Abstract
NF-κB (nuclear factor-κB) transcription factors have multiple critical roles in the regulation of immune responses. In unstimulated cells, NF-κB proteins are sequestered in the cytoplasm by IκB inhibitory proteins. Various immune stimuli induce the IκB kinase (IKK) to phosphorylate IκBs, triggering their ubiquitination and proteasomal degradation, which permits nuclear translocation of associated NF-κB subunits and activation of NF-κB target genes. Recent studies have highlighted the importance of dynamic ubiquitination-deubiquitination events in regulating this canonical NF-κB signaling pathway. Ubiquitination additionally plays critical roles in activation of the noncanonical pathway that regulates NF-κB via signal-induced processing of NF-κB2 p100. New research has also identified several novel regulatory proteins that control the transcriptional activity of nuclear NF-κB.
Introduction
NF-κB (nuclear factor-κB) regulates genes involved at multiple stages of immune responses, including innate immune cell activation, inflammation, dendritic cell maturation and lymphocyte activation. NF-κB activation is tightly controlled by canonical and atypical pathways that regulate proteolysis of IκB (an inhibitor of NF-κB) and IκB-related proteins (Box 1; Figure 1). Because such basic knowledge of NF-κB has been discussed previously [1,2], this review will instead focus on new insights gained through recent studies.
Box 1. Nuclaear factor-κB family and its regulation: canonical and atypical pathways.
NF-κB (nuclear factor-κB) represents a family of dimeric transcription factors, which in mammals comprises RelA (p65), RelB, c-Rel, NF-κB1 (p50) and NF-κB2 (p52) [2]. These structurally homologous proteins form various homodimers and heterodimers via their N-terminal Rel homology domains (RHDs). In unstimulated cells, NF-κB dimers are sequestered in the cytoplasm as latent complexes by physical association of their RHDs with NF-κB inhibitory proteins, IκBs. NF-κB1 and NF-κB2 are produced as precursor proteins, p105 and p100, which share structural homology with IκBs in their C-terminal portion. Like IκBs, p105 and p100 bind to mature NF-κBs and sequester them in the cytoplasm.
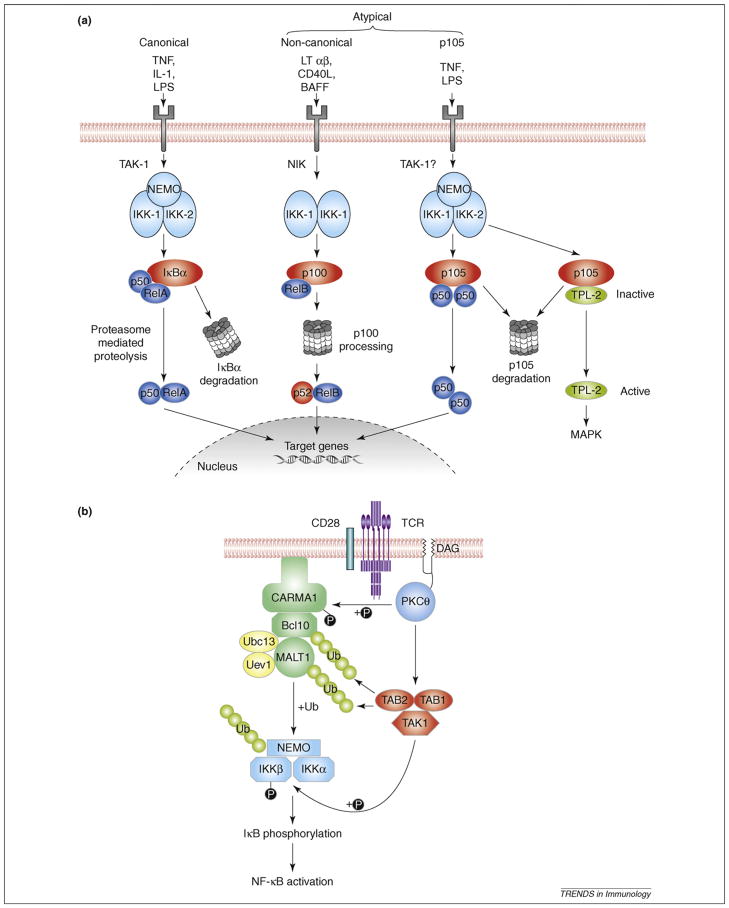
NF-κB activation is typically mediated by proteasomal degradation of the prototypical IκB member, IκBα [2]. This so-called canonical NF-κB pathway is stimulated by various immune receptors, such as the Toll-like receptors (TLRs), interleukin-1 receptor (IL-1R), tumor necrosis factor receptor (TNFR) and antigen receptors. On ligand engagement, each of these triggers signal transduction events that lead to the activation of the IκB kinase (IKK) complex, composed of two catalytic subunits (IKK1 and IKK2) and a regulatory subunit, NEMO (NF-κB essential modulator). Activated IKK phos-phorylates IκBα, predominantly via the action of IKK2, triggering its lysine-48–linked polyubiquitination and proteasomal degradation, releasing associated NF-κB subunits to translocate into the nucleus.
Atypical NF-κB pathways, involving p105 and p100, also have important immune functions [1]. The p100-mediated pathway, often called the noncanonical NF-κB pathway, is triggered by a subset of TNF family members and sequentially activates NF-κB inducing kinase (NIK) and IKK1. IKK1 phosphorylates p100, triggering its polyubiquitination and subsequent partial proteolysis by the proteasome to produce p52, which translocates into the nucleus predominantly in association with RelB [1]. Unlike the noncanonical pathway, constitutive processing of p105 to produce p50 by the proteasome is not regulated by agonist stimulation. However, p105 is phosphorylated by IKK after activation of the canonical pathway, targeting it for complete degradation by the proteasome to release associated NF-κB subunits. Because the inducible degradation of p105 regulates NF-κB as well as activation of Tpl-2 kinase [1], it is considered an atypical pathway. Tpl-2 is a mitogen-activated protein (MAP) 3-kinase that mediates TLR-stimulated activation of extra-cellular signal-regulated kinase (ERK) and production of TNF-α in macrophages, thereby regulating inflammatory responses [1].
Figure 1.
Activation of NF-κB (nuclear factor-κB) by canonical and atypical pathways. (a) NF-κB activation by Toll-like receptor (TLRs) and cytokine receptors. The canonical pathway is activated by a large number of agonists [e.g. tumor necrosis fator (TNF), interleukin 1 (IL-1) or microbial ligands such as lipopolysaccharide (LPS)] through the triggering of various cytokine receptors or TLRs and stimulate the IκB kinase (IKK) complex (IKK1-IKK2-NEMO) to phosphorylate IκBα and promote its degradation. The atypical NF-κB pathways involve stimulus-induced proteolysis of the NF-κB2 p100 (the so-called noncanonical pathway) and NF-κB1 p105 precursor proteins. A limited number of agonists induce processing of p100 to p52 via activation of NF-κB–inducing kinase (NIK) and IKK1, resulting in the nuclear translocation of p52-RelB heterodimers. By contrast, the canonical IKK complex triggers p105 proteolysis, which releases associated Rel subunits and also facilitates activation of the p105-associated TPL-2 MEK kinase, which triggers extracellular signal-regulated kinase (ERK) mitogen-activated protein (MAP) kinase activation. (b) Canonical NF-κB activation by T-cell receptor (TCR). Ligation of TCR and the costimulatory molecule CD28 triggers the association of the intermediate CBM signaling complex, composed of Carma1, Bcl10 and MALT1, with the Ubc13-Uve1 ubiquitin conjugating enzyme heterodimer. This mediates K63-linked ubiquitination of NEMO, Bcl10 and MALT1, promoting the interaction of the CBM complex with IKK and transforming growth factor β (TGF-β)-activated kinase 1 (TAK1), which is a required step for the activation of IKK. A Carma1-independent pathway, which stimulates Tak1 catalytic activity and consequently phosphorylation of the IKK2 activation loop, is also necessary for IKK activation. Ub, ubiquitin.
Regulation of canonical NF-κB signaling by ubiquitination and deubiquitination
Protein ubiquitination plays a critical role in NF-κB activation. Initially shown to mediate proteasomal degradation of IκBs and processing of NF-κB precursors, ubiquitination is now known to also serve as a nondegradative mechanism for activation of the canonical NF-κB pathway (Box 2). This latter function involves the addition of lysine 63 (K63)-linked ubiquitin chains to specific target proteins.
Box 2. Regulation of nuclear factor-κB signaling by K63-type ubiquitination.
Ubiquitination is a posttranslational modification involving covalent conjugation of ubiquitin molecules to target proteins as ubiquitin monomers (monoubiquitination) or ubiquitin chains (polyubiquiti-nation). Polyubiquitin chains can be linked via different internal lysine (K) residues, most commonly K48 and K63. K48-linked ubiquitin chains target substrates for proteasomal degradation, whereas K63-linked polyubiquitin chains mediate nondegradative functions, including activation of IκB kinase (IKK) and its upstream kinase transforming growth factor β (TGF-β)-activated kinase (TAK1) [3]. Ubiquitination is catalyzed by the coordinated actions of three enzymes: ubiquitin activating enzyme (E1), ubiquitin conjugating enzymes (ubc or E2) and ubiquitin ligases (E3). Although several E2s are involved in K48 ubiquitination, the only K63-specific E2 thus far characterized is Ubc13 in complex with a Ubc-like protein, Uev1A [3]. However, genetic evidence suggests the existence of additional K63-specific E2s, because Ubc13 deficiency blocks some, but not all, of the ubiquitination events associated with IKK activation [4]. The TNF receptor–associated factor (TRAF) family is well established to mediate K63-type ubiquitination. TRAF6 is a central component of NF-κB signaling downstream of various receptors, including Toll-like receptors (TLRs), interleukin 1 receptor (IL-1R), receptor activator of NF-κB (RANK), and CD40, whereas TRAF2 mediates NF-κB (nuclear factor-κB) activation by tumor necrosis factor receptors (TNFRs) [3]. On receptor ligation, TRAF2 and TRAF6 undergo self-ubiquitination and induce the ubiquitination of target proteins involved in IKK activation, such as NF-κB essential modulator (NEMO) and several signaling adaptors. Recently, Pellino proteins have been shown to contain a RING-like domain and possess intrinsic E3 ubiquitin ligase activity toward IRAK1 [73,74]. However, a physiological role for Pellino in NF-κB signaling has not yet been established.
Both TAK1 and IKK are dependent on K63-linked ubiquitination for their activation [3]. TAK1 is physically associated with two regulatory proteins: TAB1 and TAB2 (or a homolog, TAB3). TAB2 contains a ubiquitin-binding domain (UBD) and is responsible for recruiting TAK1 to ubiquitinated signaling molecules. Like TAB2, the IKK regulatory subunit, NEMO, contains a UBA domain that mediates recruitment of the IKK complex to ubiquitinated adaptors. In TNF-stimulated cells, TRAF2 mediates the K63 ubiquitination of an adaptor kinase, RIP1, which in turn recruits TAK1 and IKK complexes and mediates their catalytic activation. In the TLR- and IL-1R signaling pathways, ubiquitinated TRAF6 is generally considered a signaling adaptor for recruiting TAK1 and IKK. However, recent evidence suggests that IRAK1 is conjugated with K63-linked ubiquitin chains and provides a binding platform for IKK and possibly other signaling complexes [73,75,76].
Regulation of IκB kinase and mitogen-activated protein kinase activation by NF-κB essential modulator ubiquitination
Different stimuli induce ubiquitination of the IKK (IκB kinase) regulatory subunit, NEMO (NF-κB essential modulator), at distinct lysine (K) acceptor sites [3]. For example, positions K399 and K285 of NEMO are conjugated with K63-linked polyubiquitin chains in cells stimulated via the TCR (T-cell receptor) and NOD2 (nucleotide oligomerization domain), respectively. By contrast, DNA damaging agents induce NEMO monoubiquitination at positions K277 and K309. In each case, mutation of the specific ubiquitination site(s) results in attenuation of IKK activation, suggesting that ubiquitin conjugation to different residues of NEMO contributes to IKK activation. However, recent studies indicate that NEMO ubiquitination might not always be required for IKK activation [4]. Conditional knockout of Ubc13, a major K63-specific ubiquitin-conjugating enzyme (see Box 2), blocks interleukin 1 (IL-1)-stimulated ubiquitination of NEMO in mouse embryonic fibroblasts (MEFs), without affecting the activation of NF-κB [4]. Moreover, germline knockin mutation of murine NEMO at K392 (equivalent to human NEMO K399) prevents its ubiquitination after LPS (lipopolysaccharide) stimulation but has only a minor effect on IKK activation and IκB degradation [5]. Nevertheless, both Ubc13 deficiency and NEMO K392 mutation cause severe defects in antibody production and TLR (Toll-like receptor)-stimulated expression of proinflammatory genes. These findings raise the possibility that NEMO ubiquitination might regulate a signaling pathway involved in adaptive and innate immune responses that does not stimulate NF-κB. It has been suggested that activation of mitogen-activated protein (MAP) kinases [extra-cellular signal-regulated kinase (ERK), c-Jun amino-terminal kinase (JNK), and p38] is dependent on NEMO ubiquitination [4], but LPS activation of MAP kinases is not affected by NEMO K392 mutation [5]. It is also possible that NEMO ubiquitination is not completely blocked by deleting Ubc13 or mutation of a single NEMO lysine residue, and consequently, NEMO signaling function is not impaired. This cannot be ruled out because methods for detection of protein ubiquitination are currently not very sensitive. Finally, it is important to note that the lack of a prominent NF-κB signaling phenotype in conditional Ubc13 knockout mice [4] could also be caused by variations in knockout efficiency, because a more clear defect in TLR-stimulated activation of NF-κB was detected in mice heterozygous for a conventional Ubc13 knockout mutation [6].
Ubc13 and ubiquitin-dependent activation of transforming growth factor β–activated kinase 1 and IKK
In vitro studies suggest that TAK1 [transforming growth factor β (TGF-β)-activated kinase 1] is a ubiquitin-dependent IKK-activating kinase that is regulated specifically by K63-linked ubiquitination [3]. Consistently, conditional knockout of Ubc13 in thymocytes results in impaired TCR activation of TAK1 and NF-κB [7]. However, in Ubc13-deficient B cells, macrophages and MEFs, NF-κB activation by several agonists is normal [4]. Ubc13-deficient MEFs remain competent in IL-1β–stimulated ubiquitination of TRAF6 (tumor necrosis factor receptor–associated factor 6) and downstream activation of TAK1. Therefore, Ubc13 may be redundant in mediating K63-linked ubiquitination of NF-κB signaling components in certain cell types or after stimulation with particular agonists.
How ubiquitination triggers TAK1 activation and TAK1-mediated activation of IKK is incompletely understood. TNF-α (tumor necrosis factor α) stimulation of TAK1 and IKK involves recruitment of these kinases to K63-ubiquitinated RIP1 (receptor-interacting protein 1) (Box 2). TCR stimulation similarly promotes recruitment of TAK1 and IKK to the paracaspase MALT1, which is conjugated with K63-linked ubiquitin chains [8]. MALT1 is a component of a TCR intermediate signaling complex, which additionally contains the scaffold protein Carma1 and the adaptor Bcl10 [9] (Figure 1b). Within this so-called ‘CBM’ complex, Bcl10 is also conjugated with K63-linked ubiquitin chains and associates with IKK via NEMO [10]. It is generally thought that assembly of ubiquitin-dependent signaling complexes both activates TAK1 and allows TAK1 to efficiently phosphorylate and activate IKK. However, recent findings suggest that the CBM complex regulates only some of the events that lead to NF-κB activation [11]. Loss of Carma1 or Bcl10 blocks NEMO ubiquitination and IKK activation but has no effect on stimulation of TAK1 activity and activation loop phos-phorylation of IKK1 or 2. These findings indicate that activation of IKK by the TCR might involve two separate steps: a CBM-independent step mediating TAK1 activation and IKK1 or 2 phosphorylation and a CBM-dependent step mediating NEMO ubiquitination (Figure 1b). Because TCR activation of TAK1 nevertheless requires Ubc13 [7], it is likely that another Ubc13-dependent ubiquitination event triggers activation of TAK1. This may involve TAK1 itself, because transfected TAK1 undergoes K63-linked ubiquitination, which seems to be required for its autoactivation [12]. Ubiquitination of endogenous TAK1 and TAK1-associated proteins also occurs in T cells and correlates with TAK1 catalytic activation [13].
Negative regulation of NF-κB by deubiquitinases
Ubiquitination is a reversible process, with the removal of ubiquitin being mediated by a family of deubiquitinating enzymes (DUBs) [14]. Several DUBs have been demonstrated to deconjugate K63-linked ubiquitin chains and negatively regulate the activation of IKK, among which CYLD (cylindromatosis) and A20 are the best characterized [15]. The association of both of these genes with several human autoimmune diseases underscores their importance in regulating immune responses and inflammation [15].
CYLD was originally identified as a tumor suppressor involved in familial cylindromatosis, a predisposition to benign tumors of hair follicles. CYLD mutations in cylindromatosis patients occur frequently in its C-terminal portion, which contains a DUB domain belonging to the ubiquitin-specific protease (USP) subfamily. CYLD physically interacts with and deubiquitinates NEMO, thereby negatively regulating NF-κB activation [15]. CYLD also removes K63-linked ubiquitin from TRAF2, TRAF6, TRAF7, RIP1, TAK1 and Bcl-3 [15]. With the exception of Bcl-3, K63 ubiquitination of these factors contributes to the activation of IKK. Bcl-3 ubiquitination, by contrast, seems to promote its translocation into the nucleus, where it functions as a transcriptional coactivator of p50- and p52-containing complexes [16].
A20 is a zinc-finger-containing DUB that is a member of the ovarian tumor-related proteases (OTUs) subfamily and plays a critical role in terminating NF-κB activation by innate immune receptors, such as TNFR1, TLRs and NOD2 [17–20]. Like CYLD, A20 can deubiquitinate several NF-κB regulatory proteins, including TRAF6, RIP1, RIP2 and NEMO [19–23]. It has been suggested that A20 functions both as a DUB and an E3 ligase [22]. A20 removes K63-linked ubiquitin chains from RIP1 and concurrently stimulates conjugation of K48-linked ubiquitin chains to RIP1, which targets it for proteasomal degradation. A20 therefore attenuates the signaling function of RIP1 by first removing K63-linked ubiquitin chains and then promoting its proteolysis. It is not known why de-ubiquitination alone is not sufficient to turn off RIP1 signaling activity. It has been suggested that the C-terminal zinc finger domain confers E3 ligase activity to A20. However, it is uncertain whether the E3 ligase activity associated with A20 is intrinsic, because Itch E3 ligase can be recruited to A20 via TAX1BP1 (TAX1-binding protein 1), an adaptor that was originally found to associate with the Tax protein encoded by human T-cell leukemia virus type I [24].
Although A20 and CYLD target the same set of NF-κB signaling factors, knockout mouse studies indicate nonredundant roles. CYLD functions as a constitutively active DUB that prevents spontaneous ubiquitination of its targets [13,25,26]. A major phenotype associated with CYLD deficiency therefore is constitutive activation of IKK and NF-κB [13,26–28]. By contrast, A20 functions inducibly and is required for termination of signal-induced NF-κB activation [18,19]. The level of A20 is low in unstimulated cells but is strongly elevated in activated cells through NF-κB–mediated A20 gene induction [17]. The DUB catalytic activity of A20 might also be positively regulated through its phosphorylation by IKK2 [29]. Thus, CYLD and A20 seem to regulate the initial and resolving phases of NF-κB activation, respectively.
Ubiquitin-binding adaptors in the regulation of NF-κB signaling
Several ubiquitin-binding adaptors (UBAs) have been identified that contain a ubiquitin-binding domain (UBD) and have positive or negative functions in NF-κB signaling. The best-characterized UBAs with positive-signaling functions are TAK1-binding proteins 2 and 3 (TAB2 and TAB3) and the IKK regulatory subunit NEMO. As discussed above, these UBAs recruit TAK1 and IKK to upstream ubiquitinated proteins for the assembly of IKK activation complexes. The NEMO homolog optineurin contains a UBD, and gene knockdown experiments indicate that optineurin inhibits TNFα-stimulated NF-κB activation by antagonizing the binding of NEMO to ubiquiti-nated RIP1 [30]. The ABIN (A20-binding inhibitor of NF-κB) family proteins, ABIN-1, ABIN-2 and ABIN-3, also contain a UBD homologous to that of NEMO and can bind to K63 ubiquitinated proteins [31]. Each of these proteins inhibits NF-κB activation when overexpressed, and this activity is abolished when the UBD is mutated. However, a physiological role for ABIN proteins in NF-κB regulation remains to be established.
A UBA for which a physiological NF-κB inhibitory function has been clearly established is TAX1BP1 [32,33]. TAX1BP1 interacts with A20 and mediates the recruitment of A20 to K63-ubiquitinated RIP1 and TRAF6 [32,33]. Thus, in TAX1BP1-deficient cells, A20 fails to deubiquitinate RIP1 and TRAF6 or terminate the IKK activation signal, resulting in hyperproduction of proinflammatory cytokines [32,33]. The DUB function of CYLD also seems to involve UBAs. For example, p62 (also known as sequestosome 1) recruits CYLD to ubiquitinated TRAF6 [34,35] and facilitates CYLD inhibition of NF-κB activation by receptor activator of NF-κB (RANK) [34].
Noncanonical pathway of NF-κB activation
Processing of p100 and inducible degradation of p105 represent two major atypical pathways of NF-κB activation, with the former being known as the noncanonical NF-κB pathway (Figure 1a). Because p100 and p105 function as both NF-κB precursor proteins and IκB-like molecules (Box 1), their processing and degradation play important roles in mediating NF-κB signaling. The regulation of p105-specific pathway has been thoroughly discussed in a previous review [1], and this section focuses on the recent progress in our understanding of the noncanonical p100 pathway. Processing of p100 is a tightly regulated event that is induced by NF-κB–inducing kinase (NIK), a MAP kinase kinase kinase (MAP 3-kinase) that acts via activation of IKK1 [36,37]. IKK1 phosphorylates p100 at two C-terminal serines, which promotes p100 ubiquitination by the SCFβTrCP ubiquitin E3 ligase, partial proteolysis by the proteasome and subsequent nuclear translocation of p52-RelB complexes [36,38]. This noncanonical NF-κB signaling pathway is induced by a subset of TNFR superfamily members, including B cell–activating factor receptor (BAFFR), CD40, lymphotoxin β receptor (LTβR) and RANK [1] (Figure 1a).
Regulation of NIK stability by TRAF3
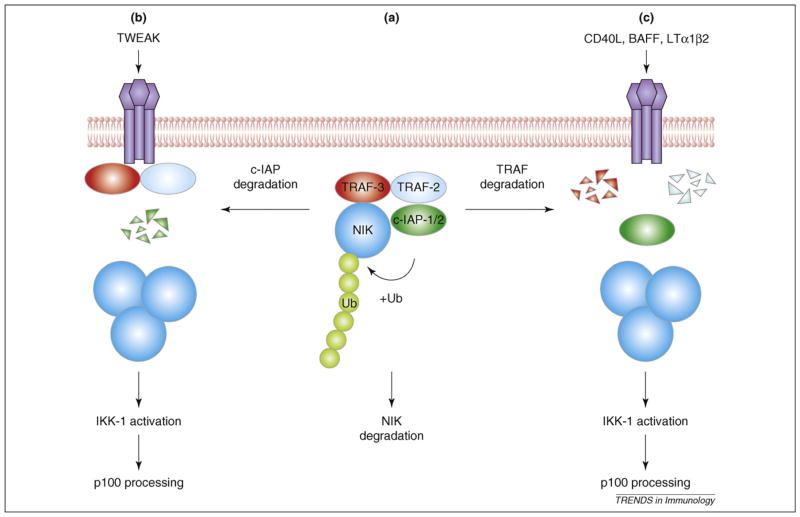
Stimulation of the noncanonical NF-κB pathway is slow and depends on protein synthesis because of the unusual mechanism of NIK activation [1]. Steady-state levels of NIK are normally very low, because of its constitutive degradation via a TRAF3-dependent mechanism (Figure 2). TRAF3 physically associates with a novel N-terminal domain of NIK and induces NIK ubiquitination and degradation [39]. Ligand stimulation induces TRAF3 degradation, resulting in NIK stabilization, thereby allowing the newly synthesized NIK to accumulate and activate IKK1. Consistently, TRAF3 deficiency results in accumulation of NIK and activation of p100 processing [39,40]. Stabilized NIK also induces delayed activation of the canonical NF-κB pathway after LTβR stimulation, resulting in upregulation of proinflammatory cytokines [41]. Thus, NIK might be able to induce the activation of both noncanonical and canonical IKK complexes.
Figure 2.
A model of NF-κB–inducing kinase (NIK) regulation and noncanonical NF-κB (nuclear factor-κB) signaling. TNF receptor–associated factor 2 (TRAF2) and TRAF3 physically associate with cellular inhibitor of apoptosis 1 (c-IAP1; or c-IAP2) and NIK, respectively, and form a NIK-degradation complex via TRAF2-TRAF3 dimerization. Within this complex, c-IAP1 or 2 functions as an E3 ubiquitin ligase that mediates NIK polyubiquitination and proteolysis, thereby preventing NIK accumulation and noncanonical NF-κB signaling (a). Induction of noncanonical NF-κB signaling by different inducers involves degradation of c-IAP1 or 2 (b) or TRAF2 and 3 (c), stabilizing NIK, which triggers activation of NF-κB.
TRAF2 and cellular inhibitors of apoptosis are components of the NIK-degradation complex
Although TRAF3 induces the ubiquitination and proteasomal degradation of NIK, it is not a direct E3 ligase for NIK [39]. Two recent studies have indicated that NIK stability is regulated by cellular inhibitor of apoptosis 1 (c-IAP1) and c-IAP2 [42,43], which function as E3 ligases because of their C-terminal RING domains. The antiapoptotic function of c-IAPs can be inhibited by a natural antagonist, the mitochondrial activator of caspases (SMAC), and by synthetic SMAC mimetics [44]. Synthetic c-IAP antagonists induce rapid degradation of c-IAP1 and c-IAP2 via the ubiquitin-proteasome pathway [42,43]. Remarkably, c-IAP antagonists also activate both the canonical and noncanonical NF-κB pathways, which seem to involve RIP1 recruitment to TNFR1 and NIK stabilization, respectively [42,43]. Transfected c-IAP1 induces NIK ubiquitination and degradation, which is dependent on the RING domain of c-IAP1 [42]. These data suggest that c-IAPs function as specific E3 ligases for NIK (Figure 2).
Interaction of c-IAP1 with NIK seems to be mediated by the adaptor function of TRAF2, because a mutant form of c-IAP1 defective in TRAF2 binding cannot associate with NIK or induce its ubiquitination. Consistently, TRAF2, like TRAF3, functions as a negative regulator of the non-canonical pathway in B cells [45]. Agonist induction of p100 processing is associated with degradation of TRAF2 and TRAF3 [39], events that are dependent on the zinc finger domain of TRAF2 [46]. Notably, simultaneous deletion of TRAF2 and TRAF3 does not cause further enhancement of noncanonical NF-κB signaling compared with individual deletions, suggesting that these TRAFs function in the same pathway [47]. Because TRAF2 dimerizes with TRAF3 and physically interacts with c-IAPs and NIK, an attractive model is that these two TRAF members bridge NIK with its ubiquitin ligases c-IAP1 and 2, and thus both serve as essential components of the NIK-degradation complex. Agonist inactivation of this complex seems to involve degradation of either c-IAPs or TRAF2 and TRAF3. For example, degradation of c-IAP1 is associated with induction of p100 processing by a TNF family member, TWEAK (TNF-like weak inducer of apoptosis) [42], whereas TRAF2 and TRAF3 proteolysis is induced by several other agonists of the noncanonical pathway [39,46] (Figure 2). It remains to be determined whether the E3 activity of c-IAPs mediates their self-degradation and the degradation of TRAF2 and TRAF3.
Regulation of NF-κB activity in the nucleus
RelA phosphorylation
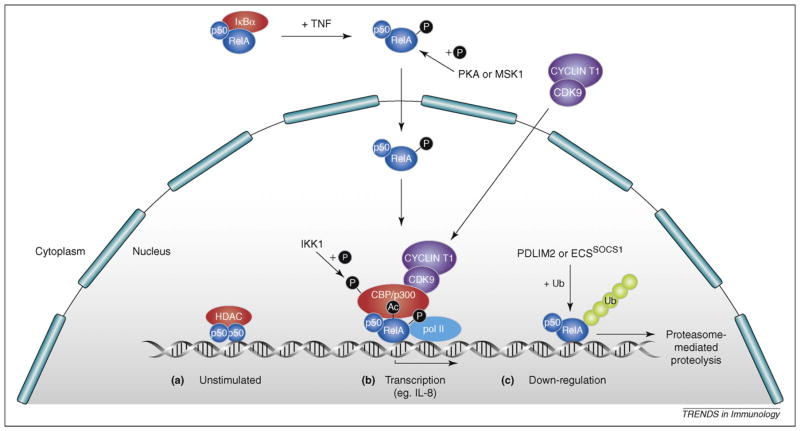
Posttranslational modification of Rel subunits and their association with other nuclear proteins are critical to regulate the transcriptional activity and specificity of NF-κB dimers (see Refs. [2,48] for recent reviews). For example, RelA phosphorylation on S276 promotes its interaction with the histone acetyltransferases CBP (CREB-binding protein) and p300 while displacing repressive histone deacetylase (HDAC) proteins [49] (Figure 3). The physiological significance of RelA S276 phosphorylation in NF-κB activation has recently been investigated by generation of a RelAS276A (serine to alanine mutation) knock-in mouse strain [50]. Remarkably, these mice die from variegated developmental abnormalities caused by recruitment of HDACs by RelAS276A into the vicinity of genes positioned near to NF-κB binding sites. Thus, unphosphorylated nuclear RelA can affect the expression of genes not directly regulated by NF-κB via epigenetic mechanisms. It is not clear whether this reflects a normal function for wild-type RelA in heterochromatin regulation [51]. Analysis of MEFs from RelAS276A knock-in mice confirmed the previous finding that S276 phosphorylation controls RelA association with CBP, but showed that this is important for expression of only a subset of NF-κB–regulated genes [50]. Interestingly, genes that are affected by the RelAS276A mutation do not correlate with their chromatin accessibility status, indicating that this is not solely determined by recruitment of CBP by RelA [52].
Figure 3.
Regulation of the transcriptional activity of NF-κB (nuclear factor-κB). (a) In unstimulated cells, homodimers of p50 and p52 actively repress transcription of NF-κB target genes to which they are bound by recruitment of histone deacetylases (HDAC). (b) After ligand stimulation [e.g. via tumor necrosis factor (TNF)], p50-RelA dimers are released from degraded IκBα to translocate into the nucleus and displace repressive p50 and p52 homodimers from NF-κB target gene promoters. Transcriptional activity of RelA is regulated by phosphorylation on multiple sites, of which serine (S) 276 is the best understood. S276 can be phosphorylated by protein kinase A (PKA) and mitogen-and stress-activated protein kinases (MSKs) 1 and 2, inducing binding to CREB-binding protein (CBP) and p300 co-activator complexes, which acetylate adjacent histones and RelA to stimulate target gene transcription. RelA S276 phosphorylation also triggers recruitment of a complex of CDK1 and cyclin T1, which promotes transcriptional elongation. IκB kinase 1 (IKK1)phosphorylation of CBP-p300 stimulates preferential binding to NF-κB complexes. (c) Downregulation of NF-κB activity involves several different mechanisms. NF-κB dimers can be transported back to the cytoplasm as a result of NF-κB–dependent IκBα resynthesis. Alternatively, RelA can be ubiquitinated by ECSSOCS1 or PDLIM2 E3 ligases, triggering its degradation by the proteasome. c-Rel–mediated transcription is also turned off by ubiquitin-dependent proteolysis in stimulated cells. Ub, ubiquitin.
ChIp (chromatin immunoprecipitation) assays have provided insight into the gene specific role of RelA S276 phosphorylation in TNF-induced expression of NF-κB target genes [53]. Binding of phospho-S276-RelA is primarily restricted to a subset of genes that are rapidly induced by TNF, such as the chemokine IL-8. Induction of such genes involves the association of phospho-S276-RelA with the P-TEFb transcriptional elongation factor, a complex of CDK-9 (cyclin-dependent kinase-9) and cyclin T1, which controls Pol II recruitment and activation. Inhibition of P-TEFb activity confirms that the phospho-S276-RelA/P-TEFb complex is required for IL-8 expression. By contrast, expression of phospho-S276-RelA–independent genes, such as IκBα, is mediated by Pol II prebound to the promoter and does not require P-TEFb activity. Thus, the phosphorylation of RelA controls its interactions with both coactivators and transcriptional elongation factors.
RelA acetylation
RelA also undergoes stimulus-induced acetylation on multiple sites [54], which seems to be mediated by associated CBP and p300 [55] (Figure 3). Acetylation of K221 increases the DNA-binding affinity of RelA for κB sites, whereas acetylation of K310 is required for full trans-activation by the NF-κB complex [54]. IKK1 directly phosphorylates and stimulates CBP acetyl transferase activity after TNF stimulation [56]. Interestingly, this promotes CBP binding to RelA while decreasing binding to p53, resulting in augmented expression of NF-κB–regulated antiapoptotic genes and reduced expression of p53-regulated proapoptotic genes. Acetylation of RelA also couples the activation of NF-κB to other signaling pathways. For example, in epithelial cells, TGF-β1 induces RelA acetylation on K221 via a Smad3 and 4–PKA–p300-dependent signaling pathway [57]. This is essential for TGF-β to synergistically enhance NF-κB-dependent transcription of TNF and IL-1β induced by Haemophilus influenzae bacteria.
Negative regulation of NF-κB in the nucleus
NF-κB–dependent expression of IκBα is one important mechanism by which NF-κB activity is switched off in a cell intrinsic manner. However, NF-κB activity can be terminated in the absence of IκBα, indicating the existence of additional regulatory mechanisms [58]. One of these involves the polyubiquitination and proteasome-mediated degradation of nuclear RelA, which is mediated by a multimeric ubiquitin ligase containing Elongins B and C, Cul2 and SOCS1 (ECSSOCS1) [59] (Figure 3). ECSSOCS1 regulation of RelA proteolysis requires COMMD1, a ubiquitously expressed inhibitor of NF-κB that is recruited to κB-responsive promoters [60,61]. COMMD1 interacts with ECSSOCS1 and increases the interaction between SOCS1 and RelA [62]. Knockdown of COMMD1 stabilizes nuclear RelA after TNF stimulation, enhancing NF-κB–dependent cellular responses. PDLIM2, a nuclear protein containing both PDZ and LIM domains, can also function as an E3 ligase for nuclear RelA [63]. PDLIM2 targets RelA to discrete intranuclear compartments, known as PML nuclear bodies, where polyubiquitinated RelA is degraded by the proteasome. PDLIM2 deficiency results in increased nuclear RelA, defective RelA ubiquitination and augmented production of proinflammatory cytokines in response to innate stimuli. It is unknown what determines whether RelA is ubiquitinated by ECSSOCS1 or PDLIM2 E3 ligases.
Studies using IKK1AA mice, which express an IKK1 mutant that lacks activation loop phosphorylation sites, suggest that IKK1 negatively regulates NF-κB–dependent transcription in macrophages after LPS stimulation [64]. It has been proposed that IKK1 turns off promoter-bound NF-κB by stimulating the ubiquitination and proteasome-mediated proteolysis of RelA and c-Rel, possibly by controlling recruitment of specific E3 ligases. However, RelA subunit stability is normal in IKK1-deficient macrophages, and the role of IKK1 in regulating Rel subunit turnover remains unclear [65]. LPS stimulation of macrophages also increases the ubiquitination and proteolysis of p50 by the proteasome [66]. This is inhibited by BCL3, which blocks p50 ubiquitination and stabilizes a p50 complex that inhibits gene transcription. Consequently, Bcl-3−/− mice and cells are hypersensitive to TLR activation. The E3 ligase that ubiquitinates p50 has not been characterized, and it is not clear whether BCL3 regulation of p50 stability can be modulated by TLR signals.
PIAS (protein inhibitor of activated STAT) family proteins are transcriptional regulators that function as SUMO E3 ligases [67]. PIAS1 inhibits binding of NF-κB and STAT1 to gene promoters and is particularly important in negative regulation of proinflammatory cytokines and chemokines. Consistently, PIAS1-deficient mice display increased protection against pathogenic infections and are hypersensitive to LPS-induced septic shock. After TNF or LPS stimulation, PIAS1 is rapidly phosphorylated on S90, which is required for PIAS1 to block the promoter binding of RelA [68]. Interestingly, PIAS1 S90 phosphorylation is mediated by IKK1, suggesting that impaired PIAS1 phosphorylation might explain the increased NF-κB–dependent transcription of IKK1AA macrophages.
New nuclear partners for NF-κB
The NF-κB transcriptional response is dependent on the interaction of NF-κB with other transcription factors and nuclear regulatory proteins [48]. In this way, the NF-κB function can be integrated with other signaling pathways, and weak DNA–transcription factor interactions can be augmented. Recent studies have led to the identification of several novel nuclear cofactors that regulate NF-κB transcriptional activity.
A partner of RelA, the ribosomal protein S3 (RPS3), was identified by affinity purification and shown to substantially increase the DNA binding activity of RelA-RelA homodimers and p50-RelA heterodimers [69]. RPS3 was recruited to the promoters of a subset of NF-κB–regulated genes whose induction after TCR stimulation was impaired by RPS3 knockdown in Jurkat T cells. Thus, RPS3 seems to be a previously unknown non-Rel subunit of NF-κB important for selection of genomic κB sites to be activated after TCR stimulation. It will clearly be important in future studies to confirm the role of RPS3 in regulating NF-κB function in primary T cells and in other cell lineages.
The second novel NF-κB cofactor, Akirin, was identified in an RNA interference screen for genes that were required for expression of the NF-κB–regulated antimicrobial peptide Attacin in Drosophila S2 cells infected with gram-negative bacteria [70]. Akirin is a nuclear protein that acts in parallel with the NF-κB transcription factor Relish, downstream of the Imd signaling pathway [71]. Mice encode two Akirin homologs, Akirin1 and Akirin2. Although Akirin1 knockout mice have no obvious phenotype, Akirin2 knockout mice are embryonic lethal, and the Akirin2-deficient MEFs are defective in the induction of a subset of NF-κB target genes in response to TLR ligands, IL-1β or TNF [70]. The precise mechanism by which Akirins modulate NF-κB activity is not known, but one possibility is that these proteins interact with chromatin or components of the transcriptional machinery.
Another novel NF-κB cofactor was identified in a genome-wide screen for RelA binding sites in LPS-stimulated monocytes [72]. An over-representation of the E2F1-binding motifs was detected among RelA-bound loci associated with expressed NF-κB target genes. RNAi knockdown revealed a requirement for E2F1 for the transcriptional activation of several LPS-inducible NF-κB target genes, including IL-1β, TNF and CCL3. E2F1 is rapidly recruited to the promoters of these genes via its physical interaction with p50-RelA heterodimers in LPS-stimulated cells. Thus E2F1 cooperates with NF-κB in the transcriptional regulation of specific LPS-responsive genes.
Concluding remarks
It has now been > 20 years since the original identification of NF-κB as a regulator of κB light chain expression in B cells. Nevertheless, NF-κB is still a major area of research and serves as a paradigm for a signaling pathway that is regulated by ubiquitination. The importance of NF-κB in inflammation, autoimmunity and cancer is clearly established, and there has been a considerable effort by the pharmaceutical industry to develop IKK inhibitors to treat these diseases. However, such inhibitors will cause global inhibition of NF-κB activation and can have many unwanted side effects. It will be important in the future to identify and generate drugs to alternative targets, which allow NF-κB activity to be modulated in a more restricted fashion. This aim should be facilitated as future research addresses several key issues that are outstanding in NF-κB research:
Establish how ubiquitination regulates TAK1 and IKK activation
Determine how the deubiquitinase activities of CYLD and A20 are dynamically regulated
Establish the physiological role of IKK-induced p105 proteolysis in NF-κB activation
Investigate the in vivo significance of Rel subunit phosphorylation and acetylation in NF-κB activation by generation of knock-in mouse strains
Determine how Rel E3 ligases such as PDLIM2, are regulated during immune responses
The answers to these questions are likely to be answered soon in this fast moving area of research and, if past experience is anything to go by, will have major implications for our understanding of signal transduction pathways in general.
Acknowledgments
Work performed in the authors’ laboratory is supported by the University of Texas MD Anderson Cancer Center, and National Institutes of Health Grants R01 AI064639, R01 AI057555, and R01 CA94922 (to S.C.S.) and UK Medical Research Council (to S.C.L.). We apologize to those investigators whose work was not cited because of space limitations.
Glossary
- ABIN
A20-binding inhibitor of NF-κB
- BAFFR
B cell–activating factor receptor
- CBP
CREB-binding protein
- CDK
cyclin-dependent kinase
- ChIp
chromatin immunoprecipitation
- c-IAP
cellular inhibitor of apoptosis
- CYLD
cylindromatosis
- DUB
deubiquitinating enzyme
- ECSSOCS1
ubiquitin ligase containing Elongins B and C, Cul2 and SOCS1
- HDAC
histone deacetylase
- IKK
IκB kinase
- IκB
inhibitor of NF-κB
- LTβR
lymphotoxin β receptor
- MAP3K
MAP kinase kinase kinase
- NF-κB
nuclear factor-κB
- NIK
NF-κB–inducing kinase
- RANK
receptor activator of NF-κB
- RIP1
receptor-interacting protein 1
- TAK1
transforming growth factor β–activated kinase 1
- TAX1BP1
TAX1-binding protein 1
- TLR
Toll-like receptor
- TNF
tumor necrosis factor
- TRAF
TNF receptor–associated factor
- UBA
ubiquitin-binding adaptors
- USP
ubiquitin-specific protease
References
- 1.Beinke S, Ley SC. Functions of NF-κB1 and NF-κB2 in immune cell biology. Biochem J. 2004;382:393–409. doi: 10.1042/BJ20040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayden MS, Ghosh S. Shared principles in NF-κB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 3.Chen ZJ. Ubiquitin signaling in the NF-κB pathway. Nat Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamamoto M, et al. Key function for the Ubc13 E2 ubiquitin-conjugating enzyme in immune receptor signaling. Nat Immunol. 2006;7:962–970. doi: 10.1038/ni1367. [DOI] [PubMed] [Google Scholar]
- 5.Ni CY, et al. K63-linked polyubiquitination of NEMO modulates TLR signaling and inflammation in vivo. J Immunol. 2008;180:7107–7111. doi: 10.4049/jimmunol.180.11.7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukushima T, et al. Ubiquitin-conjugating enzyme Ubc13 is a critical component of TNF receptor-associated factor (TRAF)-mediated inflammatory responses. Proc Natl Acad Sci U S A. 2007;104:6371–6376. doi: 10.1073/pnas.0700548104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamamoto M, et al. Cutting Edge: Pivotal function of Ubc13 in thymocyte TCR signaling. J Immunol. 2006;177:7520–7524. doi: 10.4049/jimmunol.177.11.7520. [DOI] [PubMed] [Google Scholar]
- 8.Oeckinghaus A, et al. Malt1 ubiquitination triggers NF-kB signaling upon T-cell activation. EMBO J. 2007;26:4634–4645. doi: 10.1038/sj.emboj.7601897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rawlings DJ, et al. The CARMA1 signalosome links the signaling machinery of adaptive and innate immunity in lymphocytes. Nat Rev Immunol. 2006;6:799–812. doi: 10.1038/nri1944. [DOI] [PubMed] [Google Scholar]
- 10.Wu CJ, Ashwell JD. NEMO recognition of ubiquitinated Bcl10 is required for T cell receptor-mediated NF-κB activation. Proc Natl Acad Sci U S A. 2008;105:3023–3028. doi: 10.1073/pnas.0712313105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shambharkar PB, et al. Phosphorylation and ubiquitination of the IκB kinase complex by two distinct signaling pathways. EMBO J. 2007;26:1794–1805. doi: 10.1038/sj.emboj.7601622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thiefes A, et al. The Yersinia enterocolitica effector YopP inhibits host cell signaling by inactivating the protein kinase TAK1 in the IL-1 signaling pathway. EMBO Rep. 2006;7:838–844. doi: 10.1038/sj.embor.7400754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reiley WW, et al. Deubiquitinating enzyme CYLD negatively regulates the ubiquitin-dependent kinase Tak1 and prevents abnormal T cell responses. J Exp Med. 2007;204:1475–1485. doi: 10.1084/jem.20062694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nijman SM, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Sun SC. Deubiquitylation and regulation of the immune response. Nat Rev Immunol. 2008;8:501–511. doi: 10.1038/nri2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Massoumi R, et al. CYLD inhibits tumor cell proliferation by blocking bcl-3-dependent NF-κB signaling. Cell. 2006;125:665–677. doi: 10.1016/j.cell.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 17.Beyaert R, et al. A20 and A20-binding proteins as cellular inhibitors of nuclear factor-κB-dependent gene expression and apoptosis. Biochem Pharmacol. 2000;60:1143–1151. doi: 10.1016/s0006-2952(00)00404-4. [DOI] [PubMed] [Google Scholar]
- 18.Lee EG, et al. Failure to regulate TNF-induced NF-κB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boone DL, et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 20.Hitotsumatsu O, et al. The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals. Immunity. 2008;28:381–390. doi: 10.1016/j.immuni.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evans PC, et al. Zinc-finger protein A20, a regulator of inflammation and cell survival, has de-ubiquitinating activity. Biochem J. 2004;378:727–734. doi: 10.1042/BJ20031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signaling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 23.Mauro C, et al. ABIN-1 binds to NEMO/IKKγ and co-operates with A20 in inhibiting NF-κB. J Biol Chem. 2006;281:18482–18488. doi: 10.1074/jbc.M601502200. [DOI] [PubMed] [Google Scholar]
- 24.Shembade N, et al. The E3 ligase Itch negatively regulates inflammatory signaling pathways by controlling the function of the ubiquitin-editing enzyme A20. Nat Immunol. 2008;9:254–262. doi: 10.1038/ni1563. [DOI] [PubMed] [Google Scholar]
- 25.Reiley W, et al. Regulation of the deubiquitinating enzyme CYLD by IκB kinase gamma-dependent phosphorylation. Mol Cell Biol. 2005;25:3886–3895. doi: 10.1128/MCB.25.10.3886-3895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wright A, et al. Regulation of early wave of germ cell apoptosis and spermatogenesis by deubiquitinating enzyme CYLD. Dev Cell. 2007;13:705–716. doi: 10.1016/j.devcel.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 27.Jin W, et al. Deubiquitinating enzyme CYLD regulates the peripheral development and naive phenotype maintenance of B cells. J Biol Chem. 2007;282:15884–15893. doi: 10.1074/jbc.M609952200. [DOI] [PubMed] [Google Scholar]
- 28.Hövelmeyer N, et al. Regulation of B cell homeostasis and activation by the tumor suppressor gene CYLD. J Exp Med. 2007;204:2615–2627. doi: 10.1084/jem.20070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutti JE, et al. IκB kinase β phosphorylates the K63 deubiquitinase A20 to cause feedback inhibition of the NF-κB pathway. Mol Cell Biol. 2007;27:7451–7461. doi: 10.1128/MCB.01101-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu G, et al. Optineurin negatively regulates TNFα-induced NF-κB activation by competing with NEMO for ubiquitinated RIP. Curr Biol. 2007;17:1438–1443. doi: 10.1016/j.cub.2007.07.041. [DOI] [PubMed] [Google Scholar]
- 31.Wagner S, et al. Ubiquitin binding mediates the NF-κB inhibitory potential of ABINs. Oncogene. 2008;27:3739–3745. doi: 10.1038/sj.onc.1211042. [DOI] [PubMed] [Google Scholar]
- 32.Shembade N, et al. Essential role for TAX1BP1 in the termination of TNF-alpha-, IL-1- and LPS-mediated NF-κB and JNK signaling. EMBO J. 2007;26:3910–3922. doi: 10.1038/sj.emboj.7601823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iha H, et al. Inflammatory cardiac valvulitis in TAX1BP1-deficient mice through selective NF-κB activation. EMBO J. 2008;27:629–641. doi: 10.1038/emboj.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin W, et al. Deubiquitinating enzyme CYLD regulates RANK signaling and osteoclastogenesis. J Clin Invest. 2008;118:1858–1866. doi: 10.1172/JCI34257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wooten MW, et al. Essential role of sequestosome 1/p62 in regulating accumulation of Lys63-ubiquitinated proteins. J Biol Chem. 2008;283:6783–6789. doi: 10.1074/jbc.M709496200. [DOI] [PubMed] [Google Scholar]
- 36.Xiao G, et al. NF-κB-inducing kinase regulates the processing of NF-κB2 p100. Mol Cell. 2001;7:401–409. doi: 10.1016/s1097-2765(01)00187-3. [DOI] [PubMed] [Google Scholar]
- 37.Senftleben U, et al. Activation of IKKα of a second, evolutionary conserved, NF-κB signaling pathway. Science. 2001;293:1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- 38.Liang C, et al. βTrCP binding and processing of NF-κB2/p100 involve its phosphorylation at serines 866 and 870. Cell Signal. 2006;18:1309–1317. doi: 10.1016/j.cellsig.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 39.Liao G, et al. Regulation of the NF-κB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J Biol Chem. 2004;279:26243–26250. doi: 10.1074/jbc.M403286200. [DOI] [PubMed] [Google Scholar]
- 40.He JQ, et al. Rescue of TRAF3-null mice by p100 NF-κB deficiency. J Exp Med. 2006;203:2413–2418. doi: 10.1084/jem.20061166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zarnegar B. Control of canonical NF-κB activation through the NIK-IKK complex pathway. Proc Natl Acad Sci U S A. 2008;105:3503–3508. doi: 10.1073/pnas.0707959105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Varfolomeev E, et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-κB activation, and TNFα-dependent apoptosis. Cell. 2007;131:669–681. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 43.Vince JE, et al. IAP antagonists target cIAP1 to induce TNFα-dependent apoptosis. Cell. 2007;131:682–693. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
- 44.Wu H, et al. Smac mimetics and TNFα: a dangerous liaison? Cell. 2007;131:655–658. doi: 10.1016/j.cell.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grech AP, et al. TRAF2 differentially regulates the canonical and noncanonical pathways of NF-κB activation in mature B cells. Immunity. 2004;21:629–642. doi: 10.1016/j.immuni.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 46.Moore CR, Bishop GA. Differential regulation of CD40-mediated TNF receptor-associated factor degradation in B lymphocytes. J Immunol. 2005;175:3780–3789. doi: 10.4049/jimmunol.175.6.3780. [DOI] [PubMed] [Google Scholar]
- 47.Gardam S, et al. TRAF2 and TRAF3 signal adapters act cooperatively to control the maturation and survival signals delivered to B cells by the BAFF receptor. Immunity. 2008;28:391–401. doi: 10.1016/j.immuni.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 48.Perkins ND. Integrating cell-signaling pathways with NF-κB and IKK function. Nat Rev Mol Cell Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 49.Viatour P, et al. Phosphorylation of NF-κB and IκB proteins: implications in cancer and inflammation. Trends Biochem Sci. 2005;30:43–52. doi: 10.1016/j.tibs.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 50.Dong J, et al. Repression of gene expression by unphosphorylated NF-κB p65 through epigenetic mechanisms. Genes Dev. 2008;22:1159–1173. doi: 10.1101/gad.1657408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng CS, et al. Epigenetic control: slow and global, nimble and local. Genes Dev. 2008;22:1110–1114. doi: 10.1101/gad.1677008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saccani S, et al. Two waves of nuclear factor κB recruitment to target promoters. J Exp Med. 2001;193:1351–1359. doi: 10.1084/jem.193.12.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nowak DE, et al. RelA Ser276 phosphorylation is required for activation of a subset of NF-κB dependent genes by recruiting CDK-9/Cyclin T1 complexes. Mol Cell Biol. 2008;28:3623–3638. doi: 10.1128/MCB.01152-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen LF, Greene WC. Shaping the nuclear action of NF-κB. Nat Rev Mol Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- 55.Chen LF, et al. NF-κB RelA phosphorylation regulates RelA acetylation. Mol Cell Biol. 2005;25:7966–7975. doi: 10.1128/MCB.25.18.7966-7975.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang WC, et al. Phosphorylation of CBP by IKKα promotes cell growth by switching the binding preference of CBP from p53 to NF-κB. Mol Cell. 2007;26:75–87. doi: 10.1016/j.molcel.2007.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ishinaga H, et al. TGF-β induces p65 acetylation to enhance bacteria-induced NF-κB activation. EMBO J. 2007;26:1150–1162. doi: 10.1038/sj.emboj.7601546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saccani S, et al. Degradation of promoter-bound p65/RelA is essential for the prompt termination of the nuclear factor κB response. J Exp Med. 2004;200:107–113. doi: 10.1084/jem.20040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ryo A, et al. Regulation of NF-κB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol Cell. 2003;12:1413–1426. doi: 10.1016/s1097-2765(03)00490-8. [DOI] [PubMed] [Google Scholar]
- 60.Ganesh L, et al. The gene product Murr1 restricts HIV-1 replication in resting CD4+ lymphocytes. Nature. 2003;426:853–857. doi: 10.1038/nature02171. [DOI] [PubMed] [Google Scholar]
- 61.Burstein E, et al. COMMD proteins: a novel family of structural and functional homologs of MURR1. J Biol Chem. 2005;280:22222–22232. doi: 10.1074/jbc.M501928200. [DOI] [PubMed] [Google Scholar]
- 62.Maine GN, et al. COMMD1 promotes the ubiquitination of NF-κB subunits through a cullin-containing ubiquitin ligase. EMBO J. 2007;26:436–447. doi: 10.1038/sj.emboj.7601489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tanaka T, et al. PDLIM2-mediated termination of transcription factor NF-κB activation by intranuclear sequestration and degradation of the p65 subunit. Nat Immunol. 2007;8:584–591. doi: 10.1038/ni1464. [DOI] [PubMed] [Google Scholar]
- 64.Lawrence T, et al. IKKα limits macrophage NF-κB activation and contributes to the resolution of inflammation. Nature. 2005;434:1138–1143. doi: 10.1038/nature03491. [DOI] [PubMed] [Google Scholar]
- 65.Li Q, et al. Enhanced NF-κB activation and cellular function in macrophages lacking IκB kinase 1 (IKK1) Proc Natl Acad Sci U S A. 2005;102:12425–12430. doi: 10.1073/pnas.0505997102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carmody RJ, et al. Negative regulation of toll-like receptor signaling by NF-κB p50 ubiquitination blockade. Science. 2007;317:675–678. doi: 10.1126/science.1142953. [DOI] [PubMed] [Google Scholar]
- 67.Shuai K. Regulation of cytokine signaling pathways by PIAS proteins. Cell Res. 2006;16:196–202. doi: 10.1038/sj.cr.7310027. [DOI] [PubMed] [Google Scholar]
- 68.Liu B, et al. Proinflammatory stimuli induce IKKα-mediated phosphorylation of PIAS1 to restrict inflammation and immunity. Cell. 2007;129:903–914. doi: 10.1016/j.cell.2007.03.056. [DOI] [PubMed] [Google Scholar]
- 69.Wan F, et al. Ribosomal protein S3: a KH domain subunit in NF-κB complexes that mediates selective gene regulation. Cell. 2007;131:927–939. doi: 10.1016/j.cell.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 70.Goto A, et al. Akirins are highly conserved nuclear proteins required for NF-κB-dependent gene expression in drosophila and mice. Nat Immunol. 2007;9:97–104. doi: 10.1038/ni1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hoffmann JA. The immune response of Drosophila. Nature. 2003;426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- 72.Lim CA, et al. Genome-wide mapping of RELA(p65) binding identifies E2F1 as a transcriptional activator recruited by NF-κB upon TLR4 activation. Mol Cell. 2007;27:622–635. doi: 10.1016/j.molcel.2007.06.038. [DOI] [PubMed] [Google Scholar]
- 73.Ordureau A, et al. The IRAK-catalysed activation of the E3 ligase function of Pellino isoforms induces the Lys63-linked polyubiquitination of IRAK1. Biochem J. 2008;409:43–52. doi: 10.1042/BJ20071365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schauvliege R, et al. Pellino proteins: novel players in TLR and IL-1R signaling. J Cell Mol Med. 2007;11:453–461. doi: 10.1111/j.1582-4934.2007.00040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Conze DB, et al. Lys63-linked polyubiquitination of IRAK-1 is required for interleukin-1 receptor- and toll-like receptor-mediated NF-κB activation. Mol Cell Biol. 2008;28:3538–3547. doi: 10.1128/MCB.02098-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Windheim M, et al. Interleukin-1 (IL-1) induces the Lys63-linked polyubiquitination of IL-1 receptor-associated kinase 1 to facilitate NEMO binding and the activation of IκB kinase. Mol Cell Biol. 2008;28:1783–1791. doi: 10.1128/MCB.02380-06. [DOI] [PMC free article] [PubMed] [Google Scholar]