y.C., Dong G.J., Yang Y.L., Huang L.C., Leng Y.J., Xu J., Sun C., Zhang G.H., et al. Rational design of high-yield and superior-quality rice. Nat. Plants. 2017;3:17031. doi: 10.1038/nplants.2017.31. [DOI] [PubMed] [Google Scholar]
- 77.Yuan L.P. Development of hybrid rice to ensure food security. Rice Sci. 2014;21:1–2. doi: 10.1016/S1672-6308(13)60167-5. [DOI] [Google Scholar]
- 78.Thomas H., Ougham H. The stay-green trait. J. Exp. Bot. 2014;65:3889–3900. doi: 10.1093/jxb/eru037. [DOI] [PubMed] [Google Scholar]