Abstract
Cluster of differentiation (CD)90 (Thy-1) was proposed as a marker for the liver cancer stem cells that are responsible for tumorigenic activity, however its involvement in the progression of hepatocellular carcinoma (HCC) remains unknown. The aim of the present study was to determine the effect of CD90 on the biological functions of HCC and to investigate the associated circular RNA (circRNA) involved in this process. The analysis of the in vitro data demonstrated that CD90+ cells isolated from SK-Hep-1 cells exhibited increased viability, migration and invasive abilities compared with CD90− cells. In addition, circRNA expression profiles in CD90+ and CD90− cells were screened using a microarray assay and hsa_circ_0067531 and hsa_circ_0057096 were identified to be expressed at significantly different levels. It was additionally demonstrated that the expression of hsa_circ_0067531 in HCC tissues was significantly decreased compared with normal adjacent tissues. Overall, the results of the present study suggested that CD90 may be used as a potential biomarker for HCC. Furthermore, it was demonstrated that hsa_circ_0067531 may affect the biological functions of CD90+ HCC cells and may be a promising candidate to aid in the diagnosis and therapy of HCC.
Keywords: hepatocellular carcinoma, cluster of differentiation 90, circular RNA
Introduction
Hepatocellular carcinoma (HCC) ranks as the fifth most frequently occurring neoplasm and the second leading cause of cancer-associated mortalities worldwide. A total of >745,000 fatalities occur as a result of HCC, along with 500,000 newly diagnosed cases every year (1–3). Current diagnostic standards may be inaccurate and result in missed cases, and since early diagnosis is key for successful operations, it is imperative to search for more efficient biomarkers to increase the early diagnostic rate of HCC (4,5). Cancer stem cells (CSCs) are the source of numerous solid tumor types, including HCC, and are characterized by a variety of stem cell markers (6–11). A previous study revealed that cluster of differentiation (CD)90 (Thy-1) is a potential marker for liver CSCs (12) and is expressed in a variety of cells, including T-cells, thymocytes, neurons, endothelial cells and fibroblasts. It is important in cell-to-cell and cell-to-matrix interactions, apoptosis, adhesion, migration and fibrosis (13). Previous studies have demonstrated that CD90+, however not CD90− cells, obtained from HCC cell lines, exhibit tumorigenic and metastatic capacities (12,14,15). The human HCC cell line JHH-6 demonstrates increased proliferative capacity of CD90+ compared with CD90− cells (16). Conversely, CD90 functions as a tumor suppressor in ovarian cancer (17). The specific function and mechanism by which CD90+ cells contribute to HCC progression remains to be elucidated.
Circular RNAs (circRNAs) are a novel type of endogenous noncoding RNA characterized as stable, abundant, and conserved, and usually exhibit tissue/developmental-stage specific expression (18). Numerous studies have revealed that circRNAs participate in the initiation and development of multiple diseases, including Alzheimer's, Parkinson's, atherosclerosis, and various cancers (19,20). CircRNAs bind to miRNAs, acting as miRNA sponges, and thereby regulate gene expression (21). Hsa_circ_0004018 has been reported to be involved in cancer-associated pathways in HCC via interactions with miRNAs (22). The circRNA that acts as a sponge for miR-7 is Cdr1as (ciRS-7), the knockdown of which suppresses HCC cell viability and invasion (23). This evidence suggests that circRNAs are involved in HCC progression and may be promising diagnostic or predictive biomarkers. However, the expression of circRNAs in HCC has not extensively been studied and the mechanisms of circRNAs in HCC remain to be elucidated.
The present study aimed to investigate the effect of CD90 on the cell cycle and the migration, invasion, and sphere formation abilities of HCC cells. Furthermore, differentially expressed circRNAs between CD90+ and CD90− HCC cells were identified via high throughput microarray assay.
Materials and methods
Ethics statement
The present study was approved by the Ethics Committee of Sun Yat-Sen University (Guangzhou, China), and written informed consent was obtained from all patients in Sun Yat-Sen Memorial Hospital of Sun Yat-Sen University.
Specimen collection and cell culture
A total of eight pairs of HCC tumor tissues and corresponding non-tumor tissues were collected from the Department of Hepatopancreatobiliary Surgery, Sun Yat-Sen Memorial Hospital of Sun Yat-Sen University. The present study included 8 patients with HCC, 5 males and 3 females, aged 35–66 years with a mean age of 55.9±9.5 years. Exclusion criteria were the following: i) Patients with moderate or tense ascites, ii) patients refusing liver biopsy, iii) patients with previous treatment for HCC and iv) patients with a history of severe trauma. None of the recruited participants accepted adjunctive treatment prior to the surgery. All tissue samples were immediately preserved at −80°C following washing with sterile phosphate-buffered saline (PBS). HCC cell lines (LM3, Huh7, MHCC 97L and SK-Hep-1) were purchased from Shanghai Cell Center (Shanghai, China) and cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal calf serum (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and 1% penicillin-streptomycin G (Invitrogen; Thermo Fisher Scientific, Inc.). Cells were incubated at 37°C in a humidified atmosphere containing 5% CO2.
Flow cytometry
The quantitation of CD90 expression in the cells (Huh7, MHCC 97L and SK-Hep-1) was determined using flow cytometry. A total of 2×105 cells were transferred into a centrifuge tube and suspended in PBS buffer. Following centrifugation at 1,000 × g for 5 min at 4°C, the cells were fixed with fresh fixative (4% paraformaldehyde; Thermo Fisher Scientific Inc.) at room temperature for 15 min, and then preincubated in 200 ml blocking solution (10% goat serum containing 0.05% Tween-20; Thermo Fisher Scientific Inc.) for 1 h at room temperature. The cells were incubated with fluorescein isothiocyanate (FITC)-conjugated human anti-CD90 (catalog no. 85-11-0909-41; 1:100 dilution; eBiosciences; Thermo Fisher Scientific Inc.) for 1 h at 4°C, then the cells were washed in PBS and incubated with FITC-conjugated goat anti-mouse IgG secondary antibody (catalog no. ab6785; 1:1,000 dilution; Abcam, Cambridge, MA, USA). The standard was established by isotype match control (FITC-conjugated mouse IgG1 isotype control, catalog no. 85-11-4714-81; 1:100 dilution; eBiosciences; Thermo Fisher Scientific Inc.) for 1 h at 4°C. A total of 1×106 FITC-labeled cells were measured using BD AccuriC6 Flow Cytometer with Cell Quest software version 5.1 (BD Biosciences, Franklin Lakes, NJ, USA). Each experiment was performed in triplicate. The cell cycle of SK-Hep-1 CD90+ and CD90− cells was additionally determined with flow cytometry. Briefly, cells were resuspended in PBS twice prior to fixation by dropwise addition into 95% pre-cooled ethanol for 15 min at room temperature, and then centrifuged at 3,000 × g for 10 min at 4°C, resuspended twice in PBS, and stained with propidium iodide (PI) containing 50 µg/ml RNaseA (Takara Bio, Inc., Otsu, Japan) in the dark for 30 min at room temperature. The DNA content was analyzed using BD AccuriC6 Flow Cytometer with Cell Quest software, version 5.1 (BD Biosciences).
Cell separation via magnetic sorting
Separation of CD90+ cells from SK-Hep-1 cells was performed using MACS magnetic cell sorting (Miltenyi Biotech GmbH, Bergisch Gladbach, Germany) according to the manufacturer's protocol. SK-Hep-1 cells were incubated with 50 µl of FcR blocking agent (Miltenyi Biotech GmbH) and CD90-biotin immunomagnetic beads (Miltenyi Biotech GmbH) for 10 min at 4°C. Goat anti-mouse IgG magnetic beads (Miltenyi Biotech GmbH) were used as the control. A MACS cell separation column (MS column) was used to retain the CD90+ cells linked to the beads. Unlabeled cells (SK-Hep-1 CD90− cells) were washed with 3×500 µl buffer (containing PBS, 0.5% BSA, 2 mM EDTA) and collected. The MS column was then removed from the separator and placed on another collection tube. A total of 1 ml buffer was pipetted onto the MS column, and the labeled CD90+ cells (SK-Hep-1 CD90+ cells) were flushed out by applying the plunger supplied with the column. The labeled CD90+ cells were eventually obtained via centrifugation at 3,000 × g for 10 min at 4°C and resuspension in serum-free medium (Gibco; Thermo Fisher Scientific, Inc.). The SK-Hep-1 CD90+ and SK-Hep-1 CD90− cells were harvested and washed with PBS, and the expression of CD90 in these cells was directly investigated with flow cytometry. Following sorting, the SK-Hep-1 CD90+ cells indicated an ≥85% expression level of CD90 and the SK-Hep-1 CD90− cells revealed <5% expression level of CD90.
Western blotting
SK-Hep-1 cells were lysed in lysis buffer (Gibco; Thermo Fisher Scientific, Inc.) and the concentration of total protein was analyzed using a BCA Protein Assay Kit (Sangon Biotech Co., Ltd, Shanghai, China). A total of 30 µg protein was added into each lane of the 10% SDS-PAGE gel and 5% skim milk (BD Biosciences) was used to block the polyvinylidene membranes for 10 min at room temperature. The membranes were then incubated with the primary antibody anti-CD90/Thy1 (catalog no. ab133350; 1:100; Abcam) at 4°C overnight. GAPDH was used as the internal control (anti-GAPDH antibody; catalog no. ab9485; 1:100, Abcam). The next day, the membranes were incubated with horseradish peroxidase-conjugated anti-rabbit IgG secondary antibody for 1 h at room temperature (1:500 dilution; catalog no. ab7090; Abcam). The results were visualized using the enhanced chemiluminescence substrate kit (GE Healthcare, Chicago, IL, USA). All images were analyzed with ImageJ software, version 14.8 (National Institutes of Health, Bethesda, MD, USA).
Cell viability
Cell viability was assessed with a 3-(4,5- dim ethylthiazol-2-yl)-2,5-diphenyltrtrazolium bromide (MTT) assay. The treated cells were seeded into a 96-well plate and cultured in complete medium. When the cells adhered, 0.5 mg/ml MTT was added to each well and incubated at 37°C for 4 h. The supernatant was then carefully aspirated and 100 µl of dimethyl sulfoxide was added. The absorbance was measured at a wavelength of 490 nm using a microplate reader (Thermo Fisher Scientific, Inc.).
Migration and invasion assays
Cell migration and invasion assays were performed using Transwell chambers with 8 µm pores (BD Biosciences) according to the manufacturer's protocol. For migration assays, SK-Hep-1 CD90+ (5×104 cells/well) and SK-Hep-1 CD90− cells (5×104 cells/well) were separately seeded into the upper compartment without Matrigel, for invasion assay, the cells were seeded into the upper compartment with Matrigel (Corning Inc., Corning, NY, USA) and incubated in serum-free DMEM (Gibco; Thermo Fisher Scientific, Inc.) and the lower compartment was filled with complete medium supplemented with 10% FBS. Following 24 h, migratory and invasive cells on the bottom surface of the filters were fixed with 4% paraformaldehyde for 24 h and stained with 0.1% crystal violet solution for 10 min at room temperature. The cells in at least 10 randomly selected microscopic fields were counted under a light microscope, at ×200 magnification (Nikon Corp., Tokyo, Japan). Each experiment was performed in triplicate.
Sphere formation assay
SK-Hep-1 CD90+ and SK-Hep-1 CD90− cells were trypsinized and cultured in DMEM-F12 (Sigma-Aldrich; Merck KGaA) that was supplemented with 20 ng/ml EGF, 10 ng/ml FGF, 4 ng/ml insulin, and B27 (1:50) in Ultra-Low Attachment 6-well plates (Corning Inc.). The cells were imaged under a light microscope (magnification, ×200; Olympus Corp., Tokyo, Japan) following 20 days of incubation.
Microarray hybridization
Total RNA was treated with Rnase R (Epicentre Illumina, Inc., San Diego, CA, USA) to remove linear RNA, and was then amplified and transcribed into fluorescent cRNA using Quick Amp Labeling kit (Takara Bio, Inc.) according to the manufacturer's protocol. The cRNA was then purified with the RNeasy Mini Kit (Qiagen GmbH, Hilden, Germany) and hybridized to the circRNA expression microarray according to the manufacturer's protocols. The microarray hybridization and the data collection were performed by Forevergen Bio-tech Co. (Guangzhou, China). For the microarray analysis, following filtering by flag signal, the raw data were processed and normalized. Differentially expressed circRNA were identified with P<0.05 and fold changes >2. In addition, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses was performed which provided more detailed information regarding the pathways in which the target genes were involved.
Specialty confirmation and Sanger sequencing
To verify the specialty of the PCR products of hsa_circ_0067531 and hsa_circ_0057096, the cDNA and gDNA PCR products that had been amplified by diverted primers were separated on a 2% agarose gel with TE running buffer. Only a single band in the gel was thought to be a specialty. The region was incised from the whole gel and purified with SanPrep Column DNA Gel Extraction Kit (Qiagen GmbH), and the PCR fragments were inserted into T vector for Sanger sequencing (Tsingke Biotech Co., Ltd., Beijing, China).
Quantitative polymerase chain reaction (qPCR)
Total RNA was isolated using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following indicated treatments. qPCR assays were performed with SYBR Premix Ex Taq (Takara Bio, Inc.), primers, and a cDNA template on the Applied Biosystems 7500 Real-time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The primers used in the assay are listed as follows: Forward, CGA ACC AAC TTC ACC AGC AA and reverse, CTG ATG CCC TCA CAC TTG AC for CD90; forward, 5′-TCC AGA CCA GTA CGT TCG AG-3′ and reverse, 5′-TCA CAC AGT TGA GAC AAG GGA T-3′ for cir_0067531; forward, 5′-AGG CAA AGG AAG TCC ATC TCA T-3′ and reverse, 5′-TCA ATC ACA CCC TGG GCC AT-3′ for circ_0057096 and forward, 5′-CCG AGA ATG GGA AGC TTG TC-3′ and reverse, 5′-AAG CAC CAA CAG AGG AGA A-3′ for GAPDH. PCR was conducted in a 50 µl reaction system, including 1 µl forward and 1 µl reverse primers, 0.5 µl 20× SYBR-Green I, 25 µl 2×PCR buffer, 2 µl cDNA, and 20.5 µl DEPC. The qPCR program was 94°C for 4 min, 30 cycles of 94°C for 20 sec, 60°C for 30 sec, 72°C for 30 sec, and 72°C for 5 min. Each individual sample was run in triplicate and the expression level was quantified with the comparative cycle threshold (Cq) method. Results were normalized to GAPDH expression and RNA enrichments were calculated using the 2−ΔΔCq method (24).
Statistical analysis
Quantile normalization and subsequent data processing were performed using the GeneSpring GX software, version 11.5.1 package (Agilent Technologies, Inc., Santa Clara, CA, USA). Other data were analyzed with the two-tail Student's t-test and one-way analysis of variance followed by Tukey's test. SPSS software, version 19.0 (IBM SPSS, Armonk, NY, USA). Each experiment was repeated at least three times. All results were summarized and are presented as the mean ± standard deviation. P<0.05 was considered to indicate a statistically significant difference.
Results
Distribution of CD90 cells in human HCC cell lines
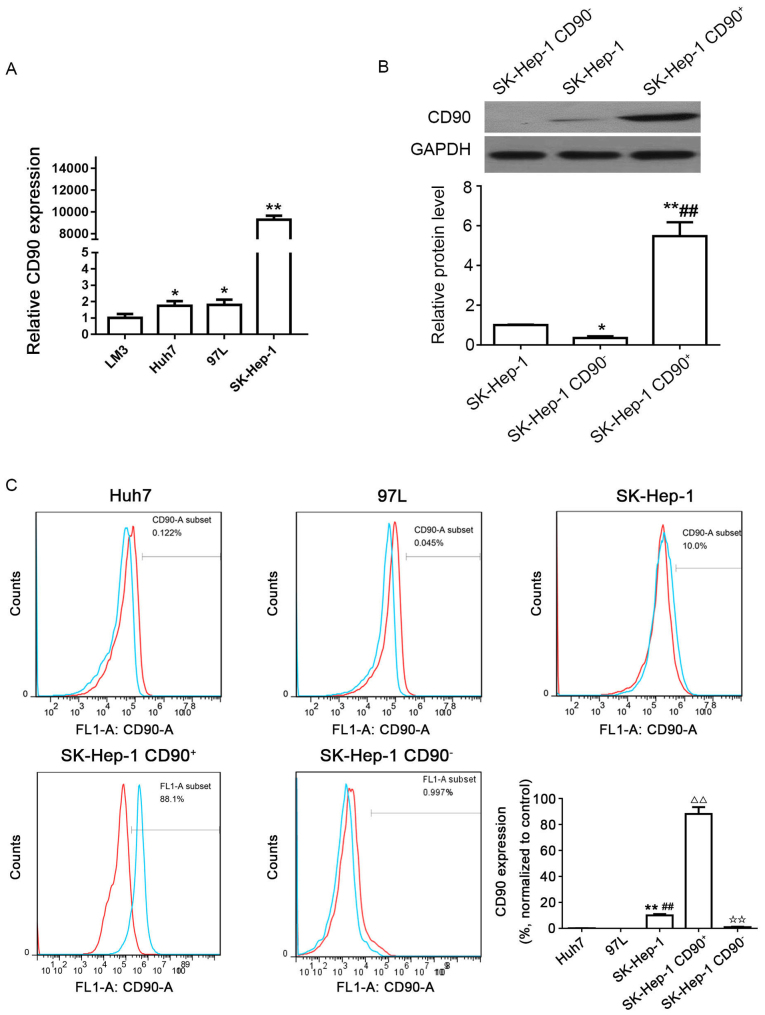
The expression levels of CD90 in the 4 different HCC cell lines were determined by qPCR; results of which demonstrated that the expression of CD90 was increased in the LM3, Huh7, and MHCC 97L cell lines, and particularly in the SK-Hep-1 cell line (Fig. 1A). The western blotting results demonstrated that the protein level of CD90 in SK-Hep-1 CD90+ cells was significantly increased compared with SK-Hep-1 and SK-Hep-1 CD90− cells. However, CD90 expression in SK-Hep-1 CD90− cells was significantly decreased compared with SK-Hep-1 cells (Fig. 1B). In addition, the expression of CD90 (blue line) and control IgG (red line) were verified in the Huh7, MHCC 97L, and SK-Hep-1 cell lines by flow cytometry. It was demonstrated that the relative expression level of CD90 in SK-Hep-1 cells (10.0%) was significantly increased compared with Huh7 (0.122%) and MHCC 97L (0.045%) cell lines (Fig. 1C). Consequently, the SK-Hep-1 cell line was selected for the ensuing experiments. The CD90+ cells were sorted from the SK-Hep-1 cell line with an expression of CD90 (88.1%) ≥85.0%, whereas CD90− cells were sorted from the SK-Hep-1 cell line with an expression of CD90 (0.997%) ≤5.0% (Fig. 1C).
Figure 1.
Distribution of CD90 cells in human HCC cell lines. (A) Expression levels of CD90 in 4 different HCC cell lines (LM3, Huh7, MHCC 97L and SK-Hep-1) were determined by quantitative polymerase chain reaction. ✩P<0.05; ✩✩P<0.01 vs. LM3. (B) Protein expression levels of CD90 in SK-Hep-1, SK-Hep-1 CD90−, and SK-Hep-1 CD90+ cells were determined by western blotting. GAPDH was used as the internal control. ✩P<0.05; ✩✩P<0.01 vs. SK-Hep-1; ##P<0.01 vs. SK-Hep-1 CD90−. (C) Expression levels of CD90 in 3 different HCC cell lines (Huh7, MHCC 97L, and SK-Hep-1), SK-Hep-1 CD90+ cells and SK-Hep-1 CD90− cells were examined with flow cytometry. Each experiment was performed in triplicate. IgG was used for control (red line). The blue line reveals the expression of CD90. The SK-Hep-1 CD90+ cells revealed an expression of 88.1% CD90, whereas CD90− cells exhibited a CD90 expression of 0.997%. ✩✩P<0.01 vs. Huh7; ##P<0.01 vs. MHCC 97L; △△P<0.01 vs. SK-Hep-1; ✩✩P<0.01 vs. SK-Hep-1 CD90+. CD, cluster of differentiation; HCC, hepatocellular carcinoma.
Effect of CD90 on the cell cycle and viability of SK-Hep-1 cells
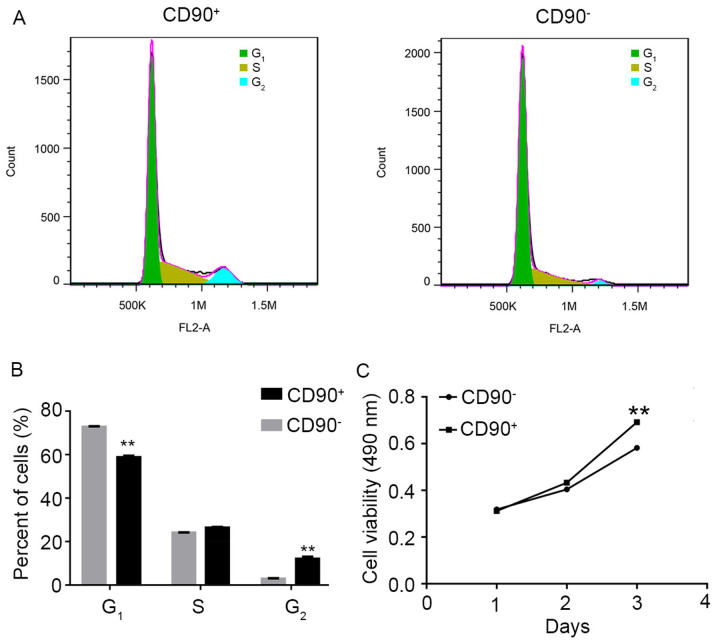
Flow cytometry analysis and MTT assay were performed to investigate the effect of CD90 on the cell cycle and viability in SK-Hep-1 cells. The raw FACScan data is presented in Fig. 2A. The numerical conversion of the cell population in G1, S, and G2 phases is presented in Fig. 2B. The results indicated that the percentage of CD90+ cells was significantly decreased in the G1 phase, compared with CD90− cells. Conversely, the percentage of CD90+ cells was significantly increased in the G2 phase, compared with CD90− cells. These results suggested that the G1 phase of CD90+ cells was shorter compared with CD90− cells. In addition, the MTT assay results demonstrated that the cell viability of CD90+ cells was significantly increased compared with CD90− cells at day 3 (Fig. 2C).
Figure 2.
Effect of SK-Hep-1 CD90+ cells on cell cycle and viability. (A) Representative image and (B) quantitative analysis of flow cytometry assay to analyze the cell cycle of CD90+ and CD90− cells. Data are presented as the mean ± standard deviation of three independent experiments. (C) MTT assay used to determine the viability of CD90+ and CD90− cells. The results demonstrated that the viability of CD90+ cells was significantly increased compared with CD90− cells. **P<0.01 vs. CD90−. CD, cluster of differentiation.
Migration and invasion abilities increase in CD90+ cells
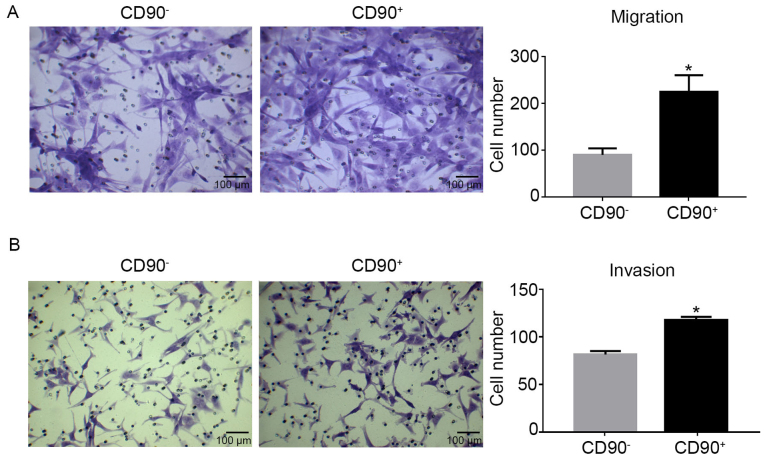
Transwell assays were used to determine the migration and invasion abilities of CD90+ and CD90− cells. The migration assay revealed that there were significantly more CD90+ cells compared with CD90− cells (Fig. 3A). Similarly, the invasion assay demonstrated the same results as the migration assay (Fig. 3B). Consequently, it was demonstrated that CD90+ cells exhibited increased migration and invasion capabilities compared with CD90− cells.
Figure 3.
Migration and invasion ability of CD90+ and CD90− cells. (A) A Transwell assay was used to determine the migration ability of CD90+ and CD90− cells. (B) A Matrigel Transwell assay was used to determine the invasion ability of CD90+ and CD90− cells. The results demonstrated that the migration and invasion abilities of CD90+ cells were significantly increased compared with CD90− cells. *P<0.05 vs. CD90−. Magnification, ×200; Scale bar, 100 µm. CD, cluster of differentiation.
CD90+ induces sphere formation
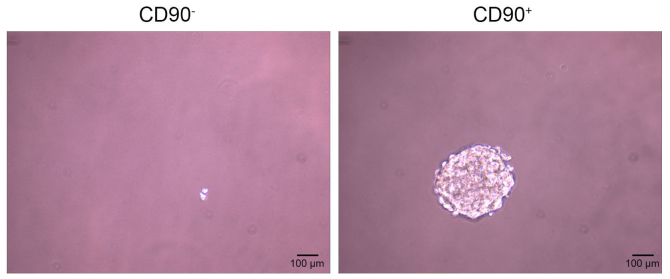
The sphere formation assay demonstrated that CD90− cells did not form spheres following 20 days of incubation, however the CD90+ cells did (Fig. 4).
Figure 4.
Sphere-forming ability of CD90+ and CD90− cells. Spheroid formation assay. CD90− and CD90+ cells were cultured on 6-well plates or as cell spheres. The cells were imaged under a light microscope (magnification, ×200) following 20 days of incubation. Each experiment was repeated three times. Scale bar, 100 µm.
Identification of differentially expressed circRNA profiles
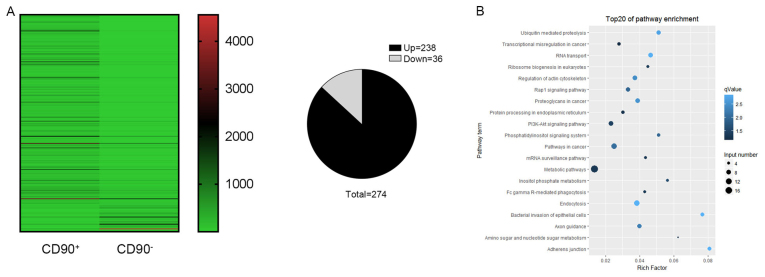
A high throughput microarray assay was used to identify the differing expressions of circRNAs in CD90+ and CD90− cells. In the present study, 274 dysregulated circRNAs were identified, including 238 upregulated and 36 downregulated circRNAs (Fig. 5A). The upregulated circRNAs were more frequently occurring compared with downregulated circRNAs. In order to further understand the biological functions of the differentially expressed circRNAs, KEGG pathway analyses was performed which provided more detailed information regarding the pathways in which the target genes were involved. It was revealed that the key signaling pathways were the metabolic pathway, pathways in cancer, and the phosphoinositide 3-kinase (P13K)-RAC-α serine/threonine-protein kinase (AKT) signaling pathway (Fig. 5B).
Figure 5.
Identification of differentially expressed circRNA profiles. Differentially expressed circRNAs between CD90+ and CD90− cells were analyzed using a high throughput microarray assay. (A) The heat map revealed 274 distinguishable (238 upregulated and 36 downregulated) circRNAs with differential expression between CD90+ and CD90− cells. (B) The key signaling pathways for these circRNAs were identified by Kyoto Encyclopedia of Genes and Genomes enrichment analyses and the top 20 are listed; the top three signaling pathways were the metabolic pathway, pathways in cancer, and the P13K-AKT signaling pathway. CD, cluster of differentiation; circRNA, circular RNA; P13K-AKT, phosphoinositide 3-kinase (P13K)-RAC-α serine/threonine-protein kinase.
Characterization of hsa_circ_0067531 and hsa_circ_0057096 in HCC
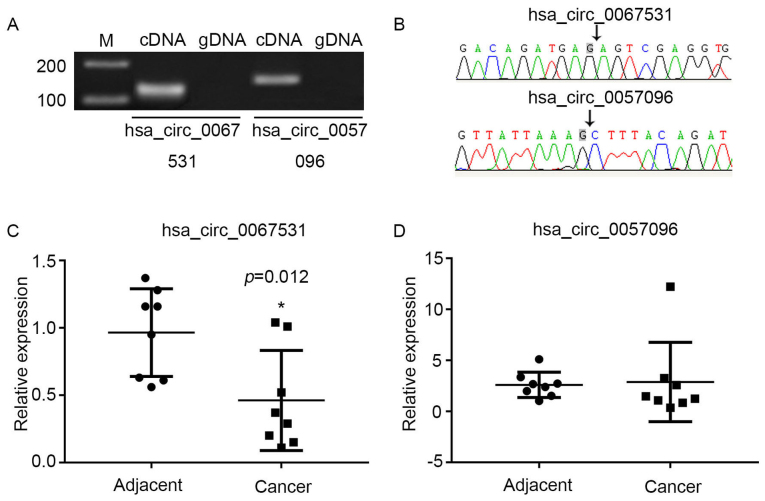
To verify the microarray data, two circRNAs with differential expression were selected from the P13K-AKT signaling pathway; hsa_circ_0067531 and hsa_ circ_0057096. The sequences of these two circRNAs from pyruvate dehydrogenase kinase (PDK)1 and phosphatidylino-sitol-4,5-bisphosphate 3-kinase catalytic subunit β (PIK3CB) are listed in Table I. Divergent primers amplify circRNAs in cDNA, however not genomic DNA (gDNA) (Fig. 6A). The Sanger sequencing verified head-to-tail splicing (Fig. 6B). The present study validated the expression levels via qPCR in eight pairs of HCC and adjacent normal tissues. It was demonstrated that the expression of hsa_circ_0067531 was significantly decreased in the HCC tissues, compared with the normal tissues (P=0.012) (Fig. 6C), however there was no significant difference in hsa_circ_0057096 expression between adjacent normal tissues and HCC tissues (Fig. 6D). Therefore, it was hypothesized that hsa_circ_0067531 may be a promising target for HCC diagnosis and therapy in the future.
Table I.
Sequences of two differentiated expression circular RNAs from PDK1 and PIK3CB.
| circBASE ID | Gene | Sequence (5′-3′) |
|---|---|---|
| hsa_circ_0057096 | PDK1 C | TTTACAGATACTGTGATACGGATCAGAAACCGACACAATGATGTCATTCCCACA ATGGCCCAGGGTGTGATTGAATACAAGGAGAGCTTTGGGGTGGATCCTGTCACC AGCCAGAATGTTCAGTACTTTTTGGATCGATTCTACATGAGTCGCATTTCAATTAG AATGTTACTCAATCAGCACTCTTTATTGTTTGGTGGAAAAGGCAAAGGAAGTCCA TCTCATCGAAAACACATTGGAAGCATAAATCCAAACTGCAAT GTACTTGAAGTTA TTAAAG |
| hsa_circ_0067531 | PIK3CB | AGTCGAGGTGGAAAAAAGTTTCTTCCTGTATTGAAAGAAATCTTGGACAGGGATCCC TTGTCTCAACTGTGTGAAAATGAAATGGATCTTATTTGGACTTTGCGACAAG ACTGCCGAGAGATTTTCCCACAATCACTGCCAAAATTACTGCTGTCAATCAAGTGG AATAAACTTGAGGATGTTGCTCAGCTTCAGGCGCTGCTTCAGATTTGGCCTAAAC TGCCCCCCCGGGAGGCCCTAGAGCTTCTGGATTTCAACTATCCAGACCAGTACG TTCGAGAATATGCTGTAGGCTGCCTGCGACAGATGAG |
PDK1, pyruvate dehydrogenase kinase; PIK3CB, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit β.
Figure 6.
Characterization of hsa_circ_0067531 and hsa_circ_0057096 in HCC tissues. (A) Head-to-tail splicing was assayed by a qPCR reaction, with divergent primers and (B) Sanger sequencing. (C) hsa_circ_0067531 and (D) hsa_circ_0057096 expressions were examined in eight pairs of the HCC tissues and in the corresponding non-tumor tissues using qPCR. The expression of hsa_circ_0067531 in HCC tissues was significantly decreased compared with adjacent normal tissues (P=0.012), however there was no significant difference in hsa_circ_0057096 expression between adjacent normal tissues and HCC tissues. *P<0.05 vs. adjacent. HCC, hepatocellular carcinoma; qPCR, quantitative polymerase chain reaction.
Discussion
circRNAs have previously emerged as novel and crucial layers of gene regulation and have demonstrated the potential to be ideal biomarkers in the diagnosis of various cancers (25,26). Certain circRNAs have already been demonstrated to exhibit important roles in cancer, including hsa_circ_002059 in gastric cancer, Cdr1as in hepatocellular carcinoma, and cir-ITCH in esophageal squamous cancer (23,27,28). Previous studies have revealed that circRNAs may be involved in the progression of cancer (20). Bachmayr-Heyda, et al (29) identified that circRNAs are significantly downregulated in colorectal cancer (CRC) tissues compared with the normal colon mucosa. Li et al (28) demonstrated that hsa_circ_002059 expression is downregulated in gastric cancer and may be a potential biomarker for its diagnosis (28). Huang et al reported that the cir-ITCH expression is decreased in esophageal squamous cell carcinoma (ESCC) and CRC, and may have an inhibitory effect on ESCC and CRC (30). The mechanisms of circRNAs in HCC remain to be elucidated.
The primary outcome of the present study was that CD90 promoted cell migration, viability and sphere-forming abilities in HCC, and the secondary outcome was that hsa_circ_0067531 and the PI3K pathway may potentially be involved in the aforementioned process. CD90 is a 25-37 kDa glycophosphatidylinositolanchored protein that operates as an important regulator of cell-to-cell and cell-to-matrix interactions in cancer (13). CD90 expression has been suggested to be associated with poor HCC prognosis (31–33) and CD90+ CSCs, however CD90− CSCs from HCC cell lines, tumor tissues, or peripheral blood, have not been reported to exhibit tumorigenic and metastatic capabilities (12,14,15). The present study first examined the expression of CD90 in the 4 human HCC cell lines, and demonstrated that CD90 expression was significantly upregulated in the HCC cell line SK-Hep-1. In addition, the data demonstrated that CD90+ cells isolated from the SK-Hep-1 cell line exhibited increased viability, migration and invasive capabilities compared with CD90− cells. The results were consistent with previous research which suggests that CD90+ cells isolated from HCC tumor tissues have the capacity to generate tumor nodules in immunodeficient mice, whereas CD90− cells do not (12,34,35).
circRNAs have previously been demonstrated to exhibit important roles in HCC cancer, including Circular RNA MTO1, Hsa_circ_0001649, and circZKSCAN1 (36–38). In the present study, 274 differentially expressed circRNAs (including upregulated and downregulated genes) were identified in CD90+ HCC cells compared with CD90− HCC cells via a high throughput microarray assay. Furthermore, KEGG pathway analyses was performed in order to further understand the biological functions of the differentially expressed circRNA. The results demonstrated that the key signaling pathways were the metabolic pathway, pathways in cancer, and the PI3K-AKT pathway. Of the significantly enriched pathways in the KEGG pathway analysis, PI3K signaling was of interest as it exhibits an important role in HCC cell cycle progression and viability (39,40). Deregulation of the PI3K/AKT signaling pathway has previously been identified in HCC (41), and Rab31, a member of the Ras superfamily, has been reported to have a role in tumor development and progression (42). The PI3K/AKT pathway was revealed to be involved in the Rab31 promotion of HCC progression (43). Previous research has revealed that inhibiting the activation of the PI3K pathway blocks the carcinogenesis and progression of HCC cells (44,45). The present study therefore selected two differentially expressed circRNAs, shsa_circ_0067531 and hsa_circ_0057096, from PDK1 and PIK3CB, respectively. It was demonstrated that hsa_circ_0067531 was markedly downregulated in HCC tissues compared with adjacent normal tissues.
In conclusion, the results of the present study suggested that CD90 may be used as a potential biomarker for HCC. It was revealed that CD90 promoted cell migration, viability and sphere-forming abilities of HCC. In addition, the expression of hsa_circ_0067531 was significantly decreased in HCC compared with adjacent normal tissues, and this suggested that hsa_circ_0067531 may be involved in the development of HCC, at least in part, through the PI3K pathway. However, the present study does not functionally verify how hsa_circ_0067531 affects HCC cell biological functions, and did not verify the involvement of the metabolic and cancer pathways in tumorigenesis of CD90+ HCC. The authors aim to investigate the mechanism underlying the modulation of hsa_circ_0067531 on HCC stem cell properties, and investigate the functional roles of the metabolic and cancer pathways in HCC development in future studies.
Acknowledgments
The present study was supported by the Major Projects on Collaborative Innovation of Industry, Guangzhou (grant no. 201508020076). The authors gratefully acknowledge the assistance of the Department of Hepatopancreatobiliary Surgery for their help in collecting medical records. In addition, the authors would like to thank all the participants of the study, without whom the study would not have been possible.
References
- 1.Asia-Pacific Working Party on Prevention of Hepatocellular Carcinoma Prevention of hepatocellular carcinoma in the Asia-Pacific region: Consensus statements. J Gastroenterol Hepatol. 2010;25:657–663. doi: 10.1111/j.1440-1746.2009.06167.x. [DOI] [PubMed] [Google Scholar]
- 2.Li Z, Zhang C, Lou C, Yan F, Mao Y, Hong X, Zhang Y. Comparison of percutaneous cryosurgery and surgical resection for the treatment of small hepatocellular carcinoma. Oncol Lett. 2013;6:239–245. doi: 10.3892/ol.2013.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 4.Bruix J, Reig M, Sherman M. Evidence-based diagnosis, staging, and treatment of patients with hepatocellular carcinoma. Gastroenterology. 2016;150:835–853. doi: 10.1053/j.gastro.2015.12.041. [DOI] [PubMed] [Google Scholar]
- 5.Kinoshita A, Koike K, Nishino H. Clinical features and prognosis of elderly patients with hepatocellular carcinoma not indicated for surgical resection. Geriatr Gerontol Int. 2017;17:189–201. doi: 10.1111/ggi.12747. [DOI] [PubMed] [Google Scholar]
- 6.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 8.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 9.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 10.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 11.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 12.Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, Chu PW, Lam CT, Poon RT, Fan ST. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153–166. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 13.Rege TA, Hagood JS. Thy-1 as a regulator of cell-cell and cell-matrix interactions in axon regeneration, apoptosis, adhesion, migration, cancer, and fibrosis. FASEB J. 2006;20:1045–1054. doi: 10.1096/fj.05-5460rev. [DOI] [PubMed] [Google Scholar]
- 14.Yamashita T, Honda M, Nakamoto Y, Baba M, Nio K, Hara Y, Zeng SS, Hayashi T, Kondo M, Takatori H, et al. Discrete nature of EpCAM+ and CD90+ cancer stem cells in human hepatocellular carcinoma. Hepatology. 2013;57:1484–1497. doi: 10.1002/hep.26168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang ZF, Ngai P, Ho DW, Yu WC, Ng MN, Lau CK, Li ML, Tam KH, Lam CT, Poon RT, Fan ST. Identification of local and circulating cancer stem cells in human liver cancer. Hepatology. 2008;47:919–928. doi: 10.1002/hep.22082. [DOI] [PubMed] [Google Scholar]
- 16.Sukowati CH, Anfuso B, Torre G, Francalanci P, Crocè LS, Tiribelli C. The expression of CD90/Thy-1 in hepatocellular carcinoma: An in vivo and in vitro study. PLoS One. 2013;8:e76830. doi: 10.1371/journal.pone.0076830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen WC, Hsu HP, Li CY, Yang YJ, Hung YH, Cho CY, Wang CY, Weng TY, Lai MD. Cancer stem cell marker CD90 inhibits ovarian cancer formation via β3 integrin. Int J Oncol. 2016;49:1881–1889. doi: 10.3892/ijo.2016.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang F, Nazarali AJ, Ji S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am J Cancer Res. 2016;6:1167–1176. [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Yang J, Zhou P, Le Y, Zhou C, Wang S, Xu D, Lin HK, Gong Z. Circular RNAs in cancer: Novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015;5:472–480. [PMC free article] [PubMed] [Google Scholar]
- 20.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51. doi: 10.1016/j.jbiotec.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 22.Fu L, Yao T, Chen Q, Mo X, Hu Y, Guo J. Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget. 2017;8:58405–58416. doi: 10.18632/oncotarget.16881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu L, Gong X, Sun L, Zhou Q, Lu B, Zhu L. The Circular RNA Cdr1as Act as an Oncogene in Hepatocellular carcinoma through targeting miR-7 expression. PLoS One. 2016;11:e0158347. doi: 10.1371/journal.pone.0158347. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Schmittgen TD. Real-time quantitative PCR. Methods. 2001;25:383–385. doi: 10.1006/meth.2001.1260. [DOI] [PubMed] [Google Scholar]
- 25.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 27.Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6:6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 29.Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D. Correlation of circular RNA abundance with viability-exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang G, Zhu H, Shi Y, Wu W, Cai H, Chen X. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/β-catenin pathway. PLoS One. 2015;10:e0131225. doi: 10.1371/journal.pone.0131225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lingala S, Cui YY, Chen X, Ruebner BH, Qian XF, Zern MA, Wu J. Immunohistochemical staining of cancer stem cell markers in hepatocellular carcinoma. Exp Mol Pathol. 2010;89:27–35. doi: 10.1016/j.yexmp.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu JW, Chang JG, Yeh KT, Chen RM, Tsai JJ, Hu RM. Overexpression of Thy1/CD90 in human hepatocellular carcinoma is associated with HBV infection and poor prognosis. Acta Histochem. 2011;113:833–838. doi: 10.1016/j.acthis.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 33.Yu XH, Xu LB, Liu C, Zhang R, Wang J. Clinicopathological characteristics of 20 cases of hepatocellular carcinoma with bile duct tumor thrombi. Dig Dis Sci. 2011;56:252–259. doi: 10.1007/s10620-010-1256-8. [DOI] [PubMed] [Google Scholar]
- 34.Ho DW, Yang ZF, Yi K, Lam CT, Ng MN, Yu WC, Lau J, Wan T, Wang X, Yan Z, et al. Gene expression profiling of liver cancer stem cells by RNA-sequencing. PLoS One. 2012;7:e37159. doi: 10.1371/journal.pone.0037159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang R, An LY, Miao QF, Li FM, Han Y, Wang HX, Liu DP, Chen R, Tang SQ. Effective elimination of liver cancer stem-like cells by CD90 antibody targeted thermosensitive magnetoliposomes. Oncotarget. 2016;7:35894–35916. doi: 10.18632/oncotarget.9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han D, Li J, Wang H, Su X, Hou J, Gu Y, Qian C, Lin Y, Liu X, Huang M, et al. Circular RNA circMTO1 acts as the sponge of miR-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 37.Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z, Yang J, Fan J, Liu L, Qin W. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16:161–169. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 38.Yao Z, Luo J, Hu K, Lin J, Huang H, Wang Q, Zhang P, Xiong Z, He C, Huang Z, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11:422–437. doi: 10.1002/1878-0261.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fulda S. Modulation of mitochondrial apoptosis by PI3K inhibitors. Mitochondrion. 2013;13:195–198. doi: 10.1016/j.mito.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 40.Fulda S. Synthetic lethality by co-targeting mitochondrial apoptosis and PI3K/Akt/mTOR signaling. Mitochondrion. 2014;19:85–87. doi: 10.1016/j.mito.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 41.Gedaly R, Angulo P, Hundley J, Daily MF, Chen C, Evers BM. PKI-587 and sorafenib targeting PI3K/AKT/mTOR and Ras/Raf/MAPK pathways synergistically inhibit HCC cell viability. J Surg Res. 2012;176:542–548. doi: 10.1016/j.jss.2011.10.045. [DOI] [PubMed] [Google Scholar]
- 42.Pan Y, Zhang Y, Chen L, Liu Y, Feng Y, Yan J. The critical role of Rab31 in cell viability and apoptosis in cancer progression. Mol Neurobiol. 2016;53:4431–4437. doi: 10.1007/s12035-015-9378-9. [DOI] [PubMed] [Google Scholar]
- 43.Sui Y, Zheng X, Zhao D. Rab p31 promoted hepatocellular carcinoma (HCC) progression via inhibition of cell apoptosis induced by PI3K/AKT/Bcl-2/BAX pathway. Tumor Biol. 2015;36:8661–8670. doi: 10.1007/s13277-015-3626-5. [DOI] [PubMed] [Google Scholar]
- 44.Liang M, Liu J, Ji H, Chen M, Zhao Y, Li S, Zhang X, Li J. A Aconitum coreanum polysaccharide fraction induces apoptosis of hepatocellular carcinoma (HCC) cells via pituitary tumor transforming gene 1 (PTTG1)-mediated suppression of the P13K/Akt and activation of p38 MAPK signaling pathway and displays antitumor activity in vivo. Tumour Biol. 2015;36:7085–7091. doi: 10.1007/s13277-015-3420-4. [DOI] [PubMed] [Google Scholar]
- 45.Zheng J, Li C, Wu X, Liu M, Sun X, Yang Y, Hao M, Sheng S, Sun Y, Zhang H, et al. Astrocyte elevated gene-1 (AEG-1) shRNA sensitizes Huaier polysaccharide (HP)-induced anti-metastatic potency via inactivating downstream P13K/Akt pathway as well as augmenting cell-mediated immune response. Tumour Biol. 2014;35:4219–4224. doi: 10.1007/s13277-013-1552-y. [DOI] [PubMed] [Google Scholar]