Abstract
Strain Marseille-P2645T was isolated in a colon sample from a Frenchwoman who underwent a colonoscopy. Bacterial cells were Gram negative, non–spore forming, mobile and strictly anaerobic. The genome of strain Marseille-P2645T is 3 950 441 bp long and contains 3374 protein-coding genes. The DNA G+C content is of 51.66 mol%. Strain Marseille-P2645T exhibited a 92.9% sequence similarity with Bacteroides helcogenes strain P36-108T (GenBank accession no. CP002352), the phylogenetically closest species with standing in nomenclature. Strain Marseille-P2645T (= CSUR P2645 = DSM 103034) is therefore a candidate as a type species of a new genus belonging to the Bacteroidaceae family, for which the name of Mediterranea massiliensis gen. nov., sp. nov., is proposed.
Keywords: Colon; culturomics; genome; Mediterranea massiliensis gen. nov., sp. nov.; taxonogenomics
Introduction
The concept of culturomics [1], [2], [3], developed in our laboratory since 2010, aims at deciphering the living microbial diversity in any milieu, notably microbes that live with humans. In contrast to metagenomics, it focuses on culture, and especially the characterization of new species become available to the scientific and medical community. Indeed, culture is the first step before experimental studies, multispecies probiotics or selection of strains for specific microbiotherapy.
The combination of endoscopic sampling and microbial culturomics allowed us to isolate a new bacterial genus from the colon lavage of a 58-year-old woman without medical history, who underwent a colonoscopy because of a positive screening test for a colorectal cancer. Strain Marseille-P2645T is the type strain of Mediterranea massiliensis gen. nov., sp. nov., the first species of the genus Mediterranea, a new member of the Bacteroidaceae family. To describe strain Marseille-P2645T, we report here the characterization of a new bacterial species using a new taxonomic strategy called taxonogenomics [4]. Taxonogenomics integrates proteomic information obtained by matrix-assisted desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) and genomic tests to describe new bacterial species [5], [6]. This polyphasic approach overcomes limitations of conventional methods for new species description (genetic, phenotypic and chemotaxonomic characteristics for new species description) [7], [8]. It combines phenotypic characteristics, analyses and comparison of the complete genome sequence. Bacteroides helcogenes strain P36-108T (GenBank accession no. CP002352) was the phylogenetically closest species with standing in nomenclature [9]. Study of the phenotypic, phylogenetic and genomic characteristics of strain Marseille-P2645T revealed that M. massiliensis was sufficiently different from Bacteroides helcogenes type strain (P36-108T) to be classified as a new genus of bacteria of the Bacteroides family [10]. We previously reported the ribosomal 16S sequence and the MALDI-TOF MS spectrum [11]. Since the first report, a new strain was isolated confirming the new species. Here we present the complete description of strain Marseille-P2645T (= CSUR P2645 = DSM 103034), together with the description of the complete genomic sequencing and annotation.
Materials and Methods
Sample collection
Strain Marseille-P2645T was first isolated in April 2016 in a liquid colon sample from a 58-year-old woman without medical history, who underwent a colonoscopy because of a positive screening test for a colorectal cancer in the gastroenterology department of Hopital Nord, Marseille, France. Signed informed consent was collected from the patient, and the study obtained approval from the ethics committee of the Institut Fédératif de Recherche IFR48 under number 2016-010.
Strain isolation and identification by MALDI-TOF MS
After collection using sterilized devices, the colonic sample was immediately placed in an antioxidant-enriched liquid medium [12] and transported to our laboratory (Institut Hospitalo-Universitaire Méditerranée Infection, Marseille, France). Once in the laboratory, the sample was placed under a laminar flow cabinet before being diluted in phosphate-buffered saline and seeded on different solid and liquid culture media. The initial growth of strain Marseille-P2645T was achieved after 1 day at 37°C on 5% sheep's blood–enriched Columbia agar (bioMérieux, Marcy l’Etoile, France) in anaerobic atmosphere (anaeroGEN; Oxoid, Dardilly, France). After isolation on pure culture, the identification of all different bacterial colonies was performed using MALDI-TOF MS using a Microflex spectrometer (Bruker Daltonics, Bremen, Germany), as previously reported [13], [14]. Each colony was deposited in duplicate on a MALDI-TOF MS target to be analysed. A matrix solution of 1.5 μL (saturated solution of cyano-4-hydroxycinnamic acid diluted in 50% acetonitrile and 2.5% trifluoroacetic acid, completed with high-performance liquid chromatography water) was deposited on each spot. After the reading of the plate, the obtained protein spectra were compared by MALDI Biotyper software with those of the Bruker database (continuously updated with our recent data). When the resulting score was >2, the identification was made at the species level, while a score of <1.7 did not enable any identification.
16S rRNA sequencing and phylogeny
After 3 failed MALDI-TOF MS identifications, a standard 16S rRNA PCR was performed using universal primers pair fD1 and rP2 in a GeneAmp PCR System 2720 thermal cycler (Thermo Fisher Scientific, Waltham, MA, USA). The amplified DNA was revealed by electrophoresis on 1.5% agarose gel. Once validated, the PCR product was purified and sequenced using the Big Dye Terminator Sequencing Kit and the following internal primers: 536F, 536R, 800F, 800R, 1050F, 1050R, 357F and 357R, as previously described [1]. Sequences were corrected using the Codon Code Aligner software (http://www.codoncode.com), and then a BLAST search (Basic Local Alignment Search Tool) was performed against the GenBank nucleotide collection (http://blast.ncbi.nlm.nih.gov/Blast.cgi). A strain was considered as a candidate novel genus if the percentage of 16S rRNA similarity with the closest species with standing in nomenclature was <95% [15]. We performed a phylogenetic analysis based on the 16S rRNA of our isolate to identify its phylogenetic affiliations. Sequences were aligned using Muscle 3.8.31 [16], and phylogenetic inferences were obtained using the approximately maximum-likelihood method within FastTree software [17]. Only bootstrap values of >95% are shown, and the numbers at the nodes are the computed local values [18].
Phenotype characteristics
Different growth conditions were tested on a 5% sheep's blood–enriched Columbia agar (bioMérieux) for strain Marseille-P2645T. Five temperatures (room temperature, 28, 37, 45 and 55°C) and three atmospheres—anaerobic (anaeroGen Compact; Oxoid), microaerophilic (campyGen Compact; Oxoid) and aerobic (in a plastic pouch to maintain a humid atmosphere)—were evaluated. Tolerance of this strain to salt was tested using 5%, 7.5%, 10%, 15% and 20% of NaCl, and the pH tolerance (5, 5.5, 6, 6.5, 7, 7.5 and 8) was also tested. Individual cells of strain Marseille-P2645T were visualized using a Tecnai G20 electron microscope (FEI Company, Limeil-Brevannes, France). Gram staining was performed and observed using a photonic microscope Leica DM2500 (Leica, Wetzlar, Germany) with a 100× oil-immersion objective. Motility testing was performed by observation of a fresh colony between the blades and slats using a DM1000 photonic microscope (Leica) at 40×. To check the ability to sporulate, strain Marseille-P2645T was grown on 5% sheep's blood–enriched Columbia agar (bioMérieux) for 2 days, and then a heat-shock test (20 minutes at 80°C) was performed.
Biochemical characterization and antibiotic susceptibility
The commercially available API ZYM, API 50CH and API 20A strips (bioMérieux) were used for biochemical tests according to the manufacturer's instructions. Catalase (bioMérieux) and oxidase (Becton Dickinson, Le Pont de Claix, France) activities were also tested.
Cellular fatty acid methyl ester (FAME) analysis was performed by gas chromatography/mass spectrometry (GC/MS). Two samples were prepared with approximately 60 mg of bacterial biomass per tube collected from several culture plates. FAMEs were prepared as described by Sasser [19]. GC/MS analyses were carried out as described by Dione et al. [20] in genome analysis and description of Anaerosalibacter massiliensis. Briefly, FAMEs were separated using an Elite 5-MS column and monitored by mass spectrometry (Clarus 500-SQ 8 S; Perkin Elmer, Courtaboeuf, France). Spectral database search was performed using MS Search 2.0 operated with the Standard Reference Database 1A (National Institute of Standards and Technology, Gaithersburg, MD, USA) and the FAMEs mass spectral database (Wiley, Chichester, UK).
Sensitivity to antibiotics was determined using a disc diffusion method on Mueller-Hinton E agar (bioMérieux). The following antibiotics were tested using Sirscan discs (i2a, Montpellier, France): doxycycline, rifampicin, vancomycin, amoxicillin, ceftriaxone, penicillin, trimethoprim/sulfamethoxazole, imipenem, oxacillin, erythromycin, tobramycin, cefotaxime, amoxicillin/clavulanic acid, gentamicin, fosfomycin and metronidazole. Inhibition diameters were measured using the Scan1200 scanner (Interscience, Saint-Nom-La Bretêche, France).
Genome sequencing and assembly
After pretreatment by a lysozyme incubation at 37°C for 2 hours, DNA of strain Marseille-P2645T was extracted on the EZ1 biorobot (Qiagen, Germantown, MD, USA) with a EZ1 DNA tissue kit. The elution volume was 50 μL. Genomic DNA was quantified by a Qubit assay with the high-sensitivity kit (Life Technologies, Carlsbad, CA, USA) to 80.7 ng/μl.
Genomic DNA of strain Marseille-P2645T was sequenced on the MiSeq Technology (Illumina, San Diego, CA, USA) with the mate-pair strategy. The genomic DNA was barcoded in order to be mixed with 11 other projects with the Nextera Mate Pair sample prep kit (Illumina). The mate-pair library was prepared with 1.5 μg of genomic DNA using the Nextera mate pair Illumina guide. The genomic DNA sample was simultaneously fragmented and tagged with a mate-pair junction adapter. The pattern of the fragmentation was validated on an Agilent 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA, USA) with a DNA 7500 labchip. The DNA fragments ranged in size from 1.5 to 11 kb, with an optimal size at 8.44 kb. No size selection was performed, and 600 ng of tagmented fragments were circularized. The circularized DNA was mechanically sheared to small fragments with an optimal at 916 bp on a Covaris device S2 in T6 tubes (Covaris, Woburn, MA, USA). The library profile was visualized on a High Sensitivity Bioanalyzer LabChip (Agilent Technologies), and the final concentration library was measured at 81.95 nmol/L. The libraries were normalized at 2 nM and pooled. After a denaturation step and dilution at 15 pM, the pool of libraries was loaded onto the reagent cartridge and then onto the instrument along with the flow cell. Automated cluster generation and sequencing run were performed in a single 39-hour run at a 2 × 151 bp read length.
Total information of 5.3 Gb was obtained from a 559K/mm2 cluster density, with a cluster passing quality control filters of 96.3% (10 450 000 passed filter clusters). Within this run, the index representation for strain Marseille-P2645T was determined to be 10.02%. The 1 047 418 paired-end reads were filtered according to the read qualities.
Genome annotation and comparisons
Prodigal was used for open reading frame (ORFs) prediction [21] with default parameters, but the predicted ORFs were excluded if they spanned a sequencing gap region (containing N). The predicted bacterial protein sequences were searched against the GenBank database [22] and the Clusters of Orthologous Groups database (COGs) using BLASTP (E value 1e-03, coverage 0.7 and identity percentage 30%). If no hit was found, a search was performed against the NR database using BLASTP with an E value of 1e-03, coverage of 0.7 and identity percentage of 30%. If the sequence lengths were smaller than 80 aa, we used an E value of 1e-05. Both tRNAs and rRNAs were predicted using the tRNAScan-SE [23] and RNAmmer [24] tools, respectively. Lipoprotein signal peptides and the number of transmembrane helices were predicted using SignalP [25] and TMHMM [26], respectively. ORFans were identified when their BLASTP E value was lower than 1e-03 for an alignment length greater than 80 aa. If alignment lengths were smaller than 80 aa, we used an E value of 1e-05. Such parameter thresholds have already been used in previous works to define ORFans. Artemis [27] and DNA Plotter [28] were used for data management and for visualization of genomic features, respectively. To estimate the mean level of nucleotide sequence similarity at the genome level between strain Marseille-P2645T and other bacteria, we used the average genomic identity of orthologous gene sequences (AGIOS) homemade software [29]. This software combines the Proteinortho software for detecting orthologous proteins between genomes compared two by two [30], then retrieves the corresponding genes and determines the mean percentage of nucleotide sequence identity among orthologous ORFs using the Needleman-Wunsch global alignment algorithm. The genome of strain Marseille-P2645T was compared with other Bacteroidetes: Bacteroides fragilis strain NCTC 9343, Bacteroides finegoldii strain DSM 17565, Bacteroides thetaiotaomicron strain VPI 8452, Bacteroides coprocola DSM 17136, Bacteroides plebeius DSM 17135, Bacteroides vulgatus ATCC 8482, Bacteroides massiliensis DSM 17679, Bacteroides fluxus YIT 12057 and Bacteroides rodentium JCM 16496. Annotation and comparison processes were performed by the multiagent software system DAGOBAH [31], which includes Figenix [32] libraries that provided pipeline analysis. We also performed genome-to-genome distance calculator (GGDC) analysis using the GGDC Web server, as previously reported [33], [34].
Results
Phylogenetic analysis
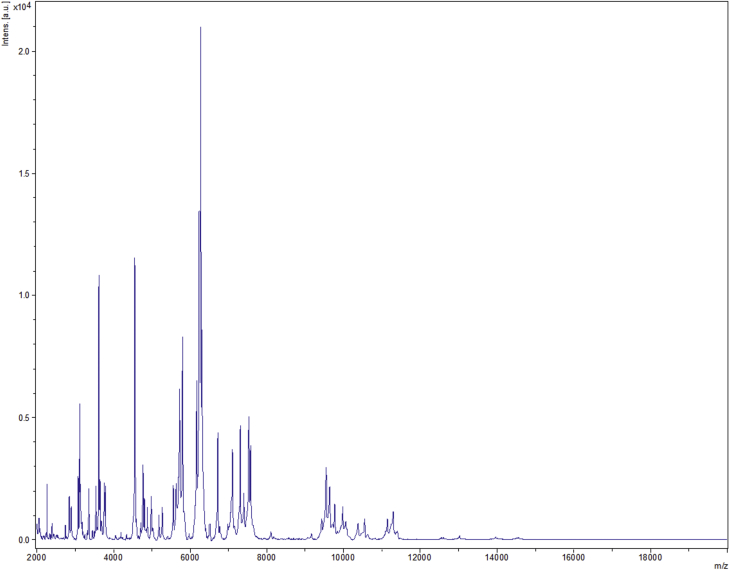
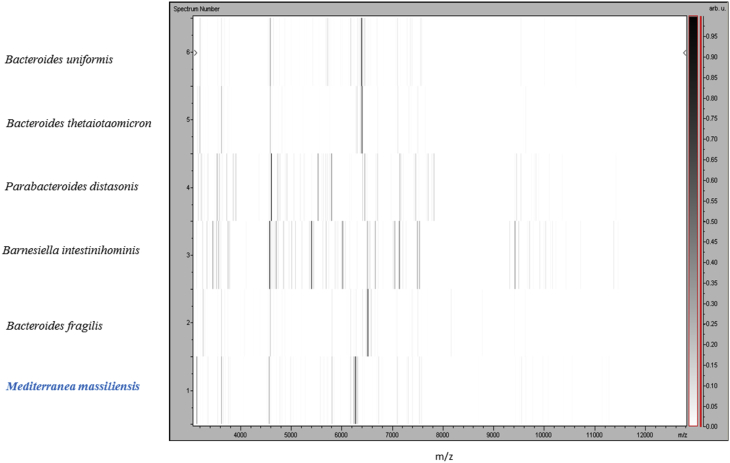
Strain Marseille-P2645T was first isolated after a 1-day anaerobic incubation period of cultivation of the sample seeded on Colombia agar with 5% sheep's blood. MALDI-TOF MS spectrum of P2645T (Fig. 1) did not match anything in our database or in Brucker's database. Phylogenetic analysis based on the 16S RNA showed that strain Marseille-P2645T exhibited a 92.9% sequence similarity [10] with Bacteroides helcogenes strain P36-108T (GenBank accession no. CP002352), the phylogenetically closest species with standing in nomenclature (Fig. 2) [9]. Bacteroides coprophilus strain CB42T (AB260026) [35] was also a close validated species of strain Marseille-P2645T; this strain is a Gram-negative anaerobic bacterium isolated from human faeces in 2007. Because of a 16S rRNA gene sequence divergence of >5% from its phylogenetically closest species with standing in nomenclature, we propose that strain Marseille-P2645T be a member of a novel genus in the family of Bacteroidaceae in the phylum of Bacteroidetes, for which we propose the name Mediterranea gen. nov. The 16S RNA of strain Marseille-P2645T was deposited in GenBank under accession number LT558847.3 (Table 1). A gel view was performed in order to observe spectra differences between M. massiliensis and other close bacteria (Fig. 3).
Fig. 1.
Reference mass spectrum from Mediterranea massiliensis strain Marseille-P2645.
Fig. 2.
Phylogenetic tree highlighting phylogenetic position of Mediterranea massiliensis strain Marseille-P2645T relative to other phylogenetically close members of family Bacteroidaceae. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using maximum likelihood method within MEGA7 software. Numbers at nodes are percentages of bootstrap values obtained by repeating analysis 500 times to generate majority consensus tree. Only bootstraps scores ≥95% were retained. Scale bar indicates 1% nucleotide sequence divergence.
Table 1.
Percentage of 16S rRNA similarities between strain Marseille-P2645T and Bacteroides genus
| Mediterranea massiliensis | Bacteroides helcogeness | Bacteroides coprophilus | Bacteroides plebeius | Bacteroides coprocola | Bacteroides massiliensis | |
|---|---|---|---|---|---|---|
| M. massiliensis | 100 | 94.90 | 94.11 | 92.68 | 93.00 | 91.63 |
| B. helcogenes | 100 | 93.39 | 92.98 | 92.97 | 93.12 | |
| B. coprophilus | 100 | 93.86 | 93.05 | 92.38 | ||
| B. plebeius | 100 | 94.55 | 93.34 | |||
| B. coprocola | 100 | 92.58 | ||||
| B. massiliensis | 100 |
Fig. 3.
Gel view comparing Mediterranea massiliensis strain Marseille-P2645T to other members of family Bacteroidaceae. Gel view displays raw spectra of loaded spectrum files arranged in pseudo–gel-like look. x-axis records m/z value. Left y-axis displays running spectrum number originating from subsequent spectra loading. Peak intensity is expressed as greyscale. Colour bar and right axis indicate intensity of each matrix-assisted desorption ionization–time of flight mass spectrometry peak and peak intensity in arbitrary units.
Phenotypic and biochemical characterizations
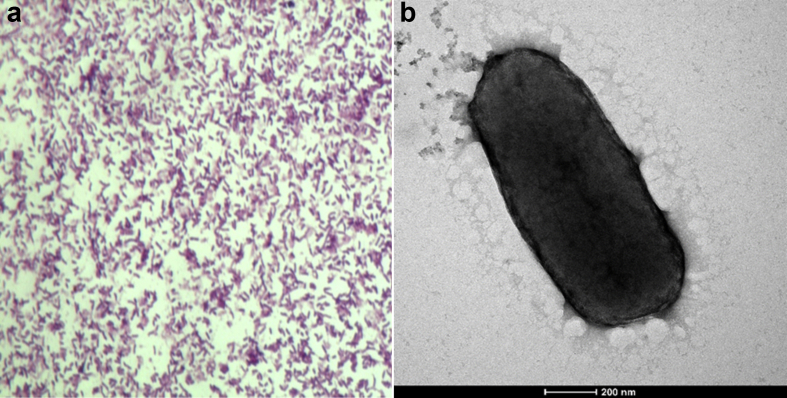
The optimum growth of Strain Marseille-P2645T was obtained after 1 day of culture at 37°C in anaerobic conditions (anaeroGEN, Oxoid). Colonies grown on 5% sheep's blood–enriched Columbia agar medium were translucent and slightly haemolytic, with a diameter of 0.5 mm. Bacterial cells were Gram-negative bacilli (Fig. 4a), 1 μm in length and 0.6 μm in diameter (Fig. 4b). This strain is non–spore-forming, peritrichous and motile. Catalase and oxidase reactions were negative. Marseille-P2645T was able to grow at 28°C and 45°C. It is strictly anaerobic and unable to grow under microaerophilic or aerobic conditions. The range of salinity used shows that strain Marseille-P2645T does not tolerate salt. Strain Marseille-P2645T survives from pH 5 to 8 with an optimal pH at 7. Classification and general features are summarized in Table 2. Using API ZYM (bioMérieux), positive reactions were obtained for alkaline phosphatase, acid phosphatase, α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, α-fucosidase and N-acetyl β-glucosaminidase, but negative reactions for α-mannosidase, α-chymotrypsin, trypsin and lipase-C14. In the API 20A (bioMérieux), the strain showed gelatin production but no indole or esculin production (ferric citrate). Using API 50CH system, positive reactions were obtained for d-glucose, mannose, starch and potassium 5-ketogluconate, whereas negative reactions were observed for l-arabinose, d-ribose, d-fructose, d-maltose, d-lactose and other constituents. Phenotypic characteristics were compared to other members of the genus Bacteroides (Table 3). Among the tested antibiotics, cells were susceptible to amoxicillin, rifampicin, imipenem, metronidazole and amoxicillin/clavulanic acid, but resistant to vancomycin, cefotaxime, gentamicin, penicillin, clindamycin, oxacillin, tobramycin, fosfomycin, ceftriaxone, doxycycline, erythromycin and trimethoprim/sulfamethoxazole. Analysis of the total cellular fatty acid composition of Mediterranea massiliensis demonstrated that the fatty acids detected are mainly saturated. The major fatty acids found for this strain were 15:0 iso (42%) and 15:0 anteiso (20%). Several specific 3-hydroxy fatty acids were observed: 17:0 3-OH iso, 16:0 3-OH and 15:0 3-OH iso (Table 4).
Fig. 4.
(a) Gram staining of Mediterranea massiliensis strain Marseille-P2645T. (b) Transmission electron microscopy of M. massiliensis strain Marseille-P2645T with Tecnai G20 electron microscope. Scale bar = 200 nm.
Table 2.
Classification and general features of Mediterranea massiliensis strain Marseille-P2645T
| Property | Term |
|---|---|
| Current classification | Domain: Bacteria |
| Phylum: Bacteroidetes | |
| Class: Bacteroidia | |
| Order: Bacteroidales | |
| Family: Bacteroidaceae | |
| Genus: Mediterranea | |
| Species: Mediterranea massiliensis | |
| Type: Strain Marseille-P2645T | |
| Gram stain | Negative |
| Cell shape | Rod |
| Motility | Motile |
| Sporulation | No sporulating |
| Temperature range | 28–45°C |
| Optimum temperature | 37°C |
| pH range: optimum | 7 |
| Salinity | 0g/L |
| Oxygen requirement | Anaerobic |
| Carbone source | Unknown |
| Habitat | Colon |
| Biotic relationship | Free-living |
| Pathogenicity | Unknown |
Table 3.
Differential characteristics of Mediterranea massiliensis strain Marseille-P2645 compared to other close bacteria of family Bacteroidaceae
| Property | Mediterranea massiliensis | Bacteroides finegoldii | Bacteroides vulgatus | Bacteroides plebeius | Bacteroides fragilis | Bacteroides thetaiotaomicron | Bacteroides massiliensis | Bacteroides rodentium | Bacteroides coprocola | Bacteroides fluxus |
|---|---|---|---|---|---|---|---|---|---|---|
| Cell diameter (μm) | 0.6 | 1–2 | 0.5–0.8 | 0.8 | 1.3 | 0.7–2 | 0.8–1.4 | 0.5–2 | 0.8 | 1.0–1.4 |
| Oxygen requirement | Anaerobic | Anaerobic | Anaerobic | Anaerobic | Anaerobic | Anaerobic | Anaerobic | Anaerobic | Anaerobic | Anaerobic |
| Shape | Bacilli | Bacilli | Bacilli | Bacilli | Bacilli | Bacilli | Bacilli | Bacilli | Bacilli | Bacilli |
| Gram stain | − | − | − | − | − | − | − | − | − | − |
| Motility | + | − | − | − | − | − | − | − | − | − |
| Indole | − | − | − | − | − | + | − | − | − | + |
| Production of: | ||||||||||
| Alkaline phosphatase | + | + | + | NA | NA | + | NA | NA | NA | + |
| Catalase | − | NA | − | NA | + | + | − | NA | NA | NA |
| Oxidase | − | NA | NA | NA | + | NA | NA | NA | NA | NA |
| Nitrate reductase | NA | − | − | NA | NA | + | NA | NA | NA | − |
| Urease | − | − | NA | NA | NA | − | NA | NA | NA | − |
| β-Galactosidase | + | + | − | NA | + | + | NA | NA | NA | + |
| N-acetyl glucosamine | − | + | NA | NA | + | + | NA | NA | NA | NA |
| Acid from: | NA | NA | NA | NA | ||||||
| l-Arabinose | − | + | + | + | − | + | − | + | − | + |
| Ribose | − | NA | + | NA | − | NA | − | NA | NA | NA |
| Mannose | + | + | + | NA | + | + | NA | NA | NA | + |
| Mannitol | − | − | − | NA | − | − | NA | NA | NA | + |
| Sucrose | NA | + | + | NA | + | + | NA | NA | NA | + |
| d-Glucose | + | + | + | NA | + | + | NA | NA | NA | + |
| d-Fructose | − | NA | + | NA | + | + | NA | NA | NA | + |
| d-Maltose | − | + | + | NA | + | + | NA | NA | NA | + |
| d-Lactose | − | + | + | NA | + | + | NA | NA | NA | + |
| G+C content (%) | 51.66 | 42.93 | 42.2 | 44.31 | 43.11 | 42.86 | 42.69 | 47.05 | 41.87 | 45.57 |
| Habitat | Human colon | Human gut | Human gut | Human gut | Human gut | Human gut | Human gut | Human gut | Human gut | Human gut |
+, positive result; −, negative result; NA, data not available.
Table 4.
Total cellular fatty acid composition of Mediterranea massiliensis strain Marseille-P2645T
| Fatty acid | Name | Mean relative %a |
|---|---|---|
| 15:0 iso | 13-methyl-Tetradecanoic acid | 41.7 ± 0.9 |
| 15:0 anteiso | 12-methyl-Tetradecanoic acid | 19.7 ± 0.2 |
| 16:00 | Hexadecanoic acid | 6.7 ± 0.3 |
| 14:00 | Tetradecanoic acid | 6.0 ± 0.4 |
| 18:1n9 | 9-Octadecenoic acid | 5.8 ± 0.3 |
| 18:2n6 | 9,12-Octadecadienoic acid | 4.5 ± 0.3 |
| 17:0 3-OH iso | 3-hydroxy-15-methyl-Hexadecanoic acid | 4.1 ± 0.1 |
| 16:0 3-OH | 3-hydroxy-Hexadecanoic acid | 3.8 ± 0.1 |
| 15:00 | Pentadecanoic acid | 1.9 ± 0.1 |
| 18:00 | Octadecanoic acid | 1.5 ± 0.1 |
| 13:0 iso | 11-methyl-Dodecanoic acid | 1.4 ± 0.1 |
| 5:0 anteiso | 2-methyl-Butanoic acid | 1.0 ± 0.1 |
| 17:0 iso | 15-methyl-Hexadecanoic acid | TR |
| 14:0 iso | 12-methyl-Tridecanoic acid | TR |
| 18:1n6 | 12-Octadecenoic acid | TR |
| 15:0 3-OH iso | 3-hydroxy-13-methyl-Tetradecanoic acid | TR |
| 20:4n6 | 5, 8, 11,14-Eicosatetraenoic acid | TR |
| 17:00 | Heptadecanoic acid | TR |
| 17:0 anteiso | 14-methyl-Hexadecanoic acid | TR |
| 13:0 anteiso | 10-methyl-Dodecanoic acid | TR |
| 13:00 | Tridecanoic acid | TR |
| 16:0 iso | 14-methyl-Pentadecanoic acid | TR |
| 16:1n7 | 9-Hexadecenoic acid | TR |
TR, trace amounts < 1%.
Mean peak area percentage.
Genome properties
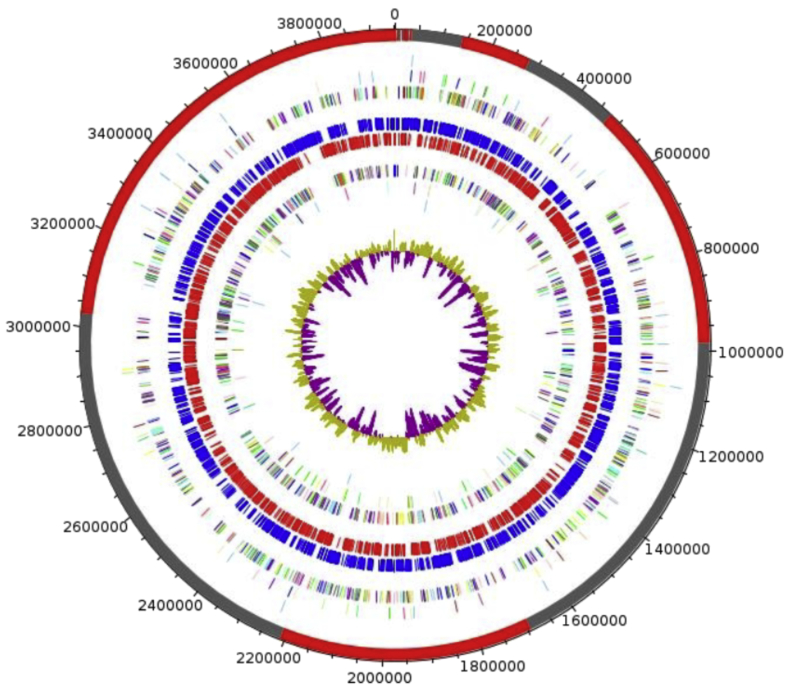
The genome of strain Marseille-P2645T is 3 950 441 bp long with 51.66 mol% G+C content (Fig. 5). It is composed of 25 scaffolds (composed of 34 contigs). Of the 3439 predicted genes, 3374 were protein-coding genes and 65 were RNA genes (three genes are 5S rRNA, one gene is 16S rRNA, one gene is 23S rRNA and 60 genes are tRNA genes). A total of 2772 genes (82.16%) were assigned a putative function (by COGs or by NR BLAST), and 92 genes were identified as ORFans (2.73%). The remaining genes were annotated as hypothetical proteins (449 genes, 13.31%). Genome statistics are summarized in Table 5, and the distribution of the genes into COGs functional categories is presented in Table 6.
Fig. 5.
Graphical circular map of genome. From outside to centre: contigs (red/grey), COGs category of genes on forward strand (three circles), genes on forward strand (blue circle), genes on reverse strand (red circle), COGs category on reverse strand (three circles), GC content. COGs, Clusters of Orthologous Groups database.
Table 5.
Nucleotide content and gene count level of genome of strain Marseille-P2645T
| Attribute | Genome (total) |
|
|---|---|---|
| Value | % of totala | |
| Size (bp) | 3 950 441 | 100 |
| No. of GC | 2 039 331 | 51.66 |
| Coding region | 3 499 039 | 88.57 |
| Total genes | 3439 | 100 |
| RNA genes | 65 | 1.890084386 |
| Protein-coding genes | 3374 | 100 |
| Genes with function prediction | 2772 | 82.16 |
| Genes assigned to COGs | 1601 | 47.45 |
| Genes with peptides signal | 835 | 24.75 |
| Genes with Pfam-A domains | 2999 | 87 |
| Genes with transmembrane helices | 646 | 19.1464138 |
| Genes associated to PKS or NRPS | 9 | 0.27 |
| Genes associated to virulence | 588 | 17.43 |
| Proteins associated to ORFans | 92 | 2.726733923 |
COGs, Clusters of Orthologous Groups database; NRPS, nonribosomal peptide synthase; ORF, open reading frame; PKS, polyketide synthase.
Total is based on either size of genome in base pairs or total number of protein-coding genes in annotated genome.
Table 6.
Number of genes associated with 25 general COGs functional categories of strain Marseille-P2645T.
| Code | Value | % of totala | Description |
|---|---|---|---|
| J | 178 | 5.275637 | Translation |
| A | 0 | 0 | RNA processing and modification |
| K | 90 | 2.667457 | Transcription |
| L | 124 | 3.6751633 | Replication, recombination and repair |
| B | 0 | 0 | Chromatin structure and dynamics |
| D | 25 | 0.7409603 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0 | Nuclear structure |
| V | 75 | 2.2228808 | Defense mechanisms |
| T | 72 | 2.1339657 | Signal transduction mechanisms |
| M | 150 | 4.4457617 | Cell wall/membrane biogenesis |
| N | 11 | 0.32602254 | Cell motility |
| Z | 0 | 0 | Cytoskeleton |
| W | 0 | 0 | Extracellular structures |
| U | 32 | 0.9484292 | Intracellular trafficking and secretion |
| O | 66 | 1.9561353 | Posttanslational modification, protein turnover, chaperones |
| X | 33 | 0.97806764 | Mobilome: prophages, transposons |
| C | 97 | 2.8749259 | Energy production and conversion |
| G | 146 | 4.327208 | Carbohydrate transport and metabolism |
| E | 122 | 3.6158864 | Amino acid transport and metabolism |
| F | 63 | 1.86722 | Nucleotide transport and metabolism |
| H | 94 | 2.7860107 | Coenzyme transport and metabolism |
| I | 58 | 1.719028 | Lipid transport and metabolism |
| P | 102 | 3.023118 | Inorganic ion transport and metabolism |
| Q | 19 | 0.56312984 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 141 | 4.179016 | General function prediction only |
| S | 61 | 1.8079432 | Function unknown |
| — | 1773 | 52.548904 | Not in COGs |
COGs, Clusters of Orthologous Groups database.
Total is based on total number of protein-coding genes in annotated genome.
Genome comparison
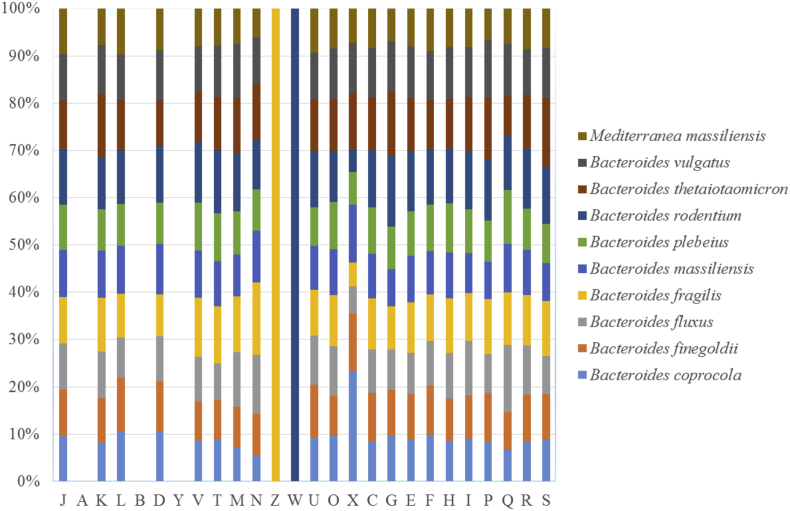
The draft genome sequence of strain Marseille-P2645T (3.95 Mb) is smaller than the Bacteroides genome analysed, and particularly that of Bacteroides finegoldii (4.89Mb), Bacteroides vulgatus (5.16), Bacteroides rodentium (4.88), Bacteroides plebeius (4.42), Bacteroides fragilis (5.24), Bacteroides fluxus (4.33), Bacteroides thetaiotaomicron (6.29), Bacteroides coprocola (4.30) and Bacteroides massiliensis (4.64). The G+C content of Mediterranea massiliensis (51.66%) is much higher (widely >1%) than that of Bacteroides finegoldii (42.93%), Bacteroides vulgatus (42.2%), Bacteroides plebeius (44.31%), Bacteroides fragilis (43.11%), Bacteroides thetaiotaomicron (42.86%), Bacteroides massiliensis (42.69%), Bacteroides rodentium (47.05%), Bacteroides coprocola (41.87%) and Bacteroides fluxus (45.57%). The gene content of Mediterranea massiliensis (3374 genes) is smaller than that of Bacteroides finegoldii (4485), Bacteroides vulgatus (4065), Bacteroides plebeius (3933), Bacteroides fragilis (4236), Bacteroides thetaiotaomicron (4825), Bacteroides massiliensis (3935), Bacteroides rodentium (4993), Bacteroides coprocola (4291) and Bacteroides fluxus (3921). Distribution of functional classes of predicted genes according to the clusters of orthologous groups of proteins between M. massiliensis and other members of the genus of Bacteroides was reported (Fig. 6). In addition, strain Marseille-P2645T shared 4485, 4065, 3933, 4236, 4825, 3935, 4993, 4291 and 3921 orthologous genes with Bacteroides finegoldii strain DSM 17565, Bacteroides vulgatus strain ATCC 8482, Bacteroides plebeius strain DSM 17135, Bacteroides fragilis strain NCTC 9343, Bacteroides thetaiotaomicron strain VPI 5482, Bacteroides massiliensis strain B84634, Bacteroides rodentium strain JCM 16496, Bacteroides coprocola strain DSM 17136 and Bacteroides fluxus strain YIT 12057. The average nucleotide sequence identity ranged from 58.89% to 66.13% between strain Marseille-P2645T, as well as the nine other members of the family Bacteroidaceae (Table 7). Results from pairwise comparison obtained for the analysis of the digital DNA-DNA hybridization (dDDH) using GGDC software are reported in Table 8. The probability of error when saying that this strain is a new species (DDH >70%) compared to nine other member of the genus Bacteroides was 0.02% (GGDC2.1 formula 2; DSMZ, http://ggdc.dsmz.de/distcalc2.php). When comparing strain Marseille-P2645Twith other strains, dDDH values ranged from 21.60% ± 4.7 for Bacteroides fluxus strain YIT 12057 to 28.10% ± 4.9 for Bacteroides coprocola strain M16 (DSM 17136).
Fig. 6.
Distribution of functional classes of predicted genes according to clusters of orthologous groups of proteins of Mediterranea Massiliensis strain Marseille-P2645.
Table 7.
Number of orthologous proteins shared between genomes (upper right), average percentage similarity of nucleotides corresponding to orthologous protein shared between genomes (lower left), and number of proteins per genome (in bold)
| Bacteroides massiliensis | Mediterranea massiliensis | Bacteroides coprocola | Bacteroides vulgatus | Bacteroides fluxus | Bacteroides rodentium | Bacteroides finegoldii | Bacteroides plebeius | Bacteroides thetaiotaomicron | Bacteroides fragilis | |
|---|---|---|---|---|---|---|---|---|---|---|
| B. massiliensis | 3935 | 1623 | 1627 | 1866 | 1744 | 1429 | 1738 | 1711 | 1787 | 1795 |
| Mediterranea massiliensis | 57.96% | 3374 | 1543 | 1554 | 1665 | 1399 | 1603 | 1630 | 1633 | 1594 |
| B. coprocola | 61.03% | 58.89% | 4291 | 1589 | 1583 | 1324 | 1662 | 1767 | 1635 | 1562 |
| B. vulgatus | 70.47% | 57.83% | 61.28% | 4065 | 1708 | 1482 | 1737 | 1697 | 1912 | 1821 |
| B. fluxus | 60.98% | 62.46% | 60.63% | 60.44% | 3921 | 1546 | 1756 | 1659 | 1822 | 1814 |
| B. rodentium | 58.71% | 74.68% | 59.73% | 59.03% | 65.13% | 4993 | 1451 | 1386 | 1591 | 1531 |
| B. finegoldii | 60.29% | 60.03% | 60.04% | 59.18% | 60.21% | 60.75% | 4485 | 1674 | 1978 | 1779 |
| B. plebeius | 61.51% | 59.80% | 63.84% | 61.40% | 58.94% | 59.89% | 59.99% | 3933 | 1721 | 1643 |
| B. thetaiotaomicron | 61.30% | 58.97% | 58.95% | 61.11% | 58.49% | 59.64% | 66.51% | 59.37% | 4825 | 2014 |
| B. fragilis | 61.21% | 58.10% | 58.62% | 61.12% | 60.41% | 59.26% | 62.81% | 59.18% | 66.13% | 4236 |
Table 8.
Pairwise comparison of Mediterranea massiliensis strain Marseille-P2645T with other species of Bacteroides genus using GGDC formula 2 (DDH estimates based on identities/HSP length)a
| Mediterranea massiliensis | Bacteroides coprocola | Bacteroides finegoldii | Bacteroides fluxus | Bacteroides fragilis | Bacteroides massiliensis | Bacteroides plebeius | Bacteroides rodentium | Bacteroides thetaiotaomicron | Bacteroides vulgatus | |
|---|---|---|---|---|---|---|---|---|---|---|
| M. massiliensis | 100% ± 00 | 28.10% ± 4.9 | 27.40% ± 4.8 | 21.60% ± 4.7 | 22.30% ± 4.8 | 27.40% ± 4.8 | 22.90% ± 4.7 | 23.10% ± 4.8 | 22.20% ± 4.8 | 24.70% ± 4.8 |
| B. coprocola | 100% ± 00 | 34.30% ± 5 | 21.70% ± 4.7 | 23.70% ± 4.7 | 25.50% ± 4.8 | 22.10% ± 4.7 | 29.30% ± 4.9 | 22.60% ± 4.7 | 22.40% ± 4.7 | |
| B. finegoldii | 100% ± 00 | 20.50% ± 4.6 | 21.10% ± 4.7 | 24.90% ± 4.8 | 25.90% ± 4.8 | 22.20% ± 4.7 | 25.70% ± 4.8 | 24.20% ± 4.8 | ||
| B. fluxus | 100% ± 00 | 22.00% ± 4.7 | 23.50% ± 4.8 | 29.10% ± 4.9 | 24.70% ± 4.8 | 20.60% ± 4.6 | 21.60% ± 4.7 | |||
| B. fragilis | 100% ± 00 | 20.20% ± 4.6 | 23.30% ± 4.8 | 21.90% ± 4.7 | 21.70% ± 4.7 | 22.30% ± 4.7 | ||||
| B. massiliensis | 100% ± 00 | 23.40% ± 4.5 | 26.20% ± 4.8 | 26.70% ± 4.8 | 25.70% ± 4.8 | |||||
| B. plebeius | 100% ± 00 | 27.40% ± 4.8 | 25.80% ± 4.8 | 21.20% ± 4.7 | ||||||
| B. rodentium | 100% ± 00 | 21.50% ± 4.7 | 26.70% ± 4.8 | |||||||
| B. thetaiotaomicron | 100% ± 00 | 26.40% ± 4.9 | ||||||||
| B. vulgatus | 100% ± 00 |
DDH, DNA-DNA hybridization; GGDC, Genome-to-Genome Distance Calculator; HSP, high-scoring segment pairs.
Confidence intervals indicate inherent uncertainty in estimating DDH values from intergenomic distances based on models derived from empirical test data sets (which are always limited in size). These results are in accordance with 16S rRNA and phylogenomic analyses as well as GGDC results.
Taxonomical and nomenclatural proposals
Description of Mediterranea gen. nov.
The genus name is Mediterranea (me.di.ter.ra.nea', N.L. fem. n., from mediterraneum, the Latin name of the Mediterranean Sea), as this strain was first isolated in Marseille, on the banks of the Mediterranean sea. This is a Gram-negative, non–spore forming, nonmotile and preferentially anaerobic bacilli. Cells do not show catalase or oxydase activity. On the basis of 16S rRNA gene sequence analysis, Mediterranea is a member of the family Bacteroidaceae. Its type species is Mediterranea massiliensis strain Marseille-P2645T.
Description of Mediterranea massiliensis gen. nov., sp. nov.
Mediterranea massiliensis gen. nov., sp. nov. (mas.si.li.en'sis, L. fem. adj., from massiliensis, ‘of Massilia,’ the Latin name for Marseille, where the strain was first cultivated) is classified as a member of the family Bacteroidaceae in the phylum Bacteroidetes. It is a Gram-negative and motile bacillus whose cells measure 1 μm in length and 0.6 μm in diameter. Optimal growth conditions are obtained at 37°C, and it is strictly anaerobic. Colonies were translucent and slightly haemolytic, with a diameter of 0.5 mm after growth on 5% sheep's blood–enriched agar. Using API ZYM, strain Marseille-P2645T exhibited alkaline phosphatase, acid phosphatase, α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase α-fucosidase and N-acetyl β-glucosaminidase activities. Using API 20A (bioMérieux), Mediterranea massiliensis exhibited gelatin production but no indole or esculin activity. Strain Marseille-P2645T exhibits d-glucose, mannose, starch and potassium 5-ketogluconate activities and does not metabolize l-arabinose, ribose, mannitol, d-fructose, d-maltose and d-lactose assay. The bacterium is susceptible to amoxicillin, rifampicin, imipenem, metronidazole and amoxicillin/clavulanic acid. Predominant cellular fatty acids are 13-methyl-tetradecanoic acid (15:0 iso), 12-methyl-tetradecanoic acid (15:0 anteiso), hexadecanoic acid (16:00), tetradecanoic acid (14:00), 9-octadecenoic acid (18:1n9), 9,12-octadecadienoic acid (18:2n6), 3-hydroxy-15-methyl-hexadecanoic acid (17:0 3-OH iso) and 3-hydroxy-hexadecanoic acid (16:0 3-OH). The genome of Mediterranea massiliensis strain Marseille-P2645T is 3 950 441 bp long with 51.66% G+C content. The genome and 16S rRNA sequences from strain Marseille-P2645T were deposited in GenBank under accession numbers FQSB00000000 and LT558847, respectively. The type strain Marseille-P2645T was deposited in CSUR and DSMZ collections under numbers CSUR P2645 and DSM 103034, and was cultivated from the colon lavage of a 58-year-old woman without medical history, who underwent a colonoscopy because of a positive screening test for a colorectal cancer.
Discussion
Currently, the development of -omics technologies has provided an overview of the complexity of microbial communities living outside and inside our organism. These considerable technologic advances have led to an exponential progression in the knowledge of the human gut microbiota. However, we have recently developed a methodology for the dynamic description of new bacteria, including new species and genera of the digestive flora, using the original approach of culturomics to explore microbial diversity of clinical and environmental samples. We report here the isolation and culture of a new bacterial genus from the colon lavage of a 58-year-old patient from Marseille, France. The isolation of this bacterium was confirmed by its deposit in two different collections: DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen) and the CSUR (Collection de Souches de l’Unité des Rickettsies). Thus, the use of an antioxidant-enriched transport medium before being seeded on a solid medium composed of 5% Columbia agar enriched with sheep's blood has allowed the isolation a new genus of bacteria called Mediterranea with the species massiliensis.
Phylogenetically, Mediterranea massiliensis is close to bacteria of the genus Bacteroides, which are Gram-negative and anaerobic bacilli colonizing mainly the intestinal flora. Here, Bacteroides helcogenes is the phylogenetically closest species of strain Marseille-P2645T. As for Bacteroides, Mediterranea massiliensis does not form spores. The main differences with closest neighbours are the motility (positive for M. Massiliensis but negative for Bacteroides species) and sequence divergence of 16S rRNA gene (>5%) compared to Bacteroides helcogenes P36-108T, the type strain of the closest phylogenetic neighbour and difference in G+C content (51.66% for strain Marseille-P2645T vs. 41% to 47% for compared Bacteroides species). These differences confirm the classification of Mediterranea massiliensis as a distinct species. Furthermore, the values of the AGIOS and dDDH of Mediterranea massiliensis compared to all other known species confirm its new genus and species status.
Conclusion
Microbial culturomics continue to significantly expand the repertoire of isolated bacteria in the digestive tract. Application of culturomics to the duodenum, ileum and colon would improve the understanding of microbiota involvement in human health and diseases. Thus, our study may contribute to a better knowledge of the associated human microorganisms, and could help to better understand physiologic functioning in health and disease. On the basis of phenotypic, genomic and phylogenetic analyses, we formally propose the creation of Mediterranea massiliensis gen. nov., sp. nov., represented by the strain Marseille-P2645T (= CSUR P2645 = DSM 103034).
Acknowledgements
The authors thank the Xegen Company (www.xegen.fr/) for automating the genomic annotation process. This study was funded by the Fondation Méditerranée Infection.
Conflict of interest
None declared.
References
- 1.Lagier J.C., Armougom F., Million M., Hugon P., Pagnier I., Robert C. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 2.Lagier J.C., Hugon P., Khelaifia S., Fournier P.E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagier J.C., Khelaifia S., Alou M.T., Ndongo S., Dione N., Hugon P. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Fournier P.E., Lagier J.C., Dubourg G., Raoult D. From culturomics to taxonomogenomics: a need to change the taxonomy of prokaryotes in clinical microbiology. Anaerobe. 2015;36:73–78. doi: 10.1016/j.anaerobe.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 5.Sentausa E., Fournier P.E. Advantages and limitations of genomics in prokaryotic taxonomy. Clin Microbiol Infect. 2013;19:790–795. doi: 10.1111/1469-0691.12181. [DOI] [PubMed] [Google Scholar]
- 6.Pagani I., Liolios K., Jansson J., Chen I.M.A., Smirnova T., Nosrat B. The Genomes OnLine Database (GOLD) v.4: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2012;40:D571–D579. doi: 10.1093/nar/gkr1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vandamme P., Pot B., Gillis M., de Vos P., Kersters K., Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stackebrandt E. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;6:152–155. [Google Scholar]
- 9.Benno Y., Watabe J., Mitsuoka T. Bacteroides pyogenes sp. nov., Bacteroides suis sp. nov., and Bacteroides helcogenes sp. nov., new species from abscesses and feces of pigs. Syst Appl Microbiol. 1983;4:396–407. doi: 10.1016/S0723-2020(83)80024-1. [DOI] [PubMed] [Google Scholar]
- 10.Kim M., Oh H.S., Park S.C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 11.Mailhe M., Ricaboni D., Benezech A., Saber K., Fournier P.E., Raoult D. ‘Mediterranea massiliensis’ gen. nov., sp. nov., a new human-associated bacterium isolated from the right and left colon lavage of a 58-year-old patient. New Microbe New Infect. 2016;13:54–55. doi: 10.1016/j.nmni.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.La Scola B., Khelaifia S., Lagier J.C., Raoult D. Aerobic culture of anaerobic bacteria using antioxidants: a preliminary report. Eur J Clin Microbiol Infect Dis. 2014;33:1781–1783. doi: 10.1007/s10096-014-2137-4. [DOI] [PubMed] [Google Scholar]
- 13.Seng P., Drancourt M., Gouriet F., La Scola B., Fournier P.E., Rolain J.M. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 14.Seng P., Rolain J.M., Fournier P.E., La Scola B., Drancourt M., Raoult D. MALDI-TOF–mass spectrometry applications in clinical microbiology. Future Microbiol. 2010;5:1733–1754. doi: 10.2217/fmb.10.127. [DOI] [PubMed] [Google Scholar]
- 15.Sankar S.A., Lagier J.C., Pontarotti P., Raoult D., Fournier P.E. The human gut microbiome, a taxonomic conundrum. Syst Appl Microbiol. 2015;38:276–286. doi: 10.1016/j.syapm.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price M.N., Dehal P.S., Arkin A.P. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimodaira H., Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- 19.Sasser M. Microbial ID; Newark, NY: 2006. Bacterial identification by gas chromatographic analysis of fatty acids methyl esters (GC-FAME) [Google Scholar]
- 20.Dione N., Sankar S.A., Lagier J.C., Khelaifia S., Michele C., Armstrong N. Genome sequence and description of Anaerosalibacter massiliensis sp. nov. New Microbe New Infect. 2016;10:66–76. doi: 10.1016/j.nmni.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyatt D., Chen G.L., Locascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J. GenBank. Nucleic Acids Res. 2017;45:D37–D42. doi: 10.1093/nar/gkw1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lagesen K., Hallin P., Rødland E.A., Stærfeldt H.H., Rognes T., Ussery D.W. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dyrløv Bendtsen J., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 26.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 27.Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M.A. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 28.Carver T., Thomson N., Bleasby A., Berriman M., Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramasamy D., Mishra A.K., Lagier J.C., Padhmanabhan R., Rossi M., Sentausa E. A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int J Syst Evol Microbiol. 2014;64:384–391. doi: 10.1099/ijs.0.057091-0. [DOI] [PubMed] [Google Scholar]
- 30.Lechner M., Findeiß S., Steiner L., Marz M., Stadler P.F., Prohaska S.J. Proteinortho: cetection of (co-)orthologs in large-scale analysis. BMC Bioinform. 2011;12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gouret P., Paganini J., Dainat J., Louati D., Darbo E., Pontarotti P. Integration of evolutionary biology concepts for functional annotation and automation of complex research in evolution: the multi-agent software system DAGOBAH. In: Pontarotti P., editor. Evolutionary biology—concepts, biodiversity, macroevolution and genome evolution. Springer-Verlag; Berlin: 2011. pp. 71–87. [Google Scholar]
- 32.Gouret P., Vitiello V., Balandraud N., Gilles A., Pontarotti P., Danchin E.G. FIGENIX: intelligent automation of genomic annotation: expertise integration in a new software platform. BMC Bioinform. 2005;6:198. doi: 10.1186/1471-2105-6-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meier-Kolthoff J.P., Auch A.F., Klenk H.P., Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Auch A.F., von Jan M., Klenk H.P., Göker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci. 2010;2:117. doi: 10.4056/sigs.531120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayashi H., Shibata K., Bakir M.A., Sakamoto M., Tomita S., Benno Y. Bacteroides coprophilus sp. nov., isolated from human faeces. Int J Syst Evol Microbiol. 2007;57:1323–1326. doi: 10.1099/ijs.0.64979-0. [DOI] [PubMed] [Google Scholar]