Abstract
Maternal obesity is a global health problem that increases offspring obesity risk. The metabolic pathways underlying early developmental programming in human infants at risk for obesity remain poorly understood, largely due to barriers in fetal/infant tissue sampling. Utilizing umbilical cord–derived mesenchymal stem cells (uMSC) from offspring of normal weight and obese mothers, we tested whether energy metabolism and gene expression differ in differentiating uMSC myocytes and adipocytes, in relation to maternal obesity exposures and/or neonatal adiposity. Biomarkers of incomplete β-oxidation were uniquely positively correlated with infant adiposity and maternal lipid levels in uMSC myocytes from offspring of obese mothers only. Metabolic and biosynthetic processes were enriched in differential gene expression analysis related to maternal obesity. In uMSC adipocytes, maternal obesity and lipids were associated with downregulation in multiple insulin-dependent energy-sensing pathways including PI3K and AMPK. Maternal lipids correlated with uMSC adipocyte upregulation of the mitochondrial respiratory chain but downregulation of mitochondrial biogenesis. Overall, our data revealed cell-specific alterations in metabolism and gene expression that correlated with maternal obesity and adiposity of their offspring, suggesting tissue-specific metabolic and regulatory changes in these newborn cells. We provide important insight into potential developmental programming mechanisms of increased obesity risk in offspring of obese mothers.
Keywords: Cell Biology, Metabolism
Keywords: Fatty acid oxidation, Mitochondria, Obesity
Umbilical cord-derived mesenchymal stem cell-specific alterations in metabolism and gene expression correlate with maternal obesity and neonatal adiposity.
Introduction
The Developmental Origins of Health and Disease (DOHaD) hypothesis, now substantiated by extensive animal and human research, suggests that both maternal nutrient deficiency as well as nutrient excess in utero and in early infancy results in an acquired susceptibility to metabolic disease later in life (1, 2) and across generations (3). Maternal obesity increases the risk for pediatric obesity and impacts multiple organs and metabolic systems underlying a broad array of chronic illnesses — including diabetes, nonalcoholic fatty liver disease, and cardiovascular disease — that are becoming increasingly common at younger ages (4). Alarmingly, although the rate of preadolescent obesity has stabilized over the last decade, 1 in 10 infants and toddlers are obese (OB), and 1 in 5 youth are both OB and at-risk for metabolic syndrome prior to the onset of puberty (4).
Our understanding of the early biochemical networks responsible for obesity in tissues and plasma from infants and young children at risk for development of obesity is lacking, due to the difficulty in obtaining tissue samples and performing intensive cellular metabolic studies in this population. Metabolomic analysis to date in newborns has been limited to umbilical cord blood (5) and umbilical cord endothelial cells (6). However, cord blood may reflect immediate events surrounding delivery rather than the offspring’s underlying steady state physiology. Cord endothelial cells, while exposed to similar nutrients as the fetus in utero, may only reflect changes in vascular endothelium and not in energy metabolizing tissues such as muscle and fat. Still, changes demonstrated in these umbilical-derived sources have made valuable connections between maternal obesity and mitochondria-associated gene expression, suggesting that there may be fundamental changes in mitochondrial metabolism in infants born to OB mothers. A recent small study of umbilical-derived mesenchymal stem cells (uMSC), which are multipotent and considered fetal in origin (7), from newborns from mothers with gestational diabetes (n = 4), has demonstrated a reduction in mitochondrial respiration (8). Maternal obesity effects on metabolomic and transcriptomic pathways, in differentiated myocytes or adipocytes in infants of OB mothers, and any relationship to infant adiposity is not currently known. Given that cellular metabolic processes are critical for the programming of stem cells (9–11), it raises the potential that maternal factors present in the OB intrauterine environment that affect energy regulation or differentiation may have significant impacts on the long-term programming and function of these cells, which exist in bone, muscle, and mature adipose tissue.
Metabolomic studies in adults with obesity have shown marked and consistent changes in nutrient handling and energy metabolism, suggestive of changes in mitochondrial metabolic fuel handling in established obesity. Increased circulating plasma short- and long-chain acylcarnitines (LCAC) (12–14) and multiple amino acids including branched chain, aromatic (e.g., phenylalanine and tyrosine), 1-carbon (e.g., methionine, cysteine, and glycine), and glutaminolytic (e.g., aspartate, alanine, and glutamine) are associated with insulin resistance and established obesity. Amino acid changes are thought to be largely due to anaplerosis, or the use of amino acid oxidative catabolism to replenish tricarboxylic acid cycle (TCA cycle) intermediates in the setting of incomplete β-oxidation (12–16). In myocytes from OB adults, there is reduced fatty acid oxidation, leading to elevations in long- and medium-chain acylcarnitines and anaplerosis, and decreased oxidative phosphorylation, indicating abnormalities in key energy pathways (13, 17–19). Similarly, in adipocytes from OB adults, there is decreased fatty acid oxidation, increased lipogenesis, and spared amino acid catabolism (13, 20, 21). Biomarkers for these processes can be observed in plasma of OB school-aged children (16, 22, 23), suggesting early changes in mitochondria metabolism in established obesity.
Despite the well-established independent risk factor of increased maternal BMI on infant birth weight and adiposity at birth (24, 25), the metabolic pathways in the infant that contribute to long-term obesity risk remain complex and unresolved. The Healthy Start Study at the University of Colorado (ClinicalTrials.gov; NCT02273297) is a longitudinal prospective observational prebirth cohort of 1,400 maternal-infant pairs beginning in early pregnancy that has revealed important associations between maternal phenotype and infant adiposity at birth (24, 26). To investigate the Biology of intraUterine Metabolic Programming (BUMP), umbilical cord tissue was obtained at birth from a convenience sample of 165 Healthy Start infants and used to culture uMSC as an ancillary part of the Healthy Start Study (Baby BUMP Project). In a subsample of these infants, we have recently shown that differentiating uMSC myocytes and adipocytes were physiologically distinct (as demonstrated by differential expression of PAX7, myogenin, myosin heavy chain, FABP4, CEBPa, and PPARγ), despite coming from the same original precursor (27). Differentiating uMSC adipocytes from offspring of OB women, compared with normal weight (NW) matched controls, had greater adipogenic potential (higher PPARγ), and uMSC lipid accumulation in the adipogenic condition was correlated with adiposity in the neonate (27). However, data demonstrating altered metabolic networks in the infant with increased obesity risk based on maternal phenotype have yet to be defined. Here, we report that in uMSC myocytes and uMSC adipocytes from infants born to OB mothers have metabolic and transcriptomic differences that correlate with maternal BMI, as well as adiposity at birth and infant exposure to excess maternal free fatty acids (FFA), particularly in offspring of OB mothers. Our results suggest that these progenitor cells are sensitive to developmental insults that correlate with neonatal adiposity and portend potential risk for future obesity.
Results
Maternal obesity phenotype correlates with offspring neonatal adiposity
Mothers in this study were classified as NW or OB based on maternal prepregnancy BMI, as shown in Table 1. For selection criteria of OB (n = 12) and NW (n = 12) groups, please refer to Methods. Briefly, these were all otherwise healthy, term pregnancies without gestational diabetes, complicating medical factors, or preeclampsia. OB mothers were selected based on the limited number available in our Baby BUMP uMSC sample. NW mothers were selected to match for age, gestational age at delivery, and infant sex. Of note, sex-stratified metabolomic analysis can be found in Supplemental Results (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.94200DS1), and are mentioned in Results when relevant.
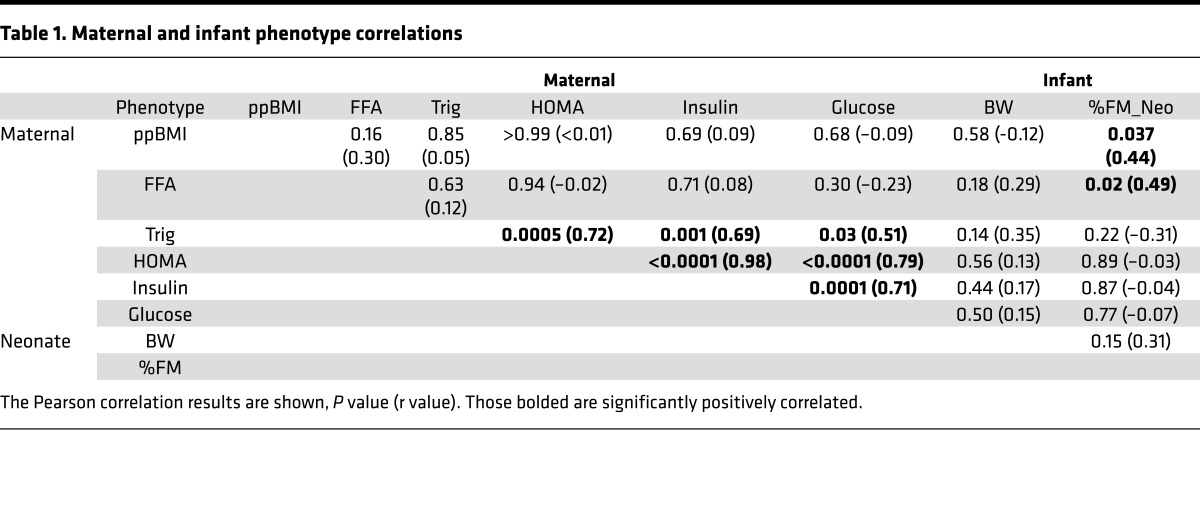
Table 1. Maternal and infant phenotype correlations.
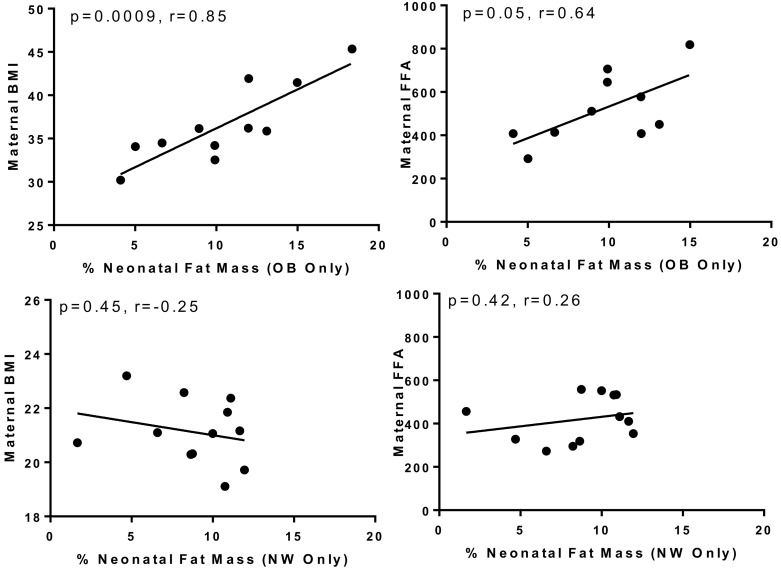
In our sample, the infants of OB mothers did not have statistically significant higher percent fat mass (%FM) compared with NW offspring (P = 0.28; Supplemental Table 1). Maternal prepregnancy BMI did correlate with neonatal fat mass in regression analyses (P = 0.04), as shown here in Table 1, primarily due to highly significant correlation between maternal BMI and neonatal %FM only in offspring of OB, but not NW, mothers (P = 0.0009, r = 0.85 vs. P = 0.45, r = –0.25, Figure 1). Therefore, the OB group had a different relationship between maternal BMI and %FM than did the NW group. Further, in this sample, there were significant correlations between maternal FFA and neonatal %FM (P = 0.02, r = 0.48) in the total group, but again significant in the OB but not NW group (P = 0.05, r = 0.68 vs. P = 0.42, r = 0.16, Figure 1). These correlations were not found in the larger Healthy Start Study. Similar correlations between maternal second trimester triglycerides, glucose, and insulin were not found in the OB or NW group. Given correlative differences between OB and NW groups for both BMI vs. %FM and FFA vs. %FM, in addition to defining uMSC differences based on categorical maternal BMI, we tested if neonatal %FM and maternal FFA (specifically in the OB but not NW group) were related to metabolomic and transcriptional differences in offspring uMSC.
Figure 1. Pearson correlation between maternal prepregnancy BMI (kg/m2), maternal free fatty acids (FFA, mg/dL), and neonatal % fat mass.
Positive correlations were found in the offspring of obese (OB, n = 12), but not normal weight (NW, n = 12) mothers.
Incomplete β-oxidation and altered oxidative metabolites correlate with infant adiposity at birth from offspring of OB but not NW mothers in uMSC myocytes
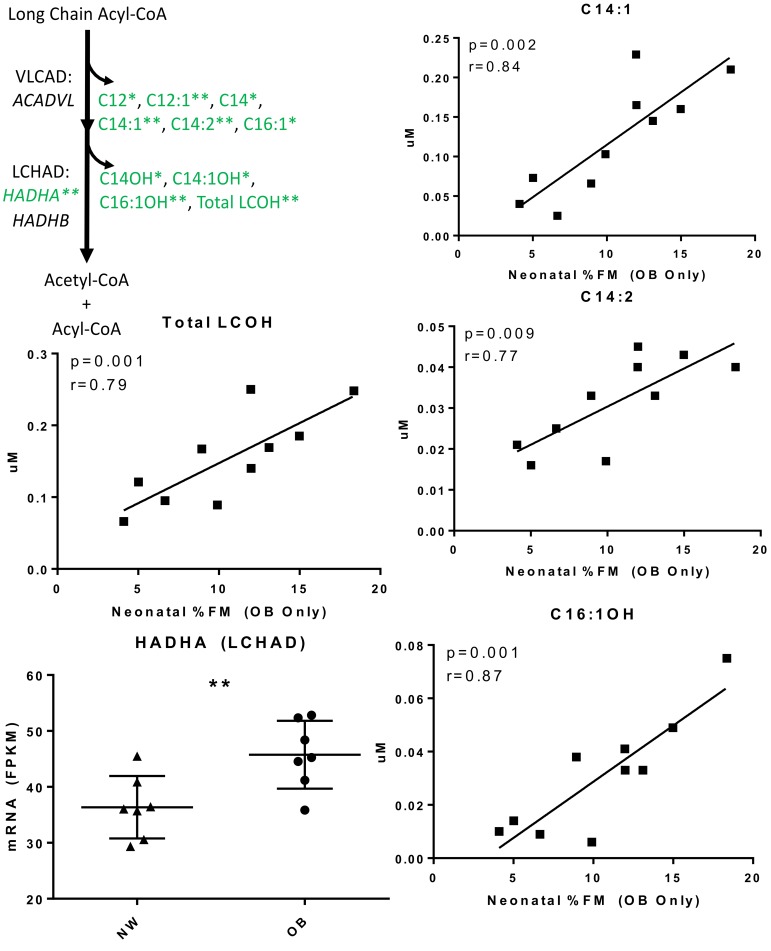
Incompleteβ-oxidation of long-chain fatty acids. To test whether maternal obesity phenotype was associated with alterations in energy metabolism in infant uMSC adipocytes and myocytes, we measured 70 primary analytes in the spent media from lipid-treated uMSC cells during differentiation, from infants born to NW (n = 12) and OB (n = 12) mothers. Very few analytes differed cross-sectionally by t test according to group assignment OB vs. NW (Supplemental Table 2; prepregnancy BMI [ppBMI]). However, there were strong, significant positive correlations between neonatal %FM and accumulation of multiple LCAC (specifically C12, C12:1, C14, C14:1, C14:2, C16:1, C18:2, and total LCAC) and long-chain hydroxy acylcarnitine species (LCOH, specifically C14-OH, C14:1-OH, C16:1-OH, and total LCOH) in the OB, but not NW, group (all P < 0.05, r > 0.60, FDR < 0.05, Figure 2 and Supplemental Table 2). These acylcarnitine signatures have been related to deficiencies in, and are substrates for, the very long–chain acyl-coa dehydrogenase (VLCAD) and long-chain 3-hydroxyacyl-CoA dehydrogenase (LCHAD) enzymatic steps of β-oxidation (28). Similar increases in LCAC and LCOH have been reported previously in plasma of adults with established obesity, and they serve as biomarkers of incomplete β-oxidation (14, 18, 29).
Figure 2. Acylcarnitine Analysis for uMSC myocytes.
Metabolomic and targeted gene expression analysis in uMSC myocytes suggesting dysfunctional β-oxidation; *P < 0.05, **P < 0.01, all FDR < 0.05 (See also Supplemental Table 2). Metabolomic data was analyzed in n = 10 offspring of obese (OB) mothers who had data available. Transcriptional analysis was performed in offspring of obese (n = 7 )and normal weight (NW, n = 7) mothers. Continuous variables were analyzed using linear regression with r values calculated by Pearson correlation. Categorical variables were analyzed using 2-tailed student t test (bar represents the mean ± SD).
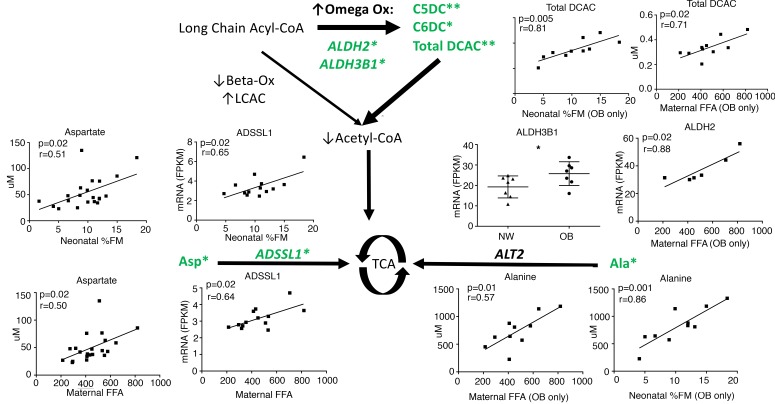
Compensatoryω-oxidation and intermediary amino acid metabolism. Increased ω-oxidation of fatty acids is a compensatory mechanism in the setting of incomplete β-oxidation and is characterized by an accumulation of dicarboxylic acylcarnitines (DCAC) in tissue, plasma, and urine. In uMSC myocytes, there were positive correlations in multiple DCAC in relation to both maternal FFA and neonatal %FM including glutarylcarnitine (C5DC, P = 0.009, r = 0.77 and 0.004, 0.81), adipylcarnitine (C6DC, P = 0.006, r = 0.79 and 0.04, 0.64), and total DCAC (P = 0.02, r = 0.71 and 0.005, 0.81), specifically in the OB group (Figure 3 and Supplemental Table 2).
Figure 3. Acylcarnitine and Amino Acid Analysis for uMSC myocytes.
Metabolomic and gene expression analysis in uMSC myocytes suggests compensatory use of ω-oxidation and amino acid catabolism as alternative fuel pathways in the setting of dysfunctional β-oxidation; *P < 0.05, **P < 0.01, FDR < 0.05 (See also Supplemental Table 3). Metabolomic data was analyzed in n = 10 offspring of obese (OB) mothers who had data available. Transcriptional analysis was performed in offspring of OB (n = 7)and normal weight (NW, n = 7) mothers. Continuous variables were analyzed using linear regression, with r values calculated by Pearson correlation. Categorical variables were analyzed using 2-tailed student t test (bar represents the mean ± SD).
As illustrated in Figure 3 and Supplemental Table 2, in uMSC myocytes, there were significant correlations between neonatal %FM and glutaminolytic amino acids, including positive correlations with alanine (P = 0.01, r = 0.53) and aspartate (P = 0.02, r = 0.51) and negative correlations with glutamine (P = 0.02, r = –0.51). As was the case for findings in the β-oxidation and ω-oxidation pathways, specifically for alanine, concentrations were positively correlated with both %FM (P = 0.001, r = 0.86) and maternal FFA (P = 0.01, r = 0.57) only in the OB group. Additionally, there were sex-specific correlations between maternal FFA and 1-carbon amino acids including glycine (P = 0.003, r = –0.99), methionine (P = 0.01, r = 0.99), and cystathionine (P = 0.008, r = –0.99) only in females of OB mothers, and correlations with FFA and phenylalanine (P = 0.03, r = 0.81), tyrosine (P = 0.03, r = 0.81), and total aromatic amino acids (P = 0.03, r = 0.81) only in males of OB mothers (Supplemental Results and Discussion). There were no significant correlations with branched-chain amino acids.
Notably, accumulated biomarkers of incomplete β-oxidation, ω-oxidation, and anaplerosis were found in the uMSC myocytes were not found in the uMSC adipocytes, despite being derived from the same precursor cell from OB offspring (Supplemental Table 3) in relation to neonatal %FM or maternal FFA.
Differential gene expression patterns support altered metabolism in uMSC myocytes from offspring of OB mothers
To test the hypothesis that maternal obesity affects gene expression in response to lipid challenge in OB and NW uMSC myocytes, RNA-sequencing (RNA-Seq) was carried out in a representative subset of these cells (n = 7 NW and n = 7 OB) (workflow outlined in Supplemental Figures 1 and 2). Based on the metabolite analysis, we first looked for targeted changes in the fatty acid oxidation pathway that might account for greater fatty acid intermediates in uMSC myocytes associated with impaired fat metabolism. However, there was significant upregulated uMSC myocyte HADHA (LCHAD) relative to maternal BMI (P = 0.01, Figure 2), and we did not find correlations in expression of HADHB (also LCHAD) or ACADVL (VLCAD).
With respect to other oxidative intermediates found in OB MSC-myocytes, ALDH3B1 (P = 0.05) was upregulated in uMSC myocytes in relation to maternal BMI, and ALDH2 (P = 0.02) was upregulated in relation to FFA in uMSC myocytes in OB but not NW groups (Figure 3), corresponding with increased DCAC species. Conversely, in amino acid metabolism, the primary gene for incorporation of alanine into the TCA cycle ALT2 (or alanine aminotransferase) was not significantly correlated with %FM, FFA, or maternal BMI. ADSSL (adenylosuccinate synthase, a key step in converting aspartate to fumarate, a TCA cycle intermediate) was upregulated relative to both neonatal %FM and maternal FFA (P = 0.02 for both).
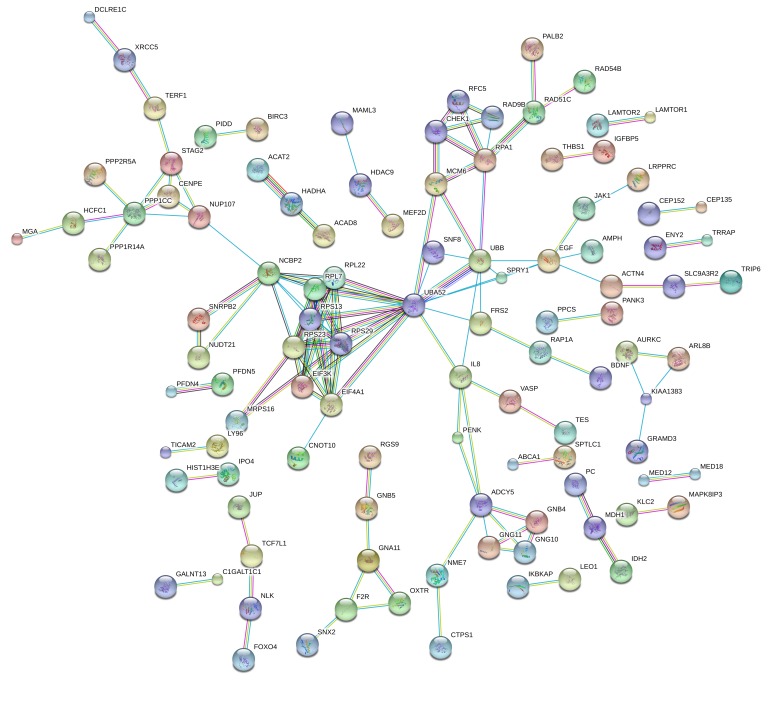
To test whether there was enrichment of gene pathways differentially expressed between OB vs. NW groups, untargeted analysis of the RNA-Seq dataset was carried out (Supplemental Figure 3). All genes with P < 0.05 (OB vs. NW, see volcano plot, Supplemental Figures 3 and 4) were analyzed by STRING online database (https://string-db.org/) (30), using the most stringent parameters for interaction score in protein encoding genes (>0.9, n = 464 genes). In addition to genes directly altered in fatty acid and amino acid metabolism, there was enrichment for multiple Gene Ontology Biological Process (GO_BP) (P = 3.6 × 10–5), including 24 of 36 processes related to metabolism and biosynthesis (Figure 4 and Supplemental Table 4). In particular, the largest enriched GO process was “Metabolic Process” (GO: 0008152, 231 total genes, FDR = 0.03) including metabolism of organic aromatic and nitrogen-containing compounds (FDR = 6.9 × 10-3 and 0.01, respectively). Among the interactions with the highest enrichment score were HADHA-ACAT2-ACAD8 (interaction score 0.997), which participate in lipid and amino acid catabolism, as well as IDH2-MDH1-PC (interaction score 0.962), genes in the TCA cycle. These genes are differentially upregulated in uMSC myocytes from the OB vs. NW group. There were also multiple GO processes involving gene expression and gene expression regulation (FDR = 0.01 and 6.9 × 10-3, respectively).
Figure 4. Differential gene expression in uMSC myocytes in offspring of obese (OB) vs.
normal weight (NW) mothers. Significantly differentially expressed genes (as calculated by 2-tailed student t test, n = 7 [NW] vs. n = 7 [OB]) were tested for relatedness using STRING online database. Network enrichment is shown. Enrichment for GO Biological Processes can be found as Supplemental Table 4.
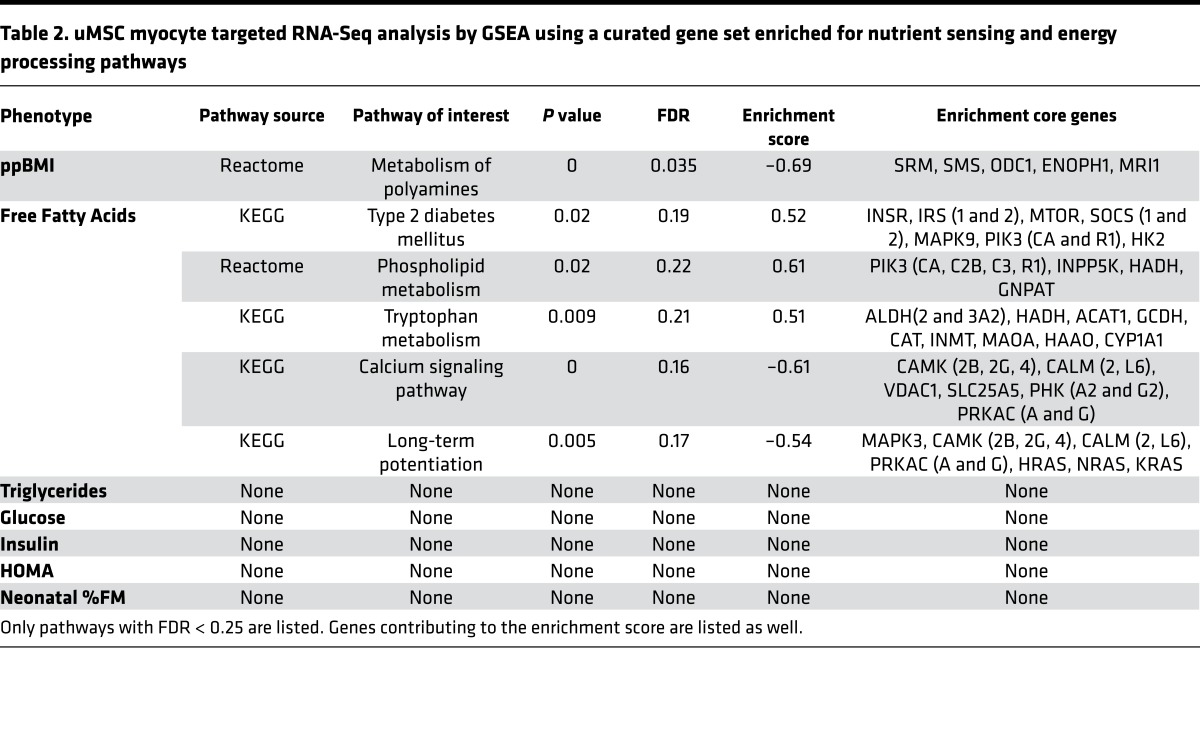
Utilizing a hand-curated, the Kyoto Encyclopedia of Genes and Genomes–based (KEGG-based; http://www.genome.jp/kegg/pathway.html) gene dataset, which included genes from multiple nutrient-sensing and energy metabolism pathways culled from the larger RNA-Seq dataset (see Methods for specific pathway KEGG pathway ID), we used Gene Set Enrichment Analysis software (GSEA, http://www.broad.mit.edu/gsea/; ref. 31) to assess directional enrichment of these pathways related to continuous maternal and neonatal phenotypes in uMSC myocytes. As illustrated in Table 2, maternal FFA were correlated with upregulation of KEGG pathways type 2 diabetes (P = 0.02, FDR = 0.19), tryptophan metabolism (P = 0.009, FDR = 0.21), and phospholipid metabolism (P = 0.02, FDR = 0.22). There was downregulation of calcium signaling (P < 0.001, FDR = 0.16), which is known to be involved in insulin-sensitive lipid and glucose metabolism (hsa04020 and 04910) and MAPK-dependent insulin signaling (hsa04910). Among the significant core enrichment genes, there was upregulation of PIK3CA (P = 0.04, r = 0.58), PIK3R1 (P = 0.05, r = 0.55), SOCS1 (0.04, r = 0.57), and SOCS2 (0.01, r = 0.66) — all associated with insulin signaling and central to the KEGG type 2 diabetes pathway (hsa04930). Importantly, PPARG, a key regulator of lipid metabolism and marker of adipogenicity, was also upregulated relative to maternal FFA (P = 0.03, r = 0.61).
Table 2. uMSC myocyte targeted RNA-Seq analysis by GSEA using a curated gene set enriched for nutrient sensing and energy processing pathways.
Neonatal %FM in the combined OB and NW groups did not show significant pathway enrichment; however, uMSC myocytes only from the OB, but not NW, group demonstrated downregulation of multiple pathways, including biocarta insulin (P < 0.001, FDR = 0.12) and insulin-related ERK (P = 0.006, FDR = 0.12) and RAS (P = 0.02, FDR = 0.14) signaling pathways (data not illustrated in the table). The reactome glycolysis pathway was also negatively enriched (P = 0.049, FDR = 0.18).
Maternal BMI, FFA, and neonatal %FM correlate with reduced MTOR and insulin-related gene expression in uMSC adipocytes
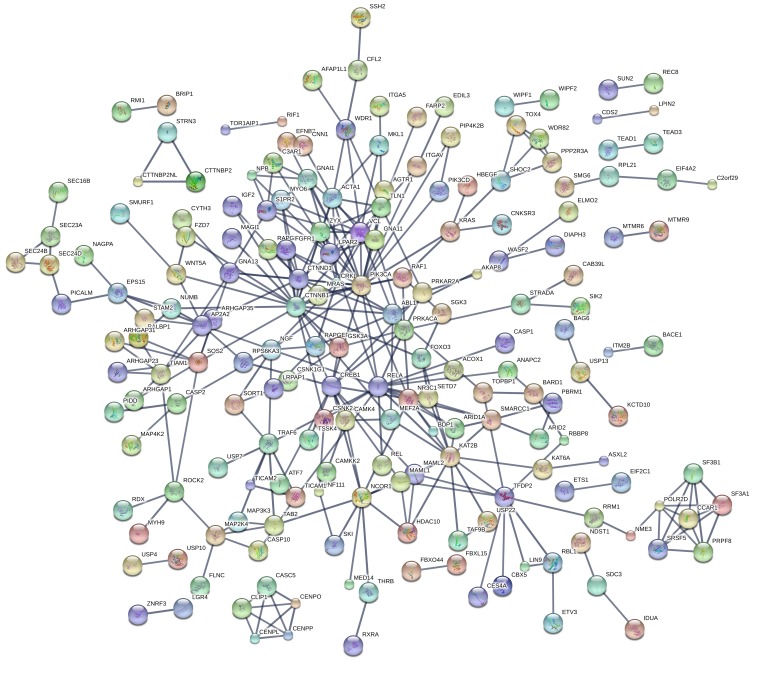
The uMSC adipocytes showed robust changes in transcriptomic analyses. We used STRING Database analysis to assess significant differential gene expression between OB vs. NW groups in uMSC adipocytes. Employing the same statistical analyses and stringent enrichment parameters as the myocyte analysis, there were a high number of gene-gene interactions related to maternal BMI (Supplemental Figure 4). Figure 4 illustrates the large number of genes enriched and the overall interaction P value of 0 (P < 1 × 10–10, n = 583 genes). This included 120 different GO biological processes, with metabolic processes again among the largest and most significant (FDR = 5.6 × 10–6). Importantly, there were 17 different KEGG pathways also enriched in OB vs. NW offspring. These are listed in Supplemental Table 5, illustrated in the larger enrichment network (Figure 5), and include MAPK (FDR = 0.005), AMPK (FDR = 0.006), RAS (FDR = 0.01), and PI3K-AKT (FDR = 0.03) signaling, which are all in the insulin-regulated signaling pathway. All were downregulated in OB vs. NW offspring cells.
Figure 5. Differential gene expression in uMSC adipocytes in offspring of obese (OB) vs.
normal weight (NW) mothers. Significantly differentially expressed genes (as calculated by 2-tailed student t test, n = 7 [NW] vs. n = 7 [OB]) were tested for relatedness using STRING online database. Network enrichment is shown. KEGG Pathway Enrichment can be found in Supplemental Table 5.
To analyze specific factors in addition to BMI that may inform the changes in gene expression, we analyzed RNA-Seq data by GSEA regression analysis using genes from our smaller, hand-curated dataset in MSC-adipocytes. Supplemental Table 6 illustrates the association between enriched pathways, core gene enrichment, maternal BMI, maternal FFA, and neonatal %FM. Maternal BMI was highly correlated with downregulation of the KEGG MTOR (P = 0, FDR = 0.003), MAPK (P = 0, FDR = 0.007), and insulin signaling pathways (P = 0, FDR = 0.004), as would be expected based on the above untargeted STRING analysis. There was further, potentially novel, downregulation in KEGG adipokine signaling (P = 0, FDR = 0.007), biocarta CREB (P = 0.002, FDR = 0.01), and KEGG WNT signaling (P = 0.008, FDR = 0.02) pathways. Maternal FFA levels correlated with downregulation of the KEGG MTOR (P = 0.04, FDR = 0.21), MAPK (P = 0.03, FDR = 0.23), AMPK (P = 0.03, FDR = 0.2), and PI3K-AKT (P = 0.02, FDR = 0.2) signaling pathways, indicating overlap and interrelatedness between maternal FFA and BMI. There was a potentially novel correlation with the reactome “transcriptional regulation of white adipocyte differentiation” (P = 0.03, FDR = 0.19), which had core enrichment for EP300 (P = 0.006, r = –0.71), CREBBP (P = 0.0001, r = –0.87), and PPARA (P = 0.0052, r = –0.72) and was specifically downregulated in the OB group only.
Similar to correlations with maternal FFA, neonatal %FM in uMSC adipocytes showed downregulation of AMPK-related MTOR signaling (P = 0.03, FDR = 0.21), PI3K signaling (P = 0.008, FDR = 0.25), and calcium-dependent signaling (P = 0.04, FDR = 0.24) pathways.
FFA and neonatal %FM correlate with increased mitochondrial respiratory chain gene expression in uMSC adipocytes
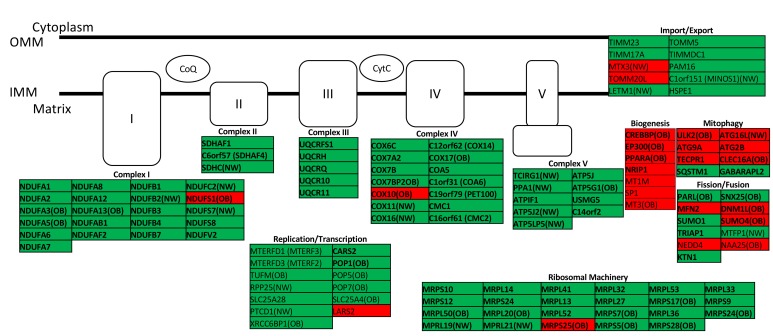
To examine whether mitochondrial gene sets in infant stem cells were affected by maternal lipid exposure or infant adiposity, we utilized a mitochondrial gene–specific hand-curated data set and tested for pathway enrichment using GSEA regression analyses. There was significant upregulation of reactome and KEGG oxidative phosphorylation pathway in relation to both maternal FFA (P = 0.02, FDR = 0.15) and neonatal %FM (P = 0.007, FDR = 0.11, Supplemental Table 6) in uMSC adipocytes. As illustrated in Figure 6, there was generalized upregulation of core gene enrichment in multiple genes in all respiratory chain complexes (I–V), mitochondrial ribosomal genes, and mitochondrial replication genes, relative to maternal FFA exposure. While both OB and NW groups demonstrated general upregulation of electron transport chain machinery, only the OB group had selectively downregulated ETC and ribosomal genes related to maternal FFA. More importantly, specific genes — include EP300, CREBBP, and PPARA (all P < 0.01, discussed above) — related to mitochondrial biogenesis were paradoxically downregulated only in the OB group in relation to FFA. There was differential regulation of mitochondrial fusion/fission genes (MFN2 and PARL) and mitochondrial mitophagy genes (NEDD4, SUMO1, TRIAP1, ATG2B, ATG9A, TECPR1, CLEC16A, and ULK2), primarily in the OB group, which directionally favors mitochondrial fission and inhibited mitophagy.
Figure 6. Mitochondrial gene expression vs.
maternal free fatty acids in the second trimester in uMSC adipocytes. Green, upregulated; red, downregulated; all P < 0.05; bold, FDR < 0.25 in targeted gene set analysis. Differential expression in obese (OB, n = 7) or normal weight (NW, n = 7) cohorts designated where applicable. Analysis was performed using linear regression. OMM, outer mitochondrial membrane; IMM, inner mitochondrial membrane; CoQ, coenzyme Q; CytC, cytochrome C)
Discussion
Our results demonstrate, for the first time to our knowledge, differences in the metabolomics and transcriptomics of stem cells in human infants born to OB women and their correlation with maternal phenotypes and neonatal adiposity. Our overall initial hypothesis that maternal BMI is an important predictor of changes in the uMSC was found to be only partially correct. Percent fat of the infant, which is the summation of all exposures throughout gestation, was a better correlate of differences in metabolism and gene expression, particularly in the offspring of OB mothers. Correlative analyses indicated clear associations in the OB group in relation to neonatal %FM and FFA exposure, suggesting the OB MSCs and NW MSCs were metabolically different. Although MSC were selected based on maternal prepregnancy BMI, there was great heterogeneic overlap in adiposity and maternal FFA concentrations between the OB and NW groups. We therefore tested relationships using both groups combined (when the phenotypic variable was continuous) and also within OB offspring specifically, as this is the group with the highest risk for metabolic derangements as children (16).
In uMSC myocytes, there was evidence for incomplete β-oxidation in relation to infant adiposity, specifically in offspring of OB but not NW mothers. Incomplete β-oxidation has recently become a topic of interest in obesity and insulin resistance, and it is central to energy metabolism in established obesity (13, 14, 20, 32). In the setting of increased lipid stress, long-chain fatty acid oxidative enzymes like VLCAD and LCHAD upregulate to meet the demand of increased fatty acid substrate (32). However, as is the case in established obesity, these steps in long-chain fatty acid metabolism cannot be upregulated enough to couple the increased substrate load with oxidative phosphorylation, resulting in a buildup of fatty acyl-CoA in the mitochondria. To preserve the CoA pool within cells, excess fatty acyl-CoAs are converted to fatty acyl-carnitines and preferentially transported out of the mitochondria, accumulating in the cytoplasm and extracellular space (33), which results in accumulation of LCAC species detectable in tissue and plasma. Here, the same markers (multiple long-chain and long-chain-hydroxy acylcarnitines) were positively correlated with neonatal adiposity, while targeted gene expression was only partially upregulated.
Biomarkers of compensatory processes associated with incomplete β-oxidation were also present in uMSC myocytes, related to neonatal adiposity in offspring of OB mothers. ω-Oxidation is characterized by elevations in DCAC and is a known compensatory process in the setting of β-oxidative dysfunction (34). In obesity and diabetes, increased ω-oxidation has been reported in the setting of dysfunctional β-oxidation in OB mice and rats (35, 36), and it has been demonstrated (as altered dicarboxylic metabolites) in limited human studies (37–39). Alterations in amino acid pools in the setting of incomplete β-oxidation may be found related to anaplerosis, a compensatory process of amino acid mobilization and oxidation to replenish the TCA cycle and maintain oxidative metabolism. Amino acid anaplerosis is found in established obesity (40). Glutaminolytic amino acids are not only important in anaplerosis, but also in biosynthetic processes (as we found enriched in our global uMSC myocyte gene expression analysis).
Our findings, taken together, suggest that incomplete β-oxidation and compensatory oxidative metabolism of fats (DCAC), and possibly amino acids, in the uMSC myoctes are associated with increased adiposity in infants born to OB mothers. This is supported by targeted and broad analysis of gene expression, with enrichment for multiple metabolic and biosynthetic processes (including oxidative metabolism of lipid, amino acids, and the TCA cycle) related to maternal obesity. While biomarkers for these processes have been previously associated with established obesity, we are the first to our knowledge to demonstrate associations in stem cells of newborn infants of OB mothers with increasing adiposity far removed from the maternal in utero environment. Our results in infant MSC-myocytes are similar to findings in fetal skeletal muscle of NHP exposed to maternal obesity, even in the absence of a Western-style diet (41), suggesting these cells may maintain a phenotype as differentiated skeletal muscle in vivo. Were these tissue-specific changes carried forward in the tissue of the child, it could favor lipid storage over oxidation upon exposure to an obesogenic environment. This would require validation, however, using muscle tissue from these same offspring, or umbilical cords from the same NHP — neither of which are available.
uMSC adipocytes did not demonstrate the same acylcarnitines or amino acid correlations found in uMSC myocytes, specifically in relation to either neonatal fat mass or maternal obesity. Differential gene expression between cells from offspring of OB vs. NW mothers revealed downregulation of nutrient-sensing pathways. AMPK, MAPK, and PI3K-AKT signaling are thought to be suppressed in established obesity (42–44). These pathways, related to insulin signaling, were downregulated in relation to maternal obesity, higher maternal FFA, and higher neonatal adiposity. This suggests a relatedness between these phenotypic variables based on uMSC physiology, but also implicates that cells have been programmed for less insulin-associated nutrient sensing in the face of maternal obesity exposures in utero.
Further, there was evidence in the uMSC adipocytes that both maternal FFA and neonatal adiposity were related to changes in oxidative phosphorylation gene expression. Specifically, maternal FFA exposure was correlated with upregulation of mitochondrial functional genes (e.g., electron transport and ribosomal machinery) but with downregulation of mitochondrial biogenesis genes including EP300, CREBBP, and PPARA. Similar patterns of upregulation in components of the electron transport chain and a downregulation of biogenesis have been reported in our NHP model in fetal offspring of OB, Western style diet–fed mothers (41). Functional effects on mitochondrial health have also been noted in fetal tissue with maternal obesity and high-fat diet exposure in rats (45). Similar alterations in mitochondrial function and quality control, including affected MFN2 protein expression, have been demonstrated across multiple generations of OB mice with high maternal fat exposure (46). The mitochondrial quality control gene ULK2 (downregulated in the OB group only) is a downstream regulator of MTORC1 (47) and of AMPK (48) via phosphorylation. Whether changes in gene expression found in this study correspond with functional changes in mitochondrial biogenesis or are related to findings of downregulated insulin–related signaling should be further investigated.
Previous research has demonstrated that obesity-prone animals exhibited increased energy efficiency and lipogenic capacity in adipose tissue and a decrease in lipolytic enzymes in muscle, resulting in greater weight gain and adiposity (49). While it is not possible to demonstrate this directly in humans generationally over time, we have shown — using a NHP model of maternal obesity and high-fat diet programming — that offspring born to an OB mother weaned to a healthy diet show permanent changes in liver metabolism, brain function, and even the microbiome differences that are maintained postnatally due to the initial exposure to maternal obesity and high-fat diet (50–52). With the combined data presented here, in both uMSC myocytes and uMSC adipocytes, this is the first evidence to our knowledge in a human fetal/neonatal cell model that maternal phenotype and underlying cellular physiology may play a role in offspring adiposity.
This study was limited by total number of subjects. However, our P values were generally robust, and patterns of analyte and gene expression were consistent within themselves such that interpretation of results was based on multiple analytes and/or genes within any given pathway. Findings in our cohort vs. the larger Healthy Start Cohort, especially in relationships of maternal BMI and FFA to infant adiposity, may reflect the intentional selection bias of choosing an equal number of OB and NW mothers vs. the ~20% of OB mothers in the larger Healthy Start Study (53). This does not negate the possible biological relevance but warrants caution when applying findings to a larger, more general population. Similarly, analyzing continuous variables like FFA across the entire group may be influenced by initial selection by maternal BMI; however, with so much overlap in FFA concentrations between the 2 groups, and no cross-sectional differences found between NW and OB cohorts, selection bias is less likely to influence correlative results. This analysis is limited to metabolomic and transcriptional data. Functional studies including enzyme assay or pathway flux analyses are necessary to further clarify mechanism. Further, correlation is not causation, so we interpret findings here carefully as a next step in understanding the underpinnings of fetal programming in obesity, since all cells at birth can be further programmed as they are subject to multiple nutritional and environmental exposures postnatally.
This work fits with the classical overnutrition model, in which the infant born to an OB mother is predisposed to obesity from the beginning of life, and the trigger for developing an OB phenotype is the postnatal obesogenic environment. Although several hypotheses have been proposed, including physiological signaling and responsiveness, genetic differences, and metabolic processes, the exact mechanisms underlying interindividual susceptibility to obesity remain largely unknown. We postulate that some of this predisposition may be due to epigenetic influences affecting gene expression, energy sensing, and fatty acid/amino acid metabolism that remain to be investigated.
Methods
Population. The Healthy Start Study is a longitudinal observational prebirth cohort of 1,410 mother-infant pairs. Pregnant women were recruited at less than 24 weeks gestation through the University of Colorado obstetrics clinics during 2010–2014. Exclusion criteria included preexisting type 1 or type 2 diabetes, a prior premature birth or fetal death, asthma with steroid management, serious psychiatric illness, or a current multiple pregnancy.
Mother and infant were followed with data collected including maternal ppBMI (obtained through medical record review) and serum samples for FFA, triglycerides, glucose, and insulin at 24–32 weeks gestation. Neonatal body composition (determined by serial air displacement plethysmography) was measured within 48 hours after delivery, and %FM was calculated by dividing FM (g) by the total body mass. Additionally, on a convenience sample, umbilical cord tissue was collected at birth, and uMSC were cultured and stored for later use (n = 164 total).
Data used in this report include 24 uMSC samples from offspring of prepregnancy OB (n = 12, BMI > 30 kg/m2) and NW (n = 12, BMI < 25 kg/m2) mothers matched for age, gestational age at delivery, and time to cell culture confluence. No mother had clinical evidence of insulin resistance, and none had preeclampsia. Maternal and infant phenotypes in the larger population have been previously described (24, 26).
uMSC growth and conditions. Our methods in growth and differentiation of uMSC have been described previously for myocytes and adipocytes (27). Undifferentiated uMSC were grown to confluence and then exposed to lipid enriched (oleate [C18:1, 133 μM], palmitate [C16:0, 67 μM], and carnitine [1 mM]) myocyte and adipocyte differentiation media. Spent media (for metabolite) and cell lysate (for RNA) were harvested on day 21 of differentiation. For further details, please see Supplemental Experimental Procedures.
Media metabolomic analysis. In media, amino acid analysis was accomplished using the Biochrom 30+ Amino Acid Analyzer through established protocols and an experienced clinical biochemical laboratory (Children’s Hospital Colorado, Aurora, Colorado). Acylcarnitine analysis was accomplished using a modified electrospray Ionization (ESI) MS/MS protocol (54). Analytes were placed in the context of known amino acid and lipid metabolism pathways in KEGG (see Supplemental Experimental Procedures for pathway details).
RNA-Seq analysis. In cell lysate, high throughput RNA-Seq analysis was performed using an Illumina High Seq 2000. Data was normalized and expression was quantified in fragments per kilobase of transcript per million mapped reads (FPKM) using Cufflinks. Data is deposited to dbGaP (accession number ***). For further details regarding harvest, mapping, and quality control, please see Supplemental Experimental Procedures. Genes from the above KEGG pathways, as well as mitochondrial biogenesis (KO03029, BRITE hierarchy gene set [see http://www.genome.jp/kegg/brite.html]), AMPK signaling (map04152), and insulin signaling (map04910) pathways were culled to form the hand-curated transcriptomic dataset, based on our hypothesis of deranged energy sensing and metabolism, and results from whole RNA-Seq dataset STRING analysis.
Statistics. Analysis workflow is illustrated in Supplemental Figures 1 and 2. Quantitative amino acid, acylcarnitine, and gene-specific RNA-Seq data were analyzed using GraphPad 6.0 software with multiple univariate Pearson correlation model for each analyte and phenotypic outcome if they were continuous (including %FM and FFA). Two-tailed student t test was used the categorical variable of maternal BMI in particular, due to the intentional selection of mothers with a BMI above 30 kg/m2 or below 25 kg/m2. Analyte correlation P values and correlation coefficients were mapped to known pathway contexts as described above. FDR was determined using the Benjamini-Hochberg procedure (55), grouping analytes relative to known pathways or single enzyme complexes when possible. Significance in statistical analysis was given to nominal P values of ≤ 0.05 with FDR ≤ 0.05. Given the potential novelty of this model and how little is known about the effects of maternal obesity in these cells, correlations with significant nominal P values and FDR between 0.05 and 0.25 were considered of marginal significance and potentially useful for hypothesis generation. STRING Database 10.0 (30) and GSEA (31) were used to analyze the categorical variable of maternal BMI and other continuous phenotype (respectively) vs. gene expression variables. For further detail, please refer to Supplemental Experimental Procedures.
Study approval. All mothers in the Healthy Start Study provided written informed consent, and the study was approved by the Colorado Multiple IRB.
Author Contributions
PRB was responsible for study conceptualization, funding acquisition, investigation, visualization, original draft. ZP, ALBS, and BADLH participated in cell culture, review, and editing. MW was responsible for data analysis and methodology. KEB contributed to methodology, review, and editing. LV assisted in data analysis, review, and editing. DD provided resources and contributed to review and editing. JEF was responsible for conceptualization, supervision, resources, review, and editing.
Supplementary Material
Acknowledgments
The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. PB was supported by the NICHD and University of Colorado Department of Pediatrics Child Health Research Career Development Award K12-HD068372 (principal investigator [PI]: PB), Children’s Hospital Colorado Research Institute Research Scholar Award (PI: PB), Nutritional and Obesity Research Center (NORC) Pilot and Feasibility grant P30DK048520 (PI: PB), and Colorado Clinical and Translational Science Institute (CCTSI) pilot grant (PI: PB); the Obesity Society (KEB) and the Building Interdisciplinary Research Careers in Women’s Health (BIRCWH; NICHD [PI: KEB]); the Healthy Start Baby BUMP Project is supported by grants from the American Heart Association (Predoctoral Fellowship 14PRE18230008, PI: AS) and the Colorado Nutrition and Obesity Research Center (DK048520, PI: JEF); and by the parent Healthy Start study (PI: DD). The Healthy Start study is supported by the National Institute of Diabetes and Digestive and Kidney Disease (DK076648, PI: DD) and NIH/NCATS Colorado CTSA grant UL1 TR001082. We also would like to thank the Colorado Biostatistics Consortium and the Children’s Hospital Colorado Clinical Biochemical Lab.
Version 1. 11/02/2017
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2017;2(21):e94200. https://doi.org/10.1172/jci.insight.94200.
Contributor Information
Zachary Patinkin, Email: zach.patinkin@gmail.com.
Michael Woontner, Email: michael.woontner@childrenscolorado.org.
Lauren Vanderlinden, Email: LAUREN.VANDERLINDEN@ucdenver.edu.
Dana Dabelea, Email: dana.dabelea@ucdenver.edu.
Jacob E. Friedman, Email: jed.friedman@ucdenver.edu.
References
- 1.Dabelea D, et al. Intrauterine exposure to diabetes conveys risks for type 2 diabetes and obesity: a study of discordant sibships. Diabetes. 2000;49(12):2208–2211. doi: 10.2337/diabetes.49.12.2208. [DOI] [PubMed] [Google Scholar]
- 2.Friedman JE. Obesity and Gestational Diabetes Mellitus Pathways for Programming in Mouse, Monkey, and Man—Where Do We Go Next? The 2014 Norbert Freinkel Award Lecture. Diabetes Care. 2015;38(8):1402–1411. doi: 10.2337/dc15-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Derraik JG, Ahlsson F, Diderholm B, Lundgren M. Obesity rates in two generations of Swedish women entering pregnancy, and associated obesity risk among adult daughters. Sci Rep. 2015;5:16692. doi: 10.1038/srep16692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ogden CL, Carroll MD, Kit BK, Flegal KM. Prevalence of childhood and adult obesity in the United States, 2011-2012. JAMA. 2014;311(8):806–814. doi: 10.1001/jama.2014.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isganaitis E, et al. Developmental programming by maternal insulin resistance: hyperinsulinemia, glucose intolerance, and dysregulated lipid metabolism in male offspring of insulin-resistant mice. Diabetes. 2014;63(2):688–700. doi: 10.2337/db13-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isganaitis E, et al. Associations of cord blood metabolites with early childhood obesity risk. Int J Obes (Lond) 2015;39(7):1041–1048. doi: 10.1038/ijo.2015.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jo CH, et al. Fetal mesenchymal stem cells derived from human umbilical cord sustain primitive characteristics during extensive expansion. Cell Tissue Res. 2008;334(3):423–433. doi: 10.1007/s00441-008-0696-3. [DOI] [PubMed] [Google Scholar]
- 8.Kim J, et al. Umbilical cord mesenchymal stromal cells affected by gestational diabetes mellitus display premature aging and mitochondrial dysfunction. Stem Cells Dev. 2015;24(5):575–586. doi: 10.1089/scd.2014.0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang J, et al. Dynamic Control of Enhancer Repertoires Drives Lineage and Stage-Specific Transcription during Hematopoiesis. Dev Cell. 2016;36(1):9–23. doi: 10.1016/j.devcel.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu JS, Cui W. Proliferation, survival and metabolism: the role of PI3K/AKT/mTOR signalling in pluripotency and cell fate determination. Development. 2016;143(17):3050–3060. doi: 10.1242/dev.137075. [DOI] [PubMed] [Google Scholar]
- 11.Ito K, et al. A PML–PPAR-δ pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat Med. 2012;18(9):1350–1358. doi: 10.1038/nm.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie B, Waters MJ, Schirra HJ. Investigating potential mechanisms of obesity by metabolomics. J Biomed Biotechnol. 2012;2012:805683. doi: 10.1155/2012/805683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newgard CB, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9(4):311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams SH, et al. Plasma acylcarnitine profiles suggest incomplete long-chain fatty acid beta-oxidation and altered tricarboxylic acid cycle activity in type 2 diabetic African-American women. J Nutr. 2009;139(6):1073–1081. doi: 10.3945/jn.108.103754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Owen OE, Kalhan SC, Hanson RW. The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem. 2002;277(34):30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- 16.Perng W, et al. Metabolomic profiles and childhood obesity. Obesity (Silver Spring) 2014;22(12):2570–2578. doi: 10.1002/oby.20901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyle KE, et al. A high-fat diet elicits differential responses in genes coordinating oxidative metabolism in skeletal muscle of lean and obese individuals. J Clin Endocrinol Metab. 2011;96(3):775–781. doi: 10.1210/jc.2010-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker PR, et al. Metabolomic analysis reveals altered skeletal muscle amino acid and fatty acid handling in obese humans. Obesity (Silver Spring) 2015;23(5):981–988. doi: 10.1002/oby.21046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lerin C, et al. Defects in muscle branched-chain amino acid oxidation contribute to impaired lipid metabolism. Mol Metab. 2016;5(10):926–936. doi: 10.1016/j.molmet.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lackey DE, et al. Regulation of adipose branched-chain amino acid catabolism enzyme expression and cross-adipose amino acid flux in human obesity. Am J Physiol Endocrinol Metab. 2013;304(11):E1175–E1187. doi: 10.1152/ajpendo.00630.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.She P, Van Horn C, Reid T, Hutson SM, Cooney RN, Lynch CJ. Obesity-related elevations in plasma leucine are associated with alterations in enzymes involved in branched-chain amino acid metabolism. Am J Physiol Endocrinol Metab. 2007;293(6):E1552–E1563. doi: 10.1152/ajpendo.00134.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wahl S, et al. Childhood obesity is associated with changes in the serum metabolite profile. Obes Facts. 2012;5(5):660–670. doi: 10.1159/000343204. [DOI] [PubMed] [Google Scholar]
- 23.McCormack SE, et al. Circulating branched-chain amino acid concentrations are associated with obesity and future insulin resistance in children and adolescents. Pediatr Obes. 2013;8(1):52–61. doi: 10.1111/j.2047-6310.2012.00087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Starling AP, et al. Associations of maternal BMI and gestational weight gain with neonatal adiposity in the Healthy Start study. Am J Clin Nutr. 2015;101(2):302–309. doi: 10.3945/ajcn.114.094946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Catalano PM, Roman-Drago NM, Amini SB, Sims EA. Longitudinal changes in body composition and energy balance in lean women with normal and abnormal glucose tolerance during pregnancy. Am J Obstet Gynecol. 1998;179(1):156–165. doi: 10.1016/S0002-9378(98)70267-4. [DOI] [PubMed] [Google Scholar]
- 26.Shapiro AL, et al. Testing the fuel-mediated hypothesis: maternal insulin resistance and glucose mediate the association between maternal and neonatal adiposity, the Healthy Start study. Diabetologia. 2015;58(5):937–941. doi: 10.1007/s00125-015-3505-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyle KE, Patinkin ZW, Shapiro AL, Baker PR, Dabelea D, Friedman JE. Mesenchymal Stem Cells From Infants Born to Obese Mothers Exhibit Greater Potential for Adipogenesis: The Healthy Start BabyBUMP Project. Diabetes. 2016;65(3):647–659. doi: 10.2337/db15-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rinaldo P, Cowan TM, Matern D. Acylcarnitine profile analysis. Genet Med. 2008;10(2):151–156. doi: 10.1097/GIM.0b013e3181614289. [DOI] [PubMed] [Google Scholar]
- 29.Mihalik SJ, et al. Increased levels of plasma acylcarnitines in obesity and type 2 diabetes and identification of a marker of glucolipotoxicity. Obesity (Silver Spring) 2010;18(9):1695–1700. doi: 10.1038/oby.2009.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szklarczyk D, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams SH. Emerging perspectives on essential amino acid metabolism in obesity and the insulin-resistant state. Adv Nutr. 2011;2(6):445–456. doi: 10.3945/an.111.000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCoin CS, Knotts TA, Adams SH. Acylcarnitines--old actors auditioning for new roles in metabolic physiology. Nat Rev Endocrinol. 2015;11(10):617–625. doi: 10.1038/nrendo.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wanders RJ, Komen J, Kemp S. Fatty acid omega-oxidation as a rescue pathway for fatty acid oxidation disorders in humans. FEBS J. 2011;278(2):182–194. doi: 10.1111/j.1742-4658.2010.07947.x. [DOI] [PubMed] [Google Scholar]
- 35.Lai RK, Goldman P. Urinary organic acid profiles in obese (ob/ob) mice: indications for the impaired omega-oxidation of fatty acids. Metab Clin Exp. 1992;41(1):97–105. doi: 10.1016/0026-0495(92)90197-I. [DOI] [PubMed] [Google Scholar]
- 36.Fiamoncini J, et al. Medium-chain dicarboxylic acylcarnitines as markers of n-3 PUFA-induced peroxisomal oxidation of fatty acids. Mol Nutr Food Res. 2015;59(8):1573–1583. doi: 10.1002/mnfr.201400743. [DOI] [PubMed] [Google Scholar]
- 37.Lippe G, Trevisan R, Nosadini R, Fabris R, Deana R. 3-Hydroxy-3-methylglutaric, adipic, and 2-oxoglutaric acids measured by HPLC in the plasma from diabetic patients. Clin Biochem. 1987;20(4):275–279. doi: 10.1016/S0009-9120(87)80012-7. [DOI] [PubMed] [Google Scholar]
- 38.Butte NF, et al. Global metabolomic profiling targeting childhood obesity in the Hispanic population. Am J Clin Nutr. 2015;102(2):256–267. doi: 10.3945/ajcn.115.111872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villarreal-Pérez JZ, et al. Plasma and urine metabolic profiles are reflective of altered beta-oxidation in non-diabetic obese subjects and patients with type 2 diabetes mellitus. Diabetol Metab Syndr. 2014;6:129. doi: 10.1186/1758-5996-6-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newgard CB. Metabolomics and Metabolic Diseases: Where Do We Stand? Cell Metab. 2017;25(1):43–56. doi: 10.1016/j.cmet.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCurdy CE, et al. Maternal obesity reduces oxidative capacity in fetal skeletal muscle of Japanese macaques. JCI Insight. 2016;1(16):e86612. doi: 10.1172/jci.insight.86612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu MJ, et al. AMP-activated protein kinase signalling pathways are down regulated and skeletal muscle development impaired in fetuses of obese, over-nourished sheep. J Physiol (Lond) 2008;586(10):2651–2664. doi: 10.1113/jphysiol.2007.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, et al. AMPK activation prevents excess nutrient-induced hepatic lipid accumulation by inhibiting mTORC1 signaling and endoplasmic reticulum stress response. Biochim Biophys Acta. 2014;1842(9):1844–1854. doi: 10.1016/j.bbadis.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCurdy CE, et al. Attenuated Pik3r1 expression prevents insulin resistance and adipose tissue macrophage accumulation in diet-induced obese mice. Diabetes. 2012;61(10):2495–2505. doi: 10.2337/db11-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mdaki KS, et al. Maternal high-fat diet impairs cardiac function in offspring of diabetic pregnancy through metabolic stress and mitochondrial dysfunction. Am J Physiol Heart Circ Physiol. 2016;310(6):H681–H692. doi: 10.1152/ajpheart.00795.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saben JL, et al. Maternal Metabolic Syndrome Programs Mitochondrial Dysfunction via Germline Changes across Three Generations. Cell Rep. 2016;16(1):1–8. doi: 10.1016/j.celrep.2016.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung CH, Seo M, Otto NM, Kim DH. ULK1 inhibits the kinase activity of mTORC1 and cell proliferation. Autophagy. 2011;7(10):1212–1221. doi: 10.4161/auto.7.10.16660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alers S, Löffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol. 2012;32(1):2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gayles EC, Pagliassotti MJ, Prach PA, Koppenhafer TA, Hill JO. Contribution of energy intake and tissue enzymatic profile to body weight gain in high-fat-fed rats. Am J Physiol. 1997;272(1 Pt 2):R188–R194. doi: 10.1152/ajpregu.1997.272.1.R188. [DOI] [PubMed] [Google Scholar]
- 50.Thorn SR, et al. Early life exposure to maternal insulin resistance has persistent effects on hepatic NAFLD in juvenile nonhuman primates. Diabetes. 2014;63(8):2702–2713. doi: 10.2337/db14-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sullivan EL, Smith MS, Grove KL. Perinatal exposure to high-fat diet programs energy balance, metabolism and behavior in adulthood. Neuroendocrinology. 2011;93(1):1–8. doi: 10.1159/000322038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma J, et al. High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model. Nat Commun. 2014;5:3889. doi: 10.1038/ncomms4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shapiro AL, et al. Maternal diet quality in pregnancy and neonatal adiposity: the Healthy Start Study. Int J Obes (Lond) 2016;40(7):1056–1062. doi: 10.1038/ijo.2016.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Blau N, Duran M, Gibson KM, eds. Laboratory guide to the methods in biochemical genetics. Berlin: Springer; 2008. [Google Scholar]
- 55.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9(7):811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.