Summary
The AbcR small RNAs (sRNAs) are a fascinating example of two highly conserved sRNAs that differ tremendously at the functional level amongst organisms. From their transcriptional activation to their regulatory capabilities, the AbcR sRNAs exhibit varying characteristics in three well-studied bacteria belonging to the Rhizobiales order: the plant symbiont Sinorhizobium meliloti, the plant pathogen Agrobacterium tumefaciens, and the animal pathogen Brucella abortus. This review outlines the similarities and differences of the AbcR sRNAs between each of these organisms, and discusses reasons as to why this group of sRNAs has diverged in their genetic organization and regulatory functions across species. In the end, this review will shed light on how regulatory systems, although seemingly conserved amongst bacteria, can vary based on the environmental niche and lifestyle of an organism.
Keywords: small RNAs, Alphaproteobacteria, AbcR1, AbcR2, Rhizobiales
Graphical abstract
The AbcR sRNAs have been described in detail in three members of the Rhizobiales: the plant symbiont Sinorhizobium, the plant pathogen Agrobacterium and the mammalian pathogen Brucella. Through the use of two 6-nucleotide motifs, the AbcR sRNAs are responsible for regulating targets involved in nutrient acquisition. Although these two sRNAs have only been characterized in three organisms thus far, it is likely the AbcR system is present and functioning in other members of the Rhizobiales.
Introduction
Small RNAs (sRNAs) are regulatory components in prokaryotes that allow organisms to respond to stimuli through rapid alteration of gene expression. Importantly, sRNAs often exert their regulatory functions by interacting imperfectly with target mRNAs, either by binding to the untranslated region (UTR) or the coding region (CDR) of the target mRNA (Storz et al., 2006; Waters and Storz, 2009; Gottesman and Storz 2011; Storz et al., 2011). The presence of regulatory sRNAs has been well documented in both Gram-positive (Johansson et al., 2002; Mann et al., 2012; Soutourina et al., 2013; Guillet et al., 2013; Miller et al., 2014) and Gram-negative (Hershberg et al., 2003; Vogel 2009; Postic et al., 2010; Bardill et al., 2012; Gómez-Lozano et al., 2012; Schiano et al., 2014; McClure et al., 2014; Baddal et al., 2015) bacteria. However, only a handful of sRNAs have been identified and characterized in Alphaproteobacteria, and in this class of bacteria, one predominant group of sRNAs is the αr15 family (del Val et al., 2012).
The first members of the αr15 family, Smr15C2 and Smr15C1, were identified in 2007 in the plant symbiont Sinorhizobium meliloti (del Val et al., 2007). αr15 sRNAs are around 100 nucleotides in length, and are commonly located within intergenic regions. The secondary structures are well conserved, generally folding into three stem loops. αr15 sRNAs are commonly trans-acting, regulating transcripts throughout the bacterial transcriptome (del Val et al., 2012). Interestingly, the αr15 family has partial homology to the SuhB non-coding RNA, which is commonly maintained in several copies on the bacterial chromosome and has a secondary structure with at least one hairpin that displays a CU-rich anti-Shine Delgarno (anti-SD) sequence (Corbino et al., 2005; del Val et al., 2012).
The AbcR (ATP-binding cassette regulator) sRNAs, AbcR1 and AbcR2, are members of the αr15 family and have been identified throughout the Rhizobiales (Ulvé et al., 2007; del Val et al., 2007; Valverde et al., 2008; Voss et al., 2009; Schlüter et al., 2010; Vercruysse et al., 2010; Wilms et al., 2011; Caswell et al., 2012; Torres-Quesada et al., 2013). S. meliloti has been predicted to have a third AbcR sRNA on the pSymA megaplasmid (del Val et al., 2012), but no experimental evidence has validated this hypothesis. The AbcR sRNAs are interesting examples of extremely conserved and structurally similar sRNAs that have minor differences between organisms, which allow them to have vastly different regulatory roles. In this MicroReview, we will focus on discussing the AbcRs in three rhizobiales: Sinorhizobium meliloti, Agrobacterium tumefaciens and Brucella abortus.
Sinorhizobium meliloti
Lifestyle
Sinorhizobium meliloti is a plant symbiont, capable of forming mutualistic associations with leguminous plants, including those from the genera Medicago, Melilotus and Trigonella (Roumiantseva et al., 2002). This bacterium lives either in soil, where it competes with organisms for essential nutrients, colonizes plants as an endophyte or resides within nodules in plant roots, where it relies on intracellular nutrients to survive and, in turn, fixes atmospheric nitrogen for the plant (Pini et al., 2012; Jones et al., 2007). S. meliloti, along with other rhizobial species, encodes around 200 ABC transport systems, allowing for the bacterium to take up nutrients, metals and other molecules that are necessary for its survival (Kaneko et al., 2000; Cheng et al., 2011). Since these transport systems are critical to the longevity of S. meliloti both extra- and intracellularly, it is of interest to understand the regulatory circuitry that governs the expression of these transporters.
Expression of the AbcR small RNAs in S. meliloti
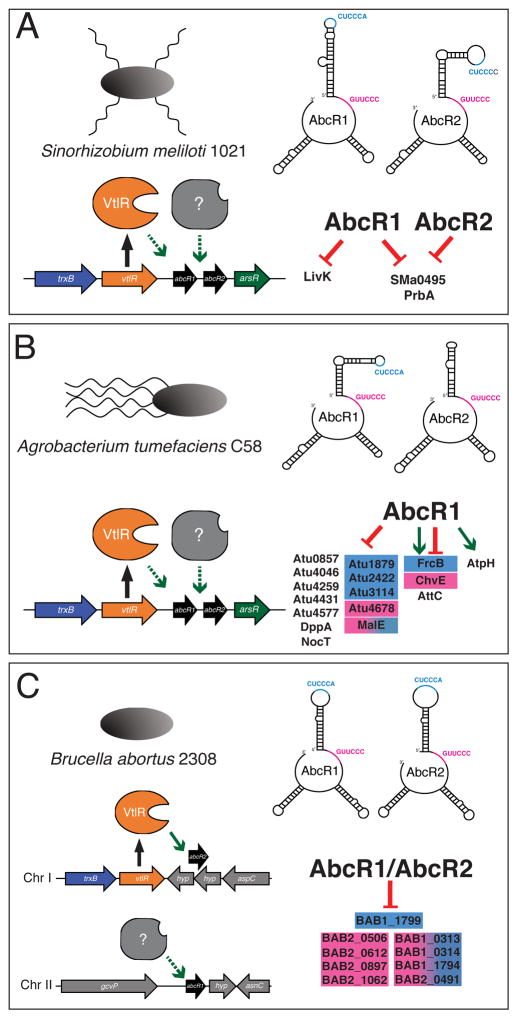
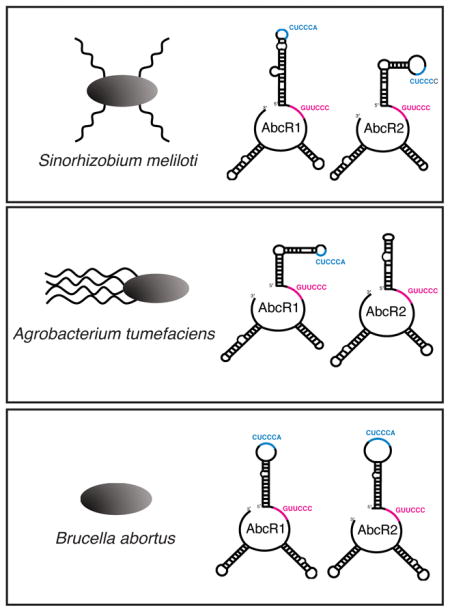
The AbcR sRNAs were among several sRNAs first identified by del Val et al. in S. meliloti (del Val, 2007). Formally referred to as Smr15C2 and Smr15C1, the sRNAs are located in tandem between smc01225, encoding the LysR-type transcriptional regulator LsrB, and smc01226, encoding an ArsR-type transcriptional regulator (Figure 1A). Smr15C2, named AbcR1, and Smr15C1, named AbcR2, are expressed independently and in response to various stimuli (Torres-Quesada et al., 2013). Specifically, AbcR1 is predominantly expressed in early phases of growth and can be detected in high abundance in mature M. sativa nodules. Conversely, AbcR2 reaches its highest expression at late stationary phase and is stress-induced (i.e., osmotic stress (NaCl), membrane stress (EtOH), low pH (5.6), and microaerobiosis). Although expression of each sRNA differs, it is interesting that the AbcR sRNAs are strikingly similar in both nucleotide sequence (85% identity) and predicted secondary structure. S. meliloti AbcR1 and AbcR2, in line with the αr15 sRNA family, are predicted to fold into 3 hairpins, where one hairpin exposes the αr15-conserved anti-SD UCCUCCC motif (Torres-Quesada et al., 2013).
Figure 1.
The AbcR sRNAs and their regulatory profiles in A) Sinorhizobium meliloti, B) Agrobacterium tumefaciens and C) Brucella abortus. The AbcR sRNAs are encoded either in tandem (S. meliloti and A. tumefaciens) or on separate chromosomes (B. abortus). VtlR binds the promoter region of abcR2, and ongoing studies in our laboratory (represented by dashed arrows) support the hypothesis that Atu2186 (VtlR) and LsrB bind the promoter region of abcR1, but not abcR2. The second transcriptional regulator of the abcRs is currently unknown. The AbcR sRNAs all fold into similar three hairpin structures. M1, colored in blue, has the sequence CUUCCA. M2, colored in fuchsia, has a sequence of GUUCCC. Experimentally confirmed AbcR-targets in each organism are listed to the right. Targets in blue are regulated by M1. Targets in fuchsia are regulated M2. Targets in blue/fuchsia are regulated by M1 and M2. Targets in black have not been experimentally confirmed to be regulated by a specific AbcR-motif.
AbcR1 and AbcR2 possess both independent and redundant regulatory functions in S. meliloti
The first target experimentally confirmed to be negatively regulated by S. meliloti AbcR1, but not AbcR2, was LivK (Torres-Quesada et al., 2013). LivK encodes for a periplasmic binding protein of an ABC transporter system, which is predicted to regulate the uptake of branched-chain amino acids. Although AbcR1 and AbcR2 possess almost identical anti-SD sequences, computational predictions of the sRNA-mRNA interactions found AbcR1 to have three additional complementary nucleotide sequences to the LivK mRNA sequence as compared to AbcR2 (Torres-Quesada et al., 2013). Although additional AbcR targets were predicted in S. meliloti, the full regulatory profile of the AbcR sRNAs remains to be elucidated.
Many sRNAs depend on the global RNA chaperon Hfq for both their own stability and successful interactions with target transcripts (reviewed in Vogel and Luisi 2011). As such, it is important to note that the AbcR sRNAs are dependent on Hfq in S. meliloti (Torres-Quesada et al., 2013). A deletion of hfq results in instability and subsequent degradation of the sRNAs, thus resulting in absence of AbcR regulation. In S. meliloti, a study utilizing RNA-seq to analyze RNA populations following the co-immunoprecipitation (Co-IP) with Hfq reveled new targets of the AbcR sRNAs (Torres-Quesada et al., 2014). In addition to the enrichment of LivK, the previously identified target of AbcR1, two additional targets were identified: SMa0495, encoding an ABC transporter amino acid binding protein; and PrbA, encoding an ABC transporter substrate-binding protein. SMa0495 and PrbA are down-regulated by both AbcR1 and AbcR2 (Torres-Quesada et al., 2014). Computational bioinformatics predicted that the sRNAs bind the 5′ UTRs, specifically in the ribosome-binding site. For interactions with PrbA, AbcR1 and AbcR2 are predicted to utilize an extended version of the previously mentioned anti-SD region, which contains a 6-nucleotide motif (CUCCCA) referred to as M1, located in the first hairpin. Conversely, the sRNAs are predicted to employ a 32-nucleotide motif, which contains a 6-nucleotide motif (GUUCCC) referred to as M2, for interaction with SMa0495 (Torres-Quesada et al., 2014). Overall, these findings shed light on how AbcR1 and AbcR2, although strikingly similar in appearance, have significant differences in their regulatory profiles. It will be interesting to further define the S. meliloti AbcR regulatory profile, and determine the extent of regulatory redundancy and/or non-redundancy that exists between AbcR1 and AbcR2 in S. meliloti.
Agrobacterium tumefaciens
Lifestyle
Agrobacterium tumefaciens, a close relative of S. meliloti, is a plant pathogen that causes tumors, known as crown galls, in dicotyledonous plants. Following association with the plant, A. tumefaciens takes advantage of the plant by injecting a short piece of its DNA, known as T-DNA, into the host cells (Moore et al., 1997). This ultimately leads to tumor formation and the production/secretion of opines, which serve as energy sources for the bacterium. As a defense mechanism, wounded plants secrete a molecule called γ-aminobutyric acid (GABA). GABA is taken up by an ABC transport system in A. tumefaciens, and can lead to disruption of bacterial cell-to-cell communication (Haudecoeur et al., 2009; Chevrot et al., 2006). Importantly, A. tumefaciens has been demonstrated to control the expression of this GABA transport system via the AbcR1 sRNA (Wilms et al., 2011).
AbcR1 regulates the ABC transport system responsible for importing GABA
Wilms et al. was the first to begin mechanistic characterization of the AbcR sRNAs in A. tumefaciens. Identical to the organization of the AbcRs in S. meliloti, the abcR1 and abcR2 genes are in tandem on the circular chromosome, located in the intergenic region of atu2186, encoding a LysR-type transcriptional regulator, and atu2187, encoding a ArsR-type transcriptional regulator (Figure 1B). In contrast to S. meliloti, AbcR1 and AbcR2 are expressed at similar levels in A. tumefaciens during different growth phases in rich medium (Wilms et al., 2011).
Deletion of abcR1 in A. tumefaciens leads to an abundance of two proteins: Atu1879 and Atu2422. Similar to the regulatory capacity of S. meliloti AbcR1, A. tumefaciens AbcR1 plays the predominant role in regulating these two targets (Wilms et al., 2011). Both Atu1879 and Atu2422 putatively encode periplasmic components of ABC transport systems. Importantly, A. tumefaciens Atu2422 is the ortholog of S. meliloti LivK, a confirmed AbcR1 target. Moreover, Atu2422 was previously shown to be a GABA and proline transporter.
To investigate the link between GABA transport and AbcR1 regulation, uptake experiments utilizing radiolabeled GABA were conducted with A. tumefaciens strains. As expected, a deletion of abcR1 resulted in increased import of GABA, due to the over-production of the Atu2422 (Wilms et al., 2011). Additionally, Atu2422 was previously demonstrated to function in the import of GABA and proline, and thus, excess amounts of unlabeled proline abolished uptake of radiolabeled GABA. Given this, it is not surprising that AbcR1 is critical to the process of GABA uptake as the sRNA down-regulates the expression of Atu2422, reducing the uptake of GABA.
AbcR1 is involved in the complex regulation of myriad transcripts in A. tumefaciens
Subsequent proteomic and bioinformatic analyses demonstrates over 60 additional targets of AbcR1, the majority of which encode components of ABC transport systems (Overlöper et al., 2014). Overall, AbcR1 regulation in A. tumefaciens is dynamic, where the sRNA is involved in both increased and decreased levels of transcripts. In some cases, AbcR regulation is governed by the growth phase of A. tumefaciens. For example, ChvE is activated by AbcR1 in exponential phase and repressed in stationary phase (Overlöper et al., 2014). ChvE is of particular interest, as this protein encodes a sugar-binding periplasmic protein apart of an ABC transport system. Importantly, ChvE is involved in A. tumefaciens pathogenesis, where it activates critical virulence cascades and transport systems (Winans et al., 1986; Huang et al., 1999; Shimoda et al., 1999; Cangelosi et al., 1999). These findings, in conjunction with AbcR1-mediated regulation of Atu2422, support the hypothesis that the AbcR system is involved in the pathogenesis of A. tumefaciens.
Through the use of CopraRNA, two binding motifs were identified in A. tumefaciens AbcR1 (Wright et al., 2013). The first motif, called M1 (CUCCCAGU), is located in the first hairpin, and the second motif, called M2 (GUUCCC), is located in a single-stranded region between the first and second hairpins. M1 was previously shown to bind the anti-SD region of Atu2422 (Wilms et al., 2011). For the additional targets identified, AbcR1 utilized M1 and/or M2 to bind the 5′ untranslated region (5′ UTR) or coding region (CDR) of transcripts (Overlöper et al., 2014). Overall, AbcR1 is a predominant regulatory AbcR sRNA, and is involved in the regulation of both ABC transport system and virulence factors.
Brucella abortus
Lifestyle
Brucella spp. are pathogenic bacteria capable of causing serious infections in a wide range of animals, such as cattle, swine, goats, sheep, and humans. In cattle, Brucella abortus results in spontaneous abortions, which can lead to substantial economic losses (Boschiroli et al., 2001; Pappas et al., 2005; Pappas et al., 2006). In humans, brucellosis, the disease caused by Brucella spp., presents as an undulating fever with common flu-like symptoms (Franco et al., 2007). Upon infection, the brucellae reside and replicate within host phagocytic cells, such as macrophages and dendritic cells. As such, eliminating the bacteria can prove to be difficult. Following entry into the host cell environment, the brucellae quickly and efficiently altering gene expression in order to adapt to the new environment by (Köhler et al., 2003; Roop et al., 2004). A common mechanism to shift their transcriptome to one that is favorable for their intracellular survival is through regulatory sRNAs.
AbcR1 and AbcR2 are functionally redundant in B. abortus
Unlike the chromosomal organization in S. meliloti and A. tumefaciens, the B. abortus AbcR sRNAs are encoded separately, with abcR1 on chromosome II and abcR2 on chromosome I. abcR1 is located between bab2_0515, encoding a glycine decarboxylase, and bab2_0516, encoding a small hypothetical protein. abcR2 is positioned on the opposite strand from bab1_1515 and bab1_1516, both of which encode small hypothetical proteins. abcR2 is flanked by bab1_1514, encoding an aspartate aminotransferase, and bab1_1517, encoding the LysR-type transcriptional regulator VtlR (Figure 1C). VtlR, for virulence-associated transcriptional LysR-family regulator, is the bona fide transcriptional activator of abcR2, but not abcR1 (Sheehan et al., 2015). Although the genetic organization of the B. abortus sRNAs differs from other bacteria, a homolog of VtlR is found upstream of abcR1 and abcR2 in both A. tumefaciens and S. meliloti.
Similar to S. meliloti and A. tumefaciens, AbcR1 and AbcR2 are dependent on the RNA chaperone Hfq in B. abortus. In contrast to A. tumefaciens, B. abortus AbcR1 and AbcR2 are redundant in function, where only a double deletion of abcR1 and abcR2 results in reduced virulence (Caswell et al., 2012). Given that AbcR1 and AbcR2 are functionally redundant, much emphasis has been directed to defining the molecular mechanism(s) of the redundancy between AbcR1 and AbcR2 in B. abortus.
B. abortus AbcR1 and AbcR2 share the same regulatory roles
Microarray and quantitative proteomic analyses of a B. abortus abcR1 abcR2 double mutant found that the AbcR sRNAs negatively regulate over 20 transcripts, the majority of which encode components of ABC transport systems (Caswell et al., 2012). Therefore, the AbcR regulatory profiles in S. meliloti, A. tumefaciens and B. abortus overlap, where the sRNAs largely regulate periplasmic proteins of ABC transport systems. In fact, one target, BAB1_1794 in B. abortus, is orthologous to LivK and Atu2422 in S. meliloti and A. tumefaciens, respectively.
Quantitative reverse-transcriptase PCR with the B. abortus single and double abcR mutants found that the sRNAs are truly redundant, with the isogenic abcR mutants resulting in gene expression levels comparable to the wild-type strain, and a double abcR mutant leading to over-expression of target genes (Sheehan and Caswell, 2017). Furthermore, AbcR1 and AbcR2 both physically bind to the 5′ UTR of one of the targets, BAB2_0879, confirming the redundant roles these sRNAs have in Brucella. Thus far, it appears this redundancy in both regulation and function is restricted to the Brucella AbcR sRNAs.
AbcR1 and AbcR2 utilize two motifs, M1 and M2, to regulate transcripts
After determining the redundancy of the B. abortus AbcR sRNAs; two motifs, named M1 and M2, were identified in both AbcR1 and AbcR2 (Sheehan and Caswell, 2017). Similar to the organization in A. tumefaciens AbcR1, M1 (CUUCCC) is located in the first hairpin, and M2 (GUUCCC) is located in a single-stranded region between the first and second hairpin (Figure 1C). Chromosomal mutations of either one or both motifs in the AbcR sRNAs found that the AbcRs utilize both motifs for their regulatory activity. Interestingly, the majority of transcripts tested were either regulated solely by M2, or by both M1 and M2. Only one target transcript, BAB1_1799, encoding a putative ABC transporter permease, was regulated by M1 alone. Conversely, four targets were regulated exclusively by M2, three of which encode putative ABC transporter periplasmic binding proteins (Sheehan and Caswell, 2017). Altogether, it seems that the AbcR sRNAs have retained almost identical motifs to carry out their regulatory roles in the Rhizobiales examined to date.
A B. abortus abcR1 abcR2 double mutant has a severe virulence defect in macrophages and experimentally infected mice (Caswell et al., 2012). Due to the role M1 and M2 play in regulation of transcripts, the single and double abcR motif mutants were tested to assess the role of these regulatory motifs in Brucella virulence. Surprisingly, while the abcR-M1 mutant exhibited wild-type infection levels, the abcR-M2 and abcR-M1/2 mutant strains were attenuated in cells and mice (Sheehan and Caswell, 2017). This result was interesting, as both motifs in the sRNAs are necessary for the proper regulation of transcripts in Brucella. However, it stands to reason that one motif, M2, may be responsible for regulating targets that are directly or indirectly involved in pathogenesis. One of these M2-regulated targets, BAB2_0612, was found to be involved in Brucella pathogenesis, strengthening this hypothesis (Sheehan and Caswell, 2017), and will be interesting to determine if any other M2 targets are linked to Brucella pathogenesis.
The AbcR sRNA system in other members of the Rhizobiales
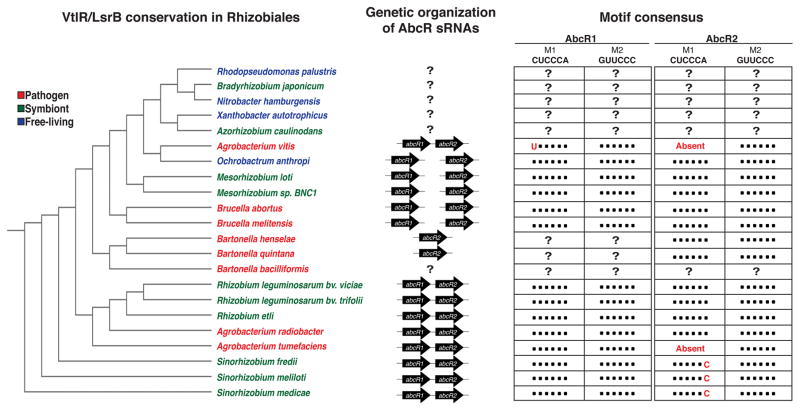
Although the AbcRs have been characterized extensively in three organisms, it is plausible that this sRNA family is conserved and functional in other bacteria in the Rhizobiales. A bioinformatics approach revealed the AbcR sRNA system in several other members of the Rhizobiales (del Val et al., 2012; Figure 2). In addition to the presence of one or both sRNAs, several of the AbcR targets and complementary binding motifs are found in several organisms including Ochrobacterium anthropi, Bartonella henselae and Mesorhizobium loti. Altogether, the AbcR system, from the LysR-type transcriptional regulator to the ABC-type transport systems, is very well conserved in this order of bacteria. The majority of ABC-type transporters regulated by the AbcR sRNAs are involved in nutrient acquisition, specifically by uptake of amino acids and polyamines (Wilms et al., 2011; Caswell et al., 2012; Torres-Quesada et al., 2013; Torres-Quesada et al., 2014; Overlöper et al., 2014). It is likely this is a necessary mechanism for rhizobiales to quickly and efficiently turn-on or turn-off transporters when in certain environments (i.e. nutrient-rich vs nutrient-limited).
Figure 2.
Conservation of the AbcR system in Rhizobiales. (Left) Phylogenetic tree analyzing the conservation of a LysR-type transcription regulator commonly found upstream of one or both abcR sRNAs. This regulator, called VtlR for virulence-associated transcriptional LysR-family regulator, has been previously shown to positively regulate abcR2 in B. abortus (Sheehan et al., 2015). Preliminary evidence has found S. meliloti LsrB (LysR-type symbiosis regulator) and A. tumefaciens VtlR to positively regulate abcR1. (Middle) The AbcR sRNAs can be found in tandem or separated in the Rhizobiales. For R. palustris, B. japonicum, N. hamburensis, X. autotrophicus, A. caulinodans, B. henselae, B. quintana, and B. bacilliformis, bioinformatics failed to identify the location(s) of one or both of the AbcRs. (Right) The AbcR sRNAs contain two regulatory motifs called M1 (CUCCCA) and M2 (GUUCCC). Black circles represent consensus with these motifs. Differences are denoted by red text.
ABC transport systems have been shown to play important roles in the formation of symbiotic relationships, specifically between R. leguminosarum bv. viciae and its pea plant host (Lodwig et al., 2003). Mutations in two ABC-type broad range amino acid transporters result in defective symbiosis and the inability of the bacteriod to fix nitrogen. However, mutations of these two ABC transporters in S. meliloti do not have any effect on nitrogen fixation or symbiosis with alfalfa plants (Prell et al., 2010). This brings up an interesting point regarding the AbcR-regulated transport systems in rhizobiales. Although deletion of the abcR sRNAs, and subsequent alternation of the expression of ABC transport systems, in S. meliloti does not result in a symbiotic defect (Torres-Quesada et al., 2013), this may not hold true in other symbionts such as R. leguminosarum bv. viciae. In this case, disregulation of these ABC transport systems, which may play roles in proper nitrogen fixation, may lead to the inability of the bacterium to form a symbiotic relationship with its host. It will be interesting to see how the AbcR sRNAs effect nitrogen-fixation and subsequent symbiosis in various rhizobiales since there are obvious differences in the role ABC transporters play in their respective organism.
Another widely conserved sRNA that has been well studied and is involved in regulating nutrient acquisition systems is GcvB. First identified in E. coli, GcvB is a posttranscriptional sRNA regulator responsible for regulating genes involved in amino acid transport and metabolism (Urbanowski et al., 2000). GcvB is an important regulatory component in many Gram-negative bacteria, tightly-controlling energy expenditure linked to the transport and biosynthesis of amino acids (McArthur et al., 2006; Sharma et al., 2007; Sharma et al., 2011; Miyakoshi et al., 2015). It seems likely that the AbcR sRNAs and GcvB are functional homologs, since both sRNAs are involved in regulating nutrient uptake in several bacteria. Moreover, at least one of the AbcR-targets in B. abortus, BAB1_1587, is shared with GcvB in Salmonella (Sharma et al., 2011; Caswell et al., 2012). Altogether, this conservation of sRNAs involved in the precise regulation of nutrient acquisition in Gram-negative bacteria is important for organisms to successfully adapt and survive in a variety of environment conditions.
The genetic organization of abcR1 and abcR2 in the Rhizobiales
The main difference in the genetic organization of the abcRs revolves around the location of the genes: are they in tandem or separated? Evolutionarily, this split is defined where abcR1 and abcR2 can be found in tandem in the Rhizobiaceae family (Sinorhizobium spp. and Agrobacterium spp.). In contrast, the Brucellaceae (Brucella spp. and Ochrobacterum spp.), Bartonellaceae (Bartonella spp.) and Phyllobacteriaceae (Mesorhizobium spp.) families have the abcR genes separated on their chromosomes (Figure 2). This divergence of the sRNAs may prove to be beneficial for some organisms, such as Brucella, since the sRNAs are redundant in function and regulatory capacity (Caswell et al., 2012; Sheehan and Caswell, 2017). If one AbcR sRNA is incapacitated (i.e., spontaneous mutation, loss of DNA, degradation of RNA, etc.), the second copy located in a difference genomic locus acts as a fail-safe. In contrast, full redundancy of the AbcRs is not observed in S. meliloti or A. tumefaciens (Torres-Quesada et al., 2013; Torres-Quesada et al., 2014; Wilms et al., 2011; Overlöper et al., 2014). Nonetheless, the AbcR sRNAs are critical regulatory components of gene expression in Rhizobiales, allowing organisms to adapt to and survive in specific environmental niches.
A LysR-type transcriptional regulator is involved in the regulation of the AbcR sRNAs
Taken together, the AbcR sRNAs have been retained by multiple members of the Rhizobiales, but have diverged in their genomic location and regulation. VtlR, the activator of abcR2 in Brucella abortus, is highly conserved amongst the Alphaproteobacteria (Sheehan et al., 2015), and in the bacteria examined in this review, VtlR is always found upstream of either one or both sRNAs (Figure 1; Figure 2). It is likely that this LysR-type regulator is involved in the regulation of either abcR1 or abcR2 across the Rhizobiales. In S. meliloti, LsrB (for LysR-type symbiosis regulator), the ortholog of VtlR, is critical for proper symbiosis between the bacterium and its plant host (Luo et al., 2005). LsrB has been shown to be the transcriptional regulator of several genes involved in LPS biosynthesis, which is required for proper nitrogen fixation in alfalfa plants (Tang et al., 2014). Analysis of the S. meliloti abcR promoter region revealed a putative LsrB-binding site, specifically upstream of abcR1. Along with preliminary evidence from our laboratory, it is likely LsrB is the transcriptional activator of abcR1 in S. meliloti. However, since both sRNAs are transcribed individually and, in some cases, have differential expression in their respective organisms, what is the other abcR transcriptional regulator?
Unfortunately, this question remains to be elucidated. Analysis of the abcR2 promoter regions in S. meliloti and A. tumefaciens revealed little nucleotide conservation, except in their -10 and -35 sites. These promoter regions, including B. abortus abcR1, share -10 and -35 motifs that resemble a σ32 promoter sequence, which is designated RpoH, and interestingly, RpoH is known to be involved in the regulation of sRNAs in S. meliloti (Gruber and Gross 2003; Barnett et al., 2012). If the sRNAs are truly dependent on this heat-shock sigma factor, this may add to the working model that the AbcRs are involved in aiding the bacterium during adaptive response to different environmental conditions. However, this observation is based solely on bioinformatics, and it will be interesting to experimentally define the transcriptional regulation of the abcR genes in each bacterium. Overall, the genetic organization and regulation of the AbcR sRNAs are well conserved, and major differences in the sRNAs underscore their diverse regulation in each bacterium.
The AbcR sRNAs have diverged immensely in their regulatory capacity
ABC transport systems are critical for organisms to import essential nutrients and export excess and/or toxic components (Davidson et al., 2008; Wilkens 2015), and the AbcR sRNAs are largely involved in the regulation of ABC transporters in S. meliloti, A. tumefaciens and B. abortus (Wilms et al., 2011; Caswell et al., 2012; Torres-Quesada et al., 2013; Torres-Quesada et al., 2014; Overlöper et al., 2014). A handful of these AbcR target systems are conserved between two of the three bacteria. Only one of the AbcR targets, encoding an ABC transporter periplasmic binding protein (i.e., S. meliloti LivK, A. tumefaciens Atu2422, and B. abortus BAB1_1794) is conserved in all three bacteria. Moreover, the complementary AbcR M1 motif is also conserved. Another AbcR target, encoding a putative ABC transporter amino acid-binding protein, is conserved in A. tumefaciens (Atu1879) and B. abortus (BAB2_0612). However, AbcR regulation of this target has diverged. In A. tumefaciens, AbcR1 utilizes the M1 regulatory motif to bind Atu1879. In contrast, B. abortus AbcR1 and AbcR2 utilize the M2 regulatory motif to bind BAB2_0612. Although the AbcRs share similar targets amongst bacteria, they have diverged in how they mechanistically regulate these transcripts.
Aside from the different AbcR-regulated targets, there are also differences in their regulatory capacity. In B. abortus and S. meliloti, AbcR1 and AbcR2 are involved in the negative regulation of targets (Caswell et al., 2012; Torres-Quesada et al., 2013; Torres-Quesada et al., 2013). In contrast, A. tumefaciens AbcR1 positively and/or negative regulates targets (i.e., FrcB and ChvE) (Overlöper et al., 2014). In some cases, this regulation is dependent on the phase of growth of A. tumefaciens. This growth-phase dependent regulation would prove to be beneficial, as AbcR1 can turn on and off targets at precise times, such as in soil vs. during infection. In Brucella, it is hypothesized that some macrophage-derived signal drives the expression of the AbcR sRNAs, potentially directly through VtlR (Sheehan et al., 2015). Going forward, it will be interesting to fully elucidate AbcR regulation in S. meliloti, as only three targets have been experimentally confirmed to date. Altogether, the AbcR sRNAs are involved in the regulation of numerous ABC transport systems that, in turn, allows the bacteria to precisely control the uptake of essential nutrients.
AbcR1 as the primary AbcR sRNA in the Rhizobiaceae
The most interesting molecular differences between the AbcRs are the two regulatory motifs, M1 and M2 (Figure 1). These motifs were first identified in A. tumefaciens, where AbcR1 contains M1 (CUCCCA) in the first hairpin and M2 (GUUCCC) in the single-stranded region between the first and second hairpin (Wilms et al., 2011; Overlöper et al., 2014). Interestingly, A. tumefaciens AbcR2 is not involved in the regulation of transcripts, and while AbcR2 does contain an intact M2 motif, M1 is absent. This small difference may explain why AbcR2 is non-functional in A. tumefaciens. It will be interesting to learn if M1 could be incorporated into AbcR2 and restore its regulatory activity.
In S. meliloti, AbcR1 and AbcR2 both have regulatory capabilities, as both sRNAs contain M1 and M2 in their respective locations (Torres-Quesada et al., 2013; Torres-Quesada et al., 2014). However, M1 in AbcR2 has one slight difference: CUCCCA is changed to CUCCCC. Although small, this may explain why AbcR1 is the major regulatory AbcR in S. meliloti. Identification of additional AbcR-regulated targets, and subsequent examination of putative complementary motif sequences would reveal if this M1 mutation in AbcR2 is hindering the activity of the sRNA in S. meliloti.
Overall, the AbcR sRNAs are major players in the regulation of ABC-type transport systems and virulence factors (Wilms et al., 2011; Caswell et al., 2012; Torres-Quesada et al., 2013; Torres-Quesada et al., 2014; Overlöper et al., 2014). Although similar in nucleotide sequences and secondary structures, they have diverged amongst the rhizobiales to allow organisms to adapt to their specific environmental niches. The AbcR sRNAs are a perfect example of how seemingly conserved molecular systems have evolved greatly based on the lifestyle of an organism.
Acknowledgments
Work in the Caswell laboratory is supported by the American Heart Association (15SDG23280044), the National Institute of Allergy and Infectious Diseases (AI117648), the Virginia-Maryland College of Veterinary Medicine, and the Fralin Life Sciences Institute at Virginia Tech. The authors would like to thank James A. Budnick for critically evaluating the review, and Dr. Birgit Scharf for assistance with reviewing and providing constructive comments on the manuscript.
References
- 1.Baddal B, Muzzi A, Censini S, Calogero RA, Torricelli G, Guidotti S, et al. Dual RNA-seq of nontypeable Haemophilus influenzae and host cell transcriptomes reveals novel insights into host-pathogen cross talk. MBio. 2015;6:e01765–15. doi: 10.1128/mBio.01765-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardill JP, Hammer BK. Non-coding sRNAs regulate virulence in the bacterial pathogen Vibrio cholerae. RNA Biol. 2012;9:392–401. doi: 10.4161/rna.19975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett MJ, Bittner AN, Toman CJ, Oke V, Long SR. Dual RpoH sigma factors and transcriptional plasticity in a symbiotic bacterium. J Bacteriol. 2012;194:4983–4994. doi: 10.1128/JB.00449-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boschiroli ML, Foulongne V, O’Callaghan D. Brucellosis: a worldwide zoonosis. Curr Opin Microbiol. 2001;4:58–64. doi: 10.1016/s1369-5274(00)00165-x. [DOI] [PubMed] [Google Scholar]
- 5.Cangelosi GA, Ankenbauer RG, Nester EW. Sugars induce the Agrobacterium virulence genes through a periplasmic binding protein and a transmembrane signal protein. Proc Natl Acad Sci U S A. 1990;87:6708–6712. doi: 10.1073/pnas.87.17.6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caswell CC, Gaines JM, Ciborowski P, Smith D, Borchers CH, Roux CM, et al. Identification of two small regulatory RNAs linked to virulence in Brucella abortus 2308. Mol Microbiol. 2012;85:345–360. doi: 10.1111/j.1365-2958.2012.08117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng J, Poduska B, Morton RA, Finan TM. An ABC-type cobalt transport system is essential for growth of Sinorhizobium meliloti at trace metal concentrations. J Bacteriol. 2011;193:4405–4416. doi: 10.1128/JB.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chevrot R, Rosen R, Haudecoeur E, Cirou A, Shelp BJ, Ron E, Faure D. GABA controls the level of quorum-sensing signal in Agrobacterium tumefaciens. Proc Natl Acad Sci U S A. 2006;103:7460–7464. doi: 10.1073/pnas.0600313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corbino KA, Barrick JE, Lim J, Welz R, Tucker BJ, Puskarz I, et al. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in Alphaproteobacteria. Genome Biol. 2005;6:R70. doi: 10.1186/gb-2005-6-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev. 2008;72:317–364. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.del Val C, Rivas E, Torres-Quesada O, Toro N, Jiménez-Zurdo JI. Identification of differentially expressed small non-coding RNAs in the legume endosymbiont Sinorhizobium meliloti by comparative genomics. Mol Microbiol. 2007;66:1080–1091. doi: 10.1111/j.1365-2958.2007.05978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.del Val C, Romero-Zaliz R, Torres-Quesada O, Peregrina A, Toro N, Jiménez-Zurdo JI. A survey of sRNA families in Alphaproteobacteria. RNA Biol. 2012;9:119–129. doi: 10.4161/rna.18643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franco MP, Mulder M, Gilman RH, Smits HL. Human brucellosis. Lancet Infect Dis. 2007;12:775–786. doi: 10.1016/S1473-3099(07)70286-4. [DOI] [PubMed] [Google Scholar]
- 14.Gómez-Lozano M, Marvig RL, Molin S, Long KS. Genome-wide identification of novel small RNAs in Pseudomonas aeruginosa. Environ Microbiol. 2012;14:2006–2016. doi: 10.1111/j.1462-2920.2012.02759.x. [DOI] [PubMed] [Google Scholar]
- 15.Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol. 2011:3. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 17.Guillet J, Hallier M, Felden B. Emerging functions for the Staphylococcus aureus RNome. PLoS Pathog. 2013;9:e1003767. doi: 10.1371/journal.ppat.1003767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haudecoeur E, Planamente S, Cirou A, Tannières M, Shelp BJ, Moréra S, Faure D. Proline antagonizes GABA-induced quenching of quorum-sensing in Agrobacterium tumefaciens. Proc Natl Acad Sci U S A. 2009;106:14587–14592. doi: 10.1073/pnas.0808005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hershberg R, Altuvia S, Margalit H. A survey of small RNA-encoding genes in Escherichia coli. Nucleic Acids Res. 2003;31:1813–1820. doi: 10.1093/nar/gkg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang ML, Cangelosi GA, Halperin W, Nester EW. A chromosomal Agrobacterium tumefaciens gene required for effective plant signal transduction. J Bacteriol. 1990;172:1814–1822. doi: 10.1128/jb.172.4.1814-1822.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002;110:551–561. doi: 10.1016/s0092-8674(02)00905-4. [DOI] [PubMed] [Google Scholar]
- 22.Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol. 2007;5:619–633. doi: 10.1038/nrmicro1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneko T, Nakamura Y, Sato S, Asamizu E, Kato T, Sasamoto S, et al. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 2000;7:331–338. doi: 10.1093/dnares/7.6.331. [DOI] [PubMed] [Google Scholar]
- 24.Köhler S, Michaux-Charachon S, Porte F, Ramuz M, Liautard JP. What is the nature of the replicative niche of a stealthy bug named Brucella? Trends Microbiol. 2003;5:215–219. doi: 10.1016/s0966-842x(03)00078-7. [DOI] [PubMed] [Google Scholar]
- 25.Logwig EM, Hosie AHF, Bourdès A, Findlay K, Allaway D, Karunakaran R, et al. Amino-acid cycling drives nitrogen fixation in the legume-Rhizobium symbiosis. Nature. 2003;422:722–726. doi: 10.1038/nature01527. [DOI] [PubMed] [Google Scholar]
- 26.Luo L, Yao Y, Becker A, Ruberg S, Yu GQ, Zhu JB, Cheng HP. Two new Sinorhizobium meliloti LysR-type transcriptional regulators required for nodulation. J Bacteriol. 2005;187:4562–4572. doi: 10.1128/JB.187.13.4562-4572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mann B, van Opijnen T, Wang J, Obert C, Wang YD, Carter R, et al. Control of virulence by small RNAs in Streptococcus pneumoniae. PLoS Pathog. 2012;8:e1002788. doi: 10.1371/journal.ppat.1002788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McArthur SD, Pulvermacher SC, Stauffer GV. The Yersinia pestis gcvB gene encodes two small regulatory RNA molecules. BMC Microbiol. 2006;6:52. doi: 10.1186/1471-2180-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McClure R, Tjaden B, Genco C. Identification of sRNAs expressed by the human pathogen Neisseria gonorrhoeae under disparate growth conditions. Front Microbiol. 2014;5:456. doi: 10.3389/fmicb.2014.00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller EW, Cao TN, Pflughoeft KJ, Sumby P. RNA-mediated regulation in Gram-positive pathogens: an overview punctuated with examples from the group A Streptococcus. Mol Microbiol. 2014;94:9–20. doi: 10.1111/mmi.12742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyakoshi M, Chao Y, Vogel J. Cross talk between ABC transporter mRNAs via a target mRNA-derived sponge of the GcvB small RNA. EMBO J. 34:1478–1492. doi: 10.15252/embj.201490546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore LW, Chilton WS, Canfield ML. Diversity of opines and opine-catabolizing bacteria isolated from naturally occurring crown gall tumors. Appl Environ Microbiol. 1997;63:201–207. doi: 10.1128/aem.63.1.201-207.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Overlöper A, Kraus A, Gurski R, Wright PR, Georg J, Hess WR, Narberhaus F. Two separate modules of the conserved regulatory RNA AbcR1 address multiple target mRNAs in and outside of the translation initiation region. RNA Biol. 2014;11:624–640. doi: 10.4161/rna.29145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pappas G, Papadimitriou P, Akritidis N, Christou L, Tsianos E. The new global map of human brucellosis. Lancet Infect Dis. 2006;2:91–99. doi: 10.1016/S1473-3099(06)70382-6. [DOI] [PubMed] [Google Scholar]
- 35.Pappas G, Akritidis N, Bosilkovski M, Tsianos E. Brucellosis. N Engl J Med. 2005;22:2325–2336. doi: 10.1056/NEJMra050570. [DOI] [PubMed] [Google Scholar]
- 36.Pini F, Frascella A, Santopolo L, Bazzicalupo M, Biondi E, Scotti C, Mengoni A. Exploring the plant-associated bacterial communities in Medicago sativa L. BMC Microbiol. 2012;12:78. doi: 10.1186/1471-2180-12-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Postic G, Frapy E, Dupuis M, Dubail I, Livny J, Charbit A, Meibom KL. Identification of small RNAs in Francisella tularensis. BMC Genomics. 2010;11:625. doi: 10.1186/1471-2164-11-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prell J, Bourdes A, Kumar S, Lodwig E, Hosie A, Kinghorn S, et al. Role of symbiotic auxotrophy in the Rhizobium-legume symbiosis. PLoS One. 2010;5:e13933. doi: 10.1371/journal.pone.0013933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roop RM, II, Bellaire BH, Valderas MW, Cardelli JA. Adaptation of brucellae to their intracellular niche. Mol Microbiol. 2004;3:621–630. doi: 10.1111/j.1365-2958.2004.04017.x. [DOI] [PubMed] [Google Scholar]
- 40.Roumiantseva ML, Andronov EE, Sharypova LA, Dammann-Kalinowski T, Keller M, Young JP, Simarov BV. Diversity of Sinorhizobium meliloti from the Central Asian Alfalfa Gene Center. Appl Environ Microbiol. 2002;68:4694–4697. doi: 10.1128/AEM.68.9.4694-4697.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schiano CA, Koo JT, Schipma MJ, Caulfield AJ, Jafari N, Lathem WW. Genome-wide analysis of small RNAs expressed by Yersinia pestis identifies a regulator of the Yop-Ysc type III secretion system. J Bacteriol. 2014;196:1659–1670. doi: 10.1128/JB.01456-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlüter JP, Reinkensmeier J, Daschkey S, Evguenieva-Hackenberg E, Janssen S, Jänicke S, et al. A genome-wide survey of sRNAs in the symbiotic nitrogen-fixing Alphaproteobacterium Sinorhizobium meliloti. BMC Genomics. 2010;11:245. doi: 10.1186/1471-2164-11-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma CM, Darfeuille F, Plantinga TH, Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007;21:2804–2817. doi: 10.1101/gad.447207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma CM, Papenfort K, Pernitzsch SR, Mollenkopf HJ, Hinton JC, Vogel J. Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol Microbiol. 81:1144–1165. doi: 10.1111/j.1365-2958.2011.07751.x. [DOI] [PubMed] [Google Scholar]
- 45.Sheehan LM, Budnick JA, Blanchard C, Dunman PM, Caswell CC. A LysR-family transcriptional regulator required for virulence in Brucella abortus is highly conserved among the Alphaproteobacteria. Mol Microbiol. 2015;98:318–328. doi: 10.1111/mmi.13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheehan LM, Caswell CC. A 6-nucleotide regulatory motif within the AbcR small RNAs of Brucella abortus mediates host-pathogen interactions. MBio. 2017:8. doi: 10.1128/mBio.00473-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shimoda N, Toyoda-Yamamoto A, Nagamine J, Usami S, Katayama M, Sakagami Y, Machida Y. Control of expression of Agrobacterium vir genes by synergistic actions of phenolic signal molecules and monosaccharides. Proc Natl Acad Sci U S A. 1990;87:6684–6688. doi: 10.1073/pnas.87.17.6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soutourina OA, Monot M, Boudry P, Saujet L, Pichon C, Sismeiro O, et al. Genome-wide identification of regulatory RNAs in the human pathogen Clostridium difficile. PLoS Genet. 2013;9:e1003493. doi: 10.1371/journal.pgen.1003493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Storz G, Opdyke JA, Wassarman KM. Regulating bacterial transcription with small RNAs. Cold Spring Harb Symp Quant Biol. 2006;71:269–273. doi: 10.1101/sqb.2006.71.033. [DOI] [PubMed] [Google Scholar]
- 51.Tang G, Wang Y, Luo L. Transcriptional regulator LsrB of Sinorhizobium meliloti positively regulates the expression of genes involved in lipopolysaccharide biosynthesis. Appl Environ Microbiol. 2014;80:5265–5273. doi: 10.1128/AEM.01393-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Torres-Quesada O, Millán V, Nisa-Martínez R, Bardou F, Crespi M, Toro N, Jiménez-Zurdo JI. Independent activity of the homologous small regulatory RNAs AbcR1 and AbcR2 in the legume symbiont Sinorhizobium meliloti. PLoS One. 2013;8:e68147. doi: 10.1371/journal.pone.0068147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Torres-Quesada O, Reinkensmeier J, Schlüter JP, Robledo M, Peregrina A, Giegerich R, et al. Genome-wide profiling of Hfq-binding RNAs uncovers extensive post-transcriptional rewiring of major stress response and symbiotic regulons in Sinorhizobium meliloti. RNA Biol. 2014;11:563–579. doi: 10.4161/rna.28239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ulvé VM, Sevin EW, Chéron A, Barloy-Hubler F. Identification of chromosomal Alphaproteobacterial small RNAs by comparative genome analysis and detection in Sinorhizobium meliloti strain 1021. BMC Genomics. 2007;8:467. doi: 10.1186/1471-2164-8-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Urbanowski ML, Stauffer LT, Stauffer GV. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol Microbiol. 2000;37:856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- 56.Valverde C, Livny J, Schlüter JP, Reinkensmeier J, Becker A, Parisi G. Prediction of Sinorhizobium meliloti sRNA genes and experimental detection in strain 2011. BMC Genomics. 2008;9:416. doi: 10.1186/1471-2164-9-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vercruysse M, Fauvart M, Cloots L, Engelen K, Thijs IM, Marchal K, Michiels J. Genome-wide detection of predicted non-coding RNAs in Rhizobium etli expressed during free-living and host-associated growth using a high-resolution tiling array. BMC Genomics. 2010;11:53. doi: 10.1186/1471-2164-11-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Voss B, Hölscher M, Baumgarth B, Kalbfleisch A, Kaya C, Hess WR, et al. Expression of small RNAs in Rhizobiales and protection of a small RNA and its degradation products by Hfq in Sinorhizobium meliloti. Biochem Biophys Res Commun. 2009;390:331–336. doi: 10.1016/j.bbrc.2009.09.125. [DOI] [PubMed] [Google Scholar]
- 59.Vogel J. A rough guide to the non-coding RNA world of Salmonella. Mol Microbiol. 2009;71:1–11. doi: 10.1111/j.1365-2958.2008.06505.x. [DOI] [PubMed] [Google Scholar]
- 60.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilkens S. Structure and mechanism of ABC transporters. F1000Prime Rep. 2015;7:14. doi: 10.12703/P7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilms I, Voss B, Hess WR, Leichert LI, Narberhaus F. Small RNA-mediated control of the Agrobacterium tumefaciens GABA binding protein. Mol Microbiol. 2011;80:492–506. doi: 10.1111/j.1365-2958.2011.07589.x. [DOI] [PubMed] [Google Scholar]
- 64.Winans SC, Ebert PR, Stachel SE, Gordon MP, Nester EW. A gene essential for Agrobacterium virulence is homologous to a family of positive regulatory loci. Proc Natl Acad Sci U S A. 1986;83:8278–8282. doi: 10.1073/pnas.83.21.8278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wright PR, Richter AS, Papenfort K, Mann M, Vogel J, Hess WR, et al. Comparative genomics boosts target prediction for bacterial small RNAs. Proc Natl Acad Sci U S A. 2013;110:E3487–3496. doi: 10.1073/pnas.1303248110. [DOI] [PMC free article] [PubMed] [Google Scholar]