Abstract
Alzheimer's disease (AD) is the most common cause of adult dementia. Yet the complete set of molecular changes accompanying this inexorable, neurodegenerative disease remains elusive. Here we adopted an unbiased lipidomics and metabolomics approach to surveying frozen frontal cortex samples from clinically characterized AD patients (n=21) and age-matched controls (n=19), revealing marked molecular differences between them. Then, by means of metabolomic pathway analysis, we incorporated the novel molecular information into the known biochemical pathways and compared it with the results of a metabolomics meta-analysis of previously published AD research. We found six metabolic pathways of the central metabolism as well as glycerophospholipid metabolism predominantly altered in AD brains. Using targeted metabolomics approaches and MS imaging, we confirmed a marked dysregulation of the mitochondrial aspartate metabolism. The altered metabolic pathways were further integrated with clinical data, showing various degrees of correlation with parameters of dementia and AD pathology. Our study highlights specific, altered biochemical pathways in the brains of individuals with AD compared with those of control subjects, emphasizing a dysregulation of the mitochondrial aspartate metabolism and supporting future venues of investigation.
Keywords: Lipidomics, Alzheimer’s disease, Metabolomics, Lipids, Metabolic pathways
Introduction
Alzheimer’s disease (AD) is the most common cause of adult-onset dementia. Rising life expectancy is associated with increased prevalence and incidence of dementia. A recent report estimates that one in every three people born in the UK in 2015 will suffer from this debilitating mental illness during their lifetime1. The need to identify novel molecular targets for AD to use in diagnostics and therapeutics is acute. Although a growing number of clinical research laboratories use mass spectrometry (MS) applications to investigate proteomic changes in AD2–6, insufficient effort has been dedicated to studying alterations in levels of small molecules, metabolites, and lipids2, 7–9. To effectively address any therapeutic challenge in AD, however, requires detailed information about its underlining molecular pathology.
Previous studies by our group and others have examined the profound biochemical alterations in the AD brain2, 10–18. Most of those studies adopted targeted analytical approaches that, based on prior hypotheses, focused on a particular group of lipids or metabolites, belonging, for example, to a specific metabolic pathway. Changes in selected lipids or metabolites in AD brains, it is speculated, result from a cascade of cellular events involving abnormal beta-amyloid protein metabolism, tau phosphorylation, oxidative stress, mitochondrial or peroxisomal dysfunctions, inflammation, neurotransmitter changes, membrane lipid dysregulation, apoptosis, or changes in other proteins/chemicals19, 20. According to a targeted analytical approach, however, the specific lipids or metabolites that undergo analysis are selected according to the questions asked, which might hinder the discovery of unexpected metabolic dysregulations in AD brains.
The use of untargeted metabolomics and lipidomics approaches, on the other hand, can offer an unbiased view of the molecular changes occurring in AD brains21, 22. Untargeted approaches are hypothesis-generating and exploratory in nature, their purpose being to obtain profiles of as many metabolites and lipids as possible in biological samples. By comparing metabolite and lipid profiles, we can thus determine patterns of variations between control and AD subjects, and we can generate novel hypotheses or support old venues of investigations. According to an untargeted analytical approach, features of interest are selected on the basis of statistical analyses and are then identified and eventually quantified using more targeted approaches. Yet, most untargeted studies are focused on biomarker discovery and usually provide only a list of individual molecular species that are altered, while missing to highlight the functional connectivity among different biochemical pathways in AD brains.
Recent technological advances in the field of MS and bioinformatics allows us to combine untargeted and targeted analytical approaches in a single analysis to provide simultaneously a bird's-eye view of the interconnected metabolic changes, as well as a magnified view on selected biochemical pathways of interest, providing novel opportunities to investigate AD brains. Furthermore, while traditional approaches generally involve processing and extracting the samples before MS analysis, which may alter the chemical composition of the molecules under study, novel analytical approaches such as MS imaging allows to determine the spatial localization of lipids or metabolites in their natural environment. By scanning through tissues, MS imaging allows us to generate topographical maps of molecular distribution in the AD brain23, 24.
In this study, we used a combination of state-of-the-art untargeted and targeted lipidomics and metabolomics, as well as MS imaging, to profile postmortem brains from AD subjects and non-demented controls. Using innovative biostatistical and bioinformatical approaches we highlighted specific biochemical pathways altered in AD, which were then correlated with clinical records such as degrees of dementia and pathology. Finally, we compared our results with a meta-analysis of previously published research on AD, supporting venues of investigations in AD research.
Experimental Section
Chemicals
All chemicals and polar metabolites used as standards were purchased from Sigma-Aldrich® (Seelze, Germany) and were of analytical-grade purity or higher. Lipid standards were purchased from Avanti® Polar Lipids (Alabaster, Alabama USA) and Cayman Chemicals (Ann Arbor, MI, USA).
Tissue Procurement
From a cohort of 19 control subjects (12:7, males to females) and 21 subjects with AD (9:12, males to females) frozen, human samples were obtained from the Banner Sun Health Research Institute (Sun City, AZ, USA) (Table 1). The samples were matched for age: 83.5 ±6.4 years for the control subjects and 82.4 ± 6.7 years for subjects with AD (mean ± SD). The samples were also matched for postmortem interval: 3.6 ± 1.7 hours for control subjects and 3.6 ± 1.4 hours for subjects with AD (mean ± SD) (Table 1). AD cases met the criteria of the National Institute on Aging–Reagan Institute for intermediate or high likelihood of Alzheimer's disease. All subjects or, where appropriate, their caregivers provided written, informed consent for both the clinical examination as well as for brain donation to the Banner Sun Health Research Institute Brain and Body Donation Program. The protocols and informed consent are approved by the Banner Health Institutional Review Board.
Table 1.
Demographic and neuropathological features of the subjects used in this study.
| AD subjects | Control subjects | |
|---|---|---|
| Total number | 21 | 19 |
| Sex (male/female) | 9/12 | 12/7 |
| Age (years) | 82.4±6.7 | 83.5±6.4 |
| Post-mortem interval (hours) | 3.6±1.4; | 3.6±1.7 |
| Tangle score (Braak and Braak) | V (n=11); VI (n=10); | I (n=2); II (n=4); III (n=7); IV (n=6); |
| Plaque Density | Frequent (20); Moderate (1) | Zero (7); Sparse (2); Moderate (4); Frequent (6) |
| MMSE score | 13.4±7.9 | 28.0±1.7 |
| BMI | 24.7±3.8 | 25.0±3.9 |
Plus and minus values are means ± SD. BMI, body-mass index; MMSE, Mini-Mental Status Examination
Sample preparation
Frozen brain samples were rapidly weighed (20 mg) and homogenized in ice-cold methanol (1 vol) containing the following internal standards: d8 arachidonic acid, cholesterol-d7, C19:0-cholesteryl ester, trinonadecenoin, 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine, 1,2-dimyristoyl-sn-glycero-3-phosphocholine, 1-heptadecenoyl-2-hydroxy-sn-glycero-3-phosphocholine, phenylalanine d2, succinate d4, glucose 13C6, carnitine d9, glutamic acid d5, lysine d4, alanine d4, AMP 13C1015N5. Metabolites were extracted by adding chloroform and water (2:1, vol/vol) and centrifuged at 10,000 xg for 10 min at 4°C. The bottom phases (mostly lipids) were dried under nitrogen, reconstituted in isopropanol/acetonitrile/water (4:3:1, vol:vol:vol; 0.1 mL) and subjected to LC/MS analysis. The upper phases (polar metabolites) were dried under vacuum, reconstituted in water/acetonitrile (1:1, vol:vol; 0.2 mL) and subjected to LC/MS analysis. A small amount (10 μL) from each sample was pooled, for use as quality controls for LC/MS analyses.
Metabolomic analyses
Chromatographic separation was achieved using an ACQUITY UPLC system (Waters® Corporation, Milford, Massachusetts USA) and hydrophilic-interaction liquid chromatography (HILIC) using a 1.7 µm (2.1 × 150 mm) ACQUITY amide column (Waters)25–27. A Synapt G2 mass spectrometer (Waters) was coupled with the UPLC system and operated in data-independent mode (MSE) 28. Pooled samples were analyzed in high definition MSE mode (HDMSE)23, 29. In positive electrospray mode, the capillary and cone voltage were 1.5 kV and 30 V, respectively. The source temperature and desolvation temperature were 120 °C and 500°C, respectively, and the flow rate of desolvation gas was 800 L/hr. During MSE experiments, the collision energy in the trap cell was 4 eV (Function 1), and in the transfer cell, it ranged from 15 eV to 30 eV (Function 2).
Samples were analyzed three times using UPLC-HILIC-MSE, once in positive ionization mode and twice in negative ionization mode, using acidic and basic chromatographic conditions, respectively25, 30. In positive mode and in negative acidic conditions, mobile phase A was 100% acetonitrile, and B was 100% H2O, with both containing 0.1% formic acid. The following elution gradient was used: 0 min, 99% A; 6 min, 40% A; 8 min, 99% A; 10 min, 99% A. In negative-mode basic conditions, mobile-phase A contained acetonitrile/sodium bicarbonate, 10 mM (95:5), and mobile-phase B contained acetonitrile/sodium bicarbonate, 10 mM (5:95). The following elution gradient was used: 0 min, 99% A; 5 min, 42% A; 6 min, 70% A; 7 min, 99%; 10 min, 99 % A. In all conditions, the flow rate was 0.4 mL/min, the column temperature 45°C, and the injection volume 3.5 µL.
Lipidomic analyses
Lipidomic analyses of the brain samples were performed using an ionKey/MS system comprised of the ACQUITY UPLC M-Class, ionKey source, and an iKey CSH C18 130 Å (1.7µm particle size) 150 μm × 100 mm column (Waters) coupled to a Synapt G2-Si (Waters). Analyses were conducted in both positive and negative electrospray in MSE mode. Pooled samples were analyzed in high definition MSE mode (HDMSE)23, 24, 31. The capillary voltage was 2.8 kV and the source temperature 110 ºC. Injections were 0.5 μL, in the partial-loop mode, with a column temperature of 55 ºC and flow rate of 3 µL/min. Mobile phase A consisted of acetonitrile/water (60:40) with 10 mM ammonium formate + 0.1% formic acid. Mobile phase B consisted of isopropanol/acetonitrile (90:10) with 10 mM ammonium formate + 0.1% formic acid. The gradient was programmed as follows: 0.0–2.0 min, from 40% B to 43% B; 2.0-2.1 min, to 50% B; 2.1–12.0 min, to 99% B; 12.0–12.1 min, to 40% B; and 12.1-14.0 min, at 40% B.
Laser Ablation Electrospray Ionization MS (LAESI-MS) imaging
Frozen, human brain samples were serially sectioned in a cryostat, to allow alternate microscopic and MS-imaging analyses. Sections were placed on standard microscope slides and kept frozen at -15°C throughout the analysis using the Peltier cooling stage of laser ablation electrospray ionization (LAESI®). Sections were analyzed using the LAESI DP-1000 Direct Ionization System (Protea® Bioscience, Morgantown, West Virginia, USA) coupled with a Synapt G2-S mass spectrometer operated in HDMS mode. The electrospray solution for LAESI was methanol/water (50:50, v:v) with 0.1% acetic acid. LAESI parameters consisted of 10 laser pulses per pixel, at 5 Hz and 800 uJ of laser energy. Data were collected in both negative- and positive-ion mode using a mass range of m/z 50 to 1500 for MS scans as well as for ion-mobility-MS scans. Identifications were made according to accurate-mass and collision cross sections (CCS) values29. Selected drift-time regions were extracted using DriftScope™ software (Waters). Ion distribution maps were created for mass values of interest using ProteaPlot v2.0.3.8 (Protea Bioscience).
Data Processing and Analysis
Data processing and analysis was conducted using Progenesis QI Informatics (Nonlinear Dynamics®, Newcastle, UK)29. Each UPLC-MS run was imported as an ion-intensity map, including m/z and retention time. These ion maps were then aligned in the retention-time direction. From the aligned runs, an aggregate run representing the compounds in all samples was used for peak picking. This aggregate was then compared with all runs, to ensure that the same ions are detected in every run. Isotope and adduct deconvolution was applied, to reduce the number of features detected. Data were normalized according to total ion intensity. A combination of analysis of the variance (ANOVA) and multivariate statistics, including principal-component analysis (PCA) and orthogonal partial least-squares discriminant analysis (OPLS-DA), identified features most responsible for differences between sample groups. Before multivariate analysis data was scaled using pareto and log trasformed. PCA and OPLS-DA were performed by using SIMCA-P (Umetrics, Umea, Sweden), Metabolites were identified by database searches against their accurate masses using publicly available databases, including the LIPID MAPS32 database and the Human Metabolome database (HMDB)33 as well as by fragmentation patterns, retention times and CCSs, when available. Pathway analysis, which consisted of enrichment analysis and pathway topological analysis, were conducted using Metabolomics Pathway Analysis (MetPA) feature within MetaboAnalyst34. TargetLynx™ software (Waters) was used for targeted analysis of selected features using internal standards for each class as previously reported25, 30. Correlation analysis was performed by using MetaboAnalyst34. Pearson test (r) was used to measure the correlation.
Metabolite cluster enrichment analysis
Enrichment analysis of changed metabolite classes, subclasses and LIPID MAPS®35 subclasses was performed according to the methods previously used by Watschinger et al.,36 with some modifications. Briefly, a distance matrix between metabolite profiles was generated by hierarchical clustering of normalized profiles (Z-scores) and then used to separate the metabolites into six clusters, according to their similarities. For each cluster HMDB33 class, HMDB subclass and LIPID MAPS35 subclass enrichment analysis was performed separately. Depending on the level on which the enrichment analysis was performed, all annotated IDs from the respective databases were used. For each individual HMDB class, HMDB subclass and LIPID MAPS subclass, a hypergeometric test based on the counts of significantly altered compounds versus all possible identifications was performed. We selected an ANOVA p-value of 0.01 and a minimum number of 3 or more compounds per category as significance criteria. Every enrichment analysis was performed separately for the complete data set as well as for all six clusters identified on the basis of profile similarity (as described above).
Metabolomics Pathway Meta-Analysis
The PubMed® database was cross searched (in March 2015) for the term “Alzheimer” and each metabolite present in the HMDB in all fields (i.e., title, abstract, keywords). Information about the human metabolome was downloaded from the HMDB at the web address www.hmdb.ca/download. Each metabolite entry was saved as an extensible markup language (XML®) file called MetaboCard. Each MetaboCard entry contains more than 110 data fields with two thirds of the information devoted to chemical and physical data and the remaining third to enzymatic or biochemical data. Many data fields are linked to other databases (KEGG®, PubChem®, MetaCyc, ChEBI, PDB, UniProt®, and GENBANK)33, 37, 38. R software,39 running on scripts developed in-house, was used to gain access to the MetaboCard entries, to obtain information about each metabolite, its synonyms, and to query the PubMed database in all fields (i.e., title, abstract, keywords). Because in the literature the same metabolite can be present under different names, each query to PubMed was parsed as follows: “Alzheimer AND (synonym#1 OR synonym#2 OR synonym#3 OR synonym#4)”. The function xmlTreeParse and xmlValue of the R-package XML were used to analyze the output from PubMed and to count the numbers of papers for each query. A metabolite-set enrichment analysis (MSEA®) was performed using over-representation analysis (ORA) using the Web tool freely available at the Internet address http://www.msea.ca/MSEA/.
Results
Table 1 shows the demographic and neuropathological characteristics of the subjects with AD and the control subjects used in this study. Notably, in order to limit artifacts due to aging or tissue degradation, the groups were closely matched for age and postmortem interval, which in average was kept under 4 hours.
Unbiased metabolomic and lipidomic analyses
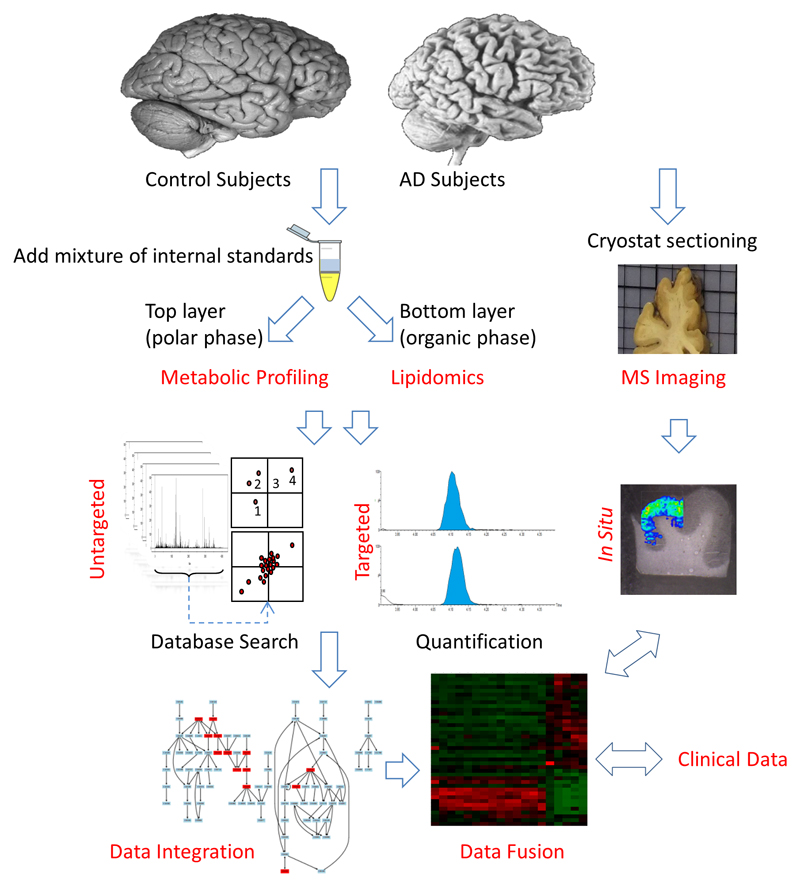
To extract metabolites and lipids from the same brain tissue samples, we used a biphasic liquid/liquid extraction method. The procedure permitted the extraction of selectively polar metabolites in the aqueous phase while lipids were isolated in the organic phase. We then applied, separately, our untargeted metabolomic and lipidomic workflows to these samples. Additional brain samples were also prepared for MS imaging analyses (Figure 1).
Figure 1. Experimental Workflow.
Frontal cortex samples from AD subjects (n = 21) and non demented controls (n = 19) were extracted using a biphasic system (CHCl3/CH3OH/H2O, 2:1:1; vol:vol:vol). Top layer was analyzed using a metabolomics approach for polar metabolites; bottom layer was analyzed using a lipidomics approach for lipid. Initial discovery data were further investigated using targeted metabolic-profiling approaches and MS imaging. The results were then fused and integrated with clinical parameters.
To determine whether the disease induced molecular differences in the AD brains, we first used multivariate statistical approaches as an initial step for data visualization. PCA and OPLS-DA showed a clear clustering of the control and AD brains for both lipidomic (Figure S1a, supporting information) and metabolomic (Figure S1b, supporting information) analyses. Integration of lipidomic and metabolomic information showed that 82% of metabolite alterations presented a joint variation with lipids, whereas 20% of the lipids evidence a unique variation (Figure S1c, supporting information). These observations indicate that AD brains sustained profound molecular changes that affected both lipids and other metabolic pathways in an interconnected fashion.
Enrichment analyses
To determine the chemical classes of lipids and metabolites altered in AD brains, we performed a hierarchical clustering analysis of all the potential molecular identifications (Figure 2a). Metabolites could be divided into six clusters, according to the distance between their individual profiles (Figure 2). Three of the clusters (Figure 2, clusters 1, 3, and 4) were characterized by a significant increase in metabolite levels in AD samples, compared with non-demented (ND) controls (Figure 2a). The remaining 3 clusters (Figure 2, clusters 2, 5, and 6) revealed the opposite effect: higher levels of metabolites in the controls (Figure 2a).
Figure 2. Metabolite cluster and enrichment analysis. of molecular species affected by AD.
a) Unsupervised hierarchical clustering analysis of all the potential molecular identifications according to their co-regulation in AD brains. The clustered profiles were divided into six different groups with distinctive behavior. Metabolite and lipid concentration changes are shown in the respective groups. Each box is calculated from all lipids belonging to the respective group, each with peak-area data from five biological replicates. Z scores were determined according to the mean and SD of all data for the respective metabolite. b) Enrichment analysis obtained by using the HMDB classification system. c) Lipid subclass enrichment analysis obtained by using the LIPID MAPS classification system. Only significantly enriched subclasses for AGMO modulation are shown (P value of enrichment <0.001). Circle size represents the number of lipid species matching the respective subclass. Enrichment P values are indicated by the intensity of red filling of the circles.
Although the apparent differences between the clusters with similar profile changes are small, our enrichment analysis revealed that the clusters have a distinct metabolic composition (Figure 2b and c). The HMDB classification system allowed differentiating metabolites according to their kingdom, super class, class, and subclass (Figure 2b). The LIPID MAPS classification system provided additional information. Located one layer below the HMDB subclass information, the LIPID MAPS subclass information, with an associated LIPID MAPS ID, was also included in the analysis for metabolites (Figure 2c). This analysis indicated that the lipid class of glycerophospholipids—in particular, phosphatydilcholines and phosphatidylethanolamines—appeared to be mostly altered in AD brains, compared with control brains, a finding cited in previous reports10, 20, 40–48. It is important to note that the same class of metabolites, such as glycerophospholipids, showed an increase in AD in clusters 1-3, whereas a decrease in clusters 4 and 5 (Figure 2b), which indicate a differential regulation of lipid species within the same subclass (Figure 2c). Changes in lipid composition may profoundly affect membrane structure, transmembrane signaling, and cell-to-cell communication in the brain, and they may thus underlie an important aspect of AD pathology. Further studies will aim to validate such lipid variations and their relation to AD.
Alterations in Metabolic Pathways
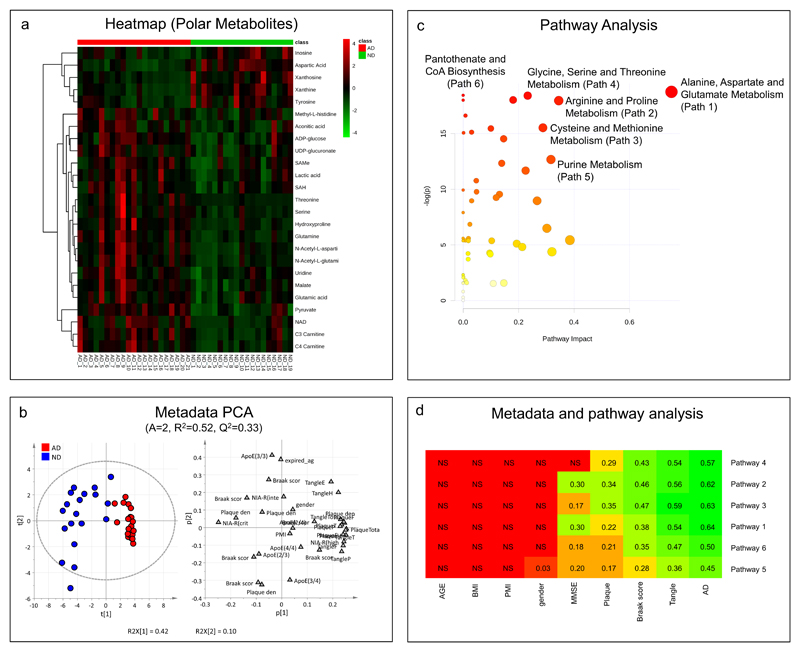
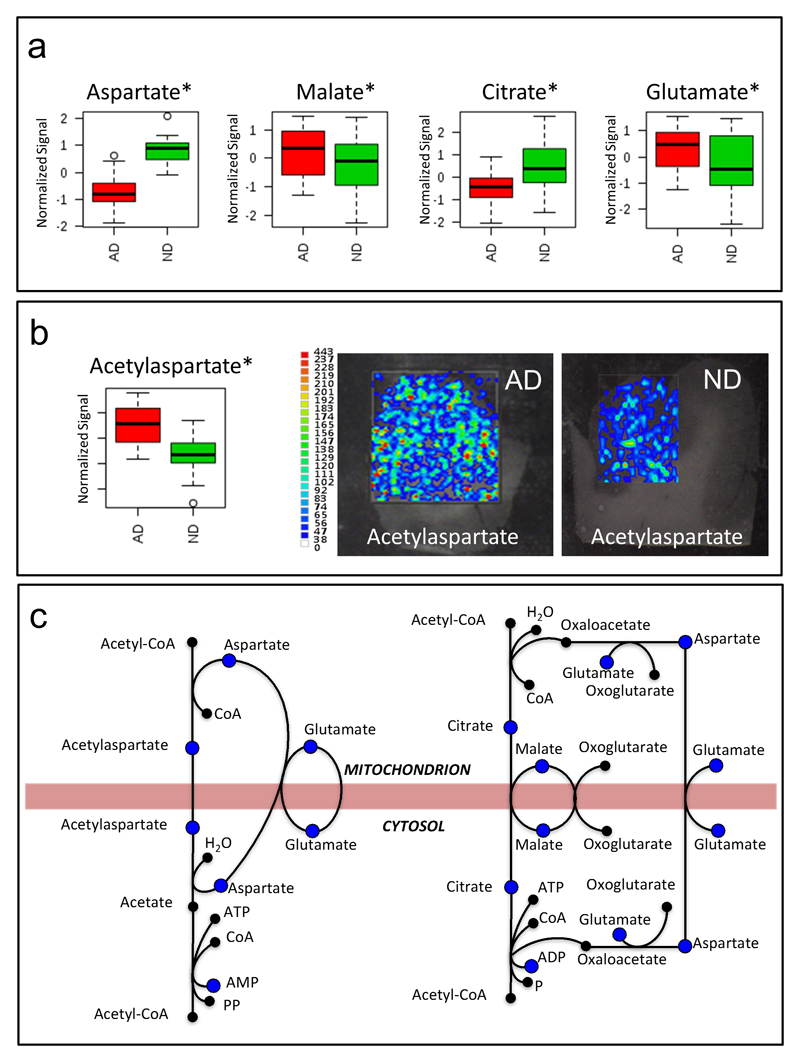
To determine the biochemical pathways mostly affected in AD brains, we identified and monitored with a targeted approach25 a subset of polar metabolite that was shown to discriminate between brains from AD patients and those from non-demented (ND) controls (Figure 3a and b). Metabolites were identified based on an in-house database24, 25, 29 and, where standards were not available, by comparing exact mass, and fragmentation spectra information with online databases. Metabolomics pathway analysis indicated six metabolic pathways affected during AD (Figure 3c, d and Table 2). In particular, the alanine, aspartate and glutamate metabolism proved one of the most affected pathways in the frontal cortices of subjects with AD, compared with those of non-demented control subjects (Figure 3c,d and Table 2). A significant dysregulation of the aspartate and glutamate metabolism was also confirmed by the MS-imaging experiment (Figure 4b). The alterations of metabolites such as aspartate, glutamate, citrate, and malate (Figure 4a) as well as the accumulation of N-acetyl aspartate (NAA) (Figure 4b), pyruvate, serine, and lactate (Figure S2, supporting information), in the frontal cortex samples of the AD subjects suggest that the transport mechanism between mitochondria and cytosol might be impaired in AD brains (Figure 4c).
Figure 3. Integration of metabolomics and clinical information.
a) Heat map of selected polar metabolites. b) Principal component analysis performed on clinical information. c) Pathway analysis performed on metabolomics results. d) Integration of pathway analysis results with clinical information. The heat map shows the relationships between clinical data and pathways; pathways and metadata are ordered according to hierarchical cluster analysis, to simplify the interpretation of the map; metadata closed each other have a similar profile with respect to the relationships with the pathways while pathways closed each other shows similar relationships with respect to the metadata; for “relationships” one means linear relationships between the concentrations of the metabolites included in the pathway and the metadata on the basis of PLS regression. The heat map was colored according to Q2CV 7-fold. “NS” means no significant PLS regression model (Q2CV 7-fold p-value> 0.05).
Table 2.
List of the most affected pathways and their metabolites in human frontal cortex of AD subjects compared to non-demented (ND) control subjects. Values are expressed as means ± SD of normalized MS intensity. HMDB, Human Metabolome Database.
| Metabolic Pathways | HMDB ID | Metabolite | AD | ND | T test p value |
|---|---|---|---|---|---|
|
Path 1: Alanine, aspartate and glutamate metabolism (p=7.19E-09) |
HMDB00812 | Acetylaspartic acid | 1.01±0.44 | 0.57±0.21 | 2.47E-04 |
| HMDB00191 | Aspartic acid | 1.50±0.78 | 3.37±1.33 | 2.67E-06 | |
| HMDB00161 | Alanine | 5.95±1.68 | 5.28±1.07 | 1.45E-01 | |
| HMDB00168 | Asparagine | 0.15±0.07 | 0.20±0.09 | 1.07E-01 | |
| HMDB00243 | Pyruvate | 0.09±0.03 | 0.05±0.03 | 6.11E-05 | |
| HMDB00148 | Glutamic acid | 24.43±8.58 | 18.31±7.06 | 1.92E-02 | |
| HMDB00641 | Glutamine | 43.09±24.72 | 16.92±10.70 | 1.28E-04 | |
| HMDB00254 | Succinic acid | 0.20±0.07 | 0.21±0.10 | 5.47E-01 | |
|
Path 2: Arginine and proline metabolism (p=1.58E-08) |
HMDB00191 | Aspartic acid | 1.50±0.78 | 3.37±1.33 | 2.67E-06 |
| HMDB00641 | Glutamine | 43.09±24.72 | 16.92±10.70 | 1.28E-04 | |
| HMDB00148 | Glutamic acid | 24.43±8.58 | 18.31±7.06 | 1.92E-02 | |
| HMDB00517 | Arginine | 12.92±6.00 | 10.34±3.81 | 1.17E-01 | |
| HMDB00162 | Proline | 0.17±0.08 | 0.13±0.06 | 6.69E-02 | |
| HMDB01185 | SAMe | 2.03±0.80 | 1.38±0.83 | 1.69E-02 | |
| HMDB00725 | Hydroxyproline | 3.21±2.02 | 1.42±0.85 | 9.91E-04 | |
| HMDB00243 | Pyruvate | 0.09±0.03 | 0.05±0.03 | 6.11E-05 | |
| HMDB01138 | Acetylglutamic acid | 17.77±8.12 | 9.81±3.66 | 3.52E-04 | |
|
Path 3: Cysteine and methionine metabolism (p=1.81E-07) |
HMDB00191 | Aspartic acid | 1.50±0.78 | 3.37±1.33 | 2.67E-06 |
| HMDB00187 | Serine | 2.67±1.61 | 1.54±0.66 | 7.31E-03 | |
| HMDB00192 | Cystine | 0.197±0.200 | 0.20±0.20 | 9.64E-01 | |
| HMDB00161 | Alanine | 5.95±1.68 | 5.28±1.07 | 1.45E-01 | |
| HMDB00696 | Methionine | 0.69±0.24 | 0.58±0.26 | 1.59E-01 | |
| HMDB00939 | SAH | 0.82±0.29 | 0.62±0.22 | 2.26E-02 | |
| HMDB01185 | SAMe | 2.03±0.80 | 1.38±0.83 | 1.69E-02 | |
| HMDB00243 | Pyruvate | 0.09±0.03 | 0.05±0.03 | 6.11E-05 | |
|
Path 4: Glycine, serine and threonine metabolism (p=1.01E-08) |
HMDB00191 | Aspartic acid | 1.50±0.78 | 3.37±1.33 | 2.67E-06 |
| HMDB00167 | Threonine | 1.52±1.70 | 0.31±0.21 | 3.86E-03 | |
| HMDB00187 | Serine | 2.67±1.61 | 1.54±0.66 | 7.31E-03 | |
| HMDB00097 | Choline | 0.59±0.26 | 0.41±0.24 | 2.73E-02 | |
| HMDB00929 | Tryptophan | 0.12±0.06 | 0.10±0.04 | 3.29E-01 | |
| HMDB00243 | Pyruvate | 0.09±0.03 | 0.05±0.03 | 6.11E-05 | |
|
Path 5: Purine metabolism (p=3.13E-06) |
HMDB00618 | Pentose 5-phosphate | 0.011±0.005 | 0.009±0.005 | 2.87E-01 |
| HMDB01178 | ADP-ribose | 0.09±0.07 | 0.06±0.06 | 1.52E-01 | |
| HMDB00641 | Glutamine | 43.09±24.72 | 16.92±10.70 | 1.28E-04 | |
| HMDB01341 | ADP | 0.016±0.013 | 0.010±0.010 | 1.45E-01 | |
| HMDB00045 | AMP | 0.32±0.26 | 0.19±0.22 | 8.95E-02 | |
| HMDB01397 | GMP | 0.07±0.04 | 0.06±0.05 | 2.97E-01 | |
| HMDB00133 | Guanosine | 2.35±1.84 | 2.35±2.04 | 9.99E-01 | |
| HMDB00175 | IMP | 0.02±0.01 | 0.01±0.01 | 2.70E-02 | |
| HMDB00289 | Uric acid | 0.80±0.43 | 1.10±0.65 | 8.90E-02 | |
| HMDB00292 | Xanthine | 1.09±0.47 | 2.29±1.08 | 4.10E-05 | |
| HMDB00299 | Xanthosine | 0.09±0.04 | 0.20±0.13 | 9.03E-04 | |
| HMDB00157 | Hypoxanthine | 25.91±9.08 | 22.68±9.34 | 2.74E-01 | |
| HMDB00195 | Inosine | 23.76±11.38 | 36.80±17.68 | 7.95E-03 | |
|
Path 6: Pantothenate and CoA biosynthesis (p=1.49E-08) |
HMDB00191 | Aspartic acid | 1.50±0.78 | 3.37±1.33 | 2.67E-06 |
| HMDB00243 | Pyruvate | 0.09±0.03 | 0.05±0.03 | 6.11E-05 | |
| HMDB00883 | Valine | 0.09±0.03 | 0.08±0.03 | 5.21E-01 | |
| HMDB00210 | Pantothenic acid | 2.19±2.18 | 1.82±0.61 | 4.77E-01 |
Figure 4. Mitochondrial shuttles.
a) Bar charts of aspartate, malate, citrate and glutamate obtained using normalized signals in AD (red) and non-demented (ND) control subjects (green). b) Bar chart of N-acetylaspartate (NAA) obtained using normalized signals in AD (red) and control subjects (green) and MS imaging of AD and control subjects brain sections. c) Mitochondrial shuttles and metabolites quantified in this experiment (blue dots). * = p < 0.05 (t-test). Targeted data used for bar charts were normalized by mean centering, scaled using unit variance and log transformed.
To determine the clinical significance of these metabolomic findings, we created scores indicative of the levels of metabolites belonging to the six metabolic pathways (Table 2) and correlated them with parameters of dementia and AD pathology (Figure 3b and d). For each pathway a PLS model was built considering the metabolite concentrations as X and the metadata as y. Permutation test on y and stability test based on Monte-Carlo sampling and PLS VIP-based were performed to check over-fitting. The set of metabolites detected by stability test was used to calculate Q2CV 7-fold (i.e., R2 calculated by 7-fold, full, cross-validation). Robust models for all pathways were found for AD, with the alanine, aspartate and glutamate pathway having the strongest correlation. No association with age, post-mortem interval (PMI), body mass index (BMI) and gender (Figure 3 b and d), with the only exception being a minor association between pathway 5 and gender (Figure 3 d). All the pathways were strongly associated with pathological hallmarks of AD, such tangles, while a smaller association was found with plaques. One of the most promising model for explaining cognitive status as measured by mini-mental state examination (MMSE) was found using metabolites of the alanine, aspartate and glutamate pathway.
An in-depth analysis of the alanine, aspartate and glutamate pathway revealed that the levels of aspartic acid and succinic acid positively correlated with the MMSE, whereas the levels of pyruvic acid, glutamine, and NAA negatively correlated with MMSE (Figure S3, supporting information). Levels of alanine, asparagine, and glutamic acid showed only a weak correlation with the clinical data (Figure S3, supporting information).
Literature comparison
To compare our results with the literature, we performed a metabolomics meta-analysis of the previously published AD literature in PubMed. We cross-searched the terms “Alzheimer” and the complete list of the metabolites and their synonyms, as reported in HMDB (41,514 entries)33. We then used the number of publications as an index of correlation between AD and the metabolite. A metabolite-set enrichment analysis38 helped identify patterns of metabolites most studied in AD research. It showed the aspartate and glutamate pathway as historically one of the most studied, agreeing with the outcome of our metabolomics investigation (Figure S4, supporting information).
Discussion
In this study we used a combination of innovative analytical approaches to conduct an unbiased metabolomics and lipidomics investigation of the biochemical pathways that are affected in AD brains.
As for all metabolomics and lipidomics analyses, study design, such as sample size, post-mortem interval and genotype (e.g., ApoE), as well as standard operating procedures, such as sample preparation, can affect results. Our study had multiple key features in terms of the analytical and analyses aspects that should be taken into consideration. First, our cohort size was of 21 AD subjects and 19 non-demented controls; groups were matched by age and post-mortem interval; frontal cortices were selected as region of the brain. Second, in order to limit artifacts due to sample degradation our criteria for subject selection required a post-mortem interval average of less than 4 hours, which is remarkably short for post-mortem brains. Third, to further limit artifacts due to enzymatic reactions and oxidation, brain samples were weighted while still frozen; next they were quenched with ice cold methanol; a liquid/liquid extraction allowed to separate hydrophilic from hydrophobic compounds in a single procedure; MS imaging was also used to support and confirm traditional analyses, while avoiding the process of extracting the samples. Finally, using data-independent acquisition mode, we obviated the need to analyze the sample in untargeted MS mode and then rerun the same sample in targeted MS/MS mode, but instead we automatically acquired the fragmentation data from each precursor ion49. The simultaneous generation of high-resolution, full scan, and fragmentation spectra served as a repository of biological information that was reused after statistical analyses to semi-quantify selected biochemical pathways. This approach allowed us to combine untargeted and targeted analysis, maximizing the identification of compounds and biochemical pathways in a single analytical run.
Our study uncovered significant alterations in key mitochondrial metabolites, which correlated with symptoms of dementia and AD pathology. Notably, as highlighted from our literature meta-analysis, previous research on AD largely focused on targeted metabolites of the alanine, aspartate and glutamate metabolism, often missing a bird's-eye view of the underlying biochemical alterations and their functional metabolic connectivity. Our metabolomic approach provided a comprehensive view of key metabolites of this pathway, pointing to a specific mitochondrial dysfunction in AD brains, which affects processes involved in the transport of metabolites between mitochondria and cytosol.
Acetyl-CoA can be transported into mitochondria by means of two main shuttles after conversion into either NAA or citrate (Figure 4c). In the first mitochondria shuttle, NAA is transported in the mitochondria and then converted to acetate by aspartoylacylase and, finally, converted back to acetyl-CoA by acetyl-CoA synthetase (Figure 4c). The observation that the levels of key metabolites in this process (i.e., NAA, aspartate and glutamate) are altered in AD and correlated with dementia and pathology, suggest that this transport mechanism is affected in AD (Figure 4). In the second mitochondria shuttle, citrate is transported in the mitochondria and then converted back into acetyl-CoA by the acetyl-CoA by ATP-citrate lyase. The observation that the levels of key metabolites in this process (i.e., citrate, malate, glutamate and aspartate) were significantly changed in AD, supports an impairment in the transport mechanism. The fact that key metabolites in both mitochondria shuttles are altered could reflect a more general mitochondrial dysfunction in AD brains.
The role of mitochondria as regulators of brain energy metabolism makes it crucial to neuronal cell survival or death. Growing evidence indicates that mitochondrial dysfunction is an early event during the progression of AD and one of the key intracellular mechanisms associated with the pathogenesis of this disease50–53. Recent findings demonstrated that α-ketoglutarate dehydrogenase complex regulates mitochondrial metabolism through post-translational modification of other enzymes in mitochondria54 and that AD patients have decreased activity of this enzyme55.
Our observations agree with a recent report using 1HNMR-based 56. Altered concentrations of brain NAA, as measured by magnetic resonance (MR), have been commonly associated with neurotoxicity and its levels have been used to assess in vivo neuronal loss and neurodegeneration in AD57–59. However, NAA concentration in the brain varies according to the brain region investigated. Indeed it has been shown that the concentration of NAA selectively decreases in specific brain areas such as hippocampus and the amygdala but not in the frontal cortex60.
We cannot exclude that the levels of NAA and other metabolites are consequence of dietary regimens or pharmacological treatments of the AD subjects. Previous evidence indicates, for example, that the levels of NAA increase during cholinergic treatment in AD61, 62 and could be reversed after other therapeutic interventions63, 64. Further investigations are needed to better understand the role of the mitochondrial aspartate metabolism in AD.
Conclusions
AD is a neurodegenerative disorder that is characterized by the loss of cognitive abilities and the appearance of pathological hallmarks in the brain, such as extra neuronal plaques and neurofibrillary tangles. In this study, we used an innovative analytical approach based on a combination of untargeted and targeted lipidomic and metabolomics, as well as MS imaging to investigate the metabolic alterations occurring in the AD brain. We explored the biochemical significance of the observed metabolic alterations according to known metabolic pathways. We then used novel bioinformatics and biostatistical approaches to integrate the novel acquired metabolic knowledge of AD brains with clinical records and literature meta-data. Most notably, we uncovered a significant dysregulation in the mitochondrial aspartate metabolism in the AD brain, which correlated with dementia and AD pathology. As such, our study provides a solid rationale for future pharmacological or metabolic interventions and other functional experiments aimed to better understand the role played by the mitochondrial aspartate metabolism in the etiology and progression of the cognitive impairment and pathological hallmarks associated with AD. Our study also suggests novel venues of investigation for biomarker discovery in peripheral tissues, such as blood, related to the metabolic changes that we observed in AD brains.
Supplementary Material
Acknowledgements
This work was partially funded by the Alzheimer’s Association (NIRG-11-203674 to G.A.), the Wellcome Trust (RG 093735/Z/10/Z to M.R.) and the ERC (Starting grant 260809 to M.R.). We are indebted to Drs. Thomas Beach and Geidy Serrano for their support and also to the Sun Health Research Institute Brain and Body Donation Program of Sun City, Arizona, for providing human biological materials. Finally, we thank Ms. Manuela Magnusdottir for her technical assistance. M.R. is a Wellcome Trust Research Career Development and Wellcome-Beit Prize fellow.
References
- 1.Lewis F. Estimation of future cases of dementia from those born in 2015. Office of Health Economics; 2015. [Google Scholar]
- 2.Fonteh AN, Harrington RJ, Huhmer AF, Biringer RG, Riggins JN, Harrington MG. Identification of disease markers in human cerebrospinal fluid using lipidomic and proteomic methods. Dis Markers. 2006;22(1–2):39–64. doi: 10.1155/2006/202938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castano EM, Roher AE, Esh CL, Kokjohn TA, Beach T. Comparative proteomics of cerebrospinal fluid in neuropathologically-confirmed Alzheimer's disease and non-demented elderly subjects. Neurol Res. 2006;28(2):155–63. doi: 10.1179/016164106X98035. [DOI] [PubMed] [Google Scholar]
- 4.Huang JT, Leweke FM, Oxley D, Wang L, Harris N, Koethe D, Gerth CW, Nolden BM, Gross S, Schreiber D, Reed B, et al. Disease Biomarkers in Cerebrospinal Fluid of Patients with First-Onset Psychosis. PLoS Med. 2006;3(11) doi: 10.1371/journal.pmed.0030428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J, Goodlett DR, Quinn JF, Peskind E, Kaye JA, Zhou Y, Pan C, Yi E, Eng J, Wang Q, Aebersold RH, et al. Quantitative proteomics of cerebrospinal fluid from patients with Alzheimer disease. J Alzheimers Dis. 2005;7(2):125–33. doi: 10.3233/jad-2005-7205. discussion 173-80. [DOI] [PubMed] [Google Scholar]
- 6.Montine TJ, Woltjer RL, Pan C, Montine KS, Zhang J. Liquid chromatography with tandem mass spectrometry-based proteomic discovery in aging and Alzheimer's disease. NeuroRx. 2006;3(3):336–43. doi: 10.1016/j.nurx.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ackermann BL, Hale JE, Duffin KL. The role of mass spectrometry in biomarker discovery and measurement. Curr Drug Metab. 2006;7(5):525–39. doi: 10.2174/138920006777697918. [DOI] [PubMed] [Google Scholar]
- 8.Chace DH. Mass spectrometry in the clinical laboratory. Chem Rev. 2001;101(2):445–77. doi: 10.1021/cr990077+. [DOI] [PubMed] [Google Scholar]
- 9.Mapstone M, Cheema AK, Fiandaca MS, Zhong X, Mhyre TR, MacArthur LH, Hall WJ, Fisher SG, Peterson DR, Haley JM, Nazar MD, et al. Plasma phospholipids identify antecedent memory impairment in older adults. Nat Med. 2014;20(4):415–8. doi: 10.1038/nm.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lukiw WJ, Cui JG, Marcheselli VL, Bodker M, Botkjaer A, Gotlinger K, Serhan CN, Bazan NG. A role for docosahexaenoic acid-derived neuroprotectin D1 in neural cell survival and Alzheimer disease. J Clin Invest. 2005;115(10):2774–83. doi: 10.1172/JCI25420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cutler RG, Kelly J, Storie K, Pedersen WA, Tammara A, Hatanpaa K, Troncoso JC, Mattson MP. Involvement of oxidative stress-induced abnormalities in ceramide and cholesterol metabolism in brain aging and Alzheimer's disease. Proc Natl Acad Sci U S A. 2004;101(7):2070–5. doi: 10.1073/pnas.0305799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puglielli L, Tanzi RE, Kovacs DM. Alzheimer's disease: the cholesterol connection. Nat Neurosci. 2003;6(4):345–51. doi: 10.1038/nn0403-345. [DOI] [PubMed] [Google Scholar]
- 13.Pappolla MA, Smith MA, Bryant-Thomas T, Bazan N, Petanceska S, Perry G, Thal LJ, Sano M, Refolo LM. Cholesterol, oxidative stress, and Alzheimer's disease: expanding the horizons of pathogenesis. Free Radic Biol Med. 2002;33(2):173–81. doi: 10.1016/s0891-5849(02)00841-9. [DOI] [PubMed] [Google Scholar]
- 14.Farooqui AA, Rapoport SI, Horrocks LA. Membrane phospholipid alterations in Alzheimer's disease: deficiency of ethanolamine plasmalogens. Neurochem Res. 1997;22(4):523–7. doi: 10.1023/a:1027380331807. [DOI] [PubMed] [Google Scholar]
- 15.Wells K, Farooqui AA, Liss L, Horrocks LA. Neural membrane phospholipids in Alzheimer disease. Neurochem Res. 1995;20(11):1329–33. doi: 10.1007/BF00992508. [DOI] [PubMed] [Google Scholar]
- 16.Farooqui AA, Liss L, Horrocks LA. Neurochemical aspects of Alzheimer's disease: involvement of membrane phospholipids. Metab Brain Dis. 1988;3(1):19–35. doi: 10.1007/BF01001351. [DOI] [PubMed] [Google Scholar]
- 17.Astarita G, Jung KM, Vasilevko V, Dipatrizio NV, Martin SK, Cribbs DH, Head E, Cotman CW, Piomelli D. Elevated stearoyl-CoA desaturase in brains of patients with Alzheimer's disease. PLoS One. 2011;6(10):e24777. doi: 10.1371/journal.pone.0024777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Astarita G, Jung KM, Berchtold NC, Nguyen VQ, Gillen DL, Head E, Cotman CW, Piomelli D. Deficient liver biosynthesis of docosahexaenoic acid correlates with cognitive impairment in Alzheimer's disease. PLoS One. 2010;5(9):e12538. doi: 10.1371/journal.pone.0012538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pettegrew JW, Panchalingam K, Hamilton RL, McClure RJ. Brain membrane phospholipid alterations in Alzheimer's disease. Neurochem Res. 2001;26(7):771–82. doi: 10.1023/a:1011603916962. [DOI] [PubMed] [Google Scholar]
- 20.Prasad MR, Lovell MA, Yatin M, Dhillon H, Markesbery WR. Regional membrane phospholipid alterations in Alzheimer's disease. Neurochem Res. 1998;23(1):81–8. doi: 10.1023/a:1022457605436. [DOI] [PubMed] [Google Scholar]
- 21.Inoue K, Tsutsui H, Akatsu H, Hashizume Y, Matsukawa N, Yamamoto T, Toyo'oka T. Metabolic profiling of Alzheimer's disease brains. Sci Rep. 2013;3:2364. doi: 10.1038/srep02364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graham SF, Chevallier OP, Roberts D, Holscher C, Elliott CT, Green BD. Investigation of the human brain metabolome to identify potential markers for early diagnosis and therapeutic targets of Alzheimer's disease. Anal Chem. 2013;85(3):1803–11. doi: 10.1021/ac303163f. [DOI] [PubMed] [Google Scholar]
- 23.Paglia G, Kliman M, Claude E, Geromanos S, Astarita G. Applications of ion-mobility mass spectrometry for lipid analysis. Anal Bioanal Chem. 2015;407(17):4995–5007. doi: 10.1007/s00216-015-8664-8. [DOI] [PubMed] [Google Scholar]
- 24.Paglia G, Angel P, Williams JP, Richardson K, Olivos HJ, Thompson JW, Menikarachchi L, Lai S, Walsh C, Moseley A, Plumb RS, et al. Ion-Mobility-Derived Collision Cross Section as an Additional Measure for Lipid Fingerprinting and Identification. Anal Chem. 2015;87(2):1137–44. doi: 10.1021/ac503715v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paglia G, Sigurjonsson OE, Rolfsson O, Valgeirsdottir S, Hansen MB, Brynjolfsson S, Gudmundsson S, Palsson BO. Comprehensive metabolomic study of platelets reveals the expression of discrete metabolic phenotypes during storage. Transfusion. 2014;54(11):2911–23. doi: 10.1111/trf.12710. [DOI] [PubMed] [Google Scholar]
- 26.Paglia G, Magnusdottir M, Thorlacius S, Sigurjonsson OE, Guethmundsson S, Palsson BO, Thiele I. Intracellular metabolite profiling of platelets: evaluation of extraction processes and chromatographic strategies. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;898:111–20. doi: 10.1016/j.jchromb.2012.04.026. [DOI] [PubMed] [Google Scholar]
- 27.Paglia G, Hrafnsdottir S, Magnusdottir M, Fleming RM, Thorlacius S, Palsson BO, Thiele I. Monitoring metabolites consumption and secretion in cultured cells using ultra-performance liquid chromatography quadrupole-time of flight mass spectrometry (UPLC-Q-ToF-MS) Anal Bioanal Chem. 2012;402(3):1183–98. doi: 10.1007/s00216-011-5556-4. [DOI] [PubMed] [Google Scholar]
- 28.Fu W, Magnusdottir M, Brynjolfson S, Palsson BO, Paglia G. UPLC-UV-MS(E) analysis for quantification and identification of major carotenoid and chlorophyll species in algae. Anal Bioanal Chem. 2012;404(10):3145–54. doi: 10.1007/s00216-012-6434-4. [DOI] [PubMed] [Google Scholar]
- 29.Paglia G, Williams JP, Menikarachchi L, Thompson JW, Tyldesley-Worster R, Halldorsson S, Rolfsson O, Moseley A, Grant D, Langridge J, Palsson BO, et al. Ion mobility derived collision cross sections to support metabolomics applications. Anal Chem. 2014;86(8):3985–93. doi: 10.1021/ac500405x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paglia G, Sigurjonsson OE, Rolfsson O, Hansen MB, Brynjolfsson S, Gudmundsson S, Palsson BO. Metabolomic analysis of platelets during storage: a comparison between apheresis- and buffy coat-derived platelet concentrates. Transfusion. 2015;55(2):301–13. doi: 10.1111/trf.12834. [DOI] [PubMed] [Google Scholar]
- 31.Pacini T, Fu W, Gudmundsson S, Chiaravalle AE, Brynjolfson S, Palsson BO, Astarita G, Paglia G. Multidimensional analytical approach based on UHPLC-UV-ion mobility-MS for the screening of natural pigments. Anal Chem. 2015;87(5):2593–2599. doi: 10.1021/ac504707n. [DOI] [PubMed] [Google Scholar]
- 32.Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CR, Shimizu T, Spener F, van Meer G, Wakelam MJ, Dennis EA. Update of the LIPID MAPS comprehensive classification system for lipids. Journal of lipid research. 2009;50(Supplement):S9–S14. doi: 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, et al. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41(Database issue):D801–7. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0-making metabolomics more meaningful. Nucleic Acids Res. 2015;43(W1):W251–7. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CR, Shimizu T, Spener F, van Meer G, Wakelam MJ, Dennis EA. Update of the LIPID MAPS comprehensive classification system for lipids. J Lipid Res. 2009;50(Suppl):S9–14. doi: 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watschinger K, Keller MA, McNeill E, Alam MT, Lai S, Sailer S, Rauch V, Patel J, Hermetter A, Golderer G, Geley S, et al. Tetrahydrobiopterin and alkylglycerol monooxygenase substantially alter the murine macrophage lipidome. Proc Natl Acad Sci U S A. 2015;112(8):2431–6. doi: 10.1073/pnas.1414887112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liesenfeld DB, Habermann N, Owen RW, Scalbert A, Ulrich CM. Review of mass spectrometry-based metabolomics in cancer research. Cancer Epidemiol Biomarkers Prev. 2013;22(12):2182–201. doi: 10.1158/1055-9965.EPI-13-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia J, Wishart DS. MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010;38(Web Server issue):W71–7. doi: 10.1093/nar/gkq329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ihaka R. R: Past and future history. Dimension Reduction, Computational Complexity and Information. 1998;30:392–396. [Google Scholar]
- 40.Brooksbank BW, Martinez M. Lipid abnormalities in the brain in adult Down's syndrome and Alzheimer's disease. Mol Chem Neuropathol. 1989;11(3):157–85. doi: 10.1007/BF03160049. [DOI] [PubMed] [Google Scholar]
- 41.Soderberg M, Edlund C, Kristensson K, Dallner G. Fatty acid composition of brain phospholipids in aging and in Alzheimer's disease. Lipids. 1991;26(6):421–5. doi: 10.1007/BF02536067. [DOI] [PubMed] [Google Scholar]
- 42.Guan Z, Wang Y, Cairns NJ, Lantos PL, Dallner G, Sindelar PJ. Decrease and structural modifications of phosphatidylethanolamine plasmalogen in the brain with Alzheimer disease. J Neuropathol Exp Neurol. 1999;58(7):740–7. doi: 10.1097/00005072-199907000-00008. [DOI] [PubMed] [Google Scholar]
- 43.Skinner ER, Watt C, Besson JA, Best PV. Differences in the fatty acid composition of the grey and white matter of different regions of the brains of patients with Alzheimer's disease and control subjects. Brain. 1993;116(Pt 3):717–25. doi: 10.1093/brain/116.3.717. [DOI] [PubMed] [Google Scholar]
- 44.Corrigan FM, Horrobin DF, Skinner ER, Besson JA, Cooper MB. Abnormal content of n-6 and n-3 long-chain unsaturated fatty acids in the phosphoglycerides and cholesterol esters of parahippocampal cortex from Alzheimer's disease patients and its relationship to acetyl CoA content. Int J Biochem Cell Biol. 1998;30(2):197–207. doi: 10.1016/s1357-2725(97)00125-8. [DOI] [PubMed] [Google Scholar]
- 45.Fraser T, Tayler H, Love S. Fatty acid composition of frontal, temporal and parietal neocortex in the normal human brain and in Alzheimer's disease. Neurochem Res. 2010;35(3):503–13. doi: 10.1007/s11064-009-0087-5. [DOI] [PubMed] [Google Scholar]
- 46.Cunnane SC, Plourde M, Pifferi F, Begin M, Feart C, Barberger-Gateau P. Fish, docosahexaenoic acid and Alzheimer's disease. Prog Lipid Res. 2009;48(5):239–56. doi: 10.1016/j.plipres.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 47.Ginsberg L, Rafique S, Xuereb JH, Rapoport SI, Gershfeld NL. Disease and anatomic specificity of ethanolamine plasmalogen deficiency in Alzheimer's disease brain. Brain Res. 1995;698(1–2):223–6. doi: 10.1016/0006-8993(95)00931-f. [DOI] [PubMed] [Google Scholar]
- 48.Han X, Holtzman DM, McKeel DW., Jr Plasmalogen deficiency in early Alzheimer's disease subjects and in animal models: molecular characterization using electrospray ionization mass spectrometry. J Neurochem. 2001;77(4):1168–80. doi: 10.1046/j.1471-4159.2001.00332.x. [DOI] [PubMed] [Google Scholar]
- 49.Plumb RS, Johnson KA, Rainville P, Smith BW, Wilson ID, Castro-Perez JM, Nicholson JK. UPLC/MS(E); a new approach for generating molecular fragment information for biomarker structure elucidation. Rapid Commun Mass Spectrom. 2006;20(13):1989–94. doi: 10.1002/rcm.2550. [DOI] [PubMed] [Google Scholar]
- 50.Reddy PH, Tripathi R, Troung Q, Tirumala K, Reddy TP, Anekonda V, Shirendeb UP, Calkins MJ, Reddy AP, Mao P, Manczak M. Abnormal mitochondrial dynamics and synaptic degeneration as early events in Alzheimer's disease: implications to mitochondria-targeted antioxidant therapeutics. Biochim Biophys Acta. 2012;1822(5):639–49. doi: 10.1016/j.bbadis.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao W, Wang J, Varghese M, Ho L, Mazzola P, Haroutunian V, Katsel PL, Gibson GE, Levine S, Dubner L, Pasinetti GM. Impaired mitochondrial energy metabolism as a novel risk factor for selective onset and progression of dementia in oldest-old subjects. Neuropsychiatr Dis Treat. 2015;11:565–74. doi: 10.2147/NDT.S74898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swerdlow RH, Burns JM, Khan SM. The Alzheimer's disease mitochondrial cascade hypothesis. J Alzheimers Dis. 2010;20(Suppl 2):S265–79. doi: 10.3233/JAD-2010-100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Navarro E, Romero SD, Yaksh TL. Release of prostaglandin E2 from brain of cat: II. In vivo studies on the effects of adrenergic, cholinergic and dopaminergic agonists and antagonists. Neuropharmacology. 1988;27(10):1067–72. doi: 10.1016/0028-3908(88)90069-x. [DOI] [PubMed] [Google Scholar]
- 54.Gibson GE, Xu H, Chen HL, Chen W, Denton TT, Zhang S. Alpha-ketoglutarate dehydrogenase complex-dependent succinylation of proteins in neurons and neuronal cell lines. J Neurochem. 2015;134(1):86–96. doi: 10.1111/jnc.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gibson GE, Blass JP, Beal MF, Bunik V. The alpha-ketoglutarate-dehydrogenase complex: a mediator between mitochondria and oxidative stress in neurodegeneration. Mol Neurobiol. 2005;31(1–3):43–63. doi: 10.1385/MN:31:1-3:043. [DOI] [PubMed] [Google Scholar]
- 56.Graham SF, H C, Green BD. Metabolic signatures of human Alzheimer’s disease (AD): 1H NMR analysis of the polar metabolome of post-mortem brain tissue. Metabolomics. 2014;10(4):744–753. [Google Scholar]
- 57.Dautry C, Vaufrey F, Brouillet E, Bizat N, Henry PG, Conde F, Bloch G, Hantraye P. Early N-acetylaspartate depletion is a marker of neuronal dysfunction in rats and primates chronically treated with the mitochondrial toxin 3-nitropropionic acid. J Cereb Blood Flow Metab. 2000;20(5):789–99. doi: 10.1097/00004647-200005000-00005. [DOI] [PubMed] [Google Scholar]
- 58.Ebisu T, Rooney WD, Graham SH, Weiner MW, Maudsley AA. N-acetylaspartate as an in vivo marker of neuronal viability in kainate-induced status epilepticus: 1H magnetic resonance spectroscopic imaging. J Cereb Blood Flow Metab. 1994;14(3):373–82. doi: 10.1038/jcbfm.1994.48. [DOI] [PubMed] [Google Scholar]
- 59.Glodzik L, Sollberger M, Gass A, Gokhale A, Rusinek H, Babb JS, Hirsch JG, Amann M, Monsch AU, Gonen O. Global N-acetylaspartate in normal subjects, mild cognitive impairment and Alzheimer's disease patients. J Alzheimers Dis. 2015;43(3):939–47. doi: 10.3233/JAD-140609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jaarsma D, Veenma-van der Duin L, Korf J. N-acetylaspartate and N-acetylaspartylglutamate levels in Alzheimer's disease post-mortem brain tissue. J Neurol Sci. 1994;127(2):230–3. doi: 10.1016/0022-510x(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 61.Hampel H, Frank R, Broich K, Teipel SJ, Katz RG, Hardy J, Herholz K, Bokde AL, Jessen F, Hoessler YC, Sanhai WR, et al. Biomarkers for Alzheimer's disease: academic, industry and regulatory perspectives. Nat Rev Drug Discov. 2010;9(7):560–74. doi: 10.1038/nrd3115. [DOI] [PubMed] [Google Scholar]
- 62.Jessen F, Traeber F, Freymann K, Maier W, Schild HH, Block W. Treatment monitoring and response prediction with proton MR spectroscopy in AD. Neurology. 2006;67(3):528–30. doi: 10.1212/01.wnl.0000228218.68451.31. [DOI] [PubMed] [Google Scholar]
- 63.Modrego PJ, Pina MA, Fayed N, Diaz M. Changes in metabolite ratios after treatment with rivastigmine in Alzheimer's disease: a nonrandomised controlled trial with magnetic resonance spectroscopy. CNS Drugs. 2006;20(10):867–77. doi: 10.2165/00023210-200620100-00006. [DOI] [PubMed] [Google Scholar]
- 64.Kalra V, Mittal R. Duration of antiepileptic drug (AED) therapy. Indian J Pediatr. 1998;65(5):772–5. doi: 10.1007/BF02731068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.