Abstract
Background
The airway microbiome is a subject of great interest for the study of respiratory disease. Anterior nare samples are more accessible than samples from deeper within the nasopharynx. However, the correlation between the microbiota found in the anterior nares and the microbiota found within the nasopharynx is unknown. We assessed the anterior nares and nasopharyngeal microbiota to determine (1) the relation of the microbiota from these two upper airway sites and (2) if associations were maintained between the microbiota from these two sites and two bronchiolitis severity outcomes.
Results
Among 815 infants hospitalized at 17 US centers for bronchiolitis with optimal 16S rRNA gene sequence reads from both nasal swab and nasopharyngeal aspirate samples, there were strong intra-individual correlations in the microbial communities between the two sample types, especially relating to Haemophilus and Moraxella genera. By contrast, we found a high abundance of Staphylococcus genus in the nasal swabs—a pattern not found in the nasopharyngeal samples and not informative when predicting the dominant nasopharyngeal genera. While these disparities may have been due to sample processing differences (i.e., nasal swabs were mailed at ambient temperature to emulate processing of future parent collected swabs while nasopharyngeal aspirates were mailed on dry ice), a previously reported association between Haemophilus-dominant nasopharyngeal microbiota and the increased severity of bronchiolitis was replicated utilizing the nasal swab microbiota and the same outcome measures: intensive care use (adjusted OR 6.43; 95% CI 2.25–20.51; P < 0.001) and hospital length-of-stay (adjusted OR 4.31; 95% CI, 1.73–11.11; P = 0.002). Additionally, Moraxella-dominant nasopharyngeal microbiota was previously identified as protective against intensive care use, a result that was replicated when analyzing the nasal swab microbiota (adjusted OR 0.30; 95% CI, 0.11–0.64; P = 0.01).
Conclusions
While the microbiota of the anterior nares and the nasopharynx are distinct, there is considerable overlap between the bacterial community compositions from these two anatomic sites. Despite processing differences between the samples, these results indicate that microbiota severity associations from the nasopharynx are recapitulated in the anterior nares, suggesting that nasal swab samples not only are effective sample types, but also can be used to detect microbial risk markers.
Electronic supplementary material
The online version of this article (10.1186/s40168-017-0385-0) contains supplementary material, which is available to authorized users.
Keywords: Microbiota, Bronchiolitis, Asthma, Nasopharynx, Anterior nares
Background
The composition and function of the airway microbiota have been associated with respiratory conditions, such as pneumonia [1, 2], asthma [3–9], and chronic obstructive pulmonary disease [4, 10–12]. However, the airway is vast and extends from the nasal openings to the alveoli deep within the lungs. Ideally, samples for microbial analysis would be collected as close as possible to the cellular inflammatory responses contributing to the respiratory illness in question, which occur in the lower respiratory tract for bronchiolitis.
Unfortunately, the deeper segments of the airway (the lower respiratory tract) must be assessed through more invasive methods, such as bronchoscopy, which is not feasible for large-scale studies. Furthermore, easy sample collection is especially important for studies involving infants and young children. As a result, multiple studies in this young population have been conducted on the microbiome using nasal swabs (NSs) or nasal brush specimens, which are easier to collect and less invasive than samples from deeper within the airway [3, 6, 7, 13, 14].
Questions remain about the correlation between the microbiota within different microenvironments along the airway. Although prior studies have demonstrated strong correlations between upper and lower airway microbiology [15, 16] and virology [17], other data have suggested significant spatial variation in the composition and structure of the upper and lower airway microbiota [18, 19]. Additionally, there are emerging data about the correlation between the microenvironments in the upper airway [7, 20]. Yan et al. found in 12 healthy adults that the microbiota of the anterior nares was significantly different from that of the middle meatus and sphenoethmoidal recess [20]. Pérez-Losada et al. compared the microbiota of the inferior turbinate and nasopharynx among 30 children and adolescents with asthma [7] and found that these two regions had distinct microbial compositions. However, both of these studies were single-center studies with less than 40 participants. Moreover, while previous studies have examined compositional differences between microenvironments, no previous study has assessed if there are differences in the associations between the microbiota from different regions of the upper airway and a clinical outcome, particularly among young children and infants for whom less invasive sampling methods are imperative.
As part of the 35th Multicenter Airway Research Collaboration (MARC-35) study, site teams collected both NS and nasopharyngeal aspirate (NPA) samples from children hospitalized for bronchiolitis within 24 h of hospitalization [21]. Our two objectives in this secondary analysis of MARC-35 data were to (1) compare the microbial composition between the anterior nares and nasopharynx of infants hospitalized with bronchiolitis and (2) assess if associations between NPA microbial composition and bronchiolitis severity [21] would be replicated or enhanced using NS microbial data. We hypothesized that although systematic co-analysis of NS and NPA samples from MARC-35 participants would have shared and divergent microbial compositions, they would both be associated with severity of illness.
Methods
Study design
The 35th Multicenter Airway Research Collaboration (MARC-35) is a multicenter prospective cohort study of infants (age < 1 year) hospitalized for bronchiolitis. The study was coordinated by the Emergency Medicine Network (EMNet) [22]. MARC-35 was conducted at 17 sites across the USA during three consecutive bronchiolitis seasons (November 1 to April 30) from 2011 to 2014.
Participants consisted of infants diagnosed with bronchiolitis (as defined by the American Academy of Pediatrics) by the attending physician [23]. The exclusion criteria included previous enrollment in the study, consent to the study more than 24 h after hospitalization, transfer to a participating hospital more than 24 h after hospitalization, and known heart-lung disease, immunodeficiency, immune suppression, or gestational age less than 32 weeks. The institutional review board at each of the 17 participating hospitals approved the study, and patients were treated at the discretion of the attending physician.
Data collection
Structured interviews of the parent or guardian were performed by site investigators to determine the demographic characteristics, medical and family history, and details of the acute illness for each patient. Additional clinical details were collected via emergency department and hospital inpatient chart reviews. Reviewers at the EMNet Coordinating Center reviewed all data and inquired about discrepancies and missing data with site investigators.
Trained site investigators collected NS and NPA samples using standardized protocols [24, 25]. The site investigators collected NS samples from the anterior nares within 24 h of hospitalization. Both nares were swabbed with a single nylon, pediatric FLOQSwab (Copan, Brescia, Italy). The NS samples were placed into a vial containing 2 mL of transport media (15% glycerol in Iscove’s media) and mailed to Massachusetts General Hospital (MGH) (Boston, MA) via US mail, where they were stored at − 80°C upon receipt. NS samples were then shipped on dry ice from MGH to Baylor College of Medicine (Houston, TX) via overnight mail, where they were again stored at − 80°C. The procedure used to collect the NS emulated future parent collected nasal swabs and shipping to ensure maximum comparability of all the nasal swab specimens collected in MARC-35.
For the NPA samples, all site teams used the same collection equipment (Medline Industries, Mundelein, IL) and also collected the samples within 24 h of hospitalization. For the collection, the child was placed supine; 1 mL of normal saline was instilled into one nare, and then an 8 French suction catheter was used to remove the mucus. This procedure was performed once on each nostril. After the sample collection from both nares, 2 mL of normal saline was suctioned through the catheter to clear the tubing. Immediately after collection, the NPA sample was added to the same transport medium as the NS in a 1:1 ratio and placed on ice. Within 1 h of collection, the NPA sample was refrigerated at 4 °C. Within 24 h of collection, the sample was transferred to a − 80 °C freezer until shipped on dry ice to Baylor College of Medicine (Houston, TX), where they were again stored at − 80°C.
Microbiota community profiling
The composition of NS and NPA microbiota was characterized at the Alkek Center for Metagenomics and Microbiome Research (CMMR) at Baylor College of Medicine by sequencing the bacterial 16S rRNA gene V4 region on the Illumina MiSeq platform as described in the initial analysis of MARC-35 data. The NPA samples were additionally tested for 17 viral pathogens (e.g., rhinovirus, respiratory syncytial virus) using real-time polymerase chain reaction (PCR) assays [21]. Briefly, bacterial genomic DNA was extracted using MO BIO PowerSoil DNA Isolation Kit (MO BIO Laboratories). The 16S rDNA V4 region was amplified by PCR and sequenced in the MiSeq platform (Illumina) using the 2 × 250 bp paired-end protocol yielding pair-end reads that overlap almost completely. The primers used for amplification contain adapters for MiSeq sequencing and single-end barcodes allowing pooling and direct sequencing of PCR products [26]. Sequencing read pairs were demultiplexed based on the unique molecular barcodes, and reads were merged using USEARCH v7.0.1090 [27] allowing zero mismatches and a minimum overlap of 50 bases. Merged reads were trimmed at the first base with a Q5 quality score. We calculated the expected error after taking into account all Q-scores across all the bases of a read and the probability of an error occurring [28]. Additionally, a quality filter was applied to the resulting merged reads, and reads containing > 0.05 expected errors was discarded. Rarefaction curves of bacterial operational taxonomic units (OTUs) were constructed using sequence data for each sample to ensure coverage of the bacterial diversity present. Samples with suboptimal amounts of sequencing reads were re-sequenced to ensure that the majority of bacterial taxa were encompassed in our analyses. Positive and negative controls were included in the extraction, amplification, and sequencing processes together with the study samples for quality control and assurance purposes. The positive control consisted of a known and previously sequenced bacterial genome that is not expected to be found in the study samples. Negative controls were non-template controls composed of the reagents used in every process from sample extraction to sequencing. There was amplification on positive controls and no amplification on negative controls.
16S rRNA gene sequences were clustered into OTUs at a similarity cutoff value of 97% using the UPARSE algorithm [29]. OTUs were determined by mapping the centroids to the SILVA database [30] containing only the 16S V4 region to determine taxonomies. A custom script constructed a rarefied OTU table from the output files generated in the previous two steps for downstream analyses of alpha-diversity (e.g., Shannon index) and beta-diversity (e.g., weighted UniFrac distance matrix) [31, 32]. Shannon diversity index is a quantitative measure that takes into account not only richness but also the proportion of each bacteria (evenness) within the local community. The weighted UniFrac algorithm calculates the distance between microbial communities based on the phylogenetic relatedness of lineages and relative abundance in each sample.
Statistical analyses
The relative abundance of each OTU was calculated for each sample, and the OTUs were combined at the genus level. For each sample type, the overall abundance of each genus was calculated by taking the sum of relative abundances by genus over all subjects. The union of the 10 most abundant genera for each site defined the overall 15 top genera for comparative analyses.
To determine whether there was a significant intra-individual correlation between the two upper airway sites, a permutation test was performed on the mean Spearman correlation for the NS and NPA sample pairs. Computing the Spearman correlation matrix of the top genera abundance for NS versus NPA assessed the intra-individual correlations by genera. Due to the compositional nature of microbiota data, Spearman correlation calculations can overestimate relationships in the data [33]. To determine the significance of the co-occurrence and mutual exclusion relationships between NS and NPA, a correlation network between the two sites was constructed using the approach of Faust et al. [34]. To construct the network, all genera that appeared in at least 10% of the samples were analyzed using four different metrics: Spearman correlation, Pearson correlation, Bray-Curtis dissimilarity, and Kullback-Leibler divergence. The network includes relationships that are significant (P < 0.05) for at least two of the above methods.
For each infant, we adjoined the genus abundance measurements for the top 15 genera from the NS and NPA samples and created a composite microbiota profile. Based on this composite data, we determined common patterns of genus abundance within individuals when comparing NS and NPA sites. We then used the composite data and Bray-Curtis dissimilarity metric to cluster subjects by partitioning around medoids (PAM). The optimal number of clusters was determined using the gap statistic and average silhouette width [35, 36].
Replication analysis
To replicate the findings comparing NPA microbiota to bronchiolitis severity outcomes using the NS microbiota data [21], the NS samples were clustered separately using the weighted UniFrac distance and PAM clustering. The optimal number of clusters was determined by the average silhouette width. We then determined associations between the NS microbiota profiles and clinical covariates using chi-squared and Kruskal-Wallis tests as appropriate. Parallel to our previous analysis, a fixed-effects logistic regression model and a mixed-effects logistic regression model adjusting for 11 clinical variables were constructed using the NS clusters for each of the severity outcomes [21] (i.e., intensive care use [i.e., admission to intensive care unit and/or use of continuous positive airway pressure and/or intubation during inpatient stay, regardless of location] and hospital length of stay). For the intensive care use outcome, these models were then repeated on isolated microbiota profiles using membership in the profile of interest (e.g., Haemophilus-dominant profile versus all other subjects) as the independent variable.
Results
Out of 921 subjects in the MARC-35 longitudinal cohort, site teams collected one NS and one NPA from 920 of these infants. We obtained optimal reads for 819 NS samples and 914 NPA samples. There were 815 subjects with optimal reads of both NS and NPA samples, and this group comprised the analytic cohort. Within this cohort, the median age at hospitalization was 3 months (IQR 2–6 months), the median weight at hospitalization was 6 kg (IQR 5–8 kg), and 31% of the infants had used antibiotics prior to hospitalization.
We found 15 unique genera for comparative analysis after examining the top 10 abundant genera from the NS and NPA (Fig. 1a). The NS microbiota was dominated by the Staphylococcus genus (40.8%). The next most abundant genera were Corynebacterium (10.4%), Moraxella (9.3%), Haemophilus (7.4%), Dolosigranulum (5.2%), Streptococcus (5.0%), and Enterobacter (4.7%), which together with Staphylococcus accounted for 80% of the NS microbiota. The NPA microbiota was dominated by Moraxella (30.7%), Streptococcus (30.5%), and Haemophilus (19.7%) genera, which comprised over 80% of the microbiota. The abundances of the dominant genera in both sample types show high variability (Table 1). Additionally, the NS microbiota showed less bacterial richness and lower Shannon diversity index scores than the NPA microbiota, corresponding to the predominance of Staphylococcus in the NS samples. However, there were several NS (6.3%) and NPA (0.5%) samples that were completely dominated by one genus, indicating that this low diversity state may be a feature of a small percentage of infant upper airway microbiota at the time of hospitalization.
Fig. 1.
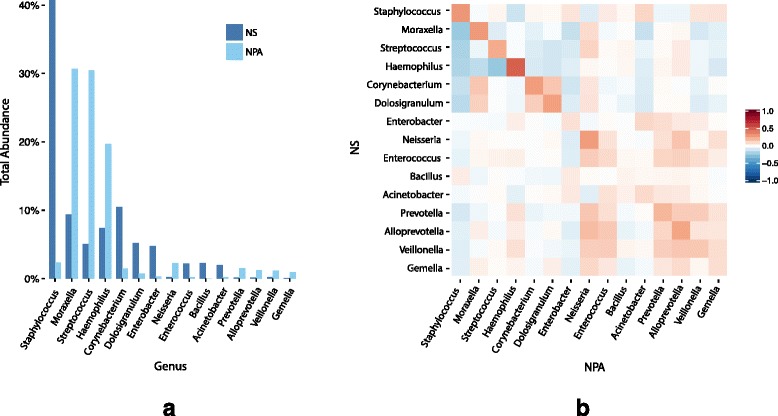
Comparison of nasal swab and nasopharyngeal microbiota. The genera abundances for the nasal swab and nasopharyngeal samples were calculated by taking the sum of all samples over each genus for each sample type. Combining the top 10 genera for each sample type gave 15 unique overall top genera. a The abundances of each of the top genera were calculated for both of the sample types. b For the top 15 genera, the Spearman correlations between the two anatomic sites are shown. The correlation heat map exhibits asymmetric behavior because it is comparing between the two sample types
Table 1.
Richness, alpha-diversity, and abundance by microbiota sample type
| Nasal swab | Nasopharyngeal aspirate | |||
|---|---|---|---|---|
| Richness, median (IQR) | ||||
| Number of genera | 6 (3–15) | 12 (6.5–20) | ||
| Alpha-diversity, median (IQR) | ||||
| Shannon index | 0.58 (0.09–1.11) | 0.90 (0.52–1.40) | ||
| Relative abundance of 15 most common genera, mean (SD), maximum | ||||
| Staphylococcus | 0.41 (0.43) | 1.00 | 0.02 (0.10) | 1.00 |
| Moraxella | 0.09 (0.21) | 0.99 | 0.31 (0.34) | 1.00 |
| Streptococcus | 0.05 (0.14) | 0.98 | 0.30 (0.30) | 1.00 |
| Haemophilus | 0.07 (0.20) | 1.00 | 0.20 (0.31) | 1.00 |
| Corynebacterium | 0.10 (0.22) | 1.00 | 0.01 (0.06) | 0.95 |
| Dolosigranulum | 0.05 (0.14) | 1.00 | 0.01 (0.04 | 0.70 |
| Enterobacter | 0.05 (0.18) | 1.00 | 0.00 (0.03) | 0.48 |
| Neisseria | 0.00 (0.01) | 0.19 | 0.02 (0.07) | 0.75 |
| Enterococcus | 0.02 (0.12) | 1.00 | 0.00 (0.01) | 0.23 |
| Bacillus | 0.02 (0.14) | 1.00 | 0.00 (0.00) | 0.02 |
| Acinetobacter | 0.02 (0.12) | 1.00 | 0.00 (0.02) | 0.25 |
| Prevotella | 0.00 (0.01) | 0.29 | 0.02 (0.05) | 0.58 |
| Alloprevotella | 0.00 (0.01) | 0.18 | 0.01 (0.04) | 0.56 |
| Veillonella | 0.00 (0.01) | 0.10 | 0.01 (0.03) | 0.27 |
| Gemella | 0.00 (0.00) | 0.06 | 0.01 (0.03) | 0.52 |
Comparison of the richness, alpha-diversity (Shannon index), and genus abundances for the nasal swab and nasopharyngeal aspirate samples. The most common genera were determined by the union of the 10 most abundant genera from each sample type and are listed in order from most to least abundant
Correlations between NS and NPA microbiota
A permutation test demonstrated that the within-individual mean correlation of 0.36 between the two upper airway sites while low was significantly higher than the randomly paired sample distribution (10,000 permutations, P = 0), which had a mean of 0.28 (SD = 0.004). The Spearman correlations between the top 15 genera from the NS and NPA are shown in Fig. 1b. Haemophilus has the greatest correlation between anatomic sites (ρ = 0.50). Additionally, there is a negative correlation between all of the top genera in NS (except Bacillus) and Staphylococcus in NPA, further illustrating the differences in Staphylococcus abundance between the NS and NPA samples. The between-site correlation network demonstrates that the top five genera in the NS and NPA co-occur within individuals, including Haemophilus and Moraxella (Fig. 2). Although Staphylococcus dominates the NS, the correlation network shows that Staphylococcus in NS is only significantly correlated with Staphylococcus and Acinetobacter in NPA.
Fig. 2.
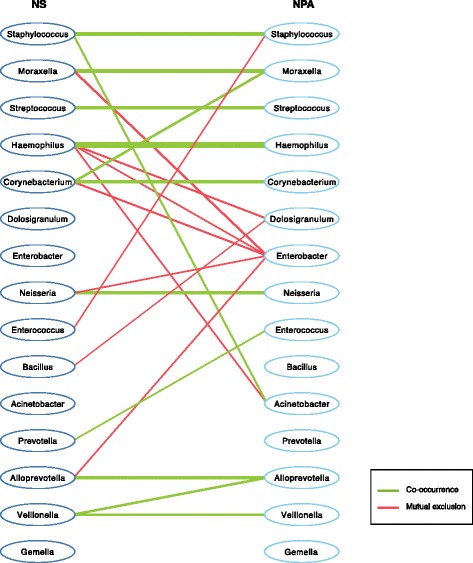
Network of significant intra-individual co-occurrence and co-exclusion associations between nasal swab and nasopharyngeal microbiota. The co-occurrence (green) and co-exclusion (red) relationships between the nasal swab (left) and nasopharyngeal (right) microbiota were assessed using four different methods: Spearman correlation, Pearson correlation, Bray-Curtis dissimilarity, and Kullback-Leibler divergence. To prevent the potential overestimation of associations that occur with the individual metrics, we retained in the network only associations between the top 15 genera that were found significant (P < 0.05) via bootstrapping for at least two of the aforementioned methods. Edges are weighted by the Spearman correlation value, with thicker lines indicating a larger correlation
Clustering the composite samples generated by adjoining the NS and NPA genus abundances for each individual demonstrated common within individual patterns between the dominant genera in the NS and NPA samples (Fig. 3). Of particular interest are the first two clusters, which show that infants with Haemophilus- and Moraxella-dominant NS samples also have Haemophilus- and Moraxella-dominant NPA samples. Additionally, the NS clusters with a high abundance of Staphylococcus correspond to high abundances of the three major genera in the NPA (i.e., Haemophilus, Moraxella, Streptococcus), indicating that high Staphylococcus samples from NS map to all of the previously identified NPA microbiota profile groups discussed below [21].
Fig. 3.
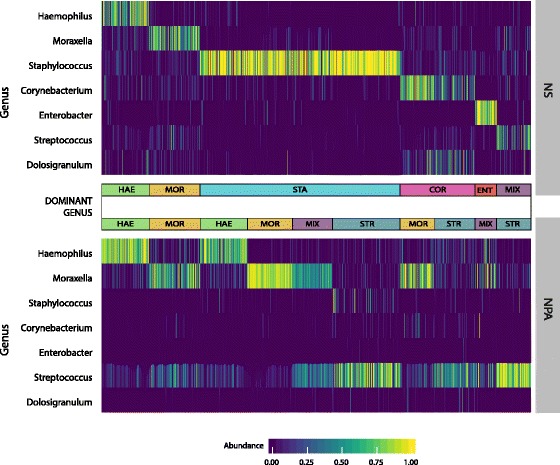
Clustering and composition of composite microbiota. For each subject in the study, adjoining the abundances of the top genera from the nasal swab and nasopharyngeal aspirate microbiotas created a composite microbiota sample. The Bray-Curtis dissimilarity was computed between each composite sample, and partitioning around medoids clustering was performed on the resulting dissimilarity values using 10 clusters. The heat map displays the abundances of the top seven genera for each of the resulting clusters, revealing common patterns of genus dominance between the nasal swab and nasopharyngeal microbiota. HAE, Haemophilus; MOR, Moraxella; STA, Staphylococcus; COR, Corynebacterium; ENT, Enterobacter; MIX, multiple genera; STR, Streptococcus
Nasal swab microbiota profiles
Using PAM clustering, we previously reported four microbiota profiles generated from the NPA samples from infants with bronchiolitis: Haemophilus-dominant, Moraxella-dominant, Streptococcus-dominant, and mixed profiles [21]. In contrast to these four NPA microbiota profiles, PAM clustering of the NS samples generated six profiles: Haemophilus-dominant (7.2%), Moraxella-dominant (13.0%), Staphylococcus-dominant (44.5%), Corynebacterium-dominant (13.4%), Enterobacter-dominant (7.5%), and mixed (14.4%) profiles (Fig. 4). This PAM analysis of the NS gives a very large and well-defined cluster of subjects with a large abundance of Staphylococcus genus. Indeed, the Staphylococcus-dominant profile consists of 78% Staphylococcus and thus shows low bacterial richness and evenness (Table 2). Moreover, the infants in the Staphylococcus-dominant profile were younger compared to other profile groups (P < 0.001), with 85% of the infants in this profile being less than 6 months of age (Table 3). The infants in the NS Haemophilus-dominant profile were older (median 5 months; IQR 3–8 months), had higher weight (median 7 kg; IQR 6–8 kg), and were more likely to have used antibiotics prior to hospitalization (50.8%) as compared to the other profiles (all P < 0.001) (Table 3), results which are all similar to the NPA findings [21].
Fig. 4.
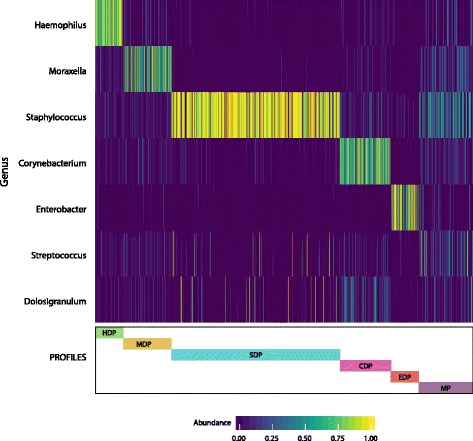
Composition of nasal swab microbiota profiles. Partitioning around medoids was performed on the weighted UniFrac distances between only the nasal swab genus abundances (independent of the nasopharyngeal aspirate genus abundances) using six clusters. The heat map shows the abundance of the top seven genera for each cluster. HDP, Haemophilus-dominant profile; MDP, Moraxella-dominant profile; SDP, Staphylococcus-dominant profile; CDP, Corynebacterium-dominant profile; EDP, Enterobacter-dominant profile; MP, mixed profile
Table 2.
Richness, alpha-diversity, and abundance by nasal swab microbiota profile
| Haemophilus-dominant profile, n = 59 (7.2%) | Moraxella-dominant profile, n = 106 (13.0%) | Staphylococcus-dominant profile, n = 363 (44.5%) | Corynebacterium-dominant profile, n = 109 (13.4%) | Enterobacter-dominant profile, n = 61 (7.5%) | Mixed profile, n = 117 (14.4%) | |
|---|---|---|---|---|---|---|
| Richness, median (IQR) | ||||||
| Number of genera | 11 (8–9) | 15 (5–23) | 4 (2–7) | 8 (5–12) | 4 (3–6) | 18 (10–27) |
| Alpha-diversity, median (IQR) | ||||||
| Shannon index | 0.77 (0.37–1.15) | 1.03 (0.50–1.39) | 0.11 (0.01–0.48) | 0.89 (0.53–1.15) | 0.37 (0.07–0.70) | 1.44 (1.11–1.80) |
| Relative abundance of 10 most abundant genera, mean (SD) | ||||||
| Staphylococcus | 0.05 (0.08) | 0.04 (0.07) | 0.78 (0.35) | 0.07 (0.13) | 0.03 (0.06) | 0.29 (0.24) |
| Corynebacterium | 0.02 (0.03) | 0.05 (0.08) | 0.02 (0.05) | 0.59 (0.27) | 0.00 (0.01) | 0.07 (0.09) |
| Moraxella | 0.06 (0.09) | 0.48 (0.33) | 0.01 (0.02) | 0.03 (0.06) | 0.01 (0.03) | 0.13 (0.13) |
| Haemophilus | 0.74 (0.20) | 0.04 (0.09) | 0.00 (0.02) | 0.03 (0.07) | 0.01 (0.03) | 0.06 (0.12) |
| Dolosigranulum | 0.02 (0.04) | 0.04 (0.07) | 0.04 (0.17) | 0.13 (0.17) | 0.01 (0.03) | 0.07 (0.14) |
| Streptococcus | 0.03 (0.06) | 0.05 (0.08) | 0.04 (0.14) | 0.04 (0.11) | 0.01 (0.02) | 0.14 (0.21) |
| Enterobacter | 0.01 (0.06) | 0.01 (0.04) | 0.00 (0.02) | 0.00 (0.01) | 0.52 (0.41) | 0.03 (0.09) |
| Bacillus | 0.00 (0.01) | 0.00 (0.00) | 0.05 (0.20) | 0.00 (0.00) | 0.00 (0.00) | 0.01 (0.08) |
| Enterococcus | 0.00 (0.02) | 0.01 (0.02) | 0.04 (0.17) | 0.01 (0.05) | 0.01 (0.05) | 0.01 (0.06) |
| Acinetobacter | 0.00 (0.00) | 0.12 (0.30) | 0.00 (0.01) | 0.00 (0.00) | 0.02 (0.09) | 0.02 (0.08) |
Summary of the richness, alpha-diversity (Shannon index), and relative abundances of the most common genera for each of the nasal swab microbiota profiles as determined by partitioning around the medoid (PAM) clustering. The 10 most abundant genera were determined by taking the sum of the abundances for each genus over all the samples and are listed in order from most to least abundant
Table 3.
Characteristics and clinical presentation of infants hospitalized for bronchiolitis by nasal swab microbiota profile
| Variables | Corynebacterium-dominant profile, n = 109 (13.4%) | Haemophilus-dominant profile, n = 59 (7.2%) | Moraxella-dominant profile, n = 106 (13.0%) | Enterobacter-dominant profile, n = 61 (7.5%) | Staphylococcus-dominant profile, n = 363 (44.5%) | Mixed profile, n = 117 (14.4%) | P value |
|---|---|---|---|---|---|---|---|
| Characteristics | |||||||
| Age (months), median (IQR) | 3 (1–6) | 5 (3–8) | 4 (2–7) | 4 (2–6) | 3 (2–4) | 4 (2–7) | < 0.001 |
| < 2 | 36 (33.0) | 7 (11.9) | 24 (22.6) | 14 (23.0) | 132 (36.4) | 23 (19.7) | < 0.001 |
| 2–5.9 | 44 (40.4) | 25 (42.4) | 48 (45.3) | 34 (55.7) | 178 (49.0) | 58 (49.6) | |
| 6–11.9 | 29 (26.6) | 27 (45.8) | 34 (32.1) | 13 (21.3) | 53 (14.6) | 36 (30.8) | |
| Male sex | 59 (54.1) | 36 (61.0) | 58 (54.7) | 37 (60.7) | 230 (63.4) | 68 (58.1) | 0.45 |
| Race/ethnicity | 0.07 | ||||||
| Non-Hispanic white | 53 (48.6) | 23 (39.0) | 49 (46.2) | 20 (32.8) | 177 (48.8) | 38 (32.5) | |
| Non-Hispanic black | 18 (16.5) | 11 (18.6) | 22 (20.8) | 22 (36.1) | 80 (22.0) | 30 (25.6) | |
| Hispanic | 34 (31.2) | 24 (40.7) | 31 (29.2) | 17 (27.9) | 93 (25.6) | 44 (37.6) | |
| Other | 4 (3.7) | 1 (1.7) | 4 (3.8) | 2 (3.3) | 13 (3.6) | 5 (4.3) | |
| Parental history of asthma | 30 (27.5) | 15 (25.4) | 42 (39.6) | 26 (42.6) | 124 (34.2) | 37 (31.6) | 0.18 |
| Maternal smoking during pregnancy | 16 (14.7) | 4 (6.8) | 14 (13.2) | 14 (23.0) | 50 (13.8) | 13 (11.1) | 0.19 |
| Mode of birth | 0.84 | ||||||
| Vaginal birth | 71 (65.1) | 37 (62.7) | 65 (61.3) | 41 (67.2) | 248 (68.3) | 75 (64.1) | |
| C-section | 38 (34.9) | 21 (35.6) | 38 (35.8) | 20 (32.8) | 111 (30.6) | 41 (35.0) | |
| Prematurity (32–37 weeks) | 17 (15.6) | 12 (20.3) | 15 (14.2) | 12 (19.7) | 60 (16.5) | 22 (18.8) | 0.87 |
| Previous breathing problems before the index hospitalization* | 23 (21.1) | 12 (20.3) | 27 (25.5) | 17 (27.9) | 62 (17.1) | 26 (22.2) | 0.26 |
| History of eczema | 17 (15.6) | 12 (20.3) | 20 (18.9) | 13 (21.3) | 61 (16.8) | 21 (17.9) | 0.92 |
| Ever attended daycare | 23 (21.1) | 17 (28.8) | 33 (31.1) | 15 (24.6) | 75 (20.7) | 27 (23.1) | 0.27 |
| Aeroallergen sensitization† | 0 (0.0) | 1 (1.7) | 2 (1.9) | 0 (0.0) | 7 (1.9) | 2 (1.7) | 0.68 |
| Food sensitization† | 21 (19.3) | 9 (15.3) | 22 (20.8) | 13 (21.3) | 64 (17.6) | 23 (19.7) | 0.92 |
| Children at home | 84 (77.1) | 48 (81.4) | 88 (83.0) | 49 (80.3) | 285 (78.5) | 92 (78.6) | 0.90 |
| Mostly breastfed for the first 3 months of age | 59 (54.1) | 31 (52.5) | 51 (48.1) | 23 (37.7) | 152 (41.9) | 45 (38.5) | 0.21 |
| Smoke exposure at home | 17 (15.6) | 5 (8.5) | 18 (17.0) | 14 (23.0) | 55 (15.2) | 18 (15.4) | 0.42 |
| Antibiotic use before index hospitalization | 29 (26.6) | 30 (50.8) | 44 (41.5) | 27 (44.3) | 96 (26.4) | 28 (23.9) | < 0.001 |
| Corticosteroid use before index hospitalization | 13 (11.9) | 13 (22.0) | 19 (17.9) | 11 (18.0) | 46 (12.7) | 19 (16.2) | 0.31 |
| Clinical presentation | |||||||
| Month of hospitalization | 0.06 | ||||||
| November | 10 (9.2) | 3 (5.1) | 10 (9.4) | 10 (16.4) | 32 (8.8) | 5 (4.3) | |
| December | 21 (19.3) | 7 (11.9) | 16 (15.1) | 6 (9.8) | 74 (20.4) | 22 (18.8) | |
| January | 31 (28.4) | 24 (40.7) | 32 (30.2) | 15 (24.6) | 97 (26.7) | 36 (30.8) | |
| February | 31 (28.4) | 14 (23.7) | 24 (22.6) | 12 (19.7) | 85 (23.4) | 32 (27.4) | |
| March | 12 (11.0) | 8 (13.6) | 21 (19.8) | 14 (23.0) | 40 (11.0) | 15 (12.8) | |
| April | 4 (3.7) | 3 (5.1) | 3 (2.8) | 4 (6.6) | 35 (9.6) | 7 (6.0) | |
| Breathing problem began < 1 day before the index hospitalization | 5 (4.6) | 2 (3.4) | 8 (7.5) | 3 (4.9) | 20 (5.5) | 10 (8.5) | 0.67 |
| Weight at presentation (kg), median (IQR) | 6 (5–8) | 7 (6–8) | 6 (5–8) | 6 (5–8) | 6 (5–7) | 7 (5–8) | < 0.001 |
| Respiratory rate at presentation (per minute), median (IQR) | 48 (40–59) | 50 (40–60) | 49 (42–60) | 50 (42–64) | 50 (40–60) | 50 (42–60) | 0.26 |
| Oxygen saturation at presentation | 0.006 | ||||||
| < 90% | 7 (6.4) | 13 (22.0) | 4 (3.8) | 7 (11.5) | 31 (8.5) | 11 (9.4) | |
| 90–93% | 23 (21.1) | 9 (15.3) | 14 (13.2) | 5 (8.2) | 50 (13.8) | 16 (13.7) | |
| ≥ 94% | 74 (67.9) | 35 (59.3) | 88 (83.0) | 46 (75.4) | 274 (75.5) | 90 (76.9) | |
| Retractions on examination | 0.32 | ||||||
| None | 18 (16.5) | 6 (10.2) | 24 (22.6) | 13 (21.3) | 58 (16.0) | 17 (14.5) | |
| Mild | 42 (38.5) | 21 (35.6) | 45 (42.5) | 23 (37.7) | 160 (44.1) | 55 (47.0) | |
| Moderate/severe | 43 (39.4) | 30 (50.8) | 33 (31.1) | 23 (37.7) | 127 (35.0) | 45 (38.5) | |
| Wheezing on examination | 63 (57.8) | 30 (50.8) | 65 (61.3) | 37 (60.7) | 214 (59.0) | 77 (65.8) | 0.31 |
| Received antibiotics during pre-hospitalization visit | 28 (25.7) | 34 (57.6) | 35 (33.0) | 25 (41.0) | 104 (28.7) | 41 (35.0) | < 0.001 |
| Received corticosteroids during pre-hospitalization visit | 6 (5.5) | 5 (8.5) | 5 (4.7) | 6 (9.8) | 34 (9.4) | 17 (14.5) | 0.15 |
| Virology | 0.11 | ||||||
| Sole RSV infection | 56 (51.4) | 30 (50.8) | 69 (65.1) | 34 (55.7) | 225 (62.0) | 60 (51.3) | |
| Sole rhinovirus infection | 7 (6.4) | 4 (6.8) | 4 (3.8) | 8 (13.1) | 17 (4.7) | 7 (6.0) | |
| RSV + rhinovirus coinfection | 14 (12.8) | 3 (5.1) | 14 (13.2) | 7 (11.5) | 44 (12.1) | 15 (12.8) | |
| RSV + non-rhinovirus pathogens | 14 (12.8) | 15 (25.4) | 9 (8.5) | 6 (9.8) | 34 (9.4) | 15 (12.8) | |
| Rhinovirus + non-RSV pathogens | 2 (1.8) | 2 (3.4) | 3 (2.8) | 2 (3.3) | 9 (2.5) | 5 (4.3) | |
| Neither RSV nor rhinovirus‡ | 11 (10.1) | 5 (8.5) | 6 (5.7) | 4 (6.6) | 26 (7.2) | 9 (7.7) | |
| No viral pathogens | 5 (4.6) | 0 (0.0) | 1 (0.9) | 0 (0.0) | 8 (2.2) | 6 (5.1) | |
| Viral genomic load (CT-value), median IQR | |||||||
| RSV | 22 (20–25) | 22 (21–26) | 22 (21–25) | 23 (21–25) | 23 (21–25) | 22 (21–27) | 0.63 |
| RV | 28 (26–31) | 29 (24–34) | 28 (27–36) | 27 (25–34) | 31 (27–36) | 30 (27–36) | 0.60 |
| Outcomes | |||||||
| Intensive care use‡ | 20 (18%) | 16 (27%) | 6 (6%) | 13 (21%) | 47 (13%) | 21 (18%) | |
| Hospital length of stay ≥ 5 days | 10 (9%) | 19 (32%) | 16 (15%) | 8 (13%) | 47 (13%) | 18 (15%) | |
| Hospital length of stay (day), median (IQR) | 2 (1–4) | 2 (2–5) | 2 (1–3) | 2 (1–3) | 2 (1–3) | 2 (1–3) | |
Data are number (%) of infants unless otherwise indicated. Percentages may not be equal to 100, because of missingness
Patient characteristics and hospital course were compared using chi-square test or Kruskal-Wallis test across the identified nasopharyngeal microbiota profiles
CT cycle threshold, IQR interquartile range, RSV respiratory syncytial virus
*Defined as an infant having cough that wakes him/her at night and/or causes emesis, or when the child has wheezing or shortness of breath without cough
†Defined by having one or more positive values for allergen-specific IgE
‡Defined as admission to intensive care unit and/or use of mechanical ventilation (continuous positive airway pressure and/or intubation during inpatient stay, regardless of location) at any time during the index hospitalization
Nasal swab microbiota profiles and Bronchiolitis severity
Using the NPA data, our group previously reported that infants with a Haemophilus-dominant NPA microbial profile had increased odds of intensive care use and risk of hospital length of stay of three or more days when compared to those with a Moraxella-dominant microbiota profile [21]. In the current analysis, we were able to replicate these severity outcome associations in NS microbiota profiles, finding that subjects with the Haemophilus-dominant NS profile were more likely to have intensive care use (unadjusted OR 6.20, P < 0.001; adjusted OR 6.43, P < 0.001) and a hospital length of stay of five or more days (unadjusted OR 3.57, P = 0.004; adjusted OR 4.31, P = 0.002) (Table 4).
Table 4.
Unadjusted and multivariate associations of nasal swab microbiota profiles with bronchiolitis severity outcomes
| Unadjusted models | Adjusted models* | |||
|---|---|---|---|---|
| Severity outcomes | OR (95% CI) | P value | OR (95% CI) | P value |
| 1) Intensive care use | ||||
| All profiles | ||||
| Haemophilus-dominant profile | 6.20 (2.38–18.28) | < 0.001 | 6.43 (2.25–20.51) | < 0.001 |
| Moraxella-dominant profile | Reference | Reference | ||
| Staphylococcus-dominant profile | 2.54 (1.14–6.78) | 0.04 | 2.17 (0.93–5.98) | 0.10 |
| Corynebacterium-dominant profile | 3.75 (1.52–10.62) | 0.007 | 4.15 (1.59–12.37) | 0.005 |
| Enterobacter-dominant profile | 4.51 (1.68–13.53) | 0.004 | 4.84 (1.67–15.46) | 0.004 |
| Mixed profile | 3.86 (1.59–10.85) | 0.005 | 3.37 (1.29–9.99) | 0.02 |
| Haemophilus vs. all others | ||||
| Combined non-Haemophilus profiles | Reference | Reference | ||
| Haemophilus-dominant profile | 2.21 (1.17–3.99) | 0.01 | 2.48 (1.19–5.03) | 0.01 |
| Moraxella vs. All others | ||||
| Combined non-Moraxella profiles | Reference | Reference | ||
| Moraxella-dominant profile | 0.30 (0.11–0.64) | 0.01 | 0.32 (0.12–0.72) | 0.01 |
| 2) Hospital length of stay ≥ 5 days | ||||
| All profiles | ||||
| Haemophilus-dominant profile | 3.57 (1.52–8.77) | 0.004 | 4.31 (1.73–11.11) | 0.002 |
| Moraxella-dominant profile | Reference | Reference | ||
| Staphylococcus-dominant profile | 1.43 (0.72–3.09) | 0.33 | 1.24 (0.61–2.75) | 0.57 |
| Corynebacterium-dominant profile | 2.03 (0.91–4.76) | 0.09 | 2.02 (0.86–4.95) | 0.11 |
| Enterobacter-dominant profile | 1.45 (0.52–3.89) | 0.46 | 1.37 (0.48–3.83) | 0.55 |
| Mixed profile | 1.75 (0.78–4.11) | 0.18 | 1.72 (0.73–4.20) | 0.22 |
Significant results of interest are in italics
Patient level variables include age at hospitalization, sex, race/ethnicity, gestational age, number of previous breathing problems (i.e., infant having cough that wakes him/her at night and/or causes emesis or when the child has wheezing or shortness of breath without cough), daycare attendance, presence of other children living in home, history of antibiotic use (i.e., infant has taken antibiotics at any time prior to hospitalization), history of corticosteroid use (i.e., infant has taken corticosteroids, inhaled or systemic, at any time prior to hospitalization), use of antibiotics during the pre-hospitalization visit (i.e., infant received antibiotics during pre-admission), and respiratory viruses detected by PCR
CI, confidence interval, OR, odds ratio
*Mixed-effects logistic regression model adjusting for 11 patient-level variables with collection site as a random effect
Analysis of the NS microbiota profiles found higher odds ratios than the NPA microbiota profiles for the severity outcomes. Isolating the Haemophilus and Moraxella NS profiles showed increased odds of intensive care use for the Haemophilus-dominant profile and a protective association of the Moraxella-dominant profile (Table 4), coincident with the patterns observed in NPA profiles of these samples. While the NS microbiota profiles did not retain the significant association with a hospital stay of three or more days, they were significantly associated with a length of stay of five or more days. Thus, the association between the Haemophilus-dominant profile and a longer hospital length of stay, as well as the protective nature of Moraxella for the hospital length of stay outcome, was maintained in the NS data.
Discussion
In this multicenter study of 815 infants with both NS and NPA samples, we found within-individual correlations between dominant genera in NS and NPA microbiota. However, the NS and NPA samples also revealed distinct compositions, with an increased abundance of Staphylococcus in the NS microbiota. Using an independent analysis of the NS microbiota, we were able to replicate a previously published association between NPA microbiota and bronchiolitis severity outcomes [21] despite sample processing differences. Our findings indicate that the associations of Haemophilus-dominant and Moraxella-dominant profiles with clinical outcomes should be consistent between NS and NPA sample types.
There was a modest but statistically significant within-individual relationship between the bacterial microbiota of the two upper airway sites. Further analysis of these correlations revealed common mappings between NS and NPA genus abundances, particularly the Haemophilus-dominant and Moraxella-dominant NS samples that were maintained in the NPA microenvironment. Beyond these two genera, the mapping between NS and NPA demonstrated inconsistency between the sample types among the dominant genera. For example, the majority of infants with dominant Staphylococcus in the NS samples had NPA samples that were dominated by Haemophilus, Moraxella, or Streptococcus. Thus, NS samples with a high abundance of Staphylococcus alone may not be informative for the corresponding most abundant genera in the NPA. This is further supported by the correlation network (Fig. 2), which shows that Staphylococcus in NS only co-occurs with Staphylococcus and Acinetobacter in NPA. We also found that some samples—particularly NS samples dominated by Staphylococcus—were almost entirely composed of a single genus. Regardless of the dominance of Staphylococcus in NS samples, the large study size allowed ample data for reliable analysis. Despite having compositional differences, the correlations between the NS and NPA microbiota confirm that NSs are a useful clinical sample type. However, researchers should be cautious of the complications imposed by the abundance of Staphylococcus in the NS samples.
Almost half of the MARC-35 infants had a Staphylococcus-dominant profile with relatively low bacterial diversity. The difference in bacterial richness and diversity between the Staphylococcus-dominant profile and the other NS profiles raises the question of whether the high abundance of Staphylococcus, commonly found in the anterior nares [20, 37–39], is associated with clinical outcomes. Although Staphylococcus is often found in the anterior nares of healthy patients [20, 39], studies have found a higher abundance of Staphylococcus in the anterior nares of patients admitted to the intensive care unit [38] and adults with chronic rhinosinusitis [37], which may imply a difference in NS Staphylococcus abundance between sick and healthy individuals. Of more relevance to infants with bronchiolitis, Staphylococcus aureus has not only been shown to enhance the replication and infectivity of rhinovirus [40] and influenza [41], but also a Staphylococcus-dominant airway microbiota is associated with an increased likelihood of severe bronchiolitis [42]. However, because the 16S rRNA gene sequencing approach is insufficient to reliably resolve data at the species level, it is unclear whether the Staphylococcus OTU in our data is in fact Staphylococcus aureus or another species of Staphylococcus. While the species of Staphyloccocus in our data may determine how it impacts other dominant genera, the presence of Staphylococcus nonetheless has potential relevance for respiratory outcomes.
In the present analysis, we have demonstrated the usefulness of this easily obtained sample type when compared with NPA samples. Specifically, we were able to recapitulate the relationship between Haemophilus-dominant nasal microbiota and bronchiolitis severity [21], as well as the protective nature of Moraxella in the airway microbiota [21, 43], with NS samples among infants with bronchiolitis (Table 4). Furthermore, the NS microbiota profiles preserved associations with age, weight, and antibiotics use (Table 3). We conclude that for associations with Haemophilus and Moraxella, NS and NPA provide similar results. However, because many Haemophilus and Moraxella dominant samples in NPA were dominated by Staphylococcus in the NS samples for the same individuals, the ability to predict bronchiolitis severity outcomes may be more limited for Staphylococcus-dominant NS samples. Additionally, the associations between other microbiota dominant profiles and clinical outcomes may differ between the anterior nares and nasopharyngeal microenvironments.
Our study has some potential limitations. First, variations between the NS and NPA microbiota may have been amplified due to differences in NS and NPA sample handling (e.g., temperature at which initially stored). Studies of fecal microbiota have found that while variations in transport media can significantly alter the microbial composition of microbiota samples, differences in storage temperature should have a relatively little effect on the resulting microbial composition [44, 45]. Although immediately freezing the NS samples would have allowed a direct comparison to the NPA samples, the NS samples were collected and processed in a manner easily replicable in outpatient clinics or in homes for community-based studies [46–50]. Mailing NS samples has been used for viral detection [47–49], but this sample collection technique has not previously been used to examine the microbiota even though both viruses, and the microbiota are associated with acute bronchiolitis severity outcomes [8, 43, 51] as well as other respiratory outcomes [41]. Although viral detection is improved when nasal samples are collected from deeper within the nasopharynx than from the anterior nares [52], the more easily accessible NS samples have been useful for detecting viruses in community studies [53]. Allowing participants to provide a mailed NS sample would remove obstacles typically faced by researchers who want to examine both viral and microbial exposures from community-based samples. In this study, despite the differences in initial storage temperature of the samples, we were able to use the NS sample data to replicate the previous association between NPA microbiota and bronchiolitis severity.
In addition, as the samples in our study were obtained in the context of acute bronchiolitis hospitalization, the within-individual correspondence we observed may not be reproducible in healthy infants. However, our study demonstrates that NS microbiota data is relevant in the context of acute illness and is associated both with the microbial composition of the nasopharynx as well as with clinically relevant outcomes. Another potential limitation is that we did not address the dynamics of the microbiota at these sites. The airway microbiota changes over time via environmental exposure and the natural progression of microbial composition in the airway [8, 43, 54]. Although our findings show the associations in infant nasal microbiota, the relationship between anterior nare samples and nasopharyngeal samples in older children is unknown. Our data should facilitate further investigations into this important domain.
Conclusions
In this multicenter cohort study of infants hospitalized with bronchiolitis, we found modest but statistically significant intra-individual correlations between NS and NPA microbiota, especially for Haemophilus and Moraxella. Given the overlap of the microbiota structure between the sample types, we were able to use NS data to replicate the previously identified associations between NPA microbiota and severity of illness (as measured by intensive care use and hospital length of stay). Our investigation also reveals important differences between NS and NPA samples—particularly the high abundance of Staphylococcus in the NS. However, despite the variations between the NS and NPA microbiota and the differential handling of the specimens, the considerable overlap of the microbiota between the anatomic sites indicates that NS can provide robust and useful samples in young children.
Acknowledgements
The following individuals were principal investigators at the 17 participating sites in the MARC-35 study. Amy D. Thompson, MD, Alfred I. DuPont Hospital for Children, Wilmington, DE; Federico R. Laham, MD, MS, Arnold Palmer Hospital for Children, Orlando, FL; Jonathan M. Mansbach, MD, MPH, Boston Children’s Hospital, Boston, MA; Vincent J. Wang, MD, MHA, Children’s Hospital of Los Angeles, Los Angeles, CA; Michelle B. Dunn, MD, Children’s Hospital of Philadelphia, Philadelphia, PA; Juan Celedon, MD, PhD, MS, Children’s Hospital of Pittsburgh, Pittsburgh, PA; Michael R. Gomez, MD, MS, Children’s Hospital at St. Francis, Tulsa, OK; Brian M. Pate, MD, The Children’s Mercy Hospital and Clinics, Kansas City, MO; Stephen J. Teach, MD, MPH, Children’s National Medical Center, Washington, DC; Richard T. Strait, MD, Cincinnati Children’s Hospital and Medical Center, Cincinnati, OH; Ilana Y. Wanik, MD, Connecticut Children’s Medical Center, Hartford, CT; Sujit S. Iyer, MD, Dell Children’s Medical Center of Central Texas, Austin, TX; Michelle D. Stevenson, MD, MS, Norton Children’s Hospital, Louisville, KY; Ari R. Cohen, MD and Wayne Shreffler, MD, PhD, Massachusetts General Hospital, Boston, MA; Anne W. Beasley, MD, Phoenix Children’s Hospital, Phoenix, AZ; Thida Ong, MD, Seattle Children’s Hospital, Seattle, WA; Charles G. Macias, MD, MPH, Texas Children’s Hospital, Houston, TX.
Funding
This study was supported by the grants U01 AI-087881 (Camargo), R01 AI-114552 (Camargo), R01 AI-108588 (Mansbach), and UG3 OD-023253 (Camargo) from the National Institutes of Health (Bethesda, MD). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Availability of data and materials
The derived results and raw sequence data have been deposited to ImmPort (http://www.immport.org) with study accession SDY1182 and are available upon request to investigators performing research in respiratory disease. The R code used to perform this analysis is included as an Additional file 1.
Abbreviations
- NPA
Nasopharyngeal aspirate
- NS
Nasal swab
- OR
Odds ratio
- OTU
Operational taxonomic unit
- PAM
Partitioning around medoid
- SD
Standard deviation
Additional file
R code for statistical analysis. (PDF 33 kb)
Authors’ contributions
PNL performed the statistical analysis and drafted the manuscript. KH developed the association analysis of severity outcomes in NPA and contributed to the critique of this manuscript. NJA performed the 16S rRNA gene sequencing and taxonomic profiling of NS samples and contributed to critique of the manuscript. JAE complied the clinical information for this study and contributed to the critique of this manuscript. DMH performed the compositional analysis of NPA data and assisted with the compilation of the dual NPA-Swab data set. JFP supervised the construction of the 16S rRNA gene data generation, helped to design the study, and contributed to the critique of this manuscript. PAP helped facilitate sample transfer and handling of both NPA and NS data and contributed to the manuscript. AFS helped design and implement the MARC-35 study and contributed to the critique of this manuscript. CAC designed and implemented the MARC-35 study and contributed to the manuscript. CAS advised PNL on statistical analysis, contributed to analysis results, and helped to write the manuscript. JMM helped design and implement the MARC-35 study, contributed significantly to the manuscript, and supervised the study design and analysis procedures. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Written informed consent was obtained from the parent or guardian of all participating infants. The institutional review board at each of the participating hospitals approved the study.
Consent for publication
Not applicable.
Competing interests
JMM has provided bronchiolitis-related consultation for Regeneron. NJA and JFP own shares at Diversigen Inc., a microbiome research company. PAP provided bronchiolitis-related consultation for Gilead, Novavax, Ablynx, and Regeneron. The remaining authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s40168-017-0385-0) contains supplementary material, which is available to authorized users.
References
- 1.Vissing NH, Chawes BL, Bisgaard H. Increased risk of pneumonia and bronchiolitis after bacterial colonization of the airways as neonates. Am J Respir Crit Care Med. 2013;188:1246–1252. doi: 10.1164/rccm.201302-0215OC. [DOI] [PubMed] [Google Scholar]
- 2.Sakwinska O, Bastic Schmid V, Berger B, Bruttin A, Keitel K, Lepage M, Moine D, Ngom Bru C, Brussow H, Gervaix A. Nasopharyngeal microbiota in healthy children and pneumonia patients. J Clin Microbiol. 2014;52:1590–1594. doi: 10.1128/JCM.03280-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castro-Nallar E, Shen Y, Freishtat RJ, Perez-Losada M, Manimaran S, Liu G, Johnson WE, Crandall KA. Integrating microbial and host transcriptomics to characterize asthma-associated microbial communities. BMC Med Genet. 2015;8:50. doi: 10.1186/s12920-015-0121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, Davies J, Ervine A, Poulter L, Pachter L, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang YJ, Nariya S, Harris JM, Lynch SV, Choy DF, Arron JR, Boushey H. The airway microbiome in patients with severe asthma: associations with disease features and severity. J Allergy Clin Immunol. 2015;136:874–884. doi: 10.1016/j.jaci.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perez-Losada M, Castro-Nallar E, Bendall ML, Freishtat RJ, Crandall KA. Dual Transcriptomic profiling of host and microbiota during health and disease in pediatric asthma. PLoS One. 2015;10:e0131819. doi: 10.1371/journal.pone.0131819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pérez-Losada M, Crandall KA, Freishtat RJ. Two sampling methods yield distinct microbial signatures in the nasopharynges of asthmatic children. Microbiome. 2016;4:1–6. doi: 10.1186/s40168-016-0170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, Holt BJ, Hales BJ, Walker ML, Hollams E, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17:704–715. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Depner M, Ege MJ, Cox MJ, Dwyer S, Walker AW, Birzele LT, Genuneit J, Horak E, Braun-Fahrlander C, Danielewicz H, et al. Bacterial microbiota of the upper respiratory tract and childhood asthma. J Allergy Clin Immunol. 2016; 10.1016/j.jaci.2016.05.050. [DOI] [PubMed]
- 10.Cabrera-Rubio R, Garcia-Nunez M, Seto L, Anto JM, Moya A, Monso E, Mira A. Microbiome diversity in the bronchial tracts of patients with chronic obstructive pulmonary disease. J Clin Microbiol. 2012;50:3562–3568. doi: 10.1128/JCM.00767-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang YJ, Kim E, Cox MJ, Brodie EL, Brown R, Wiener-Kronish JP, Lynch SV. A persistent and diverse airway microbiota present during chronic obstructive pulmonary disease exacerbations. OMICS. 2010;14:9–59. doi: 10.1089/omi.2009.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pragman AA, Kim HB, Reilly CS, Wendt C, Isaacson RE. The lung microbiome in moderate and severe chronic obstructive pulmonary disease. PLoS One. 2012;7:e47305. doi: 10.1371/journal.pone.0047305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laufer AS, Metlay JP, Gent JF, Fennie KP, Kong Y, Pettigrew MM. Microbial communities of the upper respiratory tract and otitis media in children. MBio. 2011;2:e00245–e00210. doi: 10.1128/mBio.00245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pettigrew MM, Laufer AS, Gent JF, Kong Y, Fennie KP, Metlay JP. Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl Environ Microbiol. 2012;78:6262–6270. doi: 10.1128/AEM.01051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, Bushman FD, Collman RG. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marsh RL, Kaestli M, Chang AB, Binks MJ, Pope CE, Hoffman LR, Smith-Vaughan HC. The microbiota in bronchoalveolar lavage from young children with chronic lung disease includes taxa present in both the oropharynx and nasopharynx. Microbiome. 2016;4:37. doi: 10.1186/s40168-016-0182-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mosser AG, Vrtis R, Burchell L, Lee WM, Dick CR, Weisshaar E, Bock D, Swenson CA, Cornwell RD, Meyer KC, et al. Quantitative and qualitative analysis of rhinovirus infection in bronchial tissues. Am J Respir Crit Care Med. 2005;171:645–651. doi: 10.1164/rccm.200407-970OC. [DOI] [PubMed] [Google Scholar]
- 18.Bassis CM, Erb-Downward JR, Dickson RP, Freeman CM, Schmidt TM, Young VB, Beck JM, Curtis JL, Huffnagle GB. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio. 2015;6:e00037. doi: 10.1128/mBio.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dickson RP, Huffnagle GB. The lung microbiome: new principles for respiratory bacteriology in health and disease. PLoS Pathog. 2015;11:e1004923. doi: 10.1371/journal.ppat.1004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan M, Pamp SJ, Fukuyama J, Hwang PH, Cho DY, Holmes S, Relman DA. Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. Aureus carriage. Cell Host Microbe. 2013;14:631–640. doi: 10.1016/j.chom.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasegawa K, Mansbach JM, Ajami NJ, Espinola JA, Henke DM, Petrosino JF, Piedra PA, Shaw CA, Sullivan AF, Camargo CA, Jr, et al. Association of nasopharyngeal microbiota profiles with bronchiolitis severity in infants hospitalised for bronchiolitis. Eur Respir J. 2016;48:1329–1339. doi: 10.1183/13993003.00152-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prevaes SM, de Winter-de Groot KM, Janssens HM, de Steenhuijsen Piters WA, Tramper-Stranders GA, Wyllie AL, Hasrat R, Tiddens HA, van Westreenen M, van der Ent CK, et al. Development of the nasopharyngeal microbiota in infants with cystic fibrosis. Am J Respir Crit Care Med. 2016;193:504–515. doi: 10.1164/rccm.201509-1759OC. [DOI] [PubMed] [Google Scholar]
- 23.Ralston SL, Lieberthal AS, Meissner HC, Alverson BK, Baley JE, Gadomski AM, Johnson DW, Light MJ, Maraqa NF, Mendonca EA, et al. Clinical practice guideline: the diagnosis, management, and prevention of bronchiolitis. Pediatrics. 2014;134:e1474–e1502. doi: 10.1542/peds.2014-2742. [DOI] [PubMed] [Google Scholar]
- 24.Hasegawa K, Jartti T, Mansbach JM, Laham FR, Jewell AM, Espinola JA, Piedra PA, Camargo CA., Jr Respiratory syncytial virus genomic load and disease severity among children hospitalized with bronchiolitis: multicenter cohort studies in the United States and Finland. J Infect Dis. 2015;211:1550–1559. doi: 10.1093/infdis/jiu658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lambert SB, Ware RS, Cook AL, Maguire FA, Whiley DM, Bialasiewicz S, Mackay IM, Wang D, Sloots TP, Nissen MD, et al. Observational research in childhood infectious diseases (ORChID): a dynamic birth cohort study. BMJ Open. 2012;2 [DOI] [PMC free article] [PubMed]
- 26.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 28.Edgar RC, Flyvbjerg H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics. 2015;31:3476–3482. doi: 10.1093/bioinformatics/btv401. [DOI] [PubMed] [Google Scholar]
- 29.Edgar RC. UPARSE highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 30.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011;5:169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aitchison J. The statistical analysis of compositional data; London; New York: Chapman and Hall; 1986.
- 34.Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, Huttenhower C. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol. 2012;8:e1002606. doi: 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rousseeuw PJ. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math. 1987;20:53–65. doi: 10.1016/0377-0427(87)90125-7. [DOI] [Google Scholar]
- 36.Tibshirani R, Walther G, Hastie T. Estimating the number of clusters in a data set via the gap statistic. J Royal Stat Soc Ser B Stat Methodol. 2001;63:411–423. doi: 10.1111/1467-9868.00293. [DOI] [Google Scholar]
- 37.Biswas K, Hoggard M, Jain R, Taylor MW, Douglas RG. The nasal microbiota in health and disease: variation within and between subjects. Front Microbiol. 2015;9:134. doi: 10.3389/fmicb.2015.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frank DN, Feazel LM, Bessesen MT, Price CS, Janoff EN, Pace NR. The human nasal microbiota and Staphylococcus Aureus carriage. PLoS One. 2010;5:e10598. doi: 10.1371/journal.pone.0010598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wos-Oxley ML, Plumeier I, von Eiff C, Taudien S, Platzer M, Vilchez-Vargas R, Becker K, Pieper DH. A poke into the diversity and associations within human anterior nare microbial communities. ISME J. 2010;4:839–851. doi: 10.1038/ismej.2010.15. [DOI] [PubMed] [Google Scholar]
- 40.Wang JH, Kwon HJ, Lee BJ, Jang YJ. Staphylococcal enterotoxins a and B enhance rhinovirus replication in A549 cells. Am J Rhinol. 2007;21:670–674. doi: 10.2500/ajr.2007.21.3101. [DOI] [PubMed] [Google Scholar]
- 41.Lynch SV. Viruses and microbiome alterations. Ann Am Thorac Soc. 2014;11(Suppl 1):S57–S60. doi: 10.1513/AnnalsATS.201306-158MG. [DOI] [PubMed] [Google Scholar]
- 42.Hasegawa K, Linnemann R, Mansbach J, Ajami N, Espinola J, Petrosino J, Piedra P, Stevenson M, Sullivan A, Thompson A, et al. Nasal airway microbiota profile and severe bronchiolitis in infants: a case-control study. Pediatr Infect Dis J. 2016; In press [DOI] [PMC free article] [PubMed]
- 43.Bisgaard H, Hermansen MN, Buchvald F, Loland L, Halkjaer LB, Bonnelykke K, Brasholt M, Heltberg A, Vissing NH, Thorsen SV, et al. Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med. 2007;357:1487–1495. doi: 10.1056/NEJMoa052632. [DOI] [PubMed] [Google Scholar]
- 44.Choo JM, Leong LE, Rogers GB. Sample storage conditions significantly influence faecal microbiome profiles. Sci Rep. 2015;5:16350. doi: 10.1038/srep16350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tedjo DI, Jonkers DM, Savelkoul PH, Masclee AA, van Best N, Pierik MJ, Penders J. The effect of sampling and storage on the fecal microbiota composition in healthy and diseased subjects. PLoS One. 2015;10:e0126685. doi: 10.1371/journal.pone.0126685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.uBiome: SmartGut Instructions.https://ubiome.com/smartgut-instructions. Accessed 31 May 2017.
- 47.Alsaleh AN, Whiley DM, Bialasiewicz S, Lambert SB, Ware RS, Nissen MD, Sloots TP, Grimwood K. Nasal swab samples and real-time polymerase chain reaction assays in community-based, longitudinal studies of respiratory viruses: the importance of sample integrity and quality control. BMC Infect Dis. 2014;14:15. doi: 10.1186/1471-2334-14-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fisher C, Bornstein R, Kuypers J, Boeckh M, Limaye A, Jerome KA. Prospective study comparing self-collected nasal swabs to oral washes for monitoring viral load kinetics in lung transplant recipients with respiratory virus infection. Open Forum Infect Dis. 2016;3:663.
- 49.Vargas CY, Wang L, Castellanos de Belliard Y, Morban M, Diaz H, Larson EL, LaRussa P, Saiman L, Stockwell MS. Pilot study of participant-collected nasal swabs for acute respiratory infections in a low-income, urban population. Clin Epidemiol. 2016;8:1–5. doi: 10.2147/CLEP.S95847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Cleef BA, Verkade EJM, Wulf MW, Buiting AG, Voss A, Huijsdens XW, van Pelt W, Mulders MN, Kluytmans JA. Prevalence of livestock-associated MRSA in communities with high pig-densities in The Netherlands. PLoS One. 2010;5:e9385. doi: 10.1371/journal.pone.0009385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mansbach JM, Hasegawa K, Henke DM, Ajami NJ, Petrosino JF, Shaw CA, Piedra PA, Sullivan AF, Espinola JA, Camargo CA., Jr Respiratory syncytial virus and rhinovirus severe bronchiolitis are associated with distinct nasopharyngeal microbiota. J Allergy Clin Immunol. 2016;137:1909–1913. doi: 10.1016/j.jaci.2016.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heikkinen T, Marttila J, Salmi AA, Ruuskanen O. Nasal swab versus nasopharyngeal aspirate for isolation of respiratory viruses. J Clin Microbiol. 2002;40:4337–4339. doi: 10.1128/JCM.40.11.4337-4339.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blaschke AJ, Allison MA, Meyers L, Rogatcheva M, Heyrend C, Mallin B, Carter M, Lafleur B, Barney T, Poritz MA, et al. Non-invasive sample collection for respiratory virus testing by multiplex PCR. J Clin Virol. 2011;52:210–214. doi: 10.1016/j.jcv.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Biesbroek G, Tsivtsivadze E, Sanders EA, Montijn R, Veenhoven RH, Keijser BJ, Bogaert D. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med. 2014;190:1283–1292. doi: 10.1164/rccm.201407-1240OC. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The derived results and raw sequence data have been deposited to ImmPort (http://www.immport.org) with study accession SDY1182 and are available upon request to investigators performing research in respiratory disease. The R code used to perform this analysis is included as an Additional file 1.


