Abstract
The Colorado potato beetle (Leptinotarsa decemlineata (Say)) is a significant pest of potato plants that has been controlled for more than two decades by neonicotinoid imidacloprid. L. decemlineata can develop resistance to this agent even though the molecular mechanisms underlying this resistance are not well characterized. MicroRNAs (miRNAs) are short ribonucleic acids that have been linked to response to various insecticides in several insect models. Unfortunately, the information is lacking regarding differentially expressed miRNAs following imidacloprid treatment in L. decemlineata. In this study, next-generation sequencing and quantitative real-time polymerase chain reaction (qRT-PCR) were used to identify modulated miRNAs in imidacloprid-treated versus untreated L. decemlineata. This approach identified 33 differentially expressed miRNAs between the two experimental conditions. Of interest, miR-282 and miR-989, miRNAs previously shown to be modulated by imidacloprid in other insects, and miR-100, a miRNA associated with regulation of cytochrome P450 expression, were significantly modulated in imidacloprid-treated beetles. Overall, this work presents the first report of a miRNA signature associated with imidacloprid exposure in L. decemlineata using a high-throughput approach. It also reveals interesting miRNA candidates that potentially underly imidacloprid response in this insect pest.
Keywords: microRNAs, cold hardiness, Colorado potato beetles, imidacloprid, next-generation sequencing
1. Introduction
The Colorado potato beetle (CPB) (Leptinotarsa decemlineata (Say)) is a significant insect pest harming potato crops worldwide [1]. CPBs are often considered to be the primary insect associated with potato plant defoliation [2], leading to up to 75% of foliage consumption and impacting revenues for growers [3,4]. Pest control strategies targeting CPBs typically involve pesticides, even though resistance against a variety of such compounds, including spinosad, thiamethoxam, and deltamethrin, has been observed [5,6,7]. Neonicotinoids, which target the nicotinic acetylcholine receptors (nAChRs), are a class of insecticides that are used extensively against CPBs. The neonicotinoid imidacloprid (1-(6-chloro-3-pyridylmethyl)-N-nitroimidazolidin-2-ylideneamine) was registered for such a purpose over 20 years ago, and was initially successful in managing CPB populations [6]. Unfortunately, early studies reported adult CPBs exhibiting as much as a 155-fold increase in imidacloprid resistance in select populations [8]. Efforts have been deployed in recent years to understand the molecular players associated with imidacloprid resistance in CPBs. Multiple studies have notably highlighted the differential expression of cytochrome P450s in imidacloprid-treated CPBs [9,10], positioning these enzymes as targets that could sensitize CBPs to this agent. Nevertheless, the complete molecular picture associated with imidacloprid resistance in CPBs remains incompletely characterized.
MicroRNAs (miRNAs) are short conserved non-coding RNAs capable of regulating the expression of multiple target mRNA transcripts. These molecules are involved in the response to various stresses including starvation, anoxia, and freezing [11,12]. Previous work has also revealed miRNA modulation in insects following exposure to different chemicals including pyrethroids, chlorantraniliprole and fenpropathrin [13,14,15]. In addition, recent studies have highlighted the likely regulation of pyrethroid resistance in the mosquito Culex pipiens pallens (L.) by miRNAs [16,17]. These results thus support the potential involvement of miRNAs in insecticide resistance. Unfortunately, data is currently lacking regarding a signature of differentially expressed miRNAs in response to imidacloprid treatment in CPBs.
The current study was undertaken to characterize the miRNA footprint observed in CPBs following imidacloprid exposure, and to identify novel miRNA-associated molecular leads that could be utilized in the development of alternative strategies to control CPBs using an approach relying on next-generation sequencing and qRT-PCR. The miRNA target prediction was also performed using predictive tools to better assess the likely consequences of these imidacloprid-associated miRNAs. Transcript targets linked to transcriptional regulation and glucose metabolism were observed and are further discussed.
2. Results
2.1. Small RNA Sequence Analysis
High-throughput sequencing resulted in 59,406,387 reads among all samples. Low quality reads were discarded. Reads ranging from 16 to 60 nucleotides were conserved for analysis. Small RNA libraries displayed comparable read distribution profiles in the 16 and 24 nucleotides region, with maximum peaks observed at 21 nucleotides. This approach resulted in 22,534,030 and 13,763,160 total reads for control and imidacloprid-treated insects, respectively (Table 1). Within these reads, 8,825,610 (control) and 6,211,853 (imidacloprid) unique reads were obtained. MiRBase was utilized to identify the small RNAs of interest in L. decemlineata samples using red flour beetle Tribolium castaneum reference databases [18]. The miRNAs were annotated using sRNAbench in sRNAtoolbox [19]. Small RNAs unique reads were mapped to miRNAs (26,447 for control and 24,221 for imidacloprid-treated). SnRNAs (4735 for control and 3616 for imidacloprid-treated), snRNAs (4705 for control and 2937 for imidacloprid-treated) and tRNAs (40,144 for control and 22,115 for imidacloprid-treated) were also observed.
Table 1.
Mapping statistics of reads in control versus imidacloprid-treated L. decemlineata.
| Types of RNAs | Unique Reads Control/Treated | Percent Control/Treated | Read Count Control/Treated | Percent Control/Treated |
|---|---|---|---|---|
| Total reads | 8,825,610/6,211,853 | 100%/100% | 22,534,030/13,763,160 | 100%/100% |
| miRNAs | 26,447/24,221 | 0.30%/0.39% | 1,558,719/1,511,201 | 6.92%/10.98% |
| tRNAs | 40,144/22,115 | 0.45%/0.36% | 326,281/125,085 | 1.45%/0.91% |
| snRNAs | 4705/2937 | 0.05%/0.05% | 11,161/6302 | 0.05%/0.05% |
| snoRNAs | 4735/3616 | 0.05%/0.06% | 10,813/6910 | 0.05%/0.05% |
| rRNAs | 102,238/64,189 | 1.16%/1.03% | 619,781/172,969 | 2.75%/1.26% |
2.2. miRNA Expression in Control and Imidacloprid-Treated L. decemlineata by High-Throughput Sequencing
The most frequent miRNAs detected with this approach were miR-14-3p (95,802 reads in control and 81,633 reads in imidacloprid-exposed insects) and miR-8-3p (68,314 reads in control and 79,134 reads in imidacloprid-exposed insects) (Table 2). Based on next-generation sequencing results, log2 ratios of miRNA expression in control and imidacloprid-exposed L. decemlineata demonstrated 33 differentially expressed miRNAs. A total of 14 upregulated and 19 downregulated miRNAs displaying absolute log2 fold-changes greater than 0.3 (p < 0.05, n = 3) were identified (Table 3).
Table 2.
miRNAs strongly expressed in L. decemlineata.
| miRNAs | miRNA Sequences | Normalized Expression |
|---|---|---|
| miR-14-3p | UCAGUCUUUUUCUCUCUCCUAU | 88,717.57 |
| miR-8-3p | UAAUACUGUCAGGUAAAGAUGUC | 73,723.79 |
| miR-276-3p | UAGGAACUUCAUACCGUGCUCU | 54,681.67 |
| miR-317-3p | UGAACACAGCUGGUGGUAUCUCAGU | 39,445.82 |
| miR-1-3p | UGGAAUGUAAAGAAGUAUGGAG | 16,976.97 |
| miR-2a-3p | UAUCACAGCCAGCUUUGAUGAGC | 16,697.46 |
| miR-281-5p | AAGAGAGCUAUCCGUCGACAGU | 16,606.36 |
| miR-1175-3p | UGAGAUUCAACUCCUCCAUCUC | 14,289.62 |
| miR-13b-3p | UAUCACAGCCAUUUUGACGAGU | 14,017.42 |
| bantam-3p | UGAGAUCAUUGUGAAAGCUGAUU | 13,166.75 |
Presented average normalized expressions are from all samples characterized by high-throughput sequencing.
Table 3.
Differential expression of 33 miRNAs following imidacloprid treatment in L. decemlineata.
| miRNAs | miRNA Sequences | Normalized Expression Treated/Control | Log2 Fold-Change Treated/Control | |
|---|---|---|---|---|
| miR-iab-8-3p | AGGAUACAUUCAGUAUACG | 27.77 | 53.71 | 0.95 |
| miR-252a-3p | CCUGCUGCUCAAGUGCUUAUC | 12.40 | 21.64 | 0.80 |
| miR-282-5p | UAGCCUCUCCUAGGCUUUGUCU | 13.23 | 20.32 | 0.62 |
| miR-iab-8-5p | UUACGUAUACUGAAGGUAUACCGGAC | 30.54 | 44.06 | 0.53 |
| miR-1000-5p | AUAUUGUCCUGUCACAGC | 63.29 | 86.10 | 0.44 |
| miR-3849-5p | UGACAUUUUAACCAUAGUGCU | 59.97 | 81.16 | 0.44 |
| miR-193-3p | UACUGGCCUGUUAAGUCCCAAGU | 57.31 | 77.14 | 0.43 |
| miR-7-3p | CAAGGAAUCACUAAUCAUCCCAC | 36.79 | 49.25 | 0.42 |
| miR-124-3p | UAAGGCACGCGGUGAAUGCCAAG | 183.64 | 239.11 | 0.38 |
| miR-970-3p | UCAUAAGACACACGCGGCUAU | 337.13 | 427.79 | 0.34 |
| miR-276-5p | AGCGAGGUAUAGAGUUCCUACGUG | 2120.16 | 2672.97 | 0.33 |
| let-7-5p | UGAGGUAGUAGGUUGUAUAG | 4613.21 | 5776.29 | 0.32 |
| miR-1-3p | UGGAAUGUAAAGAAGUAUGGAG | 15,177.88 | 18,776.07 | 0.31 |
| miR-133-3p | UUGGUCCCCUUCAACCAGCUGU | 1771.68 | 2187.69 | 0.30 |
| miR-263b-5p | CUUGGCACUGGAAGAAUUCAC | 95.91 | 76.77 | −0.32 |
| miR-281-5p | AAGAGAGCUAUCCGUCGACAGU | 18,510.91 | 14,701.80 | −0.33 |
| miR-2944c-3p | UAUCACAGCCAGUAGUUACC | 4468.29 | 3543.74 | −0.33 |
| miR-1175-5p | AAGUGGAGCAGUGGUCUCUUCAC | 286.32 | 224.91 | −0.35 |
| miR-92a-3p | AUUGCACUAGUCCCGGCCUA | 62.52 | 48.60 | −0.36 |
| miR-305-5p | AUUGUACUUCAUCAGGUGCUC | 7239.92 | 5605.19 | −0.37 |
| miR-iab-4-3p | CGGUAUACCUUCAGUAUACGUAAC | 24.56 | 18.65 | −0.40 |
| miR-927a-5p | UUUAGAAUUCCUACGCUUUA | 20.10 | 14.98 | −0.42 |
| miR-9c-5p | UCUUUGGUGAUCUAGCCGUGUG | 405.18 | 297.72 | −0.44 |
| miR-34-3p | CGACCACUAUCCAUACUCCCUCC | 29.11 | 21.28 | −0.45 |
| miR-316-5p | UGUCUUUUUCCGCUUUGCUGC | 11,436.58 | 8348.15 | −0.45 |
| miR-998-3p | UAGCACCAUGGGAUUCAGCUCA | 87.90 | 59.03 | −0.57 |
| miR-100-5p | AACCCGUAGAUCCGAACUUGUGGG | 5341.40 | 3528.46 | −0.60 |
| miR-750-3p | CCAGAUCUAACUCUUCCAUAUGACG | 6089.67 | 3795.84 | −0.68 |
| miR-995-3p | UAGCACCACAUGAUUCAGCUUACG | 551.70 | 340.13 | −0.70 |
| miR-2796-5p | AGGGGUUUCUUUCGGCCUCCAGCG | 41.19 | 24.70 | −0.74 |
| miR-92a-5p | AGUCCGUGAUGCGUGACAAUAU | 197.04 | 117.23 | −0.75 |
| miR-315-5p | UUUUGAUUGUUGCUCAGAAAGC | 21.80 | 12.80 | −0.77 |
| miR-989-3p | UGUGAUGUGACGUAGUGG | 4764.90 | 2748.54 | −0.79 |
2.3. qRT-PCR Quantification of Selected miRNAs
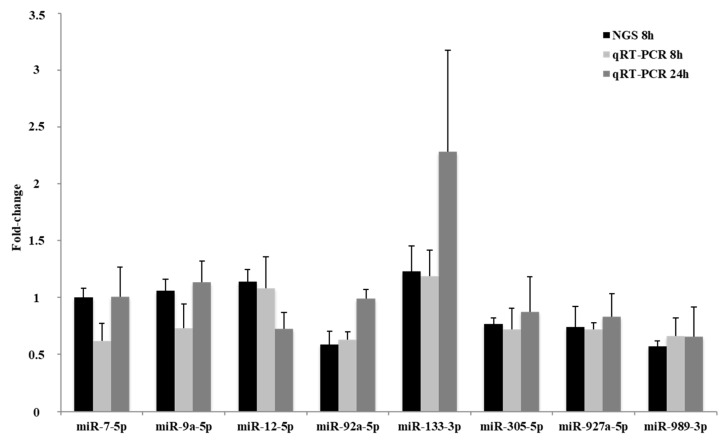
Eight miRNAs were further amplified and quantified by qRT-PCR. These miRNAs displayed reduced, elevated or stable levels in insects treated with imidacloprid for 8 h when compared with control insects using deep sequencing. Fold-changes were compared with expression results measured by next-generation sequencing (Figure 1). MiR-92a-5p, miR-133-3p, miR-305-5p, miR-927a-5p and miR-989-3p displayed substantial modulation in imidacloprid-treated versus untreated insects as measured by high-throughput sequencing, with 0.59-fold, 1.23-fold, 0.77-fold, 0.74-fold and 0.57-fold variations, respectively. These changes were comparable to the ones obtained by qRT-PCR. The miR-12-5p levels were also comparable when both methods were used to quantify its expression. MiR-7-5p and miR-9a-5p, stable miRNAs in imidacloprid-treated insects as measured by next-generation sequencing, displayed reduced levels (0.62-fold and 0.73-fold, respectively) when measured by qRT-PCR. Assessment of miRNA expression in insects treated with imidacloprid for 24 h was also performed by qRT-PCR (Figure 1). Differential expression of miR-7-5p, miR-9a-5p, miR-12-5p, miR-92a-5p, and miR-133-5p was observed between insects submitted to imidacloprid treatments of varying lengths. Levels of miR-305-5p, miR-927a-5p and miR-989-3p remained stable between the two treatment groups.
Figure 1.
Next-generation sequencing (NGS) and quantitative real-time polymerase chain reaction (qRT-PCR) expression data of select miRNAs. Expression of eight miRNAs quantified in control and imidacloprid-treated L. decemlineata. Data for qRT-PCR is normalized to transcript levels (mean ± SEM, n = 3) and NGS levels represent fold-changes in normalized read counts (mean ± SEM, n = 3).
2.4. Prediction of miRNA Targets and Functional Classification
Target prediction tools were able to identify transcripts potentially regulated by 17 L. decemlineata imidacloprid-associated miRNAs. Targets were assessed for the following miRNAs: miR-282-5p, miR-1000-5p, miR-193-3p, miR-124-3p, miR-970-3p, let-7-5p, miR-1-3p, miR-133-3p, miR-263b-5p, miR-92a-3p, miR-305-5p, miR-927a-5p, miR-9c-5p, miR-316-5p, miR-100-5p, miR-315-5p and miR-989-3p (Table 4). Common targets are underlined. Biological processes with predicted transcripts are presented in Table 5. Several target transcripts were associated with transcriptional regulation, whilst others were linked to glucose metabolism.
Table 4.
Target prediction of select L. decemlineata miRNAs with TargetScanFly and miRanda.
| miRNAs | TargetScanFly Targets | MiRanda Targets |
|---|---|---|
| miR-282-5p | Ero1L, Fus, CG14435, CG9515, CcapR | Meso18E, Rogdi, Resilin, Nkd, Kraken |
| miR-1000-5p | CG34355, Nplp1, CG10804, Net, CG13384 | Nplp1, CG10804, Kon, CG13384, Net |
| miR-193-3p | P38b, Cp7Fb, Ana, CG11041, CG11313 | CG34394, RYBP, CG6707, Pb, CG32736 |
| miR-124-3p | Sinu, Gli, CG12977, Pk92B, Axs | Pk92B, CG14299, Sinu, Gli, Cp110 |
| miR-970-3p | Btsz, CG15097, Ru, Ace, Rab6 | CG15097, Ubc-E2H, CG32372, CG42256, StmA |
| let-7-5p | Ab, CG18265, A3-3, Apt, Cpr49Aa | Ab, CG5026, CG34118, CG9548, Slam |
| miR-1-3p | CG4297, Tub, CG30457, Hmu, CG6490 | Par-6, CG18542, Pen, CG5053, Pdm2 |
| miR-133-3p | Fili, CG17193, CG2774, CG33324, CG9541 | CG30409, SkpA, Wts, Pde1c, CG32351 |
| miR-263b-5p | Qkr54B, Bx, Wls, CG32062, CG34339 | CG2371, Wls, Qkr54B, Bx, GATAe |
| miR-92a-3p | Sha, Khc-73, CrebA, CG4297, Tusp | CrebA, Cpr50Ca, CG8128, CG3077, CG14274 |
| miR-305-5p | CG33174, CG11997, CG3287, Gr98d, Nerfin-1 | CG3287, CG31855, Mi-2, NfI, Eya |
| miR-927a-5p | Gprs, CG32245, Kr-h1, CG8485, Growl | CG8485, EcR, CG9850, Aef1, Sfl |
| miR-9c-5p | Nerfin-1, Rbp9, CG11206, CG32333, Bru-2 | Sinu, CadN, CG5746, CG11206, CG9426 |
| miR-316-5p | Hr39, Rbp9, CG32121, Numb, Bsg25D | Nkd, Numb, Aret, Cib, RdgC |
| miR-100-5p | E(Pc), Gogo, CG17985, CG31772, DopR2 | CG3630, CG10979, Gogo, Pc, CG13326 |
| miR-315-5p | Eip93F, CG15465, CG32333, CG32137, CG32206 | CG12424, CG14989, CG32137, Rtnl1, CG34126 |
| miR-989-3p | Tankyrase, Fal, Lac, PGRP-SD, Qkr54B | CG12772, Chrw, CG34449, Nedd4, Kni |
Table 5.
Biological processes predicted to be modulated by select miRNAs with varying expression levels in imidacloprid-treated L. decemlineata.
| GO Biological Process | Genes Targeted | miRNAs | p-Value |
|---|---|---|---|
| Sensory perception of pain | 13 | 11 | 6.0 × 10−3 |
| Regulation of transcription, DNA-templated | 12 | 10 | 7.2 × 10−4 |
| Transcription, DNA-templated | 11 | 9 | 1.6 × 10−3 |
| Regulation of glucose metabolic process | 9 | 7 | 1.5 × 10−4 |
| Positive regulation of transcription RNA pol II promoter | 8 | 7 | 3.8 × 10−3 |
| Border follicle cell migration | 6 | 6 | 5.7 × 10−3 |
| Axon guidance | 6 | 5 | 2.3 × 10−2 |
| Imaginal disc-derived wing morphogenesis | 6 | 5 | 4.1 × 10−2 |
| Muscle organ development | 5 | 6 | 5.9 × 10−3 |
| Negative regulation of transcription, DNA-templated | 5 | 5 | 2.1 × 10−2 |
| Negative regulation of transcription RNA pol II promoter | 5 | 5 | 4.1 × 10−2 |
3. Discussion
The molecular changes underlying neonicotinoid resistance in insects have been investigated with great interest in recent years. Work conducted on various insect models treated with imidacloprid has revealed targets potentially involved in the response to this agent. These targets include select cytochrome P450s, ATP-binding cassette (ABC) transporters, and nAChRs [20,21,22]. Numerous examples exist that highlight the regulation of these imidacloprid-relevant targets by miRNAs [23,24]. While recent studies have started to associate miRNAs with insecticide resistance in various insects [14,25,26], no work so far has attempted to characterize modulated miRNAs in imidacloprid-treated CPBs. Using a high-throughput sequencing approach, the current study reveals a signature of differentially expressed miRNAs in CPBs exposed to this neonicotinoid.
The information is sparse regarding the molecular changes associated with imidacloprid response in insects. Pioneering work performed on beehives submitted to low levels of imidacloprid revealed a set of differentially expressed miRNAs in larvae gathered from imidacloprid-exposed versus unexposed hives [27]. The present study also identified similar changes in expression levels of select miRNAs, including miR-282 upregulation and miR-989 downregulation, in imidacloprid-treated CPBs. MiR-989 has been linked with key physiological processes, as Drosophila melanogaster (Meigen) miR-989 mutants notably exhibit impaired border cell migration [28], and Bombyx mori (L.) strongly upregulates miR-989 during the pupal phases [29]. It is interesting to note that levels of miR-989-3p, like miR-305-5p and miR-927a-5p, remained unchanged in insects treated with imidacloprid for different lengths of time, strengthening its potential involvement in response to this insecticide. MiR-282, on the other hand, can influence D. melanogaster viability and longevity by targeting the adenylate cyclase rutabaga [30]. While modulation of miR-282 and miR-989 in imidacloprid-treated CPBs presented here supports their probable role in response to this pesticide, additional work is required to fully delineate their functions in insects exposed to imidacloprid.
Several reports have highlighted the upregulation of cytochrome P450s in CPBs treated with imidacloprid. A high-throughput sequencing approach performed in imidacloprid-sensitive and imidacloprid-resistant CPBs revealed that transcripts for 41 cytochrome P450s were more than 2-fold upregulated in pesticide-exposed insects. In addition, cytochrome P450 activity was increased in the resistant beetles [10]. Gene expression analysis performed on CPB populations susceptible or resistant to imidacloprid reported differential cytochrome P450 levels. This further strengthens an underlying role for these enzymes in imidacloprid resistance [9]. Several studies have reported miRNA-mediated regulation of cytochrome P450 expression. Recent work conducted in the tobacco aphid Myzus persicae nicotianae (Blackman) showed that expression of CYP6CY3, a cytochrome P450 previously associated with neonicotinoid resistance [31], was regulated by miR-100 [32]. The present study highlighted downregulation of miR-100-5p levels in imidacloprid-treated CPBs, supporting a potential role for the miR-100-cytochrome P450 axis in the imidacloprid response.
Functional annotation was undertaken using the predicted targets of differentially expressed miRNAs following imidacloprid exposure in order to better characterize the potential impact of this compound in CPBs (Table 5). This approach notably highlighted gene transcription as a likely process affected by the modulated miRNAs. Some miRNAs, such as miR-92a-3p and miR-927a-5p, were predicted to impact key players involved in transcriptional regulation, including the cAMP response element binding (Creb) transcription factor and the ecdysone receptor (EcR). Previous work has reported that Creb levels were differentially expressed in honey bees (Apis mellifera carnica (Pollmann)) exposed to various neonicotinoids [33], supporting a potential role for Creb in the response to imidacloprid. Several studies have investigated the effect of imidacloprid treatment on gene expression in multiple insect models. A microarray-based approach performed in the mosquito Aedes aegypti (L.) revealed 344 and 108 differentially expressed genes in imidacloprid-resistant larvae and adults, respectively, when compared to their imidacloprid-sensitive counterparts [34]. In addition, a high-throughput sequencing approach in D. melanogaster that were sensitive or resistant to the same agent highlighted 357 transcripts that were either upregulated or downregulated in imidacloprid-resistant flies [35]. Multiple transcript targets associated with imidacloprid-modulated miRNAs were also linked to glucose metabolism. Previous reports have linked imidacloprid treatment with modulation of various targets involved in glucose homeostasis. Honey bee larvae exposed to imidacloprid exhibited increased transcript levels of multiple genes involved in the glycolytic and gluconeogenic pathways. Transcript levels of phosphoenolpyruvate carboxykinase (PEPCK), a rate-limiting enzyme for the latter cascade, were notably upregulated in imidacloprid-exposed larvae [27]. In addition, a study performed on Madagascar hissing cockroaches (Gromphadorhina portentosa (Schaum)) showed variations in glucose uptake and carbohydrate metabolism in cockroaches exposed to imidacloprid [36]. These leads generate potential processes via which the identified miRNAs can influence imidacloprid response. However, further characterization of the link that exists between these processes and imidacloprid-associated miRNAs is envisioned. Ultimately, such transcripts could be targeted by diverse means including RNA interference (RNAi)-based approaches that form part of a strategy aimed at influencing imidacloprid response in CPBs. Such approaches are currently under investigation in various pests including CPBs [37].
In conclusion, the present study revealed a set of modulated miRNAs in CPBs following exposure to the neonicotinoid imidacloprid, and is aligned with recent work aimed at better understanding the functions associated with miRNAs in these insects [38]. These results present the first example of a miRNA signature in L. decemlineata treated with this agent. Assessment of miRNA expression following different imidacloprid treatment durations also revealed varying levels of select miRNAs in short- versus long-term insecticide exposures. This warrants further investigation of early and late molecular changes associated with response to this agent. Predicted miRNA transcript targets further suggest that differential miRNA expression could impact molecular players involved in key processes. It is important to note that while the miRNA changes uncovered in this study generated valuable information regarding miRNAs underlying insect response to imidacloprid, future work is needed to investigate miRNA signatures observed in response to additional experimental conditions. These conditions include repeated topical imidacloprid exposures, usage of a diet comprised of imidacloprid-treated potato leaves, or field CPBs naturally exposed to this agent and exhibiting various degrees of tolerance against imidacloprid. Further investigations will be undertaken to examine the expression status of enzymes such as cytochrome P450s in CPB populations that do or do not respond to imidacloprid. Overall, this work provides novel information on a potential role for miRNAs in imidacloprid response and resistance, as well as contributing to the growing knowledge of the molecular levers underlying the response to this agent in Colorado potato beetles.
4. Materials and Methods
4.1. Insect Collection and Treatment
Adult Colorado potato beetles that had overwintered were collected in nursery potato fields at the Fredericton Research and Development Centre in Fredericton (NB, Canada) in June 2016. Potato fields where collection occurred were not treated with any types of insecticides. This nevertheless cannot rule out the potential exposure of beetles to insecticides in years prior to the study, nor exposure attributable to insect migration. Beetles were placed in plastic containers (50 per) containing potato leaves, and closed using a lid with insect screening for ventilation. Insects were subsequently transported to the Université de Moncton (Moncton, NB, Canada). A group of 30 insects was placed in an incubator (Thermo Fisher Scientific, Waltham, MA, USA) set at 25 °C for 5 days under 16L:8D cycles. Insects were provided with potato plants. Following acclimation, 5 µL (0.5 µg) of analytical grade imidacloprid (100 µg/mL in acetonitrile, Sigma-Aldrich, St. Louis, MO, USA) was applied topically on the abdomen of 15 beetles, as previously described [10]. A volume of 5 µL acetonitrile was applied on 15 beetles in parallel, which were used as controls. Imidacloprid-treated and control insects were returned to the incubator for 8 h. A similar experimental protocol was undertaken where insects were treated with imidacloprid for 24 h. Insects were active after the incubation period and prior to storage. A parallel bioassay performed on insects treated with increasing imidacloprid doses (0.05 µg to 5.0 µg) and not destined for high-throughput sequencing highlighted a marked impact on insect activity, albeit not viability, at maximum doses of 2.5 µg and 5.0 µg of imidacloprid (LD50 >5.0 µg/insect). All insects destined for sequencing were ultimately placed rapidly in liquid nitrogen and stored at −80 °C until RNA isolation.
4.2. Small RNA Isolation
Small RNA fractions were isolated from control and imidacloprid-treated L. decemlineata using the miRVana miRNA Isolation Kit (Thermo Fisher Scientific, Waltham, MA, USA) as per the manufacturer’s instructions and as described previously [39]. Each isolate was prepared using two L. decemlineata. RNA isolates were generated in triplicates. The quality and integrity of small RNA fractions were confirmed using the High Sensitivity RNA ScreenTape on an Agilent TapeStation 2200 instrument (Agilent Technologies (Santa Clara, CA, USA)).
4.3. Small RNA Library Construction and Sequencing
Small RNA libraries were prepared and sequenced following Ion Torrent protocols and as reported previously [40]. Samples were loaded on an Ion PI Chip v2 and sequenced with an Ion Proton Sequencer (Thermo Fisher Scientific, Waltham, MA, USA). Three control and three imidacloprid-treated libraries were constructed. FASTQ files were generated for each sample and adaptor sequences were trimmed. Reads with less than 16 and more than 60 nucleotides were discarded. Reads with Q scores of less than 20 were not included. Small RNA annotation was performed using the sRNAbench tool from sRNAtoolbox. Red flour beetle Tribolium castaneum was used as a reference [19]. The number of permitted mismatches when mapping to mature miRNAs was set to 3. Default values were used for the remaining parameters. Annotated sequences having less than 10 normalized read counts in either control or insecticide-treated samples were discarded, and log2 fold-changes were generated.
4.4. cDNA Synthesis
cDNA was synthetized as described earlier [41]. L. decemlineata miRNA sequences obtained via next-generation sequencing were used to design stem-loop primers to amplify miR-1-3p, miR-7-5p, miR-9a-5p, miR-12-5p, miR-92a-5p, miR-133-3p, miR-305-5p, miR-927a-5p and miR-989-3p (Table 6). A volume containing 0.5–1 µg of small RNA was combined with 5 μL of 300 nM of miRNA-specific stem-loop primer and the mixture was placed at 95 °C for 5 min, 60 °C for 5 min, and on ice for 1 min. Subsequently, 4 μL of 5× first strand buffer, 2 μL of 0.1 M DTT, 1 μL of 10 mM dNTPs, 2 μL of diethyl pyrocarbonate (DEPC)-treated water and 1 μL of M-MLV reverse transcriptase (Thermo Fisher Scientific, Waltham, MA, USA) were added. Mixtures were incubated at 16 °C for 30 min, 42 °C for 30 min and 85 °C for 5 min. cDNA was serially diluted and used for PCR.
Table 6.
Sequences, efficiencies, and optimal melting temperatures of primers used in this study.
| Primer | Sequence | Eff. | Temp. |
|---|---|---|---|
| miR-1-3p | 5′-ACACTCCAGCTGGGTGGAATGTAAAGAAGTA-3′ | 92.0% | 62.5 °C |
| 5′-CTCACAGTACGTTGGTATCCTTGTGATGTTCGATGCCATATTGTACTGTGAGCTCCATAC-3′ | |||
| miR-7-5p | 5′-ACACTCCAGCTGGGTGGAAGACTAGTGAT-3′ | 96.0% | 60.0 °C |
| 5′-CTCACAGTACGTTGGTATCCTTGTGATGTTCGATGCCATATTGTACTGTGAGCACAACAA-3′ | |||
| miR-9a-5p | 5′-ACACTCCAGCTGGGTCTTTGGTTATCTAG-3′ | 101.4% | 58.8 °C |
| 5′-CTCACAGTACGTTGGTATCCTTGTGATGTTCGATGCCATATTGTACTGTGAGTCATACAG-3′ | |||
| miR-12-5p | 5′-ACACTCCAGCTGGGTGAGTATTACATCAGGT-3′ | 95.6% | 64.5 °C |
| 5′-CTCACAGTACGTTGGTATCCTTGTGATGTTCGATGCCATATTGTACTGTGAGCAGTACCT-3′ | |||
| miR-92a-5p | 5′-ACACTCCAGCTGGGAGTCCGTGATGCGTGAC-3′ | 88.5% | 56.5°C |
| 5′-CTCACAGTACGTTGGTATCCTTGTGATGTTCGATGCCATATTGTACTGTGAGATATTGTC-3′ | |||
| miR-133-3p | 5′-ACACTCCAGCTGGGTTGGTCCCCTTCAACCA-3′ | 84.3% | 64.5 °C |
| 5′-CTCACAGTACGTTGGTATCCTTGTGATGTTCGATGCCATATTGTACTGTGAG-3′ | |||
| miR-305-5p | 5′-ACACTCCAGCTGGGATTGTACTTCATCAGGT-3′ | 91.3% | 63.4 °C |
| 5′-CTCACAGTACGTTGGTATCCTTGTGATGTTCGATGCCATATTGTACTGTGAGGAGCACCT-3′ | |||
| miR-927a-5p | 5′-ACACTCCAGCTGGGTTTAGAATTCCTACGCT-3′ | 105.3% | 60.9 °C |
| 5′-CTCACAGTACGTTGGTATCCTTGTGATGTTCGATGCCATATTGTACTGTGAGTAAAGCGT-3′ | |||
| miR-989-3p | 5′-ACACTCCAGCTGGGTGTGATGTGACGTAGTG-3′ | 99.5% | 56.9 °C |
| 5′-CTCACAGTACGTTGGTATCCTTGTGATGTTCGATGCCATATTGTACTGTGAGCCACTACG-3′ | |||
| Universal | 5′-CTCACAGTACGTTGGTATCCTTGTG-3′ | - | - |
Top and bottom sequences represent forward and stem-loop primers, respectively. Abbreviations: Efficiency (Eff.); Temperature (Temp.).
4.5. PCR and qRT-PCR Amplification of miRNAs
Initial PCR reactions were performed to confirm correct product amplification. Reactions consisted of 5 µL of cDNA (10−1), 5.5 µL of DEPC-treated water, 1 µL of 25 μM miRNA-specific forward primer, 1 μL of 25 μM universal reverse primer and 12.5 μL of 2× Taq FroggaMix (FroggaBio, Toronto, ON, Canada) [42]. Forward and universal primers used are presented in Table 6. An initial denaturing step at 95 °C for 5 min was performed, followed by 35 cycles at 95 °C for 15 s and at a gradient of temperatures for 1 min. Products were separated on a 2% agarose gel and visualized using a ChemiDoc MP system (Bio-Rad, Hercules, CA, USA). Products were then sequenced at the Université Laval (Quebec City, QC, Canada) and identities were confirmed using Basic Local Alignment Search Tool (BLAST).
The qRT-PCR reactions were performed by mixing 2.5 µL of diluted cDNA template with 0.5 µL of DEPC-treated water, 1 µL of 5 µM miRNA-specific forward primer, 1 µL of 5 µM universal reverse primer and 5 µL of 2× iTaq Universal SYBR Green Supermix (Bio-Rad, Hercules, CA, USA) prepared in triplicate in 96-well plates. Amplification protocol consisted of an initial denaturing step at 95 °C for 3 min, followed by 40 cycles at 95 °C for 15 s and optimal annealing temperature for each miRNAs (Table 6) for 30 s. Reactions were conducted on a CFX Connect Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). Efficiencies were calculated for each primer pair with the same protocol as above in serial cDNA dilutions. NormFinder software (Aarhus University Hospital, Aarhus, Denmark) [43] revealed miR-1-3p as the most stably expressed miRNA amongst a group of transcripts in insects exposed to imidacloprid for 8 h as determined by qRT-PCR, and was used as reference transcript.
4.6. miRNA Transcript Targets Prediction
TargetScanFly 6.2 [44] and the fruit fly miRanda algorithm [45] target prediction tools were used to identify likely mRNA candidates regulated by the imidacloprid-associated miRNAs identified by high-throughput sequencing. The top five transcript targets linked with imidacloprid-modulated miRNAs that were available in both prediction tools were obtained. A list of miRNA targets was subsequently generated and functionally annotated with the Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics resources v6.8 [46]. Biological processes associated with these targets were generated.
4.7. Quantification and Statistics
The Cq values following qRT-PCR runs were obtained using the Bio-Rad CFX Manager software (Bio-Rad, Hercules, CA, USA). miRNA levels were normalized using miR-1-3p levels amplified from the same sample. The miRNA levels were measured using the 2−ΔΔCq method [47]. Ratios of normalized miRNA levels in control CPBs to average transcript expression in imidacloprid-treated CPBs were generated. Statistical differences between control and treated conditions were assessed with the Student’s t-test. For high-throughput sequencing, annotated sequences were normalized using the trimmed mean of M-values (TMM) function in edgeR Bioconductor [48], and log2 ratios depicting miRNA expression in insecticide-treated versus control CPBs were calculated. Bioconductor “limma” package, with linear model fit and empirical Bayes statistics, was used to calculate differential expression [49]. Results for miRNAs with average normalized expression (ANE) greater than 10, log2 fold-change higher than 0.3 and p < 0.05 were investigated.
Acknowledgments
Pier Jr Morin is supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (RGPIN/402222-2012), a Research Grant (EARI 15-007) via the Enabling Agricultural and Research Innovation (EARI) program under the Canada/New Brunswick Growing Forward 2 initiative and an Emerging Project Grant (RIF 2016-036) by the New Brunswick Innovation Foundation.
Author Contributions
Mathieu D. Morin, Nicolas Crapoulet, Sébastien Boquel and Pier Jr Morin conceived and designed the experiments; Mathieu D. Morin and Pierre J. Lyons performed the experiments; Mathieu D. Morin, Pierre J. Lyons and Nicolas Crapoulet analyzed the data; Mathieu D. Morin, Sébastien Boquel, and Pier Jr Morin wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Weber D. Colorado beetle: Pest on the move. Pestic. Outlook. 2003;14:256–259. doi: 10.1039/b314847p. [DOI] [Google Scholar]
- 2.Radcliffe E.B., Lagnaoui A. Insect pests in potato: Insects. In: Vreughenhil D., Bradshaw J., Gebhardt C., Govers F., Taylor M., MacKerron D., Ross H., editors. Potato Biology and Biotechnology: Advances and Perspectives. Elsevier; Amsterdam, The Netherlands: 2007. pp. 543–567. [Google Scholar]
- 3.Hare D.J. Impact of defoliation by the Colorado potato beetle on potato yields. J. Econ. Entomol. 1980;73:369–373. doi: 10.1093/jee/73.3.369. [DOI] [Google Scholar]
- 4.Shields E.J., Wyman J.A. Effect of defoliation at specific growth stages on potato yields. J. Econ. Entomol. 1984;77:1194–1199. doi: 10.1093/jee/77.5.1194. [DOI] [Google Scholar]
- 5.Jiang W.H., Wang Z.T., Xiong M.H., Lu W.P., Liu P., Guo W.C., Li G.Q. Insecticide resistance status of Colorado Potato Beetle (Coleoptera: Chrysomelidae) adults in northern Xinjiang Uygur autonomous region. J. Econ. Entomol. 2010;103:1365–1371. doi: 10.1603/EC10031. [DOI] [PubMed] [Google Scholar]
- 6.Alyokhin A., Baker M., Mota-Sanchez D., Dively G., Grafius E. Colorado Potato Beetle Resistance to Insecticides. Am. J. Potato Res. 2008;85:395–413. doi: 10.1007/s12230-008-9052-0. [DOI] [Google Scholar]
- 7.Szendrei Z., Grafius E., Byrne A., Ziegler A. Resistance to neonicotinoid insecticides in field populations of the Colorado potato beetle (Coleoptera: Chrysomelidae) Pest Manag. Sci. 2012;68:941–946. doi: 10.1002/ps.3258. [DOI] [PubMed] [Google Scholar]
- 8.Zhao J.Z., Bishop B.A., Grafius E.J. Inheritance and synergism of resistance to imidacloprid in the Colorado potato beetle (Coleoptera: Chrysomelidae) J. Econ. Entomol. 2000;93:1508–1514. doi: 10.1603/0022-0493-93.5.1508. [DOI] [PubMed] [Google Scholar]
- 9.Clements J., Schoville S., Peterson N., Lan Q., Groves R.L. Characterizing Molecular Mechanisms of Imidacloprid Resistance in Select Populations of Leptinotarsa decemlineata in the Central Sands Region of Wisconsin. PLoS ONE. 2016;11:e0147844. doi: 10.1371/journal.pone.0147844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu F., Moural T.W., Nelson D.R., Palli S.R. A specialist herbivore pest adaptation to xenobiotics through up-regulation of multiple Cytochrome P450s. Sci. Rep. 2016;6:20421. doi: 10.1038/srep20421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barrio L., Dekanty A., Milán M. MicroRNA-mediated regulation of Dp53 in the Drosophila fat body contributes to metabolic adaptation to nutrient deprivation. Cell Rep. 2014;8:528–541. doi: 10.1016/j.celrep.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 12.Lyons P.J., Storey K.B., Morin P.J. Expression of miRNAs in response to freezing and anoxia stresses in the freeze tolerant fly Eurosta solidaginis. Cryobiology. 2015;71:97–102. doi: 10.1016/j.cryobiol.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Hong S., Guo Q., Wang W., Hu S., Fang F., Lv Y., Yu J., Zou F., Lei Z., Ma K., et al. Identification of differentially expressed microRNAs in Culex pipiens and their potential roles in pyrethroid resistance. Insect Biochem. Mol. Biol. 2014;55:39–50. doi: 10.1016/j.ibmb.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X., Guo L., Zhou X., Gao X., Liang P. miRNAs regulated overexpression of ryanodine receptor is involved in chlorantraniliprole resistance in Plutella xylostella (L.) Sci. Rep. 2015;5:14095. doi: 10.1038/srep14095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y., Xu Z., Wu Q., Peng M., Liu Y., Liu X., Shi L., Shen G., Pan Y., He L. Identification of Differentially Expressed microRNAs between the Fenpropathrin Resistant and Susceptible Strains in Tetranychus cinnabarinus. PLoS ONE. 2016;11:e0152924. doi: 10.1371/journal.pone.0152924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu B., Tian M., Guo Q., Ma L., Zhou D., Shen B., Sun Y., Zhu C. MiR-932 Regulates Pyrethroid Resistance in Culex pipiens pallens (Diptera: Culicidae) J. Med. Entomol. 2016;53:1205–1210. doi: 10.1093/jme/tjw083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian M., Liu B., Hu H., Li X., Guo Q., Zou F., Liu X., Hu M., Guo J., Ma L., et al. MiR-285 targets P450 (CYP6N23) to regulate pyrethroid resistance in Culex pipiens pallens. Parasitol. Res. 2016;115:4511–4517. doi: 10.1007/s00436-016-5238-4. [DOI] [PubMed] [Google Scholar]
- 18.Kozomara A., Griffiths-Jones S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rueda A., Barturen G., Lebrón R., Gómez-Martín C., Alganza Á., Oliver J.L., Hackenberg M. sRNAtoolbox: An integrated collection of small RNA research tools. Nucleic Acids Res. 2015;43:467–473. doi: 10.1093/nar/gkv555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X., Sun H., Zhang Y., Liu C., Liu Z. Transcriptional Changes in nAChRs, Interactive Proteins and P450s in Locusta migratoria manilensis (Orthoptera: Acrididae) CNS in Response to High and Low Oral Doses of Imidacloprid. J. Insect Sci. 2015;15:102. doi: 10.1093/jisesa/iev080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang X.L., Zhang M., Wang K., Qiao X.F., Chen M.H. Molecular cloning, expression pattern of multidrug resistance associated protein 1 (MRP1, ABCC1) gene, and the synergistic effects of verapamil on toxicity of two insecticides in the bird cherry-oat aphid. Arch. Insect Biochem. Physiol. 2016;92:65–84. doi: 10.1002/arch.21334. [DOI] [PubMed] [Google Scholar]
- 22.Sun H., Pu J., Chen F., Wang J., Han Z. Multiple ATP-binding cassette transporters are involved in insecticide resistance in the small brown planthopper, Laodelphax striatellus. Insect Mol. Biol. 2017;26:343–355. doi: 10.1111/imb.12299. [DOI] [PubMed] [Google Scholar]
- 23.Haenisch S., Werk A.N., Cascorbi I. MicroRNAs and their relevance to ABC transporters. Br. J. Clin. Pharmacol. 2014;77:587–596. doi: 10.1111/bcp.12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hogan E.M., Casserly A.P., Scofield M.D., Mou Z., Zhao-Shea R., Johnson C.W., Tapper A.R., Gardner P.D. miRNAome analysis of the mammalian neuronal nicotinic acetylcholine receptor gene family. RNA. 2014;20:1890–1899. doi: 10.1261/rna.034066.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma K., Li X., Hu H., Zhou D., Sun Y., Ma L., Zhu C., Shen B. Pyrethroid-resistance is modulated by miR-92a by targeting CpCPR4 in Culex pipiens pallens. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2017;203:20–24. doi: 10.1016/j.cbpb.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu B., Li X., Liu Y., Gao X., Liang P. Global identification of microRNAs associated with chlorantraniliprole resistance in diamondback moth Plutella xylostella (L.) Sci. Rep. 2017;7:40713. doi: 10.1038/srep40713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Derecka K., Blythe M.J., Malla S., Genereux D.P., Guffanti A., Pavan P., Moles A., Snart C., Ryder T., Ortori C.A., et al. Transient exposure to low levels of insecticide affects metabolic networks of honeybee larvae. PLoS ONE. 2013;8:e68191. doi: 10.1371/journal.pone.0068191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kugler J.M., Verma P., Chen Y.W., Weng R., Cohen S.M. miR-989 is required for border cell migration in the Drosophila ovary. PLoS ONE. 2013;8:e67075. doi: 10.1371/journal.pone.0067075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jagadeeswaran G., Zheng Y., Sumathipala N., Jiang H., Arrese E.L., Soulages J.L., Zhang W., Sunkar R. Deep sequencing of small RNA libraries reveals dynamic regulation of conserved and novel microRNAs and microRNA-stars during silkworm development. BMC Genom. 2010;11:52. doi: 10.1186/1471-2164-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vilmos P., Bujna A., Szuperák M., Havelda Z., Várallyay É., Szabad J., Kucerova L., Somogyi K., Kristó I., Lukácsovich T., et al. Viability, longevity, and egg production of Drosophila melanogaster are regulated by the miR-282 microRNA. Genetics. 2013;195:469–480. doi: 10.1534/genetics.113.153585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puinean A.M., Foster S.P., Oliphant L., Denholm I., Field L.M., Millar N.S., Williamson M.S., Bass C. Amplification of a cytochrome P450 gene is associated with resistance to neonicotinoid insecticides in the aphid Myzus persicae. PLoS Genet. 2010;6:e1000999. doi: 10.1371/journal.pgen.1000999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng T., Pan Y., Gao X., Xi J., Zhang L., Ma K., Wu Y., Zhang J., Shang Q. Reduced abundance of the CYP6CY3-targeting let-7 and miR-100 miRNAs accounts for host adaptation of Myzus persicae nicotianae. Insect Biochem. Mol. Biol. 2016;75:89–97. doi: 10.1016/j.ibmb.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 33.Christen V., Mittner F., Fent K. Molecular Effects of Neonicotinoids in HoneyBees (Apis mellifera) Environ. Sci. Technol. 2016;50:4071–4081. doi: 10.1021/acs.est.6b00678. [DOI] [PubMed] [Google Scholar]
- 34.Riaz M.A., Chandor-Proust A., Dauphin-Villemant C., Poupardin R., Jones C.M., Strode C., Régent-Kloeckner M., David J.P., Reynaud S. Molecular mechanisms associated with increased tolerance to the neonicotinoid insecticide imidacloprid in the dengue vector Aedes aegypti. Aquat. Toxicol. 2013;126:326–337. doi: 10.1016/j.aquatox.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 35.Kalajdzic P., Oehler S., Reczko M., Pavlidi N., Vontas J., Hatzigeorgiou A.G., Savakis C. Use of mutagenesis, genetic mapping and next generation transcriptomics to investigate insecticide resistance mechanisms. PLoS ONE. 2012;7:e40296. doi: 10.1371/journal.pone.0040296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawczyn T., Dolezych B., Klosok M., Augustyniak M., Stygar D., Buldak R.J., Kukla M., Michalczyk K., Karcz-Socha I., Zwirska-Korczala K. Alteration of carbohydrates metabolism and midgut glucose absorption in Gromphadorhina portentosa after subchronic exposure to imidacloprid and fenitrothion. J. Environ. Sci. Health Part A. 2012;47:1644–1651. doi: 10.1080/10934529.2012.687181. [DOI] [PubMed] [Google Scholar]
- 37.Wan P.J., Fu K.Y., Lü F.G., Wang X.X., Guo W.C., Li G.Q. Knocking down a putative Δ(1)-pyrroline-5-carboxylate dehydrogenase gene by RNA interference inhibits flight and causes adult lethality in the Colorado potato beetle Leptinotarsa decemlineata (Say) Pest Manag. Sci. 2015;71:1387–1396. doi: 10.1002/ps.3941. [DOI] [PubMed] [Google Scholar]
- 38.Morin M.D., Frigault J.J., Lyons P.J., Crapoulet N., Boquel S., Storey K.B., Morin P.J. Amplification and quantification of cold-associated microRNAs in the Colorado potato beetle (Leptinotarsa decemlineata) agricultural pest. Insect Mol. Biol. 2017;26:574–583. doi: 10.1111/imb.12320. [DOI] [PubMed] [Google Scholar]
- 39.Lyons P.J., Crapoulet N., Storey K.B., Morin P.J. Identification and profiling of miRNAs in the freeze-avoiding gall moth Epiblema scudderiana via next-generation sequencing. Mol. Cell. Biochem. 2015;410:155–163. doi: 10.1007/s11010-015-2547-3. [DOI] [PubMed] [Google Scholar]
- 40.Lyons P.J., Govaere L., Crapoulet N., Storey K.B., Morin P.J. Characterization of cold-associated microRNAs in the freeze-tolerant gall fly Eurosta solidaginis using high-throughput sequencing. Comp. Biochem. Phys. D. 2016;20:95–100. doi: 10.1016/j.cbd.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 41.Biggar K.K., Kornfield S.F., Storey K.B. Amplification and sequencing of mature microRNAs in uncharacterized animal models using stem-loop reverse transcription-polymerase chain reaction. Anal. Biochem. 2011;416:231–233. doi: 10.1016/j.ab.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 42.Lang-Ouellette D., Morin P.J. Differential expression of miRNAs with metabolic implications in hibernating thirteen-lined ground squirrels, Ictidomys tridecemlineatus. Mol. Cell. Biochem. 2014;394:291–298. doi: 10.1007/s11010-014-2105-4. [DOI] [PubMed] [Google Scholar]
- 43.Andersen C.L., Ledet-Jensen J., Ørntoft T. Normalization of real-time quantitative RT-PCR data: A model based variance estimation approach to identify genes suited for normalization, applied bladder- and colon-cancer data-sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 44.Ruby J.G., Stark A., Johnston W.K., Kellis M., Bartel D.P., Lai E.C. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Enright A.J., John B., Gaul U., Tuschl T., Sander C., Marks D.S. MicroRNA targets in Drosophila. Genome Biol. 2003;5 doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:445–447. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 47.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 48.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Biogeosciences. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:1–25. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]