Abstract
Metastatic melanoma is hallmarked by elevated glycolytic flux and alterations in cholesterol homeostasis. The contribution of cholesterol transporting receptors for the maintenance of a migratory and invasive phenotype is not well defined. Here, the scavenger receptor class B type I (SCARB1/SR-BI), a high-density lipoprotein (HDL) receptor, was identified as an estimator of melanoma progression in patients. We further aimed to identify the SR-BI controlled gene expression signature and its related cellular phenotypes. Based on whole transcriptome analysis it was found that SR-BI knockdown, but not functional inhibition of its cholesterol transporting capacity, perturbed the metastasis-associated epithelial-to-mesenchymal transition (EMT) phenotype. Furthermore, SR-BI knockdown was accompanied by decreased migration and invasion of melanoma cells and reduced xenograft tumor growth. STAT5 is an important mediator of the EMT process and loss of SR-BI resulted in decreased glycosylation, reduced DNA binding and target gene expression of STAT5. When human metastatic melanoma clinical specimens were analyzed for the abundance of SR-BI and STAT5 protein, a positive correlation was found. Finally, a novel SR-BI regulated gene profile was determined, which discriminates metastatic from non-metastatic melanoma specimens indicating that SR-BI drives gene expression contributing to growth at metastatic sites. Overall, these results demonstrate that SR-BI is a highly expressed receptor in human metastatic melanoma and is crucial for the maintenance of the metastatic phenotype.
Implications
High SR-BI expression in melanoma is linked with increased cellular glycosylation and hence is essential for a metastasis specific expression signature.
Introduction
Melanoma is an aggressive and fatal form of skin cancer. In the last decades, the worldwide prevalence of melanoma has been rising significantly (1). Patients diagnosed with tumors of early stages can be cured by surgical resection. However, patients with melanoma of advanced stages still show high mortality rates. Recently developed treatments were found to increase therapy response rates and overall survival was also increased. However, most advanced melanoma patients still succumb to the disease evidenced by extremely low cure rates (2,3). Therefore, it is of primary importance to elucidate master regulators, which enable melanoma cells to metastasize and to provide clues for curative treatment.
A key feature of cancer cells especially during metastasis is the ability to perform epithelial to mesenchymal transition (EMT), in melanoma also referred to as phenotype switching (4). The dynamic change in melanoma from a proliferative state to an invasive, EMT positive state has been shown in several studies (5–7). Intriguingly, EMT has recently been linked to metabolic reprogramming evidenced by elevated aerobic glycolysis in different cancer models (8–12). This process, termed Warburg effect, is characterized by high rates of glucose uptake which is used for increased energetic needs and cholesterol biosynthesis to boost cellular growth (13). High amounts of glucose in cancer cells do not only contribute to increased glycolysis, but also increase flux into the hexosamine biosynthetic pathway, which is needed for protein glycosylation. Therefore, altered synthesis and especially increased neo-synthesis of complex glycans is commonly observed in advanced cancer stages. EMT and increased glycosylation have recently been shown to co-occur (14–16).
A possible link between metabolic reprogramming and EMT is lipid homeostasis (17). Tumor cells frequently display increased cholesterol content, but it is unclear whether the increase originates from elevated synthesis or uptake (18). The scavenger receptor class B type I (SR-BI), also called the HDL receptor, mediates selective cholesteryl ester uptake into cells (19). Importantly, expression of SR-BI was found in many different tumor types and its expression has been linked with disease progression (20–22). Whether SR-BI plays a role in protein glycosylation or EMT remains so far unknown.
Here, we aimed to characterize the role of SR-BI expression in human metastatic melanoma. Whole transcriptome analysis revealed that presence of SR-BI protein, independent of its cholesterol transporting function, was needed to maintain the EMT phenotype. Additionally, total protein glycosylation decreased after SR-BI knockdown. We identified STAT5 as a mediator of EMT related gene expression and therefore propose a link between SR-BI induced protein glycosylation and STAT5 activation. Hence, we suggest that expression of SR-BI can drive the progression of melanoma towards a highly malignant phenotype.
Material and methods
Cell culture and pharmacological compounds
Patient-derived melanoma cells MCM1G and MCM1DLN have been characterized previously and were used 10 passages after isolation (23). Human cell lines WM793B, 1205Lu, WM35 and 451Lu were purchased from American Type Culture Collection and were used for experiments 5 passages after thawing. Cells were cultivated under standard conditions and maintained in melanoma isolation medium containing 80% MCDB153, 20% Leibovitz`s L-15, 0.5 ng/ml epidermal growth factor, 5 μg/ml human insulin, 1.68 mM calcium chloride (all from Sigma), 50 mg/l streptomycin sulfate, 30 mg/l penicillin (both from PAA), 2 μg/ml ciprofloxacin (Sigma) and supplemented with 2% fetal calf serum (FCS). Cells were screened for mycoplasmal enzymes (Lonza) every 4 weeks. Lovastatin (Sigma) was used to block cholesterol synthesis, Rapamycin (Calbiochem) to block S6 phosphorylation, MK2206 (Selleckchem) to block AKT phosphorylation and BLT-1 (Sigma) to block selective cholesteryl ester uptake. All inhibitors were dissolved in dimethyl sulfoxide (DMSO).
Cell transfection
Reagents for siRNA knockdown were obtained from Dharmacon (GE Healthcare). Cells were transfected with DharmaFECT 1 and 5 nM ON-TARGETplus SMARTpool siRNA designed against SCARB1 (L-010592), STAT5A (L-005169), STAT5B (L-010539) or non-targeting siRNA (D-001810) for 24 hours followed by either mRNA isolation or protein isolation.
Lentiviral transduction
451Lu cells were seeded in a 24-well plate and were incubated with 1:25 or 1:50 shRNA lentiviral transduction particles (stock concentration: 106 TU/ml) targeting SR-BI (SHCLNV-NM_005505; TRCN0000056963, TRCN0000056966, TRCN0000299698, TRCN0000299709, TRCN0000310483 MISSION ® Lentiviral Transduction Particles, Sigma) or control (SHC002V, MISSION® pLKO.1-puro Non-Mammalian shRNA Control Transduction Particles, Sigma). Two days after transduction cells were switched to selection medium containing 2.5 μg/ml of puromycin (Life Technologies).
RNA isolation and RT-PCR
RNA was isolated according to manufacturer's instruction (peqGOLD total RNA Kit, VWR). cDNA synthesis and PCR were performed as described previously (24). Primer pairs are listed in Supplementary table 1. Values were normalized to human ACTB mRNA. Results were quantified using the 2(-Delta Delta C(T)) Method.
Western blotting
Proteins were lysed with RIPA buffer (Cell Signaling) on ice. SDS–PAGE and Western blotting were performed according to standard protocols. Nitrocellulose membranes (Biorad) were incubated with antibodies according to Supplementary table 2. Semi-quantitative densitometric analyses of bands were performed with corresponding software from Biorad.
Whole genome expression analysis and gene set enrichment analysis
Three metastatic human melanoma cell lines (451Lu, 1205Lu, MCM1DLN) grown in MIM supplemented with 10% FSC were analyzed. Each cell line was cultured in duplicate and either treated with control siRNA + DMSO, control siRNA + BLT-1 or SR-BI siRNA + DMSO for 24 hours. RNA was isolated, pooled and hybridized to GeneChip® Human PrimeView arrays (Affymetrix). CEL files were imported to Affymetrix® Expression Console™ Software and robust multi-array average was calculated. The desktop application version of GSEA was used for gene set enrichment analysis (GSEA) (25). Following settings were taken: Expression dataset: log2 RMA normalized expression file; Collapse dataset: true; Permutation type: gene_set; Metric of ranking genes: Ratio of Classes; Collapsing mode: Median_of_probes; Normalization mode: meandiv. as ranking metric. Scatter plot was made with Affymetrix® Transcriptome Analysis Console v2.0 Software. Microarray data and description of experimental design were deposited at NCBI GEO number GSE96743.
Immunohistochemistry and immunocytochemistry
For immunohistological staining invasion assay gels were dehydrated and embedded in paraffin. 3 µm FFPE tissue sections of invasion assay gels or malignant melanoma tissue arrays (ME2082b, US Biomax Inc.) were deparaffinized followed by epitope unmasking according to standard procedure. First antibodies were incubated at 4°C over night. Biotinylated secondary antibodies were incubated at room temperature (RT) and followed by incubation of avidin biotin complexes conjugated with peroxidase at RT (Jackson Lab). Aminoethyl carbazole (AEC, Dako) was used to visualize staining and hematoxylin (Roth) was used for counterstaining. Images were taken with a Zeiss Imager Z1 microscope. For cytochemistry cells were fixed with 4% formaldehyde, washed with PBS, treated for 5 min with 1% Triton X-100, washed, blocked with 1% BSA and then stained with specific antibodies at 4°C over night (see Supplementary table 2). For WGA staining, cells were washed with PBS and incubated with fluorescent labeled WGA (5 µg/ml in HBSS) for 10 min at 37°C. Afterwards cells were fixed with 4% formaldehyde and counterstained with DAPI for 10 min at RT. Immunofluorescence images were taken with a Leica TCS SP8 microscope.
Migration and invasion assays
For in vitro cell migration assay, 3×104 cells were seeded in serum free MIM onto transwell inserts with 8 µm pore size (BD Biosciences). Attractant was MIM supplemented with 10% FCS. Cells were allowed to migrate 8 hours, fixed with 4% formaldehyde and stained with 0.1% crystal violet. Six fields of view were recorded (Zeiss Primo Vert) and cell counting was performed using ImageJ. Three dimensional gel invasion assay was performed by producing polymerized collagen I gels containing: 1.5% methylcellulose, 2 mg/ml rat-tail collagen at neutral pH. 3x105 cells were seeded on the gel at an air-liquid interface. Gels were incubated for 5 days at 37°C. Subsequently, gels were either fixed for 2 hours with 4% formaldehyde for histological analysis or mRNA was extracted.
Immunoprecipitation of glycosylated proteins
Glycoproteins were isolated according to manufacturer's instruction (Pierce™ Glycoprotein Isolation Kit, Thermo Scientific). In brief, control cells and SR-BI knockdown cells were grown in MIM supplemented with 10% FCS before protein isolation. Cells were lysed in buffer containing 20% SDS and 1.5 mg protein was loaded onto the WGA-coated column.
ELISA
Vascular endothelial growth factor (VEGF) was measured from supernatant using the Human VEGF Mini Development Kit from PeproTech®. The ABTS substrate was purchased from Sigma–Aldrich®.
EMSA
Electrophoretic mobility shift assay (EMSA) was performed as described previously (26). Briefly, cells were stimulated with IL-6 (100 ng/ml) or Oncostatin M (OSM) (50 ng/ml) (both from PeproTech) for 20 min, washed with PBS and lysed. Equal amounts of protein extracts were incubated with a 32P-labelled STAT5 (5′-3′ AGATTTCTAGGAATTCAATCC) response element for 5 min at RT before gel loading. Complexes were separated non-denaturised on 4% acrylamide gels and run in 0.25x TBE buffer. Gels were dried and binding complexes formed were analyzed with Carestream® Kodak® Biomax® MR films. Semiquantification was performed using ImageJ.
Xenograft experiments
A total of 3×106 cells were injected intradermally into flanks of six weeks old severe combined immunodeficient C.B17 female (SCID) mice (2x scrambled shRNA, 1x SR-BI shRNA clone 310483, 1x SR-BI shRNA clone 299698, 1x SR-BI shRNA clone 299709). Tumor volumes were evaluated every week by measuring the diameter with a caliper. Tumor volume was calculated using the following equation: (width*width*length)/2. All animal procedures were approved by the “Animal Care and Use Committee” of the Medical University of Vienna. All methods were carried out in accordance with the approved guidelines of the Animal Care Committee.
Patient cohort and statistical analysis
Expression intensities and survival plots were generated from GSE19234 and the SKCM-TCGA study on cutaneous melanoma (27,28). GSE8401 study was used to perform heat map analysis (29). Statistical significance was calculated by unpaired two-tailed t test and by Log-rank (Mantel-Cox) test for survival analysis. All methods involving human patient samples were approved by the institutional ethics review board from the Medical University of Vienna and carried out in accordance with the appropriate guidelines. All patient samples were obtained as anonymized tissue microarrays from US Biomax. All values are given as means±standard error of the mean (s.e.m.) analyzed with GraphPad Prism® 6. P-values are indicated as follows: ns>0.05; * P<0.05; ** P<0.01; *** P<0.001; and **** P<0.0001.
Results
SR-BI expression in melanoma patients and in melanoma cell lines
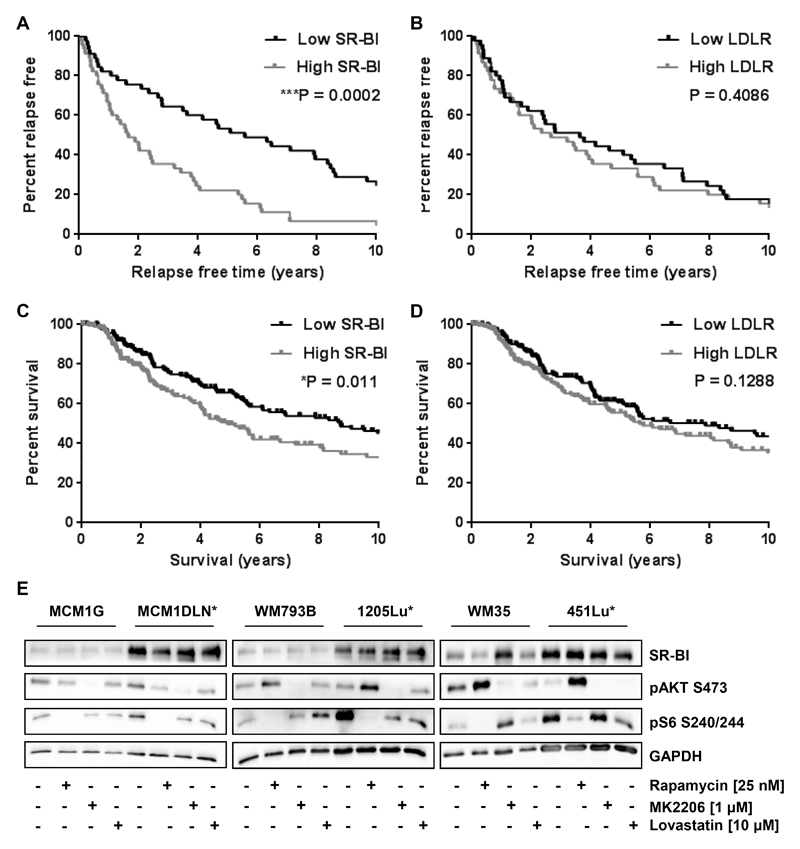
We investigated the expression of SR-BI and the low density lipoprotein receptor LDLR in human melanoma samples. Both receptors are able to bind lipoproteins and transport cholesterol into cells. We used the human melanoma dataset generated by The Cancer Genome Atlas (TCGA) and calculated the relapse free survival time according to expression above or below the median of the respective receptors. Patients with high SR-BI (Fig. 1A) displayed a significantly reduced time for re-occurrence of the tumor compared to patients with low SR-BI expression. In contrast LDLR expression did not show a significant difference (Fig. 1B). Additionally, we analyzed overall survival for SR-BI and LDLR. SR-BI levels above median revealed a significant poorer outcome after Kaplan-Meier testing in the TCGA cohort (Fig. 1C), which was confirmed in another melanoma study GSE19234 (Supplementary Fig. S1A). However, LDLR levels had no influence on overall survival (Fig. 1D, Supplementary Fig. S1B). Our analyses showed that median survival for patients expressing high SR-BI dropped from 8.6 to 4.8 years and from 10.8 to 2.6 years, respectively. Additionally, SR-BI expression according to tumor site and to tumor characteristics was analyzed. Albeit SR-BI was significantly lower expressed in the melanoma metastatic site compared to the primary site, possibly due to high SR-BI expression in pigmented cells (30,31), distant metastasis tissue showed significant higher SR-BI levels compared to regional cutaneous melanoma tissues (Supplementary Fig. S1C-E). Next, we determined SR-BI expression in six human melanoma cell lines with differing potential for malignant growth. Metastatic cells (labeled with an asterisk) showed higher SR-BI protein levels compared to non-metastatic cells (Fig. 1E). Furthermore, we tested inhibitors, which could potentially influence SR-BI expression based on their ability to disrupt cholesterol homeostasis. Rapamycin, a selective mTORC1 inhibitor, and MK2206, a selective AKT inhibitor, were used for 24 hours without altering SR-BI levels. Similarly, Lovastatin, a cholesterol synthesis inhibitor, had no effect on SR-BI expression. We conclude that SR-BI is stably expressed in non-metastatic and metastatic melanoma cells and that high levels of SR-BI correlate with poor survival outcome and metastatic potential in patients. Hence high SR-BI protein expression could play a role in melanoma metastasis.
Figure 1. High SR-BI expression indicates patients at risk for disease progression over time and robust SR-BI expression hallmarks metastatic melanoma cell lines.
(A and B) Relapse free survival time of 90 patients from the mRNA dataset SKCM-TCGA for (A) SR-BI and (B) LDLR. Grey line indicates expression levels above median, black line indicates expression levels below median. Values were calculated by log-rank testing. (C and D) Kaplan-Meier survival curves of 335 patients from the mRNA dataset SKCM-TCGA for (C) SR-BI and (D) LDLR. Grey line indicates expression levels above median, black line indicates expression levels below median. Values were calculated by log-rank testing. (E) SR-BI, phosphorylated AKT and ribosomal protein S6 protein amounts of different human melanoma cell lines without and with indicated inhibitor treatment for 24 hours. GAPDH was used as loading control. Cells able to metastasize are labeled with an asterisk.
SR-BI expression and function regulate distinct phenotypes
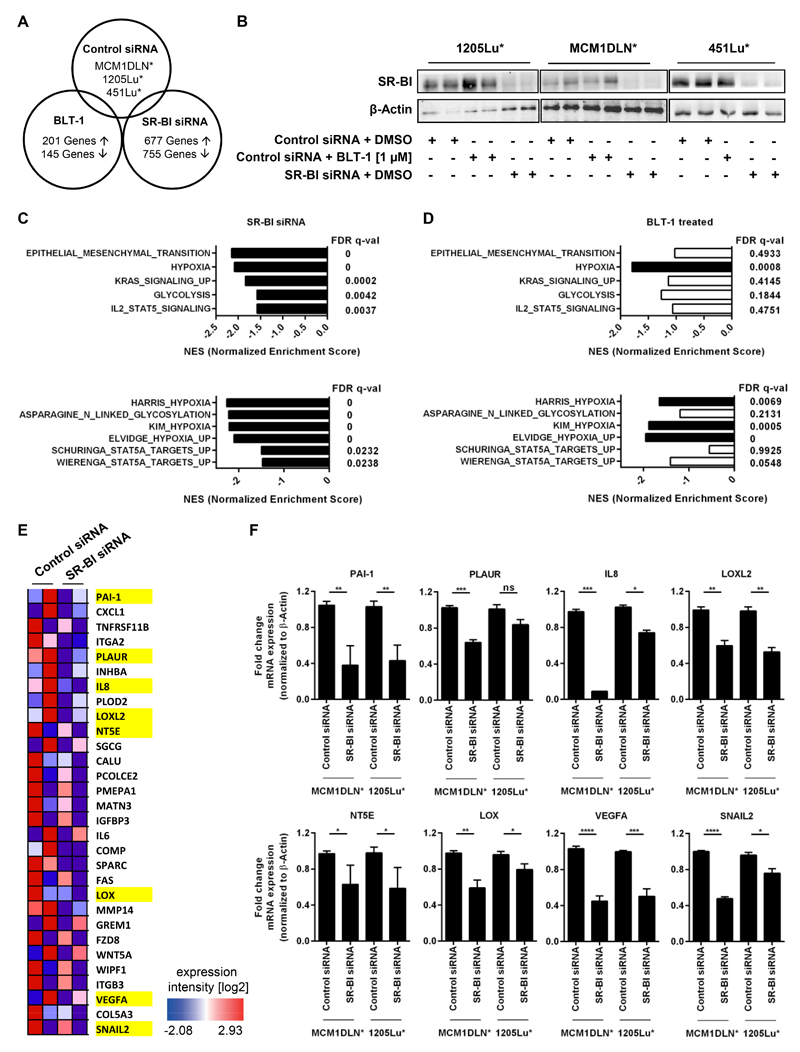
To elucidate gene expression and pathways regulated by SR-BI, independent of its cholesterol transporting function, we performed whole transcriptome analysis. Three metastatic human melanoma cell lines were treated either with siRNA targeting SR-BI or with BLT-1, an inhibitor described to specifically block SR-BI mediated lipid transport (32) (Fig. 2A). Knockdown efficiency was evaluated on protein level and, as expected, BLT-1 treated cells exhibited no alterations on SR-BI expression (Fig. 2B). Fig. 2C illustrates the strongest changes in phenotypes after SR-BI knockdown according to hallmark (upper panel) or C2CGP gene sets (lower panel). Identical phenotypes after treatment with BLT-1 are depicted in Figure 2D. Top regulated hallmark gene sets of SR-BI knockdown cell lines were the EMT pathway, followed by hypoxia, KRAS signaling, glycolysis and IL2-STAT5 signaling. In the C2CGP analysis hypoxia, glycosylation and STAT5 targets were identified. For BLT-1 treatment only hypoxia associated gene expression changes reached significance. Supplementary Fig. 2 shows enrichment plots for hallmark gene sets hypoxia, EMT and IL2-STAT5 signaling after SR-BI knockdown (upper panel) or BLT-1 treated cells (lower panel). Top 30 regulated genes of the hallmark gene set EMT after SR-BI siRNA treatment of two melanoma cell lines were illustrated as a heat map (Fig. 2E and Supplementary table 3). Characteristic genes (labeled in yellow) were validated by RT-PCR after SR-BI knockdown in two metastatic human melanoma cell lines (Fig. 2F). Here, we demonstrated that SR-BI expression, independent of its function, is important for maintaining the EMT phenotype, high cellular glycosylation and STAT5 signaling.
Figure 2. Loss of SR-BI triggers phenotypes distinct from blocking SR-BI mediated cholesterol transport.
(A) Venn diagram shows number of regulated genes after BLT-1 or SR-BI siRNA treatment of three metastatic human melanoma cell lines. (B) Western blot demonstrates SR-BI knockdown efficiency. β-Actin was used as loading control. (C and D) Gene set enrichment analysis shows normalized enrichment scores and their corresponding FDR q-value of hallmark gene sets and C2CGP study gene sets negatively enriched either after (C) SR-BI knockdown or (D) BLT-1 treatment. Black bars indicate highly significant results, white bars are not significant. (E) Heat map of top 30 regulated EMT genes of control versus SR-BI siRNA of MCM1DLN* and 1205Lu* cells. Red color displays high SR-BI expression, blue color displays low expression. Log2 expression values range from -2.08 to 2.93. Genes highlighted in yellow were evaluated in Fig. 2F. (F) RT-PCR of control versus SR-BI siRNA treated MCM1DLN* and 1205Lu* cells for indicated genes.
SR-BI depletion decreases cell migration and three dimensional invasion of cells
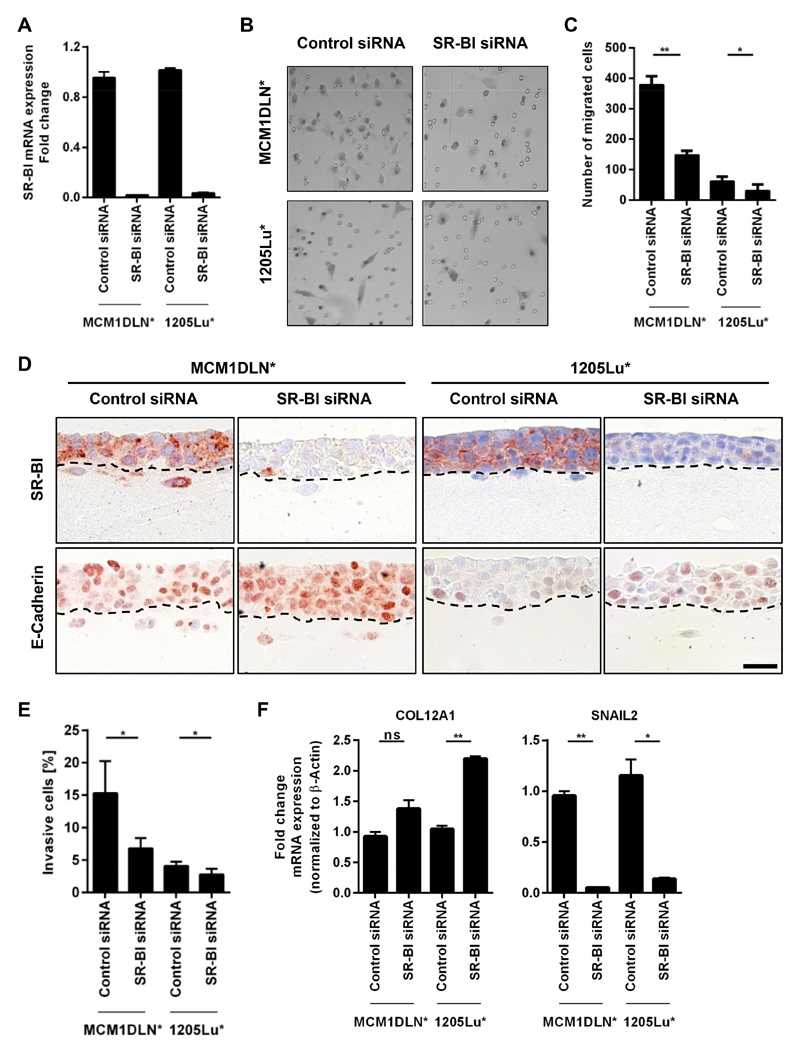
To test for functional consequences of SR-BI depletion we investigated knockdown efficiency 96 hours post transfection of MCM1DLN* and 1205Lu* by RT-PCR (Fig. 3A). Migration assays revealed a significant reduction in migration after SR-BI knockdown (Fig. 3B and Fig. 3C). Furthermore, we established in vivo-like 3D tumor structures on top of collagen I gels to test for invasive capabilities of melanoma cells. In both cell lines MCM1DLN* and 1205Lu* SR-BI loss significantly lowered invasion (Fig. 3D and Fig. 3E). Additionally, immunohistological stainings of invaded gels showed E-Cadherin, a prominent EMT marker, to be strongly increased after SR-BI knockdown (Fig. 3D). SR-BI knockdown cells displayed increased mRNA expression of COL12A1 and decreased expression of SNAIL2 (Fig. 3F). Also sphere formation assays showed significantly decreased sphere diameters, indicating stronger intercellular adhesion after SR-BI knockdown (Supplementary Fig. S3A). Cell death as a reason for smaller diameter was excluded since viability evaluation after SR-BI knockdown revealed no significant difference (Supplementary Fig. S3B). Proliferation (Supplementary Fig. S3C) and cellular cholesterol content (Supplementary Fig. S3D) were also not altered after SR-BI knockdown. These results strengthen our findings that SR-BI plays a critical role in EMT and that loss of SR-BI can induce a reversion to a less invasive phenotype.
Figure 3. SR-BI depletion decreases cell migration and invasion.
(A) SR-BI mRNA expression in control and SR-BI knockdown cells 96 hours post transfection (MCM1DLN*, 1205Lu*). (B) Transwell migration assays with representative images at 8 hours comparing control siRNA versus SR-BI siRNA. (C) Quantitation illustrates number of migrated cells (MCM1DLN*with n=3, 1205Lu* with n=4,). (D) Immunohistological stainings (anti-SR-BI and anti-E-Cadherin) of vertical invasion assays comparing control siRNA versus SR-BI siRNA. Cells below dotted line were invasive. Scale bar, 20 µm. (E) Quantitation bar chart for number of invaded cells (MCM1DLN* cells with n=4, 1205Lu* cells with n=4). (F) mRNA expression of COL12A1 and SNAIL2 of vertical invasion assays, n=3.
SR-BI facilitates protein glycosylation including STAT5
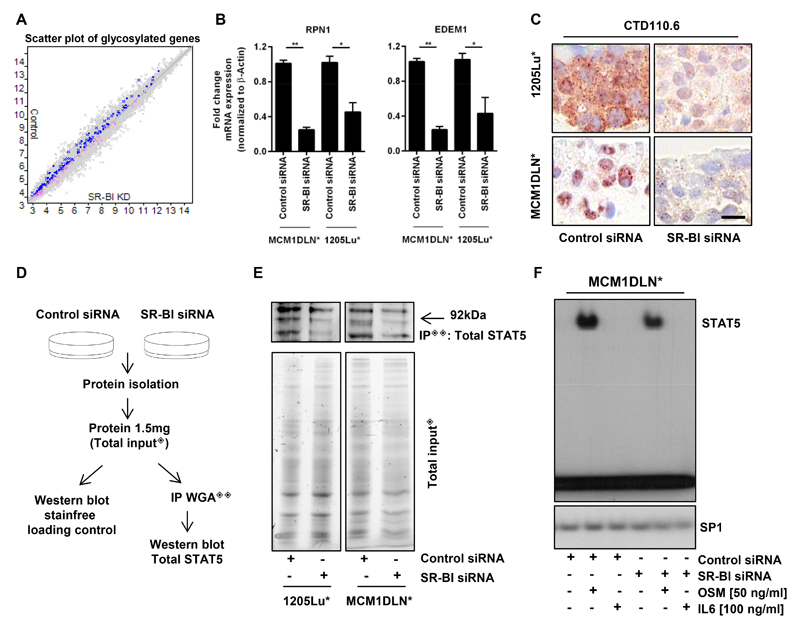
Next, we investigated expression changes in the glycosylation pathway. All genes that were part of the GO term glycosylation were plotted according to their mRNA expression comparing control versus SR-BI knockdown cells (Fig. 4A). For confirmation we analyzed RPN1 and EDEM1, two important genes for glycosylation, on mRNA level (Fig. 4B). To validate decreased glycosylation we performed an immunofluorescence with WGA (Supplementary Fig. S4A). The reduction of glycosylated proteins was also confirmed by immunohistological staining of vertical invasion gels of two metastatic human melanoma cell lines using an antibody which recognizes endogenous levels of O-GlcNAc on proteins (Fig. 4C). Since transcriptome analysis revealed changes in STAT5 controlled gene sets, we specifically tested for STAT5 glycosylation. All glycosylated proteins were isolated by WGA binding (see isolation scheme in Fig. 4D) and the amount of STAT5 was quantified by Western blotting. Equal protein input of control and siRNA treated cells revealed a vast reduction of glycosylated STAT5 in the SR-BI knockdown group. (Fig. 4E, Supplementary Fig. S4B). STAT5 phosphorylation at Tyr694 was marginally reduced (Supplementary Fig. S4C), so we tested by EMSA if DNA binding was affected after SR-BI depletion in the strongest responding cell line. Therefore, MCM1DLN* cells were treated either with control siRNA or SR-BI siRNA and additionally stimulated with OSM and as a control with IL-6. STAT5 DNA binding was significantly reduced in MCM1DLN* cells (Fig. 4F, Supplementary Fig. S4D).
Figure 4. SR-BI facilitates protein glycosylation including STAT5.
(A) Scatter plot for GO_glycosylation gene set of three metastatic human melanoma cell lines comparing control versus SR-BI knockdown. Blue dots, 147 genes significantly higher expressed in control cells; pink dots, 57 genes significantly higher expressed in SR-BI knockdown cells. (B) RT-PCR for RPN1 and EDEM1 in MCM1DLN* and 1205Lu* cells. (C) Immunohistological staining of vertical invasion assays using the antibody CTD110.6. Scale bar, 10 µm. (D) Experimental set up for WGA immunoprecipitation (IP). (E) Detection of immunoprecipitated glycosylated STAT5 (IP◈◈) by hybridization with an anti-total STAT5 antibody and visualization of total protein amounts used for IP (Total input◈). (F) Radiography of DNA bound STAT5 and SP1. SP1 was used as loading control.
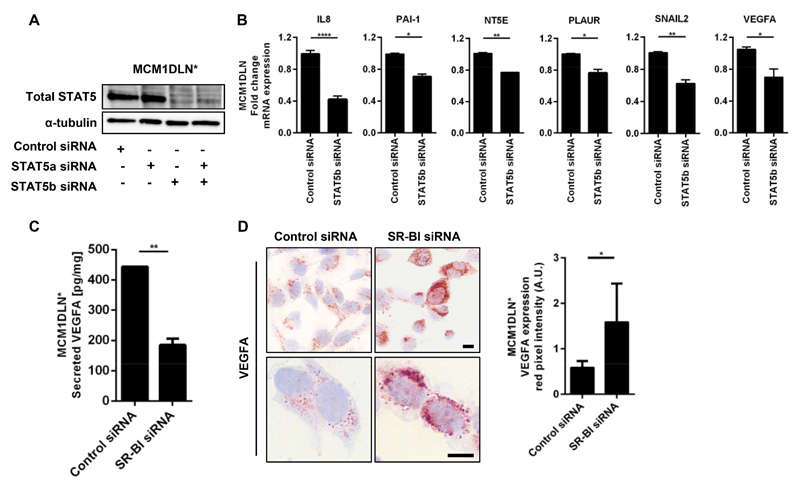
SR-BI targets are identical with STAT5 targets
To test whether STAT5 is a co-regulator for the observed EMT phenotype we performed knockdown of STAT5a and STAT5b either alone or in combination (Fig. 5A). STAT5b knockdown was sufficient to remove nearly all STAT5 protein, whereas STAT5a siRNA showed no reduction. Hence, we conclude that STAT5b was the prevalent form in this melanoma cell line, which we also confirmed on mRNA level (Supplementary Fig. 5A and 5B). STAT5 knockdown reduced expression of the SR-BI regulated genes IL8, PAI-1, NT5E, PLAUR, SNAIL2, and VEGFA (Fig. 5B). Since VEGFA is not only regulated on mRNA level, but also post-translationally by glycosylation, we investigated the amount of secreted VEGFA. We observed strongly reduced VEGFA release from cells lacking SR-BI (Fig. 5C). Additionally, VEGFA accumulated in the cytoplasm of SR-BI knockdown cells (Fig. 5D). Here we showed that STAT5 was important for maintenance of the EMT phenotype and that SR-BI knockdown as well as STAT5 knockdown led to a down regulation of identical gene sets.
Figure 5. SR-BI targets are identical with STAT5 targets.
(A) Western blot analysis of individual and combined STAT5a and STAT5b knockdown in MCM1DLN* cells. α-tubulin was used as loading control. (B) mRNA expression of selected STAT5 targets after STAT5b siRNA in MCM1DLN* cells. (C) ELISA of supernatant of secreted VEGFA of MCM1DLN* cells treated either with control siRNA or SR-BI siRNA. VEGFA was normalized to protein content, n=3. (D) Immunocytochemistry anti-VEGFA of MCM1DLN* cells comparing control versus SR-BI knockdown cells and corresponding quantitation bar chart (n=6). Scale bar, 10 µm.
SR-BI promotes melanoma progression and its signature classifies melanoma in metastatic and non-metastatic groups
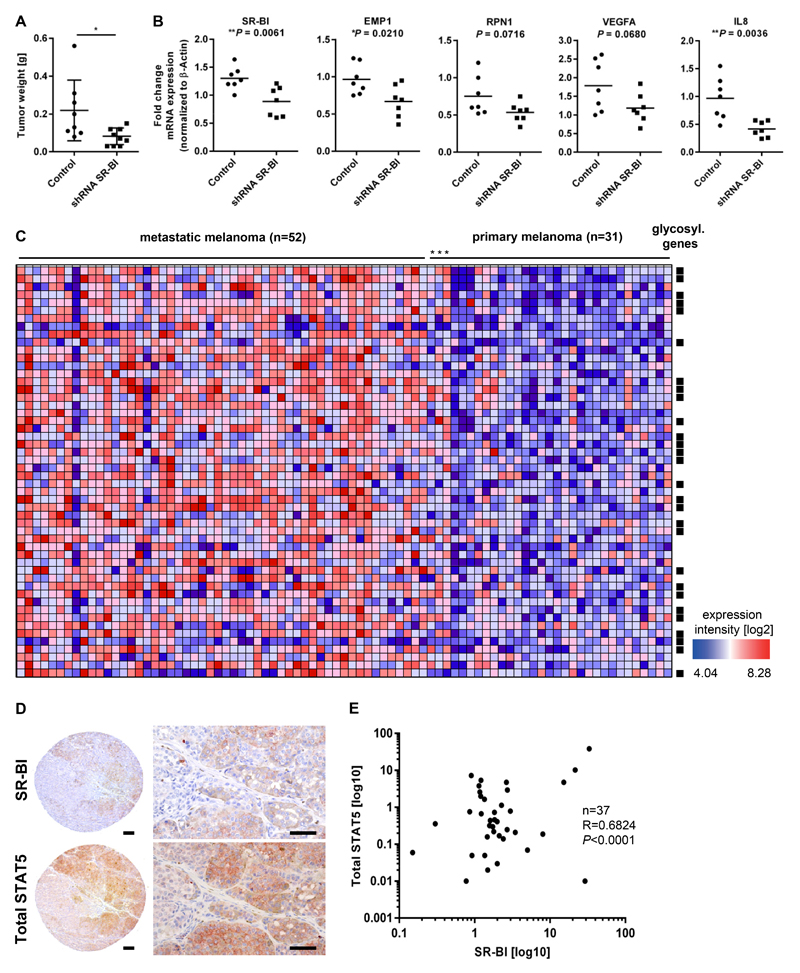
To strengthen our in vitro data, the effect of SR-BI was also analyzed in vivo. Therefore, stable SR-BI knockdown and control cells were intradermally injected into SCID mice. After 26 days, tumors were dissected and tumor weight was measured. Mice harboring SR-BI shRNA grew significantly smaller tumors (Fig. 6A). Analyzing mRNA from tumors confirmed SR-BI knockdown and down regulation of distinct target genes, like EMP1, RPN1, VEGFA and IL8, which are involved in glycosylation or the EMT process, respectively (Fig. 6B).
Figure 6. SR-BI promotes melanoma progression and its signature classifies melanoma in metastatic and non metastatic groups.
(A) Tumor weights after intradermal transplantation of 451Lu* cells either stable transduced with control shRNA (n=8) or shRNA targeting SR-BI (n=10) into SCID mice. (B) Tumor mRNA was isolated and RT-PCR of indicated genes was performed (n=7 per group). (C) Enrichment displayed as a heat map of SR-BI target genes in the patient dataset GSE8401 classifies primary and metastatic melanoma patients. Squares symbols indicate genes involved in the glycosylation pathway. (D) Immunohistological stainings of consecutive tissue from microarray slides anti-SR-BI and anti-total STAT5. Scale bar, 50µm. (E) Correlation analysis of SR-BI with STAT5 in metastatic samples derived from patient lymph nodes.
The mRNA expression dataset GSE8401 was generated during a human melanoma study characterizing primary and metastatic melanoma tissues. We used this study to perform enrichment analysis according to SR-BI target genes which we have identified. Out of 83 patients all 52 metastatic melanoma patients were hallmarked by high SR-BI target gene expression (Fig. 6C and Supplementary table 4). Only three samples out of 31 primary melanoma patients appeared as false positive. Furthermore, we found that most of the highly ranked genes are part of the glycosylation pathway, confirming our prior findings. Finally, we investigated whether SR-BI and STAT5 expression correlate in human patient biopsies. Therefore, we performed an immunohistological staining of tissue arrays, containing 37 lymph node metastases and stained two consecutive slides for SR-BI and STAT5 (Fig. 6D). Digital image quantification revealed a significant correlation of SR-BI with STAT5 (Fig. 6E). Together, these data show that SR-BI contributed to a migratory and invasive phenotype by enabling glycosylation and target gene expression of STAT5. In line, the SR-BI-derived target genes were able to classify melanoma patients according to their ability to metastasize.
Discussion
Here, we have shown that elevated expression of SR-BI can be regarded as a risk factor for patients suffering from metastasizing melanoma. Depletion of SR-BI, but not pharmacologic inhibition of its cholesterol transport function by using BLT-1, resulted in the loss of an EMT signature in melanoma. Furthermore, SR-BI knockdown reduced migration, 3D invasion and tumor growth of cells upon transplantation to SCID mice. STAT5 is a key factor for maintaining the EMT phenotype. Here we have shown that SR-BI knockdown reduces STAT5 glycosylation, STAT5 DNA binding and STAT5 target gene expression. Importantly, in human patient samples accumulation of STAT5 protein, typically co-occuring with STAT5 activation, correlated with expression of SR-BI.
SR-BI is expressed in different cell types including liver cells and steroidogenic cells, where it promotes the selective uptake of HDL cholesteryl esters. Therefore, it fulfills important functions in the reverse cholesterol transport and supplies hormone producing tissues with the required cholesterol. In breast tumor cells and in human primary endothelial cells it has been shown that HDL binding to SR-BI triggered AKT activation (33,34). Since HIF activation is linked to prominent AKT activation it is important to note that the main intracellular signaling induced by SR-BI mediated cholesterol transport is AKT phosphorylation. This we could also demonstrate when we examined gene expression changes associated with either block of SR-BI mediated lipid transport or down regulation of total cellular SR-BI expression in malignant melanoma. Despite numerous studies describing the effect on cholesterol homeostasis, little is known on lipid transport independent functions of SR-BI. Here we describe for the first time that SR-BI is important for maintaining an EMT phenotype in human melanoma and that its expression correlates with the ability to glycosylate cellular proteins including STAT5.
Since glycosylation occurs selectively at the ER and Golgi membrane network it is interesting that SR-BI has been localized to endosomal compartments and that knockout of SR-BI in primary hepatocytes led to cholesterol accumulation in the perinuclear area (35). Thus, SR-BI might play a role in intracellular membrane trafficking. Therefore, we speculate that SR-BI, which itself is highly glycosylated and palmitoylated, may serve to maintain and enhance ER and Golgi function. Recent data shows that glycosylation occurring in the ER and Golgi is enforced during melanoma progression (36). Here we could link this cell organelle specific function of SR-BI with oncogenic gene transcription.
As an important example down regulation of total protein glycosylation by SR-BI depletion also affected STAT5, which reduced its binding to DNA as well as its target gene expression including VEGFA. Interestingly, intracellular transport and release of VEGFA from the cytoplasm strictly depends on glycosylation (37,38). Hence, loss of SR-BI not only reduces VEGFA mRNA expression, but additionally it prevents VEGFA maturation and secretion. Intriguingly, SR-BI in human aortic endothelial cells is mostly localized to the cytoplasm and only expressed at the cell surface when VEGFA is available (39). Importantly, it has been demonstrated that glycosylation of STAT5 directly controls DNA binding at enhancer elements of oncogenic loci triggered by high pYSTAT5 levels as well as oligomer formation (40). These findings are also supported by the observation that an intact Golgi apparatus serves as a platform for STAT5 activation in gastrointestinal stromal tumors (41). In hematopoietic cells STAT5 could impose hypoxic signaling under normoxic conditions and recapitulated the Warburg effect in cancer cells (42). This implicates that a positive feedback loop could exist where increased glycolysis and glycosylation can lead to STAT5 stabilization and vice versa STAT5 target gene expression can lead to enforced aerobic glycolysis.
We could not observe an alteration of total STAT5 protein levels after SR-BI knockdown in melanoma cells possibly due to short term nature of our in vitro experiments. However, in human patient samples we could establish a positive correlation between SR-BI and STAT5 protein expression. In T cells STAT5 activation was paralleled by an increase in STAT5 stability (43,44). Hence, we speculate that chronic high level expression of SR-BI not only increases activation efficiency, but also stabilizes total STAT5 amounts.
In summary, our data provide evidence that SR-BI serves as a driver of an EMT-like phenotype by altering glucose related metabolic pathways feeding into STAT5 activation. High STAT5 expression has been correlated with low E-Cadherin expression in prostate cancer and reports show an involvement of STAT5 in the EMT process of hepatocellular carcinoma and head and neck cancer (45–47). Therefore, we propose that SR-BI governs intracellular processes crucial for establishing a metastatic phenotype. Moreover, we have defined SR-BI related target genes in melanoma which are enriched in metastatic melanoma patients. Hence, SR-BI presents a novel factor to estimate patient survival in melanoma and we propose it is functionally linked to increased protein glycosylation specific for metastatic tumors.
Supplementary Material
Acknowledgements
We thank Helmut Dolznig for support during 3D culturing, Jelena Brankovic and Heidemarie Huber for technical assistance. We thank the "Core Facility Genomics, Medical University of Vienna" for performing transcriptome hybridization.
Grant support: This work was supported by the Austrians Science Fund (P 25336-B13 to M. Mikula and P 25763-B13 to C. Röhrl) and a private melanoma research donation from Liechtenstein (to R. Moriggl)
Footnotes
Conception and design: K. Kinslechner, M. Mikula
Development of methodology: K. Kinslechner, D. Schörghofer, B. Schütz, M. Vallianou, B. Wingelhofer
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): W. Mikulits, C. Röhrl, M. Hengstschläger, R. Moriggl, H. Stangl, M. Mikula
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): K. Kinslechner, D. Schörghofer, B. Schütz, M. Vallianou, C. Röhrl, M. Mikula
Writing, review, and/or revision of the manuscript: K. Kinslechner, D. Schörghofer, B. Schütz, M. Vallianou, B. Wingelhofer, W. Mikulits, C. Röhrl, M. Hengstschläger, R. Moriggl, H. Stangl, M. Mikula
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): D. Schörghofer, B. Schütz, B. Wingelhofer
Study supervision: M. Hengstschläger, R. Moriggl, H. Stangl, M. Mikula
References
- 1.Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851–7. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- 2.Sanchez-Laorden B, Viros A, Girotti MR, Pedersen M, Saturno G, Zambon A, et al. BRAF Inhibitors Induce Metastasis in RAS Mutant or Inhibitor-Resistant Melanoma Cells by Reactivating MEK and ERK Signaling. Sci Signal. 2014;7 doi: 10.1126/scisignal.2004815. [DOI] [PubMed] [Google Scholar]
- 3.Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, et al. J Clin Oncol. Vol. 29. American Society of Clinical Oncology; 2011. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling; pp. 3085–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li FZ, Dhillon AS, Anderson RL, McArthur G, Ferrao PT. Front Oncol. Vol. 5. Frontiers Media SA; 2015. Phenotype switching in melanoma: implications for progression and therapy; p. 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alonso SR, Tracey L, Ortiz P, Pérez-Gómez B, Palacios J, Pollán M, et al. A High-Throughput Study in Melanoma Identifies Epithelial-Mesenchymal Transition as a Major Determinant of Metastasis. Cancer Res. 2007;67:3450–60. doi: 10.1158/0008-5472.CAN-06-3481. [DOI] [PubMed] [Google Scholar]
- 6.Hoek KS, Eichhoff OM, Schlegel NC, Dobbeling U, Kobert N, Schaerer L, et al. In vivo Switching of Human Melanoma Cells between Proliferative and Invasive States. Cancer Res. 2008;68:650–6. doi: 10.1158/0008-5472.CAN-07-2491. [DOI] [PubMed] [Google Scholar]
- 7.Caramel J, Papadogeorgakis E, Hill L, Browne GJ, Richard G, Wierinckx A, et al. A Switch in the Expression of Embryonic EMT-Inducers Drives the Development of Malignant Melanoma. Cancer Cell. 2013;24:466–80. doi: 10.1016/j.ccr.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 8.Aspuria P-JP, Lunt SY, Väremo L, Vergnes L, Gozo M, Beach JA, et al. Cancer Metab. Vol. 2. BioMed Central; 2014. Succinate dehydrogenase inhibition leads to epithelial-mesenchymal transition and reprogrammed carbon metabolism; p. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong C, Yuan T, Wu Y, Wang Y, Fan TWM, Miriyala S, et al. Loss of FBP1 by Snail-Mediated Repression Provides Metabolic Advantages in Basal-like Breast Cancer. Cancer Cell. 2013;23:316–31. doi: 10.1016/j.ccr.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W, Wei Z, Liu Y, Li H, Ren R, Tang Y. Increased 18F-FDG uptake and expression of Glut1 in the EMT transformed breast cancer cells induced by TGF-beta. Neoplasma. 2010;57:234–40. doi: 10.4149/neo_2010_03_234. [DOI] [PubMed] [Google Scholar]
- 11.Sun Y, Daemen A, Hatzivassiliou G, Arnott D, Wilson C, Zhuang G, et al. Metabolic and transcriptional profiling reveals pyruvate dehydrogenase kinase 4 as a mediator of epithelial-mesenchymal transition and drug resistance in tumor cells. Cancer Metab. 2014;2:20. doi: 10.1186/2049-3002-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu M, Quek L-E, Sultani G, Turner N. Cancer Metab. Vol. 4. BioMed Central; 2016. Epithelial-mesenchymal transition induction is associated with augmented glucose uptake and lactate production in pancreatic ductal adenocarcinoma; p. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warburg O. On the origin of cancer cells. Science. 1956;123:309–14. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 14.Du J, Hong S, Dong L, Cheng B, Lin L, Zhao B, et al. Dynamic Sialylation in Transforming Growth Factor-β (TGF-β)-induced Epithelial to Mesenchymal Transition. J Biol Chem. 2015;290:12000–13. doi: 10.1074/jbc.M115.636969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lucena MC, Carvalho-Cruz P, Donadio JL, Oliveira IA, de Queiroz RM, Marinho-Carvalho MM, et al. Epithelial mesenchymal transition induces aberrant glycosylation through hexosamine biosynthetic pathway activation. J Biol Chem. 2016;291:12917–29. doi: 10.1074/jbc.M116.729236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freire-de-Lima L, Gelfenbeyn K, Ding Y, Mandel U, Clausen H, Handa K, et al. Proc Natl Acad Sci U S A. Vol. 108. National Academy of Sciences; 2011. Involvement of O-glycosylation defining oncofetal fibronectin in epithelial-mesenchymal transition process; pp. 17690–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beloribi-Djefaflia S, Vasseur S, Guillaumond F. Oncogenesis. Vol. 5. Nature Publishing Group; 2016. Lipid metabolic reprogramming in cancer cells; p. e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuzu OF, Noory MA, Robertson GP. The Role of Cholesterol in Cancer. Cancer Res. 2016;76 doi: 10.1158/0008-5472.CAN-15-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Acton S, Rigotti A, Landschulz KT, Xu S, Hobbs HH, Krieger M. Identification of Scavenger Receptor SR-BI as a High Density Lipoprotein Receptor. Science (80-) 1996;271:518–20. doi: 10.1126/science.271.5248.518. [DOI] [PubMed] [Google Scholar]
- 20.Gutierrez-Pajares JL, Ben Hassen C, Chevalier S, Frank PG. Front Pharmacol. Vol. 7. Frontiers Media; SA: 2016. SR-BI: Linking Cholesterol and Lipoprotein Metabolism with Breast and Prostate Cancer; p. 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schörghofer D, Kinslechner K, Preitschopf A, Schütz B, Röhrl C, Hengstschläger M, et al. The HDL receptor SR-BI is associated with human prostate cancer progression and plays a possible role in establishing androgen independence. Reprod Biol Endocrinol. 2015;13:88. doi: 10.1186/s12958-015-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoekstra M, Sorci-Thomas M. Rediscovering scavenger receptor type BI. Curr Opin Lipidol. 2017;28:255–60. doi: 10.1097/MOL.0000000000000413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swoboda A, Schanab O, Tauber S, Bilban M, Berger W, Petzelbauer P, et al. MET expression in melanoma correlates with a lymphangiogenic phenotype. Hum Mol Genet. 2012;21:3387–96. doi: 10.1093/hmg/dds171. [DOI] [PubMed] [Google Scholar]
- 24.Preitschopf A, Li K, Schörghofer D, Kinslechner K, Schütz B, Thi Thanh Pham H, et al. mTORC1 is essential for early steps during Schwann cell differentiation of amniotic fluid stem cells and regulates lipogenic gene expression. PLoS One. 2014;9:e107004. doi: 10.1371/journal.pone.0107004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette Ma, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kornfeld J-W, Grebien F, Kerenyi Ma, Friedbichler K, Kovacic B, Zankl B, et al. The different functions of Stat5 and chromatin alteration through Stat5 proteins. Front Biosci. 2008;13:6237–54. doi: 10.2741/3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bogunovic D, O’Neill DW, Belitskaya-Levy I, Vacic V, Yu Y-L, Adams S, et al. Immune profile and mitotic index of metastatic melanoma lesions enhance clinical staging in predicting patient survival. Proc Natl Acad Sci U S A. 2009;106:20429–34. doi: 10.1073/pnas.0905139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tcga. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161:1681–96. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu L, Shen SS, Hoshida Y, Subramanian A, Ross K, Brunet J-P, et al. Gene Expression Changes in an Animal Melanoma Model Correlate with Aggressiveness of Human Melanoma Metastases. Mol Cancer Res. 2008;6:760–9. doi: 10.1158/1541-7786.MCR-07-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoek KS, Schlegel NC, Eichhoff OM, Widmer DS, Praetorius C, Einarsson SO, et al. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell Melanoma Res. 2008;21:665–76. doi: 10.1111/j.1755-148X.2008.00505.x. [DOI] [PubMed] [Google Scholar]
- 31.Duncan KG, Bailey KR, Kane JP, Schwartz DM. Human retinal pigment epithelial cells express scavenger receptors BI and BII. Biochem Biophys Res Commun. 2002;292:1017–22. doi: 10.1006/bbrc.2002.6756. [DOI] [PubMed] [Google Scholar]
- 32.Nieland TJF, Penman M, Dori L, Krieger M, Kirchhausen T. Discovery of chemical inhibitors of the selective transfer of lipids mediated by the HDL receptor SR-BI. Proc Natl Acad Sci U S A. 2002;99:15422–7. doi: 10.1073/pnas.222421399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danilo C, Gutierrez-Pajares JL, Mainieri MA, Mercier I, Lisanti MP, Frank PG. Scavenger receptor class B type I regulates cellular cholesterol metabolism and cell signaling associated with breast cancer development. Breast Cancer Res. 2013;15:R87. doi: 10.1186/bcr3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fruhwürth S, Krieger S, Winter K, Rosner M, Mikula M, Weichhart T, et al. Biochim Biophys Acta - Mol Cell Biol Lipids. Vol. 1841. Elsevier B.V; 2014. Inhibition of mTOR down-regulates scavenger receptor, class B, type i (SR-BI) expression, reduces endothelial cell migration and impairs nitric oxide production; pp. 944–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahras M, Naing T, Mcpherson R. Scavenger receptor class B type I localizes to a late endosomal compartment. J Lipid Res. 2008;49:1569–76. doi: 10.1194/jlr.M800055-JLR200. [DOI] [PubMed] [Google Scholar]
- 36.Agrawal P, Fontanals-Cirera B, Sokolova E, Jacob S, Vaiana CA, Argibay D, et al. A Systems Biology Approach Identifies FUT8 as a Driver of Melanoma Metastasis. Cancer Cell. 2017;31:804–819.e7. doi: 10.1016/j.ccell.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang WK, Lee MH, Kim YH, Kim MY, Kim JY. Enhanced secretion of biologically active, non-glycosylated VEGF from Saccharomyces cerevisiae. J Biotechnol. 2013;164:441–8. doi: 10.1016/j.jbiotec.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 38.Guzmán-Hernández ML, Potter G, Egervári K, Kiss JZ, Balla T. Secretion of VEGF-165 has unique characteristics, including shedding from the plasma membrane. Mol Biol Cell. 2014;25:1061–72. doi: 10.1091/mbc.E13-07-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Velagapudi S, Yalcinkaya M, Piemontese A, Meier R, Nørrelykke SF, Perisa D, et al. VEGF-A Regulates Cellular Localization of SR-BI as Well as Transendothelial Transport of HDL but Not LDL. Arterioscler Thromb Vasc Biol. 2017;37:794–803. doi: 10.1161/ATVBAHA.117.309284. [DOI] [PubMed] [Google Scholar]
- 40.Freund P, Kerenyi MA, Hager M, Wagner T, Wingelhofer B, Pham HT, et al. O-GlcNAcylation of STAT5 controls tyrosine phosphorylation and oncogenic transcription in STAT5-dependent malignancies. Leukemia. 2017:1–11. doi: 10.1038/leu.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Obata Y, Horikawa K, Takahashi T, Akieda Y, Tsujimoto M, Fletcher JA, et al. Oncogenic signaling by Kit tyrosine kinase occurs selectively on the Golgi apparatus in gastrointestinal stromal tumors. Oncogene. 2017;36:3661–72. doi: 10.1038/onc.2016.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sontakke P, Koczula KM, Jaques J, Wierenga AT, Brouwers-Vos AZ, Pruis M, et al. Hypoxia-Like Signatures Induced by BCR-ABL Potentially Alter the Glutamine Uptake for Maintaining Oxidative Phosphorylation. PLoS One. 2016;11:e0153226. doi: 10.1371/journal.pone.0153226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsiao WY, Lin YC, Liao FH, Chan YC, Huang CY. Dual-specificity phosphatase 4 regulates STAT5 protein stability and helper T cell polarization. PLoS One. 2015;10:e0145880. doi: 10.1371/journal.pone.0145880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ren Z, Aerts JL, Vandenplas H, Wang JA, Gorbenko O, Chen JP, et al. Phosphorylated STAT5 regulates p53 expression via BRCA1/BARD1-NPM1 and MDM2. Cell Death Dis. 2016;7:e2560. doi: 10.1038/cddis.2016.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benitah SA, Valeron PF, Rui H, Lacal JC. STAT5a activation mediates the epithelial to mesenchymal transition induced by oncogenic RhoA. Mol Biol Cell. 2003;14:40–53. doi: 10.1091/mbc.E02-08-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee TK, Man K, Poon RT, Lo CM, Yuen AP, Ng IO, et al. Signal transducers and activators of transcription 5b activation enhances hepatocellular carcinoma aggressiveness through induction of epithelial-mesenchymal transition. Cancer Res. 2006;66:9948–56. doi: 10.1158/0008-5472.CAN-06-1092. [DOI] [PubMed] [Google Scholar]
- 47.Koppikar P, Lui VW, Man D, Xi S, Chai RL, Nelson E, et al. Constitutive activation of signal transducer and activator of transcription 5 contributes to tumor growth, epithelial-mesenchymal transition, and resistance to epidermal growth factor receptor targeting. Clin Cancer Res. 2008;14:7682–90. doi: 10.1158/1078-0432.CCR-08-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.