Abstract
Background and aims
A 5-bp insertion-deletion (indel) polymorphism in the promoter of interferon regulatory factor 5 (IRF5) has been associated with inflammatory bowel diseases (IBD). This polymorphism generates an additional binding site for the transcription factor SP1 and has been shown to augment the expression of IRF5. Additionally, it affects a CpG dinucleotide-dense genomic region. These features of the indel suggested that it may influence the epigenetic regulation of IRF5. The aim of this study was to investigate the potential effect of the 5-bp indel on the methylation pattern of four CpG sites upstream of the polymorphism. Possible CpG site methylation differences in this region between healthy persons and individuals suffering from IBD were also tested.
Methods
Genotype was determined by 4% polyacrylamide gel electrophoresis in 33 peripheral blood leukocyte (PBL) DNA samples. DNA methylation correlates of the genotypes were measured by bisulfite pyrosequencing. IRF5 promoter methylation in association to disease state was assessed in 87 proband (49 healthy, 18 Crohn's disease, 20 ulcerative colitis) PBL samples.
Results
The polymorphism did not affect the methylation pattern of the IRF5 promoter nor could we detect significant differences in the average, low methylation of the locus between healthy persons and individuals with IBD.
Conclusions
These results implicate that epigenetic dysregulation of the IRF5 promoter is unlikely to be associated with IBD.
Keywords: IRF5, DNA methylation, Inflammatory bowel diseases, Polymorphism
Introduction
Epigenetics is the study of biological processes that can associate with mitotically heritable alterations in gene expression without a change in DNA sequence. One of the essential mechanisms of mammalian epigenetic regulation is the methylation of cytosines in cytosine-guanine dinucleotides (CpG), designated as DNA methylation. DNA methylation frequently correlates with transcriptional silencing of associated genes [1]. Importantly, correlation between DNA methylation changes and disease prevalence has been observed in a number of cases suggesting an important role for this epigenetic process in the etiology of certain human disorders [2]. Additionally, association between single nucleotide polymorphisms and allele-specific gene methylation has been detected through genome-wide assessments [3]. Not surprisingly, more and more data have emerged on correlation between genetic and epigenetic disturbances in relationship to human disorders [4–6].
Crohn's disease (CD) and ulcerative colitis (UC), frequently referred to as inflammatory bowel diseases (IBD), are immunologically mediated chronic disorders. The prevalence of IBD increased rapidly in Europe and North America in the second half of the twentieth century, and it is becoming more common in the rest of the world, especially in countries adopting a Western lifestyle [7, 8]. Such epidemiologic observations indicate that there are strong environmental influences contributing to IBD and suggest a possible influence of these on epigenetic regulatory processes. Additionally, the discordance rate in monozygotic twins for a specific disorder is thought to correlate with the potential for epigenetic influence on pathogenesis [9]. Importantly, this discordance rate is 50–80% for CD and more than 80% in UC [10]. Therefore, inflammatory bowel diseases have been recognized as disorders where epigenetic dysregulation may play a significant role [11]. Recent reports on genes with altered methylation pattern associated with IBD [12, 13] further the potential etiologic role of epigenetic dysregulation in these illnesses.
One of the genes linked to IBD among other autoimmune diseases is interferon regulatory factor 5 (IRF5) [14]. The presence of a 5-bp insertion (CGGGG) in the promoter region of the gene has been shown to confer the risk of IBD, by creating an additional binding site for transcription factor SP1 [14], and to associate with increased expression of the gene [15]. Correlation between SP1 binding and DNA methylation has been observed at other genomic regions [16]. Additionally, the particular CGGGG insertion-deletion (indel) polymorphism affects a CpG-rich area of IRF5, suggesting the possible influence of the insertion on the DNA methylation of this locus.The present study was designed to investigate the possible effects of the 5-bp indel on the methylation pattern of the locus and to examine whether there are significant differences in the methylation of this region in association with IBD.
Materials and methods
Samples
De-identified peripheral blood leukocyte (PBL) DNA samples were obtained from gene banks of the University of Pecs (Pecs, Hungary) [17] and of Yale University (New Haven, Connecticut) that were established in agreement with local and federal regulations. A total of 87 proband PBL samples from white Caucasians were analyzed. Forty-nine were healthy (28 female, 21 male), 18 with CD (nine female, nine male; four (1 female, 3 male) with history of surgery for fistulizing/stricturing disease) and 20 with UC (ten female, ten male; where two females with history of colectomy). The average age was 36–37 years in all groups without significant differences.
Bisulfite pyrosequencing
DNA was bisulfite converted according to Waterland et al. [18]. The IRF5 gene was amplified with both a universal biotinylated primer approach [19] and by traditional primer biotinylation (forward primer: GTYGTTTGGTATTTTT TTGGAGGTTTT; reverse primer: GGGACACCGCTGAT CGTTT ACCRCCCCTAAACAACTACTACTAAA— universal biotinylated primer underlined; sequencing primer: GGGGTTYGGAGTGGATT). Methylation results did not differ between the two methods (more than ten samples tested with both approaches, data not shown). A quantitative bisulfite pyrosequencing protocol was used for all methylation analyses [20] with the utilization of the Pyro Q CpG program (QIAGEN GmbH, QIAGEN Strasse 1, 40724 Hilden, Germany). The mean methylation was calculated from four CpG sites (−80, −78, −73, −68 bp from the transcription start site) upstream from the polymorphism to represent the average methylation degree of the locus. This approach is acceptable at CpG-dense regions [21]. Eighteen samples from the Hungarian cohort and 15 samples from Yale University were amplified with a Cy5-labeled primer (forward primer: CGGGGCCCGGAGTGGATTC; reverse primer: Cy5-GGGCACTTCCGCGT CTTG) and evaluated with 4% polyacrylamide gel electrophoresis to determine the genotype in relationship to the 5-bp indel in the promoter region. Standard deviation of repeat bisulfite pyrosequencing measurements of methylation at a single CpG site usually varies between 1% and 5% [22]. Therefore, methylation differences of less than 5% in group comparisons can be generally considered as nonsignificant.
CpG density calculation
CpG density in a 1,000-bp region surrounding the indel site was calculated with the CpG island searcher program (http://cpgislands.usc.edu/).
Statistical analysis
Nonpaired t test was applied for all statistical analyses in the study. Statistical significance calculations were performed by taking into account the 5% reliability limit of the pyrosequencing measurements. Error bars represent standard error of the mean. Data were plotted, and calculations for confidence intervals were done with the utilization of the GraphPad Prism software (GraphPad Software Inc., San Diego, CA, USA).
Results
The methylation of four CpG dinucleotides immediately upstream of the CGGGG indel site was examined at the IRF5 promoter in parallel with 4% polyacrylamide gel electrophoresis analysis of the insertion. The insertion introduces an additional CGGGG pentanucleotide (4×) following the already existing three repeats (3×) of the same pentanucleotide, upstream of the first IRF5 noncoding exon. Therefore, an allele without the insertion is designated as 3× (having three repeats of the CGGGG pentanucleotide, alternatively named as a deletion allele), while an allele with the insertion is designated as 4× (having four repeats of the CGGGG pentanucleotide, alternatively named as an insertion allele). 3×/3× (or homozygous deletion), hence, indicates a genotype where neither the maternal nor the paternal allele carries the CGGGG insertion at the IRF5 promoter, for example.
We first determined the genotype in a group of DNA samples with 4% polyacrylamide gel electrophoresis and found 19 with 3×/3×, six with 3×/4×, and eight with 4×/4×, respectively. Then, bisulfite pyrosequencing measurements were performed in these samples to assess genotype-DNA methylation correlation (Fig. 1a). Thereafter, the DNA methylation measurements were expanded to all of the healthy and IBD probands to determine the possible association between DNA methylation at the IRF5 promoter region and CD, or UC, respectively (Fig. 1b).
Fig. 1.
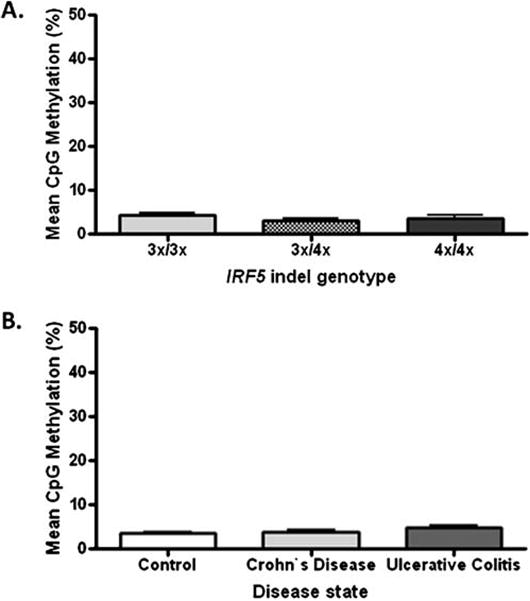
Comparison of mean CpG methylation adjacent to the IRF5 indel. a Mean methylation in relationship to genotypes. Average methylation was uniformly low (n=6–19). Confidence intervals (95%) for comparing 3×/4× (mean 3.1%) and 4×/4× (mean 3.57%) with 3×/3× (mean 4.2%) are −1.515 to 3.773 and −1.725 to 3.032, respectively. b Mean methylation in relationship to inflammatory bowel diseases (n = 18–49). Confidence intervals (95%) are −0.85% to 1.47% and −0.37% to 2.58%, respectively for comparison of CD (mean 3.74%) and UC (mean 4.9%) with healthy (mean 3.44%) groups. 3×/3× homozygous deletion, 3×/4× heterozygous insertion, 4×/4× homozygous insertion
Since the standard deviation of repeat bisulfite pyrosequencing measurements of methylation at a single CpG site usually varies between 1% and 5% [22], methylation differences of less than 5% in group comparisons can be generally considered as nonsignificant. The 95% confidence interval for a mean difference, based on variation in the measurements and sample size, estimates the range within which we can be 95% confident the true mean difference lies. Confidence intervals (95%) for comparing DNA methylation of 3×/4× and 4×/4× with 3×/3× were −1.515 to 3.773 and −1.725 to 3.032, respectively, both essentially ruling out the possibility of a difference of more than 5% (Fig. 1a). Confidence intervals (95%) of −0.85% to 1.47% and −0.37% to 2.58%, respectively, were detected for comparison of CD and UC with the healthy group (Fig. 1b). Both of these intervals provide assurance that the true mean difference is less than 5%. Further, given the variation and sample size for each of these comparisons, the power for detecting a group mean difference of 5% is greater than 90% for each. Additionally, correlation between age and the methylation level of the IRF5 promoter was not detected (n=87, r=0.14, p=0.2; data not shown).
The area of interest is a CpG-dense genomic region meeting the criteria for a dense CpG island. The mean methylation of a few CpG sites from such regions usually correlates well with regional methylation [21]. Therefore, our results indicate that the methylation pattern of the IRF5 promoter is not affected by the presence of the 5-bp insertion-deletion and does not correlate with IBD, at least in the cohort of this study.
Discussion
There is more and more evidence for interactions between genetic and epigenetic regulation in the human genome that can associate with disease. Such interactions in regards to SP1 binding sites have been specifically described [23]. Inflammatory bowel diseases have been recognized as disorders where epigenetic dysregulation may play an important etiologic role [11]. Yet, only a few definite studies have addressed this possibility thus far [12, 13]. Importantly, the 5-bp indel of the IRF5 promoter introduces an additional SP1 binding site to the region and has been associated with IBD [14]. Therefore, it appeared as an ideal candidate where epigenetic consequences of the polymorphism could arise and modulate IBD. However, in the Caucasian cohort of this study, we could not detect any significant differences in regional CpG methylation neither in relationship to the indel nor in respect of the presence of CD or UC compared to healthy individuals. Based on these findings, it is unlikely that epigenetic dysregulation of IRF5 may be a significant factor in the etiology of IBD in Caucasians. This study highlights the need for large scale epigenomic studies to identify epigenetic correlates of inflammatory bowel diseases because candidate gene approaches are less likely to yield positive results.
Acknowledgments
R.K. was supported by funding from the Broad Medical Research Program, the Broad Foundation (IBD-0252), and a young investigator joint award from the Crohn's and Colitis Foundation of America—Children's Digestive Health and Nutrition Foundation/North American Society of Pediatric Gastroenterology Hepatology and Nutrition (CCFA Ref #2426).
Contributor Information
Alfred Balasa, Section of Pediatric Gastroenterology, Baylor College of Medicine, Houston, TX, USA.
Grace Gathungu, Section of Digestive Disease, Department of Internal Medicine, Yale University, New Haven, CT, USA.
Peter Kisfali, Department of Medical Genetics, University of Pecs, Pecs, Hungary.
E O'Brian Smith, USDA Children's Nutrition Research Center, Houston, TX, USA.
Judy H. Cho, Section of Digestive Disease, Department of Internal Medicine, Yale University, New Haven, CT, USA
Bela Melegh, Department of Medical Genetics, University of Pecs, Pecs, Hungary.
Richard Kellermayer, Section of Pediatric Gastroenterology, Baylor College of Medicine, Houston, TX, USA; Section of Pediatric Gastroenterology, Hepatology & Nutrition, Baylor College of Medicine, 6621 Fannin St., CC1010.00, Houston, TX 77030-2399, USA.
References
- 1.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 2.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 3.Kerkel K, Spadola A, Yuan E, et al. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet. 2008;40:904–908. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- 4.Ligtenberg MJ, Kuiper RP, Chan TL, et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat Genet. 2009;41:112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- 5.Moser D, Ekawardhani S, Kumsta R, et al. Functional analysis of a potassium-chloride co-transporter 3 (SLC12A6) promoter polymorphism leading to an additional DNA methylation site. Neuropsychopharmacology. 2009;34:458–467. doi: 10.1038/npp.2008.77. [DOI] [PubMed] [Google Scholar]
- 6.Wang H, Ogawa M, Wood JR, et al. Genetic and epigenetic mechanisms combine to control MMP1 expression and its association with preterm premature rupture of membranes. Hum Mol Genet. 2008;17:1087–1096. doi: 10.1093/hmg/ddm381. [DOI] [PubMed] [Google Scholar]
- 7.Loftus EV., Jr Clinical epidemiology of inflammatory bowel disease: incidence, prevalence, and environmental influences. Gastroenterology. 2004;126:1504–1517. doi: 10.1053/j.gastro.2004.01.063. [DOI] [PubMed] [Google Scholar]
- 8.Sathiyasekaran M, Shivbalan S. Crohn's disease. Indian J Pediatr. 2006;73:723–729. doi: 10.1007/BF02898453. [DOI] [PubMed] [Google Scholar]
- 9.Petronis A. Epigenetics and twins: three variations on the theme. Trends Genet. 2006;22:347–350. doi: 10.1016/j.tig.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 10.Biank V, Broeckel U, Kugathasan S. Pediatric inflammatory bowel disease: clinical and molecular genetics. Inflamm Bowel Dis. 2007;13:1430–1438. doi: 10.1002/ibd.20213. [DOI] [PubMed] [Google Scholar]
- 11.Petronis A, Petroniene R. Epigenetics of inflammatory bowel disease. Gut. 2000;47:302–306. doi: 10.1136/gut.47.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tahara T, Shibata T, Nakamura M, et al. Effect of MDR1 gene promoter methylation in patients with ulcerative colitis. Int J Mol Med. 2009;23:521–527. doi: 10.3892/ijmm_00000160. [DOI] [PubMed] [Google Scholar]
- 13.Tahara T, Shibata T, Nakamura M, et al. Promoter methylation of protease-activated receptor (PAR2) is associated with severe clinical phenotypes of ulcerative colitis (UC) Clin Exp Med. 2009;9:125–130. doi: 10.1007/s10238-008-0025-x. [DOI] [PubMed] [Google Scholar]
- 14.Dideberg V, Kristjansdottir G, Milani L, et al. An insertion-deletion polymorphism in the interferon regulatory Factor 5 (IRF5) gene confers risk of inflammatory bowel diseases. Hum Mol Genet. 2007;16:3008–3016. doi: 10.1093/hmg/ddm259. [DOI] [PubMed] [Google Scholar]
- 15.Sigurdsson S, Goring HH, Kristjansdottir G, et al. Comprehensive evaluation of the genetic variants of interferon regulatory factor 5 (IRF5) reveals a novel 5 bp length polymorphism as strong risk factor for systemic lupus erythematosus. Hum Mol Genet. 2008;17:872–881. doi: 10.1093/hmg/ddm359. [DOI] [PubMed] [Google Scholar]
- 16.Brandeis M, Frank D, Keshet I, et al. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 17.Magyari L, Farago B, Bene J, et al. No association of the cytotoxic T-lymphocyte associated gene CTLA4 +49A/G polymorphisms with Crohn's disease and ulcerative colitis in Hungarian population samples. World J Gastroenterol. 2007;13:2205–2208. doi: 10.3748/wjg.v13.i15.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen L, Guo Y, Chen X, Ahmed S, Issa JP. Optimizing annealing temperature overcomes bias in bisulfite PCR methylation analysis. Biotechniques. 2007;42:48–52. doi: 10.2144/000112312. [DOI] [PubMed] [Google Scholar]
- 20.Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- 21.Bock C, Walter J, Paulsen M, Lengauer T. Inter-individual variation of DNA methylation and its implications for large-scale epigenome mapping. Nucleic Acids Res. 2008;36:e55. doi: 10.1093/nar/gkn122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mikeska T, Bock C, El-Maarri O, et al. Optimization of quantitative MGMT promoter methylation analysis using pyrosequencing and combined bisulfite restriction analysis. J Mol Diagnostics. 2007;9:368–381. doi: 10.2353/jmoldx.2007.060167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gazzoli I, Kolodner RD. Regulation of the human MSH6 gene by the Sp1 transcription factor and alteration of promoter activity and expression by polymorphisms. Mol Cell Biol. 2003;23:7992–8007. doi: 10.1128/MCB.23.22.7992-8007.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]


