Abstract
Presurgical evaluation that can precisely delineate the epileptogenic zone (EZ) is one important step for successful surgical resection treatment of refractory epilepsy patients. The noninvasive EEG-fMRI recording technique combined with general linear model (GLM) analysis is considered an important tool for estimating the EZ. However, the manual marking of interictal epileptic discharges (IEDs) needed in this analysis is challenging and time-consuming because the quality of the EEG recorded inside the scanner is greatly deteriorated compared to the usual EEG obtained outside the scanner. This is one of main impediments to the widespread use of EEG-fMRI in epilepsy. We propose a deep learning based semi-automatic IED detector that can find the candidate IEDs in the EEG recorded inside the scanner which resemble sample IEDs marked in the EEG recorded outside the scanner. The manual marking burden is greatly reduced as the expert need only edit candidate IEDs. The model is trained on data from 30 patients. Validation of IEDs detection accuracy on another 37 consecutive patients shows our method can improve the median sensitivity from 50.0% for the previously proposed template-based method to 84.2%, with false positive rate as 5 events/min. Reproducibility validation on 15 patients is applied to evaluate if our method can produce similar hemodynamic response maps compared with the manual marking ground truth results. We explore the concordance between the maximum hemodynamic response and the intracerebral EEG defined EZ and find that both methods produce similar percentage of concordance (76.9%, 10 out of 13 patients, electrode was absent in the maximum hemodynamic response in two patients). This tool will make EEG-fMRI analysis more practical for clinical usage.
Keywords: EEG-fMRI, Deep learning, IED detection, GLM, Epilepsy
Highlights
-
•
A deep learning based epileptic discharge detector for EEG-fMRI is proposed.
-
•
The burden of manually marking epileptic discharges is greatly reduced.
-
•
Our method can produce similar EEG-fMRI results compared with traditional method.
1. Introduction
Epilepsy has been ranked as one of the most common neurological disorders in the world (de Boer et al., 2008). It is characterized by repeated occurrence of epileptic seizures, defined as brief episodes of signs or symptoms due to abnormal excessive or synchronous neuronal activity in the brain (Fisher et al., 2005). Around 20 to 40% of epilepsy patients are likely to have an epilepsy refractory to antiepileptic drug therapy and those with focal epilepsy are considered surgical candidates.
Presurgical evaluation that can precisely delineate the epileptogenic zone (EZ) is one important step for successful surgical treatment. Until now, the intracranial electroencephalography recordings (icEEG) are considered the gold standard for localizing the seizure onset zone (SOZ) which in turn provides the definition of EZ (Rosenow and Luders, 2001). However, this invasive monitoring technique is not without risk (Hamer et al., 2002) and can only explore a small fraction of the brain. Furthermore, it is time consuming as the frequency of seizure occurrence is relatively low compared with (interictal epileptic discharges) IEDs. Recently, researchers have paid great attention to the noninvasive EEG-correlated functional magnetic resonance imaging (EEG-fMRI) technique as an additional tool for this work (An et al., 2013, Khoo et al., 2017, Pittau et al., 2012, Thornton et al., 2010, van Houdt et al., 2013, Zijlmans et al., 2007). Generally, IEDs are first marked by trained experts based on visual inspection of EEG simultaneously recorded with the fMRI. The general linear model (GLM) is then applied by convolving the timing of IEDs and the hemodynamic response function to estimate the hemodynamically active regions. Finally, the regions that pass a certain statistical threshold are considered as markers of the EZ.
Visual inspection and marking of IEDs can be quite challenging and time-consuming, especially in the environment of simultaneous EEG-fMRI acquisition (Gotman, 2008). Due to the strong magnetic field, the quality of the EEG is reduced in the scanner compared with EEG outside the scanner. Despite the many gradient and pulse artifact removal methods (Abreu et al., 2016, Allen et al., 2000, Allen et al., 1998, Bonmassar et al., 2002, Kim et al., 2004, Klovatch-Podlipsky et al., 2016, LeVan et al., 2013, LeVan et al., 2016, Maziero et al., 2016, Srivastava et al., 2005) which have been proposed, there is still residual artifacts affecting EEG quality. Furthermore, the marked IEDs should be accurate and consistent for acceptable statistical analysis results (Flanagan et al., 2009). Studies showed that visual inspection and marking procedure is subjective (Zijlmans et al., 2007) and requires a high level of vigilance (Nonclercq et al., 2012). Therefore, a system that can reduce the labor of manual marking and improve reproducibility is desirable.
Several studies have proposed to automatize this marking procedure and one of the well-known technique is a topography-based method (Grouiller et al., 2011). A patient-specific topographic amplitude distribution map was built by averaging IEDs recorded outside scanner and used to compute its correlation with each time frame of the EEG inside the scanner. The resulting correlation time course was used as a regressor for fMRI analysis to map the haemodynamic changes. However, IEDs are not only characterized by their topography but also by their morphology. Another study proposed a template-based method (Tousseyn et al., 2014) that computed the spatiotemporal cross-correlations between patient-specific spike-template built by IEDs recorded outside the scanner and the EEG inside the scanner. GLM analysis was performed using the time course of correlations binarized with a spike-template-specific threshold. Improved sensitivity and specificity for detecting the EZ were reported compared with the topography-based method. However, these correlation-based methods may not work well as the morphology of IEDs inside the scanner may differ from those outside the scanner because of the impact of the scanner. Furthermore, in both of these reports, only the resulting hemodynamic response maps were analyzed but there was no direct evaluation of the performance of the automatically detected IEDs compared with those detected by an expert. To build a practical IED marking tool, it is important that it could reproduce reasonable IED markings compared with those of an expert.
Inspired by the recent great progress in deep learning (LeCun et al., 2015) and its successful implementation on face recognition (Parkhi et al., 2015, Schroff et al., 2015), we propose a deep learning based IED detection method to tackle this problem. First, we train a neural network model by mapping EEG data into a space in which the same types of IEDs lie close to each other but far from other IED types or from baseline. A multi-task learning strategy is used to simultaneously classify different types of IEDs and learn a Euclidean distance embedding space. Then, we get candidate IEDs by applying our model to EEG of new studies and let the expert edit them. At last, we apply GLM analysis using fMRI images and the marked IEDs. During the validation, we first tested the accuracy of IED detection by comparing with the template based method (Tousseyn et al., 2014) and then performed an EEG-fMRI analysis using our method to see if we can reproduce the hemodynamic response maps obtained the traditional way.
2. Materials and methods
2.1. Subjects
Patients with epilepsy admitted to our epilepsy monitoring unit who have adequate number of IEDs (usually at least one IED every 2 min) on the EEG were selected for EEG/fMRI study. Between April 2004 and December 2016, 445 patients participated in EEG/fMRI studies. For training data, from the abovementioned database, we selected randomly 30 patients with focal interictal EEG findings who had > 3 IEDs on the EEG during EEG-fMRI study (An et al., 2013, Khoo et al., 2017), significant BOLD responses, and did not undergo stereo-EEG (SEEG) implantation (see rationale below). For test data, we identified 37 consecutive patients (13 male; mean age at evaluation, 28.5 ± 8.2 years, range, 16–50) with active EEG and significant BOLD responses from the database of patients who participated in EEG/fMRI studies between April 2006 (start of 3 T scanning) and December 2016, who underwent SEEG implantation for pre-surgical evaluation and with post-implantation CT or MRI (Khoo et al., 2017). The implantation, a clinical procedure, was performed independently of the experimental EEG/fMRI study. Each patient gave written informed consent for the EEG/fMRI study approved by the Research Ethics Committee of Montreal Neurological Institute and Hospital.
2.2. Acquisition and marking of EEG outside the scanner
We first used scalp EEGs recorded in the epilepsy monitoring unit. Scalp EEGs were recorded according to 10–20 system (referential FCz) with additional infero-temporal electrodes from the 10–10 system (F9, T9, P9, F10, T10 and P10), acquired using a Harmonie system (200 or 1000 Hz sampling, Stellate Harmonie, Montreal, Canada). EEGs were reviewed using referential and bipolar montages, high-pass filtered at 0.5 Hz and low-pass filtered at 50 Hz. For each subject, the trained expert first reviewed the EEG outside scanner and identified the habitual IEDs, as pointed out in the clinical reports. The IEDs were marked on a 24-h EEG recording. IEDs with the same spatial distribution but different morphology were grouped and considered as a single type. IEDs with different spatial distributions were considered as different event types. To help make the marked events of each type of IED cover the possible shape and distribution variabilities, we marked at least two events for each type. The number of marked events varied (20 ± 16) according to the availability and convenience of marking.
2.3. EEG/fMRI acquisition
The acquisition procedure of EEG/fMRI is identical to previous studies from the same lab (An et al., 2013, Moeller et al., 2009, Pittau et al., 2012). EEG was recorded inside a 3 T MRI scanner (Trio; Siemens, Erlangen, Germany) with 25 MR compatible scalp electrodes placed according to 10–20 (reference FCz) and 10–10 (F9, T9, P9, F10, T10, and P10) systems, using a Brain-Amp system (5 kHz sampling; Brain Products, Munich, Germany). Two electrodes were placed on the shoulder to record the electrocardiogram. The head of the patient was immobilized with a pillow filled with foam microspheres (Siemens, Germany) to minimize movement artifacts and for patient's comfort. Data were transmitted from a BrainAmp amplifier (Brain Products, Munich, Germany, 5 kHz sampling rate) to the EEG monitor located outside the scanner room via an optic fiber cable.
A T1-weighted anatomic image was acquired first using the following sequences: Until July 2008: 1-mm slice thickness; 256 × 256 matrix; echo time (TE), 7.4 msec; repetition time (TR), 23 msec; flip angle 30° and from July 2008: 1-mm slice thickness; 256 × 256 matrix; TE 4.18 msec; TR, 23 msec; flip angle 9°. T1 image was used for superimposition with functional images. Functional data were collected in 6-min runs lasting 60–90 min, with a T2*-weighted echo planar imaging (EPI) sequence: Until July 2008: TR, 1.75 s; TE, 30 msec; 64 × 64 matrix; 25 slices; voxel, 5 × 5 × 5 mm; flip angle 90 degrees and from July 2008: TR, 1.9 s; TE, 25 msec; 64 × 64 matrix; 33 slices; voxel, 3.7 × 3.7 × 3.7 mm; flip angle 90°.
2.4. Processing and marking of EEG inside the scanner
The EEG inside scanner was processed offline (bandpass 0.5–50 Hz), and scanner gradient artifact was removed by an averaged subtraction method (Allen et al., 2000) implemented in BrainVision Analyzer software (Brain Products, Gilching, Germany). Two ballistocardiographic (BCG) artifact removal techniques were used in our dataset. For the majority, we used the average artifact subtraction method (Allen et al., 1998) to remove the BCG artifact. For subjects in whom there was still undesirable BCG artifacts, we further applied an ICA based BCG artifact removal method (Benar et al., 2003, Srivastava et al., 2005). Both the EEG inside and outside scanner were down sampled to 200 Hz.
For each subject in the training and testing data sets, the expert marked the IEDs inside the scanner according to the marking on the EEG outside scanner. The EEG outside the scanner shows clearly the distributions and shapes of the different IED types present in a particular patient, and this is very helpful in marking correctly the degraded EEG recorded inside the scanner. This is a standard procedure in EEG-fMRI studies in epilepsy. The number of IED types was the same for EEG inside and outside the scanner: IEDs with the same distribution but different morphology were grouped; IEDs with different distributions were considered as different event types. For each IED type, the number of events ranged from 2 to 80. The average duration of EEG recording inside scanner was 50 min (range, 18–72 min).
2.5. EEG/fMRI analysis
To evaluate the reproducibility of BOLD maps, we applied the GLM method to analyze the IEDs from the manual and from the edited marking. The GLM method is similar to that used in prior studies (An et al., 2013, Fahoum et al., 2012, Moeller et al., 2009, Pittau et al., 2012). Motion correction and smoothing (6-mm full width at half maximum) are applied to the fMRI images. An autoregressive model of order 1 is used to account for the temporal autocorrelations (Worsley et al., 2002). Low frequency drifts were modeled with a third-order polynomial fitting to reach run. Timing and duration of each IED were built as a regressor and convolved with four hemodynamic response functions (HRFs) peaking at 3, 5, 7, and 9 s. Motion parameters were modeled as confounds. All regressors were included in the same general linear model. A statistic t map was created for each regressor using the other regressors as confounds for each event type. A combined t map was created by taking, at each voxel, the maximum t value from the four t maps based on the four HRFs. The single combined t map was used for comparison. To be significant, a response required five contiguous voxels having a t value > 3.1 corresponding to p < 0.01 for the individual analysis, corrected for multiple comparisons (family wise error rate), or equivalent to p < 0.05 for the combined analysis using the four HRFs (Bagshaw et al., 2004).
2.6. Spike detection method
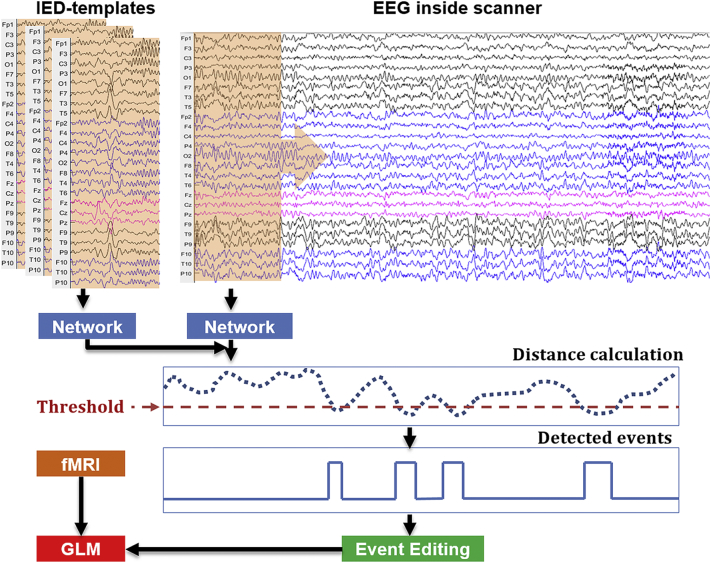
Our method contains a network that is trained on a training set. We will explain the training part in a subsequent section. Assuming that we have the trained network, here we show how to apply it on a new subject (Fig. 1). This method starts from a set of subject-specific spike-templates T ∈ {T(n) ∈ ℝC × (2R + 1)}n = 1N, where N is the number of templates, C denotes the number of channels (C = 25), and (2R + 1) the length of the template windows (range from –R to R with the spike onset in the center). We then estimate the distance between these templates and windows that slide across the EEG inside scanner. The sliding window has a size equal to the template, and a step of 32, which relates to our network architecture and reduces the computation burden (see the explanation in the supplementary section: “testing implementation”). To do this, the templates and sliding windows are first passed as inputs to a trained neural network which represents the signals in a learned spike-feature space. The distances between the sliding windows and templates in this spike-feature space are then estimated by calculating the Euclidean distance between their outputs from the network. The calculated distances represent the likelihood of having one IED type described by these templates at the time points located at the center of the corresponding sliding windows (the smaller the distance is, the more likely there is an IED). We then binarize the distances with a selected threshold H. The time points with distances lower than the threshold are treated as detected time points. Detected time points separated by < 1 s are connected together as one event. A human expert can edit the detected events to remove the false positive events. Finally, the retained events are convolved with the hemodynamic response function (HRF) and used as a regressor-of-interest in GLM for EEG-fMRI analysis.
Fig. 1.
Pipeline of our semi-automatic spike detector.
Distance calculation: The dash blue line indicates the distance between the network output of subject-specific spike-template and that of sliding windows from EEG inside the scanner. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.7. Training of the neural-network
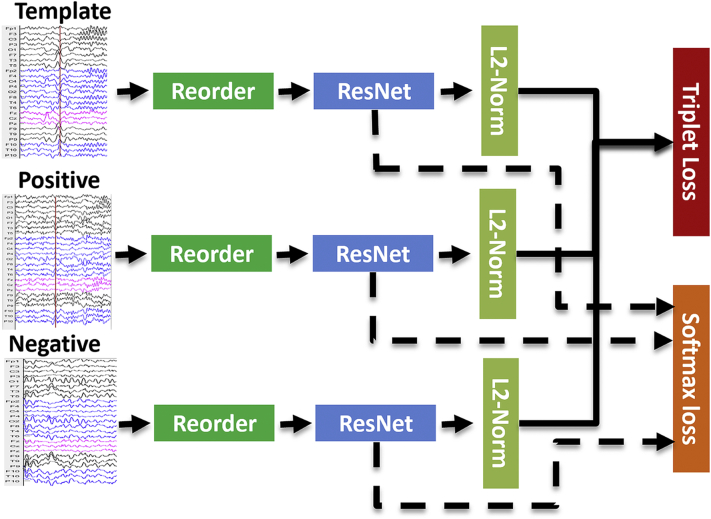
The purpose of this training step is to build a general neuro-network model that can use the IEDs outside the scanner as template to find similar IEDs inside the scanner. Fig. 2 shows our learning network architecture. During each training iteration, we first randomly select three sets of samples: (i) template sample with a window containing one type of IED extracted from EEG outside the scanner, (ii) positive sample with a window containing one IED of the same type extracted from EEG from inside the scanner; (iii) negative sample with a window extracted from the baseline of EEG inside the scanner. We use the term sample to refer to window extracted from EEG inside or outside scanner. The last two sets of samples are only necessary for subjects in the training set. After training, for IED detection in new subjects, our model does not need the expert to read the full EEG from inside scanner; only template samples from outside the scanner are needed. We then reorder the EEG channels according to channel similarities to make our algorithm invariant with respect to channel location. Samples are then fed into a deep residual network (ResNet (He et al., 2015)) and changed from 2D arrays to vectors by a fully connected layer. Finally, we train our model based on two objective functions, one being the softmax loss function for multi-class classification of different IED types, and another being the triplet loss function (Schroff et al., 2015) mapping IEDs into a space in which the Euclidean distance between the same types of IEDs is smaller than the distance between IEDs and baseline. The idea of choosing these two objective functions is mainly inspired by their successful application in face recognition. State-of-art studies in face recognition (Schroff et al., 2015) used triplet loss function. The softmax loss function is a widely used objective function for multi-class classification, it is helpful for the training of triplet loss function (Parkhi et al., 2015). In the following subsections, we explain in detail the reordering, ResNet architecture, and objective functions.
Fig. 2.
Training procedures of the neural-network.
Three sets of samples (Template, Positive and Negative) were fed as input to a deep residual network (ResNet) model, after a reordering procedure is applied to the channels to make our algorithm invariant with respect to channel location; L2 normalization (L2-Norm) is applied to the output of the network to constrain the projection to live on a hypersphere; finally, the model is trained based on two objective functions (softmax loss and triplet loss). See detailed description in the main text.
Notes: Template: template sample with a window containing one type of IED extracted from EEG outside the scanner, Positive: positive sample with a window containing one IED of the same type extracted from EEG inside the scanner, Negative: negative sample with a window extracted from the baseline inside the scanner.
2.8. Reordering
The spatial distributions of IEDs are different across different IED types and different patients: the spikes from EZs located in different lobes will appear in different EEG channels. But IEDs also have local channel similarities, spikes being visible in spatially adjacent channels. The purpose of the reordering is to make our algorithm invariant with respect to channel location and at the same time preserve local channel similarities.
We denote the EEG inside scanner of one subject as E ∈ ℝC × L, where (C = 25) is the number of channels and L is the total number of time points in the EEG. For each channel i, we set it as reference channel and get one reordered EEG as by ranking all other channels according their similarities with i using Pearson correlation. In this way, the reorder sequence is determined by both the selected reference channel and the similarity between channel signals. By selecting a different reference channel, we can get a different reorder sequence. Then, we can get the C reordered EEGs by selecting each channel as reference channel. As the reorder sequences are computed based on EEG inside scanner signals of the subject, we need to apply the same reorder operation on the subject's templates (recorded outside the scanner) to make their reordered results consistent with that of the EEG from inside the scanner. To do this, the reorder sequences for all the combinations are saved and can be applied to the spike-templates T of the same subject to get. The reorder sequences are calculated once for each subject and can apply to different types of the spike-templates.
2.9. Network architecture
Residual learning (He et al., 2015, He et al., 2016) is a recently proposed breakthrough framework for training deep neural networks. It has been shown that these networks are easier to optimize and can gain persistent improvement of accuracy in networks with > 20 layers.
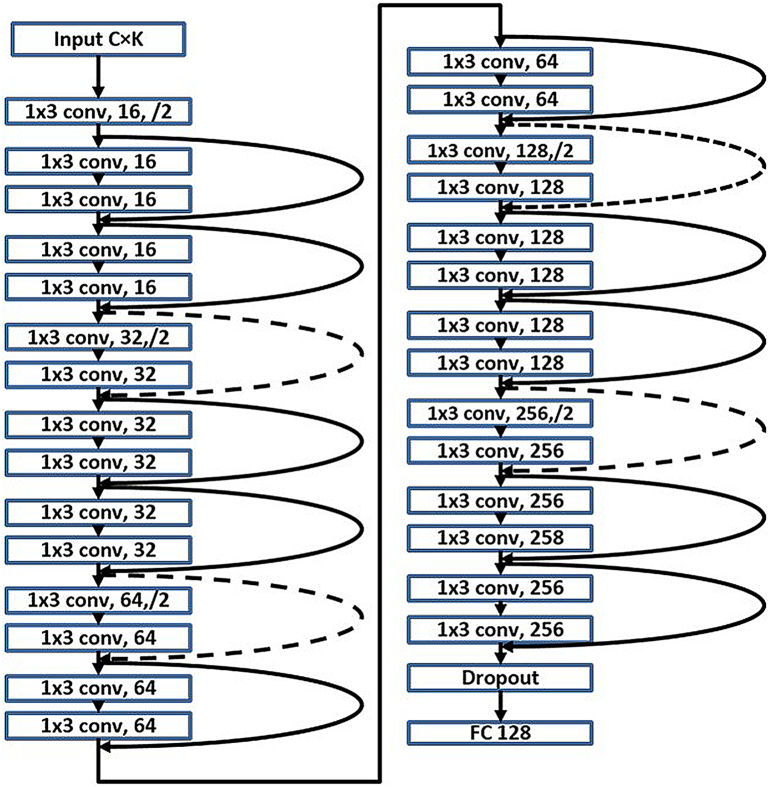
Inspired by this idea, we design our network based on a revised version of ResNet (Fig. 3). The inputs of the network can be templates from outside the scanner or sliding windows from inside the scanner. Their size is C × K with C as the number of channels and K as the number of time frames. We treat the input as C one dimensional data with length K. Parameters concerning the design of network architecture (such as the number of layers, feature map size and output size) are chosen based on the typical duration of spikes, computation time and complexity. Twenty-nine convolutional layers (in which the output of neurons are connected to local regions in the input) with filter size 1 × 3 are involved in our model. The rectified linear unit (ReLU) (Nair and Hinton, 2010) is used as the non-linear function of each layer. Following the philosophy of visual geometry group (VGG) nets (Simonyan and Zisserman, 2014), two design rules are employed for the convolutional layers (He et al., 2015): (i) the layers have the same number of filters if their output feature map sizes are the same, and (ii) the number of filters is doubled if the feature map size is halved. Downsampling is performed by the convolutional layers with a stride of 2. Identity shortcut connections are used when the input and output have the same dimensions (shown as solid shortcuts lines in Fig. 3), and Projection shortcuts (He et al., 2015) are used when the dimension increases (shown as dashed shortcut lines in Fig. 3). By using these shortcut connections, the residual networks allow the information from the input or earlier layers to flow more easily to deep layers. The feature map size goes from 16 to 256 as the layers go deeper. We end the network with a fully connected layer (FC) of size 128. Before that, dropout with coefficient value 0.5 (Baldi and Sadowski, 2013) showed 0.5 dropout can result in the maximum amount of regularization is used to improve the generalization of our model (Srivastava et al., 2014). The field of view (FOV) of our model is 417, which means our model can take into consideration ~ 2 s (as the EEG sampling rate is 200 Hz) along the time dimension. The total number of training parameters in this model is 999,920.
Fig. 3.
Network architecture.
This network contains one input layer, twenty-nine convolutional (conv) layers, one dropout layer and one full connection (FC) output layer. The conv layer computes the output of neurons that are connected to local regions in the input by using K (with value ranging from 16 to 256 as denoted by the second parameter of corresponding layer) filters of size 1 × 3, the number of filters K is doubled if the feature map size is halved (demonstrated with/2 for the third parameter of corresponding layer). Following the idea of residual learning, Identity shortcut connections (when the input and output have same dimensions, solid shortcuts line) and projection shortcut (dashed shortcuts lines) are used here to allow the information from the input or earlier layers to flow more easily to deep layers. Please see a more detailed description in the text.
2.10. Objective function
By applying the mentioned network, each template from outside the scanner or each sliding window from inside the scanner can be projected onto a vector. Our objective function contains two parts: one softmax loss function for classifying the EEG baseline and different IED types as different classes, and another triplet loss function for embedding the IEDs into a space where same type of IEDs can be easily identified. The softmax is one kind of loss function for multi-class classification. The input samples (as shown in Fig. 2) are assigned to different classes. The negative samples extracted from baseline are treated as one class. Template or positive samples containing the IED with the same reorder sequence, of the same type and from the same patient are treated as one class. This loss function can help the network learn to extract patterns that can distinguish not only IEDs from baseline but also different type of IEDs. The aim of the triplet loss function is to improve the performance of spike detection by learning the desired distance scores. This is inspired from metric learning, which generally tries to learn a reduced dimension projection that is discriminative and compact at the same time. This function helps bring the input template sample closer to the same type of IED inside the scanner than to any other window inside the scanner, such as baseline or a different type of IED. A more detailed mathematical description can be found in the supplementary section: “objective function in detail”. In the supplementary section: “training procedure”, we also explain our training procedure as well as parameter setting and training details.
2.11. Evaluation
Two evaluations are used to test the performance of our method. The first compares the accuracy of IEDs detected by different methods, and the second test if we can reproduce the EEG-fMRI analysis results by using our detector as an assistant.
2.11.1. IED detection accuracy
After training our model based on 30 patients with 70 studies, we use another 37 patients with 78 studies to test the model's accuracy. For each study, using the IEDs (at least 2) marked in EEG outside the scanner as templates, we calculate the distance between the network output of the templates and that of each window of EEG inside the scanner. By choosing one threshold, the set of distances with value lower than the threshold are marked as IED frames. This threshold is adaptively set (see description in section “Reproducibility of hemodynamic response maps” below). We then define IED frames separated by less than 1 s as belonging to the same event (we use the similar procedure for manual IED marking) and get a set of IED events. Finally, we compare the detected events with the manually marked ground truth events. If one event has overlap with one of the ground truth events, we define it as a true positive; otherwise, we define it as a false positive. Two values are computed to define performance. One is sensitivity, defined as number of true positive events divided by the number of ground truth events. The other is the false positive rate, defined as the number of false positive events per minute. This pair of values can be seen as one position in the modified Receiver Operating Characteristic (ROC) curve. By choosing different thresholds and performing the same analysis, we can draw the ROC curves and compare different methods. For our method, as the reorder sequences are determined by both the selected reference channel and the similarity between channel signals, there are 25 kinds of reorders (see more detailed description in reordering section). We can use the average results of multiple combinations of these to do the detection. We tested 5 configurations with 5, 10,15,20,25 combinations (denoted by L-c5, L-c10, L-c15, L-c20, and L-c25) here. We also test the template-based method (Tousseyn et al., 2014) which directly calculates cross-correlation between the template and spatial-temporal windows of EEG inside scanner. But one modification is made here from the original paper (Tousseyn et al., 2014): instead of using an adaptively and visually determined template size which can change in duration, we use a predefined fixed duration. In their paper, they only considered one type of IED (i.e. spike) as they defined the border of IED template that starts from the onset of the spike at baseline until the negative peak of the slow wave that follows. In our experiment, we considered all possible types of IED, such as spike and wave, burst of spike and wave, repetitive spikes, poly-spikes, which makes this procedure unusable as the durations of different events from the same IED type can be different. However, we set the template duration as one parameter r and tested 5 configurations with windows of 250 ms, 500 ms, 1000 ms, 1500 ms, 2000 ms (denoted by C-r250, C-r500, C-r1000, C-r1500, and C-r2000), representing 50, 100, 200, 300, 400 time points respectively. The ROC analysis helps us compare different methods at the subject level. We then compared the performance at the group level using the sensitivity values corresponding to the false positive rate set as 5 events/min. This value is chosen based on the need to achieve a sufficient sensitivity (with average around 80%) at a reasonable cost of false positive rate so it does not take too much time to edit the candidate events (see results in section IED detection accuracy).
2.11.2. Factors affecting detection accuracy
We are also interested in finding which factors affect the performance across studies. We select five factors: IED types with or without duration, distribution of brain regions related to the IED types, IED event rate, the length of movement artifacts marked by the expert, and the EEG quality inside the scanner (rated as good, moderate and bad by expert), and perform univariate statistical analysis with the sensitivity values (with false positive rate set as 5 events/min) using ANOVA (for categorical factors) or Pearson correlation (for factors with continuous value).
2.11.3. Reproducibility of hemodynamic response maps
To test if we can reproduce the EEG-fMRI analysis results by using the detector as an assistant, we randomly selected 15 subjects and edited the events for each study by displaying only the candidate frames marked as IEDs by our detector (i.e. the proposed human editing step in Fig. 1). During the generation of the candidate IEDs, we choose a threshold that leads to at least 5 event/min being detected. This value is based on our IEDs detection accuracy analysis that we can achieve median sensitivity of 84.2% with the false positive rate set as 5 events/min in our method (see Results below). It is true that the sensitivity may be lower as the true positives are also included in the 5 events per minute, but the number of true events is low (with median value as 0.9 event/min in our test dataset), so the average false detections/min should be still around 4–5. It should also be noted that the 5 events/min is the lower limit of the setting, during the implementation, we gradually reduced the threshold and stopped when at least 5 events/min were detected, slight change of threshold can sometimes introduce important variations in the number of new events. The time for editing each study was recorded. We then generated a hemodynamic response map for each study by performing GLM using the manually marked ground truth events and the detected/edited events to create the regressors-of-interest. Finally, we analyze the results by checking the editing time, similarities between two hemodynamic response maps and their concordance with SEEG results.
3. Results
3.1. IED detection accuracy
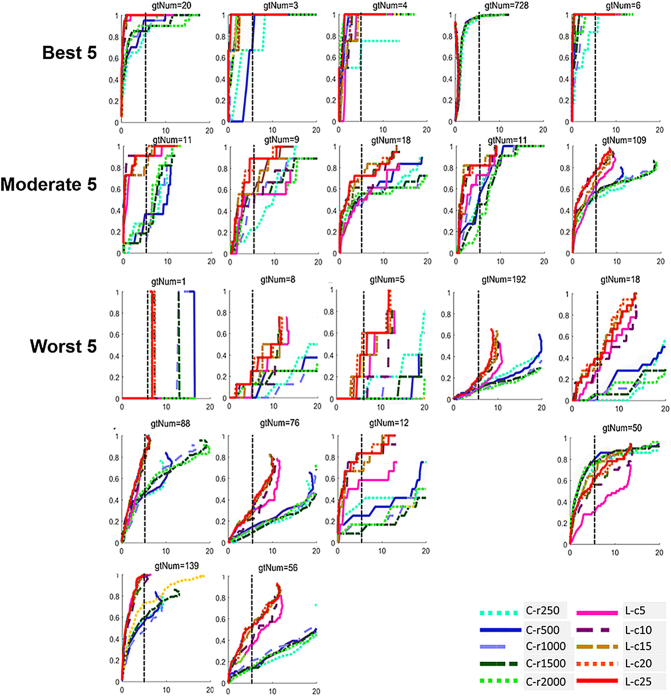
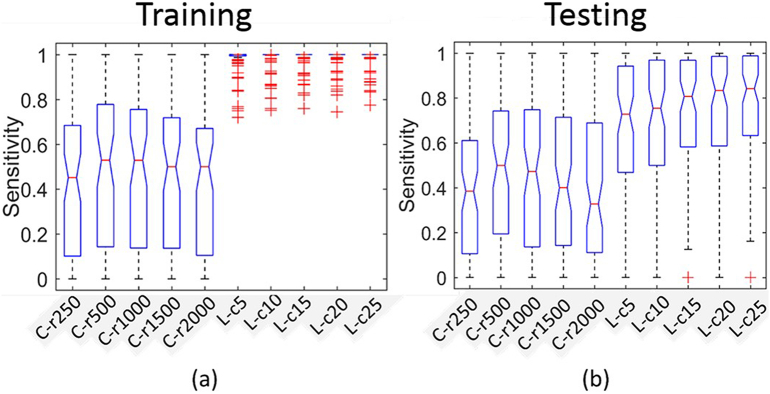
For evaluating IED detection accuracy, we compared the template-based method and our method in a test dataset of 37 patients with 78 studies. To give an intuitive impression, we show sample studies having the best, moderate and worst performance in the first three rows of Fig. 4 (We first sort all the studies by the value of average area under ROC across different methods and select the first, middle and last 5 studies for analysis). From the ROC curves, our method performed consistently better than the template based method. Our learning based method with 25 combinations (L-c25) performed best. For the best 5 studies, the sensitivities can achieve 100% and the false positive rates be < 3 events/min. For the moderate 5 studies, the sensitivities are around 80% for false positive rate at 5 events/min. Based on the above observations, we used 5 events/min as a threshold for determining the number of candidate events to detect for expert editing. This does not add too much burden for expert editing and at the same time can generally cover 80% of the ground truth events. For the worst 5 studies, the sensitivities were < 40% when the false positive rate was 5 events/min. In four of these five studies, the IEDs were not the dominant IED types of the patients. The dominant IED types for the same patients had better performance. ROC curves of other IED types from the same subject are shown in the same column in rows 4 and 5 of Fig. 4.
Fig. 4.
Examples of ROC curves comparing different methods.
For each ROC plot, the x axis corresponds to false positive rate (events/min), while the y axis corresponds to sensitivity. ROC curves in the first three rows show the best 5, moderate 5 and worst 5 studies. For the subjects corresponding to the worst 5, we also show the ROC curves of different IED types of the same subject in rows 4 and 5 at the same column if there is more than one IED type. For the ROC curves in our plots, the sensitivity does not necessarily increase monotonically with increasing false positive rate as different events may merge as we decrease the threshold for detecting events. Black vertical line shows the position at 5 events/min. C-rN represents the correlation template-based methods with template duration as N ms, using cold colors; L-cN represents our learning-based method with N as the number of reorder combinations, using warm colors.
To make a group level comparison, we found the sensitivity values with false positive rate set at 5 events/min for each study and show the box-plots for the training and testing sets in Fig. 5a and Fig. 5b. We can observe that (i) our learning-based method is consistently better than the template-based method, (ii) for the training set, the highest median sensitivity for the template based method is 52.6% for C-r500 and the highest median sensitivity for our learning-based method is 100.0% for L-c25. Post-hoc paired t-test showed that the best (L-c25) of our learning methods is significantly better than the best (C-r500) template based method (with p = 5.41e-20), (iii) for the testing set, the highest median sensitivity for the template based method is 50.0% for C-r500 and the highest median sensitivity for our learning based method is 84.2% for L-c25. Post-hoc paired t-test showed that the best (L-c25) of our learning methods is significantly better than the best (C-r500) template based method (with p = 1.39e-14).
Fig. 5.
Sensitivity comparisons across different methods.
a) Box-plot of sensitivity in the training set across different methods with false positive rate set as 5 events/min. Post-hoc paired t-test showed the best one (L-c25) of our learning methods is significantly better than the best one (C-r500) of template based methods (p = 5.41e-20). b) Box-plot of sensitivity in the testing set across different methods with false positive rate set as 5 events/min. Post-hoc paired t-test showed the best one (L-c25) of our learning methods is significantly better than the best one (C-r500) of template based methods (p = 1.39e-14). C-rN represents correlation template-based methods with template size along time dimension as N ms; L-cN represents our learning-based methods with N as the number of reorder combinations.
3.2. Factors affecting detection accuracy
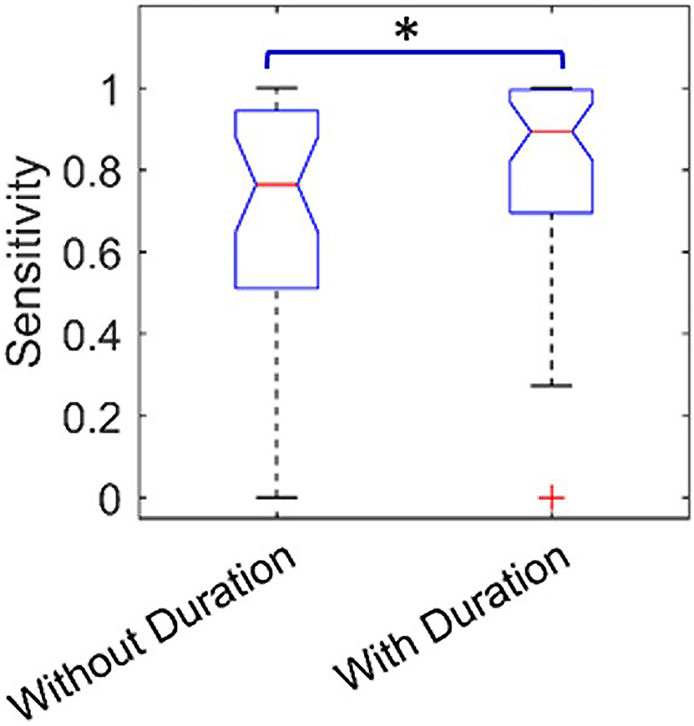
Table 1 shows the statistical analysis results of factors affecting detection accuracy. From the 5 tested factors, only one (IED types with or without duration) showed statistical significant impact on the detection accuracy: the sensitivity of IED types with duration is higher than that of IED types without duration (p = 0.0477) (Fig. 6).
Table 1.
Statistical analysis of factors affects detection accuracy of studies (sensitivity with false positive rate set as 5 envents/min).
For factors with categorical values (IED type with or without duration, distribution of brain regions related to the IED types and the EEG quality), one way ANOVA is used and its F statistical results and p value are reported. For factors with continuous values (IED event rate and the length of movement artifacts), Pearson correlation is used and its corresponding r and p values are reported. Factors showing results with statistical significant impact on detection accuracy are marked as bold.
| IEDs duration | Brain regions | Event rate | Movement artifacts | EEG quality | |
|---|---|---|---|---|---|
| ANOVA(F)/Correlation(r) | 4.05(F(1,76)) | 0.95(F(11,66)) | 0.21(r) | -5.8e-2(r) | 0.44(F(2,75)) |
| p-value | 4.77e-2 | 0.50 | 0.07 | 0.61 | 0.65 |
Fig. 6.
Boxplots show the factor (Events of IED types with or without duration) that have statistical significant differences (p = 4.77e-2) impact on the sensitivity of studies, with false positive rate set as 5 events/min.
3.3. Reproducibility of hemodynamic response maps
Table 2 shows the results of manual and edited marking on 15 patients with 21 studies. We calculated the length ratio of edited marking, which is the ratio between the frame number of candidate IEDs detected by our method and the total number of frames; the expert need to look at only 11% of the total original frames during edited marking, which is a great reduction of manual marking labor. It took 4.53 ± 3.13 min/run to mark using edited marking. We did not record the time for manual marking because they were performed before we initiated this project. But from our previous marking experience, it takes on average 3 h to mark a 10-run recording and this would be reduced to around 45 min by our method, a 75% reduction. The time taken to mark the outside-scanner EEG was not taken into account in our time calculation. But in our algorithm, we don't need to mark all the IEDs in the clinical EEG. The procedure is similar to clinical routine that one need only to quickly review the EEG segments and mark at least two events for each type. The marked clinical events will be treated as template for IED detection in our method. By comparing the number of IEDs between manual marking ground truth (GT) results and edited marking results, we found the edited marking had more IEDs in 13 of 21 studies.
Table 2.
Comparing results of ground-truth manual marking and edited marking on 15 patients.
In the column of studies, Pa_Tb denotes patient a with IED type b. Sensitivity values correspond to values as false positive rate set as 5 events/min. In the column of sign of activation, A means the max absolute t value of hemodynamic response is an activation, B means it is a deactivation. n/a means no significant activation is found for that IED type. – in the last column means absence of stereo-EEG electrode at the maximum hemodynamic response. Detailed results of IED-related BOLD response (primary cluster) and SEEG findings were listed in Table S1.
In the column of studies, Pa_Tb denotes patient a with IED type b. Sensitivity values correspond to values as false positive rate set as 5 events/min. In the column of sign of activation, A means the max absolute t value of hemodynamic response is an activation, B means it is a deactivation. n/a means no significant activation is found for that IED type. – in the last column means absence of stereo-EEG electrode at the maximum hemodynamic response. Detailed results of IED-related BOLD response (primary cluster) and SEEG findings were listed in Table S1.
| Studies | Number of runs | Editing time (min/run) | Edit length ratio | Number of IEDs (GT/Edited) | Sensitivity | Sign of activation | Same anatomical location? | Same lobe? | SEEG concordant (GT/Edited) |
|---|---|---|---|---|---|---|---|---|---|
| P1_T1 | 9 | 4.67 | 0.08 | 23/83 | 82.6 | A | N | N | N/Y |
| P2_T1 | 12 | 5.50 | 0.15 | 30/62 | 100 | A | N | Y | Y/Y |
| P3_T1 | 8 | 5.63 | 0.15 | 29/35 | 55.2 | A | Y | Y | Y/Y |
| P4_T1 | 6 | 4.83 | 0.09 | 334/179 | 88.6 | A | Y | Y | Y/Y |
| P5_T1 | 4 | 0.75 | 0.15 | 11/17 | 100 | D | Y | Y | N/N |
| P6_T1 | 11 | 1.45 | 0.02 | 35/48 | 100 | D | Y | Y | Y/Y |
| P7_T1 | 9 | 3.33 | 0.16 | 91/77 | 96.7 | A | Y | Y | Y/Y |
| P7_T2 | 9 | 2.22 | 0.17 | 3/11 | 100 | A | Y | Y | Y/Y |
| P7_T3 | 9 | 1.11 | 0.07 | 5/4 | 0 | A | N | Y | N/Y |
| P8_T1 | 11 | 7.45 | 0.14 | 192/126 | 16.2 | A | Y | Y | Y/Y |
| P9_T1 | 10 | 3.2 | 0.12 | 45/65 | 100 | A | Y | Y | N/N |
| P10_T1 | 10 | 2.8 | 0.07 | 2/16 | 100 | n/a | n/a | n/a | n/a |
| P10_T2 | 10 | 6.4 | 0.08 | 202/101 | 48.5 | D | Y | Y | Y/Y |
| P10_T3 | 10 | 13.1 | 0.11 | 326/476 | 96.3 | D | Y | Y | -/N |
| P11_T1 | 3 | 1.17 | 0.03 | 15/11 | 100 | A | Y | Y | Y/Y |
| P12_T1 | 8 | 8.38 | 0.15 | 10/41 | 40 | A | N | Y | -/N |
| P13_T1 | 6 | 6.00 | 0.21 | 18/39 | 88.9 | n/a | n/a | n/a | n/a |
| P13_T2 | 6 | 6.67 | 0.12 | 1/31 | 100 | A | N | N | Y/− |
| P14_T1 | 7 | 7.57 | 0.14 | 74/177 | 85.1 | A | Y | Y | −/− |
| P15_T1 | 10 | 1.60 | 0.08 | 36/26 | 94.4 | A | Y | Y | Y/Y |
| P15_T2 | 10 | 1.30 | 0.05 | 109/72 | 73.4 | A | Y | Y | Y/N |
After the GLM analysis, we compared the similarities between hemodynamic response maps of the GT results and edited marking results by looking at the positions of clusters with maximal absolute t-value (primary cluster) at the anatomical structure and lobe levels. Two in the 21 studies did not have significant activation clusters for both methods. In the remaining 19 studies, the primary cluster was found at the same anatomical locations in 14 studies (73.7%) and at the same lobe in 17 studies (89.5%).
As the subjects had undergone SEEG, we examined the primary cluster in relation to SEEG electrodes location by co-registering the hemodynamic response maps to the post-implantation anatomical imaging. We considered the results as concordant if the primary clusters contain SEEG electrode contacts involved at seizure onset, within 2 cm of the peak (Khoo et al., 2017). From the analysis of 19 studies, we got 12 concordant studies, 4 discordant studies and 3 studies without electrode presence for the GT results, and we got 12 concordant studies, 5 discordant studies and 2 studies without electrode presence for the edited marking results. If a subject has at least one event type that is concordant with the SEEG result, we call this subject concordant. Then, at the subject level, we also found similar percentage of concordance (76.9%, 10 out of 13 patients, 2 patients without electrode presence) for both GT results and edited marking results.
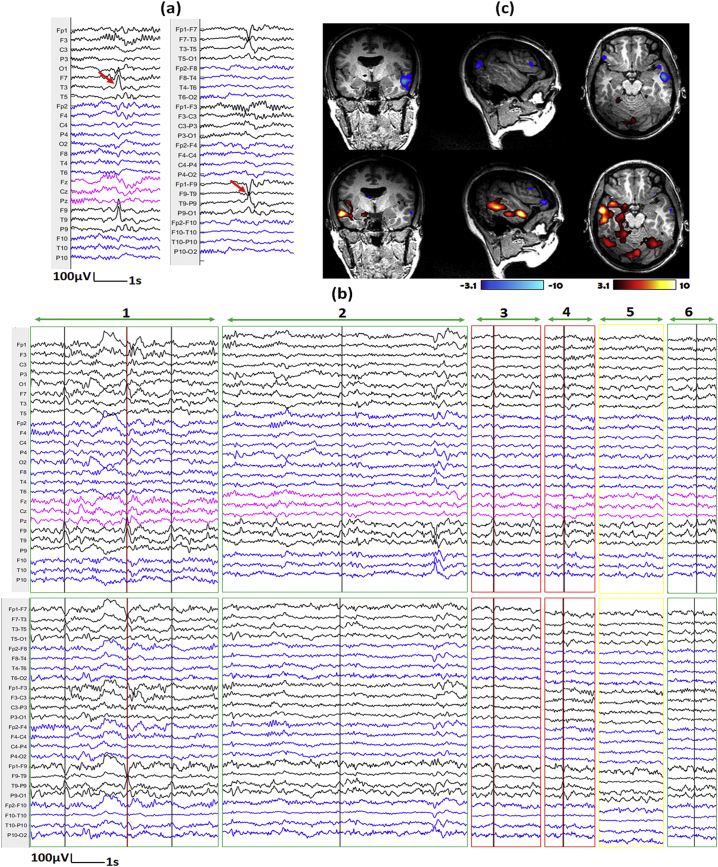
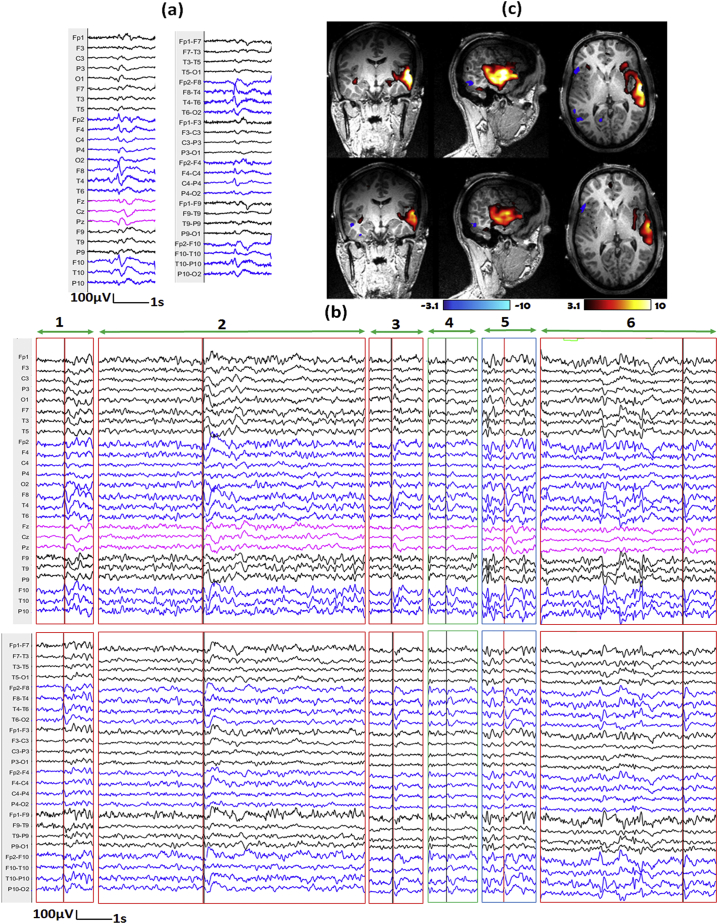
We illustrate three representative examples in Fig. 7, Fig. 8, Fig. 9. For each, we show the IED template marked outside the scanner (subfigure a), the top 6 candidate events provided by our method (subfigure b) and corresponding hemodynamic response maps from ground truth manual markers and edited makers (subfigure c). The first example comes from IED type 1 of patient 1 (Fig. 7), the number of events after editing is much larger than that of GT results (83 vs 23). The primary cluster of edited marking result is concordant with SEEG-defined SOZ, while we cannot find such concordance from hemodynamic response map of GT results. The second example comes from IED type 1 of patient 2 (Fig. 8), the number of events from edited marking results is also much larger than that of GT results (62 v.s. 30). The hemodynamic response map of GT results and edited marking results look similar and both of their primary clusters are concordant with SEEG-defined SOZ. The third example comes from IED type 2 of patient 15 (Fig. 9), the number of events from edited marking results is smaller than that of GT results (72 v.s. 109). Only the primary cluster of GT result is concordant with SEEG-defined SOZ, but we can see that the activation regions of the two results look similar, and the primary cluster of the edited marking result is very close to the primary cluster of the GT result. For detailed comparisons for all the hemodynamic response maps, please see the supplementary section: “comparisons of hemodynamic response maps”.
Fig. 7.
Demonstration of marked events and the hemodynamic response maps for IED type 1 of patient 1.
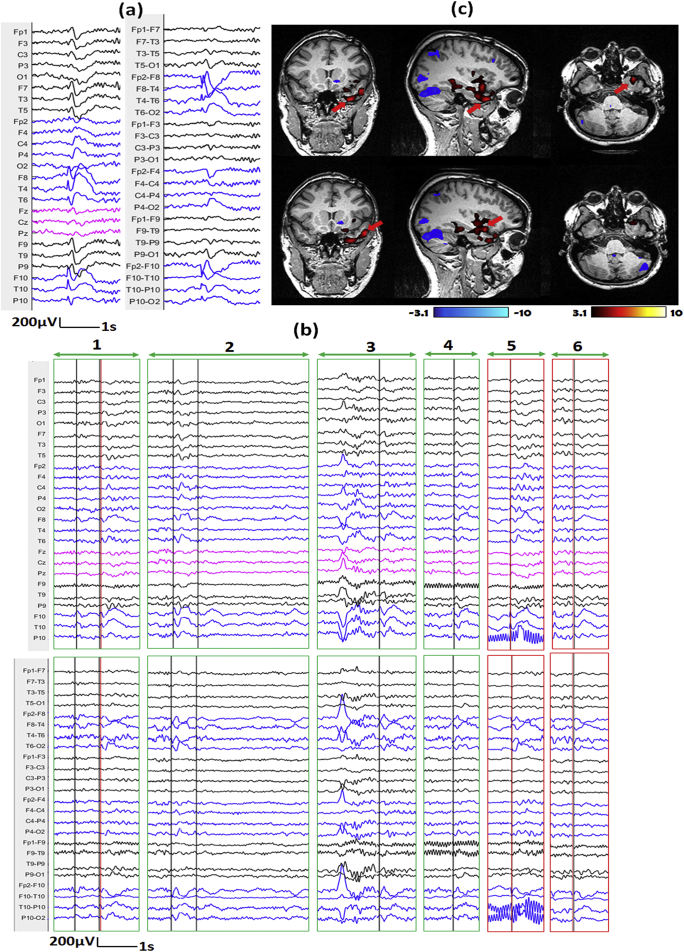
a) One example of IED template marked outside of scanner shown on average montage (left) and bipolar montage (right). The IED contains spikes with maximum at T3 and F9-T9. b) Top 6 candidate events proposed by our method for EEG inside scanner (Top: average montage, bottom: bipolar montage; Note that the candidate events provide by our method contain periods of varying length). Red lines show the positions of IEDs from the ground truth (GT) results, while black lines show the positions of IEDs from the edited results. The events were classified into different situations by marking the outlines with different colors, Green: IEDs from edited results missed in GT-results, Red: IEDs from both GT and edited results, yellow: No IED is found from either results. c) Corresponding hemodynamic response maps from ground truth manual markers (top) and edited makers (bottom). The primary cluster of edited result is concordant with SEEG-defined SOZ, while we cannot find such concordance from hemodynamic response map of GT results. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 8.
Demonstration of marked events and the hemodynamic response maps for IED type 1 of patient 2.
a) One example of IED template marked outside of scanner shown on average montage (left) and bipolar montage (right). The IED contains spikes with or without slow waves with maximum at F8, T4, T10 and P10. b) Top 6 candidate events proposed by our method for EEG inside scanner (Top: average montage, bottom: bipolar montage; Note that the candidate events provide by our method contain periods of varying length). Red lines show the positions of IEDs from the GT results, while black lines show the positions of IEDs from the edited results. The events were classified into different situations by marking the outlines with different colors, Green: IEDs from edited results that were missed in GT-results, Red: IEDs from both GT and edited results, Blue: IEDs from GT-results that were missed in edited results. c) Corresponding hemodynamic response maps from GT manual markers (top) and edited makers (bottom). The hemodynamic response map of GT results and edited marking results look similar and both of their primary clusters are concordant with SEEG-defined SOZ. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 9.
Demonstration of marked events and the hemodynamic response maps for IED type 2 of patient 15.
a) One example of IED template marked outside of scanner shown on average montage (left) and bipolar montage (right). The IED contains spikes with or without slow waves with maximum at F10, T10, F8 and T4. b) Top 6 candidate events proposed by our method for EEG inside scanner (Top: average montage, bottom: bipolar montage; Note that the candidate events provide by our method contain periods of varying length). Red lines show the positions of IEDs from the GT results, while black lines show the positions of IEDs from the edited results. The events were classified into different situations by marking the outlines with different colors, Green: IEDs from edited results that were missed in GT-results, Red: IEDs from both GT and edited results. c) Corresponding hemodynamic response maps from GT manual markers (top) and edited makers (bottom). Red arrows denote the primary cluster hemodynamic response map. The primary cluster of GT result is concordant with SEEG-defined SOZ; the activation regions of the two results look similar and the primary cluster of the edited marking result is very close to the primary cluster of the GT result. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
EEG/fMRI analysis is a useful tool for predicting the EZ for patients with refractory focal epilepsy (An et al., 2013, Khoo et al., 2017) and has attracted attention in research and clinical practice. However, one disadvantage that prevents its practical usage is that it heavily relies on manual marking of the IEDs from the EEG inside scanner. This procedure is difficult and time consuming.
The topography-based method (Grouiller et al., 2011) and the template-based method (Tousseyn et al., 2014) are trying to reduce the manual marking burden by automating this procedure. Both methods rely on correlation to find similar events using templates from EEG outside the scanner. As shown in our experiment, the template-based method achieved median sensitivity of 50.0% with false positive rate of 5 events/min. This means that many positive events will be missed. The reason may be that the correlation measurement is sensitive to artifacts features introduce by the scanner.
We introduced a data-driven deep learning-based method to overcome the above problem by learning a model using a relatively large dataset (30 patients with 70 studies). We rely on this model to find the subtle similarity between the same type of IEDs from inside and outside the scanner. Validation on another large testing dataset (37 patients with 78 studies) showed that our method could achieve median sensitivity of 84.2% with false positive rate at 5 events/min, which is significantly higher than that (50.0%) of the template-based method.
We have also found one factor (the duration property of the IEDs) statistically significantly affecting the detection accuracy. We found that the sensitivity of IEDs with duration tends to be higher compared with that of IEDs without duration (p = 0.0477). This could be explained by the fact that IEDs with duration contain more fluctuating signals that make them more easily separable from baseline.
Before performing the GLM analysis, we decided to let the expert edit the EEG based on the detected events to remove false positive events. The duration of detected events in our method is only 11% ± 5% to the duration of the original recording. We think it is necessary to add this human editing procedure as it does not add too much time, especially if we compare to marking the full recording.
Evaluation on 15 patients for reproducibility of hemodynamic response maps showed that our method got results similar to GT results in 73.7% of studies at anatomical level and in 89.5% of studies at lobar level. Comparisons with SEEG results showed both the GT results and edited marking results achieved similar concordance rate in 76.9% of the subjects. Furthermore, detailed inspection on the discordant studies showed that the edited marking results produced hemodynamic response maps with more reasonable activations (Please see the description in supplementary section: “comparisons of hemodynamic response maps”, we have systemically compared the fMRI detection results between our method and the GT method to their agreement with the SOZ results of intracranial EEG (SEEG)). We concluded that our method can reproduce comparable and even more reasonable hemodynamic response maps compared to the GT results.
To generate candidate events for human editing, we decided to select a threshold that allow detection of at least 5 events/min. We chose this value based on our evaluation that we can generally achieve around 80% sensitivity when the false positive rate is 5 events/min and also it does not add too much manual editing burden. In the template-based method (Tousseyn et al., 2014), the authors proposed to adaptively select the threshold by using a set of healthy subjects. They claimed that the adaptively selected threshold is more favorable to control false-positive detection and to avoid the need for visual verification. However, the false positive detection is not only related to the threshold but also related to other factors, such as the separability between the IEDs and the baseline, or the method used. Even if we chose a very strict threshold, there may still be false positive events as they can be mixed with the true events. So we think that visual inspection is indispensable and we rely on the human expert to deal with false positive events.
Another interesting finding is that the event numbers are not necessarily the same between GT and edited marking. It is true that in our current implementation we will miss some true positive events as the editing is based on our candidate events. However, from the validation section it seems that this does not matter too much as we can reproduce similar hemodynamic response maps. We also find that sometimes the edited marking results in more IEDs (in 13 out 21 of the studies) than the GT one. One reason maybe that the expert missed some events in the original marking as it is a time-consuming and difficult work. This is an advantage of our method as the expert only needs to mark a set of candidate events, which is only a small portion of the original recording. It should also be noted that the manual marking of IEDs is a subjective procedure. This may have a negative impact on the training of our detector. How to tackle this problem and to further improve the performance of our method is an open question and deserve further exploration.
As noted in the “EEG/fMRI acquisition” section, there has an updating of fMRI configuration at July 2018. We checked our dataset and found that subjects scanned before July 2008 were not representative in our dataset (only 4 subjects from our 37 test subjects). No significant correlation with detection accuracy was found associated with this factor. It is difficult to analyze the effect as many different factors may affect the results, such as those listed in Table 1. The effects were also implicitly handled by our training procedure as our training set contains subjects scanned both before and after the change of MR configuration. The change in MR specs may have an effect on gradient and ballistocardiographic (BCG) artifacts for the EEG data; it might be interesting to analyze its influence on artifact removal methods.
To the best of our knowledge, there has no research that study the interrater agreement on marking inside-scanner EEG yet. This is probably for two reasons: first, it is difficult to find several trained and experienced raters as one need to practice a lot to get used to the marking; second, the traditional marking is time-consuming. The lack of interrater agreement comparison is one limitation of our study. In this study, we focused on reproducing similar results with one experienced rater. As our assistant method can provide recommended events and can greatly reduce marking labor, we would hope our method can also improve the interrater reproducibility. As this research direction deserve deliberate and systematic investigation, we would investigate this topic in our future work.
5. Conclusions
We propose a deep learning-based semi-automatic spike detector for EEG-fMRI. Comparisons with expert marked IEDs showed that our method is superior to the template based method and has a sufficient performance to be used as an assistant IED marking tool for reducing the manual marking burden and improving reproducibility. This will render the application of EEG-fMRI to epilepsy more practical.
The following are the supplementary data related to this article.
IED-related BOLD response (primary cluster) and SEEG findings (N = 15).
Acknowledgements
This research was supported by grant FDN 143208 from the Canadian Institutes of Health Research. Training computations were made on the supercomputer Guillimin from McGill University, managed by Calcul Québec and Compute Canada. The operation of this supercomputer is funded by the Canada Foundation for Innovation (CFI), NanoQuébec, RMGA and the Fonds de recherche du Québec - Nature et technologies (FRQ-NT). Dr. Khoo was supported by Preston Robb Fellowship from the Montreal Neurological Institute, research fellowship from the Uehara Memorial Foundation, Japan Epilepsy Research Foundation, Osaka Medical Research Foundation for Intractable Diseases.
References
- Abreu R., Nunes S., Leal A., Figueiredo P. Physiological noise correction using ECG-derived respiratory signals for enhanced mapping of spontaneous neuronal activity with simultaneous EEG-fMRI. NeuroImage. 2016;154:115–127. doi: 10.1016/j.neuroimage.2016.08.008. [DOI] [PubMed] [Google Scholar]
- Allen P.J., Polizzi G., Krakow K., Fish D.R., Lemieux L. Identification of EEG events in the MR scanner: the problem of pulse artifact and a method for its subtraction. NeuroImage. 1998;8:229–239. doi: 10.1006/nimg.1998.0361. [DOI] [PubMed] [Google Scholar]
- Allen P.J., Josephs O., Turner R. A method for removing imaging artifact from continuous EEG recorded during functional MRI. NeuroImage. 2000;12:230–239. doi: 10.1006/nimg.2000.0599. [DOI] [PubMed] [Google Scholar]
- An D., Fahoum F., Hall J., Olivier A., Gotman J., Dubeau F. Electroencephalography/functional magnetic resonance imaging responses help predict surgical outcome in focal epilepsy. Epilepsia. 2013;54:2184–2194. doi: 10.1111/epi.12434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagshaw A.P., Aghakhani Y., Benar C.G., Kobayashi E., Hawco C., Dubeau F., Pike G.B., Gotman J. EEG-fMRI of focal epileptic spikes: analysis with multiple haemodynamic functions and comparison with gadolinium-enhanced MR angiograms. Hum. Brain Mapp. 2004;22:179–192. doi: 10.1002/hbm.20024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi P., Sadowski P.J. Understanding dropout. Adv. Neural Inf. Proces. Syst. 2013:2814–2822. [Google Scholar]
- Benar C., Aghakhani Y., Wang Y., Izenberg A., Al-Asmi A., Dubeau F., Gotman J. Quality of EEG in simultaneous EEG-fMRI for epilepsy. Clin. Neurophysiol. 2003;114:569–580. doi: 10.1016/s1388-2457(02)00383-8. [DOI] [PubMed] [Google Scholar]
- de Boer H.M., Mula M., Sander J.W. The global burden and stigma of epilepsy. Epilepsy Behav. 2008;12:540–546. doi: 10.1016/j.yebeh.2007.12.019. [DOI] [PubMed] [Google Scholar]
- Bonmassar G., Purdon P.L., Jaaskelainen I.P., Chiappa K., Solo V., Brown E.N., Belliveau J.W. Motion and ballistocardiogram artifact removal for interleaved recording of EEG and EPs during MRI. NeuroImage. 2002;16:1127–1141. doi: 10.1006/nimg.2002.1125. [DOI] [PubMed] [Google Scholar]
- Fahoum F., Lopes R., Pittau F., Dubeau F., Gotman J. Widespread epileptic networks in focal epilepsies: EEG-fMRI study. Epilepsia. 2012;53:1618–1627. doi: 10.1111/j.1528-1167.2012.03533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R.S., van Emde Boas W., Blume W., Elger C., Genton P., Lee P., Engel J., Jr. Epileptic seizures and epilepsy: definitions proposed by the International League Against Epilepsy (ILAE) and the International Bureau for Epilepsy (IBE) Epilepsia. 2005;46:470–472. doi: 10.1111/j.0013-9580.2005.66104.x. [DOI] [PubMed] [Google Scholar]
- Flanagan D., Abbott D.F., Jackson G.D. How wrong can we be? The effect of inaccurate mark-up of EEG/fMRI studies in epilepsy. Clin. Neurophysiol. 2009;120:1637–1647. doi: 10.1016/j.clinph.2009.04.025. [DOI] [PubMed] [Google Scholar]
- Gotman J. Epileptic networks studied with EEG-fMRI. Epilepsia. 2008;49(Suppl. 3):42–51. doi: 10.1111/j.1528-1167.2008.01509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grouiller F., Thornton R.C., Groening K., Spinelli L., Duncan J.S., Schaller K., Siniatchkin M., Lemieux L., Seeck M., Michel C.M. With or without spikes: localization of focal epileptic activity by simultaneous electroencephalography and functional magnetic resonance imaging. Brain. 2011;134:2867–2886. doi: 10.1093/brain/awr156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer H.M., Morris H.H., Mascha E.J., Karafa M.T., Bingaman W.E., Bej M.D., Burgess R.C., Dinner D.S., Foldvary N.R., Hahn J.F., Kotagal P., Najm I., Wyllie E., Luders H.O. Complications of invasive video-EEG monitoring with subdural grid electrodes. Neurology. 2002;58:97–103. doi: 10.1212/wnl.58.1.97. [DOI] [PubMed] [Google Scholar]
- He K., Zhang X., Ren S., Sun J. 2015. Deep Residual Learning for Image Recognition. (arXiv preprint arXiv:1512.03385) [Google Scholar]
- He K.M., Zhang X.Y., Ren S.Q., Sun J. 2016. Identity Mappings in Deep Residual Networks. Computer Vision - Eccv 2016, Pt Iv 9908; pp. 630–645. [Google Scholar]
- van Houdt P.J., de Munck J.C., Leijten F.S., Huiskamp G.J., Colon A.J., Boon P.A., Ossenblok P.P. EEG-fMRI correlation patterns in the presurgical evaluation of focal epilepsy: a comparison with electrocorticographic data and surgical outcome measures. NeuroImage. 2013;75:238–248. doi: 10.1016/j.neuroimage.2013.02.033. [DOI] [PubMed] [Google Scholar]
- Khoo H.M., Hao Y., von Ellenrieder N., Zazubovits N., Hall J., Olivier A., Dubeau F., Gotman J. The hemodynamic response to interictal epileptic discharges localizes the seizure-onset zone. Epilepsia. 2017;58:811–823. doi: 10.1111/epi.13717. [DOI] [PubMed] [Google Scholar]
- Kim K.H., Yoon H.W., Park H.W. Improved ballistocardiac artifact removal from the electroencephalogram recorded in fMRI. J. Neurosci. Methods. 2004;135:193–203. doi: 10.1016/j.jneumeth.2003.12.016. [DOI] [PubMed] [Google Scholar]
- Klovatch-Podlipsky I., Gazit T., Fahoum F., Tsirelson B., Kipervasser S., Kremer U., Ben-Zeev B., Goldberg-Stern H., Eisenstein O., Harpaz Y., Levy O., Kirschner A., Neufeld M.Y., Fried I., Hendler T., Medvedovsky M. Dual array EEG-fMRI: an approach for motion artifact suppression in EEG recorded simultaneously with fMRI. NeuroImage. 2016;142:674–686. doi: 10.1016/j.neuroimage.2016.07.014. [DOI] [PubMed] [Google Scholar]
- LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- LeVan P., Maclaren J., Herbst M., Sostheim R., Zaitsev M., Hennig J. Ballistocardiographic artifact removal from simultaneous EEG-fMRI using an optical motion-tracking system. NeuroImage. 2013;75:1–11. doi: 10.1016/j.neuroimage.2013.02.039. [DOI] [PubMed] [Google Scholar]
- LeVan P., Zhang S., Knowles B., Zaitsev M., Hennig J. EEG-fMRI gradient artifact correction by multiple motion-related templates. IEEE Trans. Biomed. Eng. 2016;63:2647–2653. doi: 10.1109/TBME.2016.2593726. [DOI] [PubMed] [Google Scholar]
- Maziero D., Velasco T.R., Hunt N., Payne E., Lemieux L., Salmon C.E., Carmichael D.W. Towards motion insensitive EEG-fMRI: correcting motion-induced voltages and gradient artefact instability in EEG using an fMRI prospective motion correction (PMC) system. NeuroImage. 2016;138:13–27. doi: 10.1016/j.neuroimage.2016.05.003. [DOI] [PubMed] [Google Scholar]
- Moeller F., Tyvaert L., Nguyen D.K., LeVan P., Bouthillier A., Kobayashi E., Tampieri D., Dubeau F., Gotman J. EEG-fMRI: adding to standard evaluations of patients with nonlesional frontal lobe epilepsy. Neurology. 2009;73:2023–2030. doi: 10.1212/WNL.0b013e3181c55d17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair V., Hinton G.E. Proceedings of the 27th International Conference on Machine Learning (ICML-10) 2010. Rectified linear units improve restricted Boltzmann machines; pp. 807–814. [Google Scholar]
- Nonclercq A., Foulon M., Verheulpen D., De Cock C., Buzatu M., Mathys P., Van Bogaert P. Cluster-based spike detection algorithm adapts to interpatient and intrapatient variation in spike morphology. J. Neurosci. Methods. 2012;210:259–265. doi: 10.1016/j.jneumeth.2012.07.015. [DOI] [PubMed] [Google Scholar]
- Parkhi O.M., Vedaldi A., Zisserman A. British Machine Vision Conference. 2015. Deep face recognition; p. 6. [Google Scholar]
- Pittau F., Dubeau F., Gotman J. Contribution of EEG/fMRI to the definition of the epileptic focus. Neurology. 2012;78:1479–1487. doi: 10.1212/WNL.0b013e3182553bf7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenow F., Luders H. Presurgical evaluation of epilepsy. Brain. 2001;124:1683–1700. doi: 10.1093/brain/124.9.1683. [DOI] [PubMed] [Google Scholar]
- Schroff F., Kalenichenko D., Philbin J. Facenet: a unified embedding for face recognition and clustering. Proc. IEEE Conf. Comput. Vis. Pattern Recognit. 2015:815–823. [Google Scholar]
- Simonyan K., Zisserman A. 2014. Very Deep Convolutional Networks for Large-Scale Image Recognition. (arXiv preprint arXiv:1409.1556) [Google Scholar]
- Srivastava G., Crottaz-Herbette S., Lau K.M., Glover G.H., Menon V. ICA-based procedures for removing ballistocardiogram artifacts from EEG data acquired in the MRI scanner. NeuroImage. 2005;24:50–60. doi: 10.1016/j.neuroimage.2004.09.041. [DOI] [PubMed] [Google Scholar]
- Srivastava N., Hinton G.E., Krizhevsky A., Sutskever I., Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014;15:1929–1958. [Google Scholar]
- Thornton R., Laufs H., Rodionov R., Cannadathu S., Carmichael D.W., Vulliemoz S., Salek-Haddadi A., McEvoy A.W., Smith S.M., Lhatoo S., Elwes R.D., Guye M., Walker M.C., Lemieux L., Duncan J.S. EEG correlated functional MRI and postoperative outcome in focal epilepsy. J. Neurol. Neurosurg. Psychiatry. 2010;81:922–927. doi: 10.1136/jnnp.2009.196253. [DOI] [PubMed] [Google Scholar]
- Tousseyn S., Dupont P., Robben D., Goffin K., Sunaert S., Van Paesschen W. A reliable and time-saving semiautomatic spike-template-based analysis of interictal EEG-fMRI. Epilepsia. 2014;55:2048–2058. doi: 10.1111/epi.12841. [DOI] [PubMed] [Google Scholar]
- Worsley K.J., Liao C.H., Aston J., Petre V., Duncan G.H., Morales F., Evans A.C. A general statistical analysis for fMRI data. NeuroImage. 2002;15:1–15. doi: 10.1006/nimg.2001.0933. [DOI] [PubMed] [Google Scholar]
- Zijlmans M., Huiskamp G., Hersevoort M., Seppenwoolde J.H., van Huffelen A.C., Leijten F.S. EEG-fMRI in the preoperative work-up for epilepsy surgery. Brain. 2007;130:2343–2353. doi: 10.1093/brain/awm141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
IED-related BOLD response (primary cluster) and SEEG findings (N = 15).