Abstract
Mutations in the CCM1 (aka KRIT1), CCM2, or CCM3 (aka PDCD10) gene cause cerebral cavernous malformation (CCM) in humans. Mouse models of CCM disease have been established by tamoxifen induced deletion of Ccm genes in postnatal animals. These mouse models provide invaluable tools to investigate molecular mechanism and therapeutic approaches for CCM disease. An accurate and quantitative method to assess lesion burden and progression is essential to harness the full value of these animal models. Here, we demonstrate the induction of CCM disease in a mouse model and the use of the contrast enhanced X-ray micro computed tomography (micro-CT) method to measure CCM lesion burden in mouse brains. At postnatal day 1 (P1), we used 4-hydroxytamoxifen (4HT) to activate Cre recombinase activity from the Cdh5-CreErt2 transgene to cleave the floxed allele of Ccm2. CCM lesions in mouse brains were analyzed at P8. For micro-CT, iodine based Lugol's solution was used to enhance contrast in brain tissue. We have optimized the scan parameters and utilized a voxel dimension of 9.5 µm, which lead to a minimum feature size of approximately 25 µm. This resolution is sufficient to measure CCM lesion volume and number globally and accurately, and provide high-quality 3-D mapping of CCM lesions in mouse brains. This method enhances the value of the established mouse models to study the molecular basis and potential therapies for CCM and other cerebrovascular diseases.
Keywords: Medicine, Issue 127, Micro computed tomography, cerebral cavernous malformation, mouse model, vascular malformation, CCM1 gene, Krit1gene
Introduction
CCM are thin walled, dilated vascular malformations in the brain with prevalence of up to 0.5% in the human population1. CCM can be inherited as a dominant disorder due to loss-of-function mutations in one of three genes: CCM1 (aka Krit1), CCM2, and CCM3 (also called PDCD10)2,3,4,5,6. These genes are present in a single signaling complex.
Various models have been developed to model human CCM disease and to understand the downstream pathways of CCM genes that are responsible for CCM7,8,9,10. The most robust model is to conditionally delete any one of the Ccm genes with tamoxifen-inducible Cdh5-CreERT2 at P1 in newborn pups8,10. These pups develop CCM lesions in the brain from P6 onward and are expected to be an ideal model for pre-clinical studies in search for mechanisms and therapeutic agents in treating CCM diseases.
CCM lesion burden in mouse brain has been measured primarily by histology-based methods, an approach that is extremely time-consuming and subject to investigator bias10,11,12. MRI based methods have been used to assess CCM lesion burden in the adult mouse model9,13. However, a highly specialized small animal MRI instrument and long scan-time of several hours to overnight is required to achieve a satisfactory resolution to identifying CCM lesions. Also, whether MRI can be used to detect CCM lesions in neonatal mice has not been reported and resolution may limit sensitivity.
We have recently developed a micro-CT technique to image and analyze CCM lesion14,15. This high-resolution, time and cost-effective method dramatically boosted the value of the CCM disease model in mechanistic and therapeutic studies. Contrast-enhancing, whole mount staining methods have been used for micro-CT imaging of soft tissues and mouse embryos16,17. Previously, we have used an osmium-based staining to enhance contrast for micro-CT imaging of CCM lesions in brain14. In this paper, we used a less toxic, non-destructive, and cost-efficient reagent, an iodine based Lugol's solution, to enhance contrast for micro-CT imaging. Iodine can diffuse throughout the brain and has a high affinity for blood18.
The detailed protocol is presented here for the induction of CCM lesions in a neonatal mouse model along with the imaging and analysis of CCM lesions with a contrast-based micro-CT. This micro-CT based method provides quantitative global measurement of CCM lesion volume, accurately identifies the number and 3-D location of CCM lesions in the mouse brain, and greatly reduces the cost and time required to phenotype these animals.
Protocol
All animal ethics and protocols were approved by The Sydney Local Health District Animal Welfare Committee and Institutional Animal Care and Use Committee (IACUC) of Tianjin Medical University. All experiments were conducted under the guidelines/regulations of Centenary Institute, University of Sydney and Tianjin Medical University
1. Induction of Cerebral Cavernous Malformations in Mouse Models
Cross Cdh5-CreErt2; Ccm2fl/fl mice with Ccm2fl/fl to generate litters with Cdh5-CreErt2; Ccm2fl/fl (Ccm2iECKO) pups and littermate controls (Ccm2fl/fl). Cdh5-CreErt2 and Ccm2fl/fl animals have been previously described19.
Dissolve 4HT in 100% ethanol and store at -80 °C in aliquots of 30 µL (4HT concentration: 10 mg/mL). On the day of use, dilute aliquoted 4HT in corn oil (0.5 mg/mL).
Intragastrically inject 50 µL of 4HT (0.5 mg/mL) to neonatal pups at P1 to induce experimental CCM lesions using an insulin syringe.
2. Sample Preparation for Micro-CT Scans
Euthanize neonatal pups at P8 via carbon dioxide asphyxiation.
Perform intra-cardiac perfusion by inserting a 29-gauge needle with 3 mL of 2% paraformaldehyde in PBS in a 10 mL syringe into the ventricle of the mouse heart. CAUTION: Paraformaldehyde is toxic; wear appropriate protection.
Detach the head from the body using a scissors. Remove all the skin off the head using a scissors and peel off the skull using forceps to dissect the whole brain.
Take images of the brain with stereomicroscope at 7.82x magnification, 1x gain, 0.6 gamma, and 20 ms exposure time.
Fix the dissected brains with 4% paraformaldehyde in PBS solution overnight. CAUTION: Paraformaldehyde is toxic; wear appropriate protection.
On the following day, detach hindbrains using a forceps and wash with PBS solution.
Incubate the hindbrains in Lugol's iodine solution for 48 h.
Following the incubation, briefly air dry the hindbrains to remove excess Lugol's iodine solution.
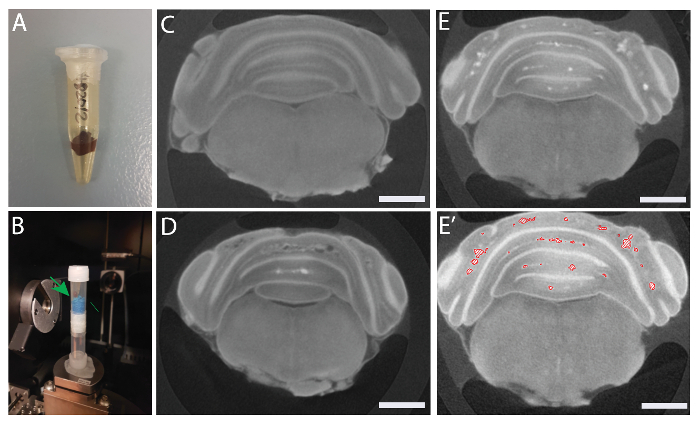
Pack the Lugol's iodine stained hindbrains in 0.65 mL microcentrifuge tubes and seal completely with plastic paraffin film to avoid tissue shrinkage (Figure 2A).
Place microcentrifuge tubes in 5 mL plastic tubes with sponges to prevent them from moving during scan (Figure 2B).
3. Micro-CT Scan of CCM in the Mouse Brain
Vertically mount the hindbrain packed in a tube on an aluminum holder in the micro-CT system.
Set the scanning parameters to 540 projections and 2 s exposure time with source conditions of 50 kV and 10 W to acquire the tomographic datasets (image resolution 9.5 μm/pixel).
Radiographs from the scan are reconstructed automatically by hardware-based projection reconstruction software supplied by the micro-CT system, producing an image series of 16-bit axial slices ("TXM" file).
4. Analysis of Micro-CT Datasets
Open the reconstructed image file ("TXM" file) in the 3-D analysis software and visualize the brain using the "volume rendering" function.
Transform the hindbrain to the desired orientation using the "bounding box" and "clipping plane" functions.
Resample the transformed image in extended mode and preserve the voxel size.
Create "edit new label field" on the transformed image (4.3) and highlight only the hindbrain using greyscale intensity.
Add the highlighted areas and run "label analysis" to obtain measurements of the whole hindbrain (i.e., total volume and area). Export measurements in an Excel file.
Visualize the hindbrain using the "isosurface rendering" function.
Create another "edit new label field" on the transformed image and highlight areas with lesions using greyscale intensity.
Add the highlighted areas and run "label" analysis to obtain measurements of the lesions within the hindbrain (i.e., total volume and area). Export measurements in an Excel file.
Visualize the lesions using the "generate surface" and "surface view" functions.
Snapshot the images of the hindbrain at desired orientations or generate a movie for 3-D visualization of the lesions.
Representative Results
A single injection of 4HT at P1 was sufficient to induce CCM lesions in the cerebellum. Lugol's iodine contrasted micro-CT sufficiently detected CCM lesions and could quantify its volume and number. Utilizing the optimized micro-CT, we imaged CCM lesions in the hindbrains of Ccm2iECKO mice. Scanned X-ray images were reconstructed to produce 3-D images of the mouse brain, which allowed visualization of entire lesions in the brain parenchyma at different depths and orientations, and assessment of the structure and 3-D location of lesions in the brain. Importantly, this image dataset can also be used for efficient and accurate quantification of lesion sizes and lesion numbers.
Figure 1 shows successful induction of CCM lesions in the cerebellum of Ccm2iECKO (B) but not in littermate controls (Ccm2fl/fl) (A) at P8.
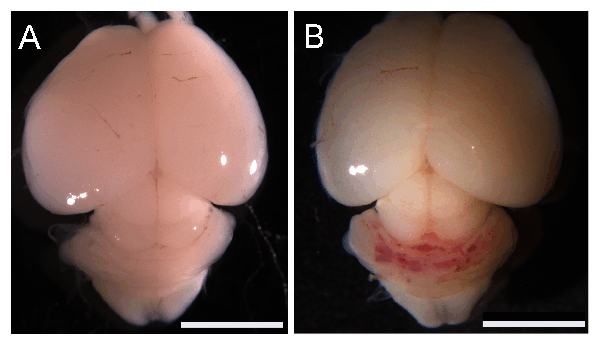
Figure 2 shows a packed sample for micro-CT scan (A-B), and representative reconstructed images from the micro-CT scan of control (Ccm2fl/fl) (C) and Ccm2iECKO (D-E). Figure 2E' shows marked lesions in Ccm2iECKO.
Figure 3 shows a rendered 3-D image of a Ccm2iECKO brain with lesions (A-B).Adjacent Table shows an example of quantitative output of the labeled individual lesions.
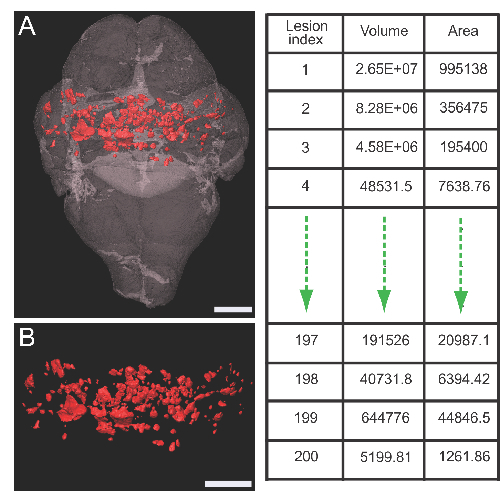
Figure 1: Induction of CCM lesions in neonatal mouse. Macroscopic images of Ccm2fl/fl (A) and Ccm2iECKO (B) brains at postnatal day 8. Scale bars (white), 5 mm. Please click here to view a larger version of this figure.
Figure 2: Sample preparation and reconstructed 2D images. Sample packed in a microcentrifuge tube (A) and 5 mL tubes (B). Green arrow shows the sample. Representative images of Ccm2fl/fl (C) and Ccm2iECKO (D-E) brains. (E') Marked lesions in Ccm2iECKO brain. Scale bars (white), 1 mm. Please click here to view a larger version of this figure.
Figure 3: Rendered 3-D images. Rendered image of Ccm2iECKO brain with lesions (A) and lesions only (B). Lesions are marked in red. Table on the right shows an example of qualitative output of individually marked lesions. Scale bars (white), 1 mm. Please click here to view a larger version of this figure.
Discussion
CCM is a common vascular malformation that affects up to 0.5% of individuals1. CCM can occur in sporadic or familial form. Patient prognosis is often unclear as CCM lesions can rupture unexpectedly to cause stroke and other neurological consequences. Currently, the only treatment option is to surgically remove lesions, which are accompanied by high risk.
Human CCM conditions have recently been reproduced in animal models7,8,9,10. These models provide invaluable tools to investigate mechanisms involved in CCM disease. An accurate, efficient, and quantitative analysis method will help to harness the full value of these models.
We have recently developed a micro-CT method to image CCM lesions in mouse models using osmium tetraoxide as a contrasting agent14. In the current study, we have subsequently used Lugol's iodine solution to enhance the contrast of brain tissue to detect lesions. Lugol's iodine solution is less toxic, non-destructive, and cheaper compared to osmium tetraoxide. Hence, the samples can be reused for other purposes, such as histology after Lugol's iodine solution is washed away.
Our micro-CT method could produce sufficient image quality to detect CCM lesions in the mouse brain. The sample preparation is straightforward and does not require highly specialized techniques. This method can provide high-resolution projections with a scan time of approximately one hour. Combining datasets from micro-CT scans and 3-D analysis software is a powerful way to generate a 3-D global view of CCM lesions in brain. The quantitative and qualitative representation of lesions relative to brain anatomy is easily achieved. Compared to histology-based 3-D reconstruction, our method is highly time- and cost-effective and produces much more accurate 3-D representations.
However, there are also limitations and critical steps to be considered. Firstly, the method requires a micro-CT system, which is highly specialized equipment and may not be readily available. Secondly, dissecting the neonatal mouse brain can be quite challenging due to the softness and size of the brain tissue. It is very important not to damage the brain during dissection in order to acquire accurate measurements and visualization of the CCM lesions in the brain. Lastly, sample preparation for micro-CT scan is critical. Lugol's iodine stained brains must be packed completely to avoid tissue shrinkage due to evaporation. Also, samples must be placed securely during the micro-CT scan. Any movement during the scan will alter the quality of the scan and hence the quantification and visualization.
We have applied our method only in fixed neonatal mouse brains. However, this method has the capacity to be used for other studies involving different samples and may serve as an exciting and novel tool for other vascular malformation research.
Disclosures
The authors have nothing to disclose.
Acknowledgments
The authors acknowledge the facilities and the scientific and technical assistance of the Sydney Microscopy & Microanalysis Research Facility (AMMRF) and the Australian Centre for Microscopy & Microanalysis (ACMM) at the University of Sydney. These studies were supported by Australian National Health and Medical Research Council (NHMRC) project grant 161558 and APP1124011 (XZ).
References
- Fischer A, Zalvide J, Faurobert E, Albiges-Rizo C, Tournier-Lasserve E. Cerebral cavernous malformations: from CCM genes to endothelial cell homeostasis. Trends Mol Med. 2013;19(5):302–308. doi: 10.1016/j.molmed.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Liquori CL, et al. Mutations in a gene encoding a novel protein containing a phosphotyrosine-binding domain cause type 2 cerebral cavernous malformations. Am J Hum Genet. 2003;73(6):1459–1464. doi: 10.1086/380314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laberge-le Couteulx S, et al. Truncating mutations in CCM1, encoding KRIT1, cause hereditary cavernous angiomas. Nat Genet. 1999;23(2):189–193. doi: 10.1038/13815. [DOI] [PubMed] [Google Scholar]
- Sahoo T, et al. Mutations in the gene encoding KRIT1, a Krev-1/rap1a binding protein, cause cerebral cavernous malformations (CCM1) Hum Mol Genet. 1999;8(12):2325–2333. doi: 10.1093/hmg/8.12.2325. [DOI] [PubMed] [Google Scholar]
- Denier C, et al. Mutations within the MGC4607 gene cause cerebral cavernous malformations. Am J Hum Genet. 2004;74(2):326–337. doi: 10.1086/381718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergametti F, et al. Mutations within the programmed cell death 10 gene cause cerebral cavernous malformations. Am J Hum Genet. 2005;76(1):42–51. doi: 10.1086/426952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald DA, et al. A novel mouse model of cerebral cavernous malformations based on the two-hit mutation hypothesis recapitulates the human disease. Hum Mol Genet. 2011;20(2):211–222. doi: 10.1093/hmg/ddq433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulday G, et al. Developmental timing of CCM2 loss influences cerebral cavernous malformations in mice. J Exp Med. 2011;208(9):1835–1847. doi: 10.1084/jem.20110571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AC, et al. Mutations in 2 distinct genetic pathways result in cerebral cavernous malformations in mice. J Clin Invest. 2011;121(5):1871–1881. doi: 10.1172/JCI44393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, et al. Cerebral cavernous malformations arise independent of the heart of glass receptor. Stroke. 2014;45(5):1505–1509. doi: 10.1161/STROKEAHA.114.004809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald DA, et al. Fasudil decreases lesion burden in a murine model of cerebral cavernous malformation disease. Stroke. 2012;43(2):571–574. doi: 10.1161/STROKEAHA.111.625467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddaluno L, et al. EndMT contributes to the onset and progression of cerebral cavernous malformations. Nature. 2013;498(7455):492–496. doi: 10.1038/nature12207. [DOI] [PubMed] [Google Scholar]
- Gibson CC, et al. Strategy for identifying repurposed drugs for the treatment of cerebral cavernous malformation. Circulation. 2015;131(3):289–299. doi: 10.1161/CIRCULATIONAHA.114.010403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JP, et al. Micro-CT Imaging Reveals Mekk3 Heterozygosity Prevents Cerebral Cavernous Malformations in Ccm2-Deficient Mice. PLoS One. 2016;11(8):0160833. doi: 10.1371/journal.pone.0160833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, et al. Cerebral cavernous malformations arise from endothelial gain of MEKK3-KLF2/4 signalling. Nature. 2016;532(7597):122–126. doi: 10.1038/nature17178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metscher BD. MicroCT for comparative morphology: simple staining methods allow high-contrast 3D imaging of diverse non-mineralized animal tissues. BMC Physiol. 2009;9:11. doi: 10.1186/1472-6793-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JT, et al. Virtual histology of transgenic mouse embryos for high-throughput phenotyping. PLoS Genet. 2006;2(4):61. doi: 10.1371/journal.pgen.0020061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R, Maga AM. A Novel Procedure for Rapid Imaging of Adult Mouse Brains with MicroCT Using Iodine-Based Contrast. PLoS One. 2015;10(11):0142974. doi: 10.1371/journal.pone.0142974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, et al. Dynamic regulation of the cerebral cavernous malformation pathway controls vascular stability and growth. Dev Cell. 2012;23(2):342–355. doi: 10.1016/j.devcel.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]


