Abstract
Lungs allografts have worse long-term survival compared with other organ transplants. This is most likely due to their unique immunoregulation that may not respond to traditional immunosuppression. For example, local NO generation by inducible NOS (iNOS) is critical for lung allograft acceptance but associates with rejection of other solid organs. The source of NO in accepting lung allografts remains unknown. Here, we report that, unlike the case for other pulmonary processes in which myeloid cells control NO generation, recipient-derived eosinophils play a critical and nonredundant role in iNOS-mediated lung allograft acceptance. Depletion of eosinophils reduces NO levels to that of recipients with global deletion of iNOS and leads to a costimulatory blockade–resistant form of rejection. Furthermore, NO production by eosinophils depends on Th1 polarization by inflammatory mediators, such as IFN-γ and TNF-α. Neutralization of such mediators abrogates eosinophil suppressive capacity. Our data point to what we believe to be a unique and previously unrecognized role of eosinophil polarization in mediating allograft tolerance and put into perspective the use of high-dose eosinophil-ablating corticosteroids after lung transplantation.
Keywords: Immunology, Transplantation
Keywords: Adaptive immunity, Respiration, Th1 response
Eosinophils are rare granulocytes with lytic capacity shown here to play a crucial role in lung graft tolerance.
Introduction
Recent work has suggested that myeloid cells may play an important role in mediating allospecific tolerance. DC-SIGN+ macrophages, for example, are necessary for heart allograft acceptance (1). Bone marrow–derived CD11b+CD115+Gr1+ monocytes, a type of myeloid-derived suppressor cells, similarly have the capacity to downregulate the adaptive immune response during tolerance induction in the heart (2). A group of DCs, described as tolerogenic DCs, can potentiate solid organ acceptance in other models as well (3). The presence of granulocytic cells, however, is generally considered deleterious for successful organ engraftment. Neutrophilia associated with ischemia/reperfusion injury has been associated with costimulatory blockade–resistant (CSB-resistant) lung allograft rejection (4), and eosinophils can contribute to rejection of multiple solid organs (5, 6). Furthermore, eosinophils are suggested to promote damaging inflammation in many other disease processes, specifically in mucosal barrier organs (7, 8).
The lung allograft is unique in its immune response after transplantation. For example, while the local production of NO correlates with rejection of kidney (9) and heart allografts (10), recent work from our laboratories has demonstrated that NO plays a critical role in amelioration of lung allograft pathology (11). Specifically, the expression of recipient-derived type II NOS, or inducible NOS (iNOS), is a critical factor for CSB-mediated lung allograft acceptance (11). Myeloid cells have the capacity to express iNOS and were thus suspected as contributing to iNOS-mediated lung allograft acceptance (12). Here, we describe the surprising observation that acceptance of allogeneic lung allografts depends on the transient presence of recipient-derived iNOS+ eosinophils. The generation of iNOS+ eosinophils is facilitated by cytokine patterns that are expressed locally within the lung during tolerance induction. Thus, our work uncovers a potentially unrecognized role for eosinophils as a mechanistic link between inflammation associated with early graft responses and tolerance. Our data further extend the notion that the immunologic environment associated with lung allograft acceptance is unique and challenge current immunosuppressive strategies for lung transplant recipients.
Results
Tolerance is associated with an influx of recipient-derived iNOS+ cells into the lung allografts.
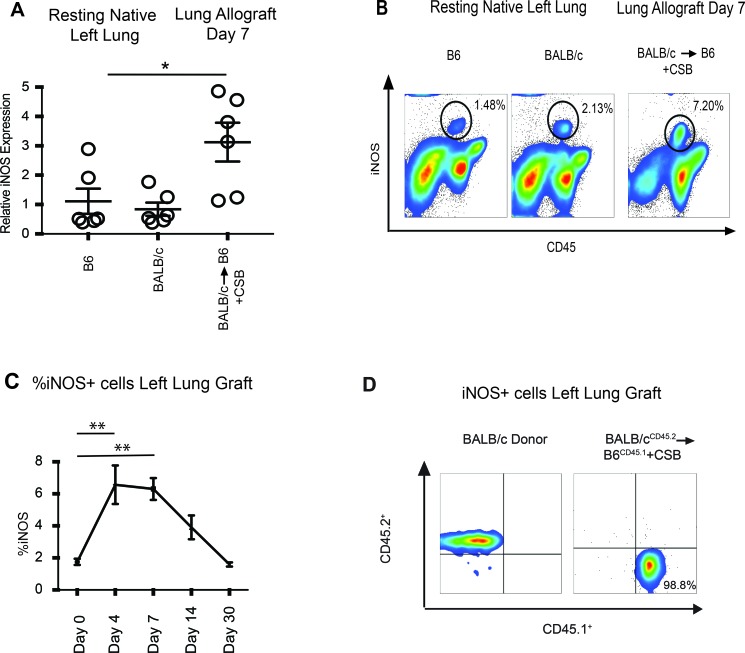
Consistent with our previous reports that recipient iNOS expression is critical for promoting lung allograft acceptance (11), we detected a relative increase in iNOS transcripts in CSB-treated lung allografts compared with resting lungs (Figure 1A). Flow cytometric analysis demonstrated a relative increase in iNOS+ hematopoietic cells within the graft (Figure 1B). We observed an early rise in this cell population, peaking on days 4–7 after transplantation, which returned to baseline by day 30 after engraftment (Figure 1C). Transplantation of CD45.2+ BALB/c lungs into CD45.1+ congenic C57BL/6 recipients (B6CD45.1) confirmed that the iNOS+ cells were exclusively of recipient origin (Figure 1D).
Figure 1. Accepting lung allografts are infiltrated by recipient-derived iNOS+ cells.
(A) Quantitative RT-PCR revealed a significant increase in iNOS transcript levels 7 days after transplantation of BALB/c lungs to B6 recipients with CSB immunosuppression. (B) Flow cytometric analysis demonstrated that lung allografts are infiltrated by iNOS+ hematopoietic cells (representative of 8 separate transplants). (C) Time-course analysis demonstrated a gradual increase in iNOS+ cells, which decrease to baseline levels 1 month after transplantation (representative of at least 4 transplants per time point). (D) Transplantation of a BALB/cCD45.2+ lung grafts to a B6CD45.1+ recipient demonstrated that all iNOS+ graft-infiltrating cells are of recipient origin (representative of 3 separate transplants). *P <.05, **P < 0.01. Comparison by Friedman’s test was used for paired data, while the Kruskal-Wallis test was used for the unpaired group of observations. Post-hoc analysis of differences and comparison of differences between pairs of data were performed with the Wilcoxon rank test and the Mann-Whitney U test for paired and unpaired observations, respectively.
Eosinophils are the dominant iNOS-expressing cells in the lung allograft.
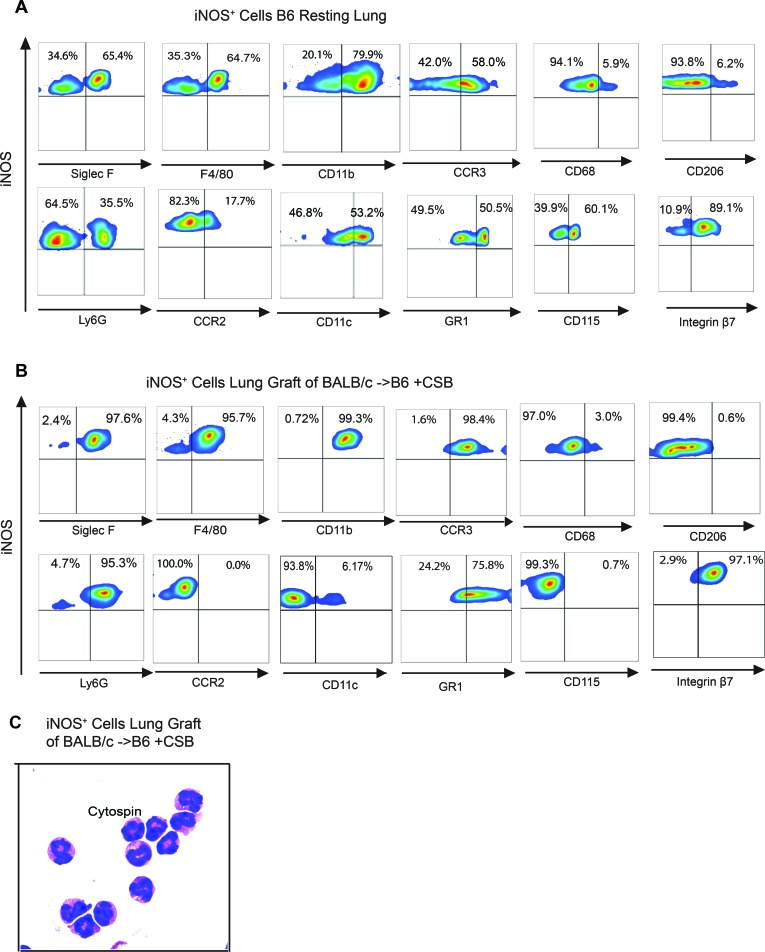
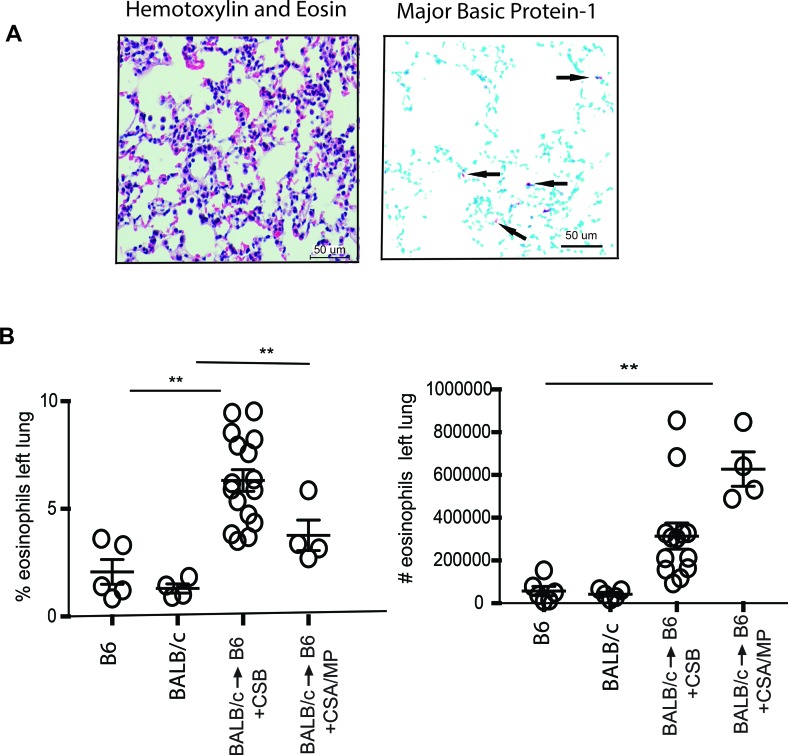
We next set out to identify the iNOS+ lung-resident cells after transplantation. In resting murine lungs, iNOS was detectable in a heterogeneous cell population containing a mixture of myeloid and granulocytic cells (Figure 2A). Within 1 week of transplantation, however, iNOS+ cells in the lung were a homogeneous population of SiglecF+F4/80+CD11b+CD11c–CCR3+CCR2–CD68–CD115–GR1+ cells, consistent with an eosinophil phenotype (Figure 2B) (13, 14). Romanowsky staining of flow cytometrically sorted cells demonstrated an intense eosinophilic cytoplasmic staining, with a characteristic eosinophil-defining ring-shaped nucleus (Figure 2C). Eosinophils from both resting lungs and accepting lung allografts (CSB-treated) expressed very low levels of CD62L, while neither of the two expressed CD101, indicating a possible regulatory capacity of the eosinophil from both models (15) (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.96455DS1). Furthermore, the accepting lung eosinophils expressed high levels of CXCL13 transcript without any significant expression of IL-27 and, therefore, might differ from the Gr-1hi eosinophils that are associated with increased CXCL13+ and IL-27 transcripts (Supplemental Figure 1B) (13). We next examined lung allograft by both histology and eosinophil major basic protein-1–based immunohistochemistry and determined that eosinophils were located primarily within the alveolar space as well as alveolar septa (Figure 3A). Quantitative analysis of accepting left lung allografts revealed an increase in both the relative percentage and total number of lung-infiltrating eosinophils over that in resting lungs (Figure 3B). Such eosinophil influx was evident, with graft acceptance induced by CSB as well as cyclosporine (CsA) and low-dose methylprednisolone (MP) (16) (Figure 3B). Thus, despite the prevailing notion that eosinophils promote deleterious inflammatory responses in the lung, eosinophils represent the dominant iNOS-producing cells in the lung allograft, and pulmonary eosinophilia is associated with allograft acceptance.
Figure 2. Phenotype of iNOS+ cells from resting lung and lung allografts.
(A) iNOS+ cells in resting B6 left lung are a heterogeneous population (representative of 7 transplants). (B) iNOS+ cells in BALB/c→B6 lung transplantation with CSB immunosuppression (representative of 7 transplants). (C) Cytospin preparation and Romanowsky staining of iNOS+ cells in accepting lung grafts, demonstrating an eosinophil phenotype. Original magnification, ×400.
Figure 3. Tissue distribution and quantification of eosinophils.
(A) Seven days after transplantation of BALB/c to B6 lung allografts with CSB immunosuppression eosinophils were distributed throughout the periphery of the lung in the alveolar space and septae, as indicated by the arrows. H&E staining and major basic protein-1 immunohistochemistry are shown. Scale bars: μm. (B) Quantitative analysis of eosinophils in resting B6 and BALB/c mice and BALB/c→B6 lung transplantation with CSB or CsA/MP immunosuppression as a percentage and total number of lung-resident cells. **P < 0.01. Comparison by Friedman’s test was used for paired data, while the Kruskal-Wallis test was used for the unpaired group of observations. Post-hoc analysis of differences and comparison of differences between pairs of data were performed with the Wilcoxon rank test and the Mann-Whitney U test for paired and unpaired observations, respectively.
Eosinophils play a critical role in NO production and lung allograft acceptance.
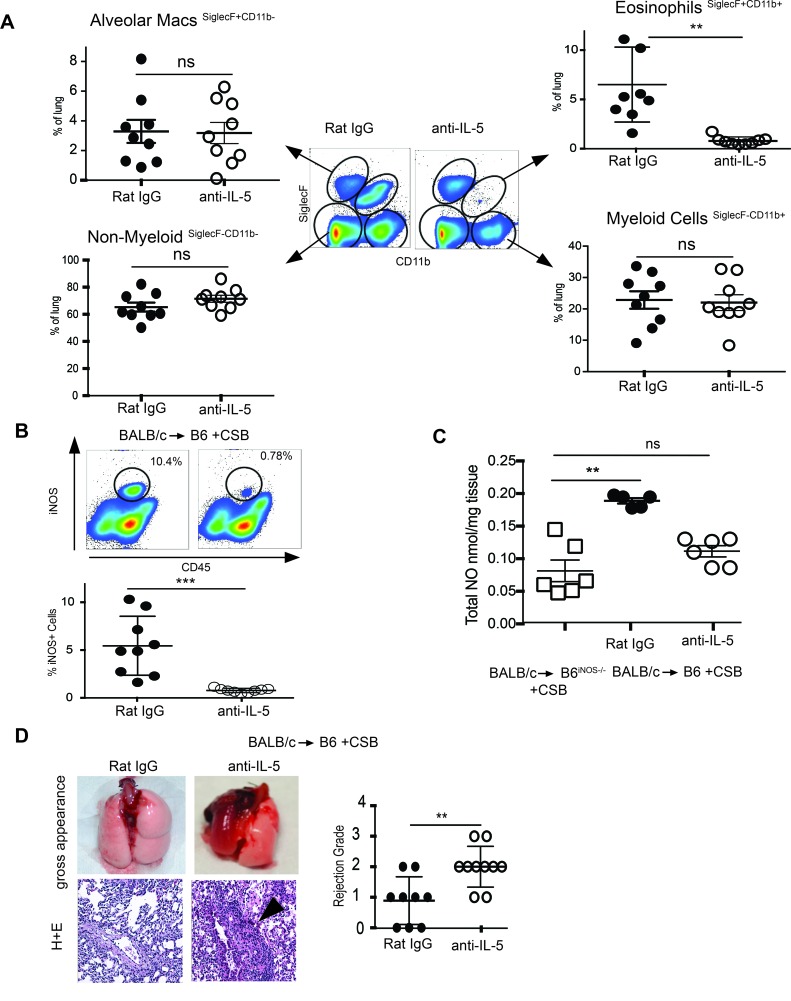
We next set out to examine whether eosinophils are necessary for tolerance induction after lung transplantation. IL-5 is the critical factor for eosinophil generation, migration, maintenance, and survival (17). Neutralization of this cytokine is a commonly accepted method for global eosinophil ablation in experimental murine models (15, 18) and therapeutic intervention for asthma (19) and hypereosinophilic patients (20). We thus neutralized IL-5 in immunosuppressed B6 recipients of BALB/c lung allografts. This treatment resulted in the near-complete elimination of graft-resident SiglecF+CD11b+ eosinophils but did not alter SiglecF+CD11b– alveolar macrophages, SiglecF–CD11b+ myeloid cells, or SiglecF–CD11b– nonmyeloid cells (Figure 4A). Depletion of eosinophils also led to the near-complete elimination of iNOS+ cells in the lung allograft (Figure 4B) and reduced graft NO production to levels comparable to those of B6iNOS–/– recipients (Figure 4C). Most importantly, lung allografts were acutely rejected after IL-5 neutralization despite CSB (Figure 4D). These data demonstrate that eosinophils are the dominant NO-producing cells in the lung after transplantation, and, in their absence, other cells are not able to produce sufficient NO to promote tolerance. To evaluate these results in an independent model of eosinophil deficiency, we transplanted B6 lung allografts to either wild-type BALB/c or ΔdblGATA eosinophil mice on a BALB/c background with CSB. We noted a significant increase in graft rejection in ΔdblGATA recipient mice, with near-complete graft destruction and severe rejection by day 7 after transplantation compared with BALB/c controls (Supplemental Figure 1C). Taken together with data described above, we can conclude that eosinophils play a crucial role in lung allograft acceptance.
Figure 4. Eosinophil depletion eliminates NO production and potentiates a CSB-resistant form of lung allograft rejection.
(A) Recipient IL-5 neutralization virtually eliminates all eosinophils in the lung allograft with minimal effect on other cell types (8–9 transplants per group). (B) IL-5 neutralization also eliminates iNOS-expressing graft-resident cells and (C) reduces lung graft NO production to levels statistically comparable to B6iNOS–/– recipients. (D) Depletion of eosinophils results in lung allograft rejection, as measured by gross changes, histologic evidence of inflammation, and ISHLT grade. Original magnification, ×100. ns, P > 0.05; **P < 0.01, ***P < 0.001. Comparison by Friedman’s test was used for paired data, while the Kruskal-Wallis test was used for the unpaired group of observations. Post-hoc analysis of differences and comparison of differences between pairs of data were performed with the Wilcoxon rank test and the Mann-Whitney U test for paired and unpaired observations, respectively.
Accepting lung allografts demonstrate Th1 polarization.
We have previously identified that environmental polarization can alter the phenotype and function of eosinophils in asthma models, which are Th2 (IL-33, GM-CSF, IL-4) cytokine producing (21). In fact, eosinophils can undergo either Th1- or Th2-induced polarization based on environmental cytokine milieu (22). While we and others have previously noted that IFN-γ and TNF-α, Th1-related cytokines, induce iNOS in several experimental models (11, 23, 24), very limited data exist on iNOS induction in eosinophils (25). In fact, some have suggested that Th2-polarizing cytokines, such as IL-4, may induce iNOS in this cell population (26). We thus considered the possibility that the cytokine milieu in the lung allograft may generate iNOS-expressing eosinophils with regulatory potential after transplantation.
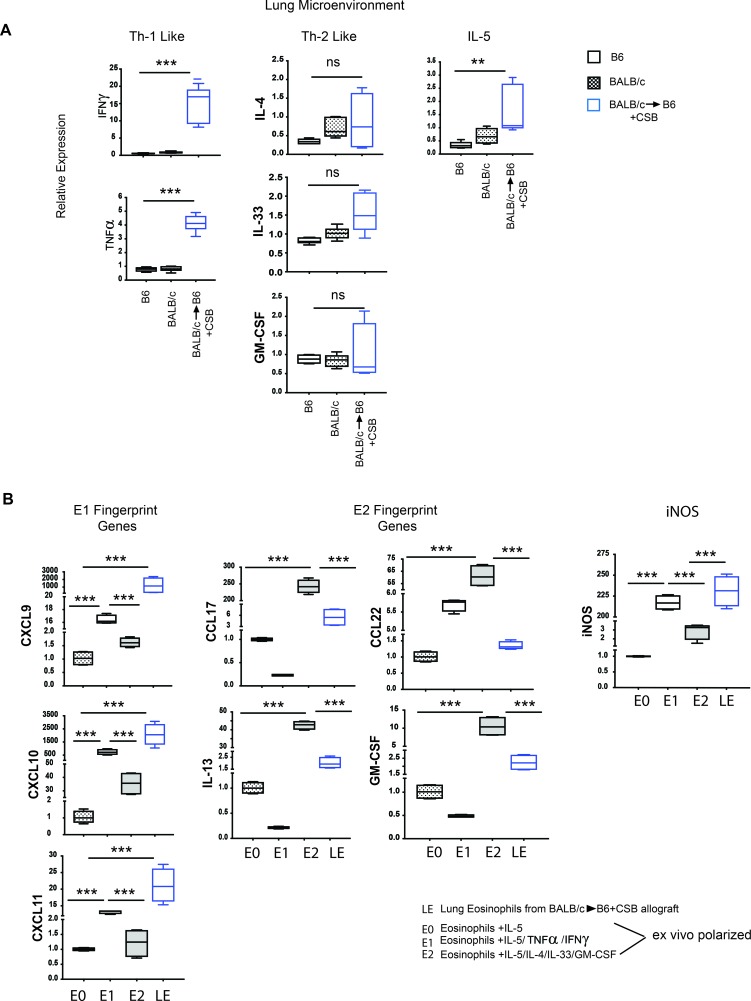
We thus evaluated the cytokine profile of lungs from resting BALB/c, B6, and CSB-treated BALB/c→B6 lung allografts 4 days after engraftment. We noted a significant increase in Th1-like cytokines, such as IFN-γ and TNF-α, as well as the eosinophil survival factor IL-5 in accepting lungs. Th2-specific cytokines, such as IL-4, IL-33, and GM-CSF, did not significantly increase after transplantation (Figure 5A). Next, we flow cytometrically sorted graft-infiltrating eosinophils from CSB-treated BALB/c→B6 lungs and compared their gene expression profiles to peripheral blood-derived eosinophils polarized by overnight exposure to a Th1 (IFN-γ and TNF-α), Th2 (IL-4, IL-33, and GM-CSF) or IL-5 alone (Th0) cytokine milieu, as previously described (21, 22). The gene expression profile of lung graft-infiltrating eosinophils closely resembled the profile of eosinophils that were exposed to IFN-γ and TNF-α (designated as E1), with high levels of CXCL9, CXCL10, and CXCL11 and with lower levels of E2 signature genes, such as CCL17, IL-13, CCL22, and GM-CSF (Figure 5B). Similar to eosinophils polarized toward an E1 phenotype, ex vivo eosinophils isolated from accepting lung grafts expressed high levels of iNOS mRNA (Figure 5B), consistent with their role as the dominant NO-producing cell after transplantation (as demonstrated in Figure 4, B and C).
Figure 5. Lung allograft acceptance is associated with Th1-like polarization.
(A) Gene expression analysis of lungs demonstrated a Th1-like polarization 4 days after transplantation (representative of 6 separate transplants, with BALB/c→B6 allografts outlined in blue, BALB/c resting lungs outlined in black with black dots, and B6 resting lungs outlined in black). (B) Eosinophils flow cytometrically sorted from BALB/c→B6 lung grafts 7 days after transplantation with CSB immunosuppression demonstrated an E1-like gene fingerprint pattern that resembles that of eosinophils exposed to IFN-γ and TNF-α ex vivo (data are from 5 separate transplants). **P < 0.01, ***P < 0.001. Comparison by Friedman’s test was used for paired data, while the Kruskal-Wallis test was used for the unpaired group of observations. Post-hoc analysis of differences and comparison of differences between pairs of data were performed with the Wilcoxon rank test and the Mann-Whitney U test for paired and unpaired observations, respectively. For the box plots, the bottom and top of the boxes represent the lower and upper quartiles, respectively; the dark band inside the box represents the median; and the top and bottom whiskers represent the maximum and minimum observed values, respectively.
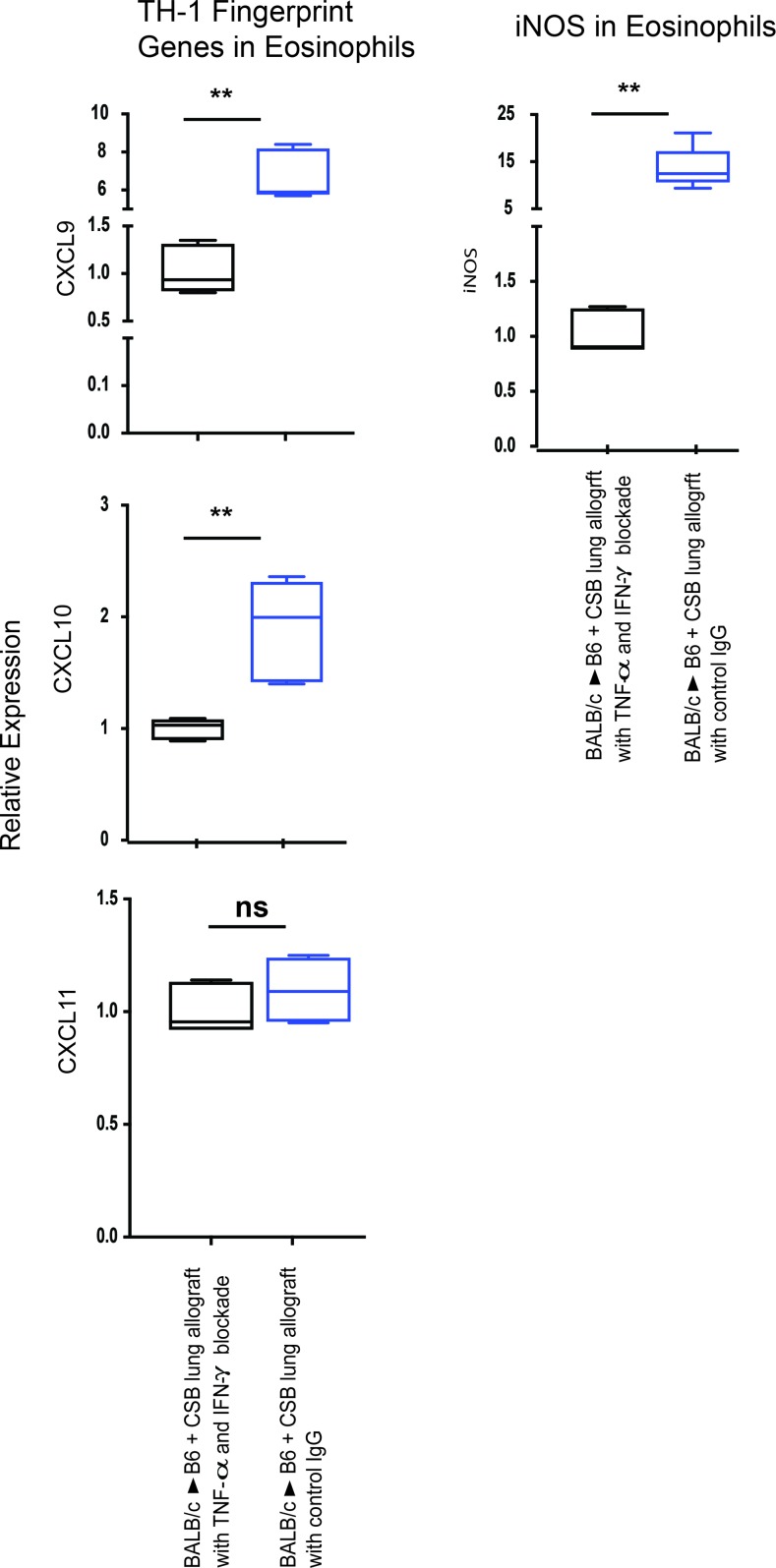
To further delineate whether Th1-polarizing cytokines contribute to iNOS production in eosinophils, we immunophenotyped this cell population from graft recipients in which IFN-γ and TNF-α were neutralized by antibody blockade. Lung-resident eosinophils from grafts where IFN-γ and TNF-α were neutralized produced less iNOS and expressed lower levels of Th1-like transcripts compared with IgG-treated control mice (Figure 6). Furthermore, we have previously described that such Th1 cytokine neutralization results in graft rejection despite immunosuppression (11). Thus, data further strengthened our assumption that E1 polarization contributes to iNOS upregulation in eosinophils and is thus responsible for downregulating alloreactivity in the lung allograft.
Figure 6. Modulation of Th1 signature genes in lung graft eosinophils.
Expression of Th1 signature genes in lung graft eosinophils after IFN-γ and TNF-α blockade. Blue boxes represent eosinophils isolated from BALB/c→B6 transplanted lung grafts with CSB, and black boxes represent eosinophils isolated from BALB/c→B6 transplanted lung grafts with CSB in the presence of IFN-γ and TNF-α neutralization (representative of a minimum of 4 transplants per group). ns, P > 0.05; **P < 0.01. Comparison by Friedman’s test was used for paired data, while the Kruskal-Wallis test was used for the unpaired group of observations. Post-hoc analysis of differences and comparison of differences between pairs of data were performed with the Wilcoxon rank test and the Mann-Whitney U test for paired and unpaired observations, respectively. For the box plots, the bottom and top of the boxes represent the lower and upper quartiles, respectively; the dark band inside the box represents the median; and the top and bottom whiskers represent the maximum and minimum observed values, respectively.
E1 polarization of eosinophils contributes to the downregulation of alloreactivity.
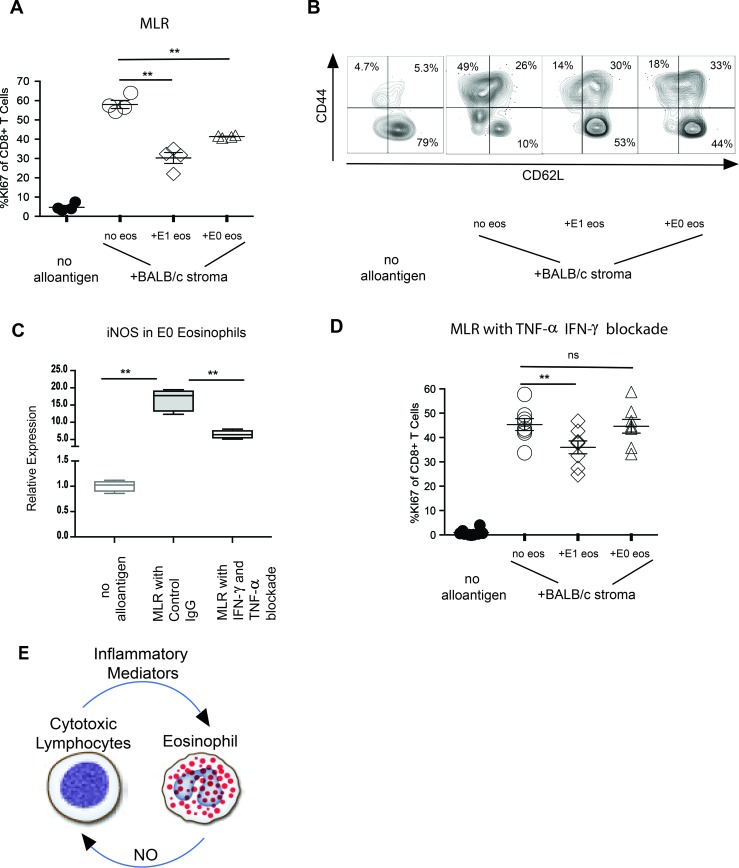
To evaluate this assumption in a more stringent fashion, we used a reductionist approach, relying on mixed lymphocyte reactions (MLRs) of alloreactive T cell activation. We thus purified eosinophils from the peripheral blood of mice, which are of the E0 phenotype (21), and polarized them toward an E1 phenotype by overnight exposure to IFN-γ, TNF-α, and IL-5 or left them in the resting E0 state by exposure to IL-5 alone. Such eosinophils were then added as regulators to MLRs consisting of BALB/cCD45.2+ splenocyte stimulators and B6CD45.1+ congenic CD8+ T cell responders. T lymphocyte proliferation was measured by Ki67 expression and the activation evaluated by surface expression of CD44 and CD62L. While resting T cells showed minimal proliferation and maintained mostly a naive CD62LhiCD44lo phenotype, exposure to BALB/c allogenic antigen-presenting cells resulted in Ki67 upregulation in the majority of the cells and activation to the CD62LloCD44hi effector phenotype in a large portion of CD8+ T cells (Figure 7, A and B). Addition of E1-polarized eosinophils as regulators to the MLR significantly inhibited CD8+ T cell proliferation and differentiation. Surprisingly, E0 eosinophils also suppressed the alloimmune response, albeit at a slightly lower rate than E1 (Figure 7A). We found this somewhat perplexing, as we have demonstrated that NO elaboration directly influences downregulation of alloreactivity (11) and E0 eosinophils express very low levels of iNOS (Figure 5B). We thus considered the possibility that Th1 cytokines produced during the MLRs might alter the polarization of resting eosinophils and upregulate iNOS. To this end, we evaluated iNOS levels in E0 eosinophils prior to, as well as after, their addition to MLRs. Indeed, eosinophils upregulated iNOS after coculture with T cells, which was suggestive of E1 polarization during the MLR. Similar to in vivo studies described above, such polarization was inhibited by IFN-γ and TNF-α blockade (Figure 7C). Consistent with these data, IFN-γ and TNF-α neutralization during the MLR abrogated the suppressive capacity of E0 eosinophils, while eosinophils polarized toward the E1 phenotype prior to the MLR maintained their capacity to suppress CD8+ T cell proliferation (Figure 7D). Taken together, our data suggest that a feedback loop exists between the inflammatory mediators that are produced within the allograft early after implantation and downregulation of alloreactivity through the tolerogenic polarization of graft-resident eosinophils (Figure 7E).
Figure 7. Modulation of CD8+ T cell proliferation and differentiation by E1-polarized eosinophil.
(A) Proliferation, as measured by Ki67 expression among B6 CD45.1+ CD8+ T cells activated with BALB/c allogenic splenocytes, with or without the addition of E1 or E0 eosinophils. Data represent 1 of 2 experiments with 4 separate replicates per experiment. (B) In vitro differentiation, as measured by CD44 and CD62L expression on B6CD45.1+ CD8+ T cells stimulated by BALB/c allogenic splenocytes, with or without the addition of E1 or E0 eosinophils. (C) iNOS expression in CD8+ T cells activated with BALB/c allogenic splenocytes, with or without the addition of IFN-γ– and TNF-α–blocking antibodies (representative experiment of 4 separate MLRs). (D) Modulation of CD8+ T cell proliferation by E0 and E1 eosinophils in MLR of BALB/c allogenic splenocyte stimulators with IFN-γ and TNF-α blockade. (E) Schematic representation of the regulatory feedback loop between alloreactive lymphocytes and eosinophils in lung grafts. ns, P > 0.05; **P < 0.01. Comparison by Friedman’s test was used for paired data, while the Kruskal-Wallis test was used for the unpaired group of observations. Post-hoc analysis of differences and comparison of differences between pairs of data were performed with the Wilcoxon rank test and the Mann-Whitney U test for paired and unpaired observations, respectively. For the box plot, the bottom and top of the boxes represent the lower and upper quartiles, respectively; the dark band inside the box represents the median; and the top and bottom whiskers represent the maximum and minimum observed values, respectively.
Discussion
The role of eosinophils in health and disease is often summarized by pervasive consensus opinions that their activity is primarily destructive and cytocidal in nature (27). Thus, traditional therapeutic strategies have focused on the destruction of this cell population for modulation of inflammatory disease processes. However, the wealth of recently available studies investigating the role(s) of eosinophils in both health and disease demonstrate that the activities of these granulocytes are far more expansive than previously appreciated (14, 17, 28–32). Masterson and colleagues recently demonstrated a protective role of eosinophils in downregulating colonic inflammation through the production of antiinflammatory lipid mediators (33), while Mesnil and colleagues identified a distinct regulatory eosinophil subset with key homeostatic function in the lung (15). Our group has suggested that eosinophils are important regulators of local immunity and/or remodeling/repair in both health and disease i.e., the LIAR hypothesis (34). In this paradigm, eosinophils are neither singularly destructive nor penultimate regulatory. Instead, they mediate activities whose effects are wide in scope. Consistent with this notion, we now demonstrate that eosinophils are participants in the immune responses leading to allograft tolerance.
Unlike most solid organs, the lungs are continuously exposed to the external environment and thus have developed mechanisms to downregulate aberrant inflammation induced by inflammatory lymphocytes (35, 36). Based on data presented here, and those described in previous experimental models (11), we can conclude that such negative feedback loops are critical for tolerance induction in lungs. This notion is supported by experimental models demonstrating that depletion of CD8+ T cells, and neutralization of Th1-type cytokines, actually prevents rather than facilitates lung allograft tolerance (11, 37). We now demonstrate that the E1-polarized eosinophil is a critical mediator in the feedback loop downregulating graft-deleterious inflammatory responses.
Unlike the case for T lymphocytes and macrophages, the functional significance of environmental polarization of eosinophils is poorly described. The finding that the Th1-like pulmonary environment and E1 polarization of eosinophils play a critical role in lung allograft tolerance is further surprising based on the association of Th2 polarization with CSB-induced tolerance in heart and kidney allografts (38, 39). Such differences further point to the unique immunologic environment of the lung. Nevertheless, these data help explain the critical role of undifferentiated CD44hiCCR7hiCD62Lhi central memory T cells in lung allograft tolerance (11). Unlike CD44hiCCR7loCD62Llo effector CD8+ T cells, which are able to destroy cellular targets via direct cytotoxicity, central memory CD8+ T cells elaborate proinflammatory cytokines but fail to mediate direct cytotoxicity. Thus, this cell population can initiate the tolerogenic eosinophil-iNOS–mediated feedback loop described above but is unable to directly destroy allogeneic grafts.
While limited data exist regarding NO production by eosinophils, this cell population has been demonstrated to contribute to asthma-related protein nitration (40, 41). Along those lines, our data demonstrate that the unique NO dependence of the lung allograft for tolerance induction takes advantage of this iNOS-dependent aspect of eosinophil biology. It is important to point out that local NO production may be important during the initial phases of tolerance induction but may not be as critical for long-term maintenance. This is evidenced by the gradual reduction of iNOS-expressing cells in the accepted graft (Figure 1C), despite the fact that local immunoregulatory circuits facilitate acceptance upon retransplantation into a secondary, nonimmunosuppressed host (42). It is possible that eosinophil-dependent NO production is important in the early perioperative period, while other suppressive cell populations, such as regulatory T cells, contribute to maintenance of tolerance. Nevertheless, our data strongly suggest that clinical interventions targeting all alloreactive cytotoxic lymphocytes or eosinophils, such as through the use of high-dose corticosteroids, may have unpredictable and hidden deleterious effects for the lung allograft’s long-term survival.
Methods
Animals.
C57BL/6(B6)CD45.2+ (H2b), B6.SJL/BoyJB6CD45.1+ (H2b), and BALB/c (H2d) mice were obtained from The Jackson Laboratory. ΔdblGATA (C.129S1(B6)-Gata1tm6Sho/J) GATA mutant mice on a BALB/C background were purchased from The Jackson Laboratory as well but were then bred and expanded at the University of Virginia along with BALB/c wild-type control mice. NJ1638 IL-5–overexpressing hypereosinophilic transgenic mice were developed in our laboratory at Mayo Clinic (43) and breed in the same laboratory or colonies were bred and maintained at the vivarium facility at the University of Virginia School of Medicine. The animals passed all necessary genotyping screening or quarantine serological tests for those that were transported from one university to the other, before they were used in this study. All mice used in this study were between 8 and 10 weeks old.
Surgery and transplantation.
Orthotopic transplantation of a left lung allograft was carried out according to our previous reports (4, 11, 42, 44). To achieve allograft acceptance, mice were treated either with double CSB of the CD28/B7 and the CD40/CD40L pathways as previously described (11, 45) or with high-dose cyclosporine A and low-dose methylprednisolone as previously described (16).
Histology.
Lungs were fixed in formaldehyde, sectioned, and stained with H&E. A lung pathologist blinded to the experimental conditions graded graft rejection using standard criteria (International Society for Heart and Lung Transplantation [ISHLT] A Grade) developed by the Lung Rejection Study Group (46).
Preparation of lung cells.
Lung tissue derived from resting or graft recipient mice was well minced with scissors and digested by placing them into RPMI 1640 medium (Thermo Fisher) containing 0.5 mg/ml collagenase II (Worthington Biochemical Corporation) and 5 U/ml DNAse (MilliporeSigma) for 60 minutes at 37°C in a shaker. The digested lung tissue was passed through a 70-μm cell strainer and treated with ACK lysing buffer (Lonza) to remove red cell contamination. This digestion methods have been previously described (11).
Flow cytometry.
All antibodies for flow cytometry were primarily fluorochrome-conjugated rat anti-mouse monoclonal antibodies. Thus, staining of samples was by direct immunofluorescence. Intracellular staining was performed as previously described (44).The following antibodies were purchased from BD Biosciences: anti-iNOS FITC, anti-Ly6G (clone IA8) APC, anti-integrin β7 PE, anti-SiglecF PE or PerCPCy5.5, anti-Gr-1 PE or APC, and anti-CD127 APC. Antibodies purchased from Biolegend include anti-Ly6C PE-Cy7, anti-CCR3 PE or PE-Cy7, and anti-CD49d APC, while the following antibodies were purchased from Thermo Fisher–eBiosciences: anti-CD45 eFluor-450, anti-CD45.2 eFluor-450 or eluor-506, anti-CD45.1 APC-eFluor-780, anti-CD11b PerCPCy5.5 or APC-eFluor-780, anti-CD8a FITC or APC-eFluor-780, anti-CD90.2 FITC or APC-eFluor-780, anti-CD11c PerCPCy5.5 or APC-eFluor-780, anti-F4/80 PE or PerCPCy5.5, anti-CD206 PE, anti-CD68 PE, anti-CD38 PE, anti-CD31 APC, anti-CD4 APC or eFluor 506, anti-CD44 PerCPCy5.5, anti-CD62L PE-Cy7, and anti-Ki-67 PE. All fluorochrome-conjugated antibodies were matched with the corresponding rat IgG isotypes as antibody controls. In addition, FMO controls were also used to separate the negative and positive populations. Dead cells were excluded with Live/Dead Fixable Stain (Thermo Fisher). To block nonspecific binding to Fc receptors, we used anti-CD16/CD32 (Thermo Fisher). Cells expressing various markers of interest were acquired in a BD Canto II, equipped with 3 lasers for 10-parameter detection; or in BD LSR Fortessa, equipped with 4 lasers for 18-parameter detection (BD Biosciences); or in FACS Scan, upgraded with 3 lasers for 10-parameter detection capacity (Cytek Development). Quality controls were performed daily on the flow cytometers according to the manufacturers’ instructions.
FACS for lung eosinophil.
Eosinophils were sorted as CD45+CD11b+Siglec–F+CD11c– cells following the staining of digested lungs with a panel of fluorochrome-conjugated antibodies similar to the ones used for flow cytometry. Sorting was done using the BD Influx (BD Biosciences) or Sony SY3200 “Synergy” Cell Sorter (Sony Biotechnology).
Differential staining for leukocytes.
Cytospin slides of sorted CD45+CD11b+Siglec–F+CD11c–recipient mouse lung graft cells were stained using the Hema 3 staining kit (Fischer Scientific); Hema 3 is a Romanowsky stain variant. Staining was done according to the manufacturer’s instruction.
NO measurement.
The Nitric Oxide Assay Kit (Colorimetric) (Abcam), based on the Greiss reagent, was used for the measurement of NO concentration in the lung. Lung tissues were homogenized using a GentleMacs Dissociator (Miltenyi Biotec), and other sample preparation procedures were according to the manufacturer’s recommendations. Prepared samples were analyzed in a iMark Microplate Absorbance Reader (Bio-Rad).
Isolation, culture, and polarization of mouse peripheral blood eosinophil.
Eosinophils were isolated from peripheral blood of hypereosinophilic IL-5–transgenic mice (NJ.1638) after density-dependent separation of the eosinophil-rich white blood cells using a combination of Histopaque 1119 and 1083 (MilliporeSigma) at a ratio of 1:9, followed by negative selection of a >98% pure eosinophil population after incubation of white blood cells with immunomagnetic beads conjugated with CD45R/B220 and CD90.2/Thy.2 antibodies (Miltenyi Biotech). The purity of eosinophils was confirmed by flow cytometry (CD45+CD11b+SiglecF+CCR3+ cells). Details of the cell preparation and eosinophil purification protocol are as previously described (21). To obtain polarized eosinophils, purified eosinophils were cultured at a 5 × 105/ml concentration for 18–24 hours in RMPI-1640 media (containing glutamine and 25 mM HEPES) (Thermo Fisher) supplemented with 10% FBS, 10 U/ml penicillin, 10 μg/ml streptomycin, 29.2 μg/ml L-glutamine, and 55 μM β-mercaptoethanol; in addition, E0 culture also included 10 ng/ml IL-5, E1 included 10 ng/ml IL-5, 15 ng/ml IFN-γ, and 15 ng/ml TNF-α, while E2 culture also had IL-5 10 ng/ml, IL-33 30 ng/ml, IL-4 10 ng/ml, and GM-CSF 10 ng/ml. All cytokines and growth factors used for eosinophil polarization culture were purchased from Peprotech.
In vitro MLR.
Stromal cells from BALB/cJ mice (CD45.2) and CD90.2-positive cells from B6.SJL/BoyJ mice (CD45.1) were obtained by negative and positive selection, respectively, by manual activated cell separation using CD90.2 MicroBeads (Miltenyi Biotech). Labeled or unlabeled CD45.1+CD90.2+ cells were cultured together with the CD45.2+ stromal cells in a round-bottomed 96-well plate at a ratio of 1:3 (CD90.2+ cells: stromal cells). In some cases, CD45.2+ eosinophils isolated from IL-5–transgenic (NJ. 1638) mice were also added to the culture at the same number as the CD90.2+ cells isolated from B6.SJL/BoyJ (CD45.1+) mice. Proliferation of the CD90.2+ cells were analyzed flow cytometrically by Ki-67 expression after 5 days of culture.
In vivo antibody-mediated cytokine neutralization.
All neutralization antibodies are of rat origin and were purchased from BioXcell. For the targeted depletion of eosinophils in allograft recipients, 200 μg anti-IL5 (clone TRFK5) was administered to each mouse on days –2, –1, +1, and 2 after transplantation. For the neutralization of IFN-γ and TNF-α, 500 μg each of the blocking antibodies, clones XMG1.2 and XT3.11, respectively, were administered, together in a cocktail twice every other day before transplant and twice every other day after transplant for a total of 4 doses. Each control animal for all the antibody-mediated cytokine neutralization experiments received an equivalent concentration of rat IgG control (clone HRPN).
In vitro antibody-mediated cytokine neutralization.
IFN-γ and TNF-α were neutralized in vitro using 10 μg/ml and 25 μg/ml, respectively, of the same blocking antibodies used for in vivo neutralization experiments. The antibodies were added twice on the culture setup days and on day 3 after setups.
Quantitative PCR.
RNA was extracted from lung digests and eosinophils were isolated from the lung or in vitro–polarized eosinophils using the TRIzol-based technique according to the manufacturer’s guidelines (Thermo Fisher). cDNA was reverse transcribed from RNA samples using the High-Capacity cDNA Reverse Transcription Kit in accordance with the manufacturer’s instructions (Thermo Fisher, Applied Biosystem). Quantitative PCR was run on the cDNA samples using Power Syber Green PCR Master Mix (Thermo Fisher, Applied Biosystem) in a CFX-96 Real-Time PCR Detection System (Bio-Rad). Cycling and reaction conditions were as provided by the manufacturer. The primers used are provided as follows (forward [FW], reverse [RV]): IFN-γ FW: ATGAACGCTACACACTGCATC, RV: CCATCCTTTTGCCAGTTCCTC; TNF-α FW: CCCTCACACTCAGATCATCTTCT, RV: GCTACGACGTGGGCTACAG; IL-5 FW: CTCTGTTGACAAGCAATGAGACG, RV: TCTTCAGTATGTCTAGCCCCTG; IL-33 FW: TCCAACTCCAAGATTTCCCCG, RV: CATGCAGTAGACATGGCAGAA; IL-4 FW: GGTCTCAACCCCCAGCTAGT, RV: GCCGATGATCTCTCTCAAGTGAT; GM-CSF FW: GGCCTTGGAAGCATGTAGAGG, RV: GGAGAACTCGTTAGAGACGACTT; iNOS FW: GTTCTCAGCCCAACAATACAAGA, RV: GTGGACGGGTCGATGTCAC; B-ACTIN FW: CGTGCGTGACATCAAAGAG, RV: TGCCACAGGATTCCATAC; CCL17 FW: TACCATGAGGTCACTTCAGATGC, RV: GCACTCTCGGCCTACATTGG; IL-13 FW: CCTGGCTCTTGCTTGCCTT, RV: GGTCTTGTGTGATGTTGCTCA; CXCL9 FW: GGAGTTCGAGGAACCCTAGTG, RV: GGGATTTGTAGTGGATCGTGC; CXCL10 FW: CCAAGTGCTGCCGTCATTTTC, RV: GGCTCGCAGGGATGATTTCAA; CXCL11 FW: GGCTTCCTTATGTTCAAACAGGG, RV: GCCGTTACTCGGGTAAATTACA; and CCL22 FW: AGGTCCCTATGGTGCCAATGT, RV: CGGCAGGATTTTGAGGTCCA.
Statistics.
Friedman’s test was used for paired data, while the Kruskal-Wallis test was used for the unpaired group of observations. Post-hoc analysis of differences and comparison of differences between pairs of data were performed with the Wilcoxon rank test and the Mann-Whitney U test for paired and unpaired observations, respectively. Differences were considered significant at P < 0.05. The primary outcome measures are presented in scatter or dot plots. For the scatter plots, the inner dark line represents the mean value, while the whiskers above and below the mean represent the SEM. For the box plots, the bottom and top of the boxes represent the lower and upper quartiles, respectively; the dark band inside the box represents the median; and the top and bottom whiskers represent the maximum and minimum observed values, respectively. Data analysis and preparation of figures were done with GraphPad Prism 7.0b software.
Study approval.
All animal procedures were approved by the Animal Studies Committee at the University of Virginia School of Medicine, Charlottesville, Virginia, USA.
Author contributions
OOO planned and conducted the immunology experiments with assistance from QW and CDN; YG planned and performed animal surgeries with assistance from XW and WL; OOO, ASK, CL, and KL analyzed data; MHS did the pathological scoring of tissue slides for rejection grading; DK, AEG, and JJL assisted with study design; and EAJ and ASK oversaw data interpretation and manuscript preparation.
Supplementary Material
Acknowledgments
This work was supported by NIH grants PO1 AI116501, 1I01BX002299-01, R01HL113436-01A1, R01HL121218-01, R21AI119506, R01 HL065228, and R01HL113931.
Version 1. 12/21/2017
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2017;2(24):e96455. https://doi.org/10.1172/jci.insight.96455.
Contributor Information
Yizhan Guo, Email: YG2J@hscmail.mcc.virginia.edu.
Qing Wang, Email: QW5D@hscmail.mcc.virginia.edu.
Kang Li, Email: KL6HS@hscmail.mcc.virginia.edu.
Christopher Daniel Nazaroff, Email: Nazaroff.Christopher@mayo.edu.
Xingan Wang, Email: wangx17@upmc.edu.
Wenjun Li, Email: liw@wudosis.wustl.edu.
Daniel Kreisel, Email: kreiseld@wudosis.wustl.edu.
Elizabeth A. Jacobsen, Email: jacobsen.elizabeth@mayo.edu.
References
- 1.Conde P, et al. DC-SIGN(+) macrophages control the induction of transplantation tolerance. Immunity. 2015;42(6):1143–1158. doi: 10.1016/j.immuni.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia MR, et al. Monocytic suppressive cells mediate cardiovascular transplantation tolerance in mice. J Clin Invest. 2010;120(7):2486–2496. doi: 10.1172/JCI41628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li H, Shi B. Tolerogenic dendritic cells and their applications in transplantation. Cell Mol Immunol. 2015;12(1):24–30. doi: 10.1038/cmi.2014.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kreisel D, et al. Emergency granulopoiesis promotes neutrophil-dendritic cell encounters that prevent mouse lung allograft acceptance. Blood. 2011;118(23):6172–6182. doi: 10.1182/blood-2011-04-347823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez OM, et al. Evidence for a nonclassical pathway of graft rejection involving interleukin 5 and eosinophils. Transplantation. 1993;55(4):909–918. doi: 10.1097/00007890-199304000-00041. [DOI] [PubMed] [Google Scholar]
- 6.Nagral A, Ben-Ari Z, Dhillon AP, Burroughs AK. Eosinophils in acute cellular rejection in liver allografts. Liver Transpl Surg. 1998;4(5):355–362. doi: 10.1002/lt.500040503. [DOI] [PubMed] [Google Scholar]
- 7.Griseri T, et al. Granulocyte macrophage colony-stimulating factor-activated eosinophils promote interleukin-23 driven chronic colitis. Immunity. 2015;43(1):187–199. doi: 10.1016/j.immuni.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carr TF, Berdnikovs S, Simon HU, Bochner BS, Rosenwasser LJ. Eosinophilic bioactivities in severe asthma. World Allergy Organ J. 2016;9:21. doi: 10.1186/s40413-016-0112-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vos IH, et al. Inhibition of inducible nitric oxide synthase improves graft function and reduces tubulointerstitial injury in renal allograft rejection. Eur J Pharmacol. 2000;391(1-2):31–38. doi: 10.1016/S0014-2999(00)00021-2. [DOI] [PubMed] [Google Scholar]
- 10.Szabolcs MJ, et al. Acute cardiac allograft rejection in nitric oxide synthase-2(-/-) and nitric oxide synthase-2(+/+) mice: effects of cellular chimeras on myocardial inflammation and cardiomyocyte damage and apoptosis. Circulation. 2001;103(20):2514–2520. doi: 10.1161/01.CIR.103.20.2514. [DOI] [PubMed] [Google Scholar]
- 11.Krupnick AS, et al. Central memory CD8+ T lymphocytes mediate lung allograft acceptance. J Clin Invest. 2014;124(3):1130–1143. doi: 10.1172/JCI71359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chong SZ, et al. Human CD8+ T cells drive Th1 responses through the differentiation of TNF/iNOS-producing dendritic cells. Eur J Immunol. 2011;41(6):1639–1651. doi: 10.1002/eji.201041022. [DOI] [PubMed] [Google Scholar]
- 13.Percopo CM, et al. SiglecF+Gr1hi eosinophils are a distinct subpopulation within the lungs of allergen-challenged mice. J Leukoc Biol. 2017;101(1):321–328. doi: 10.1189/jlb.3A0416-166R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JJ, et al. Human versus mouse eosinophils: “that which we call an eosinophil, by any other name would stain as red”. J Allergy Clin Immunol. 2012;130(3):572–584. doi: 10.1016/j.jaci.2012.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mesnil C, et al. Lung-resident eosinophils represent a distinct regulatory eosinophil subset. J Clin Invest. 2016;126(9):3279–3295. doi: 10.1172/JCI85664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen DL, et al. Increased T cell glucose uptake reflects acute rejection in lung grafts. Am J Transplant. 2013;13(10):2540–2549. doi: 10.1111/ajt.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fulkerson PC, Rothenberg ME. Targeting eosinophils in allergy, inflammation and beyond. Nat Rev Drug Discov. 2013;12(2):117–129. doi: 10.1038/nrd3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J, et al. Depletion of eosinophil infiltration by anti-IL-5 monoclonal antibody (TRFK-5) accelerates open skin wound epithelial closure. Am J Pathol. 1997;151(3):813–819. [PMC free article] [PubMed] [Google Scholar]
- 19.Nair P, et al. Mepolizumab for prednisone-dependent asthma with sputum eosinophilia. N Engl J Med. 2009;360(10):985–993. doi: 10.1056/NEJMoa0805435. [DOI] [PubMed] [Google Scholar]
- 20.Roufosse FE, et al. Long-term safety of mepolizumab for the treatment of hypereosinophilic syndromes. J Allergy Clin Immunol. 2013;131(2):461–7.e1. doi: 10.1016/j.jaci.2012.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobsen EA, et al. Differential activation of airway eosinophils induces IL-13-mediated allergic Th2 pulmonary responses in mice. Allergy. 2015;70(9):1148–1159. doi: 10.1111/all.12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu LY, Bates ME, Jarjour NN, Busse WW, Bertics PJ, Kelly EA. Generation of Th1 and Th2 chemokines by human eosinophils: evidence for a critical role of TNF-alpha. J Immunol. 2007;179(7):4840–4848. doi: 10.4049/jimmunol.179.7.4840. [DOI] [PubMed] [Google Scholar]
- 23.Fonseca SG, et al. TNF-alpha mediates the induction of nitric oxide synthase in macrophages but not in neutrophils in experimental cutaneous leishmaniasis. Eur J Immunol. 2003;33(8):2297–2306. doi: 10.1002/eji.200320335. [DOI] [PubMed] [Google Scholar]
- 24.Morikawa A, et al. Augmentation of nitric oxide production by gamma interferon in a mouse vascular endothelial cell line and its modulation by tumor necrosis factor alpha and lipopolysaccharide. Infect Immun. 2000;68(11):6209–6214. doi: 10.1128/IAI.68.11.6209-6214.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drake MG, et al. Human and mouse eosinophils have antiviral activity against parainfluenza virus. Am J Respir Cell Mol Biol. 2016;55(3):387–394. doi: 10.1165/rcmb.2015-0405OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paoliello-Paschoalato AB, Oliveira SH, Cunha FQ. Interleukin 4 induces the expression of inducible nitric oxide synthase in eosinophils. Cytokine. 2005;30(3):116–124. doi: 10.1016/j.cyto.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Watson ML, Smith D, Bourne AD, Thompson RC, Westwick J. Cytokines contribute to airway dysfunction in antigen-challenged guinea pigs: inhibition of airway hyperreactivity, pulmonary eosinophil accumulation, and tumor necrosis factor generation by pretreatment with an interleukin-1 receptor antagonist. Am J Respir Cell Mol Biol. 1993;8(4):365–369. doi: 10.1165/ajrcmb/8.4.365. [DOI] [PubMed] [Google Scholar]
- 28.Jacobsen EA, Helmers RA, Lee JJ, Lee NA. The expanding role(s) of eosinophils in health and disease. Blood. 2012;120(19):3882–3890. doi: 10.1182/blood-2012-06-330845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobsen EA, Lee NA, Lee JJ. Re-defining the unique roles for eosinophils in allergic respiratory inflammation. Clin Exp Allergy. 2014;44(9):1119–1136. doi: 10.1111/cea.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobsen EA, Taranova AG, Lee NA, Lee JJ. Eosinophils: singularly destructive effector cells or purveyors of immunoregulation? J Allergy Clin Immunol. 2007;119(6):1313–1320. doi: 10.1016/j.jaci.2007.03.043. [DOI] [PubMed] [Google Scholar]
- 31.Rosenberg HF, Dyer KD, Foster PS. Eosinophils: changing perspectives in health and disease. Nat Rev Immunol. 2013;13(1):9–22. doi: 10.1038/nri3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wegmann M. Targeting eosinophil biology in asthma therapy. Am J Respir Cell Mol Biol. 2011;45(4):667–674. doi: 10.1165/rcmb.2011-0013TR. [DOI] [PubMed] [Google Scholar]
- 33.Masterson JC, et al. Eosinophil-mediated signalling attenuates inflammatory responses in experimental colitis. Gut. 2015;64(8):1236–1247. doi: 10.1136/gutjnl-2014-306998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JJ, Jacobsen EA, McGarry MP, Schleimer RP, Lee NA. Eosinophils in health and disease: the LIAR hypothesis. Clin Exp Allergy. 2010;40(4):563–575. doi: 10.1111/j.1365-2222.2010.03484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Budden KF, et al. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol. 2017;15(1):55–63. doi: 10.1038/nrmicro.2016.142. [DOI] [PubMed] [Google Scholar]
- 36.Galli SJ, Tsai M, Piliponsky AM. The development of allergic inflammation. Nature. 2008;454(7203):445–454. doi: 10.1038/nature07204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kishimoto K, et al. Th1 cytokines, programmed cell death, and alloreactive T cell clone size in transplant tolerance. J Clin Invest. 2002;109(11):1471–1479. doi: 10.1172/JCI14947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sayegh MH, et al. CD28-B7 blockade after alloantigenic challenge in vivo inhibits Th1 cytokines but spares Th2. J Exp Med. 1995;181(5):1869–1874. doi: 10.1084/jem.181.5.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takeuchi T, Lowry RP, Konieczny B. Heart allografts in murine systems. The differential activation of Th2-like effector cells in peripheral tolerance. Transplantation. 1992;53(6):1281–1294. doi: 10.1097/00007890-199206000-00023. [DOI] [PubMed] [Google Scholar]
- 40.Iijima H, Duguet A, Eum SY, Hamid Q, Eidelman DH. Nitric oxide and protein nitration are eosinophil dependent in allergen-challenged mice. Am J Respir Crit Care Med. 2001;163(5):1233–1240. doi: 10.1164/ajrccm.163.5.2003145. [DOI] [PubMed] [Google Scholar]
- 41.MacPherson JC, et al. Eosinophils are a major source of nitric oxide-derived oxidants in severe asthma: characterization of pathways available to eosinophils for generating reactive nitrogen species. J Immunol. 2001;166(9):5763–5772. doi: 10.4049/jimmunol.166.9.5763. [DOI] [PubMed] [Google Scholar]
- 42.Li W, et al. Lung transplant acceptance is facilitated by early events in the graft and is associated with lymphoid neogenesis. Mucosal Immunol. 2012;5(5):544–554. doi: 10.1038/mi.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee NA, McGarry MP, Larson KA, Horton MA, Kristensen AB, Lee JJ. Expression of IL-5 in thymocytes/T cells leads to the development of a massive eosinophilia, extramedullary eosinophilopoiesis, and unique histopathologies. J Immunol. 1997;158(3):1332–1344. [PubMed] [Google Scholar]
- 44.Gelman AE, et al. CD4+ T lymphocytes are not necessary for the acute rejection of vascularized mouse lung transplants. J Immunol. 2008;180(7):4754–4762. doi: 10.4049/jimmunol.180.7.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larsen CP, et al. Long-term acceptance of skin and cardiac allografts after blocking CD40 and CD28 pathways. Nature. 1996;381(6581):434–438. doi: 10.1038/381434a0. [DOI] [PubMed] [Google Scholar]
- 46.Yousem SA, et al. Revision of the 1990 working formulation for the classification of pulmonary allograft rejection: Lung Rejection Study Group. J Heart Lung Transplant. 1996;15(1 Pt 1):1–15. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.