Abstract
Although metabolic reprogramming and redox imbalance are widely reported to be involved in chemo-resistance in cancer treatment, much more attention was paid to anti-cancer drug induced effect. Our previous studies showed that cancer cells can develop P-gp overexpression-mediated intrinsic drug resistance in the formation of 3D MCF-7 multi-cellular layers (MCLs) without any drug induction. However, whether metabolic reprogramming and redox imbalance functioned during this progress remained unrevealed. In our present study, LC-Q/TOF-MS and GC-MS were used in combination for analysing intracellular metabolites. The contribution of pentose phosphate pathway (PPP) and its related redox status were checked by chemical interfering and silencing/over-expression of glucose-6-phosphate dehydrogenase (G6PD). The downstream products of G6PD were assayed by quantitative real-time PCR, western blot and flow cytometry. Results showed that not only G6PD expression but also G6PD activity was significantly lowered along with 3D MCF-7 cells culture time. Impaired PPP disturbed redox-cycling, generated reactive oxygen species (ROS), which triggered cell cycle arrest and caused the switch to Chk2/p53/NF-κB pathway-mediated P-gp induction. Our results provided a new attempt to associate intrinsic small molecule metabolites (impaired PPP) communicating with cell signalling pathways through disturbed intracellular redox status to elucidate multi-cellular resistance (MCR) in 3D MCF-7 cells, which improved the understanding of the mechanisms of P-gp up-regulation in MCR with metabolomic and related redox status support.
Abbreviations: NADP+, (Nicotinamide adenine dinucleotide phosphate); NADPH, (Reduced form of Nicotiamide adenine dinucleotide phosphate); G-6-P, (Glucose-6-phosphate); F-6-P, (Fructose-6-phosphate); 6-PG, (6-Phosphoglycerate); R-5-P, (Ribose-5-phosphate); Ribu-5-P, (Ribulose-5-phosphate); Xylu-5-P, (Xylulose-5-phosphate); S-7-P, (Sedoheptulose-7-phosphate); GSH, (Reduced Glutathione); GSSG, (Oxidized Glutathione); NAC, (N-acetyl cysteine); G6PD, (Glucose-6-phosphate dehydrogenase); Mena, (Menadione); DOX, (Doxorubicin)
Chemical compounds studied in this article: Myristic-1,2-13C2 acid (PubChem CID: 16213483); Menadione (PubChem CID: 4055); PFT-α (PubChem CID:9929138); DMSO (PubChem CID: 679); N-acetylcysteine (PubChem CID: 12035)
Keywords: 3D cell culture, Cellular metabolomics, Pentose phosphate pathway, Redox-status, P-gp regulation
Highlights
-
•
Impaired pentose phosphate pathway (PPP) in the development of 3D MCF-7 cells.
-
•
PPP mediated intracellular redox disturbance and multi-cellular resistance (MCR).
-
•
P-gp up-regulation in MCR with metabolomics and related redox status support.
-
•
Intrinsic small molecule metabolites communicating with cell signalling pathway.
1. Introduction
Solid tumours are more difficult to be cured than haematological tumours due to their three-dimension (3D) structure. Therefore, the conventional anti-tumour agents shown to be effective in a two-dimension (2D) cell culture system in vitro often become less or even not effective when applied to the tumour mass; this is called multi-cellular resistance (MCR) [1], [2]. The most significant features that distinguish MCR from multi-drug resistance (MDR) could be attributed to the spatial structure and micro-environment of multi-cellular clusters rather than drug induction. For this reason, a large number of 3D cell culture models in vitro have been developed to mimic the tumour mass in vivo for more accurate evaluations of new drugs and mechanistic explorations of new drug targets [3], [4]. Until recently, many factors have been identified as responsible for MCR, including poor penetration caused by ATP-binding cassette (ABC) transporters [5], anti-apoptosis gene expression [6], abnormal endogenous oxidative stress status [7], secretion of cytokines and proteins through tumour/stromal cell-cell interactions [8], etc.
However, above mentioned researches only dealt with MCR from individual perspective. It is still unknown whether these factors work separately or in cascade for MCR, and the relationships between these factors have not been elucidated clearly. Since MCR arises without exogenous drug induction, intrinsic alteration or disturbance caused by spatial structure might be the headstream. Intracellular redox status has attracted much attention in cancer treatment. On one hand, many anticancer agents are designed to exert over production of reactive oxygen species (ROS) to disturb the redox homeostasis to kill cancer cells [9], [10]. And any adaption of cancer cells to encounter ROS is believed to cause drug resistance. On the other hand, cancer cells themselves could rearrange intracellular metabolism (metabolic reprogramming) to satisfy their rapid proliferation, and they might gradually adapt to such disorder including high oxidative stress environments and exhibit drug resistance [11]. Therefore, the relationship between redox status and drug resistance in cancer cells was not in consistence, and most studies were performed at 2D cell level [12], [13]. For MCR at 3D cell level, how endogenous oxidative stress status was disturbed, where intrinsic ROS was produced, and whether redox status triggered cell signalling pathway and conferred drug resistance, all these issues need to be addressed in the formation of 3D cancer cells.
In this study, we combined cellular metabolomics and 3D cell culture to answering why intracellular redox disturbance and MCR occur without drug induction and how to overcome it in cancer therapy with new potential target. Nevertheless, there were only a few papers describing this joint field, and most cellular metabolomic investigations of tumours still relied on 2D cell culture [14], [15]. Recently, we have successfully developed a multi-cellular layer (MCL) 3D cell culture model for MCF-7 breast cancer cells and revealed the possible MCR mechanisms that up-regulate P-gp expression through the Chk2/p53/NF-κB pathway [16]. However, it is still unknown what occurs prior to cell cycle arrest and Chk2 activation, and the potential metabolomic mechanisms and related redox-status need further elucidation. Therefore, in our present study, cellular metabolomics and related redox-status in the formation of 3D cell culture (MCLs and spheroids) were analysed and were further correlated with P-gp expression.
2. Materials and methods
2.1. Materials
Myristic-1,2-13C2 acid, methoxamine hydrochloride and menadione (Mena) were purchased from Sigma-Aldrich (USA). 5-13C-glutamine was purchased from Camabridge Isotope Laboratories, Inc. (USA). N-methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA) and chlorotrimethylsilane (TMCS) were purchased from Pierce Chemical Co. (USA). Pifithrin-α (PFT-α) was purchased from Selleckchem (USA). HPLC-grade acetonitrile, methanol and n-heptane were purchased from Merck (Germany). Deionized water was prepared by a Milli-Q system (Millipore, USA). N-acetyl cysteine (NAC), glutathione (GSH) and an oxidized glutathione (GSSG) Test Kit were purchased from Beyotime Institute of Biotechnology (Jiangsu, China). NADP/NADPH Quantitation Colorimetric Kit was purchased from BioVision, Inc. (USA). The primers for glucose-6-phosphate dehydrogenase (G6PD), mdr1, p53 and β-actin for qPCR analysis were synthesized by Invitrogen (Life Technologies, USA). Monoclonal antibodies against Chk2, p-Chk2, p53, p65, histone, and horseradish peroxidase-conjugated goat anti-mouse/rabbit IgG secondary antibodies were purchased from Cell Signalling Technology (Danvers, MA, USA). The antibody for GAPDH was purchased from Bioworld Technology (Dublin, Ohio, USA).
2.2. MCF-7 cell culture
MCF-7 human breast carcinoma cells were purchased from the American Type Culture Collection. The cells (passage 10–20) were cultured in RPMI 1640 medium supplemented with 10% foetal bovine serum, and 100 U ml−1 penicillin and streptomycin (Invitrogen, Carlsbad, CA) at 37 °C with 5% CO2, and the medium was changed every other day.
2.3. Multi-cellular layer culture
The culture of MCF-7 MCLs was performed as we described previously [16]. Briefly, the MCF-7 cells were re-suspended with pre-chilled serum-free medium containing 6% matrigel and then seeded on a 6-well plate. After incubating at 37 °C for 1 h, culture medium was added to cells in each well. The cells were cultured for 3, 5, 8, 10 and 14 days, and the medium was changed every other day (n=5).
2.4. Spheroid cell culture
MCF-7 cells were resuspended with 10% RPMI 1640 medium, and then seeded on a PrimeSurface 96U 3D cell culture plate (Sumitomo Bakelite, Japan). The cells were cultured for 3, 5, 8 days, and the medium was changed every other day (n=5).
2.5. Cell viability assays
MCF-7 MCLs (3, 5, 8, 10 and 14 days) and spheroids (3, 5 and 8 days) were exposed to a series of concentrations of doxorubicin (DOX) for 72 h at 37 °C with 5% CO2. After treatments, cells were incubated with 3-(4,5-dimethylthiazol-2-yl)−2,5-diphenyltetrazolium bromide (MTT) solution (0.5 mg/ml) for 4 h, and then, precipitated MTT was dissolved with DMSO for 30 min. Absorbance was measured at 490 nm. The IC50 values were calculated from inhibition curves using Graphpad Prism 6.
2.6. Animal and Xenograft study
All animal care and experimental procedures were conducted according to the National Research Council's Guidelines for the care and use of laboratory animals and were approved by the SPF Animal Laboratory of China Pharmaceutical University (Animal authorization reference number: SYXK2016-0011). Every effort was made to minimize animal pain, suffering and distress and to reduce the number of animals used.
Healthy female BALB/c nude mice (18–22 g and 8–10 weeks old) were obtained from Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai, China). The mice were maintained in air-conditioned rooms under controlled light (12 h light: 12 h dark) and temperature (23 ± 2 °C, 55 ± 5% humidity), fed with standard laboratory food and water ad libitum. Before MCF-7 cells were transplanted into the animal, a 2-mg E2 pellet was placed subcutaneously in the interscapular region of each mouse. Then, tumours were generated by subcutaneous injections of 5 × 106 exponentially growing MCF-7 cells into the right flank regions of the nude mice. Tumour volume (TV) was measured with micrometer calipers, and calculated using the following formula: TV (mm3) = d2 × D/2, where d and D means the shortest and the longest diameters, respectively. The maximum tumour size during the experiments was allowed by Institutional Animal Care and Use Committee. As designed, when the tumours grown to 500, 1000, and 2000 mm3 (five mice per group), the mice were killed by CO2 asphyxiation and cervical dislocation, and tumours were collected for further assessment.
2.7. GC-MS-based metabolomics assays
The samples of tumour tissues and 3D MCF-7 cells were processed as we described previously [17]. Briefly, the tissues and cells were washed and lysed by repeated freezing and thawing and homogenization, followed by adding methanol containing myristic-1,2-13C2 acid as an internal standard to extract intracellular metabolites. After centrifugation, the supernatant was evaporated to dryness, and the remaining residue underwent methoximation and trimethylsilylation subsequently. Finally, external standard methyl myristate was added before GC-MS analysis. GC-MS was performed with the SHIMADZU QP2010Ultra/SE system (Tokyo, Japan) with a 30 mm × 0.25 mm ID fused-silica capillary column chemically bonded with 0.25 µm Rxi-5MS stationary phase (Restek Corporation, PA, USA). The flow rate of helium as a carrier gas was 1.5 ml/min, and the purge flow rate was 6.0 ml/min. The gradient temperature programme was set as follows: 0–3 min, 80 °C; 3–14 min, 80–300 °C; and 14–19 min, 300 °C. Ion source temperature, 200 °C; interface temperature, 220 °C; electron beam, 70 eV; and detector voltage, −950 V. Masses were scanned from m/z 50–700 at the speed of 2500 Hz and the event time was 0.30 s. Automatic peak detection and mass spectrum deconvolution were performed with the Labsolutions (GCMS Solution software Version 2.61), as reported previously.
2.8. LC-Q/TOF-MS-based metabolomics assays
Tumour tissues and 3D MCF-7 cells were homogenized in 80% methanol solution containing 5-13C-glutamine as the internal standard. The samples were centrifuged at 30000 g for 5 min, and the supernatant was evaporated to dryness. The residue was re-dissolved and injected into an Amide XBridge HPLC column (3.5 µm; 4.6 mm × 100 mm; Waters, USA). The column temperature was set to 30 °C. A hybrid quadrupole time-of-flight tandem mass spectrometer (SCIEX TripleTOF® 5600 LC–Q/TOF-MS, Foster City, CA) was coupled with a Shimadzu Prominence HPLC system, consisting of an LC-30A binary pump, an SIL-30AC autosampler, and a CTO-30AC column oven. The mobile phase consisted of solvent A (5 mM ammonium acetate, pH=9, and 5% acetonitrile) and solvent B (acetonitrile) with the following gradient: 0–3 min 85% B, 3–6 min 85–30% B, 6–15 min 30–2% B, 15–18 min 2% B, 18–19 min 2–85% B, 19–26 min 85% B. The flow rate was 0.4 ml/min. The MS detection was performed in both positive and negative ion modes for scan analysis with a Turbo V electrospray ionization (ESI). The ESI source conditions were set as follows: TOF MS scan, m/z 50–1000; Product ion scan, m/z 50–900; Gas1, 33 psi; Gas2, 33 psi; Curtain Gas, 25 psi; Ion spray voltage, −4500/4500 V (negative/positive); Turbo spray temperature, 500 °C; DP, −93/93 V (negative/positive); and CE, −10/10 V (negative/positive). MS data acquisition was performed using Analyst TF 1.6.1 (AB SCIEX, MA, USA). The accurate mass was calibrated by Calibration Delivery System (CDS), and automatic calibration was carried out every six samples. Data exploration and peak area intergration were performed with PeakView and MultiQuant 2.0 from AB SCIEX, respectively.
2.9. Metabolites identification
Generally, all the endogenous metabolites were identified by comparing the mass spectra and retention time of the detected compounds with a reference database, and some of the metabolites were further confirmed by commercially available reference standards. For GC-MS, the following databases were used as we described previously: the National Institute of Standards and Technology (NIST) library 2.0 (2008), Wiley 9 (Wiley–VCH Verlag GmbH & Co. KGaA, Weinheim, Germany), and an in-house mass spectra library database established by the Umeå Plant Science Center, Swedish University of Agricultural Sciences (Umeå, Sweden). For LC-Q/TOF-MS, the molecular formulas of the compounds were predicted by Formula Predictor Software (AB SCIEX, Concord, ON). Meanwhile, retention time, parent ion mass spectrum information, fragmental ion mass spectrum information, as well as free online databases such as MASSBANK (http://www.massbank.jp/index-e.html), METLIN (http://metlin.scripps. edu), and MS2T (http://prime.psc.riken.jp/lcms/ms2tview/ms2tview.htm) were used in combination to identify and interpret chromatographic peaks. The peak area of each compound was weighted by internal standard and protein concentration. Data are presented as the means ± S.E.
2.10. Multivariate data analysis
Multivariate data analysis was performed with SIMCA-P 11 software (Umetrics AB, Sweden). Partial least squares discriminant analysis (PLS-DA) was employed to analyse the data as we described previously. Furthermore, a heatmap was generated with R-Project, which is available online at http://www.r-project.org/ (Vanderbilt University, Nashville Tennessee, USA). Metabolic pathway enrichment and topological analyses were elucidated by Metaboanalyst (http://www.metaboanalyst.ca), which helped to identify the most relevant pathways involved.
2.11. Detection of intracellular reactive oxygen species (ROS) levels
The 3D MCF-7 cells were incubated with reactive oxygen probe (DCFH-DA, 10 μM) in the dark at 37 °C for 45 min. After washing, an NaOH: methanol (v: v=1:1) solution was added to lyse the cells. The cell lysate was centrifuged and the supernatant was collected for determination at 535 nm (with 488 nm excitation) in a Synergy-H1 multi-mode microplate reader (Bio-Tek Instruments, Inc., USA). ROS levels were normalized to the protein levels, which were determined by BCA protein assay kit as we described previously.
2.12. Intracellular GSSG/GSH ratio assay
Intracellular GSSG and GSH were measured according to the manufacturer's instructions of a commercially available GSH and GSSG Test Kit (Beyotime Institute of Biotechnology, China), and the GSSG/GSH ratio was then calculated. In brief, the cells were lysed and any intracellular GSSG was forced to be reduced to GSH to achieve total GSH (t-GSH), which could be detected by 5,5-dithio-bis (2-nitrobenzoic) acid (DTNB). Intracellular GSSG was measured similarly, but the cell lysate was pre-excluded of intracellular GSH. The concentration of intracellular GSH was obtained by subtracting GSSG from t-GSH.
2.13. Intracellular NADP/NADPH assay
The intracellular NADP/NADPH ratio was determined with an NADP/NADPH Quantitation Colorimetric Kit (BioVision Inc., USA) based on the manufacturer's instructions. Briefly, cells lysate was prepared by repeated freezing and thawing, followed by centrifugation and ultra-filtration (10 kd). Then, total NADP/NADPH (NADPt) was measured at 450 nm with a Synergy-H1 multi-mode microplate reader (Bio-Tek Instruments, Inc., USA). For NADPH measurement, lysate was heated in a water bath at 60 °C for 30 min, and all NADP was decomposed while NADPH was still intact. NADP could be calculated by subtracting NADPH from NADPt, and the ratio could be obtained.
2.14. Cell cycle analysis
The DNA content of MCF-7 cells was determined for cell cycle analysis. Cells were fixed in 75% ethanol overnight at 4 °C, and re-suspended with a staining solution containing propidium iodide (PI) and RNase A for 30 min. After washing, the DNA content of cells was determined by flow cytometry (FACS Calibur, BD, Franklin Lakes, New Jersey, USA) and analysed with CELLQUEST software.
2.15. Knockdown and over-expression of G6PD
Reverse transfection was performed to knockdown G6PD in MCF-7 cells with Stealth Select RNAi™ siRNA, which was designed and synthesized by Invitrogen (Life Technologies, USA) to selectively target human G6PD mRNA (G6PD siRNA: 5′-ACG AGC UGA UGA AGA GAG UGG GUU U-3′). RNAi duplex-Lipofectamine® RNAiMAX (Life Technologies, USA) complexes were prepared, and then mixed with the appropriate number of MCF-7 cells to yield 30–50% confluence 24 h after plating. After incubating the cells for 72 h at 37 °C in a CO2 incubator, the cells were assayed for silence efficiency and followed by other down-stream experiments. Control siRNA, a non-targeting siRNA, was used as a negative control.
To over-express G6PD in 3D MCF-7 cells, lentivirus infection was chosen as an efficient approach. Lentivirus vector with human G6PD gene (LV-G6PD) was constructed by Shanghai Genechem Co., Ltd. (Shanghai, China), and had been fully validated before use. In brief, MCF-7 cells were infected by LV-G6PD (MOI = 20) with polybrene as an enhancer for 12 h, and then allowed to culture with normal culture medium for 72 h to achieve maximum infection efficiency. When the infected cells reached confluence, they were trypsinized, collected, and cultured into 3D forms.
2.16. G6PD activity assays
The enzyme activities of G6PD were assayed by a commercially available Glucose-6-Phosphate Dehydrogenase (G6PD) Activity Assay Kit (Cell Signalling Technology, USA). In brief, tumour tissues and MCF-7 cells were lysed into homogenate in ice-cold 1 × Cell Lysis Buffer plus 1 mM PMSF, and the samples were incubated on ice for 5 min. Then, samples were sonicated on ice and centrifuged at 14000 g for 10 min. The supernatant was added into a black 96-well plate containing 70 μl total detection solution and incubated for 30 min at 37 °C. The plate was read with excitation around 540 nm and emission around 590 nm.
2.17. Western blot
For Western blots, whole cell extracts were prepared as we previously described. After being separated on a 10% SDS-polyacrylamide gel, the proteins were transferred onto a polyvinylidene difluoride membrane (Bio-Rad, Hercules, CA, USA). Then, the membrane was blocked with 5% non-fat milk and further incubated with the primary antibodies overnight at 4 °C. Before detection, horseradish peroxidase-conjugated secondary antibody was added and treated for 1 h at 37 °C, and the signals could be detected using an enhanced chemiluminescence kit (Thermo Fisher Scientific, Waltham, MA, USA) and captured using a ChemiDoc XRS+ System (Bio-Rad, Hercules, CA, USA).
2.18. Quantitative real-time PCR assay
MCF-7 cells were suspended in a High Pure RNA Isolation Kit (RNAiso Plus, Takara Bio, Japan) and total RNA was extracted. Then, the RNA was reversely transcripted into cDNA with a PrimeScript RT Regent Kit (Takara Bio, Japan). The quantitative real-time PCR (qPCR) reactions were performed in a CFX96 real-time RT-PCR detection system (Bio-Rad, USA). The primers used in this study were as follows:
mdr1: forward primer: GCTGGGAAGATCGCTACTGA;
reverse primer: GGTACCTGCAAACTCTGAGCA;
p53: forward primer: CCATGAGCGCTGCTCAGAT;
reverse primer: CAACCTCAGGCGGCTCATA;
G6PD: forward primer: GGCAACAGATACAAGAACATGAA;
reverse primer: CCCTCATACTGGAAACCCACT;
β-actin: forward primer: GCGTGACATTAAGGAGAAG;
reverse primer: GAAGGAAGGCTGGAAGAG.
The qPCR conditions included 95 °C initial denaturation for 90 s, followed by 40 cycles of denaturation (10 s at 95 °C), annealing (30 s at 60 °C) and extension (30 s at 72 °C). A melting curve was used to monitor the specificity of the primer. The relative expressions of the detected genes were normalized to the expression of the reference gene β-actin.
2.19. P-gp expression assay
Flow cytometry was used to determine the protein expression of P-gp as we described previously. In brief, drug-treated or not treated 3D MCF-7 cells were fixed with 4% poly formaldehyde solution. After washing and blocking, the cells were incubated with FITC-conjugated anti-P-gp polyclonal antibody (BD Biosciences, USA) at 37 °C for 1 h followed by another washing. Then, the samples were loaded onto flow cytometer for analysis.
2.20. Statistical analysis
All the data are presented as the means ± S.E. of at least three independent experiments. Two-tailed Student's t-tests and one-way analysis of variances were employed for statistical analyses. Differences were considered significant at *p < 0.05, **p < 0.01, ***p < 0.001. Statistical data analysis was performed using GraphPad Prism 6.
3. Results
3.1. Time-dependent metabolic patterns of intracellular metabolites in MCF-7 MCLs and spheroids
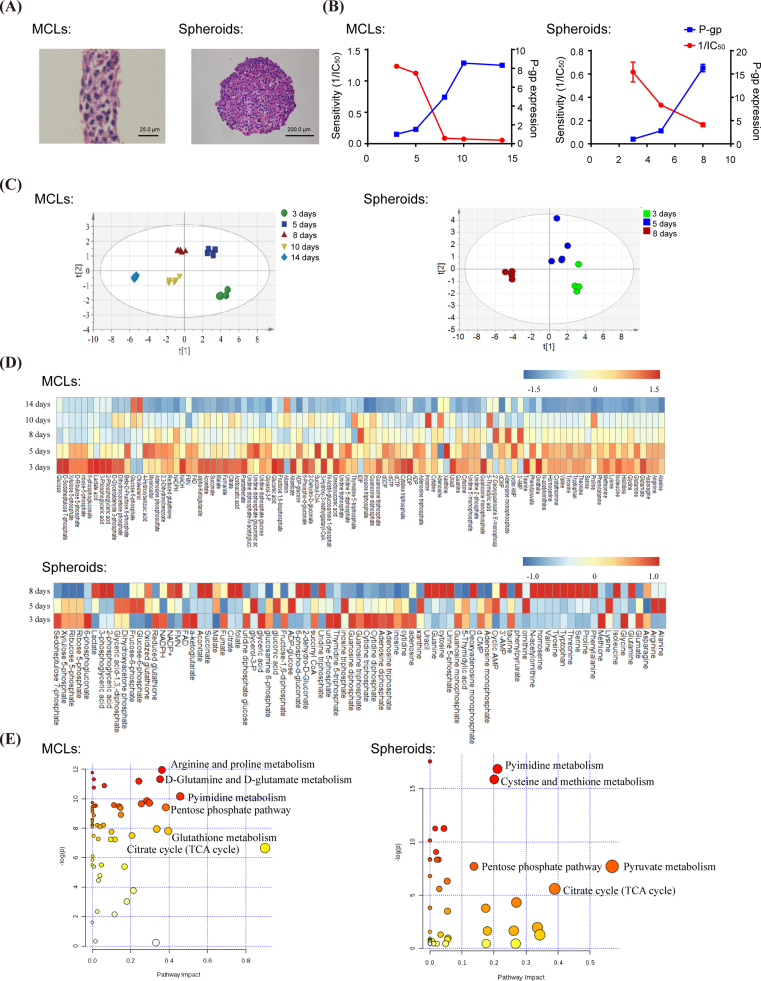
As shown in Fig. 1A, 3D MCF-7 cells were successfully developed as MCF-7 MCLs (14 days) and spheroids (8 days) two forms with haematoxylin and eosin (HE) staining. The sensitivities of 3D MCF-7 cells to anti-cancer agent doxorubicin were presented as 1/IC50, and were negatively correlated with drug resistance protein P-gp expression along with the culture time (Fig. 1B). Subsequently, the metabolic patterns in the formation of 3D MCF-7 MCLs and spheroids were analysed. As shown in Fig. 1C, all samples were scattered in the PLS-DA scores plot, and distinct separations of MCLs or spheroids samples with different culture times were observed, which moved step by step from right to left in the scores plot. Metabolite identification and statistical analysis revealed that many metabolites were severely perturbed along with the MCLs or spheroids culture time, which could be seen in the heatmaps (Fig. 1D). On this basis, a preliminary intracellular metabolite-based metabolic pathway was elucidated (Fig. 1E). By screening intracellular metabolites with similar alteration tendency in both MCLs and spheroids, pentose phosphate pathway (PPP) and its related metabolic products became our focus.
Fig. 1.
Global metabolomic profiles of MCF-7 MCLs and spheroids during the culture time. (A) MCF-7 MCLs (14 days) and spheroids (8 days) were stained by haematoxylin and eosin (HE). (B) The doxorubicin sensitivity (1/IC50) and P-gp expression of MCF-7 MCLs and spheroids during the culture time. (C) PLS-DA scores plot of MCF-7 MCLs and spheroids at designated culture time. (D) Heatmaps of the typical intracellular metabolites for MCF-7 MCLs and spheroids in response to culture time, which was generated with R-Project. Red series denote relatively higher concentrations and blue series denote relatively lower concentrations. The deeper the colour, the more significantly the metabolite changed. (E) Metabolic pathway analysis of MCF-7 MCLs and spheroids, which was elucidated by Metaboanalyst.
3.2. Inhibition of PPP in the formation of 3D MCF-7 cells
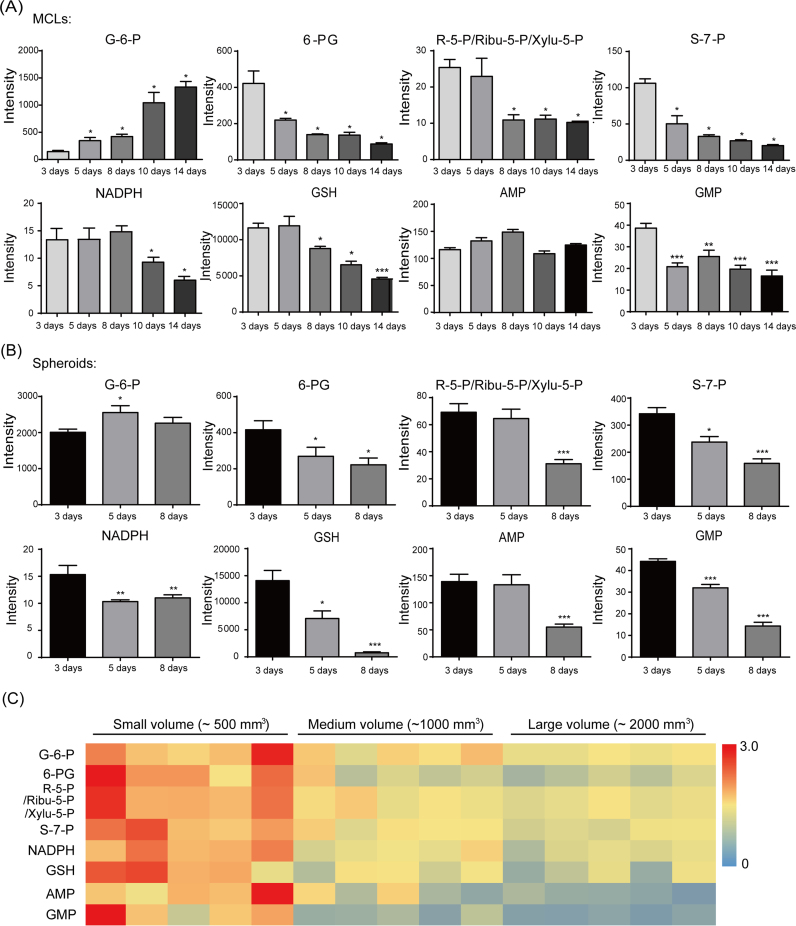
As shown in Fig. 2A, metabolic products including 6-phosphogluconate (6-PG), ribose-5-phosphate (R-5-P), ribulose-5-phosphate (Ribu-5-P), xylulose-5-phosphate (Xylu-5-P), sedoheptulose-7-phosphate (S-7-P), NADPH, GSH, adenosine monophosphate (AMP), guanosine monophosphate (GMP) in MCF-7 MCLs decreased significantly in a time-dependent manner. The same phenomenon was also observed in the formation of MCF-7 spheroids (Fig. 2B). In addition, we also determined the changes of metabolic products during the growth of MCF-7 xenografted tumours. It was shown that the major metabolic products of PPP also decreased significantly in a tumour volume-dependent manner (Fig. 2C).
Fig. 2.
Typical intracellular metabolite levels associated with PPP in MCF-7 MCLs (A), MCF-7 spheroids (B) and MCF-7 xenografted tumours (C) along with culture time. Metabolites were determined and identified by GC-MS and LC-Q/TOF-MS. For relative comparison, the peak areas were integrated and further weighted by internal standard and protein concentration, which were presented as intensity. The results are presented as the mean ± S.E. *p < 0.05, **p < 0.01, ***p < 0.001 vs the 3-day group.
3.3. Impaired G6PD in the formation of 3D MCF-7 cells
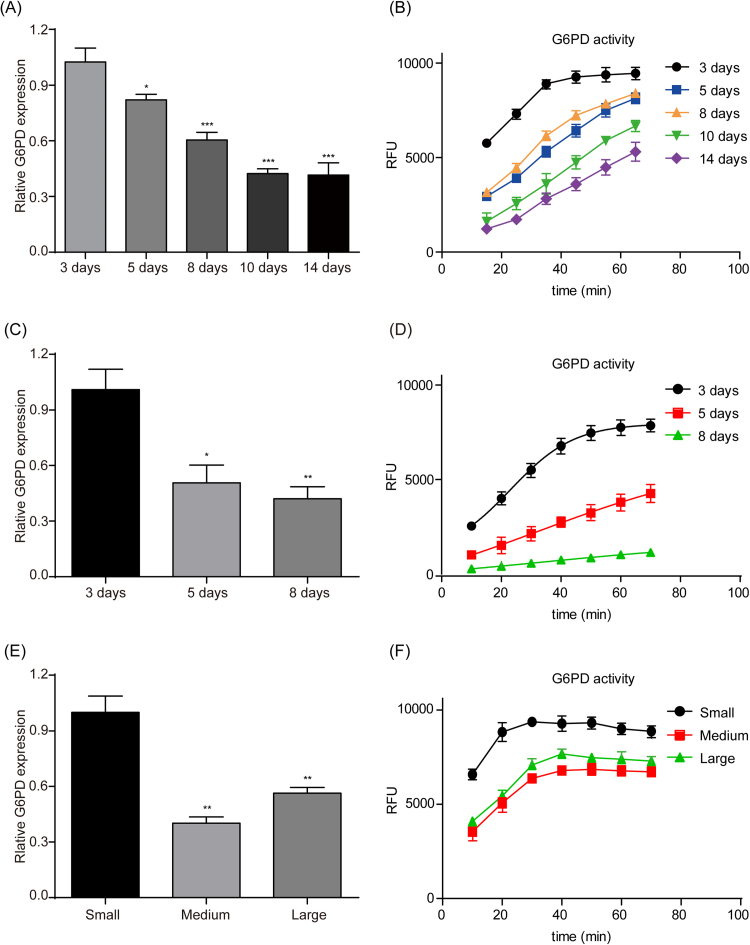
To further explore the role of PPP in the formation of 3D MCF-7 cells, we investigate the levels of G6PD, which is the key enzyme of PPP. As shown in Fig. 3A-D, both the gene expression and activity of G6PD were reduced significantly along with the culture time of 3D MCF-7 MCLs and spheroids. Furthermore, both gene expression and activity of G6PD decreased significantly during the growing progress of MCF-7 xenografted tumours (Fig. 3E-F).
Fig. 3.
Expression and activity of G6PD in MCF-7 MCLs, spheroids and xenografted tumours. Relative expressions of G6PD in the formation of MCF-7 MCLs (A) and spheroids (C) along with culture time, or in the growth of MCF-7 xenografted tumours (E) along with tumour volume were assayed by qPCR. Gene expression was normalized to the housekeeping gene β-actin. The enzyme activities of G6PD in the formation of MCF-7 MCLs (B) and spheroids (D) along with culture time, or in the growth of MCF-7 xenografted tumours (F) along with tumour volume were assayed by a Glucose-6-Phosphate Dehydrogenase Activity Assay Kit according to the manufacturer's instructions. The results are presented as the mean ± S.E. *p < 0.05, **p < 0.01, ***p < 0.001 vs the 3-day group or small volume group.
3.4. Re-activation of PPP suppressed P-gp expression with redox balance recovery
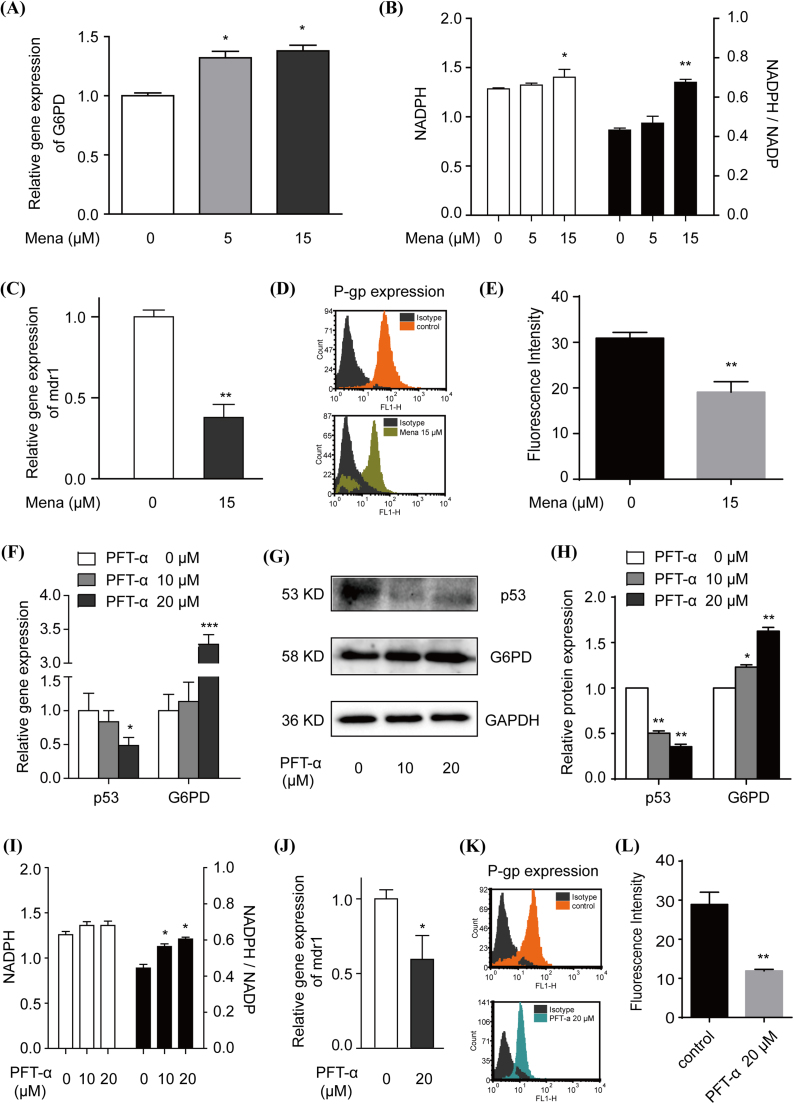
As shown in Fig. 4A and B, when 3D MCF-7 cells were treated with PPP activator Mena, G6PD expression and the NADPH/NADP+ ratio were markedly up-regulated. Subsequently, the gene and protein expression of P-gp decreased significantly (Fig. 4C–E). Similarly, when p53 inhibitor PFT-α was applied to 3D MCF-7 cells, a decrease of p53 in addition to an increase of G6PD was observed both at the gene and protein levels (Fig. 4F-H), as well as elevated NADPH/NADP+ (Fig. 4I). Further, PFT-α significantly inhibited P-gp expression (Fig. 4J-L).
Fig. 4.
The role of PPP on P-gp expression in 3D MCF-7 cells and related oxidative-reductive status. On one hand, 3D MCF-7 cells were treated with PPP activator Mena (0, 5 and 15 μM) through redox-cycling stimulation for 12 h, G6PD gene expression was assayed by qPCR (A), and intracellular NADPH level and NADPH/NADP+ ratio were detected (B). Then, mdr1 gene expression was also assayed by qPCR (C), P-gp protein levels were analysed by flow cytometry using FITC conjugated anti-P-gp antibody (D) and the mean fluorescence intensity was semi-quantified (E). On the other hand, another PPP activator PFT-α (0, 10 and 20 μM) through p53 inhibition was applied to 3D MCF-7 cells for 12 h. The gene expression (F) and protein expression (G) with semi-quantification (H) of p53 and G6PD were analysed, and the intracellular NADPH level and NADPH/NADP+ ratio were also detected (I). Subsequently, mdr1 gene expression (J) and P-gp protein expression (K) with semi-quantification (L) were assayed. Data are presented as the mean ± S.E. *p < 0.05, **p < 0.01, ***p < 0.001.
3.5. Disturbed redox balance in the formation of 3D MCF-7 cells induced P-gp expression
Previously, we found progressive G2/M cell arrest, activation of the Chk2/p53/p65 pathway, and of mdr1 expression in MCF-7 MCLs along with culture time [16]. Considering the change of PPP in the formation of 3D MCF-7 cells, we hypothesised that it may regulate P-gp expression through disturbing redox balance. As shown in Fig. 5A and B, along with the culture time, the levels of intracellular NADPH/NADP+ and GSH/GSSG in 3D MCF-7 cells decreased significantly, and the levels of intracellular ROS increased significantly. Such elevated intracellular ROS in the formation of 3D MCF-7 cells was simulated by H2O2 treatment of 2D MCF-7 cells, and it could markedly induce cell cycle arrest (Fig. 5C), stimulate Chk2/p53/p65 pathway (Fig. 5D), and increase P-gp expression (Fig. 5E). When 3D MCF-7 cells were treated with ROS scavenger NAC (10 mM), initially activated Chk2/p53/p65 pathway was suppressed (Fig. 5F), and the levels of the mdr1 gene (Fig. 5G) and P-gp protein (Fig. 5H) sharply decreased by 65% and 50%, respectively.
Fig. 5.
The relationship between intracellular oxidative-reductive status and P-gp expression in the formation of 3D MCF-7 cells. Intracellular NADPH/NADP+ ratio, GSH/GSSG ratio and ROS level in the formation of 3D MCF-7 MCLs (A) and spheroids (B) were detected by commercially available kits. Elevated ROS was further simulated by H2O2 treatment (0.2 mM, 24 h), and cell cycle (C), Chk2/p53/p65 pathway (D) and P-gp expression (E) were analysed (n = 5). Subsequently, ROS scavenger NAC (10 mM, 2 h) was added to 3D MCF-7 cells, and Chk2/p53/p65 pathway was detected by western blot (F) (n = 5), the mdr1 gene expression was assayed by qPCR (G), and P-gp protein levels were analysed by flow cytometry using FITC conjugated anti-P-gp antibody with semi-quantification (H). Data are presented as mean ± S.E. *p < 0.05, **p < 0.01, ***p < 0.001.
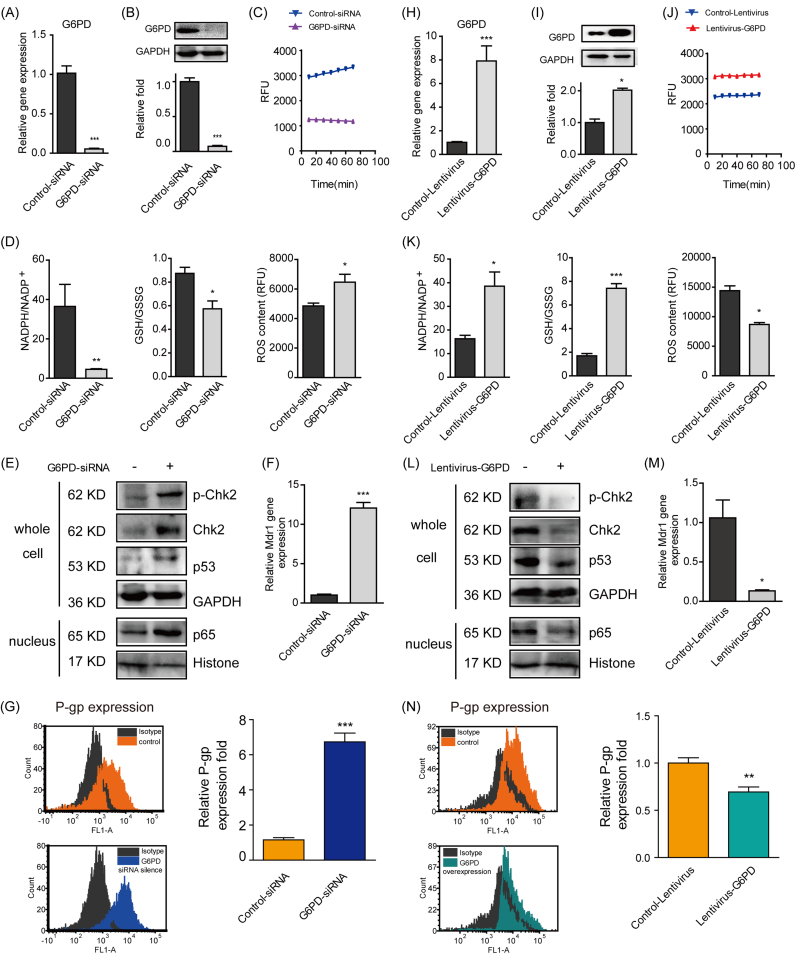
3.6. Knockdown and over-expression of G6PD negatively regulated P-gp expression
When 2D MCF-7 cells were interfered with G6PD siRNA to mimic the suppressed status of PPP in the formation of 3D MCF-7 cells, the gene, protein and activity levels of G6PD were all markedly decreased (Fig. 6A-C), demonstrating the success of G6PD knockdown. Meanwhile, the NADPH/NADP+ and GSH/GSSG ratios were also decreased in company with elevated ROS amount (Fig. 6D). With the silence of G6PD, the Chk2/p53/p65 pathway was markedly stimulated (Fig. 6E). Furthermore, the gene and protein levels of P-gp were significantly increased to 12-fold and 6-fold, respectively (Fig. 6F and G).
Fig. 6.
Knockdown and over-expression of G6PD regulated P-gp expression. 2D MCF-7 cells were transfected with G6PD siRNA to selectively knockdown G6PD. To over-express G6PD, 3D MCF-7 cells were transfected by lentivirus vector with human G6PD gene. The transfection efficiency was tested for the gene (A, H), protein (B, I) and activity (C, J) of G6PD by qPCR, western blot and enzyme activity assays, respectively. PPP related NADPH/NADP+, GSH/GSSG and ROS were also detected (D, K) by commercially available kits. The Chk2/p53/p65 pathway was determined by western blot (E, L) (n = 5). The target molecule P-gp was determined at the gene (F, M) and protein levels (G, N) by qPCR and flow cytometry (n=5), respectively. Data are presented as the mean ± S.E. *p < 0.05, **p < 0.01, ***p < 0.001.
In contrast, when 3D MCF-7 cells were infected with lentivirus-G6PD, the gene, protein and activity levels of G6PD were all markedly increased (Fig. 6H-J), indicating the over-expression of G6PD and activation of PPP. On this basis, NADPH/NADP+ and GSH/GSSG were also elevated while the ROS level was reduced (Fig. 6K). Over-expressed G6PD also suppressed Chk2/p53/p65 pathway (Fig. 6L). Importantly, the gene and protein levels of P-gp were significantly decreased to 0.13-fold and 0.76-fold, respectively (Fig. 6M and N).
Furthermore, the main metabolites of PPP, glutathione metabolism, and nucleotide metabolism were also changed correspondingly with G6PD knockdown or over-expression (Table 1).
Table 1.
Relative changes of PPP related metabolites when G6PD was knockdown or over-expressed in 2D and 3D MCF-7 cells respectively.
| 2D MCF-7 cells |
3D MCF-7 cells |
|||
|---|---|---|---|---|
| Control-siRNA | G6PD-siRNA | Control-Lentivirus | Lentivirus-G6PD | |
| G−6-P | 1.00 ± 0.32 | 0.28 ± 0.08* | 0.34±0.02 | 0.69 ± 0.03* |
| 6-PG | 1.00 ± 0.34 | 0.08 ± 0.03* | 0.47±0.06 | 7.26 ± 1.02* |
| R−5-P/ Ribu−5-P/ Xylu−5-P | 1.00 ± 0.17 | 0.38 ± 0.12* | 0.22±0.02 | 0.50 ± 0.05* |
| S−7-P | 1.00 ± 0.12 | 0.20 ± 0.04* | 0.25±0.02 | 0.46 ± 0.05* |
| NADPH | 1.00 ± 0.36 | 0.21 ± 0.15* | 2.62±0.28 | 8.81 ± 2.75* |
| GSH | 1.00 ± 0.29 | 0.04 ± 0.02* | 0.75±0.24 | 2.73 ± 0.58* |
| AMP | 1.00 ± 0.25 | 0.06 ± 0.02* | 0.68±0.09 | 6.70 ± 1.27* |
| GMP | 1.00 ± 0.30 | 0.05 ± 0.04* | 0.86±0.10 | 7.29 ± 1.19* |
4. Discussion and conclusions
Metabolic reprogramming of tumour cells has been considered one of the ten characteristics of tumours [18]. The most remarkable changes include the activation of glycolysis, increases in lipid metabolism, enhancement of mitochondrial biosynthesis and activation of the PPP [18], [19], [20], [21], [22]. It occurred not only in the process of transformation from normal cells into tumour cells but also in the development of tumour cells at the advanced stage, and it has strong relationships with the sensitivity of anti-cancer agents (both MDR and MCR) [23], [24], [25].
Previously, we found that the Chk2/p53/NF-κB pathway was responsible for the up-regulation of P-gp expression and could partially explain MCR in a 3D breast cancer cell model [16]. Therefore, this time we paid special attention to the intracellular small molecules and related redox-status in the formation of 3D cells and attempted to link the intrinsic small molecules with a cell signalling pathway through redox status disturbance.
First, global metabolomic profiles of MCF-7 MCLs and MCF-7 spheroids at designated culture times were analysed, and a dynamic migration trend in a time-dependent manner was clearly presented (Fig. 1). Given that P-gp expression in 3D MCF-7 cells also increased time-dependently in parallel to the metabolomic shift in our present and previous study [16], it was hypothesised that the intrinsic small molecules group indeed influenced and regulated P-gp expression. Hence, we subsequently identified the differential metabolites in MCF-7 MCLs or spheroids by multivariate statistical analysis and speculated on the possible involved metabolic pathways through virtual prediction. It was shown that PPP was significantly inhibited along with culture time in both MCF-7 MCLs and spheroids, while changes of glycolysis and tricarboxylic acid cycle between MCLs and spheroids were different, which was considered to be due to the culture differences of the two models. Thus, we speculated that PPP might contribute to the development of P-gp expression in 3D tumour models, and further explored the effect of PPP during this progress.
As we all know, PPP can be divided into the oxidative branch and non-oxidative branch. The oxidative branch of PPP was widely used in anti-oxidative metabolism. For example, NADPH generated from the oxidative PPP could make Saccharomyces cerevisiae tolerant to oxidizing agents [26]. Furthermore, PPP also plays an important role in the anticancer studies, such as proliferation, apoptosis, invasion, drug resistance and tumour metastasis [27]. In addition, activation of PPP was considered to contribute to drug resistance in previous reports [28], because NADPH was released when G-6-P was transformed into Ribu-5-P. Concomitantly, GSH was transformed from GSSG, and was able to scavenge ROS introduced by anti-cancer agents and thus weaken drug efficacy [28], [29]. In contrast, in our research, PPP was significantly inhibited at the point of transforming G-6-P into Ribu-5-P and presented as lowered NADPH, GSH, R-5-P and Xylu-5-P levels in both MCF-7 MCLs and spheroids in vitro and MCF-7 xenografted tumours in vivo. These data were in high agreement with our later results that ROS was increased (Fig. 5C) while purine nucleotides (AMP and GMP) were markedly decreased (Fig. 2A and B). The impaired PPP without drug induction in our study was thought to confer MCR along with culture time. To further correlate PPP with P-gp in 3D MCF-7 cells, the superoxide generator Mena was used as PPP agonist, and another p53 inhibitor, PTF-α, was also tested with the purpose of relieving the suppression effect of p53 on G6PD. Both Mena and PTF-α significantly up-regulated the levels of G6PD and the NADPH/NADP+ ratio, while the gene and protein levels of P-gp were significantly down-regulated (Fig. 4). These data suggested that the impaired PPP participated in up-regulating P-gp in the formation of 3D MCF-7 cells, and thus leaded to MCR.
One direct consequence of impaired PPP was disturbed redox status. Decreased NADPH led to reduced synthesis of GSH, which could not sufficiently counteract ROS and finally caused ROS accumulation. The redox imbalance has been shown in many types of cancer cells [30], and it might be crucial in drug resistance in cancer treatment [31], [32], [33]. Nevertheless, the contribution of metabolic pathways-induced ROS in chemoresistance was still confusing, which might depend on ROS level and cell types [34]. In our research, relatively suppressed PPP led to reduced NADPH/NADP+ and GSH/GSSG, followed by increased ROS, which might affect the expression of P-gp. Although the relationship between oxidative stress and P-gp was inconsistent in different studies [35], [36], [37], our previous studies based on 2D cell culture revealed that doxorubicin induced P-gp expression was significantly antagonized by the ROS scavenger NAC [17]. In our present study, 3D MCF-7 cells were treated with NAC to eliminate the increased ROS during the 3D structure formation. It was interesting to find that NAC could significantly inhibited activation of Chk2/p53/NF-κB, thus leaded to inhibition of P-gp expression. The effect of ROS on this pathway was further verified by H2O2 stimulation on 2D MCF-7 cells (Fig. 5C-E). These data suggested that the imbalance of oxidation-reduction by blocking PPP probably induced P-gp expression.
To further prove PPP is the headstream of Chk2/p53/NF-κB mediated P-gp expression, the key enzyme G6PD was interfered by knockdown or over-expression, and G6PD was found to negatively regulate ROS level, thus influence P-gp expression (Fig. 6). Taken together, it was indicated that excessive ROS from relatively suppressed PPP leaded to Chk2/p53/NF-κB activation, thus resulted in P-gp overexpression. In addition, the tumour suppressor p53, which acts as the key suppressor of G6PD [38], [39], may further aggravated the inhibition of PPP during the 3D culture formation.
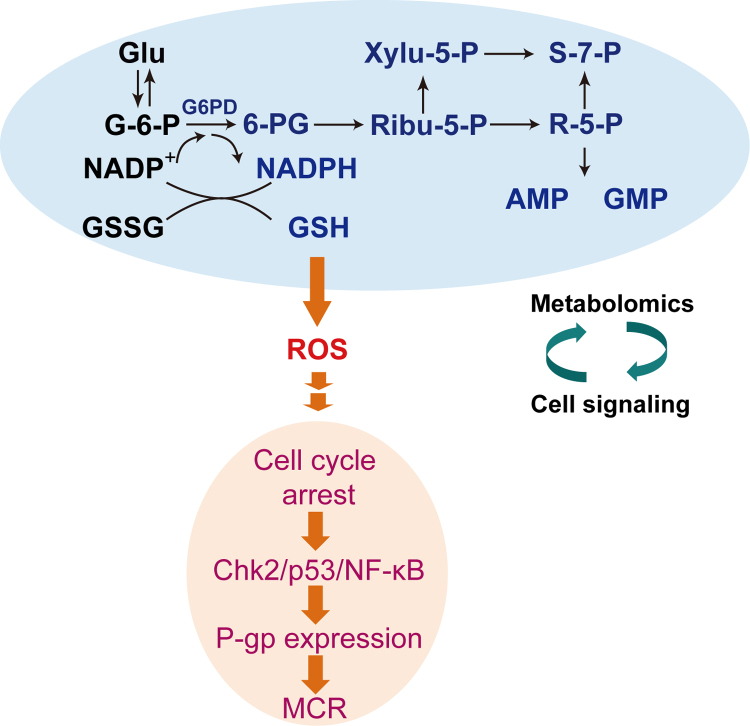
In summary, a remarkable metabolomic pattern shift was observed along with 3D MCF-7 cells culture time, and PPP was found to be crucial during this progress. Consequently, excessive intrinsic ROS generated by impaired PPP during the 3D structure formation of MCF-7 cells probably triggered cell cycle arrest, switched cells to Chk2/p53/NF-κB pathway, and finally induced P-gp expression (Fig. 7). Our research revealed the metabolic mechanism of intrinsic ROS inducing MCR, and provided a potential intervention target for cancer treatment.
Fig. 7.
A summary of our present research about the metabolomic alteration (PPP) in the formation of 3D MCF-7 cells along with culture time (red and blue series denote an increase and decrease of intracellular metabolites, respectively) and further correlated with our previous findings, both of which revealed that impaired PPP communicating with cell signalling pathways through disturbed intracellular redox status contributed to MCR in 3D MCF-7 cells.
Acknowledgements
The authors sincerely thank the post-graduates in the Key Lab of Drug Metabolism and Pharmacokinetics (China Pharmaceutical University, Nanjing, China) for their kind assistance.
This work was supported by the China National Nature Science Foundation [No. 81573494, 81703600, 81773989, 81573496, 81530098]; the Jiangsu Province Nature Science Foundation [No. BK20160076, BK20160108]; the Fundamental Research Funds for the Central Universities, Nanjing University [021414380173]; the China “Creation of New Drugs” Key Technology Projects [2015ZX09501001, 2016ZX09101031].
Acknowledgments
Conflicts of interest
The authors declare no conflicts of interest.
Authorship contribution statement
-
(1)
Study conception and design:
G.J. Wang, F. Zhou, M. Lu and J.W. Zhang
-
(2)
Acquisition, analysis and/or interpretation of data:
W.J. Wang, Q.Y. Cai, M. Lu, P. Ni, J.L. Liu and X.L. Jin
-
(3)
Drafting/revision of the work for intellectual content and context:
M. Lu, J.W. Zhang and W.J. Wang
-
(4)
Final approval and overall responsibility for the published work: G.J. Wang, M. Lu and J.W. Zhang
Contributor Information
Meng Lu, Email: ahwhlm@163.com.
Guangji Wang, Email: guangjiwang@hotmail.com.
Jingwei Zhang, Email: zhangjw_cnnj@sina.com.
References
- 1.Chitcholtan K., Sykes P.H., Evans J.J. The resistance of intracellular mediators to doxorubicin and cisplatin are distinct in 3D and 2D endometrial cancer. J. Transl. Med. 2012;10:38. doi: 10.1186/1479-5876-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Godugu C., Patel A.R., Desai U., Andey T., Sams A., Singh M. AlgiMatrix based 3D cell culture system as an in-vitro tumor model for anticancer studies. PloS One. 2013;8(1):e53708. doi: 10.1371/journal.pone.0053708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedrich J., Seidel C., Ebner R., Kunz-Schughart L.A. Spheroid-based drug screen: considerations and practical approach. Nat. Protoc. 2009;4(3):309–324. doi: 10.1038/nprot.2008.226. [DOI] [PubMed] [Google Scholar]
- 4.Susewind J., de Souza Carvalho-Wodarz C., Repnik U., Collnot E.M., Schneider-Daum N., Griffiths G.W., Lehr C.M. A 3D co-culture of three human cell lines to model the inflamed intestinal mucosa for safety testing of nanomaterials. Nanotoxicology. 2016;10(1):53–62. doi: 10.3109/17435390.2015.1008065. [DOI] [PubMed] [Google Scholar]
- 5.Chen J., Ding Z., Peng Y., Pan F., Li J., Zou L., Zhang Y., Liang H. HIF-1alpha inhibition reverses multidrug resistance in colon cancer cells via downregulation of MDR1/P-glycoprotein. PloS One. 2014;9(6):e98882. doi: 10.1371/journal.pone.0098882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakuma Y., Yamazaki Y., Nakamura Y., Yoshihara M., Matsukuma S., Koizume S., Miyagi Y. NF-kappaB signaling is activated and confers resistance to apoptosis in three-dimensionally cultured EGFR-mutant lung adenocarcinoma cells. Biochem. Biophys. Res. Commun. 2012;423(4):667–671. doi: 10.1016/j.bbrc.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 7.Wartenberg M., Fischer K., Hescheler J., Sauer H. Redox regulation of P-glycoprotein-mediated multidrug resistance in multicellular prostate tumor spheroids. Int. J. Cancer. 2000;85(2):267–274. [PubMed] [Google Scholar]
- 8.Jeong E.K., Lee S.Y., Jeon H.M., Ju M.K., Kim C.H., Kang H.S. Role of extracellular signal-regulated kinase (ERK)1/2 in multicellular resistance to docetaxel in MCF-7 cells. Int. J. Oncol. 2010;37(3):655–661. doi: 10.3892/ijo_00000714. [DOI] [PubMed] [Google Scholar]
- 9.Trachootham D., Alexandre J., Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat. Rev. Drug Discov. 2009;8(7):579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 10.Panieri E., Santoro M.M. ROS homeostasis and metabolism: a dangerous liaison in cancer cells. Cell death Dis. 2016;7(6):e2253. doi: 10.1038/cddis.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cairns R.A., Harris I.S., Mak T.W. Regulation of cancer cell metabolism. Nat. Rev. Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 12.Qiu M., Chen L., Tan G., Ke L., Zhang S., Chen H., Liu J. A reactive oxygen species activation mechanism contributes to JS-K-induced apoptosis in human bladder cancer cells. Sci. Rep. 2015;5:15104. doi: 10.1038/srep15104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar B., Koul S., Khandrika L., Meacham R.B., Koul H.K. Oxidative stress is inherent in prostate cancer cells and is required for aggressive phenotype. Cancer Res. 2008;68(6):1777–1785. doi: 10.1158/0008-5472.CAN-07-5259. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh J.C., Siegelin M.D., Vaira V., Faversani A., Tavecchio M., Chae Y.C., Lisanti S., Rampini P., Giroda M., Caino M.C., Seo J.H., Kossenkov A.V., Michalek R.D., Schultz D.C., Bosari S., Languino L.R., Altieri D.C. Adaptive mitochondrial reprogramming and resistance to PI3K therapy. J. Natl. Cancer Inst. 2015;107(3) doi: 10.1093/jnci/dju502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nevedomskaya E., Perryman R., Solanki S., Syed N., Mayboroda O.A., Keun H.C. A systems oncology approach identifies NT5E as a key metabolic regulator in tumor cells and modulator of platinum sensitivity. J. Proteome Res. 2016;15(1):280–290. doi: 10.1021/acs.jproteome.5b00793. [DOI] [PubMed] [Google Scholar]
- 16.Lu M., Zhou F., Hao K., Liu J., Chen Q., Ni P., Zhou H., Wang G., Zhang J. Alternation of adriamycin penetration kinetics in MCF-7 cells from 2D to 3D culture based on P-gp expression through the Chk2/p53/NF-kappaB pathway. Biochem. Pharmacol. 2015;93(2):210–220. doi: 10.1016/j.bcp.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 17.Cao B., Li M., Zha W., Zhao Q., Gu R., Liu L., Shi J., Zhou J., Zhou F., Wu X., Wu Z., Wang G., Aa J. Metabolomic approach to evaluating adriamycin pharmacodynamics and resistance in breast cancer cells. Metabolomics. 2013;9(5):960–973. doi: 10.1007/s11306-013-0517-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 19.DeBerardinis R.J., Lum J.J., Hatzivassiliou G., Thompson C.B. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Deberardinis R.J., Sayed N., Ditsworth D., Thompson C.B. Brick by brick: metabolism and tumor cell growth. Curr. Opin. Genet. Dev. 2008;18(1):54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phan L.M., Yeung S.C., Lee M.H. Cancer metabolic reprogramming: importance, main features, and potentials for precise targeted anti-cancer therapies. Cancer Biol. Med. 2014;11(1):1–19. doi: 10.7497/j.issn.2095-3941.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeung S.J., Pan J., Lee M.H. Roles ofp53, MYC and HIF-1 in regulating glycolysis-the seventh hallmark of cancer. Cell. Mol. life Sci.: CMLS. 2008;65(24):3981–3999. doi: 10.1007/s00018-008-8224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corazao-Rozas P., Guerreschi P., Jendoubi M., Andre F., Jonneaux A., Scalbert C., Garcon G., Malet-Martino M., Balayssac S., Rocchi S., Savina A., Formstecher P., Mortier L., Kluza J., Marchetti P. Mitochondrial oxidative stress is the Achille's heel of melanoma cells resistant to Braf-mutant inhibitor. Oncotarget. 2013;4(11):1986–1998. doi: 10.18632/oncotarget.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gopal Y.N., Rizos H., Chen G., Deng W., Frederick D.T., Cooper Z.A., Scolyer R.A., Pupo G., Komurov K., Sehgal V., Zhang J., Patel L., Pereira C.G., Broom B.M., Mills G.B., Ram P., Smith P.D., Wargo J.A., Long G.V., Davies M.A. Inhibition of mTORC1/2 overcomes resistance to MAPK pathway inhibitors mediated by PGC1alpha and oxidative phosphorylation in melanoma. Cancer Res. 2014;74(23):7037–7047. doi: 10.1158/0008-5472.CAN-14-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roesch A., Vultur A., Bogeski I., Wang H., Zimmermann K.M., Speicher D., Korbel C., Laschke M.W., Gimotty P.A., Philipp S.E., Krause E., Patzold S., Villanueva J., Krepler C., Fukunaga-Kalabis M., Hoth M., Bastian B.C., Vogt T., Herlyn M. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell. 2013;23(6):811–825. doi: 10.1016/j.ccr.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nogae I., Johnston M. Isolation and characterization of the ZWF1 gene of Saccharomyces cerevisiae, encoding glucose-6-phosphate dehydrogenase. Gene. 1990;96(2):161–169. doi: 10.1016/0378-1119(90)90248-p. [DOI] [PubMed] [Google Scholar]
- 27.Riganti C., Gazzano E., Polimeni M., Aldieri E., Ghigo D. The pentose phosphate pathway: an antioxidant defense and a crossroad in tumor cell fate. Free Radic. Biol. Med. 2012;53(3):421–436. doi: 10.1016/j.freeradbiomed.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 28.Polimeni M., Voena C., Kopecka J., Riganti C., Pescarmona G., Bosia A., Ghigo D. Modulation of doxorubicin resistance by the glucose-6-phosphate dehydrogenase activity. Biochem. J. 2011;439(1):141–149. doi: 10.1042/BJ20102016. [DOI] [PubMed] [Google Scholar]
- 29.Tai D.J., Jin W.S., Wu C.S., Si H.W., Cao X.D., Guo A.J., Chang J.C. Changes in intracellular redox status influence multidrug resistance in gastric adenocarcinoma cells. Exp. Ther. Med. 2012;4(2):291–296. doi: 10.3892/etm.2012.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawanishi S., Hiraku Y., Pinlaor S., Ma N. Oxidative and nitrative DNA damage in animals and patients with inflammatory diseases in relation to inflammation-related carcinogenesis. Biol. Chem. 2006;387(4):365–372. doi: 10.1515/BC.2006.049. [DOI] [PubMed] [Google Scholar]
- 31.Pervaiz S., Clement M.V. Tumor intracellular redox status and drug resistance--serendipity or a causal relationship? Curr. Pharm. Des. 2004;10(16):1969–1977. doi: 10.2174/1381612043384411. [DOI] [PubMed] [Google Scholar]
- 32.Sullivan R., Graham C.H. Chemosensitization of cancer by nitric oxide. Curr. Pharm. Des. 2008;14(11):1113–1123. doi: 10.2174/138161208784246225. [DOI] [PubMed] [Google Scholar]
- 33.Wise D.R., Thompson C.B. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem. Sci. 2010;35(8):427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J., Kim J., Bae J.S. ROS homeostasis and metabolism: a critical liaison for cancer therapy. Exp. Mol. Med. 2016;48(11):e269. doi: 10.1038/emm.2016.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bao Y., Guo Y., Zhuang X., Li D., Cheng B., Tan S., Zhang Z. D-alpha-tocopherol polyethylene glycol succinate-based redox-sensitive paclitaxel prodrug for overcoming multidrug resistance in cancer cells. Mol. Pharm. 2014;11(9):3196–3209. doi: 10.1021/mp500384d. [DOI] [PubMed] [Google Scholar]
- 36.Kuo M.T. Redox regulation of multidrug resistance in cancer chemotherapy: molecular mechanisms and therapeutic opportunities. Antioxid. Redox Signal. 2009;11(1):99–133. doi: 10.1089/ars.2008.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sosa V., Moline T., Somoza R., Paciucci R., Kondoh H., ME L.L. Oxidative stress and cancer: an overview. Ageing Res. Rev. 2013;12(1):376–390. doi: 10.1016/j.arr.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 38.Jiang P., Du W., Wang X., Mancuso A., Gao X., Wu M., Yang X. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat. Cell Biol. 2011;13(3):310–316. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siegl C., Prusty B.K., Karunakaran K., Wischhusen J., Rudel T. Tumor suppressor p53 alters host cell metabolism to limit Chlamydia trachomatis infection. Cell Rep. 2014;9(3):918–929. doi: 10.1016/j.celrep.2014.10.004. [DOI] [PubMed] [Google Scholar]