Abstract
Fibrinogen is a highly pleiotropic protein that is involved in the final step of the coagulation cascade, wound healing, inflammation, and angiogenesis. Heterozygous mutations in Aα, Bβ, or γ fibrinogen-chain genes (FGA, FGB, FGG) have been described as being responsible for fibrinogen deficiencies (hypofibrinogenemia, hypo-dysfibrinogenemia, dysfibrinogenemia) and for more rare conditions, such as fibrinogen storage disease and hereditary renal amyloidosis. Instead, biallelic mutations have been associated with afibrinogenemia/severe hypofibrinogenemia, i.e., the severest forms of fibrinogen deficiency, affecting approximately 1–2 cases per million people. However, the “true” prevalence for these conditions on a global scale is currently not available. Here, we defined the mutational burden of the FGA, FGB, and FGG genes, and estimated the prevalence of inherited fibrinogen disorders through a systematic analysis of exome/genome data from ~140,000 individuals belonging to the genome Aggregation Database. Our analysis showed that the world-wide prevalence for recessively-inherited fibrinogen deficiencies could be 10-fold higher than that reported so far (prevalence rates vary from 1 in 106 in East Asians to 24.5 in 106 in non-Finnish Europeans). The global prevalence for autosomal-dominant fibrinogen disorders was estimated to be ~11 in 1000 individuals, with heterozygous carriers present at a frequency varying from 3 every 1000 individuals in Finns, to 1–2 every 100 individuals among non-Finnish Europeans and Africans/African Americans. Our analysis also allowed for the identification of recurrent (i.e., FGG-p.Ala108Gly, FGG-Thr47Ile) or ethnic-specific mutations (e.g., FGB-p.Gly103Arg in Admixed Americans, FGG-p.Ser245Phe in Africans/African Americans).
Keywords: fibrinogen, afibrinogenemia, hypofibrinogenemia, dysfibrinogenemia, hepatic fibrinogen storage disease, hereditary renal amyloidosis, FGA, FGB, FGG, exome-based epidemiology
1. Introduction
Fibrinogen is a 340-kDa glycoprotein that plays a crucial role in the hemostatic cascade, being the substrate for fibrin clot formation and the support for platelet aggregation [1,2,3]. It is also essential for several other biological functions, such as wound healing, inflammation, and angiogenesis [4,5,6].
Into the circulation, thrombin cleaves fibrinopeptides A and B to convert fibrinogen into fibrin, which spontaneously polymerizes, and thus forms double-stranded protofibrils, which, in turn, assemble into fibers, ultimately leading to the fibrin clot [7]. In hepatocytes—the primary site of fibrinogen biosynthesis—the molecule is rapidly assembled in the endoplasmic reticulum (ER), and secreted as a hexamer composed of two sets of three homologous polypeptide chains (called Aα, Bβ, and γ) [8,9,10,11]. These are encoded by paralogous genes (FGA, FGB, and FGG), which are clustered in a 50-kb region on chromosome 4 (4q31.3–q32.1) [12].
Monoallelic and biallelic mutations in FGA, FGB, and FGG genes are associated with different inherited conditions, reflecting the pleiotropic function of the fibrinogen protein [13]. Congenital fibrinogen defects are conventionally classified on the basis of plasma concentration as quantitative (type I) and qualitative (type II) deficiencies [14,15,16]. Quantitative deficiencies include afibrinogenemia/severe hypofibrinogenemia (Online Mendelian Inheritance in Man (OMIM) #202400; [17]) and hypofibrinogenemia (OMIM +134820, *134830, *134850), which are characterized by the lack/extremely low or by reduced amounts of immunoreactive fibrinogen (<150–160 mg/dL), leading to hemorrhagic manifestations, which can vary from very mild to life threatening. Qualitative deficiencies comprise dysfibrinogenemia (OMIM +134820, *134830, *134850) and hypo-dysfibrinogenemia, and are characterized by a discrepancy between antigen levels and functional (abnormally low) activity. Patients that are diagnosed with these conditions are either asymptomatic, or can suffer from bleeding symptoms, thrombophilia, or even both [15].
Hypofibrinogenemia and afibrinogenemia have long been considered as different clinical entities. Indeed, they represent the phenotypic expression of the same quantitative trait (i.e., diminished plasma fibrinogen level), which is determined, respectively, by heterozygous or homozygous/combined heterozygous mutations that are affecting one of the fibrinogen genes. As for dysfibrinogenemias and hypodysfibrinogenemias, these disorders are usually inherited as an autosomal dominant trait: they are caused by a single genetic defect ultimately affecting a functional property of the fibrinogen protein, such as the release (impaired or delayed) of fibrinopeptides A and B, defective polymerization, crosslinking, or thrombin binding, as well as delayed plasmin digestion [16].
Rare hypofibrinogenemic patients can present with liver disease due to the accumulation of mutant fibrinogens within hepatocytes. This condition is called fibrinogen storage disease (FSD), and is generally caused by heterozygous mutations, leading to an impaired secretion of the abnormal fibrinogen, which however maintains its capacity for polymerization and spontaneously aggregates in hepatocellular ER. In the vast majority of cases, mutations leading to FSD are missense variants that are located in a defined region of the C-terminal γ chain (residues 284–375) [18,19]. FSD-causing mutation carriers show a great variability in the severity of liver injury, going from the lack of symptoms to severe liver fibrosis/cirrhosis; more severe manifestations can be secondary to xenobiotic intake (e.g., estrogen therapy, alcohol abuse), to viral infections, or even to cancer [18,20].
Heterozygous mutations, which are affecting a small region of the C-terminal portion of the Aα chain, have been described in patients with hereditary renal amyloidosis (HRA) (OMIM +134820). These genetic defects are associated with a mild decrease in fibrinogen levels and are supposed to destabilize the native fold of circulating Aα chain degradation peptides, so that they spontaneously aggregate into amyloid fibrils, prevalently in the glomeruli of the kidneys, and, to a lesser extent, in heart muscle, spleen, and liver. Fibrinogen amyloidosis is the most common form of HRA, and clinical symptoms include hypertension, proteinuria, and azotemia [21].
The frequency of congenital fibrinogen disorders in the general population is very low. International registries, such as those from the United States, Italy, Iran, and the United Kingdom, suggest that afibrinogenemia is one of the rarest among rare bleeding disorders, with only 1–2 cases per million people [22]; however, these registries lack prospective and systematic evaluations that could lead to incidence/prevalence determination. In addition, “true” incidence/prevalence estimates for a-, hypo-, and dysfibrinogenemia are made difficult, because many patients are asymptomatic [15,22].
With these premises, we here defined the global mutational landscape of FGA, FGB, and FGG, and tried to determine ethnic-specific prevalence of inherited fibrinogen disorders, by analyzing exome and genome data from almost 140,000 individuals available through the publicly available genome Aggregation Database (gnomAD) resource [23,24].
2. Results
2.1. Global Mutational Spectrum of the Fibrinogen Gene Cluster Using Population-Based Exome- and Genome-Sequencing Data
High-quality variants were collected from the gnomAD database in subjects of different ethnicities, i.e., Africans and African Americans (12,020 subjects), admixed Americans (17,210), Ashkenazi Jewish (5076), East Asians (9435), Finnish (12,897), non-Finnish Europeans (63,369), South Asians (15,391), and other ethnicities (population not assigned; 3234 subjects).
Among the variants reported in the repository with high confidence, we retrieved all of the null mutations (frameshifts, nonsense, and splicing variants affecting the first two and the last two intronic nucleotides), the non-synonymous variants annotated as damaging by seven out of seven prediction programs, as well as splicing defects mapping in intronic positions −3 and +3/+6 and predicted as disrupting by three out of three prediction algorithms.
We found a total of 1524 heterozygous variants (287 in FGA, 233 in FGB, 1004 in FGG) in a maximum of 277,080 different alleles, corresponding to a total of 189 unique mutations (79 in FGA, 53 in FGB, 57 in FGG) (Table 1). Among these unique mutations, 56 (29.6%) have already been reported in fibrinogen-disorder related databases. As for the 133 mutations that were not described in the literature, in Supplementary Table S1 we listed all of the null frameshift/nonsense mutations, whereas in Supplementary Table S2, we reported all of the missense/splicing/inframe-indels mutations consistently annotated as damaging by prediction programs.
Table 1.
Mutational landscape of the three fibrinogen genes.
| Gene | Type of Mutation | Number of gnomAD Mutations | Number of gnomAD Mutations Already Reported in Fibrinogen Disorder Databases 1 | |
|---|---|---|---|---|
| FGA | Frameshift | 17 | 5 | 4a, 1d/h |
| Nonsense | 20 | 11 | 9a, 2a/h | |
| Splicing | 6 | 2 | 2a | |
| Missense | 30 | 10 | 6d, 1h, 3m | |
| Inframe indels | 6 | 2 | 2a | |
| Total | 79 | 30 | ||
| FGB | Frameshift | 4 | 0 | - |
| Nonsense | 5 | 3 | 2h, 1a/h | |
| Splicing | 3 | 1 | 1h | |
| Missense | 41 | 7 | 1a, 2d, 4h | |
| Inframe indels | 0 | 0 | - | |
| Total | 53 | 11 | ||
| FGG | Frameshift | 6 | 1 | 1a |
| Nonsense | 4 | 1 | 1a | |
| Splicing | 7 | 1 | 1a | |
| Missense | 39 | 12 | 8d, 3h, 1d/h | |
| Inframe indels | 1 | 0 | - | |
| Total | 57 | 15 | ||
1 We interrogated: the Human Fibrinogen Database; the Human Gene Mutation Database (HGMD); the Expert Protein Analysis System (ExPASy); and the ClinVar resource (see Section 4). The phenotype associated with already reported mutations is also reported: a = afibrinogenemia, d = dysfibrinogenemia, h = hypofibrinogenemia, m = amyloidosis.
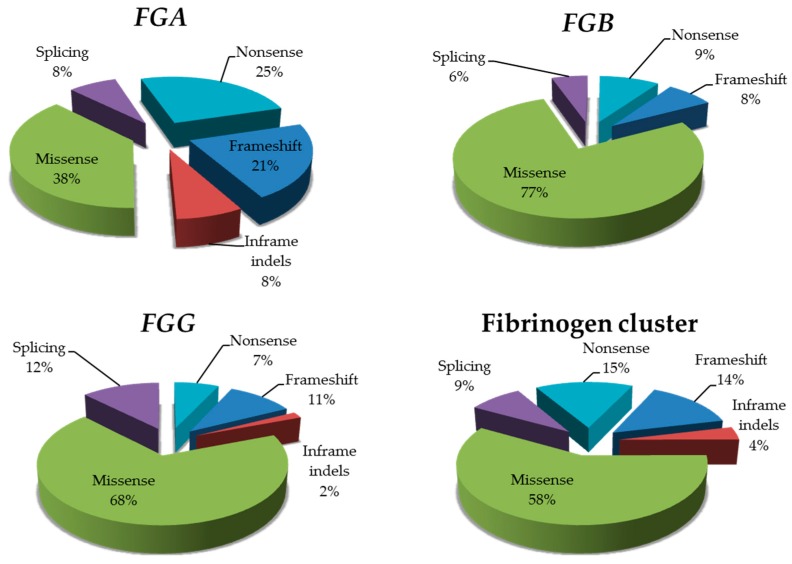
The distributions of the different types of mutations in the three genes are summarized in Figure 1: distributions were not significantly different from those reported in the Human Gene Mutation Database (HGMD) (Fisher exact test, 2-tailed p values 0.32, 0.69, and 0.83, for FGA, FGB, FGG, respectively).
Figure 1.
Distribution of point mutations in the fibrinogen genes. Pie-charts show the distribution of the different types of point mutations identified in each fibrinogen gene; an overall count is also displayed for the whole fibrinogen cluster.
2.2. Carrier Rates of Recessively-Inherited Fibrinogen Deficiencies Using Population-Based Exome- and Genome-Sequencing Data
At first, we performed prevalence calculations for the autosomal-recessive fibrinogen deficiency forms by considering separately the FGA, FGB, and FGG genes (Table 2). In general, our analysis revealed prevalence data profoundly different among the three genes, as well as among different populations. In fact, global prevalence rates increase from 0.31 per 106 individuals for the FGA gene, to 0.69 per 106 individuals for FGB, up to the remarkable prevalence of 9.54 per 106 individuals for the FGG gene. Autosomal-recessive fibrinogen deficiency appears to be virtually absent in Ashkenazi Jews (no mutations found in FGA and FGB; prevalence for FGG of 1.64 per 106 individuals), whereas, non-Finnish Europeans have a predicted exceptional rate of 23.05 per 106 individuals for the sole FGG gene.
Table 2.
Estimated prevalence of recessively-inherited fibrinogen deficiencies by ethnicity and gene.
| Gene | Population | Total Number of Alleles 1 | Total Number of Variants 2 | Collective Frequency of Variants | Heterozygote Frequency | Prevalence in 106 Individuals |
|---|---|---|---|---|---|---|
| FGA | All | 277,108 | 154 | 0.000556 | 0.00111 | 0.31 |
| Africans and African Americans | 24,030 | 9 | 0.000374 | 0.000749 | 0.14 | |
| Admixed Americans | 34,414 | 41 | 0.00119 | 0.00238 | 1.42 | |
| Ashkenazi Jewish | 10,148 | 0 | - | - | - | |
| East Asians | 18,868 | 8 | 0.000424 | 0.000848 | 0.18 | |
| Finnish | 25,794 | 0 | - | - | - | |
| Europeans (not Finnish) | 126,628 | 67 | 0.000529 | 0.00106 | 0.28 | |
| South Asians | 30,782 | 21 | 0.000682 | 0.00136 | 0.47 | |
| Other ethnicities | 6462 | 8 | 0.00124 | 0.00248 | 1.53 | |
| FGB | All | 277,144 | 231 | 0.000833 | 0.00167 | 0.69 |
| Africans and African Americans | 24,032 | 11 | 0.000458 | 0.000915 | 0.21 | |
| Admixed Americans | 34,418 | 39 | 0.00113 | 0.00226 | 1.28 | |
| Ashkenazi Jewish | 10,152 | 0 | - | - | - | |
| East Asians | 18,868 | 12 | 0.000636 | 0.00127 | 0.40 | |
| Finnish | 25,790 | 4 | 0.000155 | 0.000310 | 0.024 | |
| Europeans (not Finnish) | 126,646 | 137 | 0.00108 | 0.00216 | 1.17 | |
| South Asians | 30,782 | 23 | 0.000747 | 0.00149 | 0.56 | |
| Other ethnicities | 6466 | 5 | 0.000773 | 0.00155 | 0.60 | |
| FGG | All | 277,080 | 856 | 0.00309 | 0.00618 | 9.54 |
| Africans and African Americans | 24,034 | 64 | 0.00266 | 0.00532 | 7.09 | |
| Admixed Americans | 34,394 | 56 | 0.00163 | 0.00326 | 2.65 | |
| Ashkenazi Jewish | 10,150 | 13 | 0.00128 | 0.00256 | 1.64 | |
| East Asians | 18,854 | 13 | 0.000689 | 0.00138 | 0.47 | |
| Finnish | 25,790 | 40 | 0.00155 | 0.00310 | 2.40 | |
| Europeans (not Finnish) | 126,630 | 608 | 0.00480 | 0.00960 | 23.05 | |
| South Asians | 30,782 | 43 | 0.00140 | 0.00280 | 1.95 | |
| Other ethnicities | 6462 | 19 | 0.00294 | 0.00588 | 8.65 |
Carrier rates were estimated using the gnomAD database, including all null mutations, plus missense variants predicted to be deleterious by seven of seven prediction software, plus splicing defects located at intronic positions −3 and +3/+6 and predicted as disrupting by three of three algorithms (see Section 4). 1 Discrepancies in the number of alleles are due to slight differences in the number of individuals successfully sequenced for each specific region; 2 Mutations that were reported in fibrinogen-related databases as associated with dysfibrinogenemia, hypodysfibrinogenemia, and amyloidosis were not included.
Then, we performed prevalence calculations collapsing together the rates relative to the three genes. Table 3 shows the world-wide prevalence rate, as well as the prevalence calculated for the different populations. In particular, the world-wide prevalence was ~10 fold higher than that reported so far (i.e., 1 in 105 rather than 1 in 106); Europeans showed the highest prevalence (~2.5 in 105), whereas Ashkenazi Jews and East Asians the lowest (~1.6 in 106 and ~1.1 in 106, respectively).
Table 3.
Estimated global prevalence of recessively-inherited fibrinogen deficiencies by ethnicity.
| Population | Prevalence in 106 Individuals |
|---|---|
| All | 10.54 |
| Africans and African Americans | 7.44 |
| Admixed Americans | 5.35 |
| Ashkenazi Jewish | 1.64 |
| East Asians | 1.05 |
| Finnish | 2.42 |
| Europeans (not Finnish) | 24.5 |
| South Asians | 2.98 |
| Other ethnicities | 10.78 |
Carrier rates calculated for the three fibrinogen genes (Table 2) were summed to estimate global carrier rates in different populations.
Finally, the interrogation of the gnomAD database allowed for us to point out recurrent and ethnic-specific mutations (Table 4). Among them, two mutations that were located in FGG were extremely frequent in the analyzed populations, i.e., p.Ala108Gly and p.Thr47Ile. Both of the mutations have been described in the literature as being causative of hypofibrinogenemia and associated with a mild bleeding tendency [25,26,27]. In particular, the p.Ala108Gly variant (also known as Ala82Gly, fibrinogen Dunedin) accounts for the totality of mutated alleles in Ashkenazi Jews, and represents >70% of all mutated alleles among Europeans (for a total of 29 heterozygous individuals among 25,790 Finns, and 456 heterozygous individuals among 126,630 non-Finnish Europeans).
Table 4.
Most frequent mutations causing recessively-inherited fibrinogen deficiencies by ethnicity and gene.
| Gene | Population | RefSeqID | Genomic Position on Chr 4 1 | Type of Mutation | Consequence 2 | % of All Mutated Alleles in the Population 3 | Allele Freq in gnomAD Database |
|---|---|---|---|---|---|---|---|
| FGA | Admixed Americans | rs773619297 | 155,506,927 | frameshift | p.Gly552AlafsTer16 | 24.4 | 0.00029 |
| rs748106542 | 155,508,067 | missense | p.Asp172Asn | 29.3 | 0.00036 | ||
| Europeans (not Finnish) | rs773619297 | 155,506,927 | frameshift | p.Gly552AlafsTer16 | 11.9 | 0.000063 | |
| rs146387238 | 155,508,663 | splice donor | c.510+1G>T * | 17.9 | 0.000094 | ||
| FGB | Admixed Americans | rs774502903 | 155,487,641 | missense | p.Gly103Arg | 41.0 | 0.00047 |
| Europeans (not Finnish) | rs370703973 | 155,489,611 | missense | p.Tyr266Cys * | 51.1 | 0.00055 | |
| rs777451745 | 155,490,421 | missense | p.Ala307Val * | 10.2 | 0.00011 | ||
| South Asians | rs762523152 | 155,490,854 | missense | p.Leu383Val | 26.1 | 0.00019 | |
| rs765571602 | 155,491,770 | missense | p.Met482Val | 34.8 | 0.00026 | ||
| FGG | Africans and African Americans | rs145051028 | 155,529,735 | missense | p.Ser245Phe | 54.7 | 0.0015 |
| rs148685782 | 155,533,035 | missense | p.Ala108Gly * | 28.1 | 0.00075 | ||
| Admixed Americans | rs776288074 | 155,530,871 | missense | p.Tyr193His | 35.7 | 0.00058 | |
| rs148685782 | 155,533,035 | missense | p.Ala108Gly * | 19.6 | 0.00032 | ||
| rs138511699 | 155,533,337 | missense | p.Thr47Ile * | 23.2 | 0.00038 | ||
| Ashkenazi Jewish | rs148685782 | 155,533,035 | missense | p.Ala108Gly * | 100 | 0.0013 | |
| Finnish | rs148685782 | 155,533,035 | missense | p.Ala108Gly * | 72.5 | 0.0011 | |
| rs138511699 | 155,533,337 | missense | p.Thr47Ile * | 27.5 | 0.00043 | ||
| Europeans (not Finnish) | rs148685782 | 155,533,035 | missense | p.Ala108Gly * | 75.0 | 0.0036 | |
| rs138511699 | 155,533,337 | missense | p.Thr47Ile * | 15.0 | 0.00074 | ||
| South Asians | rs148685782 | 155,533,035 | missense | p.Ala108Gly * | 27.9 | 0.00039 | |
| rs138511699 | 155,533,337 | missense | p.Thr47Ile * | 46.5 | 0.00066 |
1 Numbering according to UCSC Genome Browser, human, February 2009 (GRCh37/hg19) assembly. 2 The consequences on the native-protein numbering are reported. Mutations reported in fibrinogen-related databases (Human Fibrinogen Database, HGMD, ExPASy, ClinVar) are indicated with an asterisk. 3 Frequencies relative to the corresponding gene. The highly-recurrent FGG p.Ala108Gly and FGG p.Thr47Ile mutations are respectively depicted in bold and underlined.
2.3. Global Carrier Rates of Autosomal-Dominant Fibrinogen Disorders Using Population-Based Exome- and Genome-Sequencing Data
Apart from afibrinogenemia/severe hypofibrinogenemia, all of the fibrinogen disorders are transmitted in an autosomal dominant fashion, and are characterized by the presence of one causative mutation involving any of the three fibrinogen-chain genes. Hence, we performed a global prevalence calculation for such disorders by clumping together the contribution of FGA, FGB, and FGG.
Our analysis estimated that heterozygous subjects should be present in the general world-wide population, at a frequency of ~1 every 100 individuals (range: 0.3 to 1.5 every 100 individuals), suggesting that fibrinogen-related disorders represents a far more frequent condition in the population than believed so far (Table 5).
Table 5.
Estimated prevalence of autosomal-dominant fibrinogen disorders by ethnicity.
| Population | Total Number of Alleles 1 | Total Number of Variants | Collective Frequency of Variants | Heterozygote Frequency | Prevalence in 103 Individuals |
|---|---|---|---|---|---|
| All | 277,144 | 1524 | 0.0055 | 0.011 | 11 |
| Africans and African Americans | 24,036 | 166 | 0.0069 | 0.014 | 14 |
| Admixed Americans | 34,418 | 176 | 0.0051 | 0.010 | 10 |
| Ashkenazi Jewish | 10,152 | 44 | 0.0043 | 0.0087 | 9 |
| East Asians | 18,868 | 35 | 0.0019 | 0.0037 | 4 |
| Finnish | 25,794 | 44 | 0.0017 | 0.0034 | 3 |
| Non-Finnish Europeans | 126,646 | 922 | 0.0073 | 0.015 | 15 |
| South Asians | 30,782 | 90 | 0.0029 | 0.0058 | 6 |
| Other ethnicities | 6466 | 47 | 0.0073 | 0.015 | 15 |
Carrier rates were estimated using the gnomAD database, including all null mutations, plus missense variants predicted to be deleterious by 7 of 7 prediction software, plus splicing defects located at intronic positions −3 and +3/+6 and predicted as disrupting by 3 of 3 algorithms (see Section 4). 1 Discrepancies in the number of alleles are due to slight differences in the number of individuals successfully sequenced for each specific region.
Finally, we specifically searched the gnomAD repository for the presence of mutations that were known to be frequently found in patients affected by dysfibrinogenemia [28], FSD [18], or HRA [29] (Table 6). No FSD-causing mutation was found in database, and only eight heterozygous individuals were carrier of the well-known mutation hotspots FGA-p.Arg35Cys/His or FGG-p.Arg301His (these mutations account for 85% of all known cases of congenital dysfibrinogenemia) [28]. As for HRA-causing mutations, we found just one heterozygous carrier of the FGA-p.Glu545Val variant (i.e., the most common HRA variant) [29], but we observed a total of 29 carriers of p.Glu545Lys, i.e., a mutation less-frequently associated with HRA, but involving the same FGA p.Glu545 residue. Interestingly, 19 of 29 p.Glu545Lys carriers were of Ashkenazi Jewish origin, so that the calculated prevalence of heterozygotes for this mutation in this population reaches the remarkable rate of 3.74 in 103 individuals.
Table 6.
World-wide frequency of dysfibrinogenemia-, fibrinogen storage disease (FSD)-, and hereditary renal amyloidosis (HRA)-causing hotspot mutations.
| Gene | Disorder | Mutation (Legacy Name) | Mutation (gnomAD) | Number of Mutated Alleles 1 |
|---|---|---|---|---|
| FGA | dysfibrinogenemia | Aα(16)Arg > Cys | p.Arg35Cys | 1 |
| dysfibrinogenemia | Aα(16)Arg > His | p.Arg35His | 5 | |
| HRA | Aα(526)Glu > Lys | p.Glu545Lys | 29 | |
| HRA | Aα(526)Glu > Val | p.Glu545Val | 1 | |
| FGG | dysfibrinogenemia | γ(275)Arg > His | p.Arg301His | 2 |
Hotspot mutations were searched in the gnomAD database. 1 The overall world-wide population was considered (277,144 total alleles).
3. Discussion
With the exception of the involvement of some peculiar molecular mechanism, such as uniparental isodisomy of the entire chromosome 4 [30], the genetic bases of fibrinogen disorders are invariably constituted by homozygous/heterozygous mutations within the fibrinogen gene cluster. Here, we took advantage of exome and genome data of ~140,000 individuals to estimate the prevalence of recessively-inherited fibrinogen deficiency, as well as the collective prevalence of all the dominantly-inherited fibrinogen disorders. Our estimates indicated that: (i) the world-wide prevalence for recessively-inherited fibrinogen deficiencies could be 10-fold higher than that reported so far; (ii) prevalence among different populations seems to be extremely different (ranging from 1 in 106 in East Asians up to 24.5 in 106 in non-Finnish Europeans); and, (iii) heterozygous carriers of mutations in the fibrinogen cluster (i.e., individuals possibly at risk to develop a form of fibrinogen disorder, as well as asymptomatic/undiagnosed subjects) should be present in the general world-wide population at a frequency of ~1 every 100 individuals.
Notably, we have to acknowledge that our estimates suffer from some important limitations. First, our prevalence calculations relied on the use of prediction programs aimed at evaluating the deleteriousness of missense/splicing mutations with unknown biological significance. In this respect, we have to notice that, although these algorithms can present limitations [31,32], their use currently represents a standard approach, especially if researchers have to deal with data from large-scale sequencing projects. Indeed, it has been demonstrated that different methods can individually show a limited overall predictive value, which, however, increases significantly when considering only concordant outputs from different software (e.g., it has been calculated an encouraging predictive value of ~90% when taking into account concordant results from four different prediction methods) [31]. We hence based our prevalence calculations on the use of seven different prediction software for missense variants, and of three programs for splicing variants. This choice proved to be quite “conservative”: for instance, excluding from prevalence calculations all of the variants that were never reported in fibrinogen-related databases, we observed a global prevalence rates for recessively-inherited fibrinogen deficiencies of 6.8 in 106 individuals (0.033 in 106 individuals for the FGA gene, 0.16 in 106 for FGB, 6.68 in 106 for FGG). Importantly, the good performance of our in-silico approach is also testified by predictions performed on fibrinogen missense variants reported as associated with fibrinogen disorders in the databases: we observed an overall prediction rate for the fibrinogen cluster of 78% (if considering all together concordant predictions from seven of seven and six of seven algorithms). The only exception pertained the nine FGA mutations that were described as cause of amyloidosis: none of them was predicted as damaging neither by 7 of 7 nor by 6 of 7 algorithms. This was probably because all these variants involve a peculiar trait of the Aα chain, i.e., the C-terminal unstructured tail of the protein.
Second, we could in theory have underestimated the total number of causative mutations for systematic bias still characterizing exome-sequencing data. For instance, promoters and intronic regions are not included by design in exome sequencing, so that variants located in such regions can easily go undetected. Insertions and deletions are not always correctly recognized by variant-calling programs, so they can go unnoticed as well. More importantly, gross deletions, and rearrangements may be not detected at all. This represents a substantial problem in the calculation of prevalence for fibrinogen deficiencies, since one of the most recurrent mutation for these disorders is the well-known 11-kb deletion, eliminating the majority of the FGA gene [16,33]. This deletion has been reported in at least eight afibrinogenemic patients [34], and, together with other two gross deletions that are described in the FGA gene (i.e., a 15-kb and a 4.1-kb deletion) [30,35], account for ~9% of all cases of afibrinogenemia characterized by mutations in FGA [34].
The high prevalence rates that we estimated from the gnomAD repository are indeed not completely unexpected. The most recent World Federation of Hemophilia (WFH) annual global survey, which was conducted in 2015, found that inherited fibrinogen deficiencies on a global scale account for 1777 of 304,362 inherited bleeding disorders (0.6%) [36], confirming fibrinogen deficiencies as quite rare disorders, and approximately 85 times less common than hemophilia A (these data are based on questionnaires sent to national hemophilia associations linked with the Federation). However, digging deeper in the WFH data, it becomes clear that prevalence data on fibrinogen deficiencies are somehow underestimated, at least in some populations. For instance, though it is not clear if data reported concern autosomal-recessive, autosomal-dominant, or both forms of the fibrinogen deficiency, it is possible to calculate exceptional prevalence rates of 8.7 and 14.3 per million people for the United Kingdom and the Slovak Republic, respectively. Concerning Italy, a country with a population that is comparable to that of the United Kingdom (~60 million people), no data are reported on fibrinogen deficiencies in the WFH 2015 survey. However, in the “Human fibrinogen database”, a total of 104 fibrinogen-deficient Italian cases are described (54 coming from our center). This figure alone is sufficient to suggest higher prevalence rates for fibrinogen-related disorders than those reported so far, especially if considering that the database has a clear bias towards published data. To have a better idea of the Italian situation, we took advantage of our whole-exome dataset [37], which was composed of exomes of 1750 healthy controls (80% males, age < 45 years, no history of thromboembolic disease). Sequence coverage of fibrinogen cluster was optimal in all individuals, and allowed us to retrieve a total of 24 variants, corresponding to eight different mutations (one frameshift mutation and seven missense damaging variants; distribution: one variant in FGA, two in FGB, 5 in FGG). Once again, the burden of mutations affecting the FGG gene appears to be the highest, with just 1 mutation (p.Ala108Gly) considerably driving the prevalence rates for recessively-inherited fibrinogen deficiencies (0.08 per 106 individuals for the FGA gene, 0.33 per 106 individuals for FGB, and an exceptional 36 per 106 individuals for the FGG gene) (Supplementary Table S3). Interestingly, the “driving effect” that was exerted by the p.Ala108Gly mutation could also be at the basis of the marked differences in prevalence rates that were observed in different populations (from 1 in 106 in East Asians to 24.5 in 106 in non-Finnish Europeans). In particular, Ivaskevicius and colleagues [38] reported that the p.Ala108Gly mutation is associated with a specific haplotype, hence denoting a single, ancestral event (founder effect). Given the observed frequencies of the mutation in the different ethnic groups (Table 4), one could speculate that the ancestral mutation event could have originated in Africans (before the major divergence of non-African populations), and that, subsequently, the mutation could have spread towards Europe and Asia following human migrations [39]. However, with the last divergence between South and East Asians, it is conceivable that the p.Ala108Gly mutation could have not reached the Far East.
The issue related to the fundamental contribution of the p.Ala108Gly mutation to prevalence estimates should be carefully kept in mind. In fact, if we do not consider this variant at all, the prevalence rates for recessively-inherited fibrinogen disorders would dramatically drop (on a global scale prevalence would be 3.2 per million people), with highest values that would be registered in Africans/Africans Americans (i.e., 4 per million people) and the lowest in Ashkenazi Jews (for this population, recessively-inherited fibrinogen deficiencies would be virtually absent). The same dramatic drop would be registered for autosomal-dominant fibrinogen disorders (Supplementary Table S4). The p.Ala108Gly mutation (legacy name γAla82Gly) was repeatedly reported as being associated with moderately-decreased fibrinogen levels and with mild bleeding tendency [25,26,34]. In addition, in a recent meta-analysis aimed at identifying loci for fibrinogen concentration, the p.Ala108Gly allele clearly emerged among the strongest predictors of decreased fibrinogen levels (β = −0.2179; p = 4.0 × 10−82) [40]. Importantly, Ivaskevicius and colleagues [38], by screening 616 blood donors, already observed the p.Ala108Gly as a common FGG variant in Caucasians, thus calculating an allele frequency of 0.0032 and a frequency for homozygous individuals of 1 in 95,000. These data well reconcile with those calculated using the gnomAD database.
Our data raise the problem of why such remarkably high prevalence can be calculated for recessively-inherited fibrinogen deficiencies. The most likely explanation could rely on the relatively-low fibrinogen levels that are associated with the highly-prevalent p.Ala108Gly mutation, and the consequent mild/absent symptomatology characterizing many carrier/homozygous individuals (which can go unnoticed) [38]. Alternatively, we can hypothesize two additional explanations.
A first possibility could be that the p.Ala108Gly allele, at the heterozygous state, confers a selective advantage, and thus, has spread throughout the gene pool. This hypothesis strongly emerges among other genetic explanations, such as a possible genetic drift (which can be excluded due to the presence of the p.Ala108Gly allele world-wide), a potential transmission distortion (which can be left out since the Hardy-Weinberg equilibrium is perfectly respected), or a high mutation rate. This last possibility can be discounted on the basis of the above-mentioned observation that the p.Ala108Gly mutation is associated with a founder effect [38]. It remains to understand why a pro-hemorrhagic allele could represent a selective advantage; in this respect, it is worth noticing that a high fibrinogen concentration has long been recognized as a strong and established predictor of cardiovascular disease outcomes (myocardial infarction, stroke, venous thromboembolism), autoimmune disorders with an inflammatory component, as well as cancer [5,41,42,43,44]. However, most of these phenotypes are late onset and therefore are predicted to have a limited effect on natural selection; moreover, the heterozygous advantage hypothesized to explain the frequency of the factor V Leiden mutation points to the opposite direction, being related to moderate hypercoagulability [45].
Conversely, it could be plausible that we do not detect in the general population the high rate of predicted homozygous/compound heterozygous individuals because of problems that are associated with pregnancy (defects in fetal implantation in afibrinogenemic/hypofibrinogenemic women and/or embryos). It is indeed well recognized that fibrinogen has a critical role in maintaining pregnancy: at six weeks of gestation, maternal endothelial cells are replaced by cytotrophoblasts, starting a remodeling process of vessels that involves an active bleeding near the cytotrophoblastic shell, followed by the formation of the Nitabuch’s layer (a fibrinoid layer). This process is highly compromised in patients with quantitative fibrinogen defects, ultimately leading to spontaneous miscarriages [46]. This notion is well supported by different studies analyzing series of fibrinogen-deficient women, all reporting miscarriage as a frequent complication [47,48,49,50]. Further corroborating this observation, a recent study—aimed at elucidating by whole-exome sequencing the genetic etiology of recurrent pregnancy loss—reported the identification of mutations in the FGA gene as having a potential role in implantation/pregnancy biology [51]. Hence, when considering the high frequency of heterozygous carriers of fibrinogen defects in the general population, we can hypothesize the fibrinogen cluster as a future “biomarker” to be screened also for recurrent pregnancy loss.
In conclusion, in this work, we have exploited the enormous potential that is provided by public-available repositories to paint a more clear landscape of the genetic burden associated with fibrinogen genes and their related disorders. From our analysis, it clearly emerges that putative disease alleles are much more frequent than expected, a trend already observed for other genes/disorders [52,53]. Caution should be placed to interpret these data, since some of the identified variants could be non-pathogenic and some others could be not fully penetrant. Nonetheless, our analysis represents the first attempt to evaluate the prevalence rates of fibrinogen disorders in populations other than those that are coming from North America and Europe, also indicating the mutations/genes to be prioritized for genetic screenings.
4. Materials and Methods
4.1. Retrieving Variants from the GnomAD Database
For calculating prevalence of fibrinogen disorders in different ethnic groups, we retrieved data from the gnomAD database (release 2.0) [23,24], which contains exome and genome data on sequence variation in 138,632 unrelated subjects of different ethnicities (123,136 exomes and 15,496 genomes) (Table 7). These individuals are part of disease-specific and population-genetic studies; subjects suffering from pediatric diseases (as well as their first-degree relatives) are not included in the database.
Table 7.
Composition of the gnomAD database.
| Population | Number of Genomes | Number of Exomes | Total |
|---|---|---|---|
| African/African American | 4368 | 7652 | 12,020 |
| Admixed American | 419 | 16,791 | 17,210 |
| Ashkenazi Jewish | 151 | 4925 | 5076 |
| East Asian | 811 | 8624 | 9435 |
| Finnish | 1747 | 11,150 | 12,897 |
| Non-Finnish European | 7509 | 55,860 | 63,369 |
| South Asian | 0 | 15,391 | 15,391 |
| Other (population not assigned) | 491 | 2743 | 3234 |
| Total | 15,496 | 123,136 | 138,632 |
Among the genetic variants that are present in the gnomAD database, we collected and considered as deleterious:
-
(i)
nonsense and frame-shift variants;
-
(ii)
disruptive splice-site variants affecting the first two or last two intronic nucleotides of the splicing site;
-
(iii)
splice-site variants located at intronic positions −3 and +3/+6 predicted as deleterious by three out of three prediction programs. These software were: Human Splicing Finder [54], NetGene2 [55], and Splice Site Prediction by Neural Network [56]. We considered as deleterious sequence variations predicted either to completely abolish the wild-type splicing site or to determine a decrease in the prediction score to less than half of the wild-type counterpart;
-
(iv)
missense variants reported as cause of fibrinogen disorders in the following databases: the Human Fibrinogen Database (release 43) [34,57]; the public release of Human Gene Mutation Database (HGMD) and its professional version (our release dates back to 2014) [58,59]; the Expert Protein Analysis System (ExPASy) [60,61]; and the ClinVar resource [62,63]; and,
-
(v)
missense variants predicted as deleterious by seven out of seven programs. These software, all included in the dbNSFP package, were: SIFT [64], PolyPhen2 (2 software) [65], MutationTaster [66], MutationAssessor [67], Likelihood Ratio Test (LRT) [68], and Functional Analysis Through Hidden Markov Model (FATHMM) [69].
Among variants present in the gnomAD database, we excluded those:
-
-
showing low-confidence level in the variant calling step (as indicated by the gnomAD server);
-
-
being mapped in FGA exon 6 and FGG exon 10. Variants in such exons affect low-abundance mRNA isoforms of the relevant chain [14], and were never clearly associated with fibrinogen disorders; and,
-
-
being mapped in introns but annotated within alternative transcripts of the fibrinogen chains of unknown significance.
4.2. Calculations of Global Prevalence for FGA, FGB, and FGG Genes
We performed prevalence calculations for
-
-
afibrinogenemia/severe hypofibriniogenmia (autosomal recessive trait); since, we expect to have two in-trans mutations affecting the same gene, in this case, we first considered each gene separately, and subsequently we summed the FGA, FGB, and FGG prevalence for estimating the global rate. For this analysis, we excluded all of the mutations that were reported in fibrinogen-related databases, as associated with dysfibrinogenemia, hypodysfibrinogenemia, and amyloidosis; and,
-
-
all the other fibrinogen disorders together (hypofibrinogenemia, dysfibrinogenemia, hypo-dysfibrinogenemia, FSD, and HRA; all autosomal dominant); in this case, we expect just one mutation in any of the three genes, hence a “cumulative” prevalence was calculated when considering the three genes together.
Abbreviations
| ER | Endoplasmic Reticulum |
| Aα | A α Polypeptide Chain |
| Bβ | B β Polypeptide Chain |
| γ | γ Polypeptide chain |
| FGA | Fibrinogen α Chain Gene |
| FGB | Fibrinogen β Chain Gene |
| FGG | Fibrinogen γ Chain Gene |
| OMIM | Online Mendelian Inheritance in Man |
| FSD | Fibrinogen Storage Disease |
| HRA | Hereditary Renal Amyloidosis |
| gnomAD | Genome Aggregation Database |
| HGMD | Human Gene Mutation Database |
| ExPASy | Expert Protein Analysis System |
| WFH | World Federation of Hemophilia |
| LRT | Likelihood Ratio Test |
| FATHMM | Functional Analysis Through Hidden Markov Model |
Supplementary Materials
Supplementary materials can be found at www.mdpi.com/1422-0067/18/12/2711/s1.
Author Contributions
Rosanna Asselta and Stefano Duga conceived and designed the study; Elvezia Maria Paraboschi performed statistical analyses; Rosanna Asselta performed statistical analyses and wrote the manuscript draft; all authors read, discussed, and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Mosesson M.W. Fibrinogen and fibrin structure and functions. J. Thromb. Haemost. 2005;3:1894–1904. doi: 10.1111/j.1538-7836.2005.01365.x. [DOI] [PubMed] [Google Scholar]
- 2.Weisel J.W., Litvinov R.I. Mechanisms of fibrin polymerization and clinical implications. Blood. 2013;121:1712–1719. doi: 10.1182/blood-2012-09-306639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Podolnikova N.P., Yakovlev S., Yakubenko V.P., Wang X., Gorkun O.V., Ugarova T.P. The interaction of integrin αIIbβ3 with fibrin occurs through multiple binding sites in the αIIb β-propeller domain. J. Biol. Chem. 2014;289:2371–2383. doi: 10.1074/jbc.M113.518126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chester D., Brown A.C. The role of biophysical properties of provisional matrix proteins in wound repair. Matrix Biol. 2017;60–61:124–140. doi: 10.1016/j.matbio.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Davalos D., Akassoglou K. Fibrinogen as a key regulator of inflammation in disease. Semin. Immunopathol. 2012;34:43–62. doi: 10.1007/s00281-011-0290-8. [DOI] [PubMed] [Google Scholar]
- 6.Feng X., Tonnesen M.G., Mousa S.A., Clark R.A. Fibrin and collagen differentially but synergistically regulate sprout angiogenesis of human dermal microvascular endothelial cells in 3-dimensional matrix. Int. J. Cell Biol. 2013;2013:231279. doi: 10.1155/2013/231279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weisel J.W. Fibrinogen and fibrin. Adv. Protein Chem. 2005;70:247–299. doi: 10.1016/S0065-323370008-5. [DOI] [PubMed] [Google Scholar]
- 8.Roy S., Sun A., Redman C. In vitro assembly of the component chains of fibrinogen requires endoplasmic reticulum factors. J. Biol. Chem. 1996;271:24544–24550. doi: 10.1074/jbc.271.40.24544. [DOI] [PubMed] [Google Scholar]
- 9.Xu W.f., Chung D.W., Davie E.W. The assembly of human fibrinogen. The role of the amino-terminal and coiled-coil regions of the three chains in the formation of the alphagamma and betagamma heterodimers and alphabetagamma half-molecules. J. Biol. Chem. 1996;271:27948–27953. doi: 10.1074/jbc.271.44.27948. [DOI] [PubMed] [Google Scholar]
- 10.Medved L., Weisel J.W. Fibrinogen and Factor XIII Subcommittee of Scientific Standardization Committee of International Society on Thrombosis and Haemostasis. Recommendations for nomenclature on fibrinogen and fibrin. J. Thromb. Haemost. 2009;7:355–359. doi: 10.1111/j.1538-7836.2008.03242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weisel J.W., Litvinov R.I. Fibrin Formation, Structure and Properties. Subcell. Biochem. 2017;82:405–456. doi: 10.1007/978-3-319-49674-0_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.University of California Santa Cruz (UCSC) Genome Browser GRCh38/hg38 Human Genome Assembly, 2013. [(accessed on 4 July 2017)]; Available online: http://genome.ucsc.edu/
- 13.Ittisoponpisan S., Alhuzimi E., Sternberg M.J., David A. Landscape of Pleiotropic Proteins Causing Human Disease: Structural and System Biology Insights. Hum. Mutat. 2017;38:289–296. doi: 10.1002/humu.23155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asselta R., Duga S., Tenchini M.L. The molecular basis of quantitative fibrinogen disorders. J. Thromb. Haemost. 2006;4:2115–2129. doi: 10.1111/j.1538-7836.2006.02094.x. [DOI] [PubMed] [Google Scholar]
- 15.Casini A., de Moerloose P., Neerman-Arbez M. Clinical Features and Management of Congenital Fibrinogen Deficiencies. Semin. Thromb. Hemost. 2016;42:366–374. doi: 10.1055/s-0036-1571339. [DOI] [PubMed] [Google Scholar]
- 16.Neerman-Arbez M., de Moerloose P., Casini A. Laboratory and Genetic Investigation of Mutations Accounting for Congenital Fibrinogen Disorders. Semin. Thromb. Hemost. 2016;42:356–365. doi: 10.1055/s-0036-1571340. [DOI] [PubMed] [Google Scholar]
- 17.Online Mendelian Inheritance in Man (OMIM) [(accessed on 4 July 2017)]; Available online: https://www.omim.org/
- 18.Asselta R., Robusto M., Braidotti P., Peyvandi F., Nastasio S., D’Antiga L., Perisic V.N., Maggiore G., Caccia S., Duga S. Hepatic fibrinogen storage disease: Identification of two novel mutations (p.Asp316Asn, fibrinogen Pisa and p.Gly366Ser, fibrinogen Beograd) impacting on the fibrinogen γ-module. J. Thromb. Haemost. 2015;13:1459–1467. doi: 10.1111/jth.13021. [DOI] [PubMed] [Google Scholar]
- 19.Lee M.J., Venick R., Bhuta S., Li X., Wang H.L. Hepatic Fibrinogen Storage Disease in a Patient with Hypofibrinogenemia: Report of a Case with a Missense Mutation of the FGA Gene. Semin. Liver Dis. 2015;35:439–443. doi: 10.1055/s-0035-1567834. [DOI] [PubMed] [Google Scholar]
- 20.Casini A., Sokollik C., Lukowski S.W., Lurz E., Rieubland C., de Moerloose P., Neerman-Arbez M. Hypofibrinogenemia and liver disease: A new case of Aguadilla fibrinogen and review of the literature. Haemophilia. 2015;21:820–827. doi: 10.1111/hae.12719. [DOI] [PubMed] [Google Scholar]
- 21.Sivalingam V., Patel B.K. Familial mutations in fibrinogen Aα (FGA) chain identified in renal amyloidosis increase in vitro amyloidogenicity of FGA fragment. Biochimie. 2016;127:44–49. doi: 10.1016/j.biochi.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 22.Peyvandi F. Epidemiology and treatment of congenital fibrinogen deficiency. Thromb. Res. 2012;130:S7–S11. doi: 10.1016/S0049-3848(13)70004-5. [DOI] [PubMed] [Google Scholar]
- 23.The Genome Aggregation Database (GnomAD) [(accessed on 15 September 2017)]; Available online: http://gnomad.broadinstitute.org/
- 24.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brennan S.O., Fellowes A.P., Faed J.M., George P.M. Hypofibrinogenemia in an individual with 2 coding (gamma82 A-->G and Bbeta235 P-->L) and 2 noncoding mutations. Blood. 2000;95:1709–1713. [PubMed] [Google Scholar]
- 26.Wyatt J., Brennan S.O., May S., George P.M. Hypofibrinogenaemia with compound heterozygosity for two gamma chain mutations-gamma 82 Ala-->Gly and an intron two GT-->AT splice site mutation. Thromb. Haemost. 2000;84:449–452. [PubMed] [Google Scholar]
- 27.Riedelová-Reicheltová Z., Riedel T., Májek P., Kotlin R., Geierová V., Suttnar J., Dyr J.E. Abnormal fibrinogen Zlín (γThr21Ile) with missense mutation causing hypofibrinogenemia. Acta Haematol. 2014;132:140–143. doi: 10.1159/000356781. [DOI] [PubMed] [Google Scholar]
- 28.Casini A., Neerman-Arbez M., Ariëns R.A., de Moerloose P. Dysfibrinogenemia: From molecular anomalies to clinical manifestations and management. J. Thromb. Haemost. 2015;13:909–919. doi: 10.1111/jth.12916. [DOI] [PubMed] [Google Scholar]
- 29.Stangou A.J., Banner N.R., Hendry B.M., Rela M., Portmann B., Wendon J., Monaghan M., Maccarthy P., Buxton-Thomas M., Mathias C.J., et al. Hereditary fibrinogen A alpha-chain amyloidosis: Phenotypic characterization of a systemic disease and the role of liver transplantation. Blood. 2010;115:2998–3007. doi: 10.1182/blood-2009-06-223792. [DOI] [PubMed] [Google Scholar]
- 30.Spena S., Duga S., Asselta R., Peyvandi F., Mahasandana C., Malcovati M., Tenchini M.L. Congenital afibrinogenaemia caused by uniparental isodisomy of chromosome 4 containing a novel 15-kb deletion involving fibrinogen Aalpha-chain gene. Eur. J. Hum. Genet. 2004;12:891–898. doi: 10.1038/sj.ejhg.5201207. [DOI] [PubMed] [Google Scholar]
- 31.Hicks S., Wheeler D.A., Plon S.E., Kimmel M. Prediction of missense mutation functionality depends on both the algorithm and sequence alignment employed. Hum. Mutat. 2011;32:661–668. doi: 10.1002/humu.21490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jian X., Boerwinkle E., Liu X. In silico tools for splicing defect prediction: A survey from the viewpoint of end users. Genet. Med. 2014;16:497–503. doi: 10.1038/gim.2013.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neerman-Arbez M., Honsberger A., Antonarakis S.E., Morris M.A. Deletion of the fibrinogen alpha-chain gene (FGA) causes congenital afibrinogenemia. J. Clin. Investig. 1999;103:215–218. doi: 10.1172/JCI5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.The Human Fibrinogen Database. [(accessed on 15 September 2017)]; Available online: http://site.geht.org/base-de-donnees-fibrinogene/
- 35.Monaldini L., Asselta R., Duga S., Peyvandi F., Karimi M., Malcovati M., Tenchini M.L. Mutational screening of six afibrinogenemic patients: Identification and characterization of four novel molecular defects. Thromb. Haemost. 2007;97:546–551. [PubMed] [Google Scholar]
- 36.World Federation of Hemophilia Annual Global Survey 2015. [(accessed on 10 October 2017)]; Available online: http://www1.wfh.org/publication/files/pdf-1669.pdf.
- 37.Do R., Stitziel N.O., Won H.H., Jørgensen A.B., Duga S., Angelica Merlini P., Kiezun A., Farrall M., Goel A., Zuk O., et al. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature. 2015;518:102–106. doi: 10.1038/nature13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ivaskevicius V., Jusciute E., Steffens M., Geisen C., Hanfland P., Wienker T.F., Seifried E., Oldenburg J. GammaAla82Gly represents a common fibrinogen gamma-chain variant in Caucasians. Blood Coagul. Fibrinolysis. 2005;16:205–208. doi: 10.1097/01.mbc.0000164430.98169.c6. [DOI] [PubMed] [Google Scholar]
- 39.Nielsen R., Akey J.M., Jakobsson M., Pritchard J.K., Tishkoff S., Willerslev E. Tracing the peopling of the world through genomics. Nature. 2017;541:302–310. doi: 10.1038/nature21347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Vries P.S., Chasman D.I., Sabater-Lleal M., Chen M.H., Huffman J.E., Steri M., Tang W., Teumer A., Marioni R.E., Grossmann V., et al. A meta-analysis of 120 246 individuals identifies 18 new loci for fibrinogen concentration. Hum. Mol. Genet. 2016;25:358–370. doi: 10.1093/hmg/ddv454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fibrinogen Studies Collaboration. Danesh J., Lewington S., Thompson S.G., Lowe G.D., Collins R., Kostis J.B., Wilson A.C., Folsom A.R., Wu K., et al. Plasma fibrinogen level and the risk of major cardiovascular diseases and nonvascular mortality: An individual participant meta-analysis. JAMA. 2005;294:1799–1809. doi: 10.1001/jama.294.14.1799. [DOI] [PubMed] [Google Scholar]
- 42.Emerging Risk Factors Collaboration. Kaptoge S., Di Angelantonio E., Pennells L., Wood A.M., White I.R., Gao P., Walker M., Thompson A., Sarwar N., et al. C-reactive protein; fibrinogen; and cardiovascular disease prediction. N. Engl. J. Med. 2012;367:1310–1320. doi: 10.1056/NEJMoa1107477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seebacher V., Polterauer S., Grimm C., Husslein H., Leipold H., Hefler-Frischmuth K., Tempfer C., Reinthaller A., Hefler L. The prognostic value of plasma fibrinogen levels in patients with endometrial cancer: A multi-centre trial. Br. J. Cancer. 2010;102:952–956. doi: 10.1038/sj.bjc.6605547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yapijakis C., Bramos A., Nixon A.M., Ragos V., Vairaktaris E. The interplay between hemostasis and malignancy: The oral cancer paradigm. Anticancer Res. 2012;32:1791–1800. [PubMed] [Google Scholar]
- 45.Van Mens T.E., Levi M., Middeldorp S. Evolution of Factor V Leiden. Thromb. Haemost. 2013;110:23–30. doi: 10.1160/TH13-02-0115. [DOI] [PubMed] [Google Scholar]
- 46.Iwaki T., Castellino F.J. Maternal fibrinogen is necessary for embryonic development. Curr. Drug Targets. 2005;6:535–539. doi: 10.2174/1389450054546006. [DOI] [PubMed] [Google Scholar]
- 47.Al-Mondhiry H., Ehmann W.C. Congenital afibrinogenemia. Am. J. Hematol. 1994;46:343–347. doi: 10.1002/ajh.2830460416. [DOI] [PubMed] [Google Scholar]
- 48.Lak M., Keihani M., Elahi F., Peyvandi F., Mannucci P.M. Bleeding and thrombosis in 55 patients with inherited afibrinogenaemia. Br. J. Haematol. 1999;107:204–206. doi: 10.1046/j.1365-2141.1999.01681.x. [DOI] [PubMed] [Google Scholar]
- 49.James A.H. More than menorrhagia: A review of the obstetric and gynaecological manifestations of bleeding disorders. Haemophilia. 2005;11:295–307. doi: 10.1111/j.1365-2516.2005.01108.x. [DOI] [PubMed] [Google Scholar]
- 50.Peyvandi F., Haertel S., Knaub S., Mannucci P.M. Incidence of bleeding symptoms in 100 patients with inherited afibrinogenemia or hypofibrinogenemia. J. Thromb. Haemost. 2006;4:1634–1637. doi: 10.1111/j.1538-7836.2006.02014.x. [DOI] [PubMed] [Google Scholar]
- 51.Quintero-Ronderos P., Mercier E., Fukuda M., González R., Suárez C.F., Patarroyo M.A., Vaiman D., Gris J.C., Laissue P. Novel genes and mutations in patients affected by recurrent pregnancy loss. PLoS ONE. 2017;12:e0186149. doi: 10.1371/journal.pone.0186149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Di Fruscio G., Garofalo A., Mutarelli M., Savarese M., Nigro V. Are all the previously reported genetic variants in limb girdle muscular dystrophy genes pathogenic? Eur. J. Hum. Genet. 2016;24:73–77. doi: 10.1038/ejhg.2015.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Asselta R., Paraboschi E.M., Rimoldi V., Menegatti M., Peyvandi F., Salomon O., Duga S. Exploring the global landscape of genetic variation in coagulation factor XI deficiency. Blood. 2017;130:e1–e6. doi: 10.1182/blood-2017-04-780148. [DOI] [PubMed] [Google Scholar]
- 54.Desmet F.O., Hamroun D., Lalande M., Collod-Béroud G., Claustres M., Béroud C. Human Splicing Finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hebsgaard S.M., Korning P.G., Tolstrup N., Engelbrecht J., Rouzé P., Brunak S. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 1996;24:3439–3452. doi: 10.1093/nar/24.17.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reese M.G., Eeckman F.H., Kulp D., Haussler D. Improved splice site detection in Genie. J. Comput. Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 57.Hanss M., Biot F. A database for human fibrinogen variants. Ann. N. Y. Acad. Sci. 2001;936:89–90. doi: 10.1111/j.1749-6632.2001.tb03495.x. [DOI] [PubMed] [Google Scholar]
- 58.The Human Gene Mutation Database. [(accessed on 15 September 2017)]; Available online: http://www.hgmd.cf.ac.uk/ac/index.php/
- 59.Stenson P.D., Ball E.V., Mort M., Phillips A.D., Shaw K., Cooper D.N. The Human Gene Mutation Database (HGMD) and its exploitation in the fields of personalized genomics and molecular evolution. Curr. Protoc. Bioinform. 2012 doi: 10.1002/0471250953.bi0113s39. [DOI] [PubMed] [Google Scholar]
- 60.The Expert Protein Analysis System. [(accessed on 15 September 2017)]; Available online: https://www.expasy.org/
- 61.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R.D., Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.The ClinVar Resource. [(accessed on 15 September 2017)]; Available online: http://www.ncbi.nlm.nih.gov/clinvar/
- 63.Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J., et al. ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 65.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwarz J.M., Rodelsperger C., Schuelke M., Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 67.Reva B., Antipin Y., Sander C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chun S., Fay J.C. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shihab H.A., Gough J., Cooper D.N., Stenson P.D., Barker G.L., Edwards K.J., Day I.N., Gaunt T.R. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum. Mutat. 2013;34:57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.