Abstract
After transcription, RNAs are always associated with RNA binding proteins (RBPs) to perform biological activities. RBPs can interact with target RNAs in sequence- and structure-dependent manner through their unique RNA binding domains. In development and progression of carcinogenesis, RBPs are aberrantly dysregulated in many human cancers with various mechanisms, such as genetic alteration, epigenetic change, noncoding RNA-mediated regulation, and post-translational modifications. Upon deregulation in cancers, RBPs influence every step in the development and progression of cancer, including sustained cell proliferation, evasion of apoptosis, avoiding immune surveillance, inducing angiogenesis, and activating metastasis. To develop therapeutic strategies targeting RBPs, RNA interference-based oligonucleotides or small molecule inhibitors have been screened based on reduced RBP-RNA interaction and changed level of target RNAs. Identification of binding RNAs with high-throughput techniques and integral analysis of multiple datasets will help us develop new therapeutic drugs or prognostic biomarkers for human cancers.
Keywords: RNA-binding proteins, Cancer, Chemoprevention, Therapeutics
INTRODUCTION
After RNA is transcribed from genome, it is not present by itself in the cell. Many proteins interact with the transcribed RNAs to make huge complex, called ribonucleoproteins (RNPs). To form RNPs, RNA binding proteins (RBPs) participate as critical regulators for RNA metabolism. RBPs can modulate the fate of binding RNAs by regulating transcription, editing, splicing, polyadenylation, translocation, and turnover.1 RBPs can also function as scaffold proteins for recruiting many factors and enzymes to modify their binding partners. By making different complexes with various combinations, RBPs can fine-tune target RNAs in a time- or space-specific manner. For exact work, RBPs are regulated by post-translational modifications (PTMs) such as acetylation, ubiquitination, and phosphorylation. For example, Src-associated protein in mitosis of 68 kDa (SAM68) is phosphorylated at tyrosine residue to mediate RNA binding activity and signal transduction.2
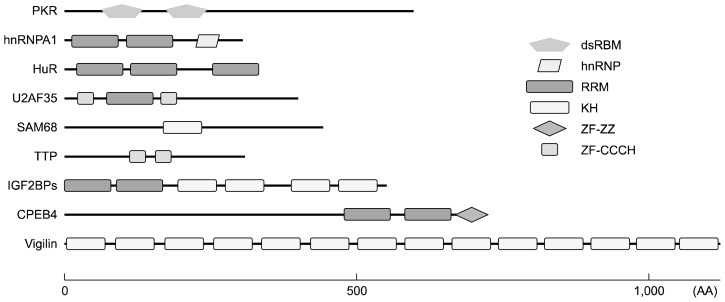
Mammalian cells contain hundreds of genes encoding RBPs that are evolutionally conserved and are transcribed into thousands of splicing variants to make RBPs. Until now, over 1,500 RBPs have been identified through high-throughput screening and are validated as members of a unique database.3,4 Due to large number of RBPs, they perform various functions to maintain the homeostasis of cellular physiology. RBPs can interact with cognate RNAs in sequence-dependent or structure-specific manner using RNA-binding domains (RBDs) containing 60–100 amino acids. RBPs can combine with different RBDs to provide specificity and affinity for binding partners (Fig. 1). However, half of known RBPs interact with RNA in the absence of specific motifs or structures.5 They may interact with RNA through concentration levels, affinity distribution, or synergistic binding with other effectors. More than 40 RBDs have been reported to be able to orchestrate the role of RBPs, through RNA recognition motif (RRM), K-homology (KH) domain, double-stranded RNA binding motif (dsRBM), Zinc finger (ZF) domain, and Piwi/Argonaute/Zwille (PAZ) domain.6 RBPs have been classified into different family members depending on compositions of RBD.
Figure 1.
Schematic diagram for various RNA binding domains of RNA binding proteins (RBPs). RBP interacts with target mRNAs using unique RNA binding domain (RBD) or a combination of different RBDs. Depending on RBDs, each RBP has specificity and affinity for its target RNAs. Each domain is schematically depicted with different shapes and colors. Each RBP is presented depending on its size. dsRBM, double-stranded RNA binding motiff; hnRNP, heterogeneous nuclear ribonucleoprotein; RRM, RNA recognition motif; KH, K-homology; ZF-ZZ, zinc finger binding with two zinc ions; ZF-CCCH, zinc finger C-x8-C-x5-C-x3-H type; PKR, protein kinase R; hnRNPA1, heterogeneous ribonucleoprotein A1; HuR, Hu-antigen R; U2AF35, U2 small nuclear RNA auxiliary factor 35; SAM68, Src-associated protein in mitosis of 68 kDa; TTP, tristetraprolin; IGF2BPs, Insulin-like growth factor 2 mRNA-binding proteins; CPEB4, cytoplasmic polyadenylation element binding protein 4; AA, amino acids.
ABERRANT EXPRESSION OF RNA BINDING PROTEINS IN CANCERS
Many reports have suggested that deregulated expression of RBP is detected in various human diseases including cancer.7–9 Altered expression of RBP causes wrong interactions with target RNAs to form incorrect RNP complex due to different affinity or concentration change. Such RNPs can affect the every post-transcriptional events in affected cells and modulate cell phenotype into pathological conditions. Neurodegenerative diseases are main representative pathological conditions caused by defected RBPs due to high expression of RBP in the brain.8,10 Loss of RBP or expression of toxic RNA is involved in the development of many neurological disorders, including fragile X syndrome, paraneoplastic neurologic syndrome, and spinal muscular atrophy.
Based on genome-wide analysis, many RBPs have been identified as key players in development and progression of cancers. They can dramatically change cell growth and proliferation.11–14 Aberrant expression of RBPs is also highly associated with survival rate of cancer patients.11,15 Unexpectedly, expression levels in fold change of validated RBPs are very low in cancers.16 Although expression levels of RBPs are not changed much in cancer cells, they can affect characteristics of cancer cells by targeting many target RNAs. Deregulation of RBPs in cancer is mainly induced by genomic alterations, epigenetic mechanism, noncoding RNA-mediated regulation, and PTMs.17,18
Although genomic alterations are hallmarks of human cancer, mutations and copy number variations have been found in only 15.2% of all RBPs.19 Due to alterations of RBPs, cancer cells can acquire differential splicing events, thus affecting various cancer hallmarks. Somatic mutations in spliceosome genes, such as serine arginine, SF3B1, U2AF1, and heterogeneous nuclear RNPs (hnRNPs) have been found in over 50% of myelodysplastic syndrome and acute myeloma leukemia patients.20,21 However, small change in one RBP can also affect global gene expression to modulate the growth of cancers. For example, germline mutation of DICER1 gene has resulted in defect in processing of precursor microRNA (miRNA) followed by abnormal expression of target RNAs.22,23 DICER1 defects are mainly found in patients of pleuropulmonary blastoma, cystic nephroma, cervical embryonal rhabdomyosarcoma, multinodular goiter, and Sertoli-Leydig cell tumors. Chromosomal translocation of the region containing RBP can also generate mutation. Fusion protein induced by rearrangement promotes transformation and progression of cancer through both RNA binding and activation domains of different proteins.24
Aberrant expression of RBPs can also be induced by post-transcriptional mechanism through noncoding RNAs or other RBPs. Mature miR-328 interacts and sequesters PCBP4 and hnRNPK to enhance the expression of oncogenic activators such MYC and PIM1 in chronic myelogenous leukemia.25 Tumor suppressor miRNAs, such as miR-34a, miR-101, miR-128, and miR-138 can suppress the expression of Musashi-1 (MSI-1) through translational repression in glioblastoma and medulloblastoma.26 Long 3′ untranslated region (UTR) containing several AU- and U-rich sequences of MSI-1 can be recognized by another RBP, Hu antigen R (HuR), thus protecting it from degradation to maintain high expression in glioblastoma.27 In cancers, the function of RBPs can be deregulated by PTMs, including phosphorylation, methylation, acetylation, and ubiquitination.28 By covalent incorporation of functional groups or proteins, the activity of RBPs is dramatically changed in many cancers. TGF-β/AKT2-dependent phosphorylation of hnRNPE1 can attenuate its suppressive activity on invasion and metastasis of breast cancer.29
ROLE OF RNA BINDING PROTEINS IN CANCER DEVELOPMENT
Because each RBP is associated with many target mRNAs and various biological processes, deregulation of RBP affects every step of cancer development, such as sustained cell proliferation, evasion of apoptosis, avoiding immune surveillance, inducing angiogenesis, and activating metastasis.17 Representative roles of RBPs in cancer development are summarized in Table 113,27–60 depending on hallmarks of cancer.
Table 1.
Role of aberrantly deregulated RBPs in human cancers
| Cancer Hallmark | RBP | Target mRNAs | Cancer types | Reference number |
|---|---|---|---|---|
| Sustained proliferation | ESRP1/2 | CCND1, CDH1, CDKN1A, MYC, PKM2, | Colon | 28, 29 |
| IGF2BP3 | MYC, CDK6 | Leukemia | 30 | |
| LIN28A/B | BMP4, HER2, HMGA1 | Breast, ovary, liver, colon | 31, 32 | |
| QKI | CDKN1B, FOS, miR-20a, NUMB | Brain, colon, lung | 33, 34 | |
| HuR | CCNA2, CCNB1 | Gastric, breast | 35, 36 | |
| SAM68 | CD44, CCND1 | Prostate, breast | 37, 38 | |
| Evading apoptosis | hnRNPH | MADD, RON | Brain | 39 |
| LARP4B | BCL2, BIK, BAX, XIAP | Brain | 40, 41 | |
| eIF4E | BCL2, BCL-XL | Lymphoma | 42 | |
| TIA1/TIAR | GADD45A, FAS, PDCD4 | Multiple | 43, 44 | |
| CELF1 | BAD, BAX, JunD | Oral | 45 | |
| Inducing angiogenesis | RBP2 | CDKI | Gastric | 46 |
| HuR | VEGF, HIF-1α, THBS1 | Breast, brain | 47, 48 | |
| LARP6 | VEGF | Breast | 49 | |
| eIF4E | VEGF, FGF-2, PDGF | Multiple | 50, 51 | |
| Activating EMT and metastasis | IGF2BPs | CD164, PDPN, CD44, IGF2, MMP9 | Breast, colon, leukemia | 52–54 |
| hnRNPE1 | DAB2, ILE1 | Breast, ovary | 27 | |
| UNR/CSDE1 | VIM, RAC1 | Melanoma | 13 | |
| RBM47 | CUL3, DKK1, MDM4, MXI1, SLK | Breast, lung | 55 | |
| Avoiding immune surveillance | IGF2BP3 | ULPB2, MICB | Leukemia | 56 |
| LIN28 | Let-7 | Leukemia | 57, 58 | |
| FXR1 | PRKCI, ECT2 | Lung | 59, 60 |
RBP, RNA binding protein; EMT, epithelial-mesenchymal transition; ESRP1/2, epithelial splicing regulatory protein 1 and 2; IGF2BP3, insulin-like growth factor 2 mRNA-binding protein 3; QKI, protein quaking; HuR, Hu antigen R; SAM68, Src-associated protein in mitosis of 68 kDa; LARP4B, La ribonucleoprotein domain family member 4B; eIF4E, eukaryotic initiation factor 4E; TIA1, T cell intracellular antigen 1; CELF1, CUGBP, elav-like family member 1; RBP2, retinol binding protein 2; UNR/CSDE1, upstream of N-Ras/Cold shock domain-containing protein E1; RBM47, RNA binding motif protein 47; FXR1, fragile X-related 1.
The main function of RBPs in cancer development is sustaining cell proliferation through suppressing or enhancing expression levels of negative or positive regulators, respectively. SAM68 regulates gene expression at post-transcriptional level through alternative splicing of pre-mRNAs. Overexpression of SAM68 has been identified in many human cancers. It modulates alternative splicing of oncogenic genes.11,39,40 For example, RAS/ERK-mediated phosphorylation of SAM68 promotes splicing of v5 exon of CD44 mRNA and stimulates cell proliferation.61 SAM68 also mediates alternative splicing of cyclin D1 into D1b isoform by recruiting spliceosomal component in prostate cancer.40 D1b isoform of cyclin D1 has higher oncogenic activity compared to D1a isoform. L1TD1, another RBP, can interact with LIN28 RNA and stabilize it to regulate the translation of OCT4.62 Upregulated OCT4 positively regulates self-renewal and proliferation of cancer cells. Insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3) interacts with 3′UTR of MYC and CDK6 mRNAs and stabilizes their transcripts to promote cell growth in B-acute lymphoblastic leukemia.32 On the other hand, QKI protein is suppressed in glioblastomas and colorectal cancer to enhance cancer progression.35,36 QKI inhibits cell growth by stabilizing p27 mRNA while degrading FOS mRNA.
Escaping apoptosis is one important mechanism to survive from extracellular stimuli and cytotoxic drugs in cancers. In this process, some RBPs play critical roles by regulating the expression of target mRNA at post-transcriptional level. Three-RRM containing HuR protein is overexpressed in many human cancers. It is closely correlated with prognosis of these patients.49,63,64 HuR promotes cell growth by stabilizing many anti-apoptotic genes, such as SIRT1, p21, MDM2, PTMA, BCL2, and MCL1.65–67 Eukaryotic initiation factor 4E (eIF4E) also regulates expression levels of proteins involved in cell survival, including BCL2 and BCL-XL.44 LARP member protein also regulates cell growth by promoting the expression of pro-survival genes such as BCL2, BIK, BAX, MDM2, and XIAP.42,43,68 CELF1 expression is increased in oral squamous cancer cells. It prevents programmed cell death and promotes cancer progression.47 This RBP interacts with 3′UTR of BAD, BAX, and JunD and destabilizes these pro-apoptotic genes. As a tumor suppressor gene, T cell intracellular antigen 1 (TIA1) protein is decreased in human cancers. It promotes apoptosis by inducing alternative splicing of FAS mRNA or increasing the stability of PDCD4 and GADD45A.45,46
Cancer tissues require generation of new blood vessels to supply nutrients and overcome hypoxia condition. In this process, histone demethylase retinoblastoma binding protein 2 (RBP2) can enhance the expression of VEGF through suppressing mRNA level of cyclin-dependent kinase inhibitor in gastric cancer.48 In triple negative breast cancer and brain tumor, HuR also modulates angiogenesis program by targeting multiple genes such as VEGF, hypoxia-inducible factor-1α, and thromospondin 1.49,50 For oncogenic activity of eIF4E, eIF4E controls the formation of blood vessel by targeting related genes, including VEGF, FGF-2, and platelet-derived growth factor.52,53,69
As a tumor promoter, RBP also enhances cancer progression by stimulating the invasion and metastasis. IGF2BPs are highly overexpressed in human cancers. It has been reported that they can stimulate the invasive activity of cancer cells through stabilization of CD44, CD164, MMP9, and podoplanin mRNAs.54–56 Their target mRNAs can facilitate the process of epithelial-mesenchymal transition (EMT) and the formation of invadopodia to enhance the infiltration and degradation of extracellular matrix. It is known that hnRNPE1 can bind to the 3′UTR of disabled-2 and interleukin-like EMT inducer mRNAs and mediate TGF-β-induced EMT and metastasis.29 UNR/CSDE1 can also promote the invasion and metastasis of melanoma by regulating VIM and RAC1 at post-transcriptional level.13 In contrast, RBM47 can suppress the metastasis of breast cancer by stabilizing transcripts of dickkopf WNT signaling pathway inhibitor 1.57
Recent report has suggested that evading immune surveillance is one important hallmark of cancer.70 IGF2BP3 induces immune escape by degrading ULPB2 and MICB, both of which are critical mediators in natural killer cells.58 Although there is no direct evidence for the involvement of LIN28 in immune surveillance of carcinogenesis, regulation of immune system by LIN28 suggests that this RBP is highly associated with cancer induction.59,60,71 Recent study also suggests that fragile X-related 1 (FXR1) protein is overexpressed in lung squamous cell carcinoma and associated with poor prognosis.72 Aberrant expression of this RBP is negatively correlated with cancer surveillance by forming complex with protein kinase C iota and epithelial cell transforming 2 mRNAs.73
THERAPEUTIC APPROACH TARGETING RNA BINDING PROTEINS
Because deregulated RBP can affect many characteristics of cancer, it might be a good therapeutic target for cancer treatment. However, regulatory mechanism of RBP is mainly originated from stabilization or degradation of target mRNA, making it difficult to modulate its activity. There are several trials for regulating RBP’s activity as cancer therapy using RNA interference-based approaches. Specific antisense oligonucleotide (ASO) against eIF4E can suppress tumor growth by repressing the translation of target mRNAs, such as VEGF, Survivin, c-Myc, Cyclin D1, and BCL-2.74–76 Most importantly, intravenous injection of ASO can significantly inhibit tumor growth without showing any side effect in a mouse model. These data support clinical trials of using eIF4E-specific ASO against human cancers. In another study, treatment with folate or transferrin receptor-targeted liposomal nanoparticle-based HuR siRNA can efficiently reduce cell viability, migration, and invasion of lung cancer.77,78 Conjugation of siRNAs against HuR with Cy3-labeled folic acid-coated DNA dendrimer nanocarrier has shown dramatic anti-oncogenic activity in ovarian cancer.79
After screening natural products against MSI-1, one group has identified (−)-gossypol from cottonseed as a potent anti-tumor agent.80 This small molecule can suppress cell growth of colon and prostate cancers by directly interacting with RNA-binding pocket of MSI-1. Because HuR interacts with adenine- and uridine-rich elements in 3′UTR of target mRNAs, it will be good strategy to disrupt this interaction for cancer therapy. Wu et al.81 has established a high-throughput screening system and identified several compounds as disruptors of HuR-mRNA interaction. These chemicals can block HuR-mRNA binding at nanomolar concentrations to inhibit the activity of HuR. Therefore, they can be used as cancer therapeutics for HuR-upregulated cancers. Interaction of LIN28 with Let-7 is also a therapeutic target for screening inhibitory chemicals using high-throughput system.82 With fluorescence resonance energy transfer-based assay, dissociation of RBP/RNA interaction can be used as a marker for screening novel drugs.
Although many researchers have attempted to develop therapeutic strategies against RBPs, success is very limited due to difficulties in direct targeting of RBPs or in specific selection of interacting mRNAs. Alternatively, we can use downstream effectors of RBPs as therapeutic target. For example, MYC is a target of many RBPs. It reversely regulates the stability of many RBPs, such as Hu-antigen R (HuR), heterogeneous ribonucleoprotein A1 (hnRNPA1), and hnRNPH to modulate cancer progression.83,84 Because MYC plays a pivotal role in many cancers, it will be a good approach to suppress the oncogenic activity of RBPs by targeting transcription or activity of MYC. As a first step, small molecule inhibitors, such as i-BET, JQ1, and MMS417 have been identified as transcriptional repressors of MYC. They can suppress cancer progression.85–87 MYC is also suppressed at post-transcriptional level by ASOs, thus decreasing its translation or splicing events.88,89 As alternative ways to block the activity of MYC, complex formation of MYC with its binding partners can be used as therapeutic target in cancer using mutant MYC or small molecule inhibitors.90–92
CONCLUSIONS
As mentioned above, RBPs play important roles in diverse biological processes. Many of them are found to be aberrantly dysregulated in various human cancers. Moreover, each RBP regulates a broad range of target mRNAs at the same time, thus leading to dramatic changes with important consequences for the development and progression of cancer. Nevertheless, expression changes of RBPs are not as strong as other cancer-related genes based on genome-wide screening and bioinformatics analysis. This might be due to technical limitations for detecting small-scale changes of differential gene expression. We can overcome these bottlenecks using systems that are more sensitive for isolating novel cancer-related RBPs, such as next generation RNA sequencing and protein mass spectrometry.4,93
As targets for cancer therapy, binding partners of RBPs are good candidates. Therefore, it is important to identify the direct target of each RBP in various cancer types. Recent progress in bioinformatics approaches and experimental techniques has improved our understanding of binding partners of RBPs involved in cancer progression. By introducing microarray and sequencing techniques, more RNAs can be identified as binding partners of proteins in diverse samples.94 After immunoprecipitation with specific antibody, bound RNAs can be isolated followed by PCR and sequencing. To identify transient or weak interaction of RBP/RNA, crosslinking methodologies can be combined with sequencing techniques, such as crosslinking and immunoprecipitation (CLIP), photoactivatable-ribonucleoside-enhanced CLIP (PAR-CLIP), and individual-nucleotide resolution CLIP (iCLIP).95–97 Another new technique called targets of RBPs identified by editing (TRIBE) has been developed to capture cell specific target of RBPs without antibody using small amounts of RNA.98
To support large-scale analysis of genome-wide data, analysis software should be developed to combine various datasets acquired from different cancers with multiple platforms. Data from genetically modified animal models or other physiological cellular models such as patient-derived tumor xenograft, tissue organoid, and microfluidic culture system can help us understand important functions of RBPs in human cancers. Then, identified RBPs or target mRNAs can be developed as therapeutic drugs for cancer therapy or are used as biomarkers for cancer progression or prognosis.
ACKNOWLEDGMENTS
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (2016-R1A2B2008007) to S Hong.
Footnotes
CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
REFERENCES
- 1.Müller-McNicoll M, Neugebauer KM. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat Rev Genet 2013;14:275–87. 10.1038/nrg3434 [DOI] [PubMed] [Google Scholar]
- 2.Lukong KE, Richard S. Sam68, the KH domain-containing super-STAR. Biochim Biophys Acta 2003;1653:73–86. [DOI] [PubMed] [Google Scholar]
- 3.Cook KB, Kazan H, Zuberi K, Morris Q, Hughes TR. RBPDB: a database of RNA-binding specificities. Nucleic Acids Res 2011;39: D301–8. 10.1093/nar/gkq1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet 2014;15:829–45. 10.1038/nrg3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jankowsky E, Harris ME. Specificity and nonspecificity in RNA-protein interactions. Nat Rev Mol Cell Biol 2015;16:533–44. 10.1038/nrm4032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol 2007;8:479–90. 10.1038/nrm2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darnell RB. RNA regulation in neurologic disease and cancer. Cancer Res Treat 2010;42:125–9. 10.4143/crt.2010.42.3.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet 2008;24:416–25. 10.1016/j.tig.2008.05.004 [DOI] [PubMed] [Google Scholar]
- 9.Neelamraju Y, Hashemikhabir S, Janga SC. The human RBPome: from genes and proteins to human disease. J Proteomics 2015;127:61–70. 10.1016/j.jprot.2015.04.031 [DOI] [PubMed] [Google Scholar]
- 10.Gabut M, Chaudhry S, Blencowe BJ. SnapShot: the splicing regulatory machinery. Cell 2008;133:192.e1. 10.1016/j.cell.2008.03.010 [DOI] [PubMed] [Google Scholar]
- 11.Busà R, Paronetto MP, Farini D, Pierantozzi E, Botti F, Angelini DF, et al. The RNA-binding protein Sam68 contributes to proliferation and survival of human prostate cancer cells. Oncogene 2007;26:4372–82. 10.1038/sj.onc.1210224 [DOI] [PubMed] [Google Scholar]
- 12.Degrauwe N, Suvà ML, Janiszewska M, Riggi N, Stamenkovic I. IMPs: an RNA-binding protein family that provides a link between stem cell maintenance in normal development and cancer. Genes Dev 2016;30:2459–74. 10.1101/gad.287540.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wurth L, Papasaikas P, Olmeda D, Bley N, Calvo GT, Guerrero S, et al. UNR/CSDE1 drives a post-transcriptional program to promote melanoma invasion and metastasis. Cancer Cell 2016;30: 694–707. 10.1016/j.ccell.2016.10.004 [DOI] [PubMed] [Google Scholar]
- 14.Dang H, Takai A, Forgues M, Pomyen Y, Mou H, Xue W, et al. Oncogenic activation of the RNA binding protein NELFE and MYC signaling in hepatocellular carcinoma. Cancer Cell 2017;32: 101–14.e8. 10.1016/j.ccell.2017.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jang HH, Lee HN, Kim SY, Hong S, Lee WS. Expression of RNA-binding motif protein 3 (RBM3) and cold-inducible RNA-binding protein (CIRP) is associated with improved clinical outcome in patients with colon cancer. Anticancer Res 2017;37:1779–85. 10.21873/anticanres.11511 [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Liu Q, Shyr Y. Dysregulated transcription across diverse cancer types reveals the importance of RNA-binding protein in carcinogenesis. BMC Genomics 2015;16 Suppl 7:S5. 10.1186/1471-2164-16-S7-S5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pereira B, Billaud M, Almeida R. RNA-binding proteins in cancer: old players and new actors. Trends Cancer 2017;3:506–28. 10.1016/j.trecan.2017.05.003 [DOI] [PubMed] [Google Scholar]
- 18.Castello A, Fischer B, Frese CK, Horos R, Alleaume AM, Foehr S, et al. Comprehensive identification of RNA-binding domains in human cells. Mol Cell 2016;63:696–710. 10.1016/j.molcel.2016.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sebestyén E, Singh B, Miñana B, Pagès A, Mateo F, Pujana MA, et al. Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks. Genome Res 2016;26:732–44. 10.1101/gr.199935.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chabot B, Shkreta L. Defective control of pre-messenger RNA splicing in human disease. J Cell Biol 2016;212:13–27. 10.1083/jcb.201510032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med 2011;365:1384–95. 10.1056/NEJMoa1103283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill DA, Ivanovich J, Priest JR, Gurnett CA, Dehner LP, Desruisseau D, et al. DICER1 mutations in familial pleuropulmonary blastoma. Science 2009;325:965. 10.1126/science.1174334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foulkes WD, Priest JR, Duchaine TF. DICER1: mutations, micro-RNAs and mechanisms. Nat Rev Cancer 2014;14:662–72. 10.1038/nrc3802 [DOI] [PubMed] [Google Scholar]
- 24.Selvanathan SP, Graham GT, Erkizan HV, Dirksen U, Natarajan TG, Dakic A, et al. Oncogenic fusion protein EWS-FLI1 is a network hub that regulates alternative splicing. Proc Natl Acad Sci U S A 2015;112:E1307–16. 10.1073/pnas.1500536112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell 2010;140:652–65. 10.1016/j.cell.2010.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vo DT, Qiao M, Smith AD, Burns SC, Brenner AJ, Penalva LO. The oncogenic RNA-binding protein Musashi1 is regulated by tumor suppressor miRNAs. RNA Biol 2011;8:817–28. 10.4161/rna.8.5.16041 [DOI] [PubMed] [Google Scholar]
- 27.Vo DT, Abdelmohsen K, Martindale JL, Qiao M, Tominaga K, Burton TL, et al. The oncogenic RNA-binding protein Musashi1 is regulated by HuR via mRNA translation and stability in glioblastoma cells. Mol Cancer Res 2012;10:143–55. 10.1158/1541-7786.MCR-11-0208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ceresa M, Mangado N, Andrews RJ, Gonzalez Ballester MA. Computational models for predicting outcomes of neuroprosthesis implantation: the case of cochlear implants. Mol Neurobiol 2015;52:934–41. 10.1007/s12035-015-9257-4 [DOI] [PubMed] [Google Scholar]
- 29.Chaudhury A, Hussey GS, Ray PS, Jin G, Fox PL, Howe PH. TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat Cell Biol 2010;12:286–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deloria AJ, Höflmayer D, Kienzl P, opatecka J, Sampl S, Klimpfinger M, et al. Epithelial splicing regulatory protein 1 and 2 paralogues correlate with splice signatures and favorable outcome in human colorectal cancer. Oncotarget 2016;7:73800–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell 2009;33:591–601. 10.1016/j.molcel.2009.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palanichamy JK, Tran TM, Howard JM, Contreras JR, Fernando TR, Sterne-Weiler T, et al. RNA-binding protein IGF2BP3 targeting of oncogenic transcripts promotes hematopoietic progenitor proliferation. J Clin Invest 2016;126:1495–511. 10.1172/JCI80046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molenaar JJ, Domingo-Fernández R, Ebus ME, Lindner S, Koster J, Drabek K, et al. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat Genet 2012;44:1199–206. 10.1038/ng.2436 [DOI] [PubMed] [Google Scholar]
- 34.Tu HC, Schwitalla S, Qian Z, LaPier GS, Yermalovich A, Ku YC, et al. LIN28 cooperates with WNT signaling to drive invasive intestinal and colorectal adenocarcinoma in mice and humans. Genes Dev 2015;29:1074–86. 10.1101/gad.256693.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen AJ, Paik JH, Zhang H, Shukla SA, Mortensen R, Hu J, et al. STAR RNA-binding protein Quaking suppresses cancer via stabilization of specific miRNA. Genes Dev 2012;26:1459–72. 10.1101/gad.189001.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji S, Ye G, Zhang J, Wang L, Wang T, Wang Z, et al. miR-574-5p negatively regulates Qki6/7 to impact β-catenin/Wnt signalling and the development of colorectal cancer. Gut 2013;62:716–26. 10.1136/gutjnl-2011-301083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang W, Caldwell MC, Lin S, Furneaux H, Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J 2000;19:2340–50. 10.1093/emboj/19.10.2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo X, Wu Y, Hartley RS. MicroRNA-125a represses cell growth by targeting HuR in breast cancer. RNA Biol 2009;6:575–83. 10.4161/rna.6.5.10079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richard S, Vogel G, Huot ME, Guo T, Muller WJ, Lukong KE. Sam68 haploinsufficiency delays onset of mammary tumorigenesis and metastasis. Oncogene 2008;27:548–56. 10.1038/sj.onc.1210652 [DOI] [PubMed] [Google Scholar]
- 40.Paronetto MP, Cappellari M, Busà R, Pedrotti S, Vitali R, Comstock C, et al. Alternative splicing of the cyclin D1 proto-oncogene is regulated by the RNA-binding protein Sam68. Cancer Res 2010; 70:229–39. 10.1158/0008-5472.CAN-09-2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lefave CV, Squatrito M, Vorlova S, Rocco GL, Brennan CW, Holland EC, et al. Splicing factor hnRNPH drives an oncogenic splicing switch in gliomas. EMBO J 2011;30:4084–97. 10.1038/emboj.2011.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trotta R, Vignudelli T, Candini O, Intine RV, Pecorari L, Guerzoni C, et al. BCR/ABL activates mdm2 mRNA translation via the La antigen. Cancer Cell 2003;3:145–60. 10.1016/S1535-6108(03)00020-5 [DOI] [PubMed] [Google Scholar]
- 43.Koso H, Yi H, Sheridan P, Miyano S, Ino Y, Todo T, et al. Identification of RNA-binding protein LARP4B as a tumor suppressor in glioma. Cancer Res 2016;76:2254–64. 10.1158/0008-5472.CAN-15-2308 [DOI] [PubMed] [Google Scholar]
- 44.Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, et al. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature 2004;428:332–7. 10.1038/nature02369 [DOI] [PubMed] [Google Scholar]
- 45.Izquierdo JM, Majós N, Bonnal S, Martínez C, Castelo R, Guigó R, et al. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol Cell 2005;19:475–84. 10.1016/j.molcel.2005.06.015 [DOI] [PubMed] [Google Scholar]
- 46.Wigington CP, Jung J, Rye EA, Belauret SL, Philpot AM, Feng Y, et al. Post-transcriptional regulation of programmed cell death 4 (PDCD4) mRNA by the RNA-binding proteins human antigen R (HuR) and T-cell intracellular antigen 1 (TIA1). J Biol Chem 2015;290:3468–87. 10.1074/jbc.M114.631937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Talwar S, Balasubramanian S, Sundaramurthy S, House R, Wilusz CJ, Kuppuswamy D, et al. Overexpression of RNA-binding protein CELF1 prevents apoptosis and destabilizes pro-apoptotic mRNAs in oral cancer cells. RNA Biol 2013;10:277–86. 10.4161/rna.23315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li L, Wang L, Song P, Geng X, Liang X, Zhou M, et al. Critical role of histone demethylase RBP2 in human gastric cancer angiogenesis. Mol Cancer 2014;13:81. 10.1186/1476-4598-13-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nabors LB, Gillespie GY, Harkins L, King PH. HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3′ untranslated regions of cytokine and angiogenic factor mRNAs. Cancer Res 2001;61:2154–61. [PubMed] [Google Scholar]
- 50.Gubin MM, Calaluce R, Davis JW, Magee JD, Strouse CS, Shaw DP, et al. Overexpression of the RNA binding protein HuR impairs tumor growth in triple negative breast cancer associated with deficient angiogenesis. Cell Cycle 2010;9:3337–46. 10.4161/cc.9.16.12711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stavraka C, Blagden S. The la-related proteins, a family with connections to cancer. Biomolecules 2015;5:2701–22. 10.3390/biom5042701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kevil C, Carter P, Hu B, DeBenedetti A. Translational enhancement of FGF-2 by eIF-4 factors, and alternate utilization of CUG and AUG codons for translation initiation. Oncogene 1995;11: 2339–48. [PubMed] [Google Scholar]
- 53.Mamane Y, Petroulakis E, Rong L, Yoshida K, Ler LW, Sonenberg N. eIF4E--from translation to transformation. Oncogene 2004;23: 3172–9. 10.1038/sj.onc.1207549 [DOI] [PubMed] [Google Scholar]
- 54.Hwang YS, Xianglan Z, Park KK, Chung WY. Functional in-vadopodia formation through stabilization of the PDPN transcript by IMP-3 and cancer-stromal crosstalk for PDPN expression. Carcinogenesis 2012;33:2135–46. 10.1093/carcin/bgs258 [DOI] [PubMed] [Google Scholar]
- 55.Samanta S, Sharma VM, Khan A, Mercurio AM. Regulation of IMP3 by EGFR signaling and repression by ERβ: implications for triple-negative breast cancer. Oncogene 2012;31:4689–97. 10.1038/onc.2011.620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vikesaa J, Hansen TV, Jønson L, Borup R, Wewer UM, Christiansen J, et al. RNA-binding IMPs promote cell adhesion and invadopodia formation. EMBO J 2006;25:1456–68. 10.1038/sj.emboj.7601039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vanharanta S, Marney CB, Shu W, Valiente M, Zou Y, Mele A, et al. Loss of the multifunctional RNA-binding protein RBM47 as a source of selectable metastatic traits in breast cancer. Elife 2014;3:e02734. 10.7554/eLife.02734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmiedel D, Tai J, Yamin R, Berhani O, Bauman Y, Mandelboim O. The RNA binding protein IMP3 facilitates tumor immune escape by downregulating the stress-induced ligands ULPB2 and MICB. Elife 2016;5:e13426. 10.7554/eLife.13426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang S, Baltimore D. RNA-binding protein Lin28 in cancer and immunity. Cancer Lett 2016;375:108–13. 10.1016/j.canlet.2016.02.050 [DOI] [PubMed] [Google Scholar]
- 60.Wang LD, Rao TN, Rowe RG, Nguyen PT, Sullivan JL, Pearson DS, et al. The role of Lin28b in myeloid and mast cell differentiation and mast cell malignancy. Leukemia 2015;29:1320–30. 10.1038/leu.2015.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matter N, Herrlich P, König H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature 2002;420:691–5. 10.1038/nature01153 [DOI] [PubMed] [Google Scholar]
- 62.Närvä E, Rahkonen N, Emani MR, Lund R, Pursiheimo JP, Nästi J, et al. RNA-binding protein L1TD1 interacts with LIN28 via RNA and is required for human embryonic stem cell self-renewal and cancer cell proliferation. Stem Cells 2012;30:452–60. 10.1002/stem.1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Durie D, Lewis SM, Liwak U, Kisilewicz M, Gorospe M, Holcik M. RNA-binding protein HuR mediates cytoprotection through stimulation of XIAP translation. Oncogene 2011;30:1460–9. 10.1038/onc.2010.527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Denkert C, Weichert W, Winzer KJ, Müller BM, Noske A, Niesporek S, et al. Expression of the ELAV-like protein HuR is associated with higher tumor grade and increased cyclooxygenase-2 expression in human breast carcinoma. Clin Cancer Res 2004;10:5580–6. 10.1158/1078-0432.CCR-04-0070 [DOI] [PubMed] [Google Scholar]
- 65.Abdelmohsen K, Lal A, Kim HH, Gorospe M. Posttranscriptional orchestration of an anti-apoptotic program by HuR. Cell Cycle 2007;6:1288–92. 10.4161/cc.6.11.4299 [DOI] [PubMed] [Google Scholar]
- 66.Lal A, Kawai T, Yang X, Mazan-Mamczarz K, Gorospe M. Antiapoptotic function of RNA-binding protein HuR effected through prothymosin alpha. EMBO J 2005;24:1852–62. 10.1038/sj.emboj.7600661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cho SJ, Zhang J, Chen X. RNPC1 modulates the RNA-binding activity of, and cooperates with, HuR to regulate p21 mRNA stability. Nucleic Acids Res 2010;38:2256–67. 10.1093/nar/gkp1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.He N, Jahchan NS, Hong E, Li Q, Bayfield MA, Maraia RJ, et al. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol Cell 2008;29:588–99. 10.1016/j.molcel.2008.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kevil CG, De Benedetti A, Payne DK, Coe LL, Laroux FS, Alexander JS. Translational regulation of vascular permeability factor by eukaryotic initiation factor 4E: implications for tumor angiogenesis. Int J Cancer 1996;65:785–90. [DOI] [PubMed] [Google Scholar]
- 70.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 71.Boldin MP, Baltimore D. MicroRNAs, new effectors and regulators of NF-κB. Immunol Rev 2012;246:205–20. 10.1111/j.1600-065X.2011.01089.x [DOI] [PubMed] [Google Scholar]
- 72.Qian J, Hassanein M, Hoeksema MD, Harris BK, Zou Y, Chen H, et al. The RNA binding protein FXR1 is a new driver in the 3q26–29 amplicon and predicts poor prognosis in human cancers. Proc Natl Acad Sci U S A 2015;112:3469–74. 10.1073/pnas.1421975112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qian J, Zou Y, Wang J, Zhang B, Massion PP. Global gene expression profiling reveals a suppressed immune response pathway associated with 3q amplification in squamous carcinoma of the lung. Genom Data 2015;5:272–4. 10.1016/j.gdata.2015.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Graff JR, Konicek BW, Vincent TM, Lynch RL, Monteith D, Weir SN, et al. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J Clin Invest 2007;117:2638–48. 10.1172/JCI32044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Graff JR, Konicek BW, Lynch RL, Dumstorf CA, Dowless MS, McNulty AM, et al. eIF4E activation is commonly elevated in advanced human prostate cancers and significantly related to reduced patient survival. Cancer Res 2009;69:3866–73. 10.1158/0008-5472.CAN-08-3472 [DOI] [PubMed] [Google Scholar]
- 76.Hsieh AC, Ruggero D. Targeting eukaryotic translation initiation factor 4E (eIF4E) in cancer. Clin Cancer Res 2010;16:4914–20. 10.1158/1078-0432.CCR-10-0433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Muralidharan R, Babu A, Amreddy N, Srivastava A, Chen A, Zhao YD, et al. Tumor-targeted nanoparticle delivery of HuR siRNA inhibits lung tumor growth in vitro and in vivo by disrupting the oncogenic activity of the RNA-binding protein HuR. Mol Cancer Ther 2017;16:1470–86. 10.1158/1535-7163.MCT-17-0134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Muralidharan R, Babu A, Amreddy N, Basalingappa K, Mehta M, Chen A, et al. Folate receptor-targeted nanoparticle delivery of HuR-RNAi suppresses lung cancer cell proliferation and migration. J Nanobiotechnol 2016;14:47. 10.1186/s12951-016-0201-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang YH, Peng W, Furuuchi N, Gerhart J, Rhodes K, Mukherjee N, et al. Delivery of therapeutics targeting the mRNA-binding protein HuR using 3DNA nanocarriers suppresses ovarian tumor growth. Cancer Res 2016;76:1549–59. 10.1158/0008-5472.CAN-15-2073 [DOI] [PubMed] [Google Scholar]
- 80.Lan L, Appelman C, Smith AR, Yu J, Larsen S, Marquez RT, et al. Natural product (−)-gossypol inhibits colon cancer cell growth by targeting RNA-binding protein Musashi-1. Mol Oncol 2015;9: 1406–20. 10.1016/j.molonc.2015.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu X, Lan L, Wilson DM, Marquez RT, Tsao WC, Gao P, et al. Identification and validation of novel small molecule disruptors of HuR-mRNA interaction. ACS Chem Biol 2015;10:1476–84. 10.1021/cb500851u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Roos M, Pradère U, Ngondo RP, Behera A, Allegrini S, Civenni G, et al. A small-molecule inhibitor of lin28. ACS Chem Biol 2016;11:2773–81. 10.1021/acschembio.6b00232 [DOI] [PubMed] [Google Scholar]
- 83.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 2010;463:364–8. 10.1038/nature08697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rounbehler RJ, Fallahi M, Yang C, Steeves MA, Li W, Doherty JR, et al. Tristetraprolin impairs myc-induced lymphoma and abolishes the malignant state. Cell 2012;150:563–74. 10.1016/j.cell.2012.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011;146:904–17. 10.1016/j.cell.2011.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A 2011;108: 16669–74. 10.1073/pnas.1108190108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 2011;478:524–8. 10.1038/nature10334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sekhon HS, London CA, Sekhon M, Iversen PL, Devi GR. c-MYC antisense phosphosphorodiamidate morpholino oligomer inhibits lung metastasis in a murine tumor model. Lung Cancer 2008;60:347–54. 10.1016/j.lungcan.2007.10.028 [DOI] [PubMed] [Google Scholar]
- 89.Iversen PL, Arora V, Acker AJ, Mason DH, Devi GR. Efficacy of antisense morpholino oligomer targeted to c-myc in prostate cancer xenograft murine model and a Phase I safety study in humans. Clin Cancer Res 2003;9:2510–9. [PubMed] [Google Scholar]
- 90.Savino M, Annibali D, Carucci N, Favuzzi E, Cole MD, Evan GI, et al. The action mechanism of the Myc inhibitor termed Omomyc may give clues on how to target Myc for cancer therapy. PLoS One 2011;6:e22284. 10.1371/journal.pone.0022284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Soucek L, Jucker R, Panacchia L, Ricordy R, Tatò F, Nasi S. Omomyc, a potential Myc dominant negative, enhances Myc-induced apoptosis. Cancer Res 2002;62:3507–10. [PubMed] [Google Scholar]
- 92.Berg T. Small-molecule modulators of c-Myc/Max and Max/Max interactions. Curr Top Microbiol Immunol 2011;348:139–49. [DOI] [PubMed] [Google Scholar]
- 93.Kwon SC, Yi H, Eichelbaum K, Föhr S, Fischer B, You KT, et al. The RNA-binding protein repertoire of embryonic stem cells. Nat Struct Mol Biol 2013;20:1122–30. 10.1038/nsmb.2638 [DOI] [PubMed] [Google Scholar]
- 94.Peritz T, Zeng F, Kannanayakal TJ, Kilk K, Eiríksdóttir E, Langel U, et al. Immunoprecipitation of mRNA-protein complexes. Nat Protoc 2006;1:577–80. 10.1038/nprot.2006.82 [DOI] [PubMed] [Google Scholar]
- 95.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010;141:129–41. 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 2010;17:909–15. 10.1038/nsmb.1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods 2005;37:376–86. 10.1016/j.ymeth.2005.07.018 [DOI] [PubMed] [Google Scholar]
- 98.McMahon AC, Rahman R, Jin H, Shen JL, Fieldsend A, Luo W, et al. TRIBE: hijacking an rna-editing enzyme to identify cell-specific targets of RNA-binding proteins. Cell 2016;165:742–53. 10.1016/j.cell.2016.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]