Abstract
Accumulation of lipid droplets and inflammatory cell infiltration is the hallmark of nonalcoholic steatohepatitis (NASH). The roles of noncoding RNAs in NASH are less known. We aim to elucidate the function of miR-141/200c in diet-induced NASH. WT and miR-141/200c–/– mice were fed a methionine and choline deficient (MCD) diet for 2 weeks to assess markers of steatosis, liver injury, and inflammation. Hepatic miR-141 and miR-200c RNA levels were highly induced in human patients with NASH fatty liver and in WT MCD mice. miR-141/200c–/– MCD mice had reduced liver weights and triglyceride (TG) levels, which was associated with increased microsomal TG transfer protein (MTTP) and PPARα but reduced SREBP1c and FAS expression. Inflammation was attenuated and F4/80 macrophage activation was suppressed in miR-141/200c–/– mice, as evidenced by decreased serum aminotransferases and IL-6 and reduced hepatic proinflammatory, neutrophil, and profibrotic genes. Treatment with LPS in BM-derived macrophages isolated from miR-200c/141–/– mice polarized macrophages toward the M2 antiinflammatory state by increasing Arg1 and IL-10 levels while decreasing the M1 marker iNOS. In addition, elevated phosphorylated AMPK (p-AMPK), p-AKT, and p-GSK3β and diminished TLR4 and p-mTOR/p-4EBP1 proteins were observed. Lipidomics and metabolomics revealed alterations of TG and phosphatidylcholine (PC) lipid species by miR-141/200c deficiency. In summary, miR-141/200c deficiency diminished NASH-associated hepatic steatosis and inflammation by reprogramming lipid and inflammation signaling pathways.
Keywords: Hepatology, Inflammation
Keywords: Fatty acid oxidation, Macrophages, Noncoding RNAs
Levels of miR-141 and miR-200c were elevated in livers of mice and humans with NASH, and deletion of miR-141/200c protected mice from developing NASH.
Introduction
The prevalence of nonalcoholic steatohepatitis (NASH) is on the rise and is estimated to affect ~5% of the general population, posing a significant risk factor for hepatic cirrhosis and hepatocellular carcinoma (1). NASH is the advanced stage of nonalcoholic fatty liver disease (NAFLD) and encompasses a continuum of hepatic disorders ranging from fatty liver or steatosis to inflammation, along with varying degrees of fibrosis and cirrhosis (2). Dysregulation of lipid metabolism is the main perpetrator in the pathogenesis of NASH, where excess triglyceride (TG) accumulation is a consequence of increased fatty acid and TG synthesis and decreased export (3). The accumulation of intrahepatic lipids provides a toxic environment for hepatocytes, leading to activation of liver resident macrophages, Kupffer cells (KCs), which in turn release proinflammatory cytokines. Thus, liver infiltration with inflammatory cells is a key feature of NASH. A growing body of evidence has reported that cell damage, mitochondrial dysfunction, oxidative stress, and inflammation play key roles in the pathogenesis of NAFLD/NASH (4).
Noncoding RNAs have emerged as critical regulators of human liver diseases (5). MicroRNAs are noncoding RNAs of 22 nucleotides in length, which regulate gene expression by binding to the 3′UTR of the target messenger RNA (6, 7). On the other hand, the transcription of miRNAs is tightly controlled by transcription factors and nuclear receptors (8–10). MicroRNAs are involved in a variety of pathological processes including growth, apoptosis, and cell migration and invasion (11, 12). The miR-200 family comprises 5 highly conserved members (miR-141, miR-200a/200b/200c, and miR-429), which are located on 2 separate chromosomal clusters (miR-141/miR-200c and miR-429/miR-200a/200b) with distinct functions, despite containing very similar seed sequences (13). Reduced miR-200b and miR-200c expression was reported in steatotic livers of mice fed a high-fat (HF) diet (14). The expression of miR-200a and miR-200b was positively correlated with the progression of liver fibrosis (15). In contrast, reduced miR-200c levels in hepatocytes contributed to IL-6–induced insulin resistance via upregulation of FOG2 expression (16). These conflicting findings suggest the need to properly investigate the in vivo role of miR-141/200c on the progression of NAFLD/NASH using specific KO mice.
There are several commonly used experimental models to study NASH. The HF or HF/high-fructose diets give rise to an obese and insulin-resistant state; however, these diets mildly produce steatosis and inflammation, and liver fibrosis was only observed with chronic long-term feeding. On the contrary, the methionine-choline deficient (MCD) diet rapidly produces the NASH phenotype of hepatic steatosis, inflammation, and fibrosis in mice within a short timeframe; however, it does not generate the obese and insulin-resistant phenotype associated with the human NASH phenotype (17). Nonetheless, feeding mice using an MCD diet is a frequently used model of NASH that induces aminotransferase elevation and hepatic histological features characterized by steatosis, focal inflammation, and fibrosis.
In the present study, we investigated the effect of loss of miR-141/200c on the development of NASH using the MCD diet, which is a robust model to induce steatohepatitis and recapitulates key features of the inflammatory and lipid profile seen in human NASH conditions, with no or minimal features of the metabolic syndrome. We report that miR-141/200c deficiency diminished hepatic steatosis, liver injury, and inflammation by altering multiple signaling pathways and reprogramming metabolic and lipid profiling. Currently, there are no effective therapies or established medical treatments for NASH; therefore, our results provide potentially novel insights into the understanding of the role of noncoding RNAs in NASH.
Results
miR-141 and miR-200c are markedly induced in NASH livers in humans and mice.
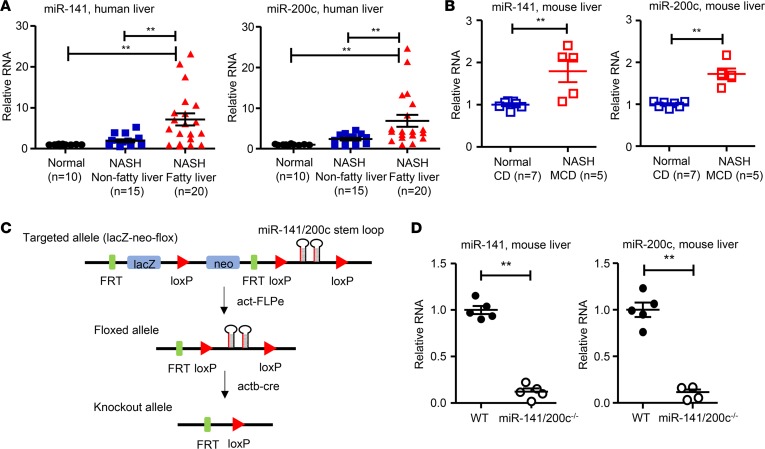
In humans, the miR-141 is located on chromosome 12 (12q13.31) together with miR-200c. To determine their involvement in NASH, we analyzed both miRNAs in 2 groups of human NASH liver specimens, i.e., NASH without fatty liver (nonfatty liver) and NASH with fatty liver (fatty liver). The liver samples without patient identification were provided by the Liver Tissue Cell Distribution System, and the clinical data sheets obtained did not provide NASH Clinical Research Network (NASH CRN) scores. As shown in Supplemental Table 2 (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.96094DS1), age was similar between NASH fatty and NASH nonfatty liver patients. Sex distribution was similar in NASH nonfatty liver, while in NASH fatty liver, more females were affected than males. Other biochemical characteristics such as model end liver disease (MELD) score, total bilirubin, albumin, aspartate aminotransferase (AST), and alkaline phosphatase (ALP) were similar between groups, while creatinine levels were significantly elevated in NASH fatty vs. NASH nonfatty groups. Compared with normal control livers, miR-141 and miR-200c expression levels were upregulated in NASH nonfatty livers, albeit not significantly, and there was a 6-fold induction of miR-141 and miR-200c RNA levels in NASH fatty livers (Figure 1A). miR-141 and miR-200c were also highly expressed in human Huh7 cells, with low levels detected in HepG2 cells (Supplemental Figure 1A). The results provide the clinical relevance of the study. In addition, liver miR-141 and miR-200c levels were significantly increased in WT mice fed the MCD diet compared with the control diet (CD) (Figure 1B). Taken together, miR-141 and miR-200c expression was upregulated in NASH in both humans and mice.
Figure 1. Levels of miR-141/200c are increased in steatotic livers of patients and mice with NASH.
(A) qPCR analysis of miR-141 and miR-200c expression in human liver specimens (n = 10–20 samples per group). (B) qPCR analysis of miR-141 and miR-200c expression in livers of WT mice fed the control diet (CD) and methionine and choline–deficient (MCD) diet (n = 5–7 mice per group). (C) Targeting strategy for generating miR-141/200c–double KO mice. (D) qPCR analysis of miR-141 and miR-200c expression in livers of miR-141/200c–/– mice and their WT littermates (n = 5 mice per group). Data are represented as mean ± SEM. Differences between 2 groups were compared using a Student’s unpaired t test. For multiple groups, differences were compared using a one-way ANOVA followed by Newman-Keuls multiple comparisons test. **P < 0.01 indicate statistical significance.
Generation of miR-141/200c–KO mouse model.
The ES clone obtained from the Knock-Out Mouse Project (KOMP) Repository at UC Davis (Davis, California, USA) was used to generate miR-141 and miR-200c double-deletion model, as KOMP was unable to delete each single miRNA due to their close proximity on chromosome 6. In brief, the miR-141/200c–double KO mouse model (miR-141/200c–/–) was generated by inserting a targeting vector upstream of the microRNA cluster stem loop and 1 loxP site immediately downstream of the microRNA stem loop (Figure 1C). The expression of miR-141 and miR-200c was mostly ablated in livers of miR-141/200c–/– mice compared with WT mice (Figure 1D), as well as in primary hepatocytes isolated from miR-141/200c–/– mice (Supplemental Figure 1B), confirming the deletion of these 2 microRNAs. The very low levels of both miRs in livers and hepatocytes of KO mice were background signals.
miR-141/200c deficiency attenuates MCD diet–induced hepatic lipid accumulation.
Body weights were similar between WT and miR-141/200c–/– mice before (data not shown) and after the 2-week MCD diet feeding regime (Figure 2A). However, absolute and relative liver weights were significantly reduced in miR-141/200c–/– MCD mice compared with WT MCD mice, while no differences were detected between groups on the CD (Figure 2A). Fasting serum glucose levels were lower in miR-141/200c–/– vs. WT mice fed the CD, which were at similar levels between WT and miR-141/200c–/– mice on the MCD diet (Supplemental Figure 2A). Fasting serum TG levels were no different between WT and miR-141/200c–/– mice on CD and MCD diets (Figure 2B). Interestingly, liver TG contents were markedly lower in miR-141/200c–/– MCD vs. WT MCD mice (Figure 2B). On the other hand, cholesterol levels were not altered in miR-141/200c–/– MCD vs. WT MCD mice (Supplemental Figure 2B).
Figure 2. MCD diet–induced steatosis is lessened by miR-141/200c deficiency.
(A) Body weight, absolute and relative liver weights in WT and miR-141/200c–/– mice fed the control diet (CD) or methionine and choline–deficient (MCD) diet (n = 4–8 mice per group). (B) Serum and liver triglyceride levels were measured by calorimetric analysis (n = 4–8 mice per group). (C) Representative images of H&E staining of liver sections from WT and miR-141/200c–/– mice fed the CD and MCD diet. (D and E) Western blot showing liver protein levels of pSREBP1, nSREPB1, FAS, MTTP, and α-TUBULIN in WT MCD and miR-141/200c–/– MCD mice. Samples were pooled from 5 individual mice for each group unless otherwise indicated. Densitometry of each blot relative to the loading control was quantified using ImageJ software. Data are represented as mean ± SEM. Differences between 2 groups were compared using a Student’s unpaired t test. For multiple groups, differences were compared using a one-way ANOVA followed by Newman-Keuls multiple comparisons test. *P < 0.05 and **P < 0.01 indicate statistical significance.
Hepatic lipid accumulation is one of the most prominent characteristic features of NASH and can induce further liver injury and inflammation. The MCD diet does not produce obesity or increased weight gain in mice, but steatosis develops within a short timeframe (18, 19). H&E staining showed no overt differences in liver architecture between WT CD and miR-141/200c–/– CD mice. However, there was noticeable decrease in lipid droplets and the infiltration of inflammatory cells in miR-141/200c–/– MCD vs. WT MCD mice (red arrows) (Figure 2C). Oil Red O staining (Supplemental Figure 2C) further revealed a marked reduction of accumulation of lipid droplets in miR-141/200c–/– MCD vs. WT MCD mice.
We next analyzed hepatic proteins and mRNAs associated with de novo lipogenesis and lipid transport. SREBP1 is a master regulator of lipogenesis (20, 21). Its precursor form (pSREBP1) was markedly increased, whereas its nuclear form SREBP1 (nSREPB1) was decreased in miR-141/200c–/– MCD vs. WT MCD mice, as shown by the quantification results (Figure 2D); this is indicative of impaired cleavage. In addition, fatty acid synthase (FAS) protein was downregulated, whereas microsomal TG transfer protein (MTTP) protein was upregulated in miR-141/200c–/– vs. WT mice (Figure 2E), suggesting decreased lipid synthesis but increased lipid transport.
miR-141/200c deficiency suppresses hepatic injury and inflammation.
The toxic effects of lipids on hepatocytes activate liver resident macrophages KCs to release inflammatory cytokines, contributing to the development of NASH. Elevated serum AST and alanine aminotransferase (ALT) levels are indicators of liver injury, which were significantly increased in WT MCD but not in miR-141/200c–/– MCD mice (Figure 3A). The reduced liver injury in miR-141/200c–/– MCD mice was accompanied by decreased serum IL-6 levels (Figure 3B). Interestingly, serum thiobarbituric acid reactive substances (TBARs), a measure of lipid peroxidation, remained unchanged between WT MCD and miR-141/200c–/– MCD mice, whereas TBAR concentration in liver homogenates was modestly reduced in miR-141/200c–/– MCD mice compared with WT MCD mice (Supplemental Figure 3A). Oxidative stress can induce inflammation and contribute to the NASH phenotype. The levels of hepatic H2O2 measured using the fluorescent probe Amplex Red was not significantly altered between WT MCD and miR-141/200c–/– MCD mice (Supplemental Figure 3B).
Figure 3. MCD diet–induced hepatic inflammation is diminished by miR-141/200c deficiency.
(A) Levels of aspartate aminotransferase (AST), alanine aminotransferase (ALT),and (B) IL-6 were measured in overnight fasted serum of WT and miR-141/200c–/– mice fed the control diet (CD) or methionine and choline–deficient (MCD) diet (n = 5–8 mice per group). (C–F) qPCR analysis of hepatic genes involved in inflammation (Saa1, Orm1, IL-1β, TNFα, IL-4, F4/80, LyG6) and fibrosis (col1a1, TGFβ) (n = 5 mice per group). (G) Representative images of Masson’s trichrome staining and F4/80 immunohistochemistry to examine fibrosis and macrophage activation in WT MCD and miR-141/200c–/– MCD. Images were quantified by digital image analysis using ImageJ software in 6 randomly chosen fields from 3 individual mice per group. (H) qPCR analysis of hepatic genes associated with M1 (iNOS) or M2 (Arg1) macrophage phenotype in WT MCD and miR-141/200c–/– MCD mice (n = 5 mice per group). Data are represented as mean ± SEM. Differences between 2 groups were compared using a Student’s unpaired t test. For multiple groups, differences were compared using a one-way ANOVA followed by Newman-Keuls multiple comparisons test. *P < 0.05 and **P < 0.01 indicate statistical significance.
To verify the inflammatory state of mice fed the MCD diet, we analyzed for hepatic genes involved in inflammation and fibrosis (Figure 3, C–F). Hepatic acute phase response genes (Saa1 and Orm1) (Figure 3C), inflammatory cytokines (IL-1β, TNFα, IL-4) (Figure 3D), macrophage and neutrophil markers (F4/80 and LyG6) (Figure 3E), and fibrotic genes (Col1a1, TGFβ) (Figure 3F) were highly induced by MCD diet in WT mice but to a lesser extent in miR-141/200c–/– mice. Additional fibrosis-related genes including MMP2, ICAM, and α-SMA did not show marked differences in WT and miR-141/200c–/– mice on either CD or MCD diet (Supplemental Figure 3C).
Masson’s trichrome staining is commonly used to visually assess collagen levels associated with diet-induced fibrosis (blue coloration). Collagen deposition was slightly lower but did not reach statistical significance in WT and miR-141/200c–/– mice (Figure 3G). In addition, the activation status of KCs was histologically examined by assessing F4/80-positive cells in liver sections (Figure 3G). Feeding an MCD diet caused recruitment of KCs into clusters in WT mice, which was markedly less abundant in miR-141/200c−/− mice (Figure 3G). Macrophages are phenotypically heterogeneous and can be characterized according to their activation or polarization state as M1 “classically activated” proinflammatory macrophages or M2 “alternatively activated” antiinflammatory macrophages (22, 23). We found that M1 polarization marker iNOS was reduced, whereas M2 polarization marker Arg1 was increased in livers of miR-141/200c–/– MCD vs. WT MCD mice (Figure 3H). Interestingly, M1 and M2 transcriptional activators HIF1α, C/EBPβ, and STAT3 remained unaltered in miR-141/200c–/– vs. WT mice (Supplemental Figure 3D). Similarly, protein expression of p-STAT3 and total STAT3 (t-STAT3) was also not altered in livers of WT and miR-141/200c–/– mice (Supplemental Figure 3E).
We also examined caspase-3, a marker of early apoptosis, and observed a modest reduction of its cleavage in miR-141/200c–/– vs. WT mice (Supplemental Figure 4A). The number of TUNEL-positive cells were lower, albeit not significant in miR-141/200c–/– vs. WT mice (Supplemental Figure 4B).
miR-141/200c–/– mice exhibit predominance of M2 macrophage phenotype when exposed to LPS.
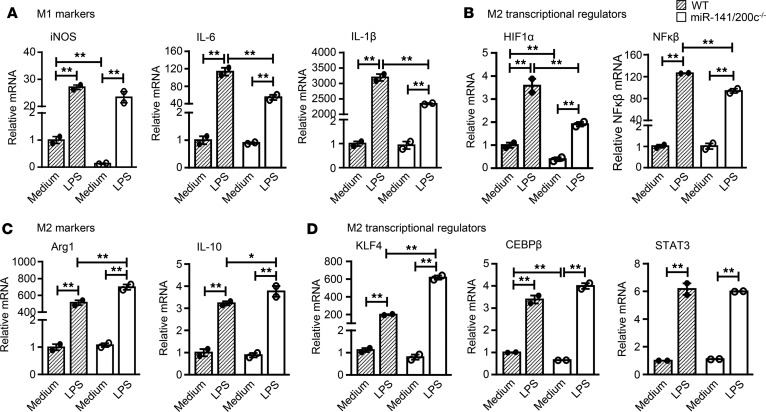
To further assess the regulatory role of miR-141/200c in inflammation, BM cells were isolated from WT and miR-141/200c–/– mice to generate BM-derived macrophages (BMDMs). BMDMs of WT mice treated with LPS (10 ng/ml) for 24 hours polarized macrophages preferentially toward a proinflammatory M1-like profile with increased expression of iNOS, IL-6, and IL-1β mRNA (Figure 4A). However, the induction of IL-6 and IL-1β mRNA expression by LPS was diminished in macrophages derived from miR-141/200c–/– mice (Figure 4A). This was accompanied by the lesser induction of M1 transcriptional regulators HIF1α and NFκβ with LPS treatment (Figure 4B). On the contrary, miR-141/200c deficiency polarized the macrophage phenotype toward the M2-like state, as evidenced by the further induction of Arg1 and IL-10 mRNA expression by LPS treatment (Figure 4C). Consistent with this, the expression of the M2 transcriptional regulator KLF4 was highly induced in miR-141/200c–/– macrophages compared with WT, while C/EBPβ and STAT3 mRNA expression was similar (Figure 4D). Taken together, miR-141/200c deficiency appears to favor macrophages toward an M2-like profile in the presence of LPS under in vitro conditions.
Figure 4. miR-141/200c deficiency polarizes macrophages toward the M2 antiinflammatory phenotype.
BM-derived macrophages (BMDMs) were generated from WT and miR-141/200c–/– mice and incubated with or without LPS (10 ng/ml) for 24 hours and then processed for RNA isolation (n = 2 independent experiments). qPCR analysis of mRNA expression of (A) M1 markers (iNOS, IL-6, and IL-1β), (B) M1 regulators (HIF1α and NF-κβ), (C) M2 markers (Arg1 and IL-10), and (D) M2 regulators (KLF4, C/EBPβ, and STAT3). Data are represented as mean ± SEM. Differences between multiple groups were compared using a one-way ANOVA followed by Newman-Keuls multiple comparisons test. *P < 0.05 and **P < 0.01 indicate statistical significance.
Multiple hepatic signaling pathways are altered by miR-141/200c deficiency.
At the cellular level, MAPKs, AMPK, PPARs, and TLR signaling pathways are major links to the pathology of NASH. Therefore, we assessed their activation status to determine the regulatory role of miR-141/200c in steatosis and inflammation (Figure 5). Nuclear receptor PPARα activates fatty acid β-oxidation, and its expression was increased in miR-141/200c–/– mice compared with WT mice (Figure 5A). In contrast, PPARγ protein expression remained unaltered. AMPK is a central regulator of energy balance, and its phosphorylation activates downstream target molecules including PPARα to promote fatty acid oxidation (24). We observed increased AMPK phosphorylation in miR-141/200c–/– mice compared with WT mice.
Figure 5. miR-141/200c deficiency affects proteins in multiple signaling pathways.
Immunoblotting analysis of proteins in (A) PPAR and AMPK; (B) ERK, SAPK/JNK, and p38; (C) AKT/GSK3β; (D) TLR4; and (E) mTOR signaling pathways in WT and miR-141/200c–/– livers fed the MCD diet. Samples were pooled from 5 individual mice for each group. Densitometry of each blot relative to the loading control was quantified using ImageJ software.
The ERKs play a major role in the cellular response induced by growth factor stimulation, and SAPK/JNK and P38 are related with stress and inflammation (25). Activation of SAPK/JNK is known to be involved in the pathogenesis of NAFLD (26), which was observed in miR-141/200c–/– mice (Figure 5B). On the contrary, there was reduced phosphorylation of ERK1/2 and P38 in miR-141/200c–/– vs. WT mice. A minor increase in AKT phosphorylation in miR-141/200c–/– vs. WT mice (Figure 5C) was consistent with studies reporting reduced AKT protein levels in NAFLD (24). Interestingly, the phosphorylated glycogen synthase kinase 3-β (GSK3β) was highly elevated (Figure 5C), resulting in inactivation of GSK3β activity to promote glycogen synthesis and prevent lipid accumulation.
Among other signaling pathway regulators, TLR4 protein was reduced in miR-141/200c–/– mice, whereas IKKβ protein was not altered (Figure 5D). On the other hand, both p-mTOR and its target protein p-4EBP1 were significantly reduced in miR-141/200c–/– vs. WT mice, whereas p70S6K was unaltered (Figure 5E). These results are in agreement with the notion that mTOR signaling can promote de novo lipogenesis through activation of SREBP1 (27), thereby contributing to the pathogenesis of NASH.
miR-141/200c deficiency disrupts hepatic TG and PC lipid species.
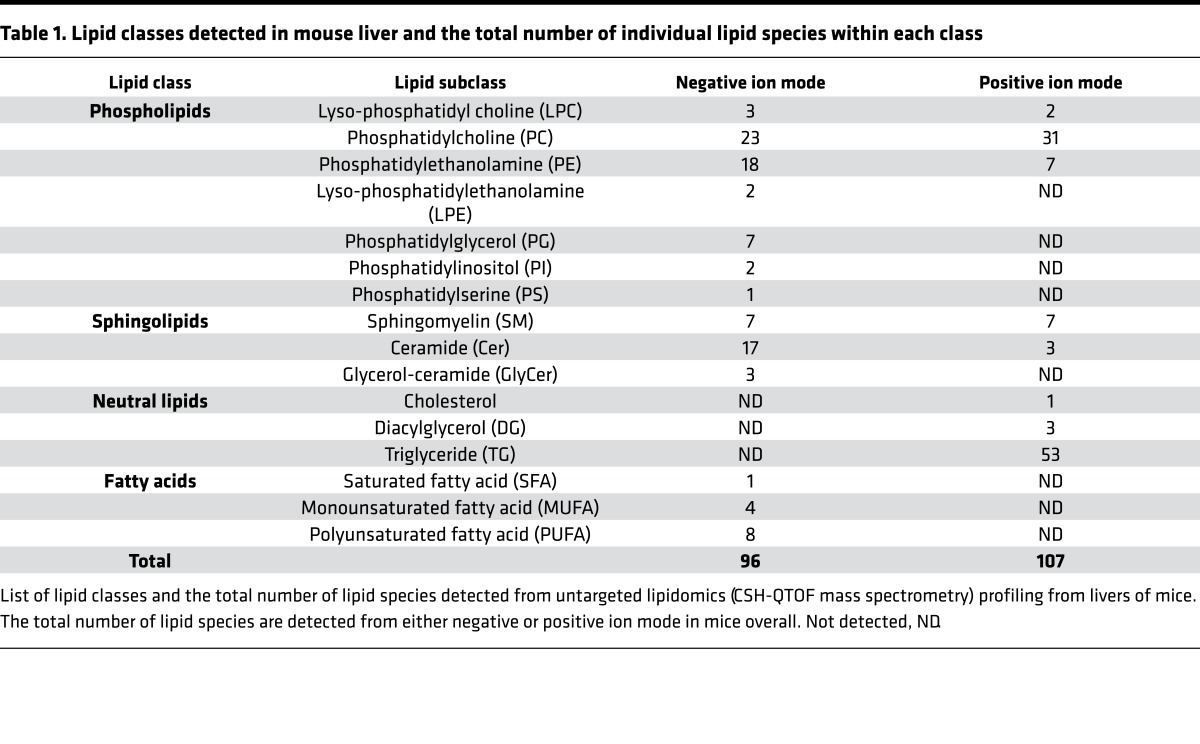
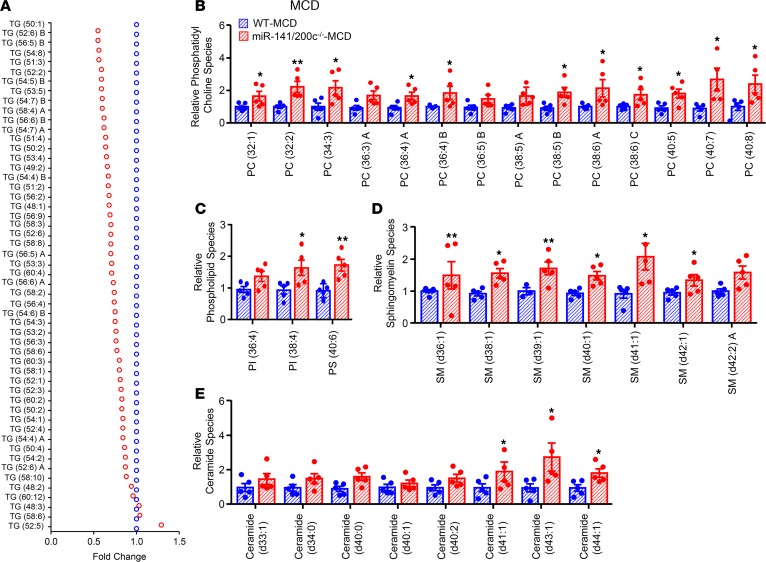
In order to comprehensively assess lipid metabolites altered by miR-141/200c–/– deficiency, we performed lipidomics analysis using CSH-QTOF mass spectrometry. We identified a total of 203 lipid molecules from 96 positive and 107 negative ions in the liver, including 96 phospholipids, 37 sphingolipids, 57 neutral lipids, and 13 free fatty acids (FFAs) (Table 1). A majority of TG lipid species, expressed as relative fold change to WT (Figure 6A) and as average ion peak height (Supplemental Figure 5, A and B), were found to be reduced in miR-141/200c–/– mice. Interestingly, a large number of phospholipids, including phosphatidylinositol (PI), phosphatidylserine (PS), and a majority of phosphatidylcholine (PC), were markedly elevated in miR-141/200c–/– vs. WT mice (Figure 6, B and C). Additionally, sphingolipids, including sphingomyelin (SM) and ceramide lipid species, were also increased in miR-141/200c–/– mice (Figure 6, D and E). No major alterations were observed in fatty acids (Supplemental Figure 6A), while many phosphatidylethanolamine (PE) species were increased in miR-141/200c–/– mice (Supplemental Figure 6B). Overall, miR-141/200c deficiency reduced hepatic neutral lipid accumulation (mainly TGs) but resulted in increases of PC, PI, and PS lipid species compared with WT mice.
Table 1. Lipid classes detected in mouse liver and the total number of individual lipid species within each class.
Figure 6. Lipidomics reveals reduced TG and increased PC lipid species in miR-141/200c–/– mice.
Livers of WT and miR-141/200c–/– mice from the methionine and choline–deficient (MCD) group were prepared and subjected to LC-MS for lipidomics analysis of various lipid classes (n = 5 mice per group). (A) Triglyceride (TG), (B) phosphatidylcholine (PC), (C) phospholipids including phosphatidylinositol (PI) and phosphatidylserine (PS), (D) sphingomyelin (SM), and (E) ceramide lipid species are expressed as the fold change relative to WT mice. Data are represented as mean ± SEM. *P < 0.05 and **P < 0.01 vs. WT by Student’s unpaired t test.
miR-141/200c deficiency alters metabolomics profiling.
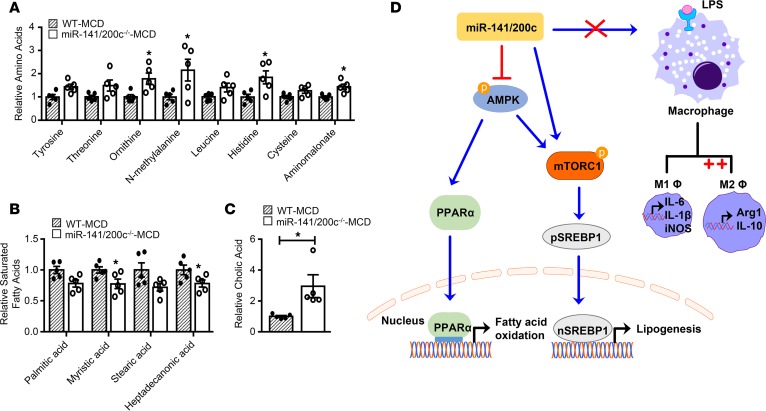
Metabolomics provides a powerful tool to depict changes in the hepatic metabolome and offers new insight into the cellular metabolic pathways regulated by miR-141/200c. We used untargeted metabolomics profiling (GC-TOF mass spectrometer) to elucidate changes in liver metabolites of WT and miR-141/200c–/– mice fed the MCD diet. Metabolomics analyses identified about 130 known metabolites in the liver belonging to major metabolic pathways such as carbohydrate and sugar complexes, fatty acids, hydroxyl acids, pentose phosphate, amino acid, and nucleotides. Of the altered metabolites between WT and miR-141/200c–/– mice, changes were mostly observed in amino acids (ornithine, N-methylalanine, histidine, leucine, and aminomalonate), saturated fatty acids (palmitic acid, myristic acid, stearic acid, and heptadecanoic acid), and cholic acid (Figure 7, A–C). Correlation matrix clustering showed subsets of metabolites with strong correlation, particularly in the amino acid pathway (Supplemental Figure 7). The data set were subjected to 2-tailed t test analysis to identify unique metabolome patterns altered between WT and miR-141/200c–/– mice. Significant changes were observed in 11 metabolites including isohexonic acid, cholic acid, glycerol, glyceric acid, myoinositol, heptadecanoic acid, myristic acid, aminomalonate, lauric acid, histidine, and ornithine (Supplemental Figure 8).
Figure 7. Metabolomics unravels distinct metabolic profiling altered by miR-141/200c deficiency.
Livers of WT and miR-141/200c–/– mice from the methionine and choline–deficient (MCD) group were prepared and subjected to LC-MS for metabolomics analysis (n = 5 mice per group). Levels of (A) amino acids, (B) saturated fatty acids, and (C) cholic acid are expressed as the fold change relative to WT mice. Data are represented as mean ± SEM. *P < 0.05 vs. WT by a Student’s unpaired t test. (D) Schematic diagram of miR-141/200c–modulated signaling pathways to control lipid metabolism and macrophage phenotype in MCD diet–induced NASH. miR-141/200c inhibits AMPK phosphorylation to reduce downstream PPARα activation and subsequently decrease fatty acid β-oxidation. At the same time, miR-141/200c targets mTOR phosphorylation to favor SREPB1 translocation to the nucleus, contributing to hepatic lipid accumulation. In the presence of LPS, miR-141/200c deficiency appears to favor antiinflammatory M2 polarization and activation as assessed by the increased expression of Arg1 and IL-10.
Discussion
microRNAs play key roles in various liver diseases, including liver cancer and fibrosis; however, limited studies have focused on the in vivo regulation of NASH development by miRNAs. Using a newly generated miR-141/200c–/– mouse model, our results demonstrate that genetic deletion of the miR-141/200c cluster significantly attenuates steatosis and inflammation in diet-induced NASH (Figure 7D). Furthermore, miR-141 and miR-200c are highly induced in human patients and mice with NASH, indicating the clinical significance of our findings.
The pathways associated with steatosis and inflammation are interconnected to form an intricate regulatory network and can mutually enhance each other, forming a vicious cycle. miR-141/200c deficiency in mice reduced TG accumulation via AMPK activation to promote PPARα-mediated fatty acid β-oxidation. Additionally, a role for mTOR signaling has been reported in patients with NAFLD (28). AMPK can negatively regulate mTOR signaling to reduce lipogenesis via downregulation of SREBP1 and its target FAS. Thus, our results are consistent with studies reporting a role for AMPK activation in the attenuation of hepatic steatosis (24). It is also worth noting that the changes in AMPK and mTOR phosphorylation may likely be a compensatory effect as a result of the decreased NASH phenotype rather than a direct role of the miR-141/200c cluster in mediating these changes.
In miR-141/200c-deficient mice fed the MCD diet, we observed reduced levels of genes associated with inflammation and fibrosis. Indeed, this could be due to the indirect effects or due to a direct loss of miR-141 and miR-200c regulation. Identification of miR-141 and miR-200c target genes warrants future investigation. To determine role of miR-141/200c in mediating inflammation, we assessed the activation status of macrophages in response to LPS. Macrophages have different functional states exhibiting a proinflammatory M1 or antiinflammatory M2 phenotype (29). The M1 phenotype is characterized by high expression of proinflammatory cytokines and toxic intermediates, while the M2 phenotype typically have increased antiinflammatory cytokine production and expression of genes involved in tissue repair, remodeling, and immune regulation (23). In both in vitro and in vivo models, we demonstrated that miR-141/200c deficiency tends to polarize macrophages away from the M1 proinflammatory state and toward the M2-like anti-inflammatory state in the presence of an M1 inducer. It is important to note that, although the precise mechanisms pertaining to this macrophage phenotype differ, it appears that miR-141/200c deficiency somehow protects against the activation of macrophages toward the M1-like phenotype. Interestingly, under in vitro conditions, the polarization toward the M2-like phenotype appears to be mediated via reduced HIF1α and NF-κβ, known transcriptional regulators of the M1 macrophage phenotype. Our findings are in accordance with studies by Wan et al., who demonstrated that reducing M1 KC polarization can reduce NASH progression and that M2 KCs promotes apoptosis of M1 KC during NAFLD progression in mice (30). Collectively, the reduced steatosis and inflammation associated with miR-141/200c deficiency may be related to, at least in part, predominance of the M2-like macrophage phenotype.
Increased levels of saturated and monounsaturated FFAs have been implicated in triggering inflammation and, thus, potentially serving as endogenous danger signals in NASH patients. Indeed, several saturated fatty acids were significantly reduced in miR-141/200c–/– mice compared with WT mice. Besides the storage of TGs, steatohepatitis is also associated with changes in phospholipid metabolism, with PCs accounting for ~70 % of the total phospholipid content (31). miR-141/200c deficiency was associated with increased PC content, which was in accordance with studies reporting that NAFLD is associated with reduced PC levels in human patients (32). Other studies have described a trend of reduced SM content and short-chain ceramide in the portal circulation of NASH subjects (33). Additionally, dietary SM supplementation in HF-fed mice has been found to reduce total hepatic TG levels (34). Thus, the increased SM and ceramide in miR-141/200c–/– mice may play a role in reducing hepatic TG accumulation. The molecular basis by which miR-141/200c regulates phospholipid and sphingolipid metabolism remains undefined and warrants further investigation.
Further studies will incorporate the use of female mice to determine whether sex differences are observed in the pathogenesis of NASH. Ideally, tissue-specific KOs would be required to bypass the limitations of traditional constitutive KO models in terms of compensatory mechanisms. Because the MCD diet has its limitation to study steatohepatitis, additional diet models would be useful to further explore the role of miR-141/200c in NASH development. Furthermore, the relative contributions of miR-141 and miR-200c need to be delineated by using siRNAs or specific inhibitors in future studies.
In summary, miR-141/200c deficiency ameliorates NASH-associated hepatic steatosis, injury, and inflammation by reprogramming multiple signaling pathways. Further studies are warranted to identify common regulatory pathways regulated by miR-141/200c in other chronic liver diseases.
Methods
Experimental animals.
miR-141/200c–/– mice and WT littermates were generated on a 129P2 background through heterozygote mating from our laboratory. The embryonic stem cells (ES cells) for double deletion of miR-141 and miR-200c used for this research project were generated by the trans-NIH KOMP and obtained from the KOMP Repository (www.komp.org). We contracted with KOMP to generate the germline-transmitted lacZ-neo-floxed mice. We then crossed the mice with a germline β-actin-Flpe to restore the functional WT allele. Conditional mice were then crossed with the β-actin-Cre transgenic mouse to generate mice deficient for both miR-141 and miR-200c (miR-141/200c–/–). Age-matched (10 weeks of age) male mice were used for all experimental procedures and were maintained on a 12-hour light-dark cycle in a temperature (22°C) and humidity controlled (45%–55%) environment with ad libitum access to food and water. miR-141/200c–/– mice and their WT littermates (n = 5–8 mice per group) were fed a CD (Harlan Teklad, TD.94149; 16.8% kcal protein, 61.3% kcal carbohydrate, 21.9% kcal fat plus L-methionine [8.2 g/kg] and choline [1.4 g/kg]) or MCD diet (Harlan Teklad, TD.90262; 15.9% kcal protein, 62.3% kcal carbohydrate, 21.8% kcal fat) for 2 weeks to induce the NASH phenotype. After 2 weeks of the feeding regime, mice were fasted overnight and then euthanized, and blood and livers were collected.
Human liver specimens.
The coded human liver specimens were obtained through the Liver Tissue Cell Distribution System funded by NIH Contract HSN276201200017C, as described previously (35–37). Three groups of liver specimens were used: control normal (n = 10), NASH nonfatty liver (n = 15), and NASH fatty liver (n = 20).
Cell lines, primary mouse hepatocyte and BM isolations, and cell culture.
The human hepatocellular carcinoma cell lines HepG2 and Huh7 (Health Science Research Resources Bank, JCRB0403) (38) were cultured in DMEM containing glucose (4.5 g/l) and 10% FBS (Gibco, Thermo Fisher Scientific) under standard conditions as described previously (35, 39). Primary mouse hepatocytes were isolated from 10-week-old WT and miR-141/200c–/– mice as previously described (39, 40). In brief, primary mouse hepatocytes were isolated from male adult mice by a 2-step collagenase perfusion method and purified by Percoll (GE Healthcare) gradient centrifugation. Approximately 5 × 105 isolated hepatocyte cells were plated onto each well of a collagen-coated 6-well plate and were maintained in DMEM containing glucose (4.5 g/l; Gibco) and 10% FBS. BM cells were isolated from the tibia and femurs of 10-week-old male WT littermates and miR-141/200c–/– mice. Cells were differentiated into mature macrophages (BMDMs) in RPMI-1641 media (Gibco) containing 10% FBS (Atlanta Biologicals), 100 U/ml penicillin (Gibco), 100 μg/ml streptomycin (Gibco), and 10 ng/ml macrophage CSF (M-CSF, R&D Systems). After 7 days in culture, BMDMs were treated with or without LPS (10 ng/ml, MilliporeSigma) for 24 hours, followed by RNA isolation.
Biochemical analysis.
TGs were extracted from liver homogenates as described previously (41). Liver and serum TGs were measured calorimetrically as described previously (21). Levels of hepatic H2O2 were measured fluorometrically using an Amplex Red reagent (Thermo Fisher Scientific). Lipid peroxidation was measured by determining levels of malondialdehyde (MDA) levels using TBARs according to the method of Ohkawa et al. (42). The absorbance of the butanol layer was measured at 532 nm using 1,1,3,3-tetraethoxypropane (MilliporeSigma) as a standard. AST, ALT, and IL-6 levels in fasted serum were measured using Infinity assay kit (Point Scientific) or an ELISA kit (Cayman Chemicals), respectively, as described previously (37). Serum glucose was measured calorimetrically (Cayman Chemicals).
Immunoblotting analysis.
Livers were lysed in radio-immunoprecipitation assay buffer (RIPA) containing phosphatase and proteasome cocktail inhibitors as previously described (43). Equal amounts of protein from 5 livers in each group (n = 5 per group) were pooled. Approximately 20 μg of protein was separated by a 10%–12% SDS-PAGE and transferred onto a PVDF membrane. The membrane was blocked in 5% milk in tris-buffered saline plus tween (TBST) and incubated in primary antibody overnight at 4°C. The specific primary antibodies used were PPARα (sc-9000, Santa Cruz Biotechnology Inc.), PPARγ (sc-7196, Santa Cruz Biotechnology Inc.), Thr172 p-AMPK (50081, Cell Signaling Technology), AMPK (5832, Cell Signaling Technology), Ser473 p-AKT (4060, Cell Signaling Technology), Thr202/Tyr204 p-ERK1/2 (4377, Cell Signaling Technology), Thr180/Tyr182 p-P38 (4511, Cell Signaling Technology), Thr183/Tyr185 p-JNK (4668, Cell Signaling Technology), TLR4 (sc-293072, Santa Cruz Biotechnology Inc.), IKKβ (8943, Cell Signaling Technology), Ser9 p-GSK3β (5558, Cell Signaling Technology), SREBP1 (sc-8984, Santa Cruz Biotechnology Inc.), FAS (sc-55580, Santa Cruz Biotechnology Inc.), α-tubulin (3873, Cell Signaling Technology), β-actin (3700, Cell Signaling Technology), MTTP (PA542391, Thermo Fisher), Ser2448 p-mTOR (5536, Cell Signaling Technology), mTOR (2983, Cell Signaling Technology), Ser 371 p-p70S6K (9208, Cell Signaling Technology), Thr37/46 p-4EBP1 (2855, Cell Signaling Technology), Ser727 p-Stat3 (sc-81523, Santa Cruz Biotechnology Inc.), Stat3 (sc-293151, Santa Cruz Biotechnology Inc.), and caspase-3 (9662, Cell Signaling Technology). The membrane was incubated in the corresponding HRP-conjugated goat anti-rabbit (170-6515, Bio-Rad) or goat anti-mouse secondary antibody (HAF007, R&D Systems) for 1 hour at room temperature. Immunodetection was determined using the Pierce ECL detection system (Thermo Fisher Scientific) according to manufacturer’s instructions. Images were obtained using the ImageQuant LAS digital analyzer (GE Healthcare Life Sciences).
Gene expression analysis by quantitative PCR.
Total RNA was extracted from mouse and human livers using Trizol reagent (MilliporeSigma) according to manufacturer’s instructions and then reverse transcribed with the iScript cDNA Synthesis Kit (Bio-Rad). Quantitative PCR (qPCR) was carried out using SYBR Green assay in Bio-Rad Cycler detection system (43). A PCR master mix containing specific primers was used, and HPRT (mouse) or U6 (human) was used as the internal control. The CT was determined, and the relative gene expression subsequently was calculated as follows: fold change 2ΔΔCT. Primer sequences are provided in Supplemental Table 1.
Liver histology.
A piece of liver was fixed in 10% formalin overnight, processed, and embedded in paraffin wax. Approximately 4-μm thick sections were deparaffinized and then stained with H&E and Masson’s trichrome (43). Livers embedded in Tissue-Tek OCT compound were cut at 10 μm for F4/80 IHC and Oil Red O staining (35). Liver sections were incubated with rat anti–mouse F4/80 (MCA497GA, Bio-Rad) overnight and incubated with HRP-conjugated goat anti–rat IgG secondary antibody (Bio-Rad) for 1 hour before detection with DAB substrate (Vector Laboratories). TUNEL-positive cells were detected using the DeadEnd Calorimetric TUNEL assay (Promega), with DAB as a peroxidase substrate, and hematoxylin was used for the counterstain. All slides were examined under an Olympus light microscope, and images were taken at ×20 magnification. Computerized image analysis using ImageJ (NIH) software was used to quantify the area stained, and results are presented as relative stained area of the total area of view. The same threshold settings were used for each analysis. About 5–10 randomly chosen fields per section were used for all analyses, and n = 3 mice were used for analysis unless otherwise stated.
Metabolomics and lipidomics analysis.
Liver samples (~50 mg) was collected from mice (n = 5 per group) and immediately snap frozen in liquid nitrogen. Metabolomics and lipidomics analyses were carried out by the West Coast University of California (UC) Davis Center for Metabolomics, as described previously (44). Raw data are processed in an untargeted (qualitative) manner using Agilent’s software MassHunter Qual. The actual data are provided as peak heights for the quantification ion (mass/charge ratio [mz] value) at the specific retention time (rt value) and are normalized to the total average peak height. Peak heights of each studied lipid or metabolite were normalized to the appropriate internal standards. The final data sets are calculated as fold change relative to WT or as average ion peak height.
Statistics.
Data are presented as mean ± SEM. A Student’s unpaired t test was used to determine differences between WT and miR-141/200c–/– mice. Differences between multiple groups were compared using a one-way ANOVA with Newman-Keuls comparisons test. P < 0.05 was considered significant.
Study approval.
All animal experimental and care procedures were approved and conducted in accordance with the guidelines of the IACUCittee of the University of Connecticut. Individual identities associated with human liver samples were not known; therefore, the IRB for Human Research at the University of Connecticut determined that the project was not research involving human subjects.
Author contributions
MT performed the experiments, analyzed the data, and wrote the manuscript. SML and DJS generated and provided miR-141/200c–/– mice. LW supervised the work and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank West Coast Metabolomics Centre, UC Davis, for lipidomics and metabolomics analyses. LW is supported by NIH R01DK104656, R01DK080440, R01ES025909, R21AA022482, R21AA024935, R01AA026322 (with DJS); VA Merit Award 1I01BX002634, P30 DK34989 (Yale Liver Center); and the National Natural Scientific Foundation of China (grant 81572443). NIH grants to Velocigene at Regeneron Inc. (U01HG004085) and the CSD Consortium (U01HG004080) funded the generation of gene-targeted ES cells for 8,500 genes in the KOMP and archived and distributed by the KOMP Repository at UC Davis and CHORI (U42RR024244).
Version 1. 11/02/2017
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2017;2(21):e96094. https://doi.org/10.1172/jci.insight.96094.
Contributor Information
Melanie Tran, Email: melanie.tran@uconn.edu.
Sang-Min Lee, Email: smlee95@unist.ac.kr.
Dong-Ju Shin, Email: dong-ju.shin@uconn.edu.
Li Wang, Email: li.wang@uconn.edu.
References
- 1.Bhala N, Younes R, Bugianesi E. Epidemiology and natural history of patients with NAFLD. Curr Pharm Des. 2013;19(29):5169–5176. doi: 10.2174/13816128113199990336. [DOI] [PubMed] [Google Scholar]
- 2.Povero D, Feldstein AE. Novel Molecular Mechanisms in the Development of Non-Alcoholic Steatohepatitis. Diabetes Metab J. 2016;40(1):1–11. doi: 10.4093/dmj.2016.40.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marra F, Gastaldelli A, Svegliati Baroni G, Tell G, Tiribelli C. Molecular basis and mechanisms of progression of non-alcoholic steatohepatitis. Trends Mol Med. 2008;14(2):72–81. doi: 10.1016/j.molmed.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Zámbó V, Simon-Szabó L, Szelényi P, Kereszturi E, Bánhegyi G, Csala M. Lipotoxicity in the liver. World J Hepatol. 2013;5(10):550–557. doi: 10.4254/wjh.v5.i10.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Z, Ross RA, Zhao S, Tu W, Liangpunsakul S, Wang L. LncRNA AK054921 and AK128652 are potential serum biomarkers and predictors of patient survival with alcoholic cirrhosis. Hepatol Commun. 2017;1:513–523. doi: 10.1002/hep4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Z, Cappello T, Wang L. Emerging role of microRNAs in lipid metabolism. Acta Pharm Sin B. 2015;5(2):145–150. doi: 10.1016/j.apsb.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song G, Wang L. Nuclear receptor SHP activates miR-206 expression via a cascade dual inhibitory mechanism. PLoS ONE. 2009;4(9):e6880. doi: 10.1371/journal.pone.0006880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Yang Z, Whitby R, Wang L. Regulation of miR-200c by nuclear receptors PPARα, LRH-1 and SHP. Biochem Biophys Res Commun. 2011;416(1-2):135–139. doi: 10.1016/j.bbrc.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song G, Wang L. MiR-433 and miR-127 arise from independent overlapping primary transcripts encoded by the miR-433-127 locus. PLoS ONE. 2008;3(10):e3574. doi: 10.1371/journal.pone.0003574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song G, Wang L. A conserved gene structure and expression regulation of miR-433 and miR-127 in mammals. PLoS ONE. 2009;4(11):e7829. doi: 10.1371/journal.pone.0007829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Z, Zhang Y, Wang L. A feedback inhibition between miRNA-127 and TGFβ/c-Jun cascade in HCC cell migration via MMP13. PLoS One. 2013;8(6):e65256. doi: 10.1371/journal.pone.0065256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song G, Zhang Y, Wang L. MicroRNA-206 targets notch3, activates apoptosis, and inhibits tumor cell migration and focus formation. J Biol Chem. 2009;284(46):31921–31927. doi: 10.1074/jbc.M109.046862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Senfter D, Madlener S, Krupitza G, Mader RM. The microRNA-200 family: still much to discover. Biomol Concepts. 2016;7(5-6):311–319. doi: 10.1515/bmc-2016-0020. [DOI] [PubMed] [Google Scholar]
- 14.Guo J, et al. Reduced miR-200b and miR-200c expression contributes to abnormal hepatic lipid accumulation by stimulating JUN expression and activating the transcription of srebp1. Oncotarget. 2016;7(24):36207–36219. doi: 10.18632/oncotarget.9183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murakami Y, et al. The progression of liver fibrosis is related with overexpression of the miR-199 and 200 families. PLoS ONE. 2011;6(1):e16081. doi: 10.1371/journal.pone.0016081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dou L, et al. miR-200s contribute to interleukin-6 (IL-6)-induced insulin resistance in hepatocytes. J Biol Chem. 2013;288(31):22596–22606. doi: 10.1074/jbc.M112.423145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rinella ME, Elias MS, Smolak RR, Fu T, Borensztajn J, Green RM. Mechanisms of hepatic steatosis in mice fed a lipogenic methionine choline-deficient diet. J Lipid Res. 2008;49(5):1068–1076. doi: 10.1194/jlr.M800042-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ibrahim SH, Hirsova P, Malhi H, Gores GJ. Animal Models of Nonalcoholic Steatohepatitis: Eat, Delete, and Inflame. Dig Dis Sci. 2016;61(5):1325–1336. doi: 10.1007/s10620-015-3977-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Machado MV, et al. Mouse models of diet-induced nonalcoholic steatohepatitis reproduce the heterogeneity of the human disease. PLoS One. 2015;10(5):e0127991. doi: 10.1371/journal.pone.0127991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee SM, Zhang Y, Tsuchiya H, Smalling R, Jetten AM, Wang L. Small heterodimer partner/neuronal PAS domain protein 2 axis regulates the oscillation of liver lipid metabolism. Hepatology. 2015;61(2):497–505. doi: 10.1002/hep.27437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang J, Tabbi-Anneni I, Gunda V, Wang L. Transcription factor Nrf2 regulates SHP and lipogenic gene expression in hepatic lipid metabolism. Am J Physiol Gastrointest Liver Physiol. 2010;299(6):G1211–G1221. doi: 10.1152/ajpgi.00322.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32(5):593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 23.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122(3):787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, et al. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab. 2011;13(4):376–388. doi: 10.1016/j.cmet.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev. 2011;75(1):50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miura K, Seki E, Ohnishi H, Brenner DA. Role of toll-like receptors and their downstream molecules in the development of nonalcoholic Fatty liver disease. Gastroenterol Res Pract. 2010;2010:362847. doi: 10.1155/2010/362847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porstmann T, et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8(3):224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sapp V, Gaffney L, EauClaire SF, Matthews RP. Fructose leads to hepatic steatosis in zebrafish that is reversed by mechanistic target of rapamycin (mTOR) inhibition. Hepatology. 2014;60(5):1581–1592. doi: 10.1002/hep.27284. [DOI] [PubMed] [Google Scholar]
- 29.Wang M, You Q, Lor K, Chen F, Gao B, Ju C. Chronic alcohol ingestion modulates hepatic macrophage populations and functions in mice. J Leukoc Biol. 2014;96(4):657–665. doi: 10.1189/jlb.6A0114-004RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wan J, et al. M2 Kupffer cells promote M1 Kupffer cell apoptosis: a protective mechanism against alcoholic and nonalcoholic fatty liver disease. Hepatology. 2014;59(1):130–142. doi: 10.1002/hep.26607. [DOI] [PubMed] [Google Scholar]
- 31.Agren JJ, Kurvinen JP, Kuksis A. Isolation of very low density lipoprotein phospholipids enriched in ethanolamine phospholipids from rats injected with Triton WR 1339. Biochim Biophys Acta. 2005;1734(1):34–43. doi: 10.1016/j.bbalip.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 32.Puri P, et al. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology. 2007;46(4):1081–1090. doi: 10.1002/hep.21763. [DOI] [PubMed] [Google Scholar]
- 33.Ilan Y. Compounds of the sphingomyelin-ceramide-glycosphingolipid pathways as secondary messenger molecules: new targets for novel therapies for fatty liver disease and insulin resistance. Am J Physiol Gastrointest Liver Physiol. 2016;310(11):G1102–G1117. doi: 10.1152/ajpgi.00095.2016. [DOI] [PubMed] [Google Scholar]
- 34.Chung RW, et al. Dietary sphingomyelin lowers hepatic lipid levels and inhibits intestinal cholesterol absorption in high-fat-fed mice. PLoS ONE. 2013;8(2):e55949. doi: 10.1371/journal.pone.0055949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, et al. Bcl2 is a critical regulator of bile acid homeostasis by dictating Shp and lncRNA H19 function. Sci Rep. 2016;6:20559. doi: 10.1038/srep20559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song Y, et al. H19 promotes cholestatic liver fibrosis by preventing ZEB1-mediated inhibition of epithelial cell adhesion molecule. Hepatology. 2017;66(4):1183–1196. doi: 10.1002/hep.29209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L, Yang Z, Trottier J, Barbier O, Wang L. Long noncoding RNA MEG3 induces cholestatic liver injury by interaction with PTBP1 to facilitate shp mRNA decay. Hepatology. 2017;65(2):604–615. doi: 10.1002/hep.28882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao Y, et al. High-mobility-group protein 2 regulated by microRNA-127 and small heterodimer partner modulates pluripotency of mouse embryonic stem cells and liver tumor initiating cells. Hepatol Commun. 2017;1(8):816–830. doi: 10.1002/hep4.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, et al. E2F1 is a novel fibrogenic gene that regulates cholestatic liver fibrosis through the Egr-1/SHP/EID1 network. Hepatology. 2014;60(3):919–930. doi: 10.1002/hep.27121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, et al. Nuclear receptor SHP, a death receptor that targets mitochondria, induces apoptosis and inhibits tumor growth. Mol Cell Biol. 2010;30(6):1341–1356. doi: 10.1128/MCB.01076-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tabbi-Anneni I, et al. Overexpression of nuclear receptor SHP in adipose tissues affects diet-induced obesity and adaptive thermogenesis. Am J Physiol Endocrinol Metab. 2010;298(5):E961–E970. doi: 10.1152/ajpendo.00655.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohkawa H, Ohishi N, Yagi K. Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction. Anal Biochem. 1979;95(2):351–358. doi: 10.1016/0003-2697(79)90738-3. [DOI] [PubMed] [Google Scholar]
- 43.Yang Z, et al. REV-ERBα Activates C/EBP Homologous Protein to Control Small Heterodimer Partner-Mediated Oscillation of Alcoholic Fatty Liver. Am J Pathol. 2016;186(11):2909–2920. doi: 10.1016/j.ajpath.2016.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tran M, Yang Z, Liangpunsakul S, Wang L. Metabolomics Analysis Revealed Distinct Cyclic Changes of Metabolites Altered by Chronic Ethanol-Plus-Binge and Shp Deficiency. Alcohol Clin Exp Res. 2016;40(12):2548–2556. doi: 10.1111/acer.13257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.