Abstract
Bipolar disorder is a common and severe mental illness with unsolved pathophysiology. A genome-wide association study (GWAS) has been used to find a number of risk genes, but it is difficult for a GWAS to find genes indirectly associated with a disease. To find core hub genes, we introduce a network analysis after the GWAS was conducted. Six thousand four hundred fifty eight single nucleotide polymorphisms (SNPs) with p < 0.01 were sifted out from Wellcome Trust Case Control Consortium (WTCCC) dataset and mapped to 2045 genes, which are then compared with the protein–protein network. One hundred twelve genes with a degree >17 were chosen as hub genes from which five significant modules and four core hub genes (FBXL13, WDFY2, bFGF, and MTHFD1L) were found. These core hub genes have not been reported to be directly associated with BD but may function by interacting with genes directly related to BD. Our method engenders new thoughts on finding genes indirectly associated with, but important for, complex diseases.
Keywords: bipolar disorder, GWAS, functional enrichment analysis, network analysis
1. Introduction
Bipolar disorder (BD) is a common and severe mental disorder characterized by alternative episodes of mania/hypomania and depression [1]. It affects 1–5% of the world’s population [2,3,4]. Genetic studies have shown that bipolar disorder is a complex genetic disease that involves the interaction of multiple genes and the environment. Genetic factors can account for up to 60–85% of the risk [5,6,7,8]. The strong genetic basis of BD inspires plenty of research focused on finding candidate genes or single nucleotide polymorphisms (SNPs) associated with this disease.
Over the past few decades, traditional family-based linkage analysis and population-based case–control association analysis have been common means of identifying bipolar disorder susceptibility genes. With the advent of the third-generation polymorphism genetic marker SNPs, genome-wide association studies (GWASs) have also been applied to large-scale scanning of new BD susceptibility gene loci and a number of genes, such as CACAN1C [9,10], ANK3 [10,11], SYNE1 [12], CSMD1 [12], ITIH1 [11], KIT [11], and DGKH [13], have been found.
GWASs have proven to be useful in finding susceptibility genes of diseases. However, when used alone, it is difficult to determine genes that have relatively high GWAS p-values but may play a role through interaction with the genes directly associated with BD. The complexity of the disease makes it even more difficult to elucidate its molecular mechanism. Therefore, although the previous study has found a lot of genetic factors with significant effects on BD, its molecular mechanism remains unresolved. In this case, a comprehensive analysis focusing on gene interactions and biological functions will provide valuable information to explore the pathogenesis of BD. It has been found that the distribution of genetic marker loci on chromosomes and the interaction between SNPs are one of the major genetic basis for complex diseases [14]. The gene network is often used to reveal complex relationships among genes.
Considering that complex mental phenotypes may be affected by many genes with small or mild effects rather than one or two genes with a major impact [15], a comprehensive analysis of the underlying genes in the pathway or network framework may provide more insights into its molecular mechanism. It will be more efficient to understand the role of genes in complex diseases using network study. Some methods have been developed in this area, but the problem is far from being solved. There is scarce known molecular interaction mechanism and systematic gene network analysis for BD. Construction of a gene interaction network can be used to explore the synergistic effect of multiple genes on BD.
In this study, we performed a GWAS to obtain BD-related genes and confirmed their function by functional enrichment analysis. To further explore the association between these genes and BD, a network was constructed using a human protein–protein interaction database, and the BD-risk genes identified in the GWAS were mapped onto the network to find core hub genes. This will provide more insight into the molecular mechanisms of BD by determining the core hub genes of the network.
2. Results
2.1. GWAS Results
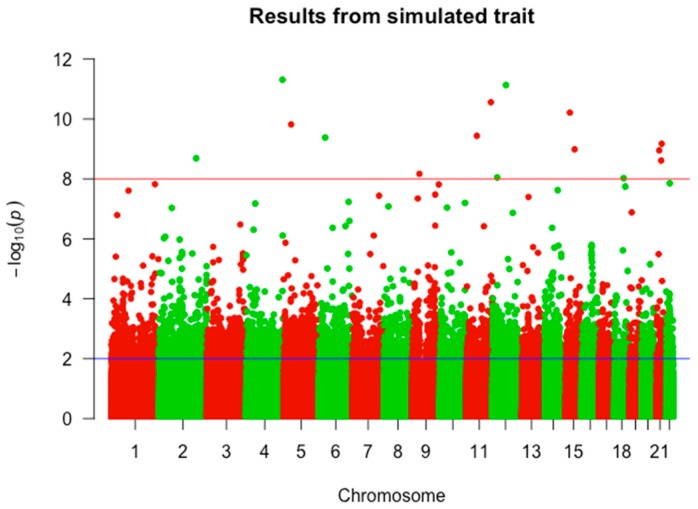
A total of 482,247 SNPs located on 22 chromosomes of 1868 BD cases and 2938 controls satisfies the quality control. The number of SNPs decreases to 354,282 after the Hardy–Weinberg equilibrium test. Finally, a total of 6458 SNPs is qualified in the GWAS where p < 0.01 and used for further analysis. The result is shown in Figure 1.
Figure 1.
Results of the genome wide association study (GWAS). The horizontal axis represents 22 chromosomes and the vertical axis represents the negative logarithm with base 10 of GWAS p-value for each SNP. Red line: canonical 5 × 10−8 cutoff. Blue line: 0.01 cutoff used in this study.
2.2. Gene Functional Analysis
A total of 2045 risk genes was obtained after mapping the 6458 SNPs onto human genes. These genes were then classified into three Gene Ontology (GO) sections: cellular components, molecular functions, and biological processes. The first 10 GO items (p < 0.01) are shown in Table 1, Table 2 and Table 3. Genes with transferase and kinase function dominate in molecular functions. In cellular components, most gene products are located in the nervous system. This coordinates with the biological process result in which most genes are involved in nervous system development.
Table 1.
Molecular functions (GO).
| Name | FDR | Gene Count |
|---|---|---|
| transferase activity, transferring phosphorus-containing groups | 5.67 × 10−5 | 118 |
| kinase activity | 5.67 × 10−5 | 103 |
| phosphotransferase activity, alcohol group as acceptor | 5.67 × 10−5 | 96 |
| protein serine/threonine kinase activity | 1.03 × 10−4 | 63 |
| protein kinase activity | 1.90 × 10−4 | 81 |
| GTPase regulator activity | 2.61 × 10−4 | 47 |
| signal transducer activity, downstream of receptor | 2.61 × 10−4 | 33 |
| GTPase activator activity | 4.57 × 10−4 | 43 |
| adenyl ribonucleotide binding | 7.16 × 10−4 | 154 |
Table 2.
Cellular components (GO).
| Name | FDR | Gene Count |
|---|---|---|
| synapse | 5.23 × 10−14 | 131 |
| postsynapse | 1.68 × 10−13 | 83 |
| synapse part | 7.30 × 10−13 | 110 |
| synaptic membrane | 1.89 × 10−12 | 62 |
| cell junction | 9.04 × 10−10 | 149 |
| postsynaptic embrane | 1.91 × 10−9 | 47 |
| neuron part | 2.94 × 10−9 | 178 |
| excitatory synapse | 7.86 × 10−9 | 49 |
| plasma membrane region | 8.87 × 10−9 | 130 |
| neuron projection | 8.89 × 10−9 | 144 |
Table 3.
Biological processes (GO).
| Name | FDR | Gene Count |
|---|---|---|
| neurogenesis | 1.28 × 10−7 | 186 |
| cell morphogenesis | 1.28 × 10−7 | 159 |
| generation of neurons | 1.28 × 10−7 | 176 |
| regulation of nervous system development | 1.28 × 10−7 | 116 |
| neuron differentiation | 3.55 × 10−7 | 162 |
| neuron development | 3.55 × 10−7 | 136 |
| cell projection morphogenesis | 3.61 × 10−7 | 116 |
| cellular component morphogenesis | 4.18 × 10−7 | 164 |
| cell projection organization | 4.63 × 10−7 | 163 |
| neuron projection morphogenesis | 6.25 × 10−7 | 88 |
2.3. Overlapped Genes in Different Mental Illnesses
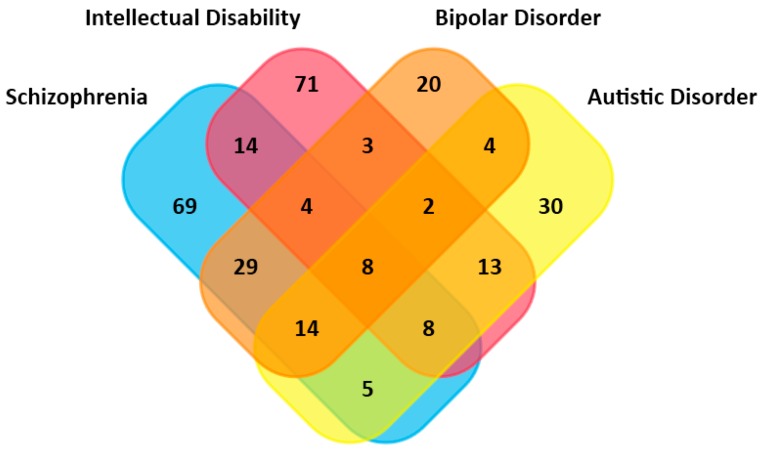
We enriched these candidate genes in BD and other three mental illnesses: schizophrenia, intellectual disability, and autistic disorder. In the total 2045 risk genes, the numbers of genes associated with schizophrenia, intellectual disability, autism, and bipolar disorder are 151, 123, 84, and 84, respectively. A Venn map of the overlap genes of these four diseases shows that, out of the 84 genes associated with bipolar disorder, 55 genes are in common with schizophrenia, 17 with intellectual disorder, and 28 with autism (Figure 2).
Figure 2.
Overlapped genes associated with four mental illnesses.
2.4. Protein Interaction Network
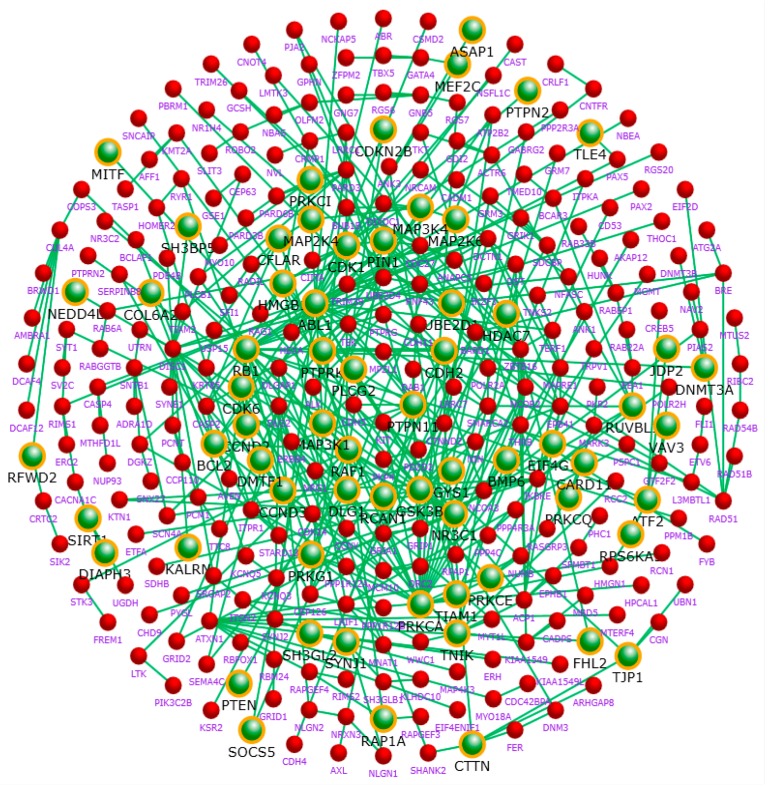
The 2045 genes from the GWAS result are mapped onto the protein–protein interaction network constructed using data from the STRING database (Figure 3).
Figure 3.
BD risk gene interaction network. Only the nodes with a degree ≥4 are shown. Green balls are BD risk genes identified in the GWAS with p < 0.01.
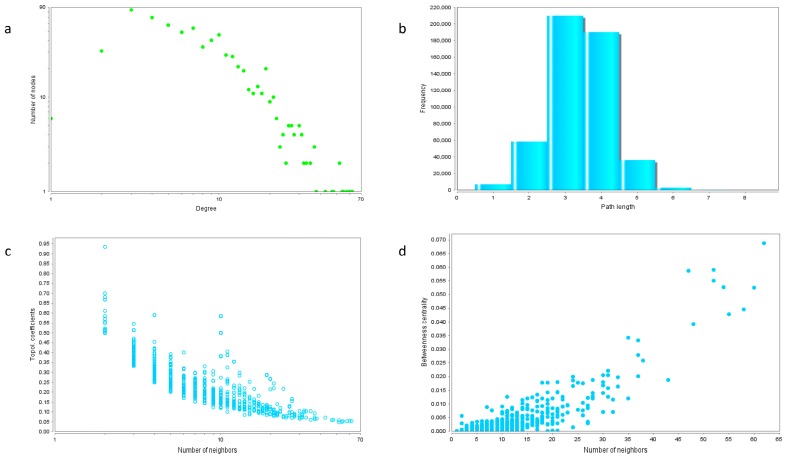
There are 1083 nodes in the network. The average node degree of the network is 7.555. The clustering coefficient is 0.232, and the characteristic path length is 3.393. The properties of the network are further analyzed and the results are shown in Figure 4. The connectivity of the network exhibits characteristic power distribution. Figure 4b shows that the shortest path with the highest frequency among the candidate genes of BD is between 3 and 4, indicating that the network is not a stochastic network but a complex network with characteristics of biological molecular network. The number of neighbors shared by the network nodes has a significant inverse relationship with its topological coefficients (Figure 4c), but shows a positive correlation with the node’s identity (Figure 4d).
Figure 4.
The topology properties of the network. (a) The distribution of number of nodes with different degrees. (b) Frequency distribution of shortest paths. (c) The relationship between topological coefficients and the number of node neighbors. (d) The relationship between betweenness and the number of node neighbors.
2.5. Hub Genes of the Network
One hundred twelve gene nodes with a degree >17 is chosen as hub genes from the network for further analysis (Table 4 and Table A1).
Table 4.
The gene nodes with a network degree >17.
| Hub Gene | Degree | Hub Gene | Degree | Hub Gene | Degree |
|---|---|---|---|---|---|
| CDK1 | 62 | PNPLA6 [16] | 27 | FARS2 | 20 |
| PTEN [17,18] | 61 | SYNJ2 | 27 | FBXO22 | 20 |
| BCL2 [19] | 60 | UBE2R2 [20] | 27 | FLT3 | 20 |
| POLR2A [21] | 55 | CACNA1D [22,23,24] | 26 | GATA4 [25] * | 20 |
| SMARCA2 [26,27] | 55 | CDK6 [28] | 26 | ITSN2 [29] | 20 |
| GSK3B [30,31,32,33] | 54 | CHRM2 [34] | 26 | KIF18A | 20 |
| ABL1 [35,36,37] | 53 | MTHFD1L [38] ** | 26 | LONRF1 | 20 |
| PRKCA [39,40] | 50 | GRIA1 [41] | 25 | NCOA3 | 20 |
| bFGF [42] ** | 48 | POLR2H | 25 | PCNT [43] | 20 |
| RB1 [44] * | 45 | TJP1 [45] * | 25 | PJA2 | 20 |
| KIT [11] | 40 | MAPRE1 [46] * | 24 | SYT1 [47] | 20 |
| RAD51 * | 38 | RUNX1 [48] | 24 | TRIM39 | 20 |
| SIRT1 [49,50,51] | 38 | UBE2D4 | 24 | WDFY2 [52] ** | 20 |
| UBE2D1 [53,54] | 37 | EHHADH | 23 | AK4 | 19 |
| DLG1 [55,56] | 36 | IQCB1 | 23 | ASB15 | 19 |
| CDC27 [57] | 35 | PPM1B [58] | 23 | ATF2 [29] | 19 |
| NEDD4L [59] | 35 | PPP4C [60] * | 23 | BUB1B | 19 |
| PRKG1 [61] * | 35 | RAD50 | 23 | DHX15 | 19 |
| RAP1A | 34 | SH3GL2 | 23 | DNM3 [62] * | 19 |
| CDH2 [63] * | 33 | DCTN1 | 22 | ETV6 | 19 |
| GNB5 [64] * | 33 | ERBB4 [65,66] | 22 | FBXL13 [67,68] ** | 19 |
| MAPK6 [69] | 33 | FBXO32 | 22 | HECW2 | 19 |
| GNG7 [70] | 32 | ITPR1 [71] * | 22 | MEF2C [72] * | 19 |
| PTPN11 [73] * | 32 | MLL [74] | 22 | NR3C1 [75] | 19 |
| ZBTB16 [76] | 32 | NCOR2 [77] | 22 | BDE4D | 19 |
| ADCY2 [78] | 31 | PRKCE [79] | 22 | RNF19B | 19 |
| DICER1 [80,81] | 31 | RAD51B | 22 | RNF217 | 19 |
| SYNJ1 [82,83,84,85] | 31 | ACTN4 | 21 | RXFP2 | 19 |
| CACNA1C [9,52,86,87] | 30 | CCND2 [88,89] | 21 | RYR1 | 19 |
| CTTN | 30 | CDH5 | 21 | THBS2 | 19 |
| DLG2 [90,91] | 30 | CUL4A [92] * | 21 | AKT3 [93] * | 18 |
| MAP3K1 [94] * | 30 | EFCAB13 | 21 | BARD1 | 18 |
| RIT2 [95] | 30 | LMO7 | 21 | CTNNA2 [11,96] | 18 |
| ANAPC5 [28] | 28 | MITF | 21 | HDAC7 | 18 |
| PLCB1 [97,98,99] | 28 | TRIM9 [100] | 21 | ITGAV | 18 |
| RAF1 [101] * | 28 | CCND3 | 20 | PARD3 [102] * | 18 |
| PARK2 [61] * | 27 | EPHB1 [103] * | 20 | PCSK2 | 18 |
| PLCG2 [13,39] | 27 |
* associated with other mental illness ** core hub genes.
Of these 112 hub genes, 45 were reported associated with BD in previous studies. Another 24 were reported associated with other mental illnesses. Gene nodes with higher degrees have a higher ratio of genes being reported associated with BD. Only five genes with a degree >23 (51 genes) are not reported associated with BD or other mental illnesses, while 24 with a degree ≤23 (61 genes) are not found reported directly associated to any mental illnesses. Obviously, risk genes with more degrees have a closer connection to BD than those with fewer degrees.
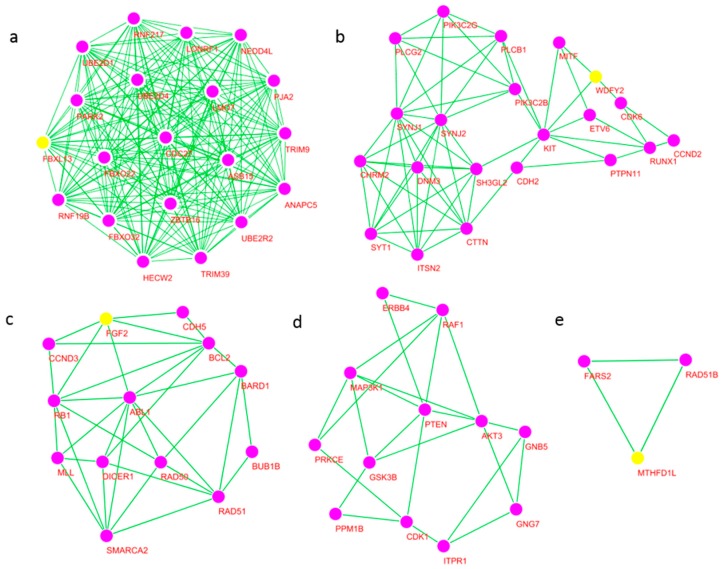
2.6. Significant Modules of the Network and Core Hub Genes
Five significant gene modules are found in the network containing 112 hub genes with Cytoscape. Four core hub genes are found in these modules: FBXL13, WDFY2, bFGF (FGF2), and MTHFD1L. No core hub gene is found for one module (Cluster 4) (Table 5, Figure 5).
Table 5.
Significant risk gene modules.
| Cluster | Score | Nodes | Edges | Node IDs |
|---|---|---|---|---|
| 1 | 20 | 20 | 190 |
ASB15, HECW2, UBE2D1, NEDD4L, ANAPC5, PJA2 TRIM39, UBE2R2, UBE2D4, CDC27, TRIM9, ZBTB16 LONRF1, PARK2, FBXL13 *, FBXO22, RNF19B, LMO7 RNF217, FBXO32 |
| 2 | 6.1 | 21 | 61 |
SYNJ1, KIT, PIK3C2G, PTPN11, PIK3C2B, SYNJ2 RUNX1, ITSN2, PLCB1, CDH2, DNM3, SYT1, CTTN WDFY2 *, CHRM2, CCND2, MITF, PLCG2, CDK6, ETV6, SH3GL2 |
| 3 | 5.5 | 13 | 33 | MLL, bFGF(FGF2) *, BUB1B, BARD1, RB1, DICER1, RAD50, RAD51, BCL2, CDH5, SMARCA2, ABL1, CCND3 |
| 4 | 4.182 | 12 | 23 |
AKT3, PTEN, ITPR1, PRKCE, GNB5, CDK1 ERBB4, GNG7, RAF1, GSK3B, PPM1B, MAP3K1 |
| 5 | 3 | 3 | 3 | FARS2, RAD51B, MTHFD1L * |
* core hub genes.
Figure 5.
Gene clusters identified with Cytoscape. Yellow nodes are core hub genes. No core hub gene is found in Cluster 4 (d). (a) Cluster 1; (b) Cluster 2; (c) Cluster 3; (d) Cluster 4; (e) Cluster 5.
3. Discussion
3.1. Most BD Risk Gene Products Are Located in the Nervous System
Our gene functional analysis of 2045 BD risk genes shows that most of their products are located in the nervous system, such as synapse and postsynapse. The result of GO biological process analysis shows most genes are involved in nervous system development. These two results verify each other and are consistent with previous studies [104]. BD risk genes may affect patients in two aspects: short-term and permanent. Environmental or internal factors may cause ectopic expression of some of the risk genes, which in turn cause episodes of BD. Some genes may work in the development of the nervous system and have a permanent effect on patients. This may explain why 60% patients will relapse into depression or mania within two years after treatment [105].
3.2. Intense Overlappings of Genes Associated with BD and Other Mental Disorders
Many symptoms and signs overlap between different mental disorders and patients often present with features of more than one disorder [106]. This may be caused by underlying genetic reasons. We compared BD risk genes with those of three other mental disorders and found intense overlaps. Similar results were also reported in other studies [28,52,86,89,106,107].
Schizophrenia and BD share the most associated genes. Previous work also found a significant correlation between a BP polygenic risk score and the clinical dimension of mania in schizophrenia patients [86]. PRKG1 was reported to be significantly associated with schizophrenia. In this study, we also find it is a hub gene in the network of BD risk genes. This gene encodes a cGMP-dependent protein kinase which acts as key mediator of the nitric oxide (NO)/cGMP signaling pathway. Another gene, SMARCA2, was also found to play a role in the pathophysiology of schizophrenia [27]. Its product is a transcription activator and involved in neuron differentiation. Many other risk genes are also found involved in signal transduction and nervous system development. This suggests that these two diseases may share some common underlying pathways.
3.3. Core Hub Genes Give New Insights of BD
We combined protein–protein network and genome wide association analysis in this study and found four core hub genes. Although genes with higher degrees are more frequently reported to be associated with BD, two core hub genes (WDFY2 and FBXL13) have relatively low degrees (20 and 19, respectively).
bFGF has not been reported to be associated with BD before, but is usually used for treatment of neurodegenerative diseases such as Alzheimer’s disease [42]. It plays an essential role in regulation of cell proliferation, differentiation, and migration. bFGF is found as a core hub gene implies the abnormal nervous development of BD patients.
There is no obvious evidence for another core hub gene, MTHFD1L, to be associated with BD, but it is thought to have an important effect on the pathophysiology of depression through rumination, and maybe via this cognitive intermediate phenotype on other mental and physical disorders [38].
WDFY2 is not directly associated with BD, but its product interacts with AKT1 [108], which has been found involved in BD and schizophrenia [109]. This result suggests that the pathophysiology of BD is even more complicated than we thought. Some genes may play a role through its interaction with genes directly associated with BD.
FBXL13 functions in the maturation of human dendritic cells [68] which are key regulators in the immune system and show mild aberrancies in bipolar disorder that can be fully restored to even activation after in vivo lithium treatment [67].
Interestingly, all the four core hub genes are not directly associated with BD. Although the role of these genes in the pathophysiology of BD requires further investigation, our method inspires new initiatives to find those genes that are important for BD but overlooked by studies using GWAS alone.
3.4. Effectiveness of GWAS Followed by Gene Network Analysis
GWAS is a successful tool for identifying human disease-associated genes. However, results of different studies often vary due to sampling even when a strict significant p-value of 5 × 10−8 is used [110]. In this study, a loose p-value threshold of 0.01 was used for the GWAS, and a gene network analysis was then used to find BD-associated genes in the GWAS result. Many resulted genes with high network degrees but relatively high GWAS p-values are reported to be associated with BD and/or other mental illnesses (Table 4), which suggests that the combination of the two methods is efficient in finding disease-related genes. It is necessary to use a loose p-value threshold in the first step to provide enough input genes for the following network analysis. A second screening using network degrees can help to make the final result more reliable.
Sklar et al. conducted a combined GWAS with 7481 BD cases and 9250 controls and identified CACNA1C and a miRNA located in the first intron of ODZ4 as BD-associated genes [87]. The calcium channel subunit coding gene CACNA1C has also been found to be associated with BD in previous studies [9,52,86] and is confirmed with a relatively high degree (30) in our results. However, the miRNA is not detected in this study, probably due to our relatively smaller sample size.
4. Materials and Methods
4.1. Bipolar Disorder Datasets
The dataset is from a study published by Wellcome Trust Case Control Consortium (WTCCC), which conducted a genome-wide scan of all SNPs of 17,000 British Caucasian loci by human SNPs genotyping chips. This dataset includes 14,000 disease samples from seven common complex diseases: bipolar disorder, bipolar depression, Crohn’s disease, hypertension, rheumatoid arthritis, type 1 diabetes, type 2 diabetes, and 3000 healthy control samples, which has been completed by more than 50 research teams [111]. The dataset is downloaded from WTCCC website [112]. This study uses the BD part of the dataset. Human SNP annotation data and human reference sequence data are downloaded from NCBI (https://www.ncbi.nlm.nih.gov/), which contain 336,843,011 SNPs on 24 human chromosomes and the start and end of genes in which they are located [113].
4.2. Screening of Risk SNPs
SNP sites that do not meet one of the following criteria are excluded for quality control: Hardy–Weinberg equilibrium test (Bonferroni corrected p < 5 × 10−7), missingness >5%, minor allele frequency <5%, and odds ratio R2 > 0.8. Risk SNPs are screened under p < 0.01. Quality control and risk gene screening are finished with Plink software [114].
4.3. Mapping Significant Risk SNPs to Genes
Risk SNPs are mapped onto human genes by comparing them with transcription start sites and stop sites. An SNP will be mapped onto its nearest gene within 5 kb if it is not located within any gene. SNPs located outside of the 5 kb of genes are removed.
4.4. Gene Function and Disease Enrichment Analysis
FunRich [115] software is used to carry out gene enrichment analysis with p < 0.01. Results are reversely ordered by FDR-values, and only the first 10 results are listed in each GO section. ToppGene [116] is used to enrich genes in four different mental illnesses.
4.5. Protein Network Analysis
STRING database [117] is used to find a protein–protein relationship and FunRich is then used to map BD risk genes to the protein–protein network. Those protein (gene) nodes with degree >17 are sifted out as hub genes, which are further analyzed with the MCODE plugin [118] of Cytoscape [119] to find out network clusters (modules) and core hub genes. The node gene with the highest MCODE node score in a cluster is designated as its core hub gene, which is crucial for the cluster.
The topological properties of a gene cluster include [120,121] the following: (1) degree, the number of genes directly connected to a gene, (2) the cluster coefficient (CC), the coincidence of the common regulatory genes between two adjacent genes, defined as
| (1) |
where represents the number of edges of the neighbors that connect to node —the mean of the clustering coefficients of all nodes is designated as the clustering coefficient of the network—(3) the shortest path, the path with the least edges between two nodes, and (4) betweenness (B(v)), the sum of the ratios of number of shortest paths connecting to a node to that of all shortest paths in a network
| (2) |
where is total number of shortest paths from node s to t, and is the number of those paths that pass through v.
Acknowledgments
This study was financially supported by the Special Project of National Science and Technology Cooperation (2014DFB30010), National Natural Science Foundation of China (61501071) and the Science and Technology Research Program of Chongqing Municipal Education Commission (KJ1704094). We thank the three anonymous reviewers for their constructive comments.
Abbreviations
| BD | bipolar disorder |
| GWAS | genome-wide association study |
| SNP | single nucleotide polymorphism |
| WTCCC | the World Healthcare Case Control Association |
Appendix A
Table A1.
Gene information of the nodes with a network degree greater than 17.
| GENE | Degree | Ensembl | UniProtKB |
|---|---|---|---|
| CDK1 | 62 | ENSG00000170312 | P06493 |
| PTEN | 61 | ENSG00000171862 | P60484 |
| BCL2 | 60 | ENSG00000171791 | P10415 |
| POLR2A | 55 | ENSG00000181222 | P24928 |
| SMARCA2 | 55 | ENSG00000080503 | P5153 |
| GSK3B | 54 | ENSG00000082701 | P49841 |
| ABL1 | 53 | ENSG00000097007 | P00519 |
| PRKCA | 50 | ENSG00000154229 | P17252 |
| FGF2 | 48 | ENSG00000138685 | P09038 |
| RB1 | 45 | ENSG00000139687 | P06400 |
| KIT | 40 | ENSG00000157404 | P10721 |
| RAD51 | 38 | ENSG00000051180 | Q06609 |
| SIRT1 | 38 | ENSG00000096717 | Q96EB6 |
| UBE2D1 | 37 | ENSG00000072401 | P51668 |
| DLG1 | 36 | ENSG00000075711 | Q12959 |
| CDC27 | 35 | ENSG00000004897 | P30260 |
| NEDD4L | 35 | ENSG00000049759 | Q96PU5 |
| PRKG1 | 35 | ENSG00000185532 | Q13976 |
| RAP1A | 34 | ENSG00000116473 | P62834 |
| CDH2 | 33 | ENSG00000170558 | P19022 |
| GNB5 | 33 | ENSG00000069966 | O14775 |
| MAPK6 | 33 | ENSG00000069956 | Q16659 |
| GNG7 | 32 | ENSG00000176533 | O60262 |
| PTPN11 | 32 | ENSG00000179295 | Q06124 |
| ZBTB16 | 32 | ENSG00000109906 | Q05516 |
| ADCY8 | 31 | ENSG00000155897 | P40145 |
| DICER1 | 31 | ENSG00000100697 | Q9UPY3 |
| SYNJ1 | 31 | ENSG00000159082 | O43426 |
| CACNA1C | 30 | ENSG00000151067 | Q13936 |
| CTTN | 30 | ENSG00000085733 | Q14247 |
| DLG2 | 30 | ENSG00000150672 | Q15700 |
| MAP3K1 | 30 | ENSG00000095015 | Q13233 |
| RIT2 | 30 | ENSG00000152214 | Q99578 |
| ANAPC5 | 28 | ENSG00000089053 | Q9UJX4 |
| PLCB1 | 28 | ENSG00000182621 | Q9NQ66 |
| RAF1 | 28 | ENSG00000132155 | P04049 |
| PARK2 | 27 | ENSG00000185345 | O60260 |
| PLCG2 | 27 | ENSG00000197943 | P16885 |
| PNPLA6 | 27 | ENSG00000032444 | Q8IY17 |
| SYNJ2 | 27 | ENSG00000078269 | O15056 |
| UBE2R2 | 27 | ENSG00000107341 | Q712K3 |
| CACNA1D | 26 | ENSG00000157388 | Q01668 |
| CDK6 | 26 | ENSG00000105810 | Q00534 |
| CHRM2 | 26 | ENSG00000181072 | P08172 |
| MTHFD1L | 26 | ENSG00000120254 | Q6UB35 |
| GRIA1 | 25 | ENSG00000155511 | P42261 |
| POLR2H | 25 | ENSG00000163882 | P52434 |
| TJP1 | 25 | ENSG00000104067 | Q07157 |
| MAPRE1 | 24 | ENSG00000101367 | Q15691 |
| RUNX1 | 24 | ENSG00000159216 | Q01196 |
| UBE2D4 | 24 | ENSG00000078967 | Q9Y2X8 |
| EHHADH | 23 | ENSG00000113790 | Q08426 |
| IQCB1 | 23 | ENSG00000173226 | Q15051 |
| PPM1B | 23 | ENSG00000138032 | O75688 |
| PPP4C | 23 | ENSG00000149923 | P60510 |
| RAD50 | 23 | ENSG00000113522 | Q92878 |
| SH3GL2 | 23 | ENSG00000107295 | Q99962 |
| DCTN1 | 22 | ENSG00000204843 | Q14203 |
| ERBB4 | 22 | ENSG00000178568 | Q15303 |
| FBXO32 | 22 | ENSG00000156804 | Q969P5 |
| ITPR1 | 22 | ENSG00000150995 | Q14643 |
| MLL | 22 | ENSG00000118058 | Q03164 |
| NCOR2 | 22 | ENSG00000196498 | Q9Y618 |
| PRKCE | 22 | ENSG00000171132 | Q02156 |
| RAD51B | 22 | ENSG00000182185 | O15315 |
| ACTN4 | 21 | ENSG00000130402 | O43707 |
| CCND2 | 21 | ENSG00000118971 | P30279 |
| CDH5 | 21 | ENSG00000179776 | P33151 |
| CUL4A | 21 | ENSG00000139842 | Q13619 |
| EFCAB13 | 21 | ENSG00000178852 | Q8IY85 |
| LMO7 | 21 | ENSG00000136153 | Q8WWI1 |
| MITF | 21 | ENSG00000187098 | O75030 |
| TRIM9 | 21 | ENSG00000100505 | Q9C026 |
| CCND3 | 20 | ENSG00000112576 | P30281 |
| EPHB1 | 20 | ENSG00000154928 | P54762 |
| FARS2 | 20 | ENSG00000145982 | O95363 |
| FBXO22 | 20 | ENSG00000167196 | Q8NEZ5 |
| FLT3 | 20 | ENSG00000122025 | P36888 |
| GATA4 | 20 | ENSG00000136574 | P43694 |
| ITSN2 | 20 | ENSG00000198399 | Q9NZM3 |
| KIF18A | 20 | ENSG00000121621 | Q8NI77 |
| LONRF1 | 20 | ENSG00000154359 | Q17RB8 |
| NCOA3 | 20 | ENSG00000124151 | Q9Y6Q9 |
| PCNT | 20 | ENSG00000160299 | O95613 |
| PJA2 | 20 | ENSG00000198961 | O43164 |
| SYT1 | 20 | ENSG00000067715 | P21579 |
| TRIM39 | 20 | ENSG00000204599 | Q9HCM9 |
| WDFY2 | 20 | ENSG00000139668 | Q96P53 |
| AK4 | 19 | ENSG00000162433 | P27144 |
| ASB15 | 19 | ENSG00000146809 | Q8WXK1 |
| ATF2 | 19 | ENSG00000115966 | P15336 |
| BUB1B | 19 | ENSG00000156970 | O60566 |
| DHX15 | 19 | ENSG00000109606 | O43143 |
| DNM3 | 19 | ENSG00000197959 | Q9UQ16 |
| ETV6 | 19 | ENSG00000139083 | P41212 |
| FBXL13 | 19 | ENSG00000161040 | Q8NEE6 |
| HECW2 | 19 | ENSG00000138411 | Q9P2P5 |
| MEF2C | 19 | ENSG00000081189 | Q06413 |
| NR3C1 | 19 | ENSG00000113580 | P04150 |
| PDE4D | 19 | ENSG00000113448 | Q08499 |
| RNF19B | 19 | ENSG00000116514 | Q6ZMZ0 |
| RNF217 | 19 | ENSG00000146373 | Q8TC41 |
| RXFP2 | 19 | ENSG00000133105 | Q8WXD0 |
| RYR1 | 19 | ENSG00000196218 | P21817 |
| THBS2 | 19 | ENSG00000186340 | P35442 |
| AKT3 | 18 | ENSG00000117020 | Q9Y243 |
| BARD1 | 18 | ENSG00000138376 | Q99728 |
| CTNNA2 | 18 | ENSG00000066032 | P26232 |
| HDAC7 | 18 | ENSG00000061273 | Q8WUI4 |
| ITGAV | 18 | ENSG00000138448 | P06756 |
| PARD3 | 18 | ENSG00000148498 | Q8TEW0 |
| PCSK2 | 18 | ENSG00000125851 | P16519 |
| PIK3C2B | 18 | ENSG00000133056 | O00750 |
| PIK3C2G | 18 | ENSG00000139144 | O75747 |
| UBQLN1 | 18 | ENSG00000135018 | Q9UMX0 |
Author Contributions
Zengyan Xie contributed literature search, study design, data interpretation, and wrote the paper and provided study supervision. Xianyan Yang contributed literature search, figures, study design, data collection, data analysis, data interpretation and wrote the paper. Xiaoya Deng contributed literature search and data checking. Mingyue Ma contributed paper revising. Kunxian Shu contributed study design and provided study supervision.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Craddock N., Sklar P. Genetics of bipolar disorder. Lancet. 2013;381:1654–1662. doi: 10.1016/S0140-6736(13)60855-7. [DOI] [PubMed] [Google Scholar]
- 2.Akiskal H.S., Bourgeois M.L., Angst J., Post R., Moller H., Hirschfeld R. Re-evaluating the prevalence of and diagnostic composition within the broad clinical spectrum of bipolar disorders. J. Affect. Disord. 2000;59:S5–S30. doi: 10.1016/S0165-0327(00)00203-2. [DOI] [PubMed] [Google Scholar]
- 3.Kessler R.C., Akiskal H.S., Ames M., Birnbaum H., Greenberg P., Hirschfeld R.M., Jin R., Merikangas K.R., Simon G.E., Wang P.S. Prevalence and effects of mood disorders on work performance in a nationally representative sample of U.S. workers. Am. J. Psychiatry. 2006;163:1561–1568. doi: 10.1176/ajp.2006.163.9.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grande I., Berk M., Birmaher B., Vieta E. Bipolar disorder. Lancet. 2016;387:1561–1572. doi: 10.1016/S0140-6736(15)00241-X. [DOI] [PubMed] [Google Scholar]
- 5.Smoller J.W., Finn C.T. Family, twin, and adoption studies of bipolar disorder. Am. J. Med. Genet. Part C Semin. Med. Genet. 2003;123C:48–58. doi: 10.1002/ajmg.c.20013. [DOI] [PubMed] [Google Scholar]
- 6.Barnett J.H., Smoller J.W. The genetics of bipolar disorder. Neuroscience. 2009;164:331–343. doi: 10.1016/j.neuroscience.2009.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan P.F., Daly M.J., O’Donovan M. Genetic architectures of psychiatric disorders: The emerging picture and its implications. Nat. Rev. Genet. 2012;13:537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lescai F., Als T.D., Li Q., Nyegaard M., Andorsdottir G., Biskopsto M., Hedemand A., Fiorentino A., O’Brien N., Jarram A., et al. Whole-exome sequencing of individuals from an isolated population implicates rare risk variants in bipolar disorder. Transl. Psychiatry. 2017;7:e1034. doi: 10.1038/tp.2017.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sklar P., Smoller J.W., Fan J., Ferreira M.A., Perlis R.H., Chambert K., Nimgaonkar V.L., McQueen M.B., Faraone S.V., Kirby A., et al. Whole-genome association study of bipolar disorder. Mol. Psychiatry. 2008;13:558–569. doi: 10.1038/sj.mp.4002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferreira M.A., O’Donovan M.C., Meng Y.A., Jones I.R., Ruderfer D.M., Jones L., Fan J., Kirov G., Perlis R.H., Green E.K., et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat. Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott L.J., Muglia P., Kong X.Q., Guan W., Flickinger M., Upmanyu R., Tozzi F., Li J.Z., Burmeister M., Absher D., et al. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proc. Natl. Acad. Sci. USA. 2009;106:7501–7506. doi: 10.1073/pnas.0813386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu W., Cohen-Woods S., Chen Q., Noor A., Knight J., Hosang G., Parikh S.V., De Luca V., Tozzi F., Muglia P., et al. Genome-wide association study of bipolar disorder in Canadian and UK populations corroborates disease loci including SYNE1 and CSMD1. BMC Med. Genet. 2014;15:2. doi: 10.1186/1471-2350-15-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baum A.E., Akula N., Cabanero M., Cardona I., Corona W., Klemens B., Schulze T.G., Cichon S., Rietschel M., Nothen M.M., et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol. Psychiatry. 2008;13:197–207. doi: 10.1038/sj.mp.4002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin N.W., Medland S.E., Verweij K.J., Lee S.H., Nyholt D.R., Madden P.A., Heath A.C., Montgomery G.W., Wright M.J., Martin N.G. Educational attainment: A genome wide association study in 9538 Australians. PLoS ONE. 2011;6:e20128. doi: 10.1371/journal.pone.0020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGuffin P., Rijsdijk F., Andrew M., Sham P., Katz R., Cardno A. The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch. Gen. Psychiatry. 2003;60:497–502. doi: 10.1001/archpsyc.60.5.497. [DOI] [PubMed] [Google Scholar]
- 16.Song Y., Wang M., Mao F., Shao M., Zhao B., Song Z., Shao C., Gong Y. Knockdown of Pnpla6 protein results in motor neuron defects in zebrafish. Dis. Models Mech. 2013;6:404–413. doi: 10.1242/dmm.009688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le-Niculescu H., Levey D.F., Ayalew M., Palmer L., Gavrin L.M., Jain N., Winiger E., Bhosrekar S., Shankar G., Radel M., et al. Discovery and validation of blood biomarkers for suicidality. Mol. Psychiatry. 2013;18:1249–1264. doi: 10.1038/mp.2013.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quintero-Rivera F., Sharifi-Hannauer P., Martinez-Agosto J.A. Autistic and psychiatric findings associated with the 3q29 microdeletion syndrome: Case report and review. Am. J. Med. Genet. Part A. 2010;152A:2459–2467. doi: 10.1002/ajmg.a.33573. [DOI] [PubMed] [Google Scholar]
- 19.Uemura T., Green M., Corson T.W., Perova T., Li P.P., Warsh J.J. Bcl-2 SNP rs956572 associates with disrupted intracellular calcium homeostasis in bipolar I disorder. Bipolar Disord. 2011;13:41–51. doi: 10.1111/j.1399-5618.2011.00897.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhernakova D.V., de Klerk E., Westra H.J., Mastrokolias A., Amini S., Ariyurek Y., Jansen R., Penninx B.W., Hottenga J.J., Willemsen G., et al. DeepSAGE reveals genetic variants associated with alternative polyadenylation and expression of coding and non-coding transcripts. PLoS Genet. 2013;9:e1003594. doi: 10.1371/annotation/296056cb-f80c-4b04-985b-180f6d3cc4ae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silberberg G., Baruch K., Navon R. Detection of stable reference genes for real-time PCR analysis in schizophrenia and bipolar disorder. Anal. Biochem. 2009;391:91–97. doi: 10.1016/j.ab.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 22.Ross J., Gedvilaite E., Badner J.A., Erdman C., Baird L., Matsunami N., Leppert M., Xing J., Byerley W. A Rare Variant in CACNA1D Segregates with 7 Bipolar I Disorder Cases in a Large Pedigree. Mol. Neuropsychiatry. 2016;2:145–150. doi: 10.1159/000448041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabir Z.D., Martinez-Rivera A., Rajadhyaksha A.M. From Gene to Behavior: L-Type Calcium Channel Mechanisms Underlying Neuropsychiatric Symptoms. Neurotherapeutics. 2017;14:588–613. doi: 10.1007/s13311-017-0532-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez-Rivera A., Hao J., Tropea T.F., Giordano T.P., Kosovsky M., Rice R.C., Lee A., Huganir R.L., Striessnig J., Addy N.A., et al. Enhancing VTA Cav1.3 L-type Ca2+ channel activity promotes cocaine and mood-related behaviors via overlapping AMPA receptor mechanisms in the nucleus accumbens. Mol. Psychiatry. 2017;22:1735–1745. doi: 10.1038/mp.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teschler S., Bartkuhn M., Kunzel N., Schmidt C., Kiehl S., Dammann G., Dammann R. Aberrant methylation of gene associated CpG sites occurs in borderline personality disorder. PLoS ONE. 2013;8:e84180. doi: 10.1371/journal.pone.0084180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sengupta S., Xiong L., Fathalli F., Benkelfat C., Tabbane K., Danics Z., Labelle A., Lal S., Krebs M.O., Rouleau G., et al. Association study of the trinucleotide repeat polymorphism within SMARCA2 and schizophrenia. BMC Genet. 2006;7:34. doi: 10.1186/1471-2156-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koga M., Ishiguro H., Yazaki S., Horiuchi Y., Arai M., Niizato K., Iritani S., Itokawa M., Inada T., Iwata N., et al. Involvement of SMARCA2/BRM in the SWI/SNF chromatin-remodeling complex in schizophrenia. Hum. Mol. Genet. 2009;18:2483–2494. doi: 10.1093/hmg/ddp166. [DOI] [PubMed] [Google Scholar]
- 28.Benes F.M., Lim B., Subburaju S. Site-specific regulation of cell cycle and DNA repair in post-mitotic GABA cells in schizophrenic versus bipolars. Proc. Natl. Acad. Sci. USA. 2009;106:11731–11736. doi: 10.1073/pnas.0903066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vine A.E., McQuillin A., Bass N.J., Pereira A., Kandaswamy R., Robinson M., Lawrence J., Anjorin A., Sklar P., Gurling H.M., et al. No evidence for excess runs of homozygosity in bipolar disorder. Psychiatr. Genet. 2009;19:165–170. doi: 10.1097/YPG.0b013e32832a4faa. [DOI] [PubMed] [Google Scholar]
- 30.Benedetti F., Serretti A., Colombo C., Lorenzi C., Tubazio V., Smeraldi E. A glycogen synthase kinase 3-beta promoter gene single nucleotide polymorphism is associated with age at onset and response to total sleep deprivation in bipolar depression. Neurosci. Lett. 2004;368:123–126. doi: 10.1016/j.neulet.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 31.Hu S., Begum A.N., Jones M.R., Oh M.S., Beech W.K., Beech B.H., Yang F., Chen P., Ubeda O.J., Kim P.C., et al. GSK3 inhibitors show benefits in an Alzheimer’s disease (AD) model of neurodegeneration but adverse effects in control animals. Neurobiol. Dis. 2009;33:193–206. doi: 10.1016/j.nbd.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X., Jope R.S. Is glycogen synthase kinase-3 a central modulator in mood regulation? Neuropsychopharmacology. 2010;35:2143–2154. doi: 10.1038/npp.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benedetti F., Bollettini I., Barberi I., Radaelli D., Poletti S., Locatelli C., Pirovano A., Lorenzi C., Falini A., Colombo C., et al. Lithium and GSK3-beta promoter gene variants influence white matter microstructure in bipolar disorder. Neuropsychopharmacology. 2013;38:313–327. doi: 10.1038/npp.2012.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cannon D.M., Klaver J.K., Gandhi S.K., Solorio G., Peck S.A., Erickson K., Akula N., Savitz J., Eckelman W.C., Furey M.L., et al. Genetic variation in cholinergic muscarinic-2 receptor gene modulates M2 receptor binding in vivo and accounts for reduced binding in bipolar disorder. Mol. Psychiatry. 2011;16:407–418. doi: 10.1038/mp.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munkholm K., Peijs L., Vinberg M., Kessing L.V. A composite peripheral blood gene expression measure as a potential diagnostic biomarker in bipolar disorder. Transl. Psychiatry. 2015;5:e614. doi: 10.1038/tp.2015.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kato T., Hayashi-Takagi A., Toyota T., Yoshikawa T., Iwamoto K. Gene expression analysis in lymphoblastoid cells as a potential biomarker of bipolar disorder. J. Hum. Genet. 2011;56:779–783. doi: 10.1038/jhg.2011.101. [DOI] [PubMed] [Google Scholar]
- 37.Munkholm K., Peijs L., Kessing L.V., Vinberg M. Reduced mRNA expression of PTGDS in peripheral blood mononuclear cells of rapid-cycling bipolar disorder patients compared with healthy control subjects. Int. J. Neuropsychopharmacol. 2014;18 doi: 10.1093/ijnp/pyu101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eszlari N., Kovacs D., Petschner P., Pap D., Gonda X., Elliott R., Anderson I.M., Deakin J.F., Bagdy G., Juhasz G. Distinct effects of folate pathway genes MTHFR and MTHFD1L on ruminative response style: A potential risk mechanism for depression. Transl. Psychiatry. 2016;6:e745. doi: 10.1038/tp.2016.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kittel-Schneider S., Lorenz C., Auer J., Weissflog L., Reif A. DGKH genetic risk variant influences gene expression in bipolar affective disorder. J. Affect. Disord. 2016;198:148–157. doi: 10.1016/j.jad.2016.03.041. [DOI] [PubMed] [Google Scholar]
- 40.Carroll L.S., Williams N.M., Moskvina V., Russell E., Norton N., Williams H.J., Peirce T., Georgieva L., Dwyer S., Grozeva D., et al. Evidence for rare and common genetic risk variants for schizophrenia at protein kinase C, alpha. Mol. Psychiatry. 2010;15:1101–1111. doi: 10.1038/mp.2009.96. [DOI] [PubMed] [Google Scholar]
- 41.Kerner B., Jasinska A.J., DeYoung J., Almonte M., Choi O.W., Freimer N.B. Polymorphisms in the GRIA1 gene region in psychotic bipolar disorder. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2009;150B:24–32. doi: 10.1002/ajmg.b.30780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng C., Zhang C., Shao X., Liu Q., Qian Y., Feng L., Chen J., Zha Y., Zhang Q., Jiang X. Enhancement of nose-to-brain delivery of basic fibroblast growth factor for improving rat memory impairments induced by co-injection of beta-amyloid and ibotenic acid into the bilateral hippocampus. Int. J. Pharm. 2012;423:226–234. doi: 10.1016/j.ijpharm.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 43.Numata S., Iga J., Nakataki M., Tayoshi S., Tanahashi T., Itakura M., Ueno S., Ohmori T. Positive association of the pericentrin (PCNT) gene with major depressive disorder in the Japanese population. J. Psychiatry Neurosci. 2009;34:195–198. [PMC free article] [PubMed] [Google Scholar]
- 44.De Ligt J., Willemsen M.H., van Bon B.W., Kleefstra T., Yntema H.G., Kroes T., Vulto-van Silfhout A.T., Koolen D.A., de Vries P., Gilissen C., et al. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 45.Iossifov I., Zheng T., Baron M., Gilliam T.C., Rzhetsky A. Genetic-linkage mapping of complex hereditary disorders to a whole-genome molecular-interaction network. Genome Res. 2008;18:1150–1162. doi: 10.1101/gr.075622.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Drago A., Crisafulli C., Sidoti A., Calabro M., Serretti A. The microtubule-associated molecular pathways may be genetically disrupted in patients with Bipolar Disorder. Insights from the molecular cascades. J. Affect. Disord. 2016;190:429–438. doi: 10.1016/j.jad.2015.10.016. [DOI] [PubMed] [Google Scholar]
- 47.Cupertino R.B., Kappel D.B., Bandeira C.E., Schuch J.B., da Silva B.S., Muller D., Bau C.H., Mota N.R. SNARE complex in developmental psychiatry: Neurotransmitter exocytosis and beyond. J. Neural Transm. 2016;123:867–883. doi: 10.1007/s00702-016-1514-9. [DOI] [PubMed] [Google Scholar]
- 48.Serretti A., Mandelli L. The genetics of bipolar disorder: Genome ‘hot regions’, genes, new potential candidates and future directions. Mol. Psychiatry. 2008;13:742–771. doi: 10.1038/mp.2008.29. [DOI] [PubMed] [Google Scholar]
- 49.Kishi T., Fukuo Y., Kitajima T., Okochi T., Yamanouchi Y., Kinoshita Y., Kawashima K., Inada T., Kunugi H., Kato T., et al. SIRT1 gene, schizophrenia and bipolar disorder in the Japanese population: An association study. Genes Brain Behav. 2011;10:257–263. doi: 10.1111/j.1601-183X.2010.00661.x. [DOI] [PubMed] [Google Scholar]
- 50.Herskovits A.Z., Guarente L. SIRT1 in neurodevelopment and brain senescence. Neuron. 2014;81:471–483. doi: 10.1016/j.neuron.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nivoli A., Porcelli S., Albani D., Forloni G., Fusco F., Colom F., Vieta E., Serretti A. Association between Sirtuin 1 Gene rs10997870 Polymorphism and Suicide Behaviors in Bipolar Disorder. Neuropsychobiology. 2016;74:1–7. doi: 10.1159/000446921. [DOI] [PubMed] [Google Scholar]
- 52.Moskvina V., Craddock N., Holmans P., Nikolov I., Pahwa J.S., Green E., Wellcome Trust Case Control Consortium. Owen M.J., O’Donovan M.C. Gene-wide analyses of genome-wide association data sets: Evidence for multiple common risk alleles for schizophrenia and bipolar disorder and for overlap in genetic risk. Mol. Psychiatry. 2009;14:252–260. doi: 10.1038/mp.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smalheiser N.R., Lugli G., Rizavi H.S., Torvik V.I., Turecki G., Dwivedi Y. MicroRNA expression is down-regulated and reorganized in prefrontal cortex of depressed suicide subjects. PLoS ONE. 2012;7:e33201. doi: 10.1371/journal.pone.0033201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dwivedi Y. Emerging role of microRNAs in major depressive disorder: Diagnosis and therapeutic implications. Dialogues Clin. Neurosci. 2014;16:43–61. doi: 10.31887/DCNS.2014.16.1/ydwivedi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uezato A., Yamamoto N., Iwayama Y., Hiraoka S., Hiraaki E., Umino A., Haramo E., Umino M., Yoshikawa T., Nishikawa T. Reduced cortical expression of a newly identified splicing variant of the DLG1 gene in patients with early-onset schizophrenia. Transl. Psychiatry. 2015;5:e654. doi: 10.1038/tp.2015.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xing J., Kimura H., Wang C., Ishizuka K., Kushima I., Arioka Y., Yoshimi A., Nakamura Y., Shiino T., Oya-Ito T., et al. Resequencing and Association Analysis of Six PSD-95-Related Genes as Possible Susceptibility Genes for Schizophrenia and Autism Spectrum Disorders. Sci. Rep. 2016;6:27491. doi: 10.1038/srep27491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hunsberger J.G., Chibane F.L., Elkahloun A.G., Henderson R., Singh R., Lawson J., Cruceanu C., Nagarajan V., Turecki G., Squassina A., et al. Novel integrative genomic tool for interrogating lithium response in bipolar disorder. Transl. Psychiatry. 2015;5:e504. doi: 10.1038/tp.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Le-Niculescu H., Patel S.D., Bhat M., Kuczenski R., Faraone S.V., Tsuang M.T., McMahon F.J., Schork N.J., Nurnberger J.I., Jr., Niculescu A.B., 3rd Convergent functional genomics of genome-wide association data for bipolar disorder: Comprehensive identification of candidate genes, pathways and mechanisms. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2009;150B:155–181. doi: 10.1002/ajmg.b.30887. [DOI] [PubMed] [Google Scholar]
- 59.Chen H., Ross C.A., Wang N., Huo Y., MacKinnon D.F., Potash J.B., Simpson S.G., McMahon F.J., DePaulo J.R., Jr., McInnis M.G. NEDD4L on human chromosome 18q21 has multiple forms of transcripts and is a homologue of the mouse Nedd4-2 gene. Eur. J. Hum. Genet. 2001;9:922–930. doi: 10.1038/sj.ejhg.5200747. [DOI] [PubMed] [Google Scholar]
- 60.Birnbaum R., Jaffe A.E., Hyde T.M., Kleinman J.E., Weinberger D.R. Prenatal expression patterns of genes associated with neuropsychiatric disorders. Am. J. Psychiatry. 2014;171:758–767. doi: 10.1176/appi.ajp.2014.13111452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao Z., Webb B.T., Jia P., Bigdeli T.B., Maher B.S., van den Oord E., Bergen S.E., Amdur R.L., O’Neill F.A., Walsh D., et al. Association study of 167 candidate genes for schizophrenia selected by a multi-domain evidence-based prioritization algorithm and neurodevelopmental hypothesis. PLoS ONE. 2013;8:e67776. doi: 10.1371/journal.pone.0067776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sakai M., Watanabe Y., Someya T., Araki K., Shibuya M., Niizato K., Oshima K., Kunii Y., Yabe H., Matsumoto J., et al. Assessment of copy number variations in the brain genome of schizophrenia patients. Mol. Cytogenet. 2015;8:46. doi: 10.1186/s13039-015-0144-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moya P.R., Dodman N.H., Timpano K.R., Rubenstein L.M., Rana Z., Fried R.L., Reichardt L.F., Heiman G.A., Tischfield J.A., King R.A., et al. Rare missense neuronal cadherin gene (CDH2) variants in specific obsessive-compulsive disorder and Tourette disorder phenotypes. Eur. J. Hum. Genet. (EJHG) 2013;21:850–854. doi: 10.1038/ejhg.2012.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lodder E.M., De Nittis P., Koopman C.D., Wiszniewski W., Moura de Souza C.F., Lahrouchi N., Guex N., Napolioni V., Tessadori F., Beekman L., et al. GNB5 Mutations Cause an Autosomal-Recessive Multisystem Syndrome with Sinus Bradycardia and Cognitive Disability. Am. J. Hum. Genet. 2016;99:704–710. doi: 10.1016/j.ajhg.2016.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen P., Chen J., Huang K., Ji W., Wang T., Li T., Wang Y., Wang H., He L., Feng G., et al. Analysis of association between common SNPs in ErbB4 and bipolar affective disorder, major depressive disorder and schizophrenia in the Han Chinese population. Prog. Neuro-psychopharmacol. Biol. Psychiatry. 2012;36:17–21. doi: 10.1016/j.pnpbp.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 66.Goes F.S., Rongione M., Chen Y.C., Karchin R., Elhaik E., Bipolar Genome S., Potash J.B. Exonic DNA sequencing of ERBB4 in bipolar disorder. PLoS ONE. 2011;6:e20242. doi: 10.1371/journal.pone.0020242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Knijff E.M., Ruwhof C., de Wit H.J., Kupka R.W., Vonk R., Akkerhuis G.W., Nolen W.A., Drexhage H.A. Monocyte-derived dendritic cells in bipolar disorder. Biol. Psychiatry. 2006;59:317–326. doi: 10.1016/j.biopsych.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 68.Ebstein F., Lange N., Urban S., Seifert U., Kruger E., Kloetzel P.M. Maturation of human dendritic cells is accompanied by functional remodelling of the ubiquitin-proteasome system. Int. J. Biochem. Cell Biol. 2009;41:1205–1215. doi: 10.1016/j.biocel.2008.10.023. [DOI] [PubMed] [Google Scholar]
- 69.Padmos R.C., Hillegers M.H., Knijff E.M., Vonk R., Bouvy A., Staal F.J., de Ridder D., Kupka R.W., Nolen W.A., Drexhage H.A. A discriminating messenger RNA signature for bipolar disorder formed by an aberrant expression of inflammatory genes in monocytes. Arch. Gen. Psychiatry. 2008;65:395–407. doi: 10.1001/archpsyc.65.4.395. [DOI] [PubMed] [Google Scholar]
- 70.Badner J.A., Koller D., Foroud T., Edenberg H., Nurnberger J.I., Jr., Zandi P.P., Willour V.L., McMahon F.J., Potash J.B., Hamshere M., et al. Genome-wide linkage analysis of 972 bipolar pedigrees using single-nucleotide polymorphisms. Mol. Psychiatry. 2012;17:818–826. doi: 10.1038/mp.2011.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsuboi D., Kuroda K., Tanaka M., Namba T., Iizuka Y., Taya S., Shinoda T., Hikita T., Muraoka S., Iizuka M., et al. Disrupted-in-schizophrenia 1 regulates transport of ITPR1 mRNA for synaptic plasticity. Nat. Neurosci. 2015;18:698–707. doi: 10.1038/nn.3984. [DOI] [PubMed] [Google Scholar]
- 72.Mitchell A.C., Javidfar B., Pothula V., Ibi D., Shen E.Y., Peter C.J., Bicks L.K., Fehr T., Jiang Y., Brennand K.J., et al. MEF2C transcription factor is associated with the genetic and epigenetic risk architecture of schizophrenia and improves cognition in mice. Mol. Psychiatry. 2017 doi: 10.1038/mp.2016.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hendriks W.J., Pulido R. Protein tyrosine phosphatase variants in human hereditary disorders and disease susceptibilities. Biochim. Biophys. Acta. 2013;1832:1673–1696. doi: 10.1016/j.bbadis.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 74.Arey R.N., Enwright J.F., 3rd, Spencer S.M., Falcon E., Ozburn A.R., Ghose S., Tamminga C., McClung C.A. An important role for cholecystokinin, a CLOCK target gene, in the development and treatment of manic-like behaviors. Mol. Psychiatry. 2014;19:342–350. doi: 10.1038/mp.2013.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Spijker A.T., van Rossum E.F., Hoencamp E., DeRijk R.H., Haffmans J., Blom M., Manenschijn L., Koper J.W., Lamberts S.W., Zitman F.G. Functional polymorphism of the glucocorticoid receptor gene associates with mania and hypomania in bipolar disorder. Bipolar Disord. 2009;11:95–101. doi: 10.1111/j.1399-5618.2008.00647.x. [DOI] [PubMed] [Google Scholar]
- 76.Spijker S., Van Zanten J.S., De Jong S., Penninx B.W., van Dyck R., Zitman F.G., Smit J.H., Ylstra B., Smit A.B., Hoogendijk W.J. Stimulated gene expression profiles as a blood marker of major depressive disorder. Biol. Psychiatry. 2010;68:179–186. doi: 10.1016/j.biopsych.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 77.Azevedo J.A., Carter B.S., Meng F., Turner D.L., Dai M., Schatzberg A.F., Barchas J.D., Jones E.G., Bunney W.E., Myers R.M., et al. The microRNA network is altered in anterior cingulate cortex of patients with unipolar and bipolar depression. J. Psychiatr. Res. 2016;82:58–67. doi: 10.1016/j.jpsychires.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Muhleisen T.W., Leber M., Schulze T.G., Strohmaier J., Degenhardt F., Treutlein J., Mattheisen M., Forstner A.J., Schumacher J., Breuer R., et al. Genome-wide association study reveals two new risk loci for bipolar disorder. Nat. Commun. 2014;5:3339. doi: 10.1038/ncomms4339. [DOI] [PubMed] [Google Scholar]
- 79.Perlis R.H., Huang J., Purcell S., Fava M., Rush A.J., Sullivan P.F., Hamilton S.P., McMahon F.J., Schulze T.G., Potash J.B., et al. Genome-wide association study of suicide attempts in mood disorder patients. Am. J. Psychiatry. 2010;167:1499–1507. doi: 10.1176/appi.ajp.2010.10040541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gouvea E.S., Ota V.K., Noto C., Santoro M.L., Spindola L.M., Moretti P.N., Carvalho C.M., Xavier G., Rios A.C., Sato J.R., et al. Gene expression alterations related to mania and psychosis in peripheral blood of patients with a first episode of psychosis. Transl. Psychiatry. 2016;6:e908. doi: 10.1038/tp.2016.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pulay A.J., Rethelyi J.M. Multimarker analysis suggests the involvement of BDNF signaling and microRNA biosynthesis in suicidal behavior. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2016;171:763–776. doi: 10.1002/ajmg.b.32433. [DOI] [PubMed] [Google Scholar]
- 82.Shelley W.B., Shelley E.D. A dermatologic diary. Portrait of a practice. Cutis. 1992;50:179–186. [PubMed] [Google Scholar]
- 83.Detera-Wadleigh S.D., Badner J.A., Berrettini W.H., Yoshikawa T., Goldin L.R., Turner G., Rollins D.Y., Moses T., Sanders A.R., Karkera J.D., et al. A high-density genome scan detects evidence for a bipolar-disorder susceptibility locus on 13q32 and other potential loci on 1q32 and 18p11.2. Proc. Natl. Acad. Sci. USA. 1999;96:5604–5609. doi: 10.1073/pnas.96.10.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Saito T., Guan F., Papolos D.F., Lau S., Klein M., Fann C.S., Lachman H.M. Mutation analysis of SYNJ1: A possible candidate gene for chromosome 21q22-linked bipolar disorder. Mol. Psychiatry. 2001;6:387–395. doi: 10.1038/sj.mp.4000871. [DOI] [PubMed] [Google Scholar]
- 85.McQuillin A., Bass N.J., Kalsi G., Lawrence J., Puri V., Choudhury K., Detera-Wadleigh S.D., Curtis D., Gurling H.M. Fine mapping of a susceptibility locus for bipolar and genetically related unipolar affective disorders, to a region containing the C21ORF29 and TRPM2 genes on chromosome 21q22.3. Mol. Psychiatry. 2006;11:134–142. doi: 10.1038/sj.mp.4001759. [DOI] [PubMed] [Google Scholar]
- 86.Ruderfer D.M., Fanous A.H., Ripke S., McQuillin A., Amdur R.L., Schizophrenia Working Group of the Psychiatric Genomics Consortium. Bipolar Disorder Working Group of the Psychiatric Genomics Consortium. Cross-Disorder Working Group of the Psychiatric Genomics Consortium. Gejman P.V., O’Donovan M.C., et al. Polygenic dissection of diagnosis and clinical dimensions of bipolar disorder and schizophrenia. Mol. Psychiatry. 2014;19:1017–1024. doi: 10.1038/mp.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Psychiatric GWAS Consortium Bipolar Disorder Working Group Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ruzicka W.B., Subburaju S., Benes F.M. Circuit- and Diagnosis-Specific DNA Methylation Changes at gamma-Aminobutyric Acid-Related Genes in Postmortem Human Hippocampus in Schizophrenia and Bipolar Disorder. JAMA Psychiatry. 2015;72:541–551. doi: 10.1001/jamapsychiatry.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Benes F.M., Lim B., Matzilevich D., Walsh J.P., Subburaju S., Minns M. Regulation of the GABA cell phenotype in hippocampus of schizophrenics and bipolars. Proc. Natl. Acad. Sci. USA. 2007;104:10164–10169. doi: 10.1073/pnas.0703806104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Noor A., Lionel A.C., Cohen-Woods S., Moghimi N., Rucker J., Fennell A., Thiruvahindrapuram B., Kaufman L., Degagne B., Wei J., et al. Copy number variant study of bipolar disorder in Canadian and UK populations implicates synaptic genes. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2014;165B:303–313. doi: 10.1002/ajmg.b.32232. [DOI] [PubMed] [Google Scholar]
- 91.MacLaren E.J., Charlesworth P., Coba M.P., Grant S.G. Knockdown of mental disorder susceptibility genes disrupts neuronal network physiology in vitro. Mol. Cell. Neurosci. 2011;47:93–99. doi: 10.1016/j.mcn.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hannah J., Zhou P. Distinct and overlapping functions of the cullin E3 ligase scaffolding proteins CUL4A and CUL4B. Gene. 2015;573:33–45. doi: 10.1016/j.gene.2015.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Howell K.R., Floyd K., Law A.J. PKBgamma/AKT3 loss-of-function causes learning and memory deficits and deregulation of AKT/mTORC2 signaling: Relevance for schizophrenia. PLoS ONE. 2017;12:e0175993. doi: 10.1371/journal.pone.0175993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hicks C., Asfour R., Pannuti A., Miele L. An integrative genomics approach to biomarker discovery in breast cancer. Cancer Inform. 2011;10:185–204. doi: 10.4137/CIN.S6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Emamalizadeh B., Jamshidi J., Movafagh A., Ohadi M., Khaniani M.S., Kazeminasab S., Biglarian A., Taghavi S., Motallebi M., Fazeli A., et al. RIT2 Polymorphisms: Is There a Differential Association? Mol. Neurobiol. 2017;54:2234–2240. doi: 10.1007/s12035-016-9815-4. [DOI] [PubMed] [Google Scholar]
- 96.Cross-Disorder Group of the Psychiatric Genomics Consortium Identification of risk loci with shared effects on five major psychiatric disorders: A genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vakalopoulos C. The effect of deficient muscarinic signaling on commonly reported biochemical effects in schizophrenia and convergence with genetic susceptibility loci in explaining symptom dimensions of psychosis. Front. Pharmacol. 2014;5:277. doi: 10.3389/fphar.2014.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lo Vasco V.R., Longo L., Polonia P. Phosphoinositide-specific Phospholipase C beta1 gene deletion in bipolar disorder affected patient. J. Cell Commun. Signal. 2013;7:25–29. doi: 10.1007/s12079-012-0182-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ban H.J., Kim S.C., Seo J., Kang H.B., Choi J.K. Genetic and metabolic characterization of insomnia. PLoS ONE. 2011;6:e18455. doi: 10.1371/journal.pone.0018455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kanazawa T., Ikeda M., Glatt S.J., Tsutsumi A., Kikuyama H., Kawamura Y., Nishida N., Miyagawa T., Hashimoto R., Takeda M., et al. Genome-wide association study of atypical psychosis. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2013;162B:679–686. doi: 10.1002/ajmg.b.32164. [DOI] [PubMed] [Google Scholar]
- 101.Yuan P., Zhou R., Wang Y., Li X., Li J., Chen G., Guitart X., Manji H.K. Altered levels of extracellular signal-regulated kinase signaling proteins in postmortem frontal cortex of individuals with mood disorders and schizophrenia. J. Affect. Disord. 2010;124:164–169. doi: 10.1016/j.jad.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kim S.K., Lee J.Y., Park H.J., Kim J.W., Chung J.H. Association study between polymorphisms of the PARD3 gene and schizophrenia. Exp. Ther. Med. 2012;3:881–885. doi: 10.3892/etm.2012.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Su L., Ling W., Jiang J., Hu J., Fan J., Guo X., Huang G., Xie X., Long J. Association of EPHB1 rs11918092 and EFNB2 rs9520087 with psychopathological symptoms of schizophrenia in Chinese Zhuang and Han populations. Asia-Pac. Psychiatry. 2016;8:306–308. doi: 10.1111/appy.12241. [DOI] [PubMed] [Google Scholar]
- 104.Chen H., Wang N., Zhao X., Ross C.A., O’Shea K.S., McInnis M.G. Gene expression alterations in bipolar disorder postmortem brains. Bipolar Disord. 2013;15:177–187. doi: 10.1111/bdi.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Geddes J.R., Miklowitz D.J. Treatment of bipolar disorder. Lancet. 2013;381:1672–1682. doi: 10.1016/S0140-6736(13)60857-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Doherty J.L., Owen M.J. Genomic insights into the overlap between psychiatric disorders: Implications for research and clinical practice. Genome Med. 2014;6:29. doi: 10.1186/gm546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lee S.A., Tsao T.T., Yang K.C., Lin H., Kuo Y.L., Hsu C.H., Lee W.K., Huang K.C., Kao C.Y. Construction and analysis of the protein–protein interaction networks for schizophrenia, bipolar disorder, and major depression. BMC Bioinform. 2011;12:S20. doi: 10.1186/1471-2105-12-S13-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fritzius T., Burkard G., Haas E., Heinrich J., Schweneker M., Bosse M., Zimmermann S., Frey A.D., Caelers A., Bachmann A.S., et al. A WD-FYVE protein binds to the kinases Akt and PKCzeta/lambda. Biochem. J. 2006;399:9–20. doi: 10.1042/BJ20060511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Karege F., Perroud N., Schurhoff F., Meary A., Marillier G., Burkhardt S., Ballmann E., Fernandez R., Jamain S., Leboyer M., et al. Association of AKT1 gene variants and protein expression in both schizophrenia and bipolar disorder. Genes Brain Behav. 2010;9:503–511. doi: 10.1111/j.1601-183X.2010.00578.x. [DOI] [PubMed] [Google Scholar]
- 110.Clarke G.M., Anderson C.A., Pettersson F.H., Cardon L.R., Morris A.P., Zondervan K.T. Basic statistical analysis in genetic case–control studies. Nat. Protoc. 2011;6:121–133. doi: 10.1038/nprot.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wellcome Trust Case Control Consortium. [(accessed on 15 December 2017)]; Available online: https://www.wtccc.org.uk/info/access_to_data_samples.html.
- 113.Brown G.R., Hem V., Katz K.S., Ovetsky M., Wallin C., Ermolaeva O., Tolstoy I., Tatusova T., Pruitt K.D., Maglott D.R., et al. Gene: A gene-centered information resource at NCBI. Nucleic Acids Res. 2015;43:D36–D42. doi: 10.1093/nar/gku1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Renteria M.E., Cortes A., Medland S.E. Using PLINK for Genome-Wide Association Studies (GWAS) and data analysis. Methods Mol. Biol. 2013;1019:193–213. doi: 10.1007/978-1-62703-447-0_8. [DOI] [PubMed] [Google Scholar]
- 115.Pathan M., Keerthikumar S., Ang C.S., Gangoda L., Quek C.Y., Williamson N.A., Mouradov D., Sieber O.M., Simpson R.J., Salim A., et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
- 116.Chen J., Bardes E.E., Aronow B.J., Jegga A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bader G.D., Hogue C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kohl M., Wiese S., Warscheid B. Cytoscape: Software for visualization and analysis of biological networks. Methods Mol. Biol. 2011;696:291–303. doi: 10.1007/978-1-60761-987-1_18. [DOI] [PubMed] [Google Scholar]
- 120.Barabasi A.L., Oltvai Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 121.Schweiger R., Linial M., Linial N. Generative probabilistic models for protein–protein interaction networks—The biclique perspective. Bioinformatics. 2011;27:i142–i148. doi: 10.1093/bioinformatics/btr201. [DOI] [PMC free article] [PubMed] [Google Scholar]