Abstract
Tissue engineering is a rapidly growing technological area for the regeneration and reconstruction of damage to the central nervous system. By combining seed cells with appropriate biomaterial scaffolds, tissue engineering has the ability to improve nerve regeneration and functional recovery. In the present study, mouse induced pluripotent stem cells (iPSCs) were generated from mouse embryonic fibroblasts (MEFs) with the non-integrating episomal vectors pCEP4-EO2S-ET2K and pCEP4-miR-302-367 cluster, and differentiated into neural stem cells (NSCs) as transplanting cells. Electrospinning was then used to fabricate randomly oriented poly(L-lactic acid) (PLLA) nanofibers and aligned PLLA nanofibers and assessed their cytocompatibility and neurite guidance effect with iPSC-derived NSCs (iNSCs). The results demonstrated that non-integrated iPSCs were effectively generated and differentiated into iNSCs. PLLA nanofiber scaffolds were able to promote the adhesion, growth, survival and proliferation of the iNSCs. Furthermore, compared with randomly oriented PLLA nanofibers, the aligned PLLA nanofibers greatly directed neurite outgrowth from the iNSCs and significantly promoted neurite growth along the nanofibrous alignment. Overall, these findings indicate the feasibility of using PLLA nanofiber scaffolds in combination with iNSCs in vitro and support their potential for use in nerve tissue engineering.
Keywords: nerve tissue engineering, poly(L-lactic acid), episomal vectors, induced pluripotent stem cells, neural stem cells, aligned nanofibers
Introduction
Spinal cord injury (SCI) is damaging to the axonal tracts and cells around the site of injury and has devastating consequences for patients' locomotor function and sensory function. Unfortunately, the mechanical barriers caused by glial scars and the inhibitory molecules secreted by astrocytes prevent axonal regeneration following SCI. Numerous efforts have been made to solve this problem, including the utilization of autografts and suturing of the gaps (1). However, the two approaches have limitations and have not proven to be feasible (2). Tissue engineering using a combination of biomaterial scaffolds and transplanted seed cells is an inspiring and progressive method of repairing and reconstructing the damaged spinal cord (3). Two important issues require consideration when attempting to bridge the injury gap with tissue engineering. One is the selection of appropriate cells for transplantation, and the other is the identification of scaffolds that are able to enhance cell survival and direct cell differentiation into neurons.
It has been reported that neural stem cells (NSCs) are promising seed cells for the treatment of SCI (4). NSCs differentiate into one of the three major neural cell types that are able to repopulate the damaged spinal cord (5,6). These cells can assist with locomotor function and sensory function recovery by promoting cell survival and the regeneration of host axons. The traditional method of obtaining NSCs, differentiation from embryonic stem cells (ESCs) (5), has ethical issues and causes allogeneic immunological rejection. Fortunately, the discovery of induced pluripotent stem cells (iPSCs) provides an improved method of obtaining NSCs (7,8). The derivation of iPSCs from patient-specific cells for use in transplantation therapies avoids the problems associated with immunological rejection and ethics. However, methods for the generation of iPSCs typically use genome-integrating retroviral or lentiviral vectors, which have safety issues due to insertional mutations and residual transgene expression during iPSC derivation, expansion and differentiation (9). To overcome these issues, integration-free iPSCs with episomal vectors pCEP4-EO2S-ET2K and pCEP4-miR-302-367 cluster have been generated. The use of the pCEP4-EO2S-ET2K vector, encoding octamer-binding transcription factor 4 (OCT4), sex determining region Y-box 2 (SOX2), Kruppel like factor 4 and simian virus 40 large T antigen, has been demonstrated to be a manufacturing practice-compliant method for the generation of integration-free iPSCs (10). The NSCs induced from iPSCs exhibit morphological features and NSC marker gene expression similar to those of wild-type NSCs. Furthermore, these induced NSCs have the potential to differentiate into neurons (11). Therefore, iPSC-derived NSCs are regarded as excellent seed cells for tissue engineering.
Numerous bioengineered scaffolds have been reported to improve neural regeneration in tissue engineering for the treatment of SCI (3). Electrospun fibers have been widely produced for neural tissue engineering scaffolds due to their ability to provide a suitable microenvironment for cell attachment, proliferation and migration that is similar to the natural extracellular matrix (12,13).
Poly(L-lactic acid) (PLLA) is an ideal bioengineered scaffold that has been approved by the United States Food and Drug Administration. PLLA has been widely used as a drug delivery, artificial catheter and tissue engineering scaffold material due to its biocompatibility, tunable degradation rate and non-immunogenicity (14,15). The aligned fibers with diameters in the nanometer range obtained by the electrospinning of natural polymers usually exhibit superior mechanical properties (16). The pore features of electrospun fibers, such as fully interconnected pore structures and a wide-open surface, facilitate the transport of water and nutrients for cell growth (17). In addition, the fibers may be aligned to provide directional guidance for axonal growth. It has been shown that electrospun nanofibers are capable of directing neurite outgrowth and glial cell migration along the aligned orientation (14,18).
In the present study, an electrospun PLLA nanofiber scaffold was synthesized as a three-dimensional (3D) system and its biocompatibility with iPSC-derived NSCs was assessed, with the aim of using the combined system to guide neurite outgrowth and build a cell delivery platform for nerve tissue engineering purposes.
Materials and methods
Ethics statement
Female BALB/c nude immune deficient mice (n=5; age, 6–7 weeks old; weight, 18–22 g) were obtained from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China) and maintained at 22°C (room temperature), 50–60% relative humidity, and a 12 h light/dark cycle in a specific pathogen-free facility. All the mice were fed a sterilized commercial diet. Pregnant BL/6 mice (n=2) carrying the OCT4-EGFP (OG2) pluripotency reporter were obtained from Guangzhou Institute of Biomedicine and Health (Guangzhou, China). All experimental procedures and animal handling were carried out in accordance with the guidelines and laws of the Animal Care and Use Committee of Sun Yat-sen University (Guangzhou, China; approval no. SYXK2012-0083), and conformed with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (8th edition, 2011).
Isolation of murine embryonic fibroblasts and iPSC generation
Mouse embryonic fibroblasts (MEFs) and wild-type NSCs were obtained from embryonic day 13.5–14.5 mouse embryos following previously published protocols (19) and cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 1 mM non-essential amino acids (NEAA) and penicillin-streptomycin (MEF medium). The oriP/EBNA1-based episomal vectors pEP4-EO2S-ET2K (7 µg; Addgene plasmid no. 20927; Addgene, Inc., Cambridge, MA, USA) and pCEP4-miR-302-367 cluster (5 µg) mixture was electroporated into 2×106 MEFs with a Nucleofector 2b Device with an Amaxa Basic Nucleofector kit (Both Lonza Group, Ltd., Basel, Switzerland) according to the manufacturer's protocol. The program used was T-020. The 1×105 cells were seeded onto plates covered with a MEF feeder layer following 2 days of culture in MEF medium. For the induction to iPSCs, the culture medium was switched to iPSC medium [knockout-DMEM supplemented with 15% knockout serum replacement (KSR; Gibco; Thermo Fisher Scientific, Inc.), 1 mM β-mercaptoethanol, 1X NEAA, 1 mM GlutaMAX, 1X penicillin-streptomycin solution, 1,000 U/ml mouse leukemia inhibitory factor (LIF; EMD Millipore, Billerica, MA, USA), 2i (0.5 µM PD0325901 and 3 µM CHIR99021), and 50 µg/ml vitamin C]. The medium was refreshed every 2 days. The colonies were counted 13–16 days after plating, and the colonies that appeared similar to mouse ESCs were selected for further cultivation. Reprogramming efficiency was calculated based on the number of ESC-like alkaline phosphatase (AP)-positive colonies normalized to the number of seeded cells that were electroporated.
Differentiation of mouse iPSCs into NSCs and neurons
iPSCs were dissociated with 0.125% trypsin-EDTA and then transferred to low-attachment dishes (19). They were cultured in iPSC medium without LIF and 2i to promote embryonic body (EB) formation. After 4 days, the EBs were treated with the same medium with additional 1 µM all-trans retinoic acid (RA; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). After another 4 days, EBs were collected by centrifugation at 300 × g for 5 min in room temperature and dissociated with Accutase (Gibco; Thermo Fisher Scientific, Inc.). For differentiation toward a neuronal lineage, the EBs were transferred to tissue culture dishes coated with 0.01% poly-L-lysine (PLL; Sigma-Aldrich; Merck KGaA) and maintained in NSC medium [DMEM/F12 supplemented with b27 and N2 (invitrogen; Thermo Fisher Scientific, Inc.) supplements, 20 ng/ml basic fibroblast growth factor (BFGF) and 20 ng/ml epidermal growth factor (EGF) (both from Peprotech, Inc., Rocky Hill, NJ, USA)]. The medium was refreshed every 2 days. After 7 days, differentiated cells were dissociated with Accutase and cultured in low-attachment dishes with NSC differentiation medium comprising DMEM/F12 supplemented with 20 ng/ml BFGF and 20 ng/ml EGF to form neurospheres. For terminal differentiation into neurons and glial cells, these cells were transferred to tissue culture dishes in NSC differentiation medium (DMEM/F12 supplemented with 5% FBS, 1 µM RA, 1 mM GlutaMAX, 1 mM sodium pyruvate, 0.1 mM NEAA and 1X penicillin/streptomycin) for another 14 days with a medium change every 2 days.
Immunofluorescence staining and AP staining
For immu-nocytochemistry, cells were fixed with 4% paraformaldehyde for 20 min in 4°C and then washed with phosphate-buffered saline (PBS) three times. After treating with 0.2% Triton X-100 and blocking solution (6% FBS; Life Technologies, Carlsbad, CA, USA) for 1 h in room temperature, the cells were incubated with primary antibodies overnight at 4°C and incubated with the respective secondary antibodies for 1 h at room temperature. Nuclei were detected by Hoechst 33342 (Sigma-Aldrich; Merck KGaA) staining in dark for 4 min at room temperature. Cells were imaged with a fluorescence microscope and confocal laser scanning microscopy (LSM 700; both Leica Microsystems, GmbH, Wetzlar, Germany). Antibodies used targeted: SOX2 (1:400; Abcam, Cambridge, MA, USA), OCT4 (1:400; EMD Millipore), homeobox protein NANOG (NANOG; 1:1,000), stage-specific embryonic antigen 1 (SSEA1; 1:1,000) and NESTiN (1:200) (all from Abcam), microtubule-associated protein 2 (MAP2; 1:800; EMD Millipore), β iii tubulin (Tuj1; 1:400; Abcam), glial fibrillary acidic protein (GFAP; 1:200), myelin basic protein (MBP; 1:200) and paired box 6 protein (PAX6; 1:200) (both from Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Alexa 488- and Alexa 555-labeled secondary antibodies were obtained from Invitrogen (Thermo Fisher Scientific, Inc.). AP staining was performed using the Alkaline Phosphatase Detection kit (EMD Millipore) according to the manufacturer's protocol. The immunofluorescence was imaged under a confocal laser scanning microscope (LSM 700; Leica Microsystems GmbH, Wetzlar, Germany). Image processing and analysis were realized using ImageJ (National Institutes of Health, Bethesda, MD, USA) and a plugin called NeuronJ (20). In this program, we used a computer mouse to select a starting point and an ending point to trace the dendrite in order to get quantification metrics. Data were organized using Excel (Microsoft, Redmond, Washington, USA).
RNA preparation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted with TRizol (invitrogen; Thermo Fisher Scientific, Inc.) and cDNA was synthesized using the Reverse Transcription kit (Takara Biotechnology, Co., Ltd., Dalian, China) following the manufacturer's protocol. RT-qPCR was conducted using Maxima™ SYBR-Green/ROX qPCR Master Mix (Thermo Fisher Scientific, Inc.) according to the instructions provided by the manufacturer, with the Stratagene Mx3000P Real-time PCR system (Agilent Technologies, Inc., Santa Clara, CA, USA). DNA was extracted from the iPS cells with Qiagen DNeasy kit (Qiagen Co., Ltd., Shanghai, China). DNA fragments were amplified by PCR with Taq DNA Polymerase (Thermo Fisher Scientific, Inc.). The PCR reaction (50 µl) included 5 µl 10X Taq Buffer, 1.5 mM MgCl2, 2 µmol bidirectional primers, 0.2 mM deoxynucleoside triphosphates (dNTPs), 1.25 U TaqDNA polymerase, 1 µg cDNA and double-distilled water. The PCR reaction conditions were as follows: denaturation at 94°C for 3 min; denaturation at 95°C for 30 sec, annealing at 58°C for 30 sec, extension at 72°C for 45 sec, 30 cycles; extension at 72°C for 10 min. PCR products were separated on a 1.2% agarose gel (with ethidium bromide staining). The resultant gel image was analyzed using the AlphaImager gel analysis system. The primer sequences used for the RT-qPCR and PCR analyses of iPSCs are listed in Table I. The sequences of the primer pairs used for the RT-qPCR analyses of NSCs are listed in Table II. Relative mRNA expression was calculated using the 2−ΔΔCT method and GAPDH was used as an internal control (21).
Table I.
Primers for the RT-qPCR and PCR detection of iPSCs.
| Names | Primer sequences |
|---|---|
| SOX2 | F: 5′-TAGAGCTAGACTCCGGGCGAT-3′ |
| R: 5′-TTGCCTTAAACAAGACCACGAAA-3′ | |
| OCT4 | R: 5′-TTGCCTTAAACAAGACCACGAAA-3′ |
| R: 5′-TGCGGGCGGACATGGGGAGATCC-3′ | |
| NANOG | F: 5′-CACAGTTTGCCTAGTTCTGAGG-3′ |
| R: 5′-GCAAGAATAGTTCTCGGGATGAA-3′ | |
| EXO-SOX2 | F: 5′-ACCAGCTCGCAGACCTACAT-3′ |
| R: 5′-CCCCCTGAACCTGAAACATA-3′ | |
| EXO-OCT4 | F: 5′-AGTGAGAGGCAACCTGGAGA-3′ |
| R: 5′-AGGAACTGCTTCCTTCACGA-3′ | |
| miR-302-367 | R: 5′-CCCCCTGAACCTGAAACATA-3′ |
| R: 5′-CTCCCAAAGAGTCCTGTTCTGTCCT-3′ | |
| EBNA | F: 5′-ATCGTCAAAGCTGCACACAG-3′ |
| R: 5′-CCCAGGAGTCCCAGTAGTCA-3′ | |
| GAPDH | F: 5′-AGGTCGGTGTGAACGGATTTG-3′ |
| R: 5′-GGGGTCGTTGATGGCAACA-3′ |
F, forward; R, reverse; PCR, polymerase chain reaction; RT-qPCR, reverse transcription-quantitative PCR; iPSC, induced pluripotent stem cell; SOX2, sex determining region Y-box 2; OCT4, octamer-binding transcription factor 4.
Table II.
Primers for the RT-qPCR detection of NSCs.
| Names | Primer sequences |
|---|---|
| NESTIN | F: 5′-CCCCTTGCCTAATACCCTTGA-3′ |
| R: 5′-GCCTCAGACATAGGTGGGATG-3′ | |
| PAX6 | F: 5′-GTTGTGTGAGTAAAATTCTGGGC-3′ |
| R: 5′-GAGTCGCCACTCTTGGCTTA-3′ | |
| BLBP | F: 5′-CGCAACCTGGAAGCTGACA-3′ |
| R: 5′-GCCCAGAGCTTTCATGTACTCA-3′ | |
| ZFP42 | F: 5′-CCGGGATGAAAGTGAGATTAGC-3′ |
| R: 5′-TCACCTCGTATGATGCACTC-3′ | |
| GAPDH | F: 5′-AGGTCGGTGTGAACGGATTTG-3′ |
| R: 5′-GGGGTCGTTGATGGCAACA-3′ |
The GAPDH primer pair used was as listed in Table I. F, forward; R, reverse; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; NSC, neural stem cell; PAX6, paired box 6; BLBP, brain lipid-binding protein.
Teratoma formation
To determine the pluripotency of the iPSCs in vivo, the iPSCs were injected into immunocom-promised mice for teratoma formation. The iPSC colonies were collected by 0.5 mM EDTA treatment, and 2×106 iPSCs were subcutaneously injected into 4-week-old BALB/c nude immune deficient mice. At 4 weeks after the cell injection, the teratomas were collected and fixed in 10% formalin at room temperature overnight. The teratomas were then dehydrated through graded ethanol, embedded in paraffin and cut into 5-µm-thick sections. After being dewaxed with ethanol, the slices were soaked with hematoxylin at room temperature for 5 min. Following rinsing with water for 10 min, the slices were stained with eosin for 3 min and dehydrated through graded ethanol. The stained tissues, which were permeabilized with xylene and mounted with resinene, could be observed under a microscope (TS100; Nikon Corporation, Tokyo, Japan).
Patch clamp analysis
iPSC-derived neurons were tested on a microscopic workbench with a patch clamp. Neurons were immersed in extracellular fluid containing 95% O2 and 5% CO2 during the whole process. Cells were visualized using an Olympus patch clamp microscope (Olympus Corporation, Tokyo, Japan). Medium-sized neurons with a bright cell margin and smooth surface were selected for patch clamp analysis. Neurons were inserted into the clamp and data were recorded using an Axon MultiClamp 700B amplifier (Axon instruments; Molecular Devices, LLC, Sunnyvale, CA, USA) and Igor 5.0 software (Wavemetrics, Inc., Lake Oswego, OR, USA).
Preparation of aligned and randomly oriented nanofibrous scaffolds by electrospinning
Polymer solution was prepared by dissolving PLLA in hexafluoroisopropanol at a concentration of 5% w/w. Electrospinning was conducted with the application of a positive voltage (15 kV) to the spinning solution with a distance between the needle tip and the collecting plate surface of 15±2 cm. The solution was placed in a syringe fitted with a 5-ml blunt-end needle and delivered through a syringe pump to control the mass flow rate at 1 ml/h. For aligned nanofibers, the collector was a motor-driven wheel. For random nanofibers, the collector was a stationary plate. The spun nanofibers were dried under vacuum at room temperature for 24 h.
Cell viability, adhesion and proliferation on PLLA nanofibers scaffolds
To investigate the biocompatibility of the PLLA nanofiber scaffolds, iNSCs were dissociated and then seeded on the nanofiber scaffolds. As a control, iNSCs were also seeded and cultured on tissue culture polystyrene (TCPs) coated with PLL, which was used as a positive control for the NSC culture experiments.
Cell survival on the PLLA fiber scaffolds and PLL-coated TCPs was evaluated using a Cell Counting kit-8 (CCK-8; Nanjing KeyGen Biotech Co., Ltd., Nanjing, China) following the manufacturer's protocol. Cells were cultured in 96-well plates for 2 days and then incubated with 100 µl CCK-8 solution per well for 2 h at 37°C, and the absorbance was measured at 450 nm (Elx800; BioTek instruments, Inc., Winooski, VT, USA). INSCs (1×104/well) were seeded onto PLLA scaffolds in a 96-well plate and PLL-coated TCPs. Following incubation for 3, 6 and 9 h, the samples were washed with PBS three times to remove the cells that did not adhere to the scaffolds. The remaining cells were collected with 0.25% trypsin-EDTA and the number of cells was counted under an inverted optical microscope (TS100; Nikon Corporation). Cell proliferation was evaluated using an CCK-8 assay. After seeding the iNSCs (1×103/well) onto PLLA and PLL-coated TCPs for 1, 3, 5, 7 and 9 days in the 96-well plates, cells were incubated with CCK- 8 solution for 2 h at 37°C. The absorbance of the solution in the 96-well plates was then measured. Finally, a proliferation curve of the iNSCs on the scaffolds was constructed.
Morphological examination using scanning electron microscopy (SEM) and nuclear staining
Samples were washed with PBS and then fixed in 2.5% glutaraldehyde overnight at 4°C. The following morning, samples were washed three times prior to dehydration with a series of 30, 50, 70 and 100% ethanol (7 min each). After air-drying, the samples were covered with gold using sputter coating (IB5 ion coater; Eiko Engineering Co., Ltd., Hitachinaka, Japan) and the cell morphologies were observed using SEM (Quanta 200; FEi; Thermo Fisher Scientific, Inc.). For nuclear staining, nuclei were detected by Hoechst 33342 staining (Sigma-Aldrich; Merck KGaA) as described above.
Statistical analysis
Data are reported as the mean ± standard deviation. Statistical analysis was conducted using Student's t-test for comparing two groups of data and analysis of variance (ANOVA) with Bonferroni comparison for three or more groups of data with SPSS 16.0 statistical software (SPSS Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Generation of mouse iPSCs and analysis of pluripotency markers
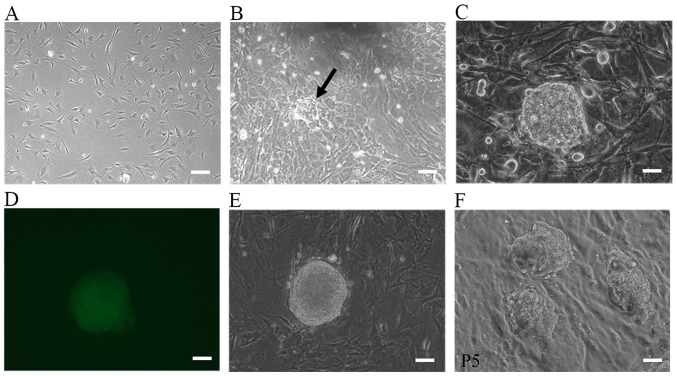
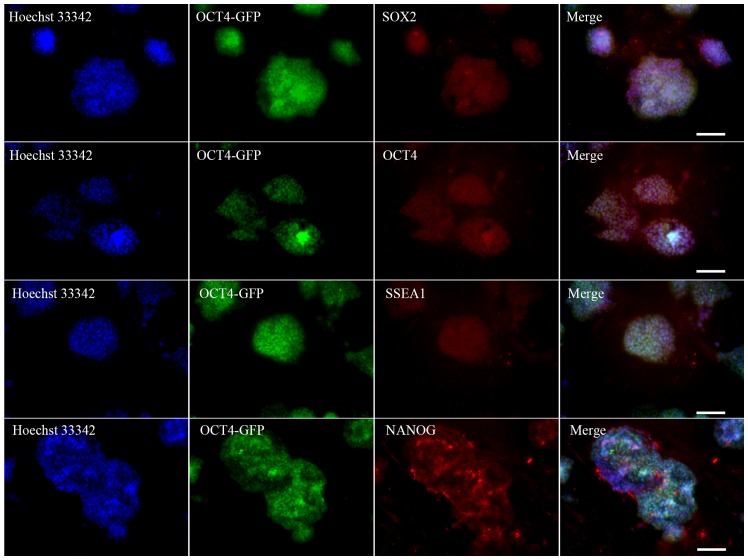
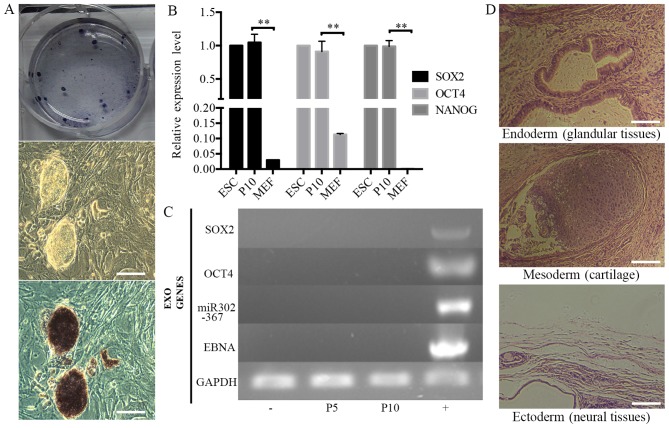
MEFs of low passage number (passage 3) were used for the induction of iPSCs. The oriP/EBNA episomal vectors were transfected into the MEFs through electroporation. ESC-like colonies began to emerge at day 7 following plasmid transfection, and by day 17, they were large enough to be selected for expansion. Fig. 1A–D shows the morphological changes during the process of iPSC generation. For the 1×105 cells routinely used to start reprogramming, the number of ESC-like colonies varied between 10 and 50 and the reprogramming efficiency was 0.01–0.05%. Colonies were manually selected and successfully expanded on MEF feeder cells. Some clones were selected for subsequent studies (Fig. 1E and F). Immunocytochemical analysis on these morphologically ESC-like cells demonstrated that they expressed the ESC markers SOX2, OCT4, SSEA1 and NANOG (Fig. 2). These colonies also stained positive for AP (Fig. 3A). The source cells, MEFs, were negative for all the aforementioned markers. The expression levels of the endogenous pluripotency marker genes SOX2, OCT4 and NANOG were analyzed by RT-qPCR and were comparable to those of mouse ESCs (Fig. 3B). Using PCR to specifically amplify the transgenes used for reprogramming, it was confirmed that the established iPSC colonies no longer harbored the exogenous reprogramming factors and episomal backbones (Fig. 3C).
Figure 1.
Representative phase contrast photographs of the iPSC generation process. An emerging iPSC colony at different time-points. The morphology of MEFs (A) prior to infection, (B) 7 days after transduction and (C) 17 days after transduction. (D) Specific excitation of the pluripotency reporter (OG2). Note that only the iPSC colonies are enhanced green fluorescent protein-positive. (E) The selected colony and (F) the established iPSC line from this colony. Scale bars, 100 µm. The arrow indicates the colony. P5, passage 5; iPSC, induced pluripotent cell; MEF, mouse embryonic fibroblast.
Figure 2.
Immunofluorescence staining of undifferentiated iPSCs, showing the morphology and staining patterns of expanded iPSC clones. Hoechst 33342 was used to stain cellular DNA. Scale bars, 100 µm. iPSC, induced pluripotent stem cell; OCT4, octamer-binding transcription factor 4; GFP, green fluorescent protein; SOX2, sex determining region Y-box 2; SSEA1, stage-specific embryonic antigen 1.
Figure 3.
Characterization of non-integrated iPSCs. (A) AP staining of iPSCs. (B) Reverse transcription-quantitative polymerase chain reaction analysis of iPSCs, using mouse ESCs and MEFs as positive and negative controls and GAPDH as an internal control. **P<0.01 as indicated. (C) Non-integration analysis of episomal DNA in the iPSCs. (-) Negative control (MEFs); (+) positive control (MEFs electroporated with pEP4-EO2S-ET2K and pCEP4-miR-302-367). P5, passage 5; P10, passage 10. (D) Hematoxylin and eosin staining of teratomas from the iPSCs. Scale bars: (A) 100 µm; (D) 200 µm. iPSCs induced pluripotent stem cell; AP, alkaline phosphatase; ESC, embryonic stem cell ; MEF, mouse embryonic fibroblast; SOX2, sex determining region Y-box 2; OCT4, octamer-binding transcription factor 4. EBNA was the backbone of pEP4-EO2S-ET2K plasmid.
Differentiation potential of iPSCs
Teratoma formation assays were performed to examine the differentiation potential of iPSCs in vivo. The iPSC colonies were collected and injected into BALB/c nude mice. Tumor formation efficiency was 100% and palpable tumor masses appeared in all cases 1 week later. The tumors were allowed to develop for another 3 weeks prior to harvesting. Histological examination was performed by a qualified pathologist and the tumors were identified as teratomas. The teratomas contained organized structures of three germ layer cell derivatives, namely glandular tissues (endoderm), cartilage (mesoderm) and neural tissue (ectoderm) (Fig. 3D).
In vitro differentiation into a neural lineage
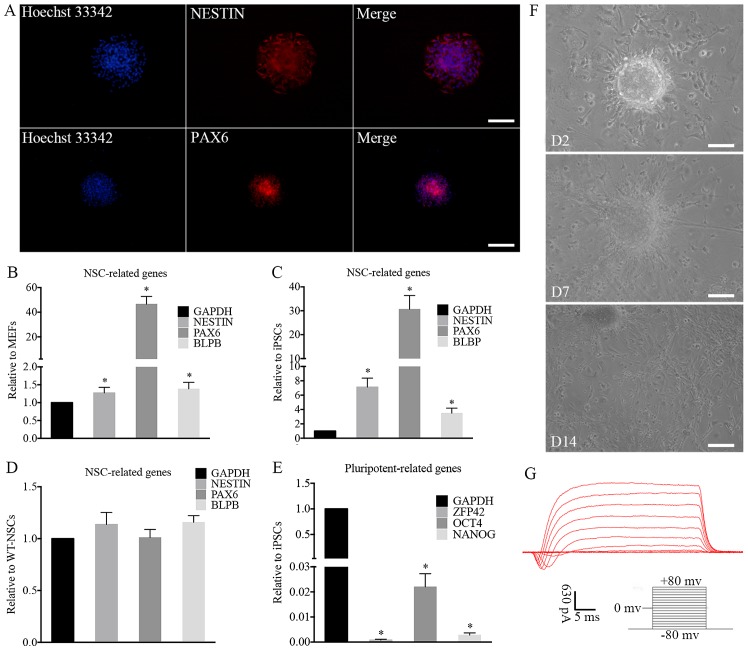
The ability of the iPSCs to differentiate into functional neurons was investigated. The differentiation potential was explored by performing immunocytochemical analysis and RT-qPCR. Immunocytochemical analysis demonstrated that the induced NSC-like cells expressed the NSC markers, NESTIN and paired box 6 (PAX6) (Fig. 4A). For further characterization, RT-qPCR revealed that the expression levels of NESTIN, PAX6 and brain lipid-binding protein (BLBP), which are typical NSC markers, were increased significantly in iNSCs compared with MEFs (Fig. 4B) and iPSCs (Fig. 4C) and the expression levels of these markers in the iNSCs were similar to those in wild-type NSCs (wt-NSCs; Fig. 4D). In addition, the iNSCs did not express significant levels of the pluripotency-related genes OCT4, NANOG and zinc finger protein 42 (ZFP42; Fig. 4E).
Figure 4.
Characterization and differentiation of iNSCs in vitro. (A) immunofluorescence of iNSCs. NSC markers were assessed by immunofluorescence, including NESTiN and PAX6. The expression of typical NSC-related genes on iNSCs significantly increased compared with that on (B) MEFs and (C) iPSCs (*P<0.05) with similar expression to (D) wt-NSCs. (E) Compared with iPSCs, the expression of pluripotency-related genes on iNSCs was negligible (*P<0.05 vs. iPSCs). (F) Differentiated cells migrated from neurospheres on D2 after adherence. Neurites and differentiated cells were observed around the adherent NSCs on D7 and D14. (G) Electrophysiological function of differentiated neurons. (A and F) Scale bars, 100 µm. iNSC, induced pluripotent stem cell-derived neural stem cell; NSC, neural stem cell; MEF, mouse embryonic fibroblast; iPSC, induced pluripotent stem cell; wt, wild-type; PAX6, paired box 6; BLBP, brain lipid-binding protein; ZFP42, zinc finger protein 42; OCT4, octamer-binding transcription factor 4; D2, day 2; D7, day 7; D14, day 14.
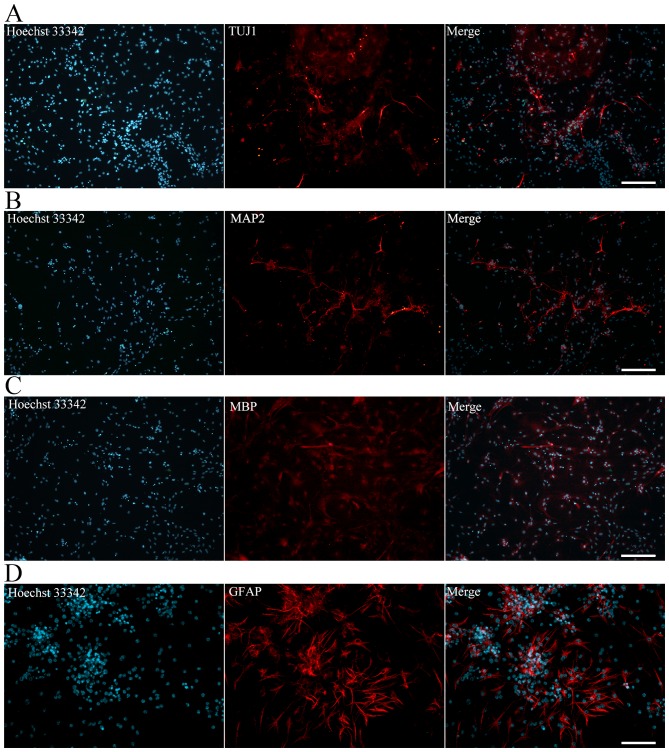
The iNSCs were further differentiated into neurons, astrocytes and oligodendrocytes. The iNSCs cultured on PLL-coated TCPs in NSC differentiation medium exhibited neural lineage cell-like morphology (Fig. 4F). Cells (63.2%) differentiated from iNSCs were detected to exhibit inward Na+ and outward K+ currents, which are the properties of mature, functional neurons (Fig. 4G). At 7 days after differentiation, immunofluorescence staining revealed Tuj1-positive neurons (Fig. 5A). MAP2-positive mature neurons (Fig. 5B), MBP-positive oligodendrocytes (Fig. 5C) and GFAP-positive astrocytes (Fig. 5D) were also observed after culturing for more 7 days. Together, these data indicate that the iNSCs were able to differentiate into neurons, astrocytes and oligodendrocytes.
Figure 5.
Immunofluorescence of differentiated induced pluripotent stem cell-derived neural stem cells. Neurons, oligodendrocytes and astrocytes were assessed by immunofluorescence for (A) TUJ1, (B) MAP2, (C) MBP and (D) GFAP. Scale bars (A-C) 200 µm; (D) 100 µm. TUJ1, β iii tubulin; MAP2, microtubule-associated protein 2; MBP, myelin basic protein; GFAP, glial fibrillary acidic protein.
iNSC survival, adhesion and process extension on PLLA nanofiber scaffolds
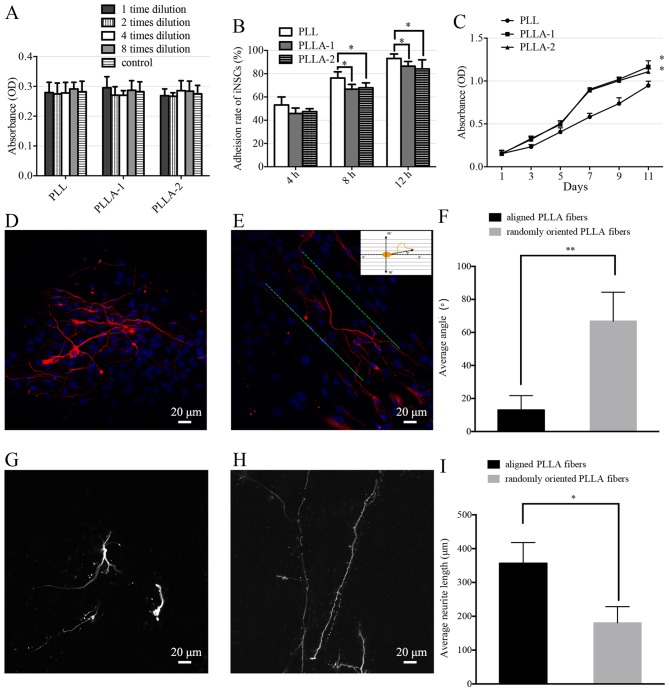
The cell viability of the iNSCs on the PLL-coated TCPs and PLLA nanofiber scaffolds did not differ significantly (Fig. 6A). The adhesion rates of iNSCs on PLL-coated TCPs were significantly higher than those of iNSCs on PLLA nanofibers at each time-point (Fig. 6B). The proliferation of iNSCs on PLLA nanofibers and PLL-coated TCPs was evaluated (Fig. 6C). The number of the cells in the PLL-coated TCPs and PLLA scaffolds increased steadily from the first day of incubation; however, the iNSCs proliferated significantly more strongly on the PLLA scaffolds compared with those on PLL-coated TCPs.
Figure 6.
Anaysis of iNSCs cultured on PLLA nanofibers as a 3D culture system. (A) Viability, (B) cell adhesion rates and (C) proliferation of iNSCs cultured on PLL-coated tissue culture polystyrene (PLL), aligned PLLA nanofibers (PLLA-1) and randomly oriented PLLA fibers (PLLA-2) are shown. *P<0.05 vs. the PLL group. (D-I) Orientation and length of neurite growth on PLLA scaffolds. (D and G) Neurites of iPSC-derived neurons on randomly oriented PLLA fibers and (E and H) aligned PLLA fibers. Neurons were dual-labeled with MAP2 (red) and Hoechst 33342 (blue). The green line shows the direction of the aligned fibers. Quantification of (F) neurite angles and (I) neurite lengths is shown. *P<0.05 and **P<0.01, as indicated. iNSC, induced pluripotent stem cell-derived neural stem cell; PLLA, poly(L-lactic acid); 3D, three-dimensional; iPSC, induced pluripotent stem cell; MAP2, microtubule-associated protein 2; OD, optical density
Analysis of neurite growth on PLLA nanofiber scaffolds as a 3D culture system
Following 14 days of iNSC culture with differentiation medium, the neurites exhibited randomly oriented growth patt erns on the randomly oriented PLLA nanofibers (Fig. 6D), and parallel growth on aligned PLLA nanofibers (Fig. 6E). Fig. 6E shows how the measurement of the neurite orientation was calculated. The average neurite angle on the randomly oriented PLLA nanofibers was calculated as 66.9±17.5°. The average neurite angle on the aligned PLLA nanofibers was 13.0±8.8°, which was significantly smaller than that on the randomly oriented nanofibers (Fig. 6F). The neurite length was measured as a linear distance between the tip of the neurite and the cell junction. The average neurite length on the randomly oriented PLLA nanofibers (Fig. 6G) was calcu lated to be 180.7±47.7 µm. The average neurite length on the aligned PLLA nanofibers (Fig. 6H) was 356.7±61.2 µm, which was significantly longer than that of the neurites on the randomly oriented nanofibers (Fig. 6I).
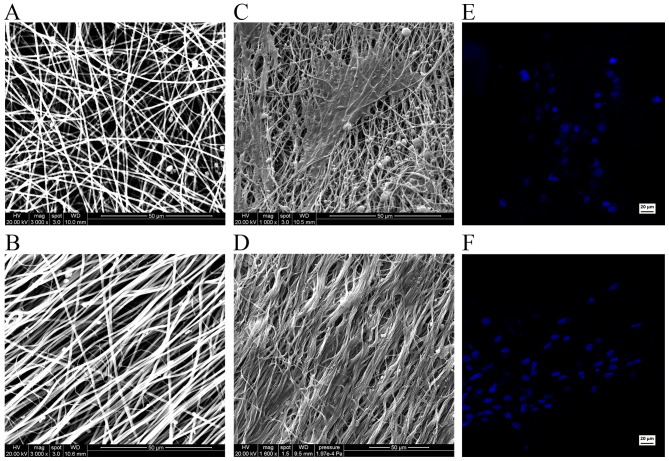
SEM images reveal the relationship between the iNSCs and the supporting nanofibers. Scanning electron micrographs demonstrated the randomly oriented PLLA fibers (Fig. 7A) and aligned PLLA fibers (Fig. 7B). On the randomly oriented PLLA nanofibers, the cells projected neurites that were randomly oriented (Fig. 7C). However, on the aligned PLLA nanofibers, the neurites were observed to be parallel to the nanofibers (Fig. 7D). Hoechst staining demonstrated the normal morphology and quality of the nuclei in the two groups (Fig. 7E and F).
Figure 7.
Morphological examination using scanning electron microscopy and nuclear staining. Representative scanning electron micrographs of template (A) randomly oriented poly(L-lactic acid) (PLLA) fibers and (B) aligned PLLA fibers. The fibers of the scaffolds were entangled to form a three-dimensional matrix. (C) Randomly oriented neurites on random fibers. (D) Aligned neurites on aligned fibers. Hoechst staining shows the normal morphology and quality of the nuclei, with no obvious cellular apoptosis or necrosis in the cells on the (E) random and (F) aligned fibers.
Discussion
Following SCI, nervous system damage causes severe physical and sensory loss that may have severe adverse effects on the quality of life. SCI often causes the death of glial cells and nerve cells local to the injury, which was considered to be unrenewable until 1981 when David and Aguayo reported the injured axons could be regenerated in the appropriate circumstances (22). Numerous studies have attempted to supplement neurons and glial cells with the aim of reestablishing the neural connections and achieving functional recovery (23). NSCs are able to differentiate into neurons, astrocytes and oligodendrocytes. The ability of transplanted NSCs to integrate into host tissue and thereby recover the function of damaged neural tissue by improving neurogenesis and axonal regrowth has been reported (24).
However, due to the multifactorial and multiphasic pathophysiology of the brain and SCI, NSC transplantation alone is likely to cause glial scar and tissue collapse, ultimately affecting the recovery of spinal cord function.
In one cell transplantation approach, a cell suspension is directly injected via fine needles or glass capillaries. Using such an approach, it has been observed that the cell suspension quickly diffuses following injection and does not distribute into the spine cord gap (25). Furthermore, with the lack of an effective support, the surrounding tissues usually collapse. One available strategy to address these issues is to combine the seeding cells with scaffolds to support the reconstruction. The ideal scaffolds should inhibit glial scar formation and provide guidance for neurite outgrowth. This encouraged the present research team to prepare an ideal scaffold for nerve tissue engineering and investigate its biocompatibility with seeding cells to promote reconstruction of the injured spinal cord.
Stem cells, such as ESCs, mesenchymal stem cells and NSCs, have been investigated in nervous system treatment (5,26). Each of these stem cells displays different degrees of regeneration and functional restoration. Among them, NSCs have been reported to be a suitable source of therapeutic cells in neural lesions with the ability to promote the survival of injured neurons and functional recovery (27). Furthermore, transplanted NSCs do not cause obvious graft rejection due to their low immunogenicity (28). These advantages indicate that NSCs are the ideal seeding cells for tissue engineering to repair SCI.
There are a number of methods by which SCs may be obtained. The primitive method is to isolate them from the hippocampus, which is not a practical approach for transplantation (29). In addition, NSCs can be differentiated from ESCs (30)and iPSCs(31). However, the ethical issues which hamper the further development of ESCs remain to be solved. In a revolutionary study, Takahashi and Yamanaka reprogrammed adult somatic cells to become ESC-like cells, termed iPSCs, which paved the way for a new era in regenerative medicine and tissue engineering (32). The iPSC line was observed to behave similarly to ESCs, and conformed to the major aspects of pluripotency, such as unlimited self-renewal, multi-lineage differentiation potential. A number of studies have demonstrated that iPSCs are able to effectively differentiate into functional neurons in vitro and in vivo (33). The generation of patient-specific iPSCs reduces the risk of immune rejection following transplantation and provides the most suitable seeding cells for regenerative medicine. However, initial attempts to generate iPSCs typically used genome-integrating retroviral or lentiviral vectors, which limits their clinical application (9). The genomic integration of transgenes creates insertional mutagenesis and the continued expression of oncogenic proteins, which increases the risk of tumor formation (34).
To overcome these obstacles, several non-integrating approaches had been reported to generate mouse and human iPSCs, including Sendai virus (35), the piggyBac system (36), episomal vectors (37) and direct protein delivery (38). The majority of these reprogramming approaches are inefficient or laborious. The direct delivery of proteins, RNA or modifying Sendai virus vectors is technically demanding, and requires the repeated delivery of the reprogramming factors (34).
In the present study, electroporation of episomal vector (pCEP4-EO2S-ET2K) was conducted to deliver the reprogramming factors into MEFs and obtain non-integrating iPSCs. In addition, pCEP4-miR-302-367 cluster (39), which greatly enhances reprogramming efficiency, was added to the transfection system.
A reprogramming efficiency of up to 0.05% was achieved, which was lower than that of the retroviral or the lentiviral infection approaches (0.1–1%) (40), but higher than that of standard episomal vectors (~0.005%) (37). Furthermore, exogenous reprogramming factors were not detectable in the reprogrammed iPSCs at passages 5 and 10, which is an important safety advantage for clinical application. In addition, plasmid vectors can be manufactured and qualified for good manufacturing practice with a relatively low cost. The capacity of the reprogramed iPSCs to differentiate into neural lineage cells was then investigated. The iNSCs exhibited the expression of the hallmark NSC markers NESTIN, PAX6 and BLBP, with similar expression levels to those in wt-NSCs. In addition, the expression of pluripotent-related genes in these cells was extremely low compared with that of iPSCs. The iNSCs were cultured for further induction in vitro and transplanted in PLLA scaffolds. Cells were observed to survive for prolonged periods and differentiate into mature neurons with the expected electrophysiological properties and glial cells.
Despite surgical interventions and entubulation, the functional recovery of SCI remains very challenging in clinical practice (23). The misdirection of regenerating neurons and the gaps between the injured spinal cord are the main issues of concern (41).
Recently, the development of tissue engineering approaches using functional cells combined with biodegradable scaffolds has shown considerable promise (3,42). The ideal scaffold is able to provide mechanical support as well as a suitable environment, similar to the natural extracellular matrix, that is able to improve cell adhesion and growth. Due to its topographic features and physical properties, PLLA has been widely studied in many fields, particularly tissue engineering (15). Furthermore, it has been reported that this scaffold construct provides a microenvironment for seeding cells that maintains the morphology and functional phenotype of the cells without causing immunological rejection (14). Aligned PLLA nanofibers have been demonstrated to promote the differentiation of pheochromocytoma 12 cells, guide neurite outgrowth and support Schwann cell migration along the aligned orientation (14,18). Furthermore, nanofibrous scaffolds have the capacity to be rolled and packed into a cylindrical shape mimicking the normal spinal cord. This may not only provide ample substrate for cell transplantation, but also support the structure to avoid collapse of the surrounding tissues (43). In addition, the ability of aligned PLLA nanofibers to conduct neonatal nerve growth along the fibrous orientation and thereby promote nerve functional recovery in vivo has been demonstrated (44).
In the present study, electrospinning was used to synthesize aligned electrospun PLLA nanofibers and randomly oriented electrospun PLLA nanofibers as a delivery platform. The biocompatibility of these nanofibers with iNSCs was then tested in vitro. The viability of the iNSCs, measured by CCK-8 assay, was similar on the two PLLA nanofiber scaffolds and on the PLL-coated TCPs. This indicates that PLLA nanofibers have no cytotoxic or inhibitory effects on iNSC survival. Even though cell adhesion rates did not exhibit an apparent improvement on PLLA nanofibrous scaffolds compared with those on PLL-coated TCPs at each time-point, the majority of the cells adhered to the PLLA as time passed. This may be due to the hydrophobic surface of the PLLA scaffolds providing less favorable adhesion conditions than the hydrophilic surface of the PLL-coated TCPs. The proliferation of iNSCs did not differ significantly on day 1 according to whether they were supported on the two PLLA nanofiber scaffolds or the PLL-coated TCPs. However, from day 3, the iNSCs proliferated more strongly on the PLLA nanofibers scaffolds compared with those on the PLL-coated TCPs. Therefore, it may be concluded that the PLLA nanofibrous scaffolds greatly improved iNSCs proliferation.
The aligned PLLA nanofibers appear to be better suited for neurite outgrowth than were the randomly oriented PLLA nanofibers, because the alignment provided guidance cues to direct axonal outgrowth. In the present study, the neurite length and angle between neurite outgrowth and PLLA nanofibers were calculated. The average length of the neurites in the differentiated iNSCs cultured on the aligned PLLA nanofibers was significantly greater than those on the randomly oriented PLLA nanofibers. The majority of the neurites from differentiated iNSCs on the aligned PLLA nanofibers were oriented within 13.0±8.8°, whereas those on those on the randomly oriented PLLA nanofibers exhibited random orientation (66.9±17.5°). Therefore, the aligned PLLA nanofibrous scaffolds exhibit a potent ability to direct differentiated iNSCs and elongate neurite growth length. SEM results also demonstrated that differentiated iNSCs tightly attached and aligned with the nanofibers. The above results demonstrate that aligned PLLA nanofibers are highly effective for neurite guidance and the extension of differentiated iNSCs.
In conclusion, iPSC-derived NSCs are promising cell candidates for the treatment of SCI as they can be generated from a wide range of somatic cells without causing immunology issues and ethical issues. Also, they can effectively develop into a neural lineage. In the present study, integration-free iPSCs were successfully generated with the episomal vectors pEP4-EO2S-ET2K and pCEP4-miR-302-367 cluster. The integration-free iPSCs were differentiated into NSCs and their cytocompatibility and neurite growth on PLLA nanofibrous scaffolds in vitro were investigated. The results indicated that PLLA nanofibers had no cytotoxic or adverse effects on iNSC survival and proliferation, and the aligned PLLA nanofibers were able to direct and elongate neurite outgrowth, supporting their potential usefulness for nerve guidance when implanted at the SCI site. The combination of iPSC-derived NSCs with PLLA scaffolds as a cell-scaffold strategy has great potential for applications in nerve tissue engineering. Studies are ongoing to investigate their feasibility for such applications in vivo.
Acknowledgments
This study was supported by the Natural Science Foundation of China (grant nos. 81772349, 31470949, 31170947 and 81472122), the Guangdong Natural Sciences Foundation of China (S2012020011099 and S2013010016413) and the Guangzhou Science and Technology Planning Project of China (2013J4100062). The authors thank Mr. Baojian Liao for the plasmids, Mr. Bin Jiang for patch clamp analysis, and Mr. Dongbo Qiu and members of the Cell-gene Therapy Translational Medicine Research Center for help and discussion.
References
- 1.Nichols CM, Brenner MJ, Fox IK, Tung TH, Hunter DA, Rickman SR, Mackinnon SE. Effects of motor versus sensory nerve grafts on peripheral nerve regeneration. Exp Neurol. 2004;190:347–355. doi: 10.1016/j.expneurol.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 2.IJkema-Paassen J, Jansen K, Gramsbergen A, Meek MF. Transection of peripheral nerves, bridging strategies and effect evaluation. Biomaterials. 2004;25:1583–1592. doi: 10.1016/S0142-9612(03)00504-0. [DOI] [PubMed] [Google Scholar]
- 3.Evans NR, Davies EM, Dare CJ, Oreffo RO. Tissue engineering strategies in spinal arthrodesis: the clinical imperative and challenges to clinical translation. Regen Med. 2013;8:49–64. doi: 10.2217/rme.12.106. [DOI] [PubMed] [Google Scholar]
- 4.Ziv Y, Avidan H, Pluchino S, Martino G, Schwartz M. Synergy between immune cells and adult neural stem/progenitor cells promotes functional recovery from spinal cord injury. Proc Natl Acad Sci USA. 2006;103:13174–13179. doi: 10.1073/pnas.0603747103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu Y, Wang MY. Neural stem cell grafts for complete spinal cord injury. Neurosurgery. 2012;71:N13–N15. doi: 10.1227/01.neu.0000423041.74441.42. [DOI] [PubMed] [Google Scholar]
- 6.Piltti KM, Salazar DL, Uchida N, Cummings BJ, Anderson AJ. Safety of human neural stem cell transplantation in chronic spinal cord injury. Stem Cells Transl Med. 2013;2:961–974. doi: 10.5966/sctm.2013-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi HW, Kim JS, Choi S, Hong YJ, Kim MJ, Seo HG, Do JT. Neural stem cells differentiated from iPS cells spontaneously regain pluripotency. Stem Cells. 2014;32:2596–2604. doi: 10.1002/stem.1757. [DOI] [PubMed] [Google Scholar]
- 8.Chen X, Gu Q, Wang X, Ma Q, Tang H, Yan X, Guo X, Yan H, Hao J, Zeng F. Directed neuronal differentiation of mouse embryonic and induced pluripotent stem cells and their gene expression profiles. Int J Mol Med. 2013;32:25–34. doi: 10.3892/ijmm.2013.1372. [DOI] [PubMed] [Google Scholar]
- 9.Okita K, Ichisaka T, Yamanaka S. Generation of germ-line-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 10.Briggs JA, Sun J, Shepherd J, Ovchinnikov DA, Chung TL, Nayler SP, Kao LP, Morrow CA, Thakar NY, Soo SY, et al. Integration-free induced pluripotent stem cells model genetic and neural developmental features of down syndrome etiology. Stem Cells. 2013;31:467–478. doi: 10.1002/stem.1297. [DOI] [PubMed] [Google Scholar]
- 11.Hoveizi E, Ebrahimi-Barough S, Tavakol S, Sanamiri K. In vitro differentiation of human iPS cells into neural like cells on a biomimetic polyurea. Mol Neurobiol. 2017;54:601–607. doi: 10.1007/s12035-015-9663-7. [DOI] [PubMed] [Google Scholar]
- 12.Pham QP, Sharma U, Mikos AG. Electrospinning of polymeric nanofibers for tissue engineering applications: a review. Tissue Eng. 2006;12:1197–1211. doi: 10.1089/ten.2006.12.1197. [DOI] [PubMed] [Google Scholar]
- 13.Ebrahimi-Barough S, Hoveizi E, Norouzi Javidan A, Ai J. Investigating the neuroglial differentiation effect of neuroblastoma conditioned medium in human endometrial stem cells cultured on 3D nanofibrous scaffold. J Biomed Mater Res A. 2015;103:2621–2627. doi: 10.1002/jbm.a.35397. [DOI] [PubMed] [Google Scholar]
- 14.Wang HB, Mullins ME, Cregg JM, McCarthy CW, Gilbert RJ. Varying the diameter of aligned electrospun fibers alters neurite outgrowth and Schwann cell migration. Acta Biomater. 2010;6:2970–2978. doi: 10.1016/j.actbio.2010.02.020. [DOI] [PubMed] [Google Scholar]
- 15.Kabiri M, Oraee-Yazdani S, Shafiee A, Hanaee-Ahvaz H, Dodel M, Vaseei M, Soleimani M. Neuroregenerative effects of olfactory ensheathing cells transplanted in a multi-layered conductive nanofibrous conduit in peripheral nerve repair in rats. J Biomed Sci. 2015;22:35. doi: 10.1186/s12929-015-0144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim SH, Mao HQ. Electrospun scaffolds for stem cell engineering. Adv Drug Deliv Rev. 2009;61:1084–1096. doi: 10.1016/j.addr.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 17.Ghasemi-Mobarakeh L, Prabhakaran MP, Morshed M, Nasr-Esfahani MH, Ramakrishna S. Electrospun poly(epsilon-caprolactone)/gelatin nanofibrous scaffolds for nerve tissue engineering. Biomaterials. 2008;29:4532–4539. doi: 10.1016/j.biomaterials.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 18.Yu Y, Lü X, Ding F. influence of poly(L-lactic acid) aligned nanofibers on C12 differentiation. J Biomed Nanotechnol. 2015;11:816–827. doi: 10.1166/jbn.2015.1973. [DOI] [PubMed] [Google Scholar]
- 19.Liu C, Huang Y, Pang M, Yang Y, Li S, Liu L, Shu T, Zhou W, Wang X, Rong L, et al. Tissue-engineered regeneration of completely transected spinal cord using induced neural stem cells and gelatin-electrospun poly (lactide-co-glycolide)/polyethylene glycol scaffolds. PLoS One. 2015;10:e0117709. doi: 10.1371/journal.pone.0117709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- 21.Livak, Schmittgen Analysis of relative gene expression data using real-time quantitative PCR and the 22(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.David S, Aguayo AJ. Axonal elongation into peripheral nervous system 'bridges' after central nervous system injury in adult rats. Science. 1981;214:931–933. doi: 10.1126/science.6171034. [DOI] [PubMed] [Google Scholar]
- 23.Johnson EO, Zoubos AB, Soucacos PN. Regeneration and repair of peripheral nerves. Injury. 2005;36(Suppl 4):S24–S29. doi: 10.1016/j.injury.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 24.Darsalia V, Kallur T, Kokaia Z. Survival, migration and neuronal differentiation of human fetal striatal and cortical neural stem cells grafted in stroke-damaged rat striatum. Eur J Neurosci. 2007;26:605–614. doi: 10.1111/j.1460-9568.2007.05702.x. [DOI] [PubMed] [Google Scholar]
- 25.Paul C, Samdani AF, Betz RR, Fischer I, Neuhuber B. Grafting of human bone marrow stromal cells into spinal cord injury: a comparison of delivery methods. Spine. 2009;34:328–334. doi: 10.1097/BRS.0b013e31819403ce. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu G, Cheng Y, Guo S, Feng Y, Li Q, Jia H, Wang Y, Tong L, Tong X. Transplantation of adipose-derived stem cells for peripheral nerve repair. Int J Mol Med. 2011;28:565–572. doi: 10.3892/ijmm.2011.725. [DOI] [PubMed] [Google Scholar]
- 27.Park Ki, Teng YD, Snyder EY. The injured brain interacts reciprocally with neural stem cells supported by scaffolds to reconstitute lost tissue. Nat Biotechnol. 2002;20:1111–1117. doi: 10.1038/nbt751. [DOI] [PubMed] [Google Scholar]
- 28.Ring KL, Tong LM, Balestra ME, Javier R, Andrews-Zwilling Y, Li G, Walker D, Zhang WR, Kreitzer AC, Huang Y. Direct reprogramming of mouse and human fibroblasts into multi-potent neural stem cells with a single factor. Cell Stem Cell. 2012;11:100–109. doi: 10.1016/j.stem.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 30.Elkabetz Y, Studer L. Human ESC-derived neural rosettes and neural stem cell progression. Cold Spring Harb Symp Quant Biol. 2008;73:377–387. doi: 10.1101/sqb.2008.73.052. [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi Y, Okada Y, Itakura G, Iwai H, Nishimura S, Yasuda A, Nori S, Hikishima K, Konomi T, Fujiyoshi K, et al. Pre-evaluated safe human iPSC-derived neural stem cells promote functional recovery after spinal cord injury in common marmoset without tumorigenicity. PloS one. 2012;7:e52787. doi: 10.1371/journal.pone.0052787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi K, Yamanaka S. induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 33.Aboody K, Capela A, Niazi N, Stern JH, Temple S. Translating stem cell studies to the clinic for CNS repair: current state of the art and the need for a Rosetta stone. Neuron. 2011;70:597–613. doi: 10.1016/j.neuron.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 34.Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, Hong H, Nakagawa M, Tanabe K, Tezuka K, et al. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 35.Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Talluri TR, Kumar D, Glage S, Garrels W, Ivics Z, Debowski K, Behr R, Kues WA. Non-viral reprogramming of fibroblasts into induced pluripotent stem cells by Sleeping Beauty and piggyBac transposons. Biochem Biophys Res Commun. 2014;450:581–587. doi: 10.1016/j.bbrc.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 37.Jia F, Wilson KD, Sun N, Gupta DM, Huang M, Li Z, Panetta NJ, Chen ZY, Robbins RC, Kay MA, et al. A nonviral minicircle vector for deriving human iPS cells. Nat Methods. 2010;7:197–199. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim D, Kim CH, Moon Ji, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu S, Wilson KD, Ghosh Z, Han L, Wang Y, Lan F, Ransohoff KJ, Burridge P, Wu JC. MicroRNA-302 increases repro gramming efficiency via repression of NR2F2. Stem Cells. 2013;31:259–268. doi: 10.1002/stem.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wrobel MR, Sundararaghavan HG. Directed migration in neural tissue engineering. Tissue Eng Part B Rev. 2014;20:93–105. doi: 10.1089/ten.teb.2013.0233. [DOI] [PubMed] [Google Scholar]
- 42.Hoveizi E, Tavakol S, Ebrahimi-Barough S. Neuroprotective effect of transplanted neural precursors embedded on PLA/CS scaffold in an animal model of multiple sclerosis. Mol Neurobiol. 2015;51:1334–1342. doi: 10.1007/s12035-014-8812-8. [DOI] [PubMed] [Google Scholar]
- 43.Li J, Shi R. Fabrication of patterned multi-walled poly-l-lactic acid conduits for nerve regeneration. J Neurosci Methods. 2007;165:257–264. doi: 10.1016/j.jneumeth.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 44.Hurtado A, Cregg JM, Wang HB, Wendell DF, Oudega M, Gilbert RJ, McDonald JW. Robust CNS regeneration after complete spinal cord transection using aligned poly-L-lactic acid microfibers. Biomaterials. 2011;32:6068–6079. doi: 10.1016/j.biomaterials.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]