Abstract
Overview
MicroRNAs (miRNAs) have been implicated in neurodegenerative diseases including Parkinson’s disease and Alzheimer’s disease (AD). Here, we evaluated the expression of miRNAs in anterior cingulate (AC; Brodmann area [BA] 24) and primary motor (MO; BA 4) cortical tissue from aged human brains in the University of Kentucky AD Center autopsy cohort, with a focus on dementia with Lewy bodies (DLB).
Methods
RNA was isolated from gray matter of brain samples with pathology-defined DLB, AD, AD+DLB, and low-pathology controls, with n=52 cases initially included (n=23 with DLB), all with low (<4hrs) postmortem intervals. RNA was profiled using Exiqon miRNA microarrays. Quantitative PCR for post-hoc replication was performed on separate cases (n=6 controls) and included RNA isolated from gray matter of MO, AC, primary somatosensory (BA 3), and dorsolateral prefrontal (BA 9) cortical regions.
Results
The miRNA expression patterns differed substantially according to anatomic location: of the relatively highly-expressed miRNAs, 150/481 (31%) showed expression that was different between AC versus MO (at p<0.05 following correction for multiple comparisons), most (79%) with higher expression in MO. A subset of these results were confirmed in qPCR validation focusing on miR-7, miR-153, miR-133b, miR-137, and miR-34a. No significant variation in miRNA expression was detected in association with either neuropathology or sex after correction for multiple comparisons.
Conclusion
A subset of miRNAs (some previously associated with α-synucleinopathy and/or directly targeting α-synuclein mRNA) were differentially expressed in AC and MO, which may help explain why these brain regions show differences in vulnerability to Lewy body pathology.
Keywords: Synuclein, noncoding, epigenetic, neuropathology, neuroanatomy, gender, miR-15/107, miR-107, miR-15, miR-16, miR-133, brain, sensory, frontal
Introduction
Neurodegenerative diseases (NDs) are devastating brain disorders with an enormous impact on public health. Among the prevalent NDs are Alzheimer’s disease (AD) and dementia with Lewy bodies (DLB), and “gold-standard” diagnoses can only be made with certainty at autopsy (Beach et al., 2012; Nelson et al., 2009b; Nelson et al., 2010a). According to consensus-based recommendations, DLB pathology includes aberrant cerebral cortical deposits of insoluble α-synuclein/Lewy bodies (McKeith et al., 2004; McKeith et al., 2017). The prevalence of Lewy body pathology is 10%–30% in most series of dementia patients (Heidebrink, 2002; Neltner et al., 2016; Rahkonen et al., 2003; Sonnen et al., 2009; Zaccai et al., 2005). It is relatively challenging to study “pure” DLB cases since most persons with cortical Lewy body pathology also have comorbid AD pathology (Irwin et al., 2017; Nelson et al., 2009b; Nelson et al., 2010a; Nelson et al., 2010b). Notably, focused duplication of the gene (SNCA) that encodes α-synuclein is enough to produce α-synuclein/Lewy body pathology (Gwinn et al., 2011; Nishioka et al., 2006; Singleton et al., 2003), so, other gene regulatory factor(s) that cause increased α-synuclein expression could potentiate the pathology.
An enigmatic characteristic of NDs is that they tend to affect specific brain areas initially, and then progress to additional brain regions in a predictable sequence. Symptoms reflect the spatiotemporal progression. In DLB, Lewy bodies are observed in the anterior cingulate gyrus (Brodmann Area [BA] 24; AC) early in the disease (Thal et al., 2004). By contrast, the primary motor cortex (BA 4; MO) and primary somatosensory (BA 3) neocortical regions are relatively resistant to pathology (Thal et al., 2004).
Brain region-specific gene expression may contribute to the initiation and/or propagation of pathology in some parts of the brain, while causing other anatomically-defined brain areas to be less vulnerable. Testing this hypothesis requires factoring in gene regulatory phenomena in addition to mRNA levels, because mRNA is an imperfect proxy for protein levels, and transcription is only one among many different nodes of gene expression regulation in the human brain (Nelson and Keller, 2007). Thus, new information about factors that affect gene regulation post-transcriptionally may be helpful to achieve better understanding of ND pathogenesis.
MicroRNAs (miRNAs) are short (~22 nucleotides in length) noncoding RNAs that have been implicated in NDs (Liu et al., 2008; Nelson et al., 2008a). Encoded by small genes that can be located practically anywhere in the genome (Londin et al., 2015), miRNAs regulate gene expression in multiple ways – predominantly by interacting with mRNA “targets” post-transcriptionally – and each miRNA has the potential to regulate dozens or even hundreds of targeted mRNAs (Kiriakidou et al., 2004; Liu et al., 2008; Wang et al., 2010). Relative to protein-coding genes, the human brain miRNA repertoire is smaller, with under 1000 moderately- or highly-expressed brain transcripts (Hebert and Nelson, 2012). However, individual miRNAs can be expressed at up to an order of magnitude higher transcript copies than highly expressed mRNAs (Hebert and Nelson, 2012). Furthermore, individual miRNAs have previously been implicated in AD and Lewy body pathology (Hebert and De Strooper, 2009; Nelson et al., 2008a; Pietrzak et al., 2016; Ubhi et al., 2014; Weinberg et al., 2015).
Fundamental characteristics of any miRNA include the cells it is expressed in and the factors that are associated with expression variance. For this reason, miRNA expression profiling is an important tool for understanding miRNA biology in health and in disease states. Although there have been prior expression profiling studies of miRNAs in human tissues, many brain areas have not been assessed thoroughly to date, and there are many extant unanswered questions. For example, the miRNA profiles in AC and MO regions have not been compared with each other and the relevance of miRNAs to DLB is unknown.
To address these issues, we evaluated the expression of miRNAs in well-characterized human brains. We focused specifically on AC and MO tissue from aged human brains that had been snap-frozen in liquid nitrogen at autopsy from the University of Kentucky Alzheimer’s Disease Center (UK-ADC) cohort. We were interested in testing whether the miRNA expression patterns varied according to anatomic region, sex, and/or neuropathology.
Results
Human brain samples were obtained at autopsies after short post-mortem interval (PMI), followed by dissection of gray matter for RNA isolation and miRNA expression profiling. The rationale for the areas of brain chosen for miRNA analyses is depicted in Fig. 1. Some aspects of RNA quality are depicted in Fig. 2. Patient characteristics, including clinical and pathological parameters, and PMI, are shown in summary form in Table 1. Provided in supplemental material are the following additional data: case-by-case clinical, pathological, PMI, and RNA Integrity Number (RIN) data (Supplemental Table 1). Note that a total of 7 microarrays failed QC due to RIN values <4.0. We also note that, as we have reported previously, “pure DLB” is relatively rare and predominantly seen in males (Nelson et al., 2009b; Nelson et al., 2010a; Nelson et al., 2010b), whereas most cases with DLB pathology have concomitant AD pathology (Irwin et al., 2017). Thus, a group of cases with AD pathology but no Lewy bodies was used as a control cohort. The total number of tested subjects with DLB pathology was 23, of which 16 had substantial comorbid AD pathology.
Figure 1. Rationale for selecting brain tissue for RNA extraction and miRNA profiling in the current study.
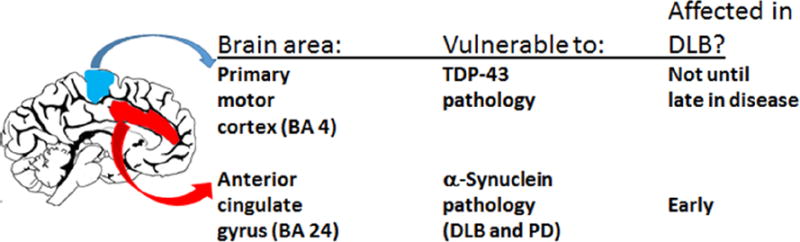
Particular brain areas show different vulnerabilities to specific brain diseases. Whereas the anterior cingulate gyrus (AC; Brodmann area 24) is affected early in DLB, the primary motor cortex (MO; Brodmann area 4) is generally not affected until late in the disease, if at all.
Figure 2. Focus on technical factors using high-quality RNA probed with state-of-the-art miRNA microarrays.
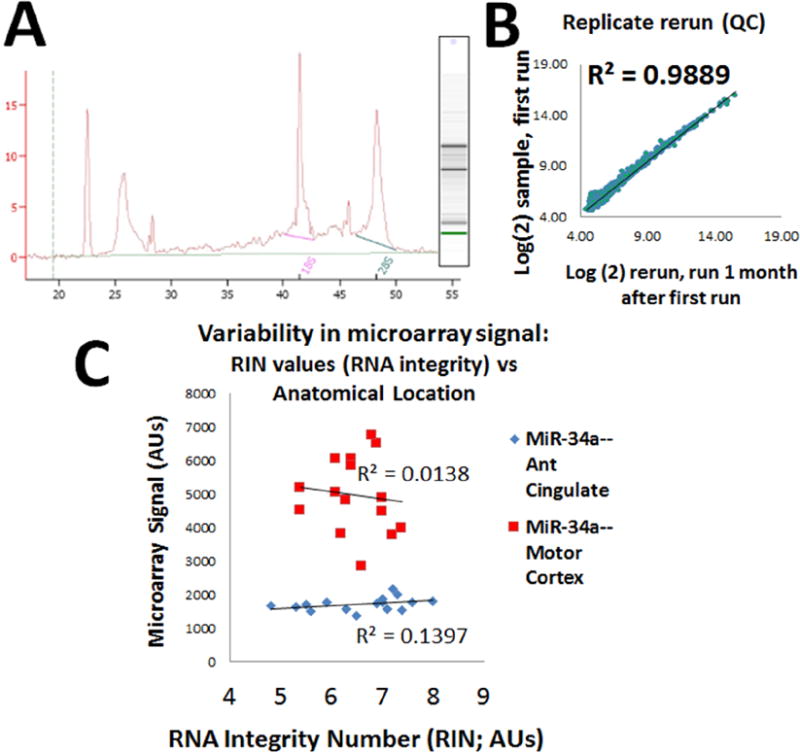
Quality control (QC) included Agilent BioAnalyzer (electropheragram, A), and the RNA Integrity Number (RIN) were mostly between 6-8, with inclusion criteria of RIN >4.0. Gray matter dissection was performed as described previously (Nelson et al., 2008b; Nelson and Wang, 2010; Wang et al., 2008c; Wang et al., 2011). For added QC, we performed extra technical replicates of six different samples a month after the primary microarrays were run and these showed within-case correlations (B is a representative correlation), demonstrating outstanding replicability. C. Variability in microarray signals correlated poorly with RIN number indicating that RNA integrity (>=4) and RNA degradation had weak impact on miRNA expression results in our preliminary samples. Shown here are results for miR-34a which are representative.
Table 1. Summary information on cases used in the current study.
For detailed information on each case, see Supplemental Table 1
| Pathology-defined case categories | n | n, F | Avg age at death | PMI (Hrs Avg) |
|---|---|---|---|---|
| Ctrl | 16 | 8 | 82.4 | 2.8 |
| DLB | 7 | 2 | 82.7 | 2.6 |
| AD | 13 | 8 | 89.8 | 2.5 |
| AD+DLB | 16 | 10 | 79.0 | 2.7 |
All the miRNA expression data for each case are provided in Supplemental Tables 2 and 3. All of the values of miRNA expression depicted in Tables 2–5 were the result of a statistical approach that enabled the use of microarray data from the two rounds of evaluations, as described in the Methods section. A total of 481 relatively high-expressing miRNAs met the inclusion criterion (log-2 expression >7 in at least one data set) and were the basis of further analyses.
Table 2. A subset of miRNAs that show differential expression in anterior cingulate gyrus and motor cortex (top 10 up-regulated in motor cortex [blue] and top 10 up-regulated in anterior cingulate [yellow] by p-value; note that 130 other differentially expressed miRNAs are presented in Supplemental Table 4).
Values shown are estimates of the average miRNA expression value for the corresponding combination of sex, brain region, and pathology adjusted for age and year.
| RNA | Adj Pval For Brain* | Anterior Cingulate | Motor Cortex | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Males | Females | Males | Females | ||||||
| DLB | Non-DLB | DLB | Non-DLB | DLB | Non-DLB | DLB | Non-DLB | ||
| hsa-miR-31-3p | 5.21E-07 | 6.6021 | 6.7391 | 6.5896 | 6.684 | 6.9523 | 6.9757 | 6.9071 | 7.0142 |
| hsa-miR-31-5p | 3.18E-06 | 7.7926 | 7.8246 | 7.82 | 7.7891 | 8.0879 | 8.1552 | 8.0402 | 8.1721 |
| hsa-miRPlus-G1246-3p | 5.90E-06 | 6.971 | 7.111 | 7.0029 | 7.0199 | 7.3388 | 7.2277 | 7.2391 | 7.2529 |
| hsa-miR-34a-5p | 6.84E-06 | 7.5044 | 7.2532 | 7.3762 | 7.1129 | 8.0542 | 8.0157 | 8.0519 | 8.1428 |
| hsa-miR-133b | 8.17E-06 | 6.0775 | 5.9338 | 6.1173 | 5.9231 | 6.6235 | 6.3554 | 6.4687 | 6.5011 |
| hsa-miR-338-3p | 8.65E-06 | 9.5422 | 9.7136 | 9.3534 | 9.577 | 9.9171 | 9.9867 | 9.9809 | 10.1153 |
| hsa-miR-584-5p | 4.56E-05 | 6.9292 | 6.9521 | 7.0193 | 6.7815 | 7.2134 | 7.1281 | 7.2317 | 7.1843 |
| hsa-miR-365a/b-3p | 4.66E-05 | 7.862 | 8.013 | 7.8536 | 7.9969 | 8.2927 | 8.1894 | 8.1764 | 8.2443 |
| hsa-miR-758-3p | 6.63E-05 | 6.2306 | 6.1672 | 6.1978 | 6.2106 | 6.049 | 5.9753 | 5.8999 | 6.0197 |
| hsa-miR-185-5p | 6.63E-05 | 8.1934 | 8.3073 | 8.187 | 8.2434 | 8.5712 | 8.4618 | 8.4916 | 8.5461 |
| hsa-miR-33b-5p | 6.63E-05 | 6.7699 | 6.778 | 6.7368 | 6.749 | 7.1108 | 7.0492 | 7.0398 | 7.0707 |
| hsa-miR-135a-5p | 7.47E-05 | 8.3028 | 8.6913 | 8.4991 | 8.7826 | 8.1154 | 8.0386 | 7.8877 | 8.0347 |
| hsa-miR-551b-3p | 7.90E-05 | 7.0652 | 7.3022 | 7.2082 | 7.3993 | 7.0217 | 6.7284 | 6.7882 | 6.7126 |
| hsa-miR-218-5p | 0.000299 | 7.6044 | 7.7582 | 7.658 | 7.773 | 7.4514 | 7.289 | 7.3475 | 7.2747 |
| hsa-miR-1185-5p | 0.001029 | 6.6961 | 6.6594 | 6.6661 | 6.7199 | 6.5659 | 6.449 | 6.4068 | 6.5388 |
| hsa-miR-154-3p | 0.001091 | 6.1423 | 6.205 | 6.1887 | 6.2029 | 6.0806 | 5.9777 | 5.8626 | 6.0123 |
| hsa-miR-543 | 0.001818 | 6.1106 | 6.1487 | 6.0874 | 6.1801 | 6.0588 | 5.8923 | 5.9416 | 5.9761 |
| hsa-miR-132-5p | 0.002194 | 6.8244 | 6.9952 | 6.8267 | 6.9137 | 6.7527 | 6.7007 | 6.5669 | 6.7445 |
| hsa-miR-137 | 0.002194 | 6.3179 | 6.8285 | 6.5601 | 6.8326 | 6.1137 | 5.7082 | 5.9383 | 5.8436 |
| hsa-miR-7-5p | 0.002496 | 10.3032 | 10.5859 | 10.3652 | 10.6905 | 10.1936 | 10.1601 | 10.0723 | 10.0952 |
-p-value is adjusted for multiple comparisons
Table 5. MiRNA expression for miRNAs previously implicated in α-synucleinopathy; those up-regulated in motor cortex are shaded blue and those up-regulated in anterior cingulate gyrus are shaded yellow.
Values shown are estimates of the average miRNA expression value for the corresponding combination of sex, brain region, and pathology adjusted for age and year.
| RNA | Anterior Cingulate | Motor Cortex | Adj p-value for brain* | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Males | Females | Males | Females | ||||||
| DLB | Non-DLB | DLB | Non-DLB | DLB | Non-DLB | DLB | Non-DLB | ||
| hsa-miR-133b | 6.0775 | 5.9338 | 6.1173 | 5.9231 | 6.6235 | 6.3554 | 6.4687 | 6.5011 | 8.17E-06 |
| hsa-miR-133a-3p | 5.848 | 5.7429 | 5.8764 | 5.802 | 6.2502 | 6.0379 | 6.1812 | 6.14 | 9.91E-05 |
| hsa-miR-34c-5p | 6.0231 | 6.0994 | 6.0893 | 5.9919 | 6.2591 | 6.2074 | 6.2372 | 6.2648 | 0.0001 |
| hsa-miR-20a-5p | 9.6119 | 9.9028 | 9.6552 | 9.8962 | 9.9988 | 10.088 | 9.904 | 10.0526 | 0.0016 |
| hsa-miR-7-5p | 10.3032 | 10.5859 | 10.3652 | 10.6905 | 10.1936 | 10.1601 | 10.0723 | 10.0952 | 0.0025 |
| hsa-miR-34b-5p | 5.6875 | 5.5041 | 5.4729 | 5.4263 | 5.805 | 5.7586 | 5.6199 | 5.7868 | 0.0048 |
| hsa-miR-153-3p | 7.3275 | 7.3024 | 7.3644 | 7.4377 | 7.2315 | 7.0217 | 6.9554 | 7.1223 | 0.0216 |
| hsa-miR-320a | 7.8979 | 8.0834 | 7.9415 | 8.0285 | 8.2249 | 8.0812 | 8.1417 | 8.1644 | 0.0267 |
| hsa-miR-7-2-3p | 6.171 | 6.0477 | 6.1892 | 5.9223 | 6.3006 | 6.1158 | 6.3905 | 6.2319 | 0.0301 |
| hsa-miR-373-3p | 5.5635 | 5.4798 | 5.5206 | 5.5004 | 5.5702 | 5.5429 | 5.6026 | 5.6076 | NS |
| hsa-miR-320b | 7.8607 | 8.0713 | 7.9686 | 8.0195 | 8.1417 | 8.0489 | 8.0914 | 8.1176 | NS |
| hsa-miR-205-5p | 6.3984 | 6.3697 | 6.3739 | 6.459 | 6.4445 | 6.408 | 6.4608 | 6.4703 | NS |
| hsa-miR-128-3p | 8.3193 | 8.4338 | 8.4448 | 8.5202 | 8.4255 | 8.2395 | 8.2868 | 8.2694 | NS |
| hsa-miR-301b | 5.5309 | 5.5481 | 5.5393 | 5.5584 | 5.5139 | 5.5696 | 5.6117 | 5.6708 | NS |
| hsa-miR-20b-3p | 6.3735 | 6.4784 | 6.4437 | 6.4443 | 6.45 | 6.4575 | 6.4907 | 6.4973 | NS |
| hsa-miR-20b-5p | 7.808 | 8.1349 | 7.875 | 8.1615 | 7.922 | 8.1938 | 7.9443 | 8.1676 | NS |
| hsa-miR-224-3p | 6.3124 | 6.399 | 6.4666 | 6.2922 | 6.4324 | 6.3306 | 6.4809 | 6.3485 | NS |
| hsa-miR-26b-5p | 9.3938 | 9.6238 | 9.4095 | 9.6125 | 9.4779 | 9.6279 | 9.4428 | 9.6327 | NS |
| hsa-miR-373-5p | 5.7349 | 5.6502 | 5.6515 | 5.672 | 5.72 | 5.6069 | 5.7453 | 5.6778 | NS |
| hsa-miR-214-3p | 6.4538 | 6.8021 | 6.9107 | 6.2816 | 6.7823 | 6.4591 | 6.6973 | 6.4082 | NS |
-p-value is adjusted for multiple comparisons
MiRNAs differentially expressed by brain area
Even after correcting for multiple comparisons, 150 of the relatively highly-expressed miRNAs were detected at different levels in the MO and AC samples across all conditions and microarray iterations. Of these, 119 (79%) were higher expressed in MO (versus AC), whereas 31 (21%) were higher expressed in AC. Table 2 shows the 10 miRNAs most highly expressed in MO and the 10 miRNAs most highly expressed in AC in terms of the statistical comparison. All of the miRNAs found to be differentially expressed at a p<0.05 level after correction for multiple comparisons are listed in Supplemental Table 4.
MiRNAs with variation that is associated with sex and DLB pathology
After correcting for multiple comparisons to decrease likelihood of false discovery, there was no miRNA that showed significant differences associated with either sex or pathology-confirmed DLB status. Shown in Table 3 are the miRNAs showing nominal association with sex or DLB status (p < 0.05, but not corrected for multiple comparisons).
Table 3. No miRNAs showed differential expression in sex by DLB after multiple comparison adjustment.
The two miRNAs presented here have unadjusted p-values less than 0.05 and may show a trend. estimates of the average miRNA expression value for the corresponding combination of sex, brain region, and pathology adjusted for age and year.
| RNA | Unadj Pval For Sex*DLB | Anterior Cingulate | Motor Cortex | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Males | Females | Males | Females | ||||||
| DLB | Non-DLB | DLB | Non-DLB | DLB | Non-DLB | DLB | Non-DLB | ||
| hsa-miR-516a-3p/hsa-miR-516b-3p | 0.022494 | 6.4476 | 6.2454 | 6.1295 | 6.2893 | 6.6242 | 6.312 | 6.4782 | 6.4181 |
| hsa-miR-4328 | 0.046346 | 6.8247 | 6.5422 | 6.7308 | 6.7731 | 6.8403 | 6.6068 | 6.6816 | 6.8325 |
MiR-15/107 family
Members of the miR-15/107 family of miRNAs, which share regulatory targets (Finnerty et al., 2010), have been linked to both AD pathology and to regulation of α-synuclein and/or synucleinopathy (Alderman and Yang, 2016; Gui et al., 2015; Li et al., 2014b; Liu et al., 2014; Liu et al., 2016; Moncini et al., 2016; Nelson and Wang, 2010; Parsi et al., 2015; Wang et al., 2008b; Yao et al., 2010; Zhang and Cheng, 2014). Some of the miR-15/107 miRNAs also are enriched in brain (Wang et al., 2014). Shown on Table 4 are microarray values of members of the miR-15/107 family: miR-15a, miR-15b, miR-16, miR-103, miR-107, miR-424, miR-497, miR-503, and miR-646. None of these miRNAs showed an association with sex and DLB pathology that was statistically significant. However, three of the four highest-expressing miR-15/107 family genes in the current sample – hsa-miR-15a-5p, hsa-miR-15b-5p, and hsa-miR-16-5p – were expressed at higher levels in MO than AC. MiR-107 showed a trend for lower expression in association with AD pathology (not shown), as expected (Nelson and Wang, 2010; Wang et al., 2008b).
Table 4. MiRNA expression for miRNAs in the miR-15/107 family; those up-regulated in motor cortex are shaded blue.
Estimates of the average miRNA expression value for the corresponding combination of sex, brain region, and pathology adjusted for age and year.
| RNA | Anterior Cingulate | Motor Cortex | Adj p-value for brain | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Males | Females | Males | Females | ||||||
| DLB | Non-DLB | DLB | Non-DLB | DLB | Non-DLB | DLB | Non-DLB | ||
| hsa-miR-16-5p | 10.7802 | 10.9397 | 10.7707 | 10.9432 | 11.0162 | 11.1346 | 10.8987 | 11.1181 | 0.0348 |
| hsa-miR-103a-3p | 10.3849 | 10.4951 | 10.4098 | 10.5276 | 10.4741 | 10.5314 | 10.3938 | 10.5017 | NS |
| hsa-miR-15a-5p | 9.7437 | 9.9063 | 9.7371 | 9.8953 | 9.9933 | 10.0579 | 9.8941 | 10.0603 | 0.0133 |
| hsa-miR-15b-5p | 9.0169 | 9.2194 | 8.9979 | 9.2099 | 9.3371 | 9.3348 | 9.2122 | 9.3747 | 0.0017 |
| hsa-miR-107 | 9.0015 | 9.2017 | 9.0256 | 9.255 | 9.0791 | 9.146 | 9.0793 | 9.1631 | NS |
| hsa-miR-424-5p | 7.1216 | 7.164 | 7.0938 | 7.1006 | 7.1603 | 6.9496 | 7.0832 | 7.0907 | NS |
| hsa-miR-497-5p | 6.2485 | 6.1855 | 6.2816 | 6.2125 | 6.3988 | 6.1022 | 6.2365 | 6.2629 | NS |
| hsa-miR-503-3p | 5.5115 | 5.4314 | 5.4414 | 5.3967 | 5.5273 | 5.4061 | 5.3884 | 5.4512 | NS |
| hsa-miR-503-5p | 5.3822 | 5.3436 | 5.4269 | 5.3724 | 5.3249 | 5.3651 | 5.3579 | 5.3747 | NS |
| hsa-miR-646 | 4.8733 | 4.7618 | 4.8528 | 4.7567 | 4.7535 | 4.764 | 4.68 | 4.8211 | NS |
-p-value is adjusted for multiple comparisons
Other miRNAs previously implicated in α-synucleinopathy
Although we found that a subset of miRNAs were differentially expressed in the AC and the MO (see Table 2), these altered miRNAs were different than those previously implicated in α-synucleinopathy: miR-7, miR-153, miR-1643, miR-34b, miR-34c, miR-214, miR-26b, miR-320, miR-20, miR-128, miR-133, miR-433, miR-205, miR-224, miR-301b, and miR-373 (Choi et al., 2014; de Mena et al., 2010; Doxakis, 2010; Fragkouli and Doxakis, 2014; Janeczek and Lewohl, 2013; Junn et al., 2009; Kabaria et al., 2015; Kim et al., 2013; Kong et al., 2015; Li et al., 2014a; Ma et al., 2013; Majidinia et al., 2016; Mouradian, 2012; Niu et al., 2016; Patil et al., 2015; Recasens et al., 2016; Tagliafierro et al., 2017; Wang et al., 2008a; Wang et al., 2015; Xie and Chen, 2016; Zhou et al., 2016). Results for these miRNAs are shown in Table 5. None of these miRNAs showed association with sex or pathology that was statistically significant. However, a subset of these – hsa-miR-133a/b-3p, hsa-miR-34c-5p, hsa-miR-20a-5p, hsa-miR-7-5p, hsa-miR-34b-5p, hsa-miR-153-3p, hsa-miR-320a, hsa-miR-7-2-3p – were expressed differentially in the AC and MO regions.
Replication (qPCR) experiment for miRNA expression variance associated with anatomical areas
Quantitative PCR for post-hoc replication was performed on six low-pathology control cases (Fig. 3). As these experiments were performed for the purpose of replication, none of these cases were among those used in the microarray experiments. In addition to evaluating RNA from MO and AC in these cases, we also analyzed RNA from primary somatosensory and dorsolateral frontal cortical regions. Average age of death of the six individuals was 80.5 years, average PMI was 3.5 hrs (See summary data on the six patients in Supplemental Table 5). Tested with TaqMan single-tube miRNA RT-qPCR were the following miRNAs: miR-133b, miR-137, miR-153, miR-34a, and miR-7. As was found with the microarrays, miR-133b and miR-34a were expressed higher in the MO than AC, while miR-7 and miR-137 were expressed at lower level in MO than AC. Intriguingly, and extending the results from the microarrays to study new brain areas, primary sensory cortex (BA 3) also had relatively high miR-133b and miR-34a and low miR-7 and miR-137. By contrast, frontal cortex (BA9) had relatively low miR-133b and miR-34a and high miR-7 and miR-137. Some of the specific predicted miRNA recognition sites on α-synuclein mRNA (with miRNAs that are expressed at relatively low levels in AC) are shown in Fig. 4. A schematic depiction of the overall hypotheses tested and our interpretation of the data are presented in Fig. 5.
Figure 3. RT-qPCR results on a replication cohort (n=6) separate from those studied with miRNA microarrays, in order to compare the expression of miRNAs in primary sensorimotor cortex (SEN—blue bars); primary motor cortex (MO–orange); dorsolateral frontal cortex (FRON–gray); and anterior cingulate gyrus (AC–yellow).
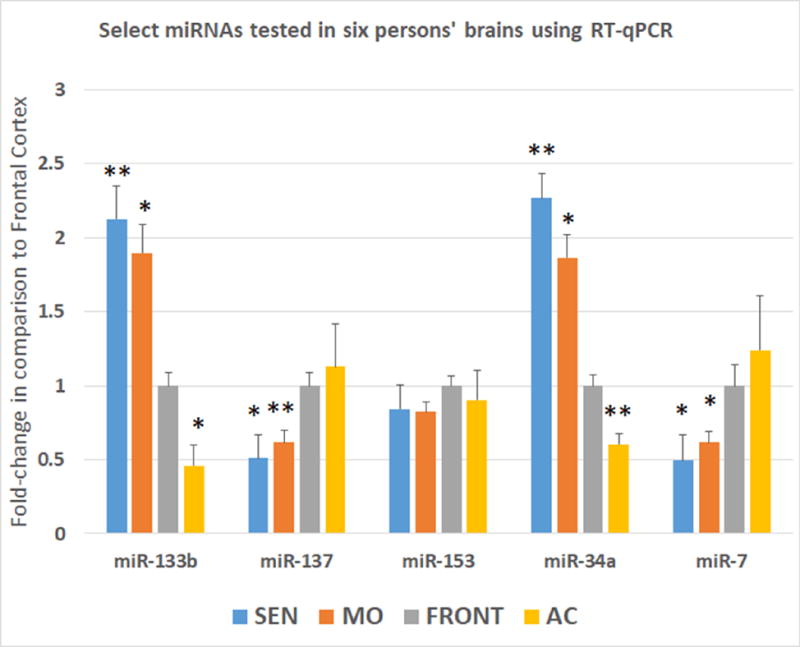
A subset of miRNAs were chosen that had differential expression in MO and AC according to the microarray studies, and had also been implicated in Lewy body disease and/or α-synuclein regulation. Note that miR-133b and miR-34a were expressed at higher levels in primary somatosensory cortex and motor cortex, whereas miR-137 and miR-7 were expressed at lower levels in these brain areas. However, miR-153 did not show differential expression in these samples. *-p<0.05; **-p<0.01 using Student’s t-test. Error bars = StDev.
Figure 4.
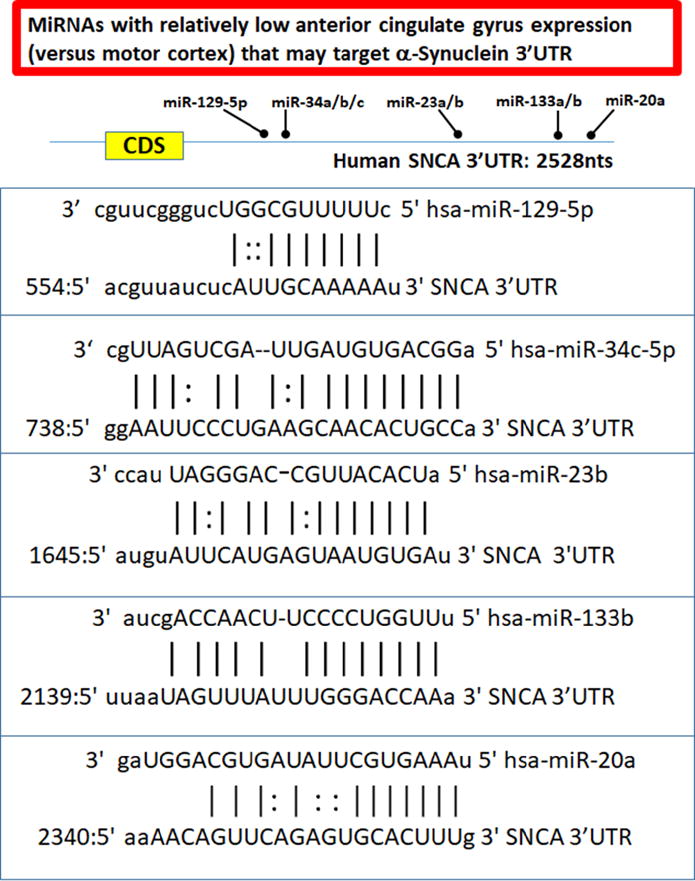
One hypothesis for why neurons in the anterior cingulate gyrus are vulnerable to α-synuclein/Lewy body pathology is that relatively low expression of miRNAs that target the 3′-untranslated region (3′UTR) of α-synuclein in the anterior cingulate gyrus could be partly responsible. The results of the current study were correlated with a miRNA web-based miRNA/mRNA target prediction algorithm (Betel et al., 2010; John et al., 2004) to highlight the potential target sites on the 3′UTR of the SNCA transcript that may be relevant to this hypothesis. These putative miRNA recognition elements all are “conserved miRNAs with good mirSVR scores” according to the website (http://www.microrna.org/microrna/faq.do).
Figure 5.

Schematic depiction of three separate hypotheses related to brain miRNA expression that could help explain differences in vulnerability to neurodegenerative diseases that are at least partially sex-specific and region-specific. HYPOTHESIS 1: the combination of underlying pathogenic mechanisms and the pathologic phenotype alter the neurochemical milieu and result in a condition-specific miRNA profile. The current study did not find support for this hypothesis with the caveat that the sample sizes were relatively small and thus statistically underpowered to correlate subtle variations in miRNA expression patterns with various pathologic and demographic parameters. HYPOTHESIS 2: there are sex-specific miRNA expression patterns that help to explain the differential vulnerability of males and females to subtypes of neurodegenerative diseases. The current study did not find evidence of robust sex-specific miRNA expression patterns in the sampled brain regions. HYPOTHESIS 3: there are neuroanatomic area-specific patterns of miRNA expression that may help cause those brain regions to have region-specific vulnerability to pathologic phenotypes. Since there were robust differences detected in miRNA patterns across brain regions, including among miRNAs that are hypothesized to target disease-relevant transcripts, we interpret the data from the current study to be compatible with this hypothesis.
Discussion
The purpose of the current study was to test whether variation in miRNA levels detected in human brain tissue was associated strongly with any of the following: DLB pathology; sex; or neuroanatomical region (AC vs MO). The hypotheses that were being tested, and our interpretation of the results, are depicted in Fig. 5. Of these factors, we only found evidence of the anatomical region being associated strongly with variation in miRNA expression: many miRNAs were differentially expressed in AC and MO. For example, miR-133b and miR-34a were expressed at relatively high levels in primary motor and somatosensory cortices, whereas miR-7 and miR-137 were expressed at higher levels in AC and frontal cortex. These data are compatible with the hypothesis that brain areas with lower vulnerability to Lewy body pathology express higher miR-133b and miR-34a, but lower levels of miR-137 and miR-7.
This study had technical and theoretical limitations. Whereas the miRNA microarray results were referred to as a proxy for “miRNA expression”, there are other factors, including miRNA stability/degradation (Sethi and Lukiw, 2009) and miRNA expression platform-related idiosyncrasies, that influenced our results. Some of the experimental conditions that affected RNA stability were at least partly mitigated by the short PMI (<4hrs), and the samples having been snap-frozen in liquid nitrogen at autopsy. However, the presence of purely technical variation introduced by the profiling platform is underscored by the different results from the two iterations of the miRNA microarray, and we have seen even greater variation between different platforms previously (Nelson et al., 2008b; Wang et al., 2008c). Microarrays, like PCR-based and “deep sequencing” methods, each have biases (Hebert and Nelson, 2012). For these reasons, we posit that there is no true gold-standard miRNA expression profiling method and the different methods must be used to complement each other, as in the current study using qPCR to validate and replicate the microarray results. Although our study represents one of the larger miRNA microarray studies to date focused on human brain, we did not have a sample size appropriate for evaluating subtle, or context-specific, associations between tested potential dependent variables and miRNA expression. This problem is exacerbated because we are testing multiple parameters and also many different miRNAs, so that substantial statistical corrections were required to account for the multiple tests.
MiRNAs have been previously linked to neurodegeneration (Delay and Hebert, 2011; Hebert and De Strooper, 2007; Nelson et al., 2008a; Rademakers et al., 2008) and implicated in α-synucleinopathic conditions (Asikainen et al., 2010; Kim et al., 2007; Lim et al., 2010; Minones-Moyano et al., 2011; Schonrock et al., 2010). This is one of the first studies published to date that has focused on DLB and miRNA expression (see (Hebert et al., 2013)). To date there is no miRNA that is specifically linked to DLB as far as we know. Correspondingly, we did not discover a strong association between DLB pathology and any particular miRNA’s expression. However, the finding of region-specific miRNA pattern may be clinically relevant to DLB and other diseases.
Lewy body pathology apparently spreads through the brain in a manner that is predictable; the AC is affected in relatively early stages of DLB, whereas the MO is generally not affected until late stages (Dickson et al., 2009; Toledo et al., 2016). This spatiotemporal sequence is inferred by cross-sectional (autopsy) studies and awaits confirmation until suitable biomarkers can be performed in longitudinal studies of living persons. The reason(s) for this common pattern of disease progression are unknown.
MiRNAs have been identified as “master regulators” of gene expression (Garofalo and Croce, 2011; Leung and Sharp, 2006) and so, theoretically, the different expressions of miRNAs in AC and MO could possibly be a factor underlying the differential vulnerability of these brain regions to Lewy body pathology. If this were true, then, the impact of a given miRNA, and a given brain area, could differ dramatically according to the circumstances—for example, whereas the MO is affected late in DLB, the same cortical region may be affected relatively early in persons showing clinical and pathological features associated with the ALS/FTLD disease spectrum (Brettschneider et al., 2013).
Prior studies of gene expression in human brains can be correlated with our results. Particular miRNAs have been shown to be potential regulators of α-synuclein expression in the brain (Choi et al., 2014; de Mena et al., 2010; Doxakis, 2010; Fragkouli and Doxakis, 2014; Janeczek and Lewohl, 2013; Junn et al., 2009; Kabaria et al., 2015; Kim et al., 2013; Kong et al., 2015; Li et al., 2014a; Ma et al., 2013; Majidinia et al., 2016; Mouradian, 2012; Niu et al., 2016; Patil et al., 2015; Recasens et al., 2016; Tagliafierro et al., 2017; Wang et al., 2008a; Wang et al., 2015; Xie and Chen, 2016; Zhou et al., 2016). Among these, the following miRNAs were shown to be differentially expressed in MO and AC in the current study: hsa-miR-133a/b-3p, hsa-miR-34c-5p, hsa-miR-20a-5p, hsa-miR-7-5p, hsa-miR-34b-5p, hsa-miR-153-3p, hsa-miR-320a, and hsa-miR-7-2-3p. Using another approach, Pietrzak et al assessed RNA expression in the AC from eight DLB subjects and 10 controls, and then applied bioinformatics to identify potential miRNA binding sites enriched in mRNAs (Pietrzak et al., 2016). This method yielded a list of candidate miRNAs that may participate in DLB pathogenesis. Among the 10 miRNA candidates identified (Pietrzak et al., 2016) – miR-25, miR-124, miR-506, miR-363, miR-82, miR-26a, miR-367, miR-1297, miR-140, miR-936 – we only found evidence that one (miR-25) was differentially expressed in AC and MO areas.
We reported previously that males are approximately three times more likely than females to die with relatively ‘pure’ neocortical Lewy body pathology (Nelson et al., 2010b). We also showed that clinicians’ knowledge of this sex-linked dimorphism in Lewy body pathology would improve clinical diagnostic accuracy (Nelson et al., 2010b). Smaller prior studies showed the same trend – males have higher DLB/cortical Lewy body risk (Barker et al., 2002; Fujishiro et al., 2008; Hishikawa et al., 2003; Kraybill et al., 2005; Weiner et al., 1996) and also higher Parkinson’s disease risk (Shulman and Bhat, 2006; Shulman, 2007). MiRNA expression is one possible explanation for sex differences; sexually dimorphic and sex hormone-regulated miRNA expression have been described (Bannister et al., 2009; Bizuayehu et al., 2012; Cutting et al., 2012; Jalava et al., 2012; Murata et al., 2010; Zhou et al., 2011) but these phenomena were previously not studied in human brain. In the present study, we could not find compelling evidence for sexually dimorphic miRNA expression in either AC or MO regions.
In conclusion, the current study was designed to detect large effect size phenomena related to variation in miRNA expression in human AC and MO. We analyzed relatively high-quality RNA derived from well characterized human brains. Using both miRNA microarrays and post-hoc studies with qPCR, we identified miRNAs that are differentially expressed – across sex and disease states – between AC and MO, and may represent part of the reason for the differential vulnerability of those two brain regions to brain pathology. It is hoped that the data we are providing with this manuscript will enable other researchers to both generate and test hypotheses related to the roles of brain miRNAs in health and disease.
Methods and materials
RNA isolation from a human cerebral cortex
Samples were derived from short-PMI autopsies, representing different NDs as defined according to pathology. All methods were in compliance with a University of Kentucky IRB protocol. Premortem clinical evaluations and pathological assessments were as described previously (Nelson et al., 2007; Schmitt et al., 2000; Wang et al., 2008c). Tissue used for pathologic evaluation was dissected from MO and AC (immediately adjacent to the tissues sampled for RNA studies), immersion-fixed in formalin and evaluated for AD- and DLB-type pathology using conventional methods (Nelson et al., 2009a). None of these cases has ever been evaluated previously using a microarray or any other high-throughput profiling method. The inclusion criteria that were applied for this convenience sample of DLB brains: PMI <4hrs; no evidence of frontotemporal dementia; no cancer in the brain parenchyma; and no large infarctions in the brain, or microinfarcts found within 3cm of the brain tissue samples. For broader inclusion/exclusion criteria of this autopsy cohort, see (Schmitt et al., 2012). Criteria of operationalizing AD was Braak NFT stage>III with Aβ plaques more than “Sparse”, whereas controls had Braak NFT stages <IV and had intact cognition in their last clinical examination.
RNA was isolated as previously described in detail (Wang et al., 2008b; Wang et al., 2008c). Briefly, RNA was extracted from the gray matter of snap-frozen cerebral cortical brain tissue. All biochemical analyses were performed blind with respect to patient information. Tissue (0.5–2 gms) that had been snap-frozen in liquid nitrogen and then transferred to a −80° C freezer was thawed in ice-cold isotonic lysis buffer with RNAsin (Promega, Madison, WI; 250 U/ml) and Complete protease inhibitor pills (Roche, Basel Switzerland). Prior to RNA extraction (as the tissue thawed), meninges and large blood vessels were removed and gray matter was carefully dissected away from white matter. Trizol LS (Invitrogen, Carlsbad, CA) was used according to manufacturer’s instructions, except an additional overnight −20°C precipitation step was included during isopropanol precipitation. RNA quality was confirmed using A260/A280 readings and also the Agilent Bioanalyzer for RIN.
Microarray methods
The quality of the total RNA after rethawing was again verified by Exiqon as part of the quality control process, using an Agilent 2100 Bioanalyzer profile. Two different batches of microarrays were analyzed, the first using the 6th generation (Product #208310) Exiqon miRNA microarray, miRBase version 17, and second round performed with 7th generation (Product #208500), miRBase version 20. Briefly, 1 μg total RNA from sample and reference was labeled with Hy3™ and Hy5™ fluorescent label, respectively, using the miRCURY™ LNA Array power labeling kit (Exiqon, Denmark) following the procedure described by the manufacturer. The Hy3™-labeled samples and a Hy5™-labeled reference RNA samples (referred to hereafter as Cy3 and Cy5) were mixed pair-wise and hybridized to the miRCURY™ LNA array version 11.0 (Exiqon, Denmark), which contains capture probes targeting all miRNAs for human, mouse or rat registered in the miRBASE version 14.0 at the Sanger Institute. The hybridization was performed according to the miRCURY™ LNA array manual using a Tecan HS4800 hybridization station. After hybridization the microarray slides were scanned and stored in an ozone free environment (ozone level below 2.0 ppb) in order to prevent potential bleaching of the fluorescent dyes. The miRCURY™ LNA array microarray slides were scanned using the Agilent G2565BA Microarray Scanner System (Agilent Technologies, Inc., USA) and the image analysis was carried out using the ImaGene 8.0 software (BioDiscovery, Inc., USA). The quantified signals were background corrected (Normexp with offset value 10 as described previously (Ritchie et al., 2007)) and normalized using the global Lowess (LOcally WEighted Scatterplot Smoothing) regression algorithm.
Analytic and statistical methods
We analyzed miRNAs from RNA with RIN greater than 4.0 and mean expression (log^2) value greater than 7.0 in at least one of the two data sets in order to avoid analyzing miRNAs that were not expressed or lacking in data quality in both experiments. To analyze the data, a mixed model was fit to each miRNA with a random effect for each individual in the study and fixed effects to adjust for age and Exiqon array iteration in miRNA expression. This model accounts for variability in miRNA expression across individuals, as well as for differences in expression due to age and Exiqon array iteration. In addition, each model included fixed effects for DLB (for cases where DLB severity is greater than 0) vs. non-DLB (cases where DLB severity is 0), sex, and brain region (AC vs. MO), plus all two-way interactions among these variables, and an interaction among DLB, sex, and brain. Each interaction allows each combination of sex, brain region, and DLB status to have a different association with miRNA expression, rather than forcing, for instance, differences in brain region to be the same in males and in females. Thus, in summary, the mixed models allow researchers to detect effects of DLB, brain region, and sex on miRNA expression, while accounting for variability across individuals and differences across age and array iteration. After the mixed models were fit, tests were performed to look for evidence of effects of interest. Specifically, tests were performed comparing the average miRNA expression across brain regions. Multiple testing corrections were made for fixed effect tests using a Benjamini-Hochberg correction. Significance was noted when the adjusted p-values for the corresponding test were less than 0.05.
RT-qPCR validation in separate cases of a subset of miRNAs
Quantitative PCR for post-hoc replication was performed on separate cases (n=6) and included RNA isolated from MO, AC, primary somatosensory, and frontal cortical regions as described above, all from the same cases. Different cases were used in comparison to those above—only controls lacking substantial DLB or AD pathology were evaluated for these replication experiments. RNA was extracted as above and evaluated with TaqMan® single-tube miRNA RT-qPCR (Thermo Fisher Scientific) according to the manufacturer’s instructions. An equal quantity (350 ng) of RNA was used in the TaqMan® single-tube assays. TaqMan® assays were designed to follow MIQE (Minimum Information for Publication of Quantitative Real-Time PCR Experiments) guidelines (Bustin et al., 2009).
Supplementary Material
Acknowledgments
The authors are deeply grateful to the study participants and clinicians who made this research possible. We thank Sonya Anderson and Ela Patel for technical support. Study funding: P30 AG028383, R21 NS085830 from the National Institutes of Health. Thanks to Ruth S. Nelson for art work. The corresponding author Dr. Peter Nelson had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- Alderman C, Yang Y. The anti-melanoma activity and oncogenic targets of hsa-miR-15a-5p. RNA Dis. 2016:3. [PMC free article] [PubMed] [Google Scholar]
- Asikainen S, et al. Global microRNA expression profiling of Caenorhabditis elegans Parkinson’s disease models. J Mol Neurosci. 2010;41:210–8. doi: 10.1007/s12031-009-9325-1. [DOI] [PubMed] [Google Scholar]
- Bannister SC, et al. Sexually dimorphic microRNA expression during chicken embryonic gonadal development. Biol Reprod. 2009;81:165–76. doi: 10.1095/biolreprod.108.074005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker WW, et al. Relative frequencies of Alzheimer disease, Lewy body, vascular and frontotemporal dementia, and hippocampal sclerosis in the State of Florida Brain Bank. Alzheimer Dis Assoc Disord. 2002;16:203–12. doi: 10.1097/00002093-200210000-00001. [DOI] [PubMed] [Google Scholar]
- Beach TG, et al. Accuracy of the clinical diagnosis of Alzheimer disease at National Institute on Aging Alzheimer Disease Centers, 2005-2010. J Neuropathol Exp Neurol. 2012;71:266–73. doi: 10.1097/NEN.0b013e31824b211b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, et al. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizuayehu TT, et al. Sex-Biased miRNA Expression in Atlantic Halibut (Hippoglossus hippoglossus) Brain and Gonads. Sex Dev. 2012:257–266. doi: 10.1159/000341378. [DOI] [PubMed] [Google Scholar]
- Brettschneider J, et al. Stages of pTDP-43 pathology in amyotrophic lateral sclerosis. Ann Neurol. 2013;74:20–38. doi: 10.1002/ana.23937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–22. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Choi DC, et al. MicroRNA-7 protects against 1-methyl-4-phenylpyridinium-induced cell death by targeting RelA. J Neurosci. 2014;34:12725–37. doi: 10.1523/JNEUROSCI.0985-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutting AD, et al. The potential role of microRNAs in regulating gonadal sex differentiation in the chicken embryo. Chromosome Res. 2012;20:201–13. doi: 10.1007/s10577-011-9263-y. [DOI] [PubMed] [Google Scholar]
- de Mena L, et al. FGF20 rs12720208 SNP and microRNA-433 variation: no association with Parkinson’s disease in Spanish patients. Neurosci Lett. 2010;479:22–5. doi: 10.1016/j.neulet.2010.05.019. [DOI] [PubMed] [Google Scholar]
- Delay C, Hebert SS. MicroRNAs and Alzheimer’s Disease Mouse Models: Current Insights and Future Research Avenues. Int J Alzheimers Dis. 2011;2011:894938. doi: 10.4061/2011/894938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson DW, et al. Neuropathological assessment of Parkinson’s disease: refining the diagnostic criteria. Lancet Neurol. 2009;8:1150–7. doi: 10.1016/S1474-4422(09)70238-8. [DOI] [PubMed] [Google Scholar]
- Doxakis E. Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153. J Biol Chem. 2010;285:12726–34. doi: 10.1074/jbc.M109.086827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnerty JR, et al. The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol. 2010;402:491–509. doi: 10.1016/j.jmb.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkouli A, Doxakis E. miR-7 and miR-153 protect neurons against MPP(+)-induced cell death via upregulation of mTOR pathway. Front Cell Neurosci. 2014;8:182. doi: 10.3389/fncel.2014.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujishiro H, et al. Validation of the neuropathologic criteria of the third consortium for dementia with Lewy bodies for prospectively diagnosed cases. J Neuropathol Exp Neurol. 2008;67:649–56. doi: 10.1097/NEN.0b013e31817d7a1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo M, Croce CM. microRNAs: Master regulators as potential therapeutics in cancer. Annu Rev Pharmacol Toxicol. 2011;51:25–43. doi: 10.1146/annurev-pharmtox-010510-100517. [DOI] [PubMed] [Google Scholar]
- Gui Y, et al. Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget. 2015;6:37043–53. doi: 10.18632/oncotarget.6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn K, et al. Clinical features, with video documentation, of the original familial lewy body parkinsonism caused by alpha-synuclein triplication (Iowa kindred) Mov Disord. 2011;26:2134–6. doi: 10.1002/mds.23776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert SS, De Strooper B. Molecular biology. miRNAs in neurodegeneration. Science. 2007;317:1179–80. doi: 10.1126/science.1148530. [DOI] [PubMed] [Google Scholar]
- Hebert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Hebert SS, Nelson PT. Studying microRNAs in the brain: technical lessons learned from the first ten years. Exp Neurol. 2012;235:397–401. doi: 10.1016/j.expneurol.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert SS, et al. A study of small RNAs from cerebral neocortex of pathology-verified Alzheimer’s disease, dementia with lewy bodies, hippocampal sclerosis, frontotemporal lobar dementia, and non-demented human controls. J Alzheimers Dis. 2013;35:335–48. doi: 10.3233/JAD-122350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidebrink JL. Is dementia with Lewy bodies the second most common cause of dementia? J Geriatr Psychiatry Neurol. 2002;15:182–7. doi: 10.1177/089198870201500402. [DOI] [PubMed] [Google Scholar]
- Hishikawa N, et al. Clinical and neuropathological correlates of Lewy body disease. Acta Neuropathol. 2003;105:341–50. doi: 10.1007/s00401-002-0651-4. [DOI] [PubMed] [Google Scholar]
- Irwin DJ, et al. Neuropathological and genetic correlates of survival and dementia onset in synucleinopathies: a retrospective analysis. Lancet Neurol. 2017;16:55–65. doi: 10.1016/S1474-4422(16)30291-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalava SE, et al. Androgen-regulated miR-32 targets BTG2 and is overexpressed in castration-resistant prostate cancer. Oncogene. 2012 doi: 10.1038/onc.2011.624. [DOI] [PubMed] [Google Scholar]
- Janeczek P, Lewohl JM. The role of alpha-synuclein in the pathophysiology of alcoholism. Neurochem Int. 2013;63:154–62. doi: 10.1016/j.neuint.2013.06.007. [DOI] [PubMed] [Google Scholar]
- John B, et al. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junn E, et al. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci U S A. 2009;106:13052–7. doi: 10.1073/pnas.0906277106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabaria S, et al. Inhibition of miR-34b and miR-34c enhances alpha-synuclein expression in Parkinson’s disease. FEBS Lett. 2015;589:319–25. doi: 10.1016/j.febslet.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, et al. A mir-153 binding site variation in SNCA in a patient with Parkinson’s disease. Mov Disord. 2013;28:1755–6. doi: 10.1002/mds.25505. [DOI] [PubMed] [Google Scholar]
- Kim J, et al. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–4. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriakidou M, et al. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–78. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, et al. High Throughput Sequencing Identifies MicroRNAs Mediating alpha-Synuclein Toxicity by Targeting Neuroactive-Ligand Receptor Interaction Pathway in Early Stage of Drosophila Parkinson’s Disease Model. PLoS One. 2015;10:e0137432. doi: 10.1371/journal.pone.0137432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraybill ML, et al. Cognitive differences in dementia patients with autopsy-verified AD, Lewy body pathology, or both. Neurology. 2005;64:2069–73. doi: 10.1212/01.WNL.0000165987.89198.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Sharp PA. Function and localization of microRNAs in mammalian cells. Cold Spring Harb Symp Quant Biol. 2006;71:29–38. doi: 10.1101/sqb.2006.71.049. [DOI] [PubMed] [Google Scholar]
- Li G, et al. Targeted suppression of chaperone-mediated autophagy by miR-320a promotes alpha-synuclein aggregation. Int J Mol Sci. 2014a;15:15845–57. doi: 10.3390/ijms150915845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, et al. MiRNA-15a mediates cell cycle arrest and potentiates apoptosis in breast cancer cells by targeting synuclein-gamma. Asian Pac J Cancer Prev. 2014b;15:6949–54. doi: 10.7314/apjcp.2014.15.16.6949. [DOI] [PubMed] [Google Scholar]
- Lim PK, et al. Neurogenesis: role for microRNAs and mesenchymal stem cells in pathological states. Curr Med Chem. 2010;17:2159–67. doi: 10.2174/092986710791299894. [DOI] [PubMed] [Google Scholar]
- Liu QY, et al. Identification of microRNAs involved in Alzheimer’s progression using a rabbit model of the disease. Am J Neurodegener Dis. 2014;3:33–44. [PMC free article] [PubMed] [Google Scholar]
- Liu W, et al. MicroRNA-107 prevents amyloid-beta induced blood-brain barrier disruption and endothelial cell dysfunction by targeting Endophilin-1. Exp Cell Res. 2016;343:248–57. doi: 10.1016/j.yexcr.2016.03.026. [DOI] [PubMed] [Google Scholar]
- Liu X, Fortin K, Mourelatos Z. MicroRNAs: biogenesis and molecular functions. Brain Pathol. 2008;18:113–21. doi: 10.1111/j.1750-3639.2007.00121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londin E, et al. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proc Natl Acad Sci U S A. 2015;112:E1106–15. doi: 10.1073/pnas.1420955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, et al. Advances with microRNAs in Parkinson’s disease research. Drug Des Devel Ther. 2013;7:1103–13. doi: 10.2147/DDDT.S48500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majidinia M, et al. The roles of non-coding RNAs in Parkinson’s disease. Mol Biol Rep. 2016;43:1193–1204. doi: 10.1007/s11033-016-4054-3. [DOI] [PubMed] [Google Scholar]
- McKeith I, et al. Dementia with Lewy bodies. Lancet Neurol. 2004;3:19–28. doi: 10.1016/s1474-4422(03)00619-7. [DOI] [PubMed] [Google Scholar]
- McKeith IG, et al. Diagnosis and management of dementia with Lewy bodies: Fourth consensus report of the DLB Consortium. Neurology. 2017;89:88–100. doi: 10.1212/WNL.0000000000004058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minones-Moyano E, et al. MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum Mol Genet. 2011;20:3067–78. doi: 10.1093/hmg/ddr210. [DOI] [PubMed] [Google Scholar]
- Moncini S, et al. The miR-15/107 Family of microRNA Genes Regulates CDK5R1/p35 with Implications for Alzheimer’s Disease Pathogenesis. Mol Neurobiol. 2016 doi: 10.1007/s12035-016-0002-4. [DOI] [PubMed] [Google Scholar]
- Mouradian MM. MicroRNAs in Parkinson’s disease. Neurobiol Dis. 2012;46:279–84. doi: 10.1016/j.nbd.2011.12.046. [DOI] [PubMed] [Google Scholar]
- Murata T, et al. miR-148a is an androgen-responsive microRNA that promotes LNCaP prostate cell growth by repressing its target CAND1 expression. Prostate Cancer Prostatic Dis. 2010;13:356–61. doi: 10.1038/pcan.2010.32. [DOI] [PubMed] [Google Scholar]
- Nelson PT, et al. Clinicopathologic correlations in a large Alzheimer disease center autopsy cohort: neuritic plaques and neurofibrillary tangles “do count” when staging disease severity. J Neuropathol Exp Neurol. 2007;66:1136–46. doi: 10.1097/nen.0b013e31815c5efb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Keller JN. RNA in brain disease: no longer just “the messenger in the middle”. J Neuropathol Exp Neurol. 2007;66:461–8. doi: 10.1097/01.jnen.0000240474.27791.f3. [DOI] [PubMed] [Google Scholar]
- Nelson PT, Wang WX, Rajeev BW. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008a;18:130–8. doi: 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, et al. Technical variables in high-throughput miRNA expression profiling: much work remains to be done. Biochim Biophys Acta. 2008b;1779:758–65. doi: 10.1016/j.bbagrm.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, et al. Alzheimer’s-type neuropathology in the precuneus is not increased relative to other areas of neocortex across a range of cognitive impairment. Neurosci Lett. 2009a;450:336–9. doi: 10.1016/j.neulet.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, et al. Relative preservation of MMSE scores in autopsy-proven dementia with Lewy bodies. Neurology. 2009b;73:1127–33. doi: 10.1212/WNL.0b013e3181bacf9e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, et al. Low sensitivity in clinical diagnoses of dementia with Lewy bodies. J Neurol. 2010a;257:359–66. doi: 10.1007/s00415-009-5324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, et al. Association between male gender and cortical Lewy body pathology in large autopsy series. J Neurol. 2010b doi: 10.1007/s00415-010-5630-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Wang WX. MiR-107 is reduced in Alzheimer’s disease brain neocortex: validation study. J Alzheimers Dis. 2010;21:75–9. doi: 10.3233/JAD-2010-091603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neltner JH, et al. Brain pathologies in extreme old age. Neurobiol Aging. 2016;37:1–11. doi: 10.1016/j.neurobiolaging.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka K, et al. Clinical heterogeneity of alpha-synuclein gene duplication in Parkinson’s disease. Ann Neurol. 2006;59:298–309. doi: 10.1002/ana.20753. [DOI] [PubMed] [Google Scholar]
- Niu M, et al. MiR-133b ameliorates axon degeneration induced by MPP(+) via targeting RhoA. Neuroscience. 2016;325:39–49. doi: 10.1016/j.neuroscience.2016.03.042. [DOI] [PubMed] [Google Scholar]
- Parsi S, et al. Preclinical Evaluation of miR-15/107 Family Members as Multifactorial Drug Targets for Alzheimer’s Disease. Mol Ther Nucleic Acids. 2015;4:e256. doi: 10.1038/mtna.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil KS, et al. A Proteomics Approach to Investigate miR-153-3p and miR-205-5p Targets in Neuroblastoma Cells. PLoS One. 2015;10:e0143969. doi: 10.1371/journal.pone.0143969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrzak M, et al. Gene expression profiling of brain samples from patients with Lewy body dementia. Biochem Biophys Res Commun. 2016;479:875–880. doi: 10.1016/j.bbrc.2016.09.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademakers R, et al. Common variation in the miR-659 binding-site of GRN is a major risk factor for TDP43-positive frontotemporal dementia. Hum Mol Genet. 2008;17:3631–42. doi: 10.1093/hmg/ddn257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahkonen T, et al. Dementia with Lewy bodies according to the consensus criteria in a general population aged 75 years or older. J Neurol Neurosurg Psychiatry. 2003;74:720–4. doi: 10.1136/jnnp.74.6.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recasens A, Perier C, Sue CM. Role of microRNAs in the Regulation of alpha-Synuclein Expression: A Systematic Review. Front Mol Neurosci. 2016;9:128. doi: 10.3389/fnmol.2016.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23:2700–7. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- Schmitt FA, et al. “Preclinical” AD revisited: neuropathology of cognitively normal older adults. Neurology. 2000;55:370–6. doi: 10.1212/wnl.55.3.370. [DOI] [PubMed] [Google Scholar]
- Schmitt FA, et al. University of Kentucky Sanders-Brown Healthy Brain Aging Volunteers: Donor Characteristics, Procedures, and Neuropathology. Curr Alzheimer Res. 2012;9:724–33. doi: 10.2174/156720512801322591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonrock N, et al. Neuronal microRNA deregulation in response to Alzheimer’s disease amyloid-beta. PLoS ONE. 2010;5:e11070. doi: 10.1371/journal.pone.0011070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi P, Lukiw WJ. Micro-RNA abundance and stability in human brain: specific alterations in Alzheimer’s disease temporal lobe neocortex. Neurosci Lett. 2009;459:100–4. doi: 10.1016/j.neulet.2009.04.052. [DOI] [PubMed] [Google Scholar]
- Shulman LM, Bhat V. Gender disparities in Parkinson’s disease. Expert Rev Neurother. 2006;6:407–16. doi: 10.1586/14737175.6.3.407. [DOI] [PubMed] [Google Scholar]
- Shulman LM. Gender differences in Parkinson’s disease. Gend Med. 2007;4:8–18. doi: 10.1016/s1550-8579(07)80003-9. [DOI] [PubMed] [Google Scholar]
- Singleton AB, et al. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- Sonnen JA, et al. Neuropathology in the adult changes in thought study: a review. J Alzheimers Dis. 2009;18:703–11. doi: 10.3233/JAD-2009-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliafierro L, et al. Genetic analysis of alpha-synuclein 3′ untranslated region and its corresponding microRNAs in relation to Parkinson’s compared to dementia with Lewy bodies. Alzheimers Dement. 2017 doi: 10.1016/j.jalz.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thal DR, Del Tredici K, Braak H. Neurodegeneration in normal brain aging and disease. Sci Aging Knowledge Environ. 2004;2004:pe26. doi: 10.1126/sageke.2004.23.pe26. [DOI] [PubMed] [Google Scholar]
- Toledo JB, et al. Pathological alpha-synuclein distribution in subjects with coincident Alzheimer’s and Lewy body pathology. Acta Neuropathol. 2016;131:393–409. doi: 10.1007/s00401-015-1526-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubhi K, et al. Widespread microRNA dysregulation in multiple system atrophy - disease-related alteration in miR-96. Eur J Neurosci. 2014;39:1026–41. doi: 10.1111/ejn.12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, et al. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am J Hum Genet. 2008a;82:283–9. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, et al. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008b;28:1213–23. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, et al. Focus on RNA isolation: obtaining RNA for microRNA (miRNA) expression profiling analyses of neural tissue. Biochim Biophys Acta. 2008c;1779:749–57. doi: 10.1016/j.bbagrm.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, et al. Individual microRNAs (miRNAs) display distinct mRNA targeting “rules”. RNA Biol. 2010;7 doi: 10.4161/rna.7.3.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, et al. Patterns of microRNA expression in normal and early Alzheimer’s disease human temporal cortex: white matter versus gray matter. Acta Neuropathol. 2011;121:193–205. doi: 10.1007/s00401-010-0756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, et al. Expression of miR-15/107 family microRNAs in human tissues and cultured rat brain cells. Genomics Proteomics Bioinformatics. 2014;12:19–30. doi: 10.1016/j.gpb.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZH, et al. MicroRNA-214 participates in the neuroprotective effect of Resveratrol via inhibiting alpha-synuclein expression in MPTP-induced Parkinson’s disease mouse. Biomed Pharmacother. 2015;74:252–6. doi: 10.1016/j.biopha.2015.08.025. [DOI] [PubMed] [Google Scholar]
- Weinberg RB, Mufson EJ, Counts SE. Evidence for a neuroprotective microRNA pathway in amnestic mild cognitive impairment. Front Neurosci. 2015;9:430. doi: 10.3389/fnins.2015.00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner MF, et al. Alzheimer’s disease and its Lewy body variant: a clinical analysis of postmortem verified cases. Am J Psychiatry. 1996;153:1269–73. doi: 10.1176/ajp.153.10.1269. [DOI] [PubMed] [Google Scholar]
- Xie Y, Chen Y. microRNAs: Emerging Targets Regulating Oxidative Stress in the Models of Parkinson’s Disease. Front Neurosci. 2016;10:298. doi: 10.3389/fnins.2016.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, et al. MicroRNA-related cofilin abnormality in Alzheimer’s disease. PLoS ONE. 2010;5:e15546. doi: 10.1371/journal.pone.0015546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccai J, McCracken C, Brayne C. A systematic review of prevalence and incidence studies of dementia with Lewy bodies. Age Ageing. 2005;34:561–6. doi: 10.1093/ageing/afi190. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Cheng Y. miR-16-1 promotes the aberrant alpha-synuclein accumulation in parkinson disease via targeting heat shock protein 70. Scientific World Journal. 2014;2014:938348. doi: 10.1155/2014/938348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, et al. Estrogen-regulated genes in rat testes and their relationship to recovery of spermatogenesis after irradiation. Biol Reprod. 2011;85:823–33. doi: 10.1095/biolreprod.111.091611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, et al. MicroRNA-7 targets Nod-like receptor protein 3 inflammasome to modulate neuroinflammation in the pathogenesis of Parkinson’s disease. Mol Neurodegener. 2016;11:28. doi: 10.1186/s13024-016-0094-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


