Abstract
Historically, progress in membrane protein research has been hindered due to solubility issues. The introduction of biomembrane mimetics has since stimulated the field’s momentum. One mimetic, the nanodisc, has proved to be an exceptional system for solubilizing membrane proteins. Herein, we critically evaluate the advantages and imperfections from employing nanodiscs in biophysical and biochemical studies. Specifically, we examine the techniques that have been modified to study membrane proteins in nanodiscs. Techniques discussed include fluorescence microscopy, solution state/solid state NMR, electron microscopy, SAXS, and several mass spectroscopy methods. Newer techniques such as SPR, charge sensitive optical detection, and scintillation proximity assays are also reviewed. Lastly, we cover nanodiscs advancing nanotechnology through nanoplasmonic biosensing, lipoprotein-nanoplatelets, and sortase-mediated labeling of nanodiscs.
Keywords: nanodisc, lipids, lipid composition, lipid bilayer, membrane bilayer, membrane protein, protein-protein interaction, POPC, POPS, DMPC, DMPG, cholesterol, protein-lipid interaction, native mass spectroscopy, electron microscopy, membrane scaffold protein (MSP) MALDI, ESI, small angle x-ray scattering, SAXS, fluorescence correlation spectroscopy FRET, solution phase and solid state NMR, isotope labeling, SPR
1. INTRODUCTION TO NANODISC TECHNOLOGY
Membrane proteins are a vital component of cells. Until recently, membrane proteins were not studied as extensively as cytosolic proteins. This is due to the propensity of membrane proteins to aggregate outside a membrane environment during their isolation from membranes using detergents. Therefore, in recent years, several lipid membrane mimics 1–3 were developed in order to facilitate interrogation of membrane proteins by rigorous biophysical and analytical techniques. In 2007, Sligar and coworkers at the University of Illinois at Urbana-Champaign developed a new technology termed as nanodiscs. Nanodiscs are essentially small nanoscale lipid bilayers that allows for the solubilization of membrane protein in a uniform, membrane-mimicking environment. Nanodiscs have several advantages over other types of lipid bilayer-mimicking technologies, including their homogeneity, control of membrane protein oligomeric state and long-term stability as has been discussed extensively in previous reviews 4, 5.
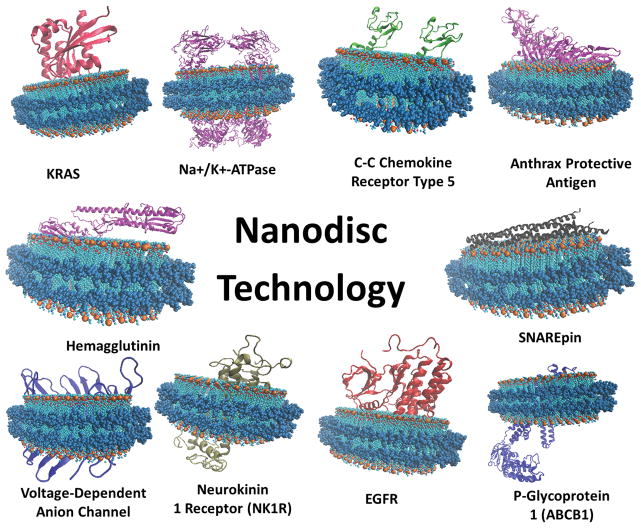
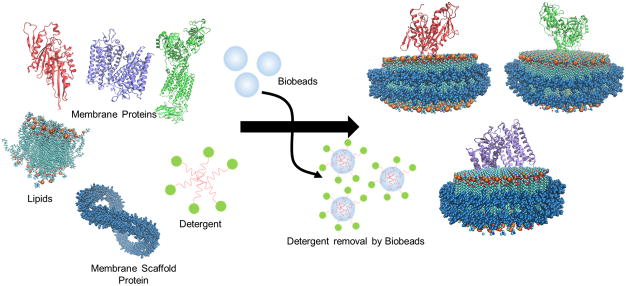
The formation of nanodiscs involves the incubation of detergent-solubilized phospholipids, target membrane protein and membrane scaffold protein (MSP) in a stoichiometric-dependent manner. The controlled ratio of these constituents is critical for proper nanodisc assembly and has been described and reviewed previously 6–8. Furthermore, the diameter of the assembled nanodiscs is dependent upon the length of the membrane scaffold protein (MSP) that is used. As such, there are various constructs of MSP that can be exploited to augment the size of the nanodiscs to fit the type of membrane protein system that is integrated. The two common constructs utilized for constructing nanodiscs are MSP1D1 and MSP1E3D1 9. MSP1D1 is used to form smaller nanodiscs with a diameter of ~9.7 nm whereas MSP1E3D1 results in larger nanodiscs with a diameter of ~12.9 nm 9. Additionally, less conventional constructs have been introduced that result in even larger nanodiscs with a diameter of 16–17 nm 10. By utilizing a carefully controlled ratio of the aforementioned constituents, the self-assembly of homogeneous nanodiscs can be initiated upon detergent removal. After the nanodiscs are assembled, size exclusion chromatography is typically utilized to separate the membrane protein nanodiscs from empty nanodiscs and other remnants of the assembly mixture. The homogenous peak consists of the membrane protein-nanodiscs that are collected and used for further biochemical studies 6–8. By capitalizing on its homogeneity, nanodisc technology has already been used to study a variety of membrane proteins (Figure 1, Table 1).
Figure 1.
Table 1.
Proteins incorporated into nanodiscs thus far and the biochemical and biophysical techniques used within each study.
| GPCRs | ||
|---|---|---|
| Protein | Techniques Utilized | Ref. |
|
| ||
| Rhodopsin | SAXS, Activity assays | 172 |
| Neurotensin receptor 1 | Dynamic light scattering (DLS), Analytical ultracentrifugation | 173 |
| μ-opioid receptor | Prism-based single-molecule total internal reflection fluorescence (TIRF), Step photobleaching analysis | 174 |
| Beta-2 adrenergic receptor | Antagonist and agonist ligand binding assays | 175 |
| ADRB2 | Dot blot assay | 176 |
| DRD1 | Dot blot assay | 176 |
| C-C chemokine receptor type 5 | SPR, Guanine nucleotide exchange assay, 1H-NMR | 177 |
| Mouse olfactory receptor | Electrophysiology characterization, Odorant vapor assay | 178 |
| A2A | SPR, SPA | 179 |
| NK1R | Dot blot assay, Electron paramagnetic resonance spectroscopy (EPR), Fluorescence correlation spectroscopy (FCS) | 176 |
|
| ||
| Transporters | ||
|
| ||
| SecYEG translocon complex | CN-PAGE, Crosslinking analysis, Analytical ultracentrifugation, FRET | 180 |
| P-Glycoprotein 1 (ABCB1) | ATPase activity assay, SPR, Nucleotide and drug binding assay | 181 |
| GLUT4 | Fluorescence quenching, Ligand binding assay | 182 |
| MalE-MalFGK2 Complex | DLS, ATPase activity assay | 183 |
| YopB | Fluorescence correlation spectroscopy (FCS) | 184 |
| Bacterial leucine transporter (LeuT) | Scintillation proximity assay (SPA), Electron microscopy (EM) | 121 |
| Anthrax protective antigen (PA) | Cryo-Electron microscopy | 139 |
| MsbA | ATPase activity assay, SPR | 185 |
| MraY | sGFP assay, Flash photolysis, Activity assays | 186 |
| FhuA (TonA) | Isothermal titration calorimetry, CN-PAGE | 187 |
| Bacteriorhodopsin (monomer/trimer) | Decay-associated spectra, Flash photolysis, Ultrafast vibrational dynamics | 188 |
| VDAC (heterodimer/heterotrimer) | TEM, TIRF | 189 |
| KvAP | 1H-NMR | 190 |
| KcsA | 1H-NMR, 31P-NMR | 191 |
| Na+/K+-ATPase | Peptide fingerprinting, Kinase activity assay, Na+/K+-ATPase activity assay | 14 |
| Vacuolar H+-ATPase | Biolayer interferometry | 160 |
| KcsA-Kv1.3 | SPR, BSI analysis, Electrophysiology characterization | 192 |
| Proteorhodopsin | sGFP assay, Single-molecule force spectroscopy (SMFS), Flash photolysis | 186 |
| Nicotinic acetylcholine receptor | ELISA, Negative stain electron microscopy, Animal studies | 193 |
|
| ||
| Cytochrome P450s | ||
|
| ||
| CYP2B4 | Atomic force microscopy (AFM), Force curve analysis | 194 |
| CYP6B1 | Thin-layer chromatography (TLC), Substrate binding assay | 195 |
| CYP73A5 | NADPH consumption assay, Spectral binding titrations | 196 |
| CYP3A4 | Redox titrations | 197 |
| CYP2J2 | Stopped flow spectroscopy, Spectral binding titrations | 47 |
| CYP5A1 | Carbon monoxide binding assay, Substrate analog binding assay | 29 |
| CYP19 | Spectral binding titrations, Substrate binding assay | 198 |
| CYP86A8 | Spectral binding titrations, Substrate binding assay | 199 |
| CYP17A1 | Radioassays, Substrate incubation assay | 200 |
|
| ||
| Other Membrane Proteins | ||
|
| ||
| Glycolipid receptor (GM1) | Surface plasmon resonance (SPR) | 201 |
| Tar (dimer) | Activity assays | 202 |
| TRPV1 | Cryo-EM | 203 |
| Cytochrome P450 reductase | AFM, Mica activity assay | 204 |
| Monoamine oxidase A (MAO-A) | Activity assays, Inhibition assays | 205 |
| Tissue factor (TF) | SPR, Substrate binding assay | 206 |
| Factor VIIa | SPR, Substrate binding assay | 206 |
| Curdlan synthase (CrdS) | SAXS, MALDI, TEM, Curdlan synthase activity assay | 207 |
| Gamma-glutamyl carboxylase (GGCX) | Hydrogen exchange mass spectrometry, GGCX carboxylase activity assay | 82 |
| Integrin-linked kinase (ILK) | SILAC | 90 |
| YidC | SILAC | 92 |
| SNAP-25 | TIRF, EPR | 208 |
| PglC | DLS, FRET | 209 |
| PgIA | DLS, FRET | 209 |
| Glycoprotein IIb/IIIa | EM | 149 |
| ZipA | Analytical ultracentrifugation, FCS | 135 |
| Anthrax lethal factor (N-terminal domain) | Cryo-Electron microscopy | 139 |
| Spider Toxin VSTx1 | 1H-NMR | 3 |
| Neurotoxin II (Cobra Venom) | 1H-NMR | 3 |
| STIM1 | DLS, EM, 31P-NMR, Heteronuclear singular quantum correlation (HSQC) | 31 |
| Annexin V | Silicon photonic microring resonator array | 37 |
| Antiamoebin I | 1H-NMR, 31P-NMR | 191 |
| CD4-Ubiquitin | SPR | 210 |
| EGF-bound EGFR (dimer) | Kinase activity assay, Negative stain electron microscopy | 211 |
| Cytochrome c oxidase | Spectral binding titrations, Electron transfer assay | 212 |
| COX-2 | Negative stain electron microscopy, Activity assays | 213 |
| K-RAS4B | 1H-NMR, 13C-NMR, BLI binding assays | 214 |
| Bcl-xl | 1H-NMR, 13C-NMR, 15N-NMR | 63 |
| C2A domain (granuphilin) | 1H-NMR, 15N-NMR, Isothermal titration calorimetry (ITC), Microscale thermophoresis (MST) | 215 |
| Ail (attachment invasion locus) | 1H-NMR, 13C-NMR, 15N-NMR, Pull-down assays, ELISA | 80 |
| OmpA | 1H-NMR, 13C-NMR, 15N-NMR, Differential scanning calorimetry, In vitro cross- linking assay | 216 |
| OmpX | 1H-NMR, 15N-NMR | 217 |
| VAMP-2 | TIRF, EPR | 208 |
| Syntaxin-1a | TIRF, EPR | 208 |
| hMGL | Peptide-level HX-MS, MALDI-Time of flight (MALDI-TOF), Fluorogenic kinetics assay | 218 |
| Rheb | 1H-NMR, 15N-NMR, Real-time NMR-based GTPase assay | 219 |
| YgaP (dimer) | 1H-NMR, 15N-NMR | 70 |
| LHCII (trimer) | Time-resolved fluorescence spectroscopy, EM, Steady-state absorbance and fluorescence spectroscopy | 220 |
| GPIb-IX | ELISA, Electron microscopy | 221 |
| Hemagglutinin (H1N1) | Animal studies, ELISA, T-cell proliferation assays | 222 |
| AgrC | Autokinase assay, phospho-transfer assay, B-lactamase assay | 223 |
| SV31 | GFP fluorescence analysis, LILBID mass spectroscopy | 224 |
Historically, nanodisc reviews have summarized the formation of nanodiscs, their structure, as well as the effect of nanodiscs on protein function 6, 11, 12. More recently, reviews have individually covered techniques that have been combined with nanodisc technology, such as NMR 13. To date, mass spectrometry procedures, fluorescence based techniques, and electron microscopy protocols have been modified to study membrane proteins in nanodiscs. Additionally, nanodiscs have recently been gaining prominence in interdisciplinary studies bridging the gap between engineering techniques and membrane protein study. Herein, we will highlight the various biophysical techniques that have been combined with nanodiscs for membrane protein interrogation from 2012–2017 focusing on fluorescence spectroscopy, solution state and solid state NMR, electron microscopy, small-angle x-ray scattering, various forms of mass spectrometry, surface plasmon resonance, charge sensitive optical detection, scintillation proximity assays, nanoplasmonic biosensing, lipoprotein-nanoplatelets, and sortase-mediated labeling of nanodiscs.
2. BIOPHYSICAL STUDIES OF MEMBRANE PROTEINS IN NANODISCS
2.1. Preparation of cell free large scale bacterial and mammalian protein nanodisc library for molecular screening
Membrane protein stabilization in nanodiscs has been largely been used for overexpressed purified proteins (Table 1) which allows the interrogation of the protein in an isolated, controlled systems without external interfering factors. However, it may lead to the loss of key protein-protein and protein-lipid interactions that might be critical for its function. In order to keep some of these interactions intact, nanodisc technology has been recently extended to form libraries of membrane proteins in nanodiscs where the membrane proteins were directly captured from the membrane environment of the cellular membranes. A schematic outlining the assembly of a nanodisc library can be seen in figure 2. Membrane protein nanodisc libraries have been prepared from membranes of simple organisms such as E. coli as well as for complex mammalian membrane systems such as synapses and cancer cells 14–16. The formation of nanodisc library systems allows for protein-protein interactions to be captured leading to the identification of interactomes while removing the need for protein overexpression and complicated purification procedures of the individual membrane proteins. Furthermore, it allows the capture of poorly expressed proteins that cannot be overexpressed in cell lines without expansive optimization.
Figure 2.
The first report of a nanodisc library was established from E. coli membrane proteins by Marty and coworkers 15. The authors optimize the nanodisc lipid to MSP ratios for a library system and quantify the incorporation efficiency to be 85% by SDS-PAGE gels. Ultimately, several proteins comprising the major bands on the SDS PAGE gel were identified by mass spectrometry. However, bacteria are simple organisms that display much less complexity in their membrane protein composition, and are specifically lacking complex proteins like GPCRs that are highly unstable in solution and are key targets for the pharmaceutical industry.
A further advance in the nanodisc library system was the incorporation of the synaptosomal membrane proteome in nanodiscs by Wilcox and coworkers 16. In this work, the authors hypothesized that the nanodisc platform can be utilized to identify the binding sites for Alzheimer’s associated Aβ oligomers. By incorporating the synaptosomal membrane proteins directly into nanodiscs, the authors exhibited binding of Aβ oligomers to proteins in the nanodisc. Furthermore, they were able to screen small molecule inhibitors in a high throughput assay, thereby providing a cell free system for screening of drug molecules to a membrane protein library created from synaptosomes.
More recently, advances toward nanodisc library systems include the incorporation of human osteosarcoma cell membrane proteins into nanodiscs by Roy and coworkers 14. This work represents an extensive characterization of the nanodisc-incorporated proteome by high throughput peptide fingerprinting mass spectrometry. This technique identified not only the specific protein species that were incorporated; it also provided a semi-quantitative amount of the proteins that were incorporated. By analysis of this data, the authors showed that changing the lipid composition could preferentially incorporate specific classes of membrane proteins. Notably, the zwitterionic 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) lipids that are typically used for nanodisc formation, also utilized for the E. coli and synaptosomal nanodisc libraries, did not incorporate the majority of proteins that are known to interact with anionic lipids. Tuning the lipid composition to include 20% anionic 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS) provided a more faithful replication of the initial membrane mixture. Furthermore, nanodiscs with three distinct lipids—POPC (72%), POPS (20%) and cholesterol (8%) were prepared that most closely mimicking the plasma membrane lipid composition. However, this nanodisc mixture, while incorporating all major protein classes, showed preferential incorporation of GPCRs indicating possible favorable interactions of GPCRs with cholesterol. Finally, the authors exhibited retention of functional activity of the incorporated proteome by measuring Na+/K+ ATPase activity and receptor tyrosine kinase activity 17.
Another recent advance has been made by Mak and colleagues, with the insertion of membrane proteins from various human cell lines such as human embryonic kidney 293 (HEK-293) cells, as well as freshly isolated human red blood cells and platelets. Interestingly, the authors eschewed traditional membrane isolation steps and generated the nanodiscs directly from cellular membrane lipids18. Whole cell lysate was combined with MSP1E3D1 and supplementary 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethyleneglycol)-2000] (PEG-PE), then incubated with BioBeads to initiate nanodisc assembly. This allowed the authors to achieve a higher percentage of membrane protein incorporation, while simultaneously retaining a native-like membrane environment 18. After purification, nanodisc formation was verified via electron microscopy, while protein incorporation was confirmed by Western blot 18. Taken together these studies show that it is possible to create a large library of membrane proteins directly from membranes and use the library to screen for small molecule binding studies that can aid in drug development. The studies also indicate the existence of specific lipid-protein interactions that help in enriching certain classes of proteins in nanodiscs with different lipid compositions.
2.2. Protein-protein interactions in nanodiscs and the challenge of conformational flip-flop
Nanodiscs can serve as an excellent technique to capture protein complexes and study protein-protein interactions. A recent study of the yeast proteome suggested that on average there are two protein-protein interactions per membrane protein 19. Hence, the co-reconstitution of multiple proteins into a single nanodisc can enable the study of membrane proteins in a stabilized system. Previous efforts into the incorporation of multiple proteins have focused on controlling the ratio of proteins to MSP to lipids in the nanodisc preparation to achieve a homogeneous mixture 20–23. However, due to the randomized nature of protein incorporation into nanodiscs, the nanodisc mixture is usually non-uniform. As a result, multiple purification steps are required to get a homogenous mixture of the desired composition, often leading to poor yields. Another major problem in the co-incorporation of proteins into nanodiscs is the bi-directionality in protein integration, where the proteins may be incorporated on opposite sides of the nanodisc, which limits the study of protein-protein interactions. In summary, the controlled co-incorporation of more than one protein type in nanodiscs with the physiologically relevant conformation has been a challenge.
A recent advance towards this controlled co-incorporation was established using modified VDAC complex 24. In this work, the authors tethered VDAC protein complexes with DNA to co-incorporate the complexes into nanodiscs, excising the linker using DNase. This made sure proteins dually incorporated into nanodiscs in the correct conformation. However, even with this technique there are drawbacks—including the insertion of the individual proteins into two nanodiscs, resulting in single protein discs after excision. This was indeed observed by TEM before excising the linker. This problem could potentially be minimized by further optimization of the linker size. However, this method provides higher yields of protein complex incorporation than expected from traditional methods. While nanodiscs can be used for interrogating protein-protein interaction but the different conformations that the proteins can adopt in relation to each other is still a challenge.
2.3. Nanodiscs with variable lipid compositions
Nanodisc technology establishes novelty in the ability to regulate the lipid composition of a native-like bilayer with precision. This ability is of importance to researchers studying membrane proteins, as the phospholipids that comprise the native plasma membrane directly modulate the functionality of membrane or membrane-associated proteins 25. Given that the conformation and function of the membrane protein is dictated by explicit molecular interactions between the protein and lipids of the phospholipid bilayer, alterations of bilayer lipid composition will result in deviations of protein activity 26. Hence, when performing in vitro studies of membrane proteins in the lipid bilayer system of nanodiscs, with the goal of simulating the native cellular environment and native protein activity, it is important to select an appropriate lipid composition for the nanodiscs assembly.
Common lipid compositions chosen for nanodiscs include: solely the zwitterionic phospholipid POPC, a mixture of POPC and the anionic phospholipid POPS, or a mixture of POPC and the anionic phospholipid POPG 27–29. In an investigation of the receptor histidine kinase AgrC, Wang et al. found that the variant AgrC-I required high anionic lipid concentrations for robust activation 30. It was also found that although AgrC-I is functional in a 1:3 mixture of DMPC and DMPG, the other variants tested—AgrC-II and AgrC-III—were not 30. All three variants were functional in nanodiscs with a 1:3 ratio of POPC to POPG 30.
Nanodiscs can have different sizes depending on the length of the MSP; these different sized nanodiscs can be used for different processes. Wang and coworkers modified MSP by deleting the last four helices, which allowed the formation of smaller nanodiscs without altering the disc like structure 31. The smaller nanodisc size allowed for better NMR spectra for STIM1 in different nanodisc environments. The authors measured 31P spectra for nanodisc containing POPC, POPE and POPC-POPE lipids. The POPC-POPE nanodiscs showed 2 different peaks, each corresponding to the individual peaks observed in POPC and POPE nanodiscs, indicating that the lipid environment is similar in the single lipid as well as the mixed lipid nanodisc. Furthermore, the authors performed 2D NMR on the incorporated protein to gain structural insights. Thus, the use of smaller nanodisc size is beneficial towards obtaining higher resolution solution state NMR spectra of proteins incorporated into nanodiscs.
Sometimes, nanodiscs must be disassembled after or during an experiment for analysis purposes. This is usually done to determine the lipid contents of a given set of nanodiscs. However, François-Martin and Pincet disrupted nanodiscs for the purpose of measuring lipid mixing 32. Older versions of this experiment require disruption with detergent to induce fluorescence dequenching. In this case, discs prepared from POPC and fluorescent DOPE lipids were joined to liposomes via SNARE fusion proteins 32. Dequenching was then used to monitor mixing of the fluorescent lipids through the newly joined membranes 32.
An important aspect of nanodisc composition is the consideration of the phase transition temperature (Tm) of the lipids employed 25. An intrinsic property of phospholipids within lipid bilayers is that they are either arranged in an ordered solid (gel) phase or a disordered liquid crystalline phase, with the former occurring at lower temperatures and the latter occurring at higher temperatures. The phase transition temperature is directly dependent upon the head group and length of the fatty acid chain. Nanodisc assembly, as well as the following experiments with assembled nanodiscs, must be conducted at temperatures that do not fall below the Tm of the lipid(s) used. This precaution should be implemented to ensure that the lipid bilayer stays in the native phase, a liquid crystalline phase. Sligar and coworkers characterized the phase transition behavior of 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and DMPC nanodiscs by creating nanodiscs consisting of either 100% DPPC or 100% DMPC and subjecting them to differential scanning calorimetry (DSC) and generalized polarization of laurdan fluorescence 33. Their findings noted that, upon incorporation into nanodiscs, the Tm of DPPC and DMPC was 3–4 °C higher than their usual value. These findings that phase transition temperatures for phospholipids are altered upon nanodisc incorporation are significant for researchers choosing a proper lipid composition; the Tm of a phospholipid that is protein-unbound can only serve as a guideline and experimental studies must be performed to determine the Tm of the nanodisc-incorporated phospholipid 33.
Nanodiscs with different lipid composition have been used to study membrane bound active peptides 3. In a recent work, the authors showed that arenicin-2, a pore-forming antimicrobial peptide, disintegrates nanodiscs containing both zwitterionic phosphatidylcholine (PC) and anionic phosphatidylglycerol (PG). However, a spider toxin VSTx1, possessing a slightly lower positive charge compared to arenicin-2, showed effective binding to the nanodiscs containing anionic lipids such as 1,2-dioleoyl-sn-glycero-3-phosphoglycerol (DOPG) and did not cause their disruption. Due to the lower charge, VSTx1 has a lower affinity to nanodiscs containing zwitterionic lipids and only weakly interacts with MSP. Finally, neurotoxin II (NTII) from cobra venom showed low affinity for nanodiscs containing anionic lipids and it did not bind to zwitterionic lipid containing nanodiscs. The authors also show that NTII interacts with the nanodisc surface in several orientations and the exchange between the different orientations is fast. As the interaction between NTII and phosphoserine is only possible in one specific orientation, the chance of a membrane disintegrating reaction is low.
Nanodisc have also been utilized in combination with cell free expression of membrane proteins for protein stabilization 34. Henrich et al. exhibited the co-translational association of synthesized membrane proteins with preformed nanodiscs, to avoid detergent contact of the protein. This is particularly useful in cases where the membrane protein is extremely susceptible to detergent contact as is the case with MraY, the protein used in this study. Furthermore, the authors show that MraY grown in gram negative bacteria showed a strong preference for anionic lipids, whereas the same protein grown in gram positive bacteria, showed no such preference. Gram negative expressed MraY did not show any activity in nanodiscs containing less than 40% 1,2-dimyristoyl-sn-glycero-3-phosphorylglycerol (DMPG). This major difference could be due to differential membrane integration patterns.
Finely-tuned lipid compositions have been utilized to achieve functionality of target membrane proteins from a multitude of protein classes. Dijkmam et al. demonstrated that the class A GPCR neurotensin receptor 1, when incorporated into nanodiscs rich in phosphatidylethanolamine, experienced the Gαi1 subunit having a 4-fold higher affinity for the GPCR 35. While various other GPCRs have been incorporated into nanodiscs to study their function (Table 1), the effect of various lipid compositions on GPCR functionality has not been thoroughly investigated. Rues and coworkers co-translationally expressed turkey β-1 adrenergic receptor modified with a GFP tag and incorporated into nanodiscs with varying lipids including 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), DMPG, POPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG), 1-stearoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (SOPG), 1-2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), DOPG, 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS), 1,2-dioleoyl-sn-glycero-3-phosphate (DOPA), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), and 1,2-dielaidoyl-sn-glycero-3-phospho-(1-rac-glycerol) (DEPG) 36. The homogeneity of various empty nanodiscs was measured. Next, for protein nanodiscs the incorporation efficiency was monitored using fluorescence and it was noted to be consistently high in all lipids, except DOPA and DOPE where a low fluorescent signal was observed. Lower incorporation was also observed with natural lipid extracts. Charged headgroups such as PG or PS supported the functional conformation and 18 carbon atom chains were preferential. The trans configuration of the double bond in DEPG membranes resulted into higher activity if DOPG membranes having cis configuration The activity of the incorporated receptor was measured using quantitative binding assays with radiolabeled alprenolol. The alprenolol binding with purified ND samples was 75% with SOPG, 50% with POPS, DOPE and DOPS, and 25% with DOPA.
The study of protein-lipid interactions has also been combined with novel analytical techniques like silicon photonic micro-ring resonator arrays 37. In this work, the authors utilize the nanodisc platform to study both protein-lipid interactions as well as membrane protein-soluble protein interactions. Nanodiscs of various compositions immobilized on the surface of a micro-ring resonator array show preferential binding when exposed to annexin, an anionic phospholipid binding protein. As the amount of POPS increases from 0% to 50% the net annexin binding also increases. Protein-lipid interactions in nanodiscs have also been investigated via gas phase native mass spectrometry 38. In this study, the authors inserted ammonium transporter AmtB and tetrameric aquaporin AqpZ into POPC and DMPC nanodiscs. By employing orbitrap mass spectrometry and optimizing the collision energy, the authors ejected the proteins with the associated lipids without losing lipids to excessive energy or drawing non-specifically bounded lipids. Deconvolution of the spectrum with UniDec and Kendrick mass defect software revealed that 9 lipids were associated with each protein. These results were further confirmed for AmtB with MD simulations.
In summary, the ability to tightly control the lipid composition of the nanodiscs is important to study the protein-lipid interactions in several membrane-bound protein and peptide systems.
3. USE OF FLUORESCENCE SPECTROSCOPY TO STUDY MEMBRANE PROTEINS IN NANODISCS
Fluorescence spectroscopy is one of the most powerful techniques to study protein folding, dynamics, protein-lipid assembly, and protein-protein interactions. Fluorescence spectroscopy has broad applicability across different proteins and in different environments. Most proteins have natural fluorophores such as tyrosine and tryptophan residues, which allow study of changes in protein conformation without any further mutation of the parent protein. Besides using the natural fluorophores, site-specific labeling with external fluorophores is also done using either mutagenesis or chemical modifications. One of the hallmarks of using fluorescence spectroscopy for membrane protein study is the requirement of a small amount of material (picomole to nanomole range) that gives a high signal-to-noise ratio. Fluorescence spectroscopy is also a convenient method to study fast protein conformational changes as the fluorescence emission lifetime is in the nanosecond range, which is relatively fast compared to most conformational transitions. In order to adapt established fluorescence measurement techniques to study membrane proteins, nanodiscs have recently been used is several applications as outlined below.
3.1. Fluorescence Correlation Spectroscopy
Fluorescence correlation spectroscopy (FCS) is a well-established technique that has been extensively used in the study of protein-protein interactions. The fundamental principal of FCS is to perform correlation analysis on the random diffusion of a fluorescent molecule in solution to determine various constants of the molecule including its diffusion constant and hydrodynamic radius 39. Additionally, data obtained through FCS can be utilized to determine the rates of chemical reactions as well as the average concentration of the molecule in solutions 40, 41. We recognize that FCS has been coupled with nanodisc technology for biophysical and biochemical studies since 2010, however for the purposes of this review we will only focus on the work performed after 2012.
FCS was used to investigate bacteriorhodopsin (BR) nanolipoprotein discs (NLPD). The NLPD is structurally and functionally similar to the nanodisc. Gao and coworkers incorporated fluorescently labeled BR protein into NLPD containing labeled lipids and performed FCS analysis. The diffusion curves for BR-NLPD were compared to empty NLPD, BR, and truncated apolipoprotein in order to determine the diameter of each species 42. This determined that the BR nanodisc insertion rate was 55%. Atomic Force Microscopy measurements were used to validate this rate of insertion42. Overall this study provided some basic characterization of BR nanodiscs and established a method to analyze membrane proteins by FCS.
Another study performing FCS on membrane proteins inserted into nanodiscs analyzed GPCRs. GPCRs are important signaling proteins that are frequently studied after detergent solubilization even though this procedure impairs protein functionality 39, 43. The nanodisc alleviates these concerns. Additionally, the nanodisc based FCS is advantageous for kinetics studies because it requires a low amount of material 43. In this study the GPCRs NK1R, ADRB2, and DRD1 were inserted into nanodiscs in order to increase the GPCRs’ solubility, and perform substrate binding studies 43. The increased solubility is another benefit of nanodisc technology. NK1R-ND and its natural substrate, Substance P, were both fluorescently labeled so binding could be analyzed by FCS. When incubating with NK1R increased substance P’s diffusion times signifying that substance P (a small peptide) was binding to NK1R (a significantly larger substance) 43. Data analysis revealed a Kd of 83 + 33 nM and this Kd was consistent with dot blotting results 43. Establishing FCS as a valid means of studying GPCR-NDs will prove beneficial for future studies because it provides high quality quantitative data and because FCS measurements can be taken rapidly 43. Gao and coworkers state that these advantages combined with FCS technique requiring low volumes of material makes FCS an ideal method for use in GPCR drug screening studies 43.
A third study on the use of FCS to study membrane proteins incorporated into nanodiscs focused on Yersinia pestis membrane proteins YopB and LcrV 39. YopB is an effector protein that plays a role in host cell invasion and YopB’s delivery into the host cell is moderated by the injectisome 39. LcrV is a cytosolic protein found in the needle tip of the injectisome and a study suggested that an interaction between these two proteins is necessary for pore formation 39, 44. Analyzing the shift in fluorescently labeled LcrV’s diffusion curve upon incubation with YopB revealed a Kd of 20.5 ± 2 nM while incubating LcrV with empty nanodiscs does not significantly alter diffusion, which suggests that LcrV requires YopB for pore formation and not just a lipid bilayer 39.
In conclusion, these studies show the viability of FCS-nanodisc coupling to study a wide variety of membrane proteins. Furthermore, FCS provides very accurate, quantitative binding data using low amounts of protein, which is ideal considering the low expression levels of membrane proteins.
3.2. Fluorescence resonance energy transfer (FRET) to study interactions within the bilayer
Membrane proteins can be classified into two broad categories integral and peripheral based on the nature of the membrane-protein interactions. Integral membrane proteins have one or more of the protein segments that are embedded in the phospholipid bilayer. Peripheral membrane proteins on the other hand are usually bound to the membrane indirectly by interactions with integral membrane proteins or direct interactions with lipid polar head groups. Overall most membrane proteins have buried protein segments that are not accessible to the aqueous solution. Fluorescence resonance energy transfer (FRET) is a method that is frequently used to study interactions that occur within lipid bilayers. Several studies have performed FRET on membrane proteins incorporated into liposomes, but nanodiscs serve as a better tool for use in these studies due to enhanced homogeneity, stability, resistance to aggregation, and a reduction in scattering affects due to aggregated proteins 21, 45–47. In these types of FRET studies a donor molecule, frequently an excitable amino acid, will be integrated into the protein at the location of interest and when excited this amino acid will emit a wavelength that then excites the acceptor, which is the measured species.
One recent study that utilized FRET analysis on membrane proteins investigated PgIC and PgIA, two of the enzymes in the Campylobacter jejuni N-Linked protein glycosylation pathway 48. This study selected the tetramethyl rhodamine (TAMRA) and Cyanine5 (Cy5) FRET-pair to probe the cofacial organization of these proteins in nanodiscs 48. PgIC was labeled in its N-Terminal or globular domain with Cy5 (the acceptor molecule) and PgIA was labeled with TAMRA (the donor molecule) 48. Hartley et al. found a greater FRET signal when PgIC was globularly labeled suggesting that the globular domains are cofacially oriented in the nanodisc 48.
We recently conducted a study using FRET to measure the insertion of Cytochrome P450 2J2 (CYP2J2) mutants into the lipid bilayer. CYP2J2 in humans is an extrahepatic CYP in the heart that epoxidizes omega-3 and omega-6 polyunsaturated fatty acids to create biologically active metabolites 49. Previous studies on CYPs suggested that CYP membrane insertion includes a partially imbedded active site and that the N-terminus and F-G Loop are significantly immersed in the membrane 46, 50. In order to study the role the F-G Loop plays in CYP2J2 membrane insertion, we mutated the F-G Loop region of a 34 AA N-Terminally truncated CYP2J2 (Δ34) to create the following constructs: Δ34-I236D, Δ34-F239H and Δ34-I236D/F239H 45. We then used stoichiometric ratios to ensure that these constructs were inserted into nanodiscs containing one and only one phospholipid with pyrene lipids 45. CYP2J2 has several naturally occurring Trp residues that can form a FRET pair with the pyrene-nanodisc and this FRET pair was used to probe the insertion of CYP2J2 into the bilayer 45. Furthermore, this FRET pair has been used to analyze the insertion CYP3A4, CYP1A2, cytochrome C, and 14-3-3γ into liposomes but nanodiscs are a better membrane mimic because of their increased homogeny and stability in aqueous solution 45–47, 51–55. Trp-Pyrene FRET analysis of the mutant 2J2 in 100% POPC nanodiscs did not demonstrate a significant variance in 2J2 membrane insertion relative to Δ34 but decreases in insertion relative to Δ34 were observed in 30% POPS:70% POPC nanodiscs and even greater decreases relative to Δ34 were observed in the 60% POPS:40% POPC nanodiscs 45. This concludes that the F-G loop mutants are inserted into the membrane less than wild type Δ34. Overall this study is another example of nanodisc technology being combined with FRET to probe membrane protein structure and their interactions with the membrane. Additionally, lipid composition was shown to have a significant effect on membrane insertion and since nanodisc lipid composition can be easily manipulated, nanodiscs are ideal mimics in studies like this where lipid composition plays an important role in the biophysical studies.
FRET was also used to further analyze the previously mentioned SecYEG translocon. This study used a FRET pair featuring a fluorophore bound to SecYEG’s substrate protopeptide and a fluorophore bound to the SecYEG translocon itself that only activates upon formation of a translocon intermediate 56, 57. Using this FRET pair to analyze SecYEG monomers in nanodiscs, they found that SecYEG is active as a monomer. This agrees with the results of a previous SecYEG study investigating SecYEG in unilamellar vesicles 56, 57. Furthermore, there was no change in FRET upon analyzing SecYEG oligomers in one nanodisc, which is a sign that oligomerization does not promote translocation 56.
Nanodiscs and FRET have been used to analyze the formation of SNAREpin complexes as part of neurotransmitter vesicle fusion and neurotransmitter release 58. The Ca2+ mediated relationship between SNAREpin, synaptogamin, and the lipid bilayer was analyzed using FRET as well 59. Furthermore, FRET has been paired with other fluorescence techniques, where in one study FRET was combined with FCS to analyze the assembly of the YidC-ribosome complex in nanodiscs 60. Kedrov and colleagues used FCS to characterize the diffusion of recombinant YidC and its binding to ribosomes 60. Then, by labeling both with AlexaFluor 488 and Cy3, they investigated the oligomerization of YidC using FRET. The YidC system was then transferred into nanodiscs where FCS was again used to measure oligomerization 60.
3.3 Luminescence Resonance Energy Transfer
Nanodisc technology has also been coupled with the technique of luminescence resonance energy transfer (LRET) to study a range of membrane proteins. In one study conducted by Zoghbi et al., LRET was combined with nanodiscs to examine the structure of the bacterial flippase protein MsbA 61. A single cysteine mutant outside the nucleotide binding domain (NBD) of the protein was labeled with Tb3+ in a thiol selective labelling. The Tb3+ is a donor for the LRET, which showed a large sensitized emission for ATP bound protein. The binding of ATP induces the NBD dimerization process which leads to increase in LRET, thus conformational changes can be monitored by LRET changes. The distance between the NBDs was monitored by measuring the LRET. It was shown that MsbA in nanodiscs undergoes conformational changes during the ATP hydrolysis event, a finding that was much smaller than the previously published results. This finding is more consistent with a closed outward facing crystal structure. Two distinct distances were detected under steady state conditions, which indicate a conformational equilibrium between two states, supporting previous results. The authors purport that the native-like state of the nanodisc-reconstituted MsbA is responsible for their results contrasting the widely open inward facing crystal structure and studies in non-physiological conditions. Within this study the LRET experiments were also conducted using liposomes as a comparative measure. It was observed that the MsbA reconstituted in nanodiscs had higher activity than in detergent-solubilized liposomes. Additionally, it was revealed while performing distance calculations that nanodiscs had more consistent LRET data than liposomes. The authors attributed the discrepancies in the liposome data to light scattering from the liposomes, conveying an advantage for the use of nanodiscs in LRET experiments.
4. NMR AND NANODISCS
4.1 Solution state NMR
Nanodiscs have been coupled to both solution state and solid-state NMR techniques. For instance, solution state NMR studies were used to study the structure of the anti-apoptotic protein BCL-XL. BCL-XL is overexpressed in tumors and found to encourage tumor formation, cell survival, and resistance to chemotherapy. 62 In the past, BCL-XL studies have evaluated truncated or detergent solubilized protein 63 and NMR analysis was performed on truncated, water soluble BCL-XL in 1996 64. Full length BCL-XL has not been structurally characterized although detergent solubilized BCL-XL was used to study how it associates with the membrane 65–67. One of these studies determined that detergents were capable of interfering with BCL-XL’s ability to bind and form protein complexes 67. Full length BCL-XL nanodiscs were analyzed via solution NMR to obtain structural information about full length BCL-XL 63. These experiments determined BCL-XL integrates into nanodiscs homogenously, BCL-XL has a globular fold when integrated, the C-Terminal tail traverses the nanodisc membrane, and BCL-XL binds to BH3-Ligand with high affinity 63. This study was able to achieve atomic resolution of BCL-XL due to the detergent-free environment, which demonstrates the utility of using nanodiscs in NMR studies 63. BCL-XL nanodiscs have also been evaluated by Yao et al. using SAXS 68. This study determined that C-terminus of BCL-XL forms α-helix and that the C-terminus disrupts the solubility of the protein 68
Solution NMR has also been used to probe the membrane interactions of a gain of function rat sarcoma ras mutant known as K-RAS4B, E. Coli Outer Membrane Protein A, and the bacterial α helical membrane protein YgaP 69–71. Additionally, solution NMR was used to probe the effect of truncating MSP on nanodisc size, and determine which MSP variant is best for analyzing beta barrel proteins, 72, 73. These studies found that MSP1D1ΔH5, which creates a smaller nanodisc with a diameter of ~9 nm, provided the best stability and NMR resolution. Chemical shift perturbation was used to analyze membrane dependent regulation of the Rheb (Ras homologue enriched in brain) GTPase 74.
Nanodiscs have also been combined with solution NMR to perform structural studies of the Cytochrome-b5 CYP2B4 complex 75. This work is notable because it is one of the first to analyze a multiple protein complex by NMR. Nanodiscs served as an easy to use platform for complex stabilization and the binding observed was much stronger than that observed in detergent micelles 75. Additionally, the detergent-free environment of nanodiscs allows for improvement in resolution over its detergent-based counterparts.
4.2 Solid State NMR
One of the challenges of analyzing proteins by solution NMR is that the larger the protein’s molecular weight the lower the resolution of the NMR structure 76. Solid State NMR have recently been used to evaluate larger protein structures 76. Solid state NMR has also been used to analyze membrane proteins inserted into lipid nanodiscs. One of these studies analyzed proteorhodopsin which is a type-1 retinal protein from marine algae that is easy to analyze due to its optical properties 77. This study used polyethylene glycol to help sediment the nanodiscs inside a MAS rotor and found that proteorhodopsisn complexes in nanodisc that are formed using DHPC lipids are more homogenous than detergent solubilized proteorhodopsin 77. Furthermore, reduced protein dynamics and increased lipid order was observed for the proteorhodopsin nanodisc samples 77. Solid State NMR has also been used to study the Yersinia pestis outer membrane protein Ail. Ail enables resistance to the host’s immune system and mediates adhesion to human cells 78. This protein crystal structure was recently determined 79. Ail in micelles has recently been studied by NMR but the detergent concentration necessary for NMR prohibited ligand binding 80. Fortunately, solid state NMR on catalytically active Ail nanodiscs was able to observe ligand binding which can be used to probe protein activity 78. Furthermore, the Ail nanodisc NMR samples were sedimented by ultracentrifugation instead of polyethylene glycol or lyophilization 78. This is advantageous because polyethylene glycol can disrupt nanodiscs of particular lipid compositions and can inactivate proteins 78, 81. This work also diluted some of the sedimented samples back to original concentrations, and solution NMR studies on these samples found that sedimentation did not disrupt the protein or lipid structures 78. Together these two studies established methods for analyzing membrane proteins by solid state NMR and pave the way for future structural and functional studies.
5. MASS SPECTROMETRY
Mass spectrometry is one of the primary methods for protein identification from complex mixtures of biological origin. This is largely attributable to the instrumental advances that allow routine analysis of small amounts (femtomoles) of peptides in complex biological mixtures along with the rapid growth in genomic databases that are amenable to searching with mass spectrometry data. There are a multitude of treatments that can be combined with mass spectrometry to study different aspects of a protein. Mass spectrometers employed in proteomic analysis use either matrix-assisted laser desorption/ionization (MALDI) or electrospray ionization (ESI). However, most of this work has focused on soluble proteins because detergent solubilization and previous membrane mimics for membrane proteins have failed to adequately provide for protein activity, proper oligomerization, and sample homogeneity 82, 83. Nanodiscs successfully alleviate the majority of these concerns by providing a homogenous environment that prevents unwanted protein oligomerization and ensures protein activity. Therefore, recent work has focused on the combination of nanodisc technology with previously developed mass spectrometric methods. These approaches include using nanodiscs in combination with Hydrogen Exchange Mass Spectrometry, Stable Isotope Labeling of Amino Acids in Cell Cultures, and Matrix Assisted Laser Desorption Mass Spectroscopy as described below.
5.1 Hydrogen Exchange Mass Spectrometry
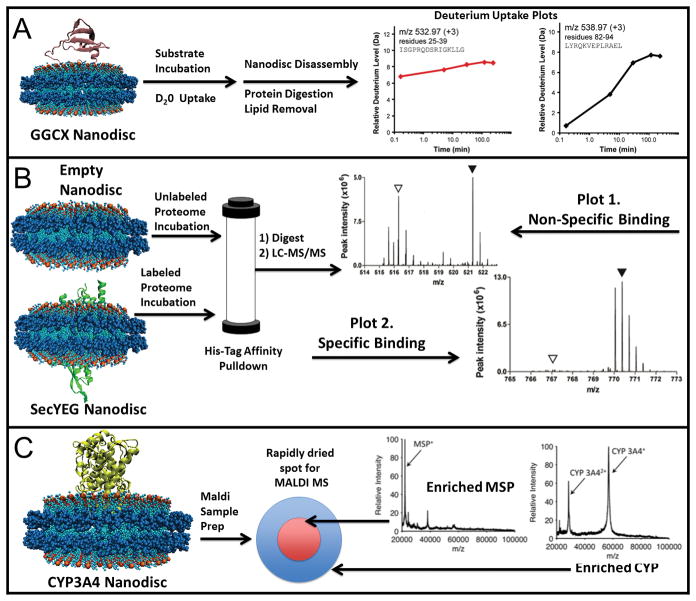
Hydrogen exchange mass spectrometry (HX-MS) has been used to investigate conformational changes of the membrane protein Gamma-Glutamyl Carboxylase (GGCX) in nanodiscs upon ligand binding. In HX-MS, deuterium is incorporated into the protein of interest (GGCX in this case) during a hydrogen exchange step 83–85. The frequency of uptake at specific amino acid residues can then be measured by mass spectrometry 86. These reactions are quenched by acidification and then cholate is added to disassemble the nanodiscs 83. The protein is then digested by pepsin, phospholipids are removed by ZrO2 resin, and the sample is analyzed by UPLC/ESI-MS in order to measure the frequency of deuterium uptake 83. The frequency of uptake reveals how accessible to solvent is to a given residue in the protein. Differences in deuteration levels between substrate bound and substrate free protein reveal conformational changes during substrate binding. A schematic of a HX-MS experiment is shown as Figure 3. Previous HX-MS membrane protein studies used liposomes and detergent micelles to stabilize the proteins, but these systems lack homogeneity 82, 83 that is circumvented by the use of nanodiscs in this case. The pairing of HX-MS and nanodisc technology elucidates information about the accessibility of individual residues while using very low amounts of protein 82, 83. This is an additional advantage of coupling HX-MS with nanodisc technology because many membrane proteins have low expression levels.
Figure 3.
GGCX post-translationally converts specific glutamic acid (Glu) residues into gamma-carboxyl glutamic acid (Gla) in many Vitamin K Dependent (VKD) proteins 82. VKD proteins have been implicated in blood clotting, signaling, and bone structure 87. This carboxylation requires Vitamin K GGCX binds to its substrates at a highly conserved 18-residue sequence known as the pro-peptide (pCon) 82, 87. Investigation into the structure of GGCX bound to the pCon domain of VKD proteins has stalled because sample preparation was leading to aggregation, oligomerization, and precipitation 82. GGCX was thus incorporated into nanodiscs to circumvent these problems. HX-MS was then performed on these nanodisc complexes 82, 83. Comparing HX-MS results of GGCX nanodiscs with GGCX nanodiscs bound to pCon provided information on conformational changes due to the binding of pCon.
As previously stated, the conformational changes of GGCX nanodiscs upon pCon binding were evaluated by comparing unbound GGCX nanodiscs to pCon bound discs and also evaluated the effect of varying the length of the deuteration reaction. Upon substrate binding less deuterium uptake was observed in many regions of GGCX suggesting increased solvent protection and hydrogen bonding 82. Furthermore, significant differences were observed in areas known to bind to pCon 82. These results were in agreement with current literature 82. A schematic of HXMS is shown as figure 3A.
As HX-MS is coupled with nanodisc technology to study additional membrane proteins, one concern is the amount of time required to digest large membrane proteins. The larger the protein, the longer the amount of time required for complete digestion. This is problematic because deuterium begins diffusing out of the protein as soon as the deuterium uptake reaction is quenched 83. This could be countered by improving the speed of protein digestion and lipid removal from the membrane protein-nanodisc system. Removing MSP contamination can also increase sequence coverage after digestion 88. Adhikary and colleagues inserted LeuT into nanodiscs and used HX-MS in concert with molecular dynamics simulations to identify conformational ensembles. Neutravidin beads were used to trap biotinylated MSP, thus improving LeuT peptide detection 88. Taken together, HX-MS coupled with nanodisc technology will continue to be a valuable tool to investigate membrane protein conformations.
5.2. Stable Isotope Labeling of Amino Acids in Cell Culture (SILAC) approach in mass spectrometry
There is a need to identify protein-protein interactions in order to understand complex cellular processes, and recent work has focused on understating the protein interactome. An interactome is all of the different proteins and molecules that a given protein interacts with. Some techniques that have been used to identify interactomes of soluble proteins include, but are not limited to, tandem affinity pulldown, stable isotope labeling of amino acids in cell cultures (SILAC), yeast two hybrid screening, chemical labeling, gel chromatography, and blue native gel electrophoresis 89, 90. Unfortunately, these methods struggle to identify membrane protein interactomes because the detergents used to solubilize membrane proteins frequently disrupt protein-protein interactions 91. Additionally, many membrane protein interactome interactions have weak affinities or are dependent on specific lipids that are associated with the proteins in vivo 92. As a result, studies of membrane protein interactomes have not been investigated as thoroughly as their soluble counterparts. Recently SILAC has been combined with nanodisc technology to determine the interactomes of target membrane proteins.
SILAC has been used to identify the interactomes of GLUT4, Integrin Kinase, and Protein Phosphatase 1 90, 93, 94. The first step in SILAC is to obtain two versions of the proteome to be evaluated. One is a heavy proteome made from deuterated amino acids and the other is the light proteome containing unlabeled amino acids 95. For a protocol capitalizing on the usage of nanodiscs, the protein being studied is purified separately and then inserted into nanodiscs. These discs are then incubated with the heavy proteome while empty nanodiscs are incubated with the light proteome. The light proteome empty nanodisc incubation functions as a control for nonspecific binding of proteins to lipids or MSP 92. The mixtures are combined and then purified through Ni-NTA affinity purification. This method of purification is important as the resin does not interact with the incorporated proteome and consequently avoids disrupting any protein-protein interactions. After elution, the protein-ND complexes are then analyzed by LC/MS/MS and identified using protein databases 92. Then, it is possible to determine the ratio heavy to light ratio for each proteomic protein and this ratio is known as the SILAC Ratio 92, 95. The higher the SILAC ratio the more specific the interaction between the protein from the proteome and the target protein. A high affinity and low affinity interaction are shown as part of figure 3B.
SILAC was first combined with nanodisc technology in an attempt to determine the interactome of membrane proteins SecYEG translocon, maltose transporter MalFGK2, and membrane insertase YidC by Zhang and coworkers. SecYEG is a channel protein known to interact with SecA, Syd, FtsY, and ribosomes 96. SILAC analysis of SecYEG-ND created with E. Coli total lipid extract found that SecYEG bound to SYD but not SecA 92. However, SecYEG-ND created with phosphatidylglycerol (PG) did bind SecA 92. This example illustrates the effects of microenvironments and surrounding lipid composition on membrane protein-protein interactions. Thus the ability to easily create nanodiscs of varying lipid compositions is critical to facilitate these studies. The SecYEG-ND did not bind FtsY and ribosomes, but this was not surprising due to these interactions requiring a ribosome-nascent chain complex to form during co-translational translocation 97, 98. Furthermore, the SILAC method observed DnaJ specifically binding to SecYEG although this binding had a much lower SILAC ratio than the others 92. The MalFGK2 experiment observed significant MalE binding 92. Although MalE is known to bind to MalFGK2 this result was remarkable because MalE only binds to detergent solubilized MalFGK2 in very specific situations 99. This is another example of the advantages of the nanodisc SILAC approach. A previous proteomics analysis of YidC did not capture any proteins that appear to interact with YidC 100. This finding was repeated by the nanodisc SILAC studies 92. Overall, the studies on these proteins agree with previous literature demonstrating the validity of the nanodisc SILAC method.
SILAC is a highly accurate quantitative experimental method that could be used to identify the interactomes of many membrane proteins as well as quantify the strength of the protein-protein interactions present within the interactomes. Furthermore, as many membrane protein interactions depend on the presence of specific lipid types, these experiments can be pursued using nanodiscs of varying lipid composition to attempt to identify new interactions similar to the SecYEG-SecA binding. Therefore, using SILAC methodologies in combination with nanodiscs, one can unearth protein-protein interactions that will lead to the discovery of the missing steps in a protein signaling pathway.
5.3. Matrix Assisted Laser Desorption Ionization Mass Spectrometry (MALDI MS) and Electrospray Ionization (ESI)
Most of the mass spectrometry methods including the previously mentioned HX-MS and SILAC analyze digested peptide fragment as opposed to full length proteins. Although the bottom-up proteomics methods provide valuable information it is important to analyze full-length proteins to obtain information regarding label attachment, post translational modifications and other applications 101–105. A previous study, used self-assembled monolayers for matrix-assisted laser desorption ionization (SAMDI) to analyze intact membrane proteins in nanodiscs but unfortunately the data featured a strong, overwhelming MSP peak that was significantly greater than the inserted protein 106. In order to correct this problem, modified MALDI methods have been developed as explained below 104, 107, 108. One of the key steps in MALDI using nanodiscs is the preparation of the sample matrix. The most common method of sample preparation is the dried drop method. In this method, the analyte is mixed with matrix and spotted on the sample plate to dry 101. This preparation was tested with CYP3A4 (cytochrome P450 3A4) and the MS data problematically contained significantly higher MSP peaks than the target CYP3A4 peaks 101. As a result, CYP3A4 nanodiscs were prepared by using the ultra-thin layer method and evaluate these nanodiscs. In the ultra-thin layer method a formic acid/water/isopropanol solution 104 was mixed with matrix, centrifuged, and the supernatant combined with protein nanodiscs 101. The solution was spotted onto an ultra-thin layer plate where the polycrystalline layer formed across the spots 101. Both α-cyano-4-hydroxycinnamic acid (4HCCA) and sinapinic acid (SA) matrixes were used to study CYP3A4 and both choices of matrixes eliminated MSP signal and enhanced CYP3A4 peaks 101. The 4HCCA matrix showed more poly cationic species than SA and this can be useful when analyzing multiple peaks to obtain a high accuracy mass value. The SA matrix generated a better signal from singly and doubly charged CYP3A4 which will be useful for simpler spectra 101. SA matrix was then used to study both Cytochrome P450 Reductase (a mostly soluble protein with a transmembrane anchor), and Rhodopsin (a protein anchored to the membrane more tightly than CYP3A4 and Cytochrome P450 Reductase). The MSP signal was removed in using the two different matrix suggesting that this effect of enhancement of the intact membrane protein peak as opposed to MSP peak is result of the ultra-thin layer plate methodology 101.
Marty et al. then investigated the means behind MSP signal elimination and protein signal amplification by further studying a CYP3A4 spot. Manually moving the laser across and depth probing the spot revealed that CYP3A4 was distributed homogenously while no MSP was observed 101. When varying the time period, crystals were allowed to form before excess sample was washed off and this resulted in the formation of a coffee ring pattern would form with no MSP on the outside layer but very concentrated MSP on the inside 101. Based on this observation, Marty and coworkers concluded that in longer crystallization times the MSP partitions into the solvent and is aspirated off with the solvent while the CYP3A4 deposits into the polycrystalline film 101 (figure 3C).
This ultra-thin layer MALDI MS method is an important development because it eliminates the MSP signal while enhancing membrane protein signal. In the future, ultra-thin layer MALDI MS will be used to analyze intact protein complexes. Ultra-thin layer MALDI can also be used to identify known protein targets within a heterogeneous mixture. Marty et al. have used native mass spectrometry to determine the molecular weight and polydispersity of intact nanodiscs, showing that the integrity of the “empty” nanodiscs can be preserved in the gas phase 109.
More recently, Campuzano and colleagues compared the use of industry standard orthogonal acceleration time-of-flight (OA-ToF) to both Orbitrap and Fourier-transform ion cyclotron resonance (FT-ICR) 110. Bacteriorhodopsin-containing and empty nanodiscs were subjected to both ESI and MALDI MS to gauge the efficiency of membrane protein liberation, as well as the lipid content of the empty discs 110. It was found that both Orbitrap and FT-ICR MS produced cleaner spectra with less protein unfolding, and that FT-ICR and isideal for membrane protein liberation 110. Future research will focus towards improving data analysis methods and spectral resolution which currently hinder the application of native mass spectrometry for studying membrane protein-nanodisc complexes111, 112. Overall, these studies have demonstrated the advantages of using nanodisc technology to alleviate problems created when analyzing detergent solubilized membrane proteins or membrane proteins incorporated into other lipid bilayer systems via mass spectrometry.
6. OTHER BIOCHEMICAL TECHNIQUES
6.1 Nanodiscs and surface plasmon resonance
Surface plasmon resonance (SPR) is a technique vastly used for studying small molecule binding to proteins. Although extensively used for soluble proteins, it has been challenging to stabilize membrane proteins for SPR analysis. However, nanodiscs present an excellent stabilization complex since it the protein embedded in the nanodisc has both its epitopes exposed on either side of the lipid bilayer. Thus, it is a valuable technique towards characterization of protein-small molecule binding. A review on the optimization of various parameters for SPR on nanodiscs including various chips and immobilization techniques has been recently published 113.
Nanodiscs have been co-employed with the cell free expression of endothelin A and B to stabilize these receptors 114. Furthermore, SPR and other techniques including radioassays and fluorescence measurements were utilized to study the binding of ligand ET, a peptide ligand of endothelin A and B. Specifically, b-ET-1 or b-ET-3 ligand was immobilized on the surface of SA biosensor chips and KD values were calculated. Additionally, the effect of lipid composition on the KD value was analyzed. By varying the lipid composition in the nanodisc, slightly lower KD values were obtained when 5% cholesterol hemisuccinate or total brain extract lipids were utilized instead of 100% PC lipids. However, the role of anionic lipids could not be analyzed due to the strong association of b-ET-1 with the anionic lipid. This work is one of the first examples of GPCR-NDs and SPR.
Recently, human adenosine A2A receptor has been incorporated into nanodiscs and binding was studied by SPR 115. For this study, the authors expressed and purified a variant of the MSP1D1—carrying an amino-terminal His tag for purification purposes that can be removed by TEV cleavage, and a C-terminal C9 tag (TETSQVAPA) to immobilize the A2A/nanodisc complex via the 1D4 antibody on the SPR chip. Alternately, A2A-GFP-His nanodiscs were also immobilized via the His tag present on the carboxy terminal tail of the GFP fused to the receptor by direct affinity coupling on nickel NTA chip. This technique allows the immobilization of empty nanodiscs to the reference channel. Furthermore, the authors found that neither immobilization technique has significant effects on the KD values as verified by radio-binding assays. Results indicate that nanodisc stabilized A2A shows increased binding activity than detergents (80% vs 52%) 78. An additional prominent feature observed in this study is that association rates for two compounds appear significantly faster when the A2A receptor is inserted in nanodiscs resulting in stronger affinities than those measured on detergent solubilized receptors. The authors hypothesize that the presence of a lipid bilayer modifies the access pathway of the ligand to the receptor. Thus, SPR technology can be used for determining kinetic parameters more accurately when combined with membrane proteins in nanodiscs.
Finally, ion channel KcsA-Kv1.3 was incorporated into nanodiscs and binding to peptide ligand was studied 116. In this study, the authors immobilize biotinylated nanodiscs with or without the ion channel incorporated into regenerable streptavidin sensorchips. The authors studied small molecule binding as well as peptide binding to the ion channel by SPR and observed that small molecule binding required a slightly higher temperature than peptide binding, possibly indicating differential binding sites. The binding site for the small molecules may require higher mobility of the lipids surrounding the channel and therefore would only be able to bind at a temperature significantly higher than the phase transition temperature of the lipid used for nanodisc assembly, DMPC. The authors also concurrently studied these binding modes with BSI or back scattering interferometry, which they found to be more sensitive towards small molecule binding to the channel than SPR. Additionally, the authors noted discrepancies between the BSI and SPR values. Altogether, a combination of these two techniques, both compatible with nanodisc complexes, provides a high throughput platform for various binding assays.
6.2. Charge Sensitive Optical Detection
In addition to SPR, the nanodisc technology has been combined with other surface based technologies like CSOD or charge sensitive optical detection 117. As the name suggests, this technique is sensitive to charge as opposed to mass change. This is very useful for detecting protein binding with small molecule ligands since the fold change in charge on binding is often more significant than the change in mass. This technique has been applied to investigate the binding of two peptides and one small molecule to KcsA-Kv1.3 channel. In this technique, an optical fiber tip made of silica, is first functionalized by APTES [(3-aminopropyl) triethoxysilane]. This further assists the attachment of the channel incorporated in nanodiscs by biotin-streptavidin interaction to immobilize the protein on the fiber tip. The fiber tip can be dipped into a well containing the ligand and the association and dissociation parameters (in buffer) can be measured by changes in charge. This technique is also sensitive in serum and 3.9% DMSO. However, this method does not allow the study of systems where binding kinetics do not result in a net change in charge.
6.3. Scintillation Proximity Assay
Transporter proteins are found all throughout cell membranes, which catalyze the transport of ions, neurotransmitters, amino acids, and other solutes across the lipid bilayer. It is important to study transporters because transporter dysfunction is associated with several diseases and as a result therapeutic and deleterious drugs target transporters 118–120. A key method used to study transporters is radioligand binding assays and these assays frequently use protein from intact membranes or protein that is detergent solubilized 121. Unfortunately, other membrane proteins interfere with analysis of transporters in intact membranes 122, 123. Additionally, detergent micelles do not adequately mimic cellular membranes for these types of studies for reasons mentioned within this review and in other studies 124. The nanodisc is the ideal system to eliminate these concerns and the Singh group recently combined nanodiscs with a radioligand binding assay known as the scintillation proximity assay (SPA) to study the membrane transporter LeuT.
The SPA is a robust radioligand binding assay that has previously been used to study membrane and detergent solubilized proteins 125–127. In this assay scintillant-incorporated beads emit light when bound to a radioligand or bound to a protein that then binds a radioligand 121. When using nanodiscs to analyze membrane proteins via SPA the protein is incorporated into nanodiscs and then the entire complex is immobilized on the bead 121. The beads are then incubated with ligand and if a ligand binds the protein it is close enough to stimulate the scillant and generates a fluorescent signal that can be measured while a non-binding radio ligand would generate no such signal 121. The strength of the fluorescence is directly proportional to the strength of the binding interaction 121, 128.
The protein analyzed by SPA, LeuT, is a robustly studied, structurally stable non-polar amino acid transporter 128. Previous literature has found that Leucine binds to LeuT 10–100 fold tighter than Alanine 128, 129. The Singh group incubated scillant bead bound LeuT nanodiscs and scillant bead bound empty nanodisc controls with varying concentrations of radiolabeled Ala and Leu. These experiments found LeuT’s Leu Kd to be 23.4 nM which is similar to detergent solubilized LeuT 121. However, LeuT bound to Ala with a Kd of 1620 nM 1.5 times greater than the detergent solubilized LeuT 121. This result points to the ability of the nanodisc lipids to increase protein activity in comparison to detergent solubilized systems.
This study paves the way for numerous future works by confirming that the scintillation proximity assay can be adjusted to feature nanodiscs. Membrane protein nanodiscs could start being used in SPA drug binding studies. Perhaps the most intriguing future use of this technology would be to vary the lipid composition of the nanodiscs. Studying LeuT in nanodiscs found that nanodisc lipids improved binding and it is known that lipid composition and cholesterol concentration influence membrane protein activity 130–132. Therefore, it is reasonable to believe that since SPA assay is sensitive enough to detect increased affinity due to lipids, the assay could discern the differences in binding caused by varying nanodisc lipid composition. Altering the nanodisc lipid composition is relatively easy and will be discussed later in this review.
6.4. Electron Microscopy
There are a multitude of electron microscopy techniques used to study proteins and determine information regarding 3D structure. Nanodiscs have enabled the use of both negative staining (2D) and cryo-electron (3D) microscopy to obtain more accurate information regarding membrane protein structures. For example, Hernandez-Rocamora et al. have improved their understanding of the ZipA-FtsZ complex by using 2D Electron microscopy. The ZipA-FtsZ complex is essential to cell division in E. Coli cells because it helps to form the bacterial division ring complex 133. A previous study discovered that ZipA’s transmembrane region is irreplaceable and necessary for biological function 134. Therefore, the ZipA-FtsZ complex and other ZipA interactions should be probed using unmodified protein in a physiological membrane mimicking environment. Hence, nanodiscs were used to analyze ZipA-FtsZ by FCS and dynamic light scattering (DLS) 135. Another study incorporated ZipA into nanodiscs constructed with E. Coli lipid extract and analyzed by negative staining electron microscopy 133. This microscopy observed ZipA-ND in a small complex conformation measuring 10–11 nm in diameter and a large conformation measuring 12–13 nm in diameter 133. This is different from the FCS and DLS experiments that identified only one species 135. However, FCS and DLS cannot resolve differences between species of such similar sizes, and as such illustrates the importance of using many different techniques when trying to elucidate properties of membrane proteins. Furthermore, ZipA-ND were observed binding to antibodies and to FtsZ 133. The results of this study demonstrate that negative staining nanodisc electron microscopy is a valid means of observing this protein complex. Additionally, negative staining electron microscopy was recently used to analyze cyclooxygenase-2 as well as the anthrax protective antigen, both of which were incorporated into nanodiscs 136, 137. Another protein that was recently analyzed by negative staining electron microscopy using nanodiscs was 5-lipoxygenase (5LO) 138. Negative staining electron microscopy and titrations determined that the binding of monomeric 5LO to nanodiscs was upregulated by calcium. This finding was expected because calcium causes 5LO to bind to the nuclear membrane 138. Dimeric 5LO was analyzed as well and it was determined that dimeric 5LO is inactive and does not bind nanodiscs 138.
As previously mentioned, the Anthrax Protecting Antigen (APA) was analyzed by negative staining electron microscopy, although the staining procedure caused some shrinkage of the protein’s outer envelope 136, 139. In order to observe a more realistic envelope and therefore obtain better insight regarding APA structure, APA-ND were analyzed by cryo-electron microscopy 139. Using this method, Gogol et al. was able to elucidate information regarding differences in APA structure upon binding to its ligand, the N-terminal domain of Anthrax Lethal Factor (NTALF) 139. The model created from the data obtained by this method had an improved correlation score over the negative staining electron microscopy model 139. Additionally, cryo-electron microsopy studies benefit significantly from using nanodiscs because the nanodiscs have a high contrast shape when viewed edge-wise, which enables high-resolution analysis 139–142. In addition, Frauenfeld et al. have analyzed how the SecYEG translocon complexes with ribosome 143. This study observed rRNA directly interacting with nanodisc lipids in the vicinity of the lateral gate and was able to create a model of a signal-anchor gated SecYEG translocon 143.
Both microscopy techniques have also been used to study platelet αIIbβ3 receptor. This integrin receptor is essential for normal hemostasis and is a target for heart attack treatment and prevention due to its role in thrombosis 144, 145. This protein has been robustly studied using multiple techniques such as cryo-electron microscopy, small angle neutron scattering (SANS), and small angle x-ray scattering (SAXS) experiments performed on detergent solubilized αIIbβ3. These studies disagree on the orientation of the receptor’s ectodomain (specifically the head region) in relation to the transmembrane domain 146–148. Analyzing αIIbβ3-ND by negative staining and cryo-electron microscopy determined that the protein’s head faces up 149. This is in agreement with the cryo-electron microscopy results of detergent solubilized αIIbβ3 146. Although the SANS experiments on detergent solubilized αIIbβ3 observe a downward facing head, the reconstruction created from the αIIbβ3-ND EM data actually fit the shape generated by the SANS model better than a head down crystal structure 149. This is one example of electron microscopy being combined with nanodiscs to resolve ambiguity created when studying detergent solubilized proteins that can exhibit an altered conformation. Another investigation focusing on vacuolar H+ ATPase (V-ATPase) used negatively stained EM images of V-ATPase nanodiscs to build a 3D model of the complex. Due to a lack of distinguishing features within the complex, calmodulin was added to the final nanodiscs to bind a peptide on subunit a of the complex and add asymmetry to the complex—thus enabling the construction of a 3D model.
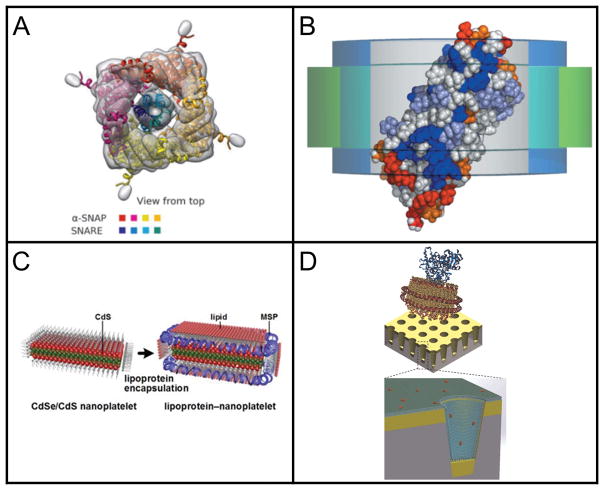
Cryo-electron microscopy and nanodiscs have also been combined to analyze SNAP-SNARE assembly 150. SNARE complexes are theorized to be the minimal machinery necessary for membrane fusion 151. After membrane fusion the SNARE complex is bound by α soluble N-ethylmaleimide-sensitive factor (NSF) attachment proteins (α-SNAPs) and NSF and this binding causes SNARE complex disassembly 150. Prior to disassembly the complex is known as the 20S complex. 20S complexes were inserted into nanodiscs and analyzed by cryo-elecron microscopy which determined that four α-SNAPS bind around a left handed SNARE helical bundle and a chock formed by conserved regions of each α SNAP protrudes into the groove of the SNARE bundle 150. Zhou and coworkers obtained new insights on the mechanism of SNARE complex disassembly from this Cryo-Electron Microscopy Data and a reconstruction of the 20S complex is shown in Figure 4A.
Figure 4.
6.5. Small-Angle X-Ray Scattering
Nanodiscs have been recognized for better preserving protein functionality upon solubilization into an aqueous environment over more traditional molecular tools, such as detergent and liposomes 152. Preservation of the native protein’s functionality and structure has opened the door for Knyde et al. to perform solution studies of proteins reconstituted in nanodisc bilayers. One such methodology that groups have employed to perform solution studies on empty nanodiscs or nanodiscs with target membrane protein incorporated, is solution-based small-angle x-ray scattering (SAXS). Studies that make use of SAXS to investigate nanodiscs allows for the gathering of structural information about the protein in its near-native conformational state in aqueous solution. Additionally, this technique has shown promising results in studies investigating molecular-level protein-lipid interactions in nanodisc-solubilized systems 153.
Notably, nanodiscs serve as a preferred solubilization method for SAXS studies on membrane proteins due to their highly homogeneous structure 154. A high degree of homogeneity of nanodisc enables resolution of membrane protein structures in nanodisc with higher resolution as compared to other solubilization techniques 28. Despite an advantageous higher resolution over other methodologies, SAXS studies containing nanodiscs incorporated with target membrane protein have not been extensively conducted. After surveying the literature, one will find that only bacteriorhodopsin (bR), cytochrome P450 3A4 (CYP3A4), and (1, 3)-β-d-glucan (curdlan) synthase have been reconstituted in nanodiscs and analyzed by small-angle x-ray scattering 153, 155, 156.
Kynde and coworkers made use of SAXS to study the structure of bacteriorhodopsin solubilized in POPC/POPG nanodiscs 153. Within their study, it was found that bR was not centered within the nanodisc, but was actually laterally displaced closer to the edge of the MSP-lipid junction 153. Furthermore, the structural study revealed that bR possessed a minor tilt with respect to the phospholipid bilayer 153. The group had also investigated the structural properties of nanodiscs, including both the lipid and protein components, and found through experimentation utilizing SAXS that the bilayer of the bR-nanodisc had a lesser thickness than empty nanodiscs 153 (Figure 4B).
In addition to nanodiscs incorporated with target protein, empty nanodiscs have also been extensively analyzed by small-angle X-ray scattering studies to obtain a greater understanding of their structural properties. Shih and coworkers combined SAXS and coarse-grained molecular dynamics simulations to examine the disassembly of nanodiscs upon adding high concentrations of sodium cholate 157. This study gained insight on the molecular movements occurring during spontaneous nanodisc assembly. Moreover, work by Sligar and coworkers have implemented SAXS methodologies in multiple works, studying empty nanodiscs, furthering the literature understanding on the intrinsic structural properties of nanodiscs. In 2005 they used SAXS to quantify lateral thermal expansion of empty nanodiscs undergoing lipid phase transitions and observed a behavior similar to nanodiscs subjected to protein incorporation; the nanodiscs decreased in thickness upon thermal increase 153, 158. Later, another SAXS study was accomplished on empty nanodiscs by Skar-Gislinge et al. that revealed the lipid packing within nanodiscs to induce an elliptical shape 154. Their reporting had matched the twisted-belt model previously theorized by Thomas et al. in 2008 159. Predating both studies, SAXS technology had been used by Denisov et al. who constructed nanodiscs sized 9.5 to 12.8 nm in diameter and showed that it is the length of the MSP that determines the diameter of a nanodisc 9.
6.6 Biolayer Interferometry
Quantifying protein-protein interactions can be challenging, especially in regards to membrane proteins. In a recent paper by Sharma and Wilkens, biolayer interferometry (BLI) is used to determine kinetic constants of the multi-subunit V-ATPase complex 160. V-ATPase is a ubiquitous ATP-dependent proton pump regulated by reversible disassembly. The authors reconstituted V-ATPase into nanodiscs containing native vacuolar lipids and tagged MSP for binding to BLI sensors, at which point the nanodiscs were treated with the inhibitor SidK to examine binding kinetics 160. Dissociation of the two main units of V-ATPase was then tested under chaotropic and inhibitory conditions 160. The successful use of BLI now offers another avenue of investigation protein-protein interactions of membrane proteins in nanodiscs.
7. NANODISC USED FOR ADVANCING NANOTECHNOLOGY
7.1 Nanoplasmonic Biosensing
Several research groups have combined nanodisc technology with nanotechnology. One example is combining the nanodisc technology with plasmonics for the purpose of nanoplasmonic biosensing. The plasmophore is a theory that fluorophores, when excited by incident light, can interact with nearby metals to create plasmons 161. This plasmon can then radiate at an observable emission that retains the fluorophore’s emission spectrum and this is known as a plasmophore 161–163. A study conducted by Zhu et al., combined the unique properties of nanodiscs and quantum dots (QDs) to analyze fluorescence and ultimately bring validation to the plasmophore concept 163. Their study created gold elliptical nanodiscs (AuND) by combining three-beam interference lithography with electron beam deposition of gold. Quantum dots emit light at specific wavelengths and an antenna molecule can enhance the observed fluorescence. Within this experiment, the AuND acts as an antenna molecule and are fluoresced by the light that emits from the quantum dots 163. When the AuNDs were brought in close proximity to the quantum dots, the emission polarization transformed from the quantum dot spectra into the antenna spectra, but the rate enhancement appeared to be polarization independent 163.
As demonstrated by Plucinski and coworkers, nanodiscs have been used to stabilize cytochrome P450s, a superfamily of heme-containing isozymes, which are heavily involved in drug metabolism, on plasmonic surfaces 164. The stabilizing effects of the nanodisc maintain protein functionality while on the metallic surface used for nanoplasmonic substrate binding assays. The group utilized a gold nanoplasmonic Lycurgus cup array sensor (nanoLCA) to detect the binding of small molecules to Cytochrome P450 2J2 164. The nanoLCA biosensor functions by taking advantage of the properties of both localized surface plasmon resonance (LSPR) and surface plasmon polariton-Bloch wave (SPP-BW) to measure spectral changes that are indicative of a drug-binding event 164. The nanoLCA biosensor, in conjunction with a protein of interest that is both immobilized and stabilized by nanodisc technology, allows for a viable methodology of performing high-throughput surface-based drug screenings 164.
7.2. Lipoprotein-Nanoplatelets
Nanoplatelets are semiconductive anisotropic nanocrystals with well-defined thicknesses residing at the atomic level 165. Current syntheses of nanoplatelets allows for them to be synthesized as planar sheets with widths of only 1.2 to 2.8 nm thick 165, 166. The precisely-defined dimensions of nanoplatelets are important because even minuscule alterations in shape will greatly affect the optical properties of the nanoparticle 165. Nanoplatelets (NPLs) possess a multitude of unique physical and electrical properties, but most notable for investigators within the life sciences is a fluorescence emission that remains stable across an extensive spectrum of wavelengths, a light collecting efficiency that is even greater than green fluorescent protein, and an emission bandwidth that is narrower than both quantum dots and organic fluorophores 167. As such, NPLs present themselves as a probable tool for investigators wishing to conduct single-molecule imaging or optical cellular tagging studies 167. However, for NPLs to be implemented within in vivo studies, they must undergo a phase transfer from the organic solvents used within syntheses to a biocompatible, aqueous buffer. Historically, the problem of stabilizing the NPLs during the phase transfer remained as a persistent complication, but nanodisc technology provides a probable solution. In a recently conducted study, researchers from Das Lab in collaboration with Smith and coworkers encapsulated NPLs within the lipoprotein of nanodiscs to construct lipoprotein-nanoplatelets (L-NPLs) 167. By use of an evaporation-encapsulation methodology outlined by the authors, they were able to successfully create L-NPLs through utilizing the nanodisc components: MSPs, phospholipids, and detergents 167. The lipoprotein-encapsulated NPLs that emerge are now biocompatible and can be used in aqueous media. The authors proceeded to characterize the L-NPLs by using a number of analytical techniques, namely through spectral analysis and transmission electron microscopy. They determined there to be an average of 7.7 MSPs per nanoplatelet and that the MSPs form as two rings, adsorbing around the edges of the top and bottom planar surfaces of the rectangular-shaped NPLs 167. Additionally, they postulated that the lipids from the lipoprotein mask the planar surfaces of the NPL that are not encapsulated by MSP 167.
As indicated by this study, nanodisc technology presents a viable technique for solubilizing and stabilizing nanoplatelets in aqueous buffers. The authors showed that the lipoprotein-nanoplatelets were stable in their aqueous environment even a month after the removal of detergent and empty nanodiscs from solution 167. Moreover, the authors note a number of future applications for nanoplatelets coupled with nanodisc technology. The L-NPLs can be utilized as a probe for single-molecule imaging studies as well as the potential for L-NPLs to serve as fluorescent cellular labels 167. These suggestions are supported by their evidence of single-particle fluorescence data which shows that L-NPLs have significantly rapid uptake into cells, which the authors claim can be attributed to both the large surface area and low curvature of the L-NPLs 167.
7.3 Sortase mediated labeling of nanodiscs
A protocol was recently developed by Petrache et al. to use Sortase A as a means of labeling nanodisc MSP with a fluorescent fluorescin-desipeptide 168. These labeled nanodiscs were then incubated with HeLA cells and were able to be observed inside the cells 168. Importantly, this group’s protocol consisted of a “retrofit approach” which allows the MSP modification to be applied to preformed nanodiscs. This approach is significant and may be applied to any number of applications that require the labeling of preformed nanodiscs.
8. CONCLUSION AND IMPROVEMENT OF NANODISC TECHNOLOGY FOR FUTURE STUDIES OF MEMBRANE PROTEINS
There has been rapid progress in the past five years in expanding the use of nanodisc technology for membrane protein studies using various biophysical and nanotechnological tools. Despite the rapid progress there is still prospect to improve this technology for membrane protein study. For instance, the nanodisc technology uses inclusion of exogenous lipids. While external lipids have proven to be excellent for the stabilization of proteins, they still do not resemble the native environment completely. From our section on protein-lipid interactions in nanodisc, it is to be noted that for several different types of membrane proteins, there is requirement of specific lipids. Thus, progress needs to be made towards inclusion of native lipids in nanodisc assembly. Moreover, this inclusion would assist in retaining the lipid-protein interactions that are key for protein functionality and activity. The diversity of the native phospholipids would create an environment different from the straight-chained aliphatic lipids typically used for nanodisc assembly. Studies are in progress where nanodiscs with native lipids are being created to study protein structure and function.
Nanodiscs are homogenous in their lipid composition and this is in stark contrast to dynamic membranes where lipid compositions vary in the outer and inner leaflets. Furthermore, the presence of flippases and floppases result in the translocation of phospholipids in the membrane over time. It is known that the asymmetric lipid bilayers are important for the study of several membrane proteins. Currently it is difficult to maintain asymmetric lipid bilayers during assembly. Therefore, in future, template based methods are needed to create asymmetric lipid bilayers in nanodiscs in order to evaluate the role of this parameter in membrane protein function.
The combination of nanodisc technology with various biochemical technologies occludes study of the section of the protein that is largely buried in the lipid bilayers. While the nanodisc technology is very useful in studying interactions at the ends of the proteins, they completely shield the interactions along the length of the molecule that is buried within the nanodisc lipids. This can be important in fluorescence studies whereby the interactions at allosteric sites buried in the membrane region cannot be completely characterized using standard techniques.
While nanodisc is a platform to study protein-protein interactions, one big challenge is controlling the conformation of the proteins with respect to each other. Hence, the two different membrane proteins can adopt multiple conformations in the nanodiscs and there is no way to control the conformation of the two protein in nanodiscs or to ensure that they form productive complex. This challenge has been partially solved by using a DNA linker for the two interacting proteins and then cleaving the linker once the proteins are in nanodiscs, however the yields are extremely low and the method requires detailed optimization and is not easy to adapt to other protein partners. Therefore more template directed assembly methods needs to be explored in order to control the conformation of the protein with respect to each other in nanodiscs.
The nanodiscs were created with the goal to facilitate functional studies of membrane proteins. Although nanodiscs have been used to perform structural studies of membrane protein with SAXS, NMR and crystallography there are several limitations 169. It is still challenging to solve crystal structures of membrane proteins in nanodiscs. It will be a major challenge to solve the structure of membrane proteins in nanodiscs. However, with the recent advances in X-ray crystallography methods, this may soon be feasible. For additional information regarding nanodiscs used in NMR studies of membrane proteins, two reviews by Puthenveetil et al. and Viegas et al. have been recently published 13, 170.
The purpose of this review was to highlight the published technologies that have been recently combined with nanodiscs to study membrane proteins from 2012–2017. This review cites over 200 publications that utilize nanodisc technology in this time period, and although we meticulously searched the literature to include as many works as possible, we acknowledge that not every paper that used nanodisc technology has been included. For further understanding of the nanodisc technology and alternative methods for lipid bilayer mimics for membrane protein study, we recommend referring to other recently published reviews on nanodiscs 4, 171.
References
- 1.Lee SC, Knowles TJ, Postis VLG, Jamshad M, Parslow RA, Lin YP, Goldman A, Sridhar P, Overduin M, Muench SP, Dafforn TR. A method for detergent-free isolation of membrane proteins in their local lipid environment. Nature protocols. 2016;11:1149–1162. doi: 10.1038/nprot.2016.070. [DOI] [PubMed] [Google Scholar]
- 2.Denisov IG, Sligar SG. Nanodiscs for structural and functional studies of membrane proteins. Nature Structural & Molecular Biology. 2016;23:481–486. doi: 10.1038/nsmb.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shenkarev ZO, Lyukmanova EN, Paramonov AS, Panteleev PV, Balandin SV, Shulepko MA, Mineev KS, Ovchinnikova TV, Kirpichnikov MP, Arseniev AS. Lipid-protein nanodiscs offer new perspectives for structural and functional studies of water-soluble membrane-active peptides. Acta Naturae. 2014;6:84–94. [PMC free article] [PubMed] [Google Scholar]
- 4.Denisov IG, Sligari SG. Nanodiscs in Membrane Biochemistry and Biophysics. Chemical Reviews. 2017;117:4669–4713. doi: 10.1021/acs.chemrev.6b00690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hein C, Henrich E, Orbán E, Dötsch V, Bernhard F. Hydrophobic supplements in cell-free systems: Designing artificial environments for membrane proteins. Engineering in Life Sciences. 2014;14:365–379. [Google Scholar]
- 6.Bayburt TH, Sligar SG. Membrane protein assembly into Nanodiscs. FEBS Letters. 2010;584:1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bayburt TH, Grinkova YV, Sligar SG. Self-Assembly of Discoidal Phospholipid Bilayer Nanoparticles with Membrane Scaffold Proteins. Nano letters. 2002;2:853–856. [Google Scholar]
- 8.Bayburt TH, Sligar SG. Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein science. 2003;12:2476–2481. doi: 10.1110/ps.03267503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J Am Chem Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 10.Grinkova YV, Denisov IG, Sligar SG. Engineering extended membrane scaffold proteins for self-assembly of soluble nanoscale lipid bilayers. Protein Eng Des Sel. 2010;23:843–848. doi: 10.1093/protein/gzq060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, Sligar SG. Chapter 11 - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Justesen BH, Günther-Pomorski T. Chromatographic and electrophoretic methods for nanodisc purification and analysis. Reviews in Analytical Chemistry. 2014;33:165–172. [Google Scholar]
- 13.Viegas A, Viennet T, Etzkorn M. The power, pitfalls and potential of the nanodisc system for NMR-based studies. Biol Chem. 2016;397:1335–1354. doi: 10.1515/hsz-2016-0224. [DOI] [PubMed] [Google Scholar]
- 14.Roy J, Pondenis H, Fan TM, Das A. Direct Capture of Functional Proteins from Mammalian Plasma Membranes into Nanodiscs. Biochemistry. 2015;54:6299–6302. doi: 10.1021/acs.biochem.5b00954. [DOI] [PubMed] [Google Scholar]
- 15.Marty MT, Wilcox KC, Klein WL, Sligar SG. Nanodisc-solubilized membrane protein library reflects the membrane proteome. Anal Bioanal Chem. 2013;405:4009–4016. doi: 10.1007/s00216-013-6790-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilcox KC, Marunde MR, Das A, Velasco PT, Kuhns BD, Marty MT, Jiang H, Luan CH, Sligar SG, Klein WL. Nanoscale Synaptic Membrane Mimetic Allows Unbiased High Throughput Screen That Targets Binding Sites for Alzheimer’s-Associated Abeta Oligomers. PLoS One. 2015;10:e0125263. doi: 10.1371/journal.pone.0125263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gregersen JL, Fedosova NU, Nissen P, Boesen T. Reconstitution of Na+,K+-ATPase in Nanodiscs. In: Bublitz M, editor. P-Type ATPases: Methods and Protocols. Springer New York; New York, NY: 2016. pp. 403–409. [DOI] [PubMed] [Google Scholar]
- 18.Mak S, Sun R, Schmalenberg M, Peters C, Luppa PB. Express incorporation of membrane proteins from various human cell types into phospholipid bilayer nanodiscs. Biochemical Journal. 2017;474:1361–1371. doi: 10.1042/BCJ20161110. [DOI] [PubMed] [Google Scholar]
- 19.Babu M, Vlasblom J, Pu S, Guo X, Graham C, Bean BD, Burston HE, Vizeacoumar FJ, Snider J, Phanse S, Fong V, Tam YY, Davey M, Hnatshak O, Bajaj N, Chandran S, Punna T, Christopolous C, Wong V, Yu A, Zhong G, Li J, Stagljar I, Conibear E, Wodak SJ, Emili A, Greenblatt JF. Interaction landscape of membrane-protein complexes in Saccharomyces cerevisiae. Nature. 2012;489:585–589. doi: 10.1038/nature11354. [DOI] [PubMed] [Google Scholar]
- 20.Denisov IG, Baas BJ, Grinkova YV, Sligar SG. Cooperativity in cytochrome P450 3A4: linkages in substrate binding, spin state, uncoupling, and product formation. J Biol Chem. 2007;282:7066–7076. doi: 10.1074/jbc.M609589200. [DOI] [PubMed] [Google Scholar]
- 21.McDougle DR, Palaria A, Magnetta E, Meling DD, Das A. Functional studies of N-terminally modified CYP2J2 epoxygenase in model lipid bilayers. Protein Sci. 2013;22:964–979. doi: 10.1002/pro.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grinkova YV, Denisov IG, Sligar SG. Functional reconstitution of monomeric CYP3A4 with multiple cytochrome P450 reductase molecules in Nanodiscs. Biochem Biophys Res Commun. 2010;398:194–198. doi: 10.1016/j.bbrc.2010.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denisov IG, Sligar SG. Cytochromes P450 in nanodiscs. Biochim Biophys Acta. 2011;1814:223–229. doi: 10.1016/j.bbapap.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raschle T, Lin C, Jungmann R, Shih WM, Wagner G. Controlled Co-reconstitution of Multiple Membrane Proteins in Lipid Bilayer Nanodiscs Using DNA as a Scaffold. ACS Chem Biol. 2015 doi: 10.1021/acschembio.5b00627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inagaki S, Ghirlando R, Grisshammer R. Biophysical characterization of membrane proteins in nanodiscs. Methods. 2013;59:287–300. doi: 10.1016/j.ymeth.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee AG. Biological membranes: the importance of molecular detail. Trends Biochem Sci. 2011;36:493–500. doi: 10.1016/j.tibs.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 27.Kim C, Ye F, Hu X, Ginsberg MH. Talin activates integrins by altering the topology of the beta transmembrane domain. J Cell Biol. 2012;197:605–611. doi: 10.1083/jcb.201112141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kynde SAR, Skar-Gislinge N, Pedersen MC, Midtgaard SR, Simonsen JB, Schweins R, Mortensen K, Arleth L. Small-angle scattering gives direct structural information about a membrane protein inside a lipid environment. Acta Crystallographica Section D-Biological Crystallography. 2014;70:371–383. doi: 10.1107/S1399004713028344. [DOI] [PubMed] [Google Scholar]
- 29.Meling DD, Zelasko S, Kambalyal A, Roy J, Das A. Functional role of the conserved i-helix residue I346 in CYP5A1-Nanodiscs. Biophys Chem. 2015;200–201:34–40. doi: 10.1016/j.bpc.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 30.Wang B, Zhao A, Xie Q, Olinares PD, Chait BT, Novick RP, Muir TW. Functional plasticity of the AgrC receptor histidine kinase required for staphylococcal virulence. Cell Chemical Biology. 2017;24:76–86. doi: 10.1016/j.chembiol.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Mu Z, Li Y, Bi Y, Wang Y. Smaller Nanodiscs are Suitable for Studying Protein Lipid Interactions by Solution NMR. Protein J. 2015;34:205–211. doi: 10.1007/s10930-015-9613-2. [DOI] [PubMed] [Google Scholar]
- 32.François-Martin C, Pincet F. Actual fusion efficiency in the lipid mixing assay - Comparison between nanodiscs and liposomes. Scientific Reports. 2017 doi: 10.1038/srep43860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaw AW, McLean MA, Sligar SG. Phospholipid phase transitions in homogeneous nanometer scale bilayer discs. FEBS Lett. 2004;556:260–264. doi: 10.1016/s0014-5793(03)01400-5. [DOI] [PubMed] [Google Scholar]
- 34.Henrich E, Ma Y, Engels I, Munch D, Otten C, Schneider T, Henrichfreise B, Sahl HG, Dotsch V, Bernhard F. Lipid Requirements for the Enzymatic Activity of MraY Translocases and in vitro Reconstitution of Lipid II Synthesis Pathway. J Biol Chem. 2015 doi: 10.1074/jbc.M115.664292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dijkman PM, Watts A. Lipid modulation of early G protein-coupled receptor signalling events. Biochim Biophys Acta. 2015;1848:2889–2897. doi: 10.1016/j.bbamem.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 36.Rues RB, Dotsch V, Bernhard F. Co-translational formation and pharmacological characterization of beta1-adrenergic receptor/nanodisc complexes with different lipid environments. Biochim Biophys Acta. 2016;1858:1306–1316. doi: 10.1016/j.bbamem.2016.02.031. [DOI] [PubMed] [Google Scholar]
- 37.Sloan CD, Marty MT, Sligar SG, Bailey RC. Interfacing lipid bilayer nanodiscs and silicon photonic sensor arrays for multiplexed protein-lipid and protein-membrane protein interaction screening. Anal Chem. 2013;85:2970–2976. doi: 10.1021/ac3037359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marty MT, Hoi KK, Gault J, Robinson CV. Probing the Lipid Annular Belt by Gas-Phase Dissociation of Membrane Proteins in Nanodiscs. Angew Chem Int Ed Engl. 2015 doi: 10.1002/anie.201508289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ly S, Bourguet F, Fischer Nicholas O, Lau Edmond Y, Coleman Matthew A, Laurence Ted A. Quantifying Interactions of a Membrane Protein Embedded in a Lipid Nanodisc using Fluorescence Correlation Spectroscopy. Biophysical Journal. 2012;106:L05–L08. doi: 10.1016/j.bpj.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwille P, Oehlenschlager F, Walter NG. Quantitative hybridization kinetics of DNA probes to RNA in solution followed by diffusional fluorescence correlation analysis. Biochemistry. 1996;35:10182–10193. doi: 10.1021/bi960517g. [DOI] [PubMed] [Google Scholar]
- 41.Rauer B, Neumann E, Widengren J, Rigler R. Fluorescence correlation spectrometry of the interaction kinetics of tetramethylrhodamin alpha-bungarotoxin with Torpedo californica acetylcholine receptor. Biophys Chem. 1996;58:3–12. doi: 10.1016/0301-4622(95)00080-1. [DOI] [PubMed] [Google Scholar]
- 42.Gao T, Blanchette CD, He W, Bourguet F, Ly S, Katzen F, Kudlicki WA, Henderson PT, Laurence TA, Huser T, Coleman MA. Characterizing diffusion dynamics of a membrane protein associated with nanolipoproteins using fluorescence correlation spectroscopy. Protein Sci. 2011;20:437–447. doi: 10.1002/pro.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao T, Petrlova J, He W, Huser T, Kudlick W, Voss J, Coleman MA. Characterization of de novo synthesized GPCRs supported in nanolipoprotein discs. PLoS One. 2012;7:e44911. doi: 10.1371/journal.pone.0044911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edqvist PJ, Aili M, Liu J, Francis MS. Minimal YopB and YopD translocator secretion by Yersinia is sufficient for Yop-effector delivery into target cells. Microbes Infect. 2007;9:224–233. doi: 10.1016/j.micinf.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 45.McDougle DR, Baylon JL, Meling DD, Kambalyal A, Grinkova YV, Hammernik J, Tajkhorshid E, Das A. Incorporation of charged residues in the CYP2J2 F-G loop disrupts CYP2J2–lipid bilayer interactions. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2015;1848:2460–2470. doi: 10.1016/j.bbamem.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bayburt TH, Sligar SG. Single-molecule height measurements on microsomal cytochrome P450 in nanometer-scale phospholipid bilayer disks. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:6725–6730. doi: 10.1073/pnas.062565599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meling DD, McDougle DR, Das A. CYP2J2 epoxygenase membrane anchor plays an important role in facilitating electron transfer from CPR. J Inorg Biochem. 2014;142C:47–53. doi: 10.1016/j.jinorgbio.2014.09.016. [DOI] [PubMed] [Google Scholar]
- 48.Hartley MD, Schneggenburger PE, Imperiali B. Lipid bilayer nanodisc platform for investigating polyprenol-dependent enzyme interactions and activities. Proc Natl Acad Sci U S A. 2013;110:20863–20870. doi: 10.1073/pnas.1320852110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arnold C, Markovic M, Blossey K, Wallukat G, Fischer R, Dechend R, Konkel A, von Schacky C, Luft FC, Muller DN, Rothe M, Schunck WH. Arachidonic acid-metabolizing cytochrome P450 enzymes are targets of {omega}-3 fatty acids. J Biol Chem. 2010;285:32720–32733. doi: 10.1074/jbc.M110.118406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berka K, Hendrychova T, Anzenbacher P, Otyepka M. Membrane position of ibuprofen agrees with suggested access path entrance to cytochrome P450 2C9 active site. J Phys Chem A. 2011;115:11248–11255. doi: 10.1021/jp204488j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim KH, Ahn T, Yun CH. Membrane properties induced by anionic phospholipids and phosphatidylethanolamine are critical for the membrane binding and catalytic activity of human cytochrome P450 3A4. Biochemistry. 2003;42:15377–15387. doi: 10.1021/bi035280k. [DOI] [PubMed] [Google Scholar]
- 52.Ahn T, Guengerich FP, Yun CH. Membrane insertion of cytochrome P450 1A2 promoted by anionic phospholipids. Biochemistry. 1998;37:12860–12866. doi: 10.1021/bi980804f. [DOI] [PubMed] [Google Scholar]
- 53.Mustonen P, Virtanen JA, Somerharju PJ, Kinnunen PK. Binding of cytochrome c to liposomes as revealed by the quenching of fluorescence from pyrene-labeled phospholipids. Biochemistry. 1987;26:2991–2997. doi: 10.1021/bi00385a006. [DOI] [PubMed] [Google Scholar]
- 54.Song Y, Yang Z, Ke Z, Yao Y, Hu X, Sun Y, Li H, Yin J, Zeng C. Expression of 14–3–3γ in patients with breast cancer: Correlation with clinicopathological features and prognosis. Cancer epidemiology. 2012;36:533–536. doi: 10.1016/j.canep.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 55.McDougle DR, Palaria A, Magnetta E, Meling DD, Das A. Functional studies of N-terminally modified CYP2J2 epoxygenase in model lipid bilayers. Protein Science. 2013;22:964–979. doi: 10.1002/pro.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taufik I, Kedrov A, Exterkate M, Driessen AJ. Monitoring the activity of single translocons. J Mol Biol. 2013;425:4145–4153. doi: 10.1016/j.jmb.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 57.Kedrov A, Kusters I, Krasnikov VV, Driessen AJM. A single copy of SecYEG is sufficient for preprotein translocation. The EMBO journal. 2011;30:4387–4397. doi: 10.1038/emboj.2011.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shin J, Lou X, Kweon DH, Shin YK. Multiple conformations of a single SNAREpin between two nanodisc membranes reveal diverse pre-fusion states. Biochem J. 2014;459:95–102. doi: 10.1042/BJ20131668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krishnakumar SS, Kummel D, Jones SJ, Radoff DT, Reinisch KM, Rothman JE. Conformational dynamics of calcium-triggered activation of fusion by synaptotagmin. Biophys J. 2013;105:2507–2516. doi: 10.1016/j.bpj.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kedrov A, Sustarsic M, de Keyzer J, Caumanns JJ, Wu ZC, Driessen AJ. Elucidating the native architecture of the YidC: ribosome complex. J Mol Biol. 2013;425:4112–4124. doi: 10.1016/j.jmb.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 61.Zoghbi ME, Cooper RS, Altenberg GA. The Lipid Bilayer Modulates the Structure and Function of an ATP-binding Cassette Exporter. J Biol Chem. 2016;291:4453–4461. doi: 10.1074/jbc.M115.698498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boise LH, Gonzalezgarcia M, Postema CE, Ding LY, Lindsten T, Turka LA, Mao XH, Nunez G, Thompson CB. Bcl-X, a Bcl-2-Related Gene That Functions as a Dominant Regulator of Apoptotic Cell-Death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 63.Yao Y, Fujimoto LM, Hirshman N, Bobkov AA, Antignani A, Youle RJ, Marassi FM. Conformation of BCL-XL upon Membrane Integration. Journal of molecular biology. 2015;427:2262–2270. doi: 10.1016/j.jmb.2015.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Muchmore SW, Sattler M, Liang H, Meadows RP, Harlan JE, Yoon HS, Nettesheim D, Chang BS, Thompson CB, Wong SL, Ng SC, Fesik SW. X-ray and NMR structure of human Bcl-x(L), an inhibitor of programmed cell death. Nature. 1996;381:335–341. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]
- 65.Chi XK, Kale J, Leber B, Andrews DW. Regulating cell death at, on, and in membranes. Biochimica Et Biophysica Acta-Molecular Cell Research. 2014;1843:2100–2113. doi: 10.1016/j.bbamcr.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 66.Kvansakul M, Hinds MG. Structural biology of the Bcl-2 family and its mimicry by viral proteins. Cell Death & Disease. 2013;4 doi: 10.1038/cddis.2013.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Losonczi JA, Olejniczak ET, Betz SF, Harlan JE, Mack J, Fesik SW. NMR studies of the anti-apoptotic protein Bcl-xL in micelles. Biochemistry. 2000;39:11024–11033. doi: 10.1021/bi000919v. [DOI] [PubMed] [Google Scholar]
- 68.Yao Y, Nisan D, Fujimoto LM, Antignani A, Barnes A, Tjandra N, Youle RJ, Marassi FM. Characterization of the membrane-inserted C-terminus of cytoprotective BCL-XL. Protein Expr Purif. 2016;122:56–63. doi: 10.1016/j.pep.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Susac L, Horst R, Wuthrich K. Solution-NMR Characterization of Outer-Membrane Protein A from E. coli in Lipid Bilayer Nanodiscs and Detergent Micelles. Chembiochem. 2014;15:995–1000. doi: 10.1002/cbic.201300729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tzitzilonis C, Eichmann C, Maslennikov I, Choe S, Riek R. Detergent/nanodisc screening for high-resolution NMR studies of an integral membrane protein containing a cytoplasmic domain. Plos One. 2013;8:e54378. doi: 10.1371/journal.pone.0054378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mazhab-Jafari MT, Marshall CB, Smith MJ, Gasmi-Seabrook GM, Stathopulos PB, Inagaki F, Kay LE, Neel BG, Ikura M. Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site. Proceedings of the National Academy of Sciences. 2015;112:6625–6630. doi: 10.1073/pnas.1419895112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang XM, Mu ZJ, Li Y, Bi YC, Wang YJ. Smaller Nanodiscs are Suitable for Studying Protein Lipid Interactions by Solution NMR. Protein Journal. 2015;34:205–211. doi: 10.1007/s10930-015-9613-2. [DOI] [PubMed] [Google Scholar]
- 73.Kucharska I, Edrington TC, Liang BY, Tamm LK. Optimizing nanodiscs and bicelles for solution NMR studies of two beta-barrel membrane proteins. Journal of Biomolecular Nmr. 2015;61:261–274. doi: 10.1007/s10858-015-9905-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mazhab-Jafari MT, Marshall CB, Stathopulos PB, Kobashigawa Y, Stambolic V, Kay LE, Inagaki F, Ikura M. Membrane-dependent modulation of the mTOR activator Rheb: NMR observations of a GTPase tethered to a lipid-bilayer nanodisc. J Am Chem Soc. 2013;135:3367–3370. doi: 10.1021/ja312508w. [DOI] [PubMed] [Google Scholar]
- 75.Zhang M, Huang R, Ackermann R, Im SC, Waskell L, Schwendeman A, Ramamoorthy A. Reconstitution of the Cytb5–CytP450 Complex in Nanodiscs for Structural Studies using NMR Spectroscopy. Angewandte Chemie International Edition. 2016 doi: 10.1002/anie.201600073. [DOI] [PubMed] [Google Scholar]
- 76.Bertini I, Luchinat C, Parigi G, Ravera E, Reif B, Turano P. Solid-state NMR of proteins sedimented by ultracentrifugation. Proc Natl Acad Sci U S A. 2011;108:10396–10399. doi: 10.1073/pnas.1103854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mors K, Roos C, Scholz F, Wachtveitl J, Dotsch V, Bernhard F, Glaubitz C. Modified lipid and protein dynamics in nanodiscs. Biochim Biophys Acta. 2013;1828:1222–1229. doi: 10.1016/j.bbamem.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 78.Ding Y, Fujimoto LM, Yao Y, Marassi FM. Solid-state NMR of the Yersinia pestis outer membrane protein Ail in lipid bilayer nanodiscs sedimented by ultracentrifugation. J Biomol Nmr. 2015;61:275–286. doi: 10.1007/s10858-014-9893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yamashita S, Lukacik P, Barnard TJ, Noinaj N, Felek S, Tsang TM, Krukonis ES, Hinnebusch BJ, Buchanan SK. Structural insights into Ail-mediated adhesion in Yersinia pestis. Structure. 2011;19:1672–1682. doi: 10.1016/j.str.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ding Y, Fujimoto LM, Yao Y, Plano GV, Marassi FM. Influence of the lipid membrane environment on structure and activity of the outer membrane protein Ail from Yersinia pestis. Biochimica et Biophysica Acta (BBA)-Biomembranes. 2015;1848:712–720. doi: 10.1016/j.bbamem.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kijac A, Shih AY, Nieuwkoop AJ, Schulten K, Sligar SG, Rienstra CM. Lipid – protein correlations in nanoscale phospholipid bilayers determined by solid-state nuclear magnetic resonance. Biochemistry. 2010;49:9190–9198. doi: 10.1021/bi1013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Parker CH, Morgan CR, Rand KD, Engen JR, Jorgenson JW, Stafford DW. A conformational investigation of propeptide binding to the integral membrane protein gamma-glutamyl carboxylase using nanodisc hydrogen exchange mass spectrometry. Biochemistry. 2014;53:1511–1520. doi: 10.1021/bi401536m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hebling CM, Morgan CR, Stafford DW, Jorgenson JW, Rand KD, Engen JR. Conformational analysis of membrane proteins in phospholipid bilayer nanodiscs by hydrogen exchange mass spectrometry. Anal Chem. 2010;82:5415–5419. doi: 10.1021/ac100962c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rand KD, Jørgensen TJ. Development of a peptide probe for the occurrence of hydrogen (1H/2H) scrambling upon gas-phase fragmentation. Analytical chemistry. 2007;79:8686–8693. doi: 10.1021/ac0710782. [DOI] [PubMed] [Google Scholar]
- 85.Rand KD, Adams CM, Zubarev RA, Jørgensen TJ. Electron capture dissociation proceeds with a low degree of intramolecular migration of peptide amide hydrogens. Journal of the American Chemical Society. 2008;130:1341–1349. doi: 10.1021/ja076448i. [DOI] [PubMed] [Google Scholar]
- 86.Marcsisin SR, Engen JR. Hydrogen exchange mass spectrometry: what is it and what can it tell us? Anal Bioanal Chem. 2010;397:967–972. doi: 10.1007/s00216-010-3556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nelsestuen GL, Shah AM, Harvey SB. Vitamin K-dependent proteins. Vitam Horm. 2000;58:355–389. doi: 10.1016/s0083-6729(00)58031-5. [DOI] [PubMed] [Google Scholar]
- 88.Adhikary S, Deredge DJ, Nagarajan A, Forrest LR, Wintrode PL, Singh SK. Conformational dynamics of a neurotransmitter: sodium symporter in a lipid bilayer. Proceedings of the National Academy of Science of the United States of America. 2017;114:E1786–E1795. doi: 10.1073/pnas.1613293114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vermeulen M, Hubner NC, Mann M. High confidence determination of specific protein–protein interactions using quantitative mass spectrometry. Current opinion in biotechnology. 2008;19:331–337. doi: 10.1016/j.copbio.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 90.Dobreva I, Fielding A, Foster LJ, Dedhar S. Mapping the Integrin-Linked Kinase Interactome Using SILAC. Journal of proteome research. 2008;7:1740–1749. doi: 10.1021/pr700852r. [DOI] [PubMed] [Google Scholar]
- 91.Reisinger V, Eichacker LA. Solubilization of membrane protein complexes for blue native PAGE. J Proteomics. 2008;71:277–283. doi: 10.1016/j.jprot.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 92.Zhang XX, Chan CS, Bao H, Fang Y, Foster LJ, Duong F. Nanodiscs and SILAC-based mass spectrometry to identify a membrane protein interactome. J Proteome Res. 2012;11:1454–1459. doi: 10.1021/pr200846y. [DOI] [PubMed] [Google Scholar]
- 93.Trinkle-Mulcahy L, Andersen J, Lam YW, Moorhead G, Mann M, Lamond AI. Repo-Man recruits PP1 gamma to chromatin and is essential for cell viability. J Cell Biol. 2006;172:679–692. doi: 10.1083/jcb.200508154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Foster LJ, Rudich A, Talior I, Patel N, Huang X, Furtado LM, Bilan PJ, Mann M, Klip A. Insulin-dependent interactions of proteins with GLUT4 revealed through stable isotope labeling by amino acids in cell culture (SILAC) J Proteome Res. 2006;5:64–75. doi: 10.1021/pr0502626. [DOI] [PubMed] [Google Scholar]
- 95.Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable Isotope Labeling by Amino Acids in Cell Culture, SILAC, as a Simple and Accurate Approach to Expression Proteomics. Molecular & Cellular Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 96.Dalal K, Duong F. The SecY complex: conducting the orchestra of protein translocation. Trends Cell Biol. 2011;21:506–514. doi: 10.1016/j.tcb.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 97.Schaffitzel C, Ban N. Generation of ribosome nascent chain complexes for structural and functional studies. J Struct Biol. 2007;158:463–471. doi: 10.1016/j.jsb.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 98.Kuhn P, Weiche B, Sturm L, Sommer E, Drepper F, Warscheid B, Sourjik V, Koch HG. The bacterial SRP receptor, SecA and the ribosome use overlapping binding sites on the SecY translocon. Traffic. 2011;12:563–578. doi: 10.1111/j.1600-0854.2011.01167.x. [DOI] [PubMed] [Google Scholar]
- 99.Sharma S, Davidson AL. Vanadate-induced trapping of nucleotides by purified maltose transport complex requires ATP hydrolysis. J Bacteriol. 2000;182:6570–6576. doi: 10.1128/jb.182.23.6570-6576.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.van Bloois E, Dekker HL, Froderberg L, Houben EN, Urbanus ML, de Koster CG, de Gier JW, Luirink J. Detection of cross-links between FtsH, YidC, HflK/C suggests a linked role for these proteins in quality control upon insertion of bacterial inner membrane proteins. FEBS Lett. 2008;582:1419–1424. doi: 10.1016/j.febslet.2008.02.082. [DOI] [PubMed] [Google Scholar]
- 101.Marty MT, Das A, Sligar SG. Ultra-thin layer MALDI mass spectrometry of membrane proteins in nanodiscs. Anal Bioanal Chem. 2012;402:721–729. doi: 10.1007/s00216-011-5512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.le Coutre J, Whitelegge JP, Gross A, Turk E, Wright EM, Kaback HR, Faull KF. Proteomics on full-length membrane proteins using mass spectrometry. Biochemistry. 2000;39:4237–4242. doi: 10.1021/bi000150m. [DOI] [PubMed] [Google Scholar]
- 103.Weinglass AB, Whitelegge JP, Hu Y, Verner GE, Faull KF, Kaback HR. Elucidation of substrate binding interactions in a membrane transport protein by mass spectrometry. EMBO J. 2003;22:1467–1477. doi: 10.1093/emboj/cdg145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gabant G, Cadene M. Mass spectrometry of full-length integral membrane proteins to define functionally relevant structural features. Methods. 2008;46:54–61. doi: 10.1016/j.ymeth.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 105.Alves ID, Sachon E, Bolbach G, Millstine L, Lavielle S, Sagan S. Analysis of an intact G-protein coupled receptor by MALDI-TOF mass spectrometry: molecular heterogeneity of the tachykinin NK-1 receptor. Anal Chem. 2007;79:2189–2198. doi: 10.1021/ac062415u. [DOI] [PubMed] [Google Scholar]
- 106.Marin VL, Bayburt TH, Sligar SG, Mrksich M. Functional assays of membrane-bound proteins with SAMDI-TOF mass spectrometry. Angew Chem Int Ed Engl. 2007;46:8796–8798. doi: 10.1002/anie.200702694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cadene M, Chait BT. A robust, detergent-friendly method for mass spectrometric analysis of integral membrane proteins. Anal Chem. 2000;72:5655–5658. doi: 10.1021/ac000811l. [DOI] [PubMed] [Google Scholar]
- 108.Fenyo D, Wang Q, DeGrasse JA, Padovan JC, Cadene M, Chait BT. MALDI sample preparation: the ultra thin layer method. J Vis Exp. 2007:192. doi: 10.3791/192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Marty MT, Zhang H, Cui W, Blankenship RE, Gross ML, Sligar SG. Native mass spectrometry characterization of intact nanodisc lipoprotein complexes. Anal Chem. 2012;84:8957–8960. doi: 10.1021/ac302663f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Campuzano IDG, Li H, Bagal D, Lippens JL, Svitel J, Kurzeja RJM, Xu H, Schnier PD, Loo JA. Native MS analysis of bacteriorhodopsin and an empty nanodisc by orthogonal acceleration time-of-flight, orbitrap and ion cyclotron resonance. Analytical Chemistry. 2016;88:12427–12436. doi: 10.1021/acs.analchem.6b03762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hoi KK, Robinson CV, Marty MT. Unraveling the Composition and Behavior of Heterogeneous Lipid Nanodiscs by Mass Spectrometry. Anal Chem. 2016;88:6199–6204. doi: 10.1021/acs.analchem.6b00851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Marty MT, Hoi KK, Robinson CV. Interfacing Membrane Mimetics with Mass Spectrometry. Acc Chem Res. 2016 doi: 10.1021/acs.accounts.6b00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Trahey M, Li MJ, Kwon H, Woodahl EL, McClary WD, Atkins WM. Applications of Lipid Nanodiscs for the Study of Membrane Proteins by Surface Plasmon Resonance. Curr Protoc Protein Sci. 2015;81:29 13 21–29 13 16. doi: 10.1002/0471140864.ps2913s81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Proverbio D, Roos C, Beyermann M, Orban E, Dotsch V, Bernhard F. Functional properties of cell-free expressed human endothelin A and endothelin B receptors in artificial membrane environments. Biochim Biophys Acta. 2013;1828:2182–2192. doi: 10.1016/j.bbamem.2013.05.031. [DOI] [PubMed] [Google Scholar]
- 115.Bocquet N, Kohler J, Hug MN, Kusznir EA, Rufer AC, Dawson RJ, Hennig M, Ruf A, Huber W, Huber S. Real-time monitoring of binding events on a thermostabilized human A2A receptor embedded in a lipid bilayer by surface plasmon resonance. Biochim Biophys Acta. 2015;1848:1224–1233. doi: 10.1016/j.bbamem.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 116.Xu H, Hill JJ, Michelsen K, Yamane H, Kurzeja RJ, Tam T, Isaacs RJ, Shen F, Tagari P. Characterization of the direct interaction between KcsA-Kv1.3 and its inhibitors. Biochim Biophys Acta. 2015;1848:1974–1980. doi: 10.1016/j.bbamem.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 117.Ma G, Guan Y, Wang S, Xu H, Tao N. Study of Small-Molecule-Membrane Protein Binding Kinetics with Nanodisc and Charge-Sensitive Optical Detection. Anal Chem. 2016;88:2375–2379. doi: 10.1021/acs.analchem.5b04366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rask-Andersen M, Masuram S, Fredriksson R, Schioth HB. Solute carriers as drug targets: current use, clinical trials and prospective. Mol Aspects Med. 2013;34:702–710. doi: 10.1016/j.mam.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 119.Gadsby DC, Vergani P, Csanady L. The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature. 2006;440:477–483. doi: 10.1038/nature04712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kristensen AS, Andersen J, Jorgensen TN, Sorensen L, Eriksen J, Loland CJ, Stromgaard K, Gether U. SLC6 neurotransmitter transporters: structure, function, and regulation. Pharmacol Rev. 2011;63:585–640. doi: 10.1124/pr.108.000869. [DOI] [PubMed] [Google Scholar]
- 121.Nasr ML, Singh SK. Radioligand binding to nanodisc-reconstituted membrane transporters assessed by the scintillation proximity assay. Biochemistry. 2014;53:4–6. doi: 10.1021/bi401412e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Quick M, Javitch JA. Monitoring the function of membrane transport proteins in detergent-solubilized form. Proc Natl Acad Sci U S A. 2007;104:3603–3608. doi: 10.1073/pnas.0609573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Harder D, Fotiadis D. Measuring substrate binding and affinity of purified membrane transport proteins using the scintillation proximity assay. Nat Protoc. 2012;7:1569–1578. doi: 10.1038/nprot.2012.090. [DOI] [PubMed] [Google Scholar]
- 124.Garavito RM, Ferguson-Miller S. Detergents as tools in membrane biochemistry. J Biol Chem. 2001;276:32403–32406. doi: 10.1074/jbc.R100031200. [DOI] [PubMed] [Google Scholar]
- 125.Sun S, Almaden J, Carlson TJ, Barker J, Gehring MR. Assay development and data analysis of receptor-ligand binding based on scintillation proximity assay. Metab Eng. 2005;7:38–44. doi: 10.1016/j.ymben.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 126.Janowski BA, Grogan MJ, Jones SA, Wisely GB, Kliewer SA, Corey EJ, Mangelsdorf DJ. Structural requirements of ligands for the oxysterol liver X receptors LXRalpha and LXRbeta. Proc Natl Acad Sci U S A. 1999;96:266–271. doi: 10.1073/pnas.96.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Reinscheid RK, Nothacker HP, Bourson A, Ardati A, Henningsen RA, Bunzow JR, Grandy DK, Langen H, Monsma FJ, Jr, Civelli O. Orphanin FQ: a neuropeptide that activates an opioidlike G protein-coupled receptor. Science. 1995;270:792–794. doi: 10.1126/science.270.5237.792. [DOI] [PubMed] [Google Scholar]
- 128.Singh SK, Piscitelli CL, Yamashita A, Gouaux E. A competitive inhibitor traps LeuT in an open-to-out conformation. Science. 2008;322:1655–1661. doi: 10.1126/science.1166777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shi L, Quick M, Zhao Y, Weinstein H, Javitch JA. The mechanism of a neurotransmitter:sodium symporter--inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol Cell. 2008;30:667–677. doi: 10.1016/j.molcel.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Divito CB, Amara SG. Close encounters of the oily kind: regulation of transporters by lipids. Mol Interv. 2009;9:252–262. doi: 10.1124/mi.9.5.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hong WC, Amara SG. Membrane cholesterol modulates the outward facing conformation of the dopamine transporter and alters cocaine binding. J Biol Chem. 2010;285:32616–32626. doi: 10.1074/jbc.M110.150565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Orlowski S, Martin S, Escargueil A. P-glycoprotein and ‘lipid rafts’: some ambiguous mutual relationships (floating on them, building them or meeting them by chance?) Cell Mol Life Sci. 2006;63:1038–1059. doi: 10.1007/s00018-005-5554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hernandez-Rocamora VM, Garcia-Montanes C, Rivas G, Llorca O. Reconstitution of the Escherichia coli cell division ZipA-FtsZ complexes in nanodiscs as revealed by electron microscopy. J Struct Biol. 2012;180:531–538. doi: 10.1016/j.jsb.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 134.Hale CA, Rhee AC, de Boer PA. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J Bacteriol. 2000;182:5153–5166. doi: 10.1128/jb.182.18.5153-5166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hernandez-Rocamora VM, Reija B, Garcia C, Natale P, Alfonso C, Minton AP, Zorrilla S, Rivas G, Vicente M. Dynamic interaction of the Escherichia coli cell division ZipA and FtsZ proteins evidenced in nanodiscs. J Biol Chem. 2012;287:30097–30104. doi: 10.1074/jbc.M112.388959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Akkaladevi N, Hinton-Chollet L, Katayama H, Mitchell J, Szerszen L, Mukherjee S, Gogol EP, Pentelute BL, Collier RJ, Fisher MT. Assembly of anthrax toxin pore: Lethal-factor complexes into lipid nanodiscs. Protein Science: A Publication of the Protein Society. 2013;22:492–501. doi: 10.1002/pro.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Orlando BJ, McDougle DR, Lucido MJ, Eng ET, Graham LA, Schneider C, Stokes DL, Das A, Malkowski MG. CYCLOOXYGENASE-2 CATALYSIS AND INHIBITION IN LIPID BILAYER NANODISCS. Archives of biochemistry and biophysics. 2014;546:33–40. doi: 10.1016/j.abb.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kumar RB, Zhu L, Idborg H, Rådmark O, Jakobsson P-J, Rinaldo-Matthis A, Hebert H, Jegerschöld C. Structural and Functional Analysis of Calcium Ion Mediated Binding of 5-Lipoxygenase to Nanodiscs. PLoS One. 2016;11:e0152116. doi: 10.1371/journal.pone.0152116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gogol EP, Akkaladevi N, Szerszen L, Mukherjee S, Chollet-Hinton L, Katayama H, Pentelute BL, Collier RJ, Fisher MT. Three dimensional structure of the anthrax toxin translocon-lethal factor complex by cryo-electron microscopy. Protein Sci. 2013;22:586–594. doi: 10.1002/pro.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lyumkis D, Moeller A, Cheng A, Herold A, Hou E, Irving C, Jacovetty EL, Lau P-W, Mulder AM, Pulokas J, Quispe JD, Voss NR, Potter CS, Carragher B. Chapter Fourteen - Automation in Single-Particle Electron Microscopy: Connecting the Pieces. In: Grant JJ, editor. Methods in enzymology. Academic Press; 2010. pp. 291–338. [DOI] [PubMed] [Google Scholar]
- 141.Zhu Y, Carragher B, Glaeser RM, Fellmann D, Bajaj C, Bern M, Mouche F, Haas Fd, Hall RJ, Kriegman DJ, Ludtke SJ, Mallick SP, Penczek PA, Roseman AM, Sigworth FJ, Volkmann N, Potter CS. Automatic particle selection: results of a comparative study. Journal of structural biology. 2004;145:3–14. doi: 10.1016/j.jsb.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 142.Carragher B, Kisseberth N, Kriegman D, Milligan RA, Potter CS, Pulokas J, Reilein A. Leginon: An Automated System for Acquisition of Images from Vitreous Ice Specimens. Journal of structural biology. 2000;132:33–45. doi: 10.1006/jsbi.2000.4314. [DOI] [PubMed] [Google Scholar]
- 143.Frauenfeld J, Gumbart J, van der Sluis EO, Funes S, Gartmann M, Beatrix B, Mielke T, Berninghausen O, Becker T, Schulten K, Beckmann R. Cryo–EM structure of the ribosome–SecYE complex in the membrane environment. Nature structural & molecular biology. 2011;18:614–621. doi: 10.1038/nsmb.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cox D, Brennan M, Moran N. Integrins as therapeutic targets: lessons and opportunities. Nat Rev Drug Discov. 2010;9:804–820. doi: 10.1038/nrd3266. [DOI] [PubMed] [Google Scholar]
- 145.Coller BS. Historical Perspective and Future Directions in Platelet Research. Journal of thrombosis and haemostasis: JTH. 2011;9:374–395. doi: 10.1111/j.1538-7836.2011.04356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Adair BD, Yeager M. Three-dimensional model of the human platelet integrin α(IIb)β(3) based on electron cryomicroscopy and x-ray crystallography. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:14059–14064. doi: 10.1073/pnas.212498199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nogales A, García C, Pérez J, Callow P, Ezquerra TA, González-Rodríguez J. Three-dimensional Model of Human Platelet Integrin αIIbβ3 in Solution Obtained by Small Angle Neutron Scattering. The Journal of biological chemistry. 2010;285:1023–1031. doi: 10.1074/jbc.M109.050039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Eng ET, Smagghe BJ, Walz T, Springer TA. Intact α(IIb)β(3) Integrin Is Extended after Activation as Measured by Solution X-ray Scattering and Electron Microscopy. The Journal of biological chemistry. 2011;286:35218–35226. doi: 10.1074/jbc.M111.275107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Choi W-S, Rice WJ, Stokes DL, Coller BS. Three-dimensional reconstruction of intact human integrin αIIbβ3: new implications for activation-dependent ligand binding. Blood. 2013;122:4165–4171. doi: 10.1182/blood-2013-04-499194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhou Q, Huang X, Sun S, Li X, Wang HW, Sui SF. Cryo-EM structure of SNAP-SNARE assembly in 20S particle. Cell Res. 2015;25:551–560. doi: 10.1038/cr.2015.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 152.Lyukmanova EN, Shenkarev ZO, Khabibullina NF, Kopeina GS, Shulepko MA, Paramonov AS, Mineev KS, Tikhonov RV, Shingarova LN, Petrovskaya LE, Dolgikh DA, Arseniev AS, Kirpichnikov MP. Lipid-protein nanodiscs for cell-free production of integral membrane proteins in a soluble and folded state: comparison with detergent micelles, bicelles and liposomes. Biochim Biophys Acta. 2012;1818:349–358. doi: 10.1016/j.bbamem.2011.10.020. [DOI] [PubMed] [Google Scholar]
- 153.Kynde SA, Skar-Gislinge N, Pedersen MC, Midtgaard SR, Simonsen JB, Schweins R, Mortensen K, Arleth L. Small-angle scattering gives direct structural information about a membrane protein inside a lipid environment. Acta Crystallogr D Biol Crystallogr. 2014;70:371–383. doi: 10.1107/S1399004713028344. [DOI] [PubMed] [Google Scholar]
- 154.Skar-Gislinge N, Simonsen JB, Mortensen K, Feidenhans’l R, Sligar SG, Lindberg Moller B, Bjornholm T, Arleth L. Elliptical structure of phospholipid bilayer nanodiscs encapsulated by scaffold proteins: casting the roles of the lipids and the protein. J Am Chem Soc. 2010;132:13713–13722. doi: 10.1021/ja1030613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Baas BJ, Denisov IG, Sligar SG. Homotropic cooperativity of monomeric cytochrome P450 3A4 in a nanoscale native bilayer environment. Arch Biochem Biophys. 2004;430:218–228. doi: 10.1016/j.abb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 156.Periasamy A, Shadiac N, Amalraj A, Garajova S, Nagarajan Y, Waters S, Mertens HD, Hrmova M. Cell-free protein synthesis of membrane (1,3)-beta-d-glucan (curdlan) synthase: co-translational insertion in liposomes and reconstitution in nanodiscs. Biochim Biophys Acta. 2013;1828:743–757. doi: 10.1016/j.bbamem.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 157.Shih AY, Freddolino PL, Sligar SG, Schulten K. Disassembly of nanodiscs with cholate. Nano Lett. 2007;7:1692–1696. doi: 10.1021/nl0706906. [DOI] [PubMed] [Google Scholar]
- 158.Denisov IG, McLean MA, Shaw AW, Grinkova YV, Sligar SG. Thermotropic phase transition in soluble nanoscale lipid bilayers. J Phys Chem B. 2005;109:15580–15588. doi: 10.1021/jp051385g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Thomas MJ, Bhat S, Sorci-Thomas MG. Three-dimensional models of HDL apoA-I: implications for its assembly and function. J Lipid Res. 2008;49:1875–1883. doi: 10.1194/jlr.R800010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sharma S, Wilkens S. Biolayer interferometry of lipid nanodisc-reconstituted yeast vacuolar H+-ATPase. Protein Science. 2017 doi: 10.1002/pro.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lakowicz JR, Ray K, Chowdhury M, Szmacinski H, Fu Y, Zhang J, Nowaczyk K. Plasmon-controlled fluorescence: a new paradigm in fluorescence spectroscopy. Analyst. 2008;133:1308–1346. doi: 10.1039/b802918k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gersten J, Kaasbjerg K, Nitzan A. Induced spin filtering in electron transmission through chiral molecular layers adsorbed on metals with strong spin-orbit coupling. J Chem Phys. 2013;139:114111. doi: 10.1063/1.4820907. [DOI] [PubMed] [Google Scholar]
- 163.Zhu Q, Zheng S, Lin S, Liu TR, Jin C. Polarization-dependent enhanced photoluminescence and polarization-independent emission rate of quantum dots on gold elliptical nanodisc arrays. Nanoscale. 2014;6:7237–7242. doi: 10.1039/c4nr01261e. [DOI] [PubMed] [Google Scholar]
- 164.Plucinski L, Ranjan Gartia M, Arnold WR, Ameen A, Chang TW, Hsiao A, Logan Liu G, Das A. Substrate binding to cytochrome P450–2J2 in Nanodiscs detected by nanoplasmonic Lycurgus cup arrays. Biosens Bioelectron. 2016;75:337–346. doi: 10.1016/j.bios.2015.07.041. [DOI] [PubMed] [Google Scholar]
- 165.Ithurria S, Tessier MD, Mahler B, Lobo RP, Dubertret B, Efros AL. Colloidal nanoplatelets with two-dimensional electronic structure. Nat Mater. 2011;10:936–941. doi: 10.1038/nmat3145. [DOI] [PubMed] [Google Scholar]
- 166.Ithurria S, Dubertret B. Quasi 2D colloidal CdSe platelets with thicknesses controlled at the atomic level. J Am Chem Soc. 2008;130:16504–16505. doi: 10.1021/ja807724e. [DOI] [PubMed] [Google Scholar]
- 167.Lim S, McDougle DR, Das A, Smith A. Lipoprotein Nanoplatelets: Fluorescent, Zwitterionic Probes for Molecular and Cellular Imaging. Journal of the American Chemical Society, Accepted with major revisions. 2015 doi: 10.1021/jacs.5b11225. [DOI] [PubMed] [Google Scholar]
- 168.Petrache AI, Machin DC, Williamson DJ, Webb ME, Beales PA. Sortase-mediated labelling of lipid nanodiscs for cellular tracing. Mol Biosyst. 2016 doi: 10.1039/c6mb00126b. [DOI] [PubMed] [Google Scholar]
- 169.Denisov IG, Sligar SG. Nanodiscs for structural and functional studies of membrane proteins. Nat Struct Mol Biol. 2016;23:481–486. doi: 10.1038/nsmb.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Puthenveetil R, Nguyen K, Vinogradova O. Nanodiscs and solution NMR: preparation, application and challenges. Nanotechnology Reviews. doi: 10.1515/ntrev-2016-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Malhotra K, Alder NN. Advances in the use of nanoscale bilayers to study membrane protein structure and function. Biotechnology and Genetic Engineering Reviews. 2014;30:79–93. doi: 10.1080/02648725.2014.921502. [DOI] [PubMed] [Google Scholar]
- 172.Bayburt TH, Leitz AJ, Xie G, Oprian DD, Sligar SG. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J Biol Chem. 2007;282:14875–14881. doi: 10.1074/jbc.M701433200. [DOI] [PubMed] [Google Scholar]
- 173.Inagaki S, Ghirlando R, White JF, Gvozdenovic-Jeremic J, Northup JK, Grisshammer R. Modulation of the Interaction between Neurotensin Receptor NTS1 and Gq Protein by Lipid. J Mol Biol. 2012;417:95–111. doi: 10.1016/j.jmb.2012.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kuszak AJ, Pitchiaya S, Anand JP, Mosberg HI, Walter NG, Sunahara RK. Purification and Functional Reconstitution of Monomeric mu-Opioid Receptors ALLOSTERIC MODULATION OF AGONIST BINDING BY G(i2) Journal of Biological Chemistry. 2009;284:26732–26741. doi: 10.1074/jbc.M109.026922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Leitz AJ, Bayburt TH, Barnakov AN, Springer BA, Sligar SG. Functional reconstitution of beta(2)-adrenergic receptors utilizing self-assembling Nanodisc technology. Biotechniques. 2006;40:601-+. doi: 10.2144/000112169. [DOI] [PubMed] [Google Scholar]
- 176.Gao TJ, Petrlova J, He W, Huser T, Kudlick W, Voss J, Coleman MA. Characterization of De Novo Synthesized GPCRs Supported in Nanolipoprotein Discs. Plos One. 2012;7 doi: 10.1371/journal.pone.0044911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yoshiura C, Kofuku Y, Ueda T, Mase Y, Yokogawa M, Osawa M, Terashima Y, Matsushima K, Shimada I. NMR Analyses of the Interaction between CCR5 and Its Ligand Using Functional Reconstitution of CCR5 in Lipid Bilayers. J Am Chem Soc. 2010;132:6768–6777. doi: 10.1021/ja100830f. [DOI] [PubMed] [Google Scholar]
- 178.Goldsmith BR, Mitala JJ, Josue J, Castro A, Lerner MB, Bayburt TH, Khamis SM, Jones RA, Brand JG, Sligar SG, Luetje CW, Gelperin A, Rhodes PA, Discher BM, Johnson ATC. Biomimetic Chemical Sensors Using Nanoelectronic Readout of Olfactory Receptor Proteins. Acs Nano. 2011;5:5408–5416. doi: 10.1021/nn200489j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bocquet N, Kohler J, Hug MN, Kusznir EA, Rufer AC, Dawson RJ, Hennig M, Ruf A, Huber W, Huber S. Real-time monitoring of binding events on a thermostabilized human A(2A) receptor embedded in a lipid bilayer by surface plasmon resonance. Bba-Biomembranes. 2015;1848:1224–1233. doi: 10.1016/j.bbamem.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 180.Johnson PJ, Halpin A, Morizumi T, Brown LS, Prokhorenko VI, Ernst OP, Miller RD. The photocycle and ultrafast vibrational dynamics of bacteriorhodopsin in lipid nanodiscs. Physical Chemistry Chemical Physics. 2014;16:21310–21320. doi: 10.1039/c4cp01826e. [DOI] [PubMed] [Google Scholar]
- 181.Ritchie TK, Kwon H, Atkins WM. Conformational analysis of human ATP-binding cassette transporter ABCB1 in lipid nanodiscs and inhibition by the antibodies MRK16 and UIC2. Journal of Biological Chemistry. 2011;286:39489–39496. doi: 10.1074/jbc.M111.284554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kraft TE, Hresko RC, Hruz PW. Expression, purification, and functional characterization of the insulin-responsive facilitative glucose transporter GLUT4. Protein Sci. 2015;24:2008–2019. doi: 10.1002/pro.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bao H, Duong F, Chan CS. A step-by-step method for the reconstitution of an ABC transporter into nanodisc lipid particles. Journal of visualized experiments: JoVE. 2012 doi: 10.3791/3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ly S, Bourguet F, Fischer NO, Lau EY, Coleman MA, Laurence TA. Quantifying interactions of a membrane protein embedded in a lipid nanodisc using fluorescence correlation spectroscopy. Biophysical journal. 2014;106:L05–L08. doi: 10.1016/j.bpj.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kawai T, Caaveiro JM, Abe R, Katagiri T, Tsumoto K. Catalytic activity of MsbA reconstituted in nanodisc particles is modulated by remote interactions with the bilayer. FEBS letters. 2011;585:3533–3537. doi: 10.1016/j.febslet.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 186.Roos C, Zocher M, Müller D, Münch D, Schneider T, Sahl HG, Scholz F, Wachtveitl J, Ma Y, Proverbio D. Characterization of co-translationally formed nanodisc complexes with small multidrug transporters, proteorhodopsin and with the E. coli MraY translocase. Biochimica et Biophysica Acta (BBA)-Biomembranes. 2012;1818:3098–3106. doi: 10.1016/j.bbamem.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 187.Mills A, Le H-T, Coulton JW, Duong F. FhuA interactions in a detergent-free nanodisc environment. Biochimica et Biophysica Acta (BBA)-Biomembranes. 2014;1838:364–371. doi: 10.1016/j.bbamem.2013.09.022. [DOI] [PubMed] [Google Scholar]
- 188.Ritchie T, Grinkova Y, Bayburt T, Denisov I, Zolnerciks J, Atkins W, Sligar S. Chapter eleven-reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods in enzymology. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Raschle T, Lin C, Jungmann R, Shih WM, Wagner G. Controlled co-reconstitution of multiple membrane proteins in lipid bilayer nanodiscs using DNA as a scaffold. ACS chemical biology. 2015;10:2448–2454. doi: 10.1021/acschembio.5b00627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Shenkarev ZO, Paramonov AS, Lyukmanova EN, Shingarova LN, Yakimov SA, Dubinnyi MA, Chupin VV, Kirpichnikov MP, Blommers MJ, Arseniev AS. NMR structural and dynamical investigation of the isolated voltage-sensing domain of the potassium channel KvAP: implications for voltage gating. J Am Chem Soc. 2010;132:5630–5637. doi: 10.1021/ja909752r. [DOI] [PubMed] [Google Scholar]
- 191.Shenkarev Z, Lyukmanova E, Solozhenkin O, Gagnidze I, Nekrasova O, Chupin V, Tagaev A, Yakimenko Z, Ovchinnikova T, Kirpichnikov M. Lipid-protein nanodiscs: possible application in high-resolution NMR investigations of membrane proteins and membrane-active peptides. Biochemistry (Moscow) 2009;74:756–765. doi: 10.1134/s0006297909070086. [DOI] [PubMed] [Google Scholar]
- 192.Xu H, Hill JJ, Michelsen K, Yamane H, Kurzeja RJ, Tam T, Isaacs RJ, Shen F, Tagari P. Characterization of the direct interaction between KcsA-Kv1. 3 and its inhibitors. Biochimica et Biophysica Acta (BBA)-Biomembranes. 2015;1848:1974–1980. doi: 10.1016/j.bbamem.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 193.Sheng JR, Grimme S, Bhattacharya P, Stowell MH, Artinger M, Prabahakar BS, Meriggioli MN. In vivo adsorption of autoantibodies in myasthenia gravis using Nanodisc-incorporated acetylcholine receptor. Experimental neurology. 2010;225:320–327. doi: 10.1016/j.expneurol.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Bayburt TH, Sligar SG. Single-molecule height measurements on microsomal cytochrome P450 in nanometer-scale phospholipid bilayer disks. Proc Natl Acad Sci U S A. 2002;99:6725–6730. doi: 10.1073/pnas.062565599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Civjan NR, Bayburt TH, Schuler MA, Sligar SG. Direct solubilization of heterologously expressed membrane proteins by incorporation into nanoscale lipid bilayers. Biotechniques. 2003;35:556–560. 562–553. doi: 10.2144/03353rr02. [DOI] [PubMed] [Google Scholar]
- 196.Duan H, Civjan NR, Sligar SG, Schuler MA. Co-incorporation of heterologously expressed Arabidopsis cytochrome P450 and P450 reductase into soluble nanoscale lipid bilayers. Arch Biochem Biophys. 2004;424:141–153. doi: 10.1016/j.abb.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 197.Das A, Grinkova YV, Sligar SG. Redox potential control by drug binding to cytochrome P450 3A4. J Am Chem Soc. 2007;129:13778–13779. doi: 10.1021/ja074864x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Grinkova YV, Denisov IG, Waterman MR, Arase M, Kagawa N, Sligar SG. The ferrous-oxy complex of human aromatase. Biochem Biophys Res Commun. 2008;372:379–382. doi: 10.1016/j.bbrc.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Duan H, Schuler MA. Heterologous expression and strategies for encapsulation of membrane-localized plant P450s. Phytochemistry Reviews. 2006;5:507–523. [Google Scholar]
- 200.Gregory MC, Denisov IG, Grinkova YV, Khatri Y, Sligar SG. Kinetic solvent isotope effect in human P450 CYP17A1-mediated androgen formation: evidence for a reactive peroxoanion intermediate. J Am Chem Soc. 2013;135:16245–16247. doi: 10.1021/ja4086403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Borch J, Torta F, Sligar SG, Roepstorff P. Nanodiscs for immobilization of lipid bilayers and membrane receptors: kinetic analysis of cholera toxin binding to a glycolipid receptor. Anal Chem. 2008;80:6245–6252. doi: 10.1021/ac8000644. [DOI] [PubMed] [Google Scholar]
- 202.Boldog T, Grimme S, Li M, Sligar SG, Hazelbauer GL. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc Natl Acad Sci U S A. 2006;103:11509–11514. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Gao Y, Cao E, Julius D, Cheng Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature. 2016 doi: 10.1038/nature17964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Bayburt TH, Carlson JW, Sligar SG. Reconstitution and imaging of a membrane protein in a nanometer-size phospholipid bilayer. J Struct Biol. 1998;123:37–44. doi: 10.1006/jsbi.1998.4007. [DOI] [PubMed] [Google Scholar]
- 205.Cruz F, Edmondson DE. Kinetic properties of recombinant MAO-A on incorporation into phospholipid nanodisks. J Neural Transm (Vienna) 2007;114:699–702. doi: 10.1007/s00702-007-0673-0. [DOI] [PubMed] [Google Scholar]
- 206.Shaw AW, Pureza VS, Sligar SG, Morrissey JH. The local phospholipid environment modulates the activation of blood clotting. J Biol Chem. 2007;282:6556–6563. doi: 10.1074/jbc.M607973200. [DOI] [PubMed] [Google Scholar]
- 207.Periasamy A, Shadiac N, Amalraj A, Garajová S, Nagarajan Y, Waters S, Mertens HD, Hrmova M. Cell-free protein synthesis of membrane (1, 3)-β-d-glucan (curdlan) synthase: Co-translational insertion in liposomes and reconstitution in nanodiscs. Biochimica et Biophysica Acta (BBA)-Biomembranes. 2013;1828:743–757. doi: 10.1016/j.bbamem.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 208.Shin J, Lou X, Kweon D-H, Shin Y-K. Multiple conformations of a single SNAREpin between two nanodisc membranes reveal diverse pre-fusion states. Biochemical Journal. 2014;459:95–102. doi: 10.1042/BJ20131668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hartley MD, Schneggenburger PE, Imperiali B. Lipid bilayer nanodisc platform for investigating polyprenol-dependent enzyme interactions and activities. P Natl Acad Sci USA. 2013;110:20863–20870. doi: 10.1073/pnas.1320852110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Glück JM, Koenig BW, Willbold D. Nanodiscs allow the use of integral membrane proteins as analytes in surface plasmon resonance studies. Analytical biochemistry. 2011;408:46–52. doi: 10.1016/j.ab.2010.08.028. [DOI] [PubMed] [Google Scholar]
- 211.Mi LZ, Grey MJ, Nishida N, Walz T, Lu C, Springer TA. Functional and structural stability of the epidermal growth factor receptor in detergent micelles and phospholipid nanodiscs. Biochemistry. 2008;47:10314–10323. doi: 10.1021/bi801006s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Nasvik Ojemyr L, von Ballmoos C, Gennis RB, Sligar SG, Brzezinski P. Reconstitution of respiratory oxidases in membrane nanodiscs for investigation of proton-coupled electron transfer. FEBS Lett. 2012;586:640–645. doi: 10.1016/j.febslet.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Orlando BJ, McDougle DR, Lucido MJ, Eng ET, Graham LA, Schneider C, Stokes DL, Das A, Malkowski MG. Cyclooxygenase-2 catalysis and inhibition in lipid bilayer nanodiscs. Arch Biochem Biophys. 2014;546:33–40. doi: 10.1016/j.abb.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Mazhab-Jafari MT, Marshall CB, Smith MJ, Gasmi-Seabrook GM, Stathopulos PB, Inagaki F, Kay LE, Neel BG, Ikura M. Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site. Proc Natl Acad Sci U S A. 2015;112:6625–6630. doi: 10.1073/pnas.1419895112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wan C, Wu B, Song Z, Zhang J, Chu H, Wang A, Liu Q, Shi Y, Li G, Wang J. Insights into the molecular recognition of the granuphilin C2A domain with PI (4, 5) P2. Chemistry and physics of lipids. 2015;186:61–67. doi: 10.1016/j.chemphyslip.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 216.Ishida H, Garcia-Herrero A, Vogel HJ. The periplasmic domain of Escherichia coli outer membrane protein A can undergo a localized temperature dependent structural transition. Biochimica et Biophysica Acta (BBA)-Biomembranes. 2014;1838:3014–3024. doi: 10.1016/j.bbamem.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 217.Bibow S, Carneiro MG, Sabo TM, Schwiegk C, Becker S, Riek R, Lee D. Measuring membrane protein bond orientations in nanodiscs via residual dipolar couplings. Protein Sci. 2014;23:851–856. doi: 10.1002/pro.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Nasr ML, Shi X, Bowman AL, Johnson M, Zvonok N, Janero DR, Vemuri VK, Wales TE, Engen JR, Makriyannis A. Membrane phospholipid bilayer as a determinant of monoacylglycerol lipase kinetic profile and conformational repertoire. Protein Science. 2013;22:774–787. doi: 10.1002/pro.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Mazhab-Jafari MT, Marshall CB, Stathopulos PB, Kobashigawa Y, Stambolic V, Kay LE, Inagaki F, Ikura M. Membrane-dependent modulation of the mTOR activator Rheb: NMR observations of a GTPase tethered to a lipid-bilayer nanodisc. J Am Chem Soc. 2013;135:3367–3370. doi: 10.1021/ja312508w. [DOI] [PubMed] [Google Scholar]
- 220.Pandit A, Shirzad-Wasei N, Wlodarczyk LM, van Roon H, Boekema EJ, Dekker JP, de Grip WJ. Assembly of the major light-harvesting complex II in lipid nanodiscs. Biophys J. 2011;101:2507–2515. doi: 10.1016/j.bpj.2011.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Yan R, Mo X, Paredes AM, Dai K, Lanza F, Cruz MA, Li R. Reconstitution of the platelet glycoprotein Ib-IX complex in phospholipid bilayer Nanodiscs. Biochemistry. 2011;50:10598–10606. doi: 10.1021/bi201351d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Bhattacharya P, Grimme S, Ganesh B, Gopisetty A, Sheng JR, Martinez O, Jayarama S, Artinger M, Meriggioli M, Prabhakar BS. Nanodisc-incorporated hemagglutinin provides protective immunity against influenza virus infection. J Virol. 2010;84:361–371. doi: 10.1128/JVI.01355-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wang B, Zhao A, Xie Q, Olinares PD, Chait BT, Novick RP, Muir TW. Functional Plasticity of the AgrC Receptor Histidine Kinase Required for Staphylococcal Virulence. Cell Chemical Biology. 2017 doi: 10.1016/j.chembiol.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Waberer L, Henrich E, Peetz O, Morgner N, Dötsch V, Bernhard F, Volknandt W. The synaptic vesicle protein SV31 assembles into a dimer and transports Zn2+ Journal of Neurochemistry. 2017;140:280–293. doi: 10.1111/jnc.13886. [DOI] [PubMed] [Google Scholar]