Abstract
Structural changes in the retina are common manifestations of ophthalmic diseases. Optical coherence tomography (OCT) enables their identification in vivo—rapidly, repetitively, and at a high resolution. This protocol describes OCT imaging in the mouse retina as a powerful tool to study optic neuropathies (OPN). The OCT system is an interferometry-based, non-invasive alternative to common post mortem histological assays. It provides a fast and accurate assessment of retinal thickness, allowing the possibility to track changes, such as retinal thinning or thickening. We present the imaging process and analysis with the example of the Opa1delTTAG mouse line. Three types of scans are proposed, with two quantification methods: standard and homemade calipers. The latter is best for use on the peripapillary retina during radial scans; being more precise, is preferable for analyzing thinner structures. All approaches described here are designed for retinal ganglion cells (RGC) but are easily adaptable to other cell populations. In conclusion, OCT is efficient in mouse model phenotyping and has the potential to be used for the reliable evaluation of therapeutic interventions.
Keywords: Neurobiology, Issue 127, optical coherence tomography, mouse, retinal imaging, retinal ganglion cells, retinal nerve fiber layer, in vivo, thickness, peripapillary
Introduction
OCT is a diagnostic tool that facilitates the examination of retinal structures1, including the optic nerve head (ONH). Over the years it has become a dependable indicator of disease progression in humans2,3, as well as in rodents4,5. It uses interferometry to create cross-sectional images of retinal layers at a 2-µm axial resolution. The innermost layer is the retinal nerve fiber layer (RNFL), containing RGC axons, which is followed by the ganglion cell layer (GCL), containing mostly RGC bodies. Next is the inner plexiform layer (IPL), where RGC dendrites meet bipolar, horizontal, and amacrine cell axons. These, together with horizontal cells, form the inner nuclear layer (INL), and their protrusions connect with photoreceptor axons in the outer plexiform layer (OPL). This is followed by the outer nuclear layer (ONL), with photoreceptor cell bodies, and is separated from the photoreceptor layer by the outer limiting membrane (OLM), also called the inner segment/outer segment (IS/OS) layer. Finally, the last observable layers in the mouse retina are the retinal pigment epithelium (RPE) and the choroid (C). The RNFL alone is normally too thin to be measured in mice; thus, analyzing the RNFL/GCL instead is preferable4,5. Another possibility is the GC complex layer, which contains the latter in addition to the IPL, making it thicker and thus even easier to measure on OCT scans4. Consequently, OCT can provide insight into the pathological status of the retina, such as in OPNs.
Alternatively, the thickness of the mouse retina is often analyzed with post mortem histology. However, this technique faces limitations relating to tissue collection, fixation, cutting, staining, mounting, etc. Hence, some defects, such as subtle thickness changes, cannot be detected. Finally, because the same mouse cannot be tested at several time points, the number of animals per study greatly increases, unlike for OCT. All in all, the non-invasiveness, high-resolution, possibility for repetition, time monitoring in time, and ease of use of the OCT technology make it the method of choice in retinal disease studies.
Mouse models are used to identify gene defects and to elucidate molecular mechanisms underlying retinopathies6. OPN is a form of retinopathy with substantial damage to the optic nerve (ON), which is made up of approximately 1.2 million RGC axons. OPN can be focused on the ON or can be secondary to other disorders, inborn or not7, leading to visual field loss and later, blindness. Characteristic traits of OPN are RGC loss and ON damage, which can be observed in human OCT as RNFL and GCL thinning2,3. Meanwhile, the pathophysiology of OPN is still poorly understood, and hence the need to test mouse retinas remains.
This manuscript describes the imaging and quantification of retinal layer thickness, using the example of the Opa1delTTAG mouse line8,9, a model of dominant optic atrophy (DOA)10. To assess RGC pathophysiology, radial, rectangular, and annular scans were quantified. This was done either with standard calipers provided by the OCT software or with a homemade macro developed for an open-source image processing program. The standard calipers are difficult to manipulate and often thicker than the RNFL/GCL, while the homemade calipers are easy to use, reproducible, and more precise. The macro performs a measurement for an automatically detected layer, in 5 points and at fixed positions, on both sides of the ONH in the peripapillary region. The goal of the presented protocol is to describe OCT scan acquisition to specify retinal positioning, with a focus on RGCs.
Protocol
The experimental protocol was approved by the Institut national de la santé et de la recherche médicale (Inserm; Montpellier, France), is consistent with the European directives, and complies with the ARVO Statement for the Use of Animals in Ophthalmic Research. It was carried out under the agreement of the Languedoc Roussillon Comity of Ethics in Animal Experimentation (CEEALR; nuCEEA-LR-12123).
1. Equipment Setup and Pre-imaging Preparation
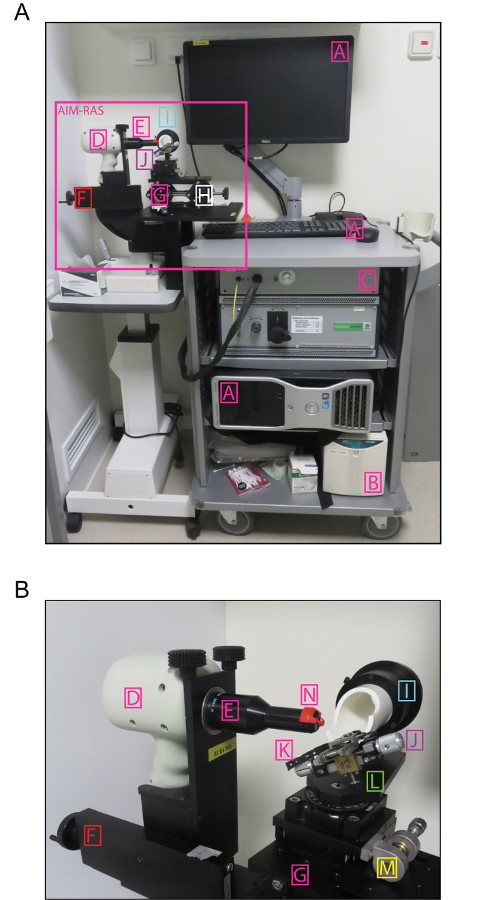
NOTE: Here, OCT was performed on mouse retinas using the spectral domain (SD) ophthalmic imaging system (Figure 1A). The SD-OCT apparatus consists of a base and an animal imaging mount (AIM) with a rodent alignment stage (RAS) (Figure 1B). The base includes the computer, the OCT engine, the SD-OCT probe, and the mouse-specific lens. The probe is mounted onto the AIM, which includes the Z-translator. The RAS is used for mouse positioning thanks to the table with the X- and Y-translator, the cassette that can be rotated and swiveled, and the removable bite bar with the nose band. The software provided by the manufacturer allows for the acquisition and analysis of OCT files, although the latter may also be done with an open-source image processing program.
Firmly place the probe in the AIM.
Connect the mouse-specific lens to the probe.
Adjust the reference arm settings for the specific lens (here, power at 086 and position at 964).
Making sure that the mouse-specific lens is sufficiently distant from the cassette, attach the bite bar with the nose band onto the cassette. NOTE: The distance between the lens and the cassette may be adjusted with the Z-translator screw, located at the back of the AIM. The nose band is a commercially available elastic band, and its tension should be adjusted depending upon the size of the mouse.
Turn on the power supply (lower right corner of the cart) and then the computer.
To launch the imaging program, double-click on the appropriate shortcut on the screen. NOTE: This depends on the type of lens; here, Mouse Retina.
- Create a new Clinical Study and add the desired Protocols. Otherwise, use the existing Clinical Study and/or Protocols.
- Add a new study by choosing the "Study Name"; "ID"; "Treatment Arm Specifications" (here, "Uncategorized"); and predefined "Scan Protocols", if they exist.
Choose an "Examiner" in the "Clinical Study" section. NOTE: For a new "Examiner", go to "Setup Examiners & Physicians" to define it.
Add a new Patient in the Patient/Exam section by click on Add Patient; entering the "ID", "name", "surname", "sex", and "date of birth" (optional). NOTE: Do this just before testing each mouse, if more convenient. Make sure that the ID is no longer than 10 characters. For the mouse, the refractive error for both eyes is 0 and the axial length is 23.0.
Click on "Add Exam" and select the desired "Protocol" by adding "Preset Scans" from the list, starting with the eye that will be measured first (here, the right eye), or by customizing the scans.
To customize a scan, either use an existing scan as a template and edit it, or create it from scratch through the "Add Custom Scan" option. After adding all the scans to the list, define a new protocol using the "Enter "New Protocol Name" option (OS: oculus sinister, left eye; OD: oculus dexter, right eye). NOTE: Here, the protocol involves three scans: (i) a radial scan, with a 1.4 mm diameter, 0 mm horizontal and vertical offsets, 1,000 lines of A-scans/B-scan, 100 B-scans/volume, 1 frame/B-scan, 80 lines of inactive A-scans/B-scan, and 1 volume; (ii) a rectangular scan, with a 1.4-mm length and width, 0° angle, 0-mm horizontal and vertical offsets, 1,000 lines of A-scans/B-scan, 100 B-scans, 1 frame/B-scan, 80 lines of inactive A-scans/B-scan, and 1 volume; and (iii) an annular scan, with a 0.4-mm minimum and 0.6-mm maximum diameter, 0-mm horizontal and vertical offsets, 1,000 lines of A-scans/B-scan, 3 B-scans/volume, 48 frames/B-scan, 80 lines of inactive A-scans/B-scan, and 1 volume.
2. Mouse Preparation
- Eye dilation and immobilization
- At least 15 min before data acquisition, instill 10% phenylephrine eye drops into the mouse eyes, remove the excess, and instill 0.5% tropicamide eye drops. NOTE: The first dilatation can be done at once for all the mice that will be tested in the session. Make sure that the liquids are at room temperature.
- Immediately after inducing general anesthesia (step 2.2), administer 0.4% oxybuprocaine hydrochloride eye drops, keeping them in place for 3 s to anesthetize and immobilize the eyes. Afterwards, wipe off the drops and repeat the instillation of 10% phenylephrine and 0.5% tropicamide to ascertain that the eyes are well dilated. NOTE: Make sure that the liquids are at room temperature and that the mouse does not swallow the oxybuprocaine.
- General anesthesia
- Prepare an anesthetic solution of ketamine (20 mg/mL) and xylazine (1.17 mg/mL) in saline. Store at 4 °C for a maximum of 2 weeks.
- Approximately 5 min before testing, intraperitoneally inject 5 - 10 µL of anesthetic solution per 1 g of body mass, depending upon the age and size of the mouse. NOTE: Young and/or thin mice need less anesthetic, take less time to anesthetize, but also awaken faster. Here, 8 µL/g (160 mg/kg of ketamine, 9.33 mg/kg of xylazine).
- Optionally, lubricate the eyes with viscous eye drops or an ophthalmic gel to avoid corneal dryness.
3. Mouse Positioning
Lubricate the eyes with viscous glycol-based eye drops to provide corneal hydration. NOTE: If the eyes seem dry during the exam, re-apply the eye drops.
Wrap the mouse in a sheet of surgical gauze to keep it warm.
Using a sponge or cotton wick, apply a thin layer of an ophthalmic gel with 0.3% hypromellose onto each eye, to minimize light refraction, avoid opacities, and secure corneal hydration. While doing so, move the eyelashes and whiskers aside.
Position the mouse in the cassette, with the head straight and pointing forward.
Gently open the bite bar clamp and place the bar in the mouth; use the nose band to secure the position. NOTE: Ensure that the bite bar is in the middle of the cassette.
Place a cotton roll under the right (left) side if the right (left) eye is being tested.
While keeping the clip-on aiming tip on the mouse-specific lens, bring it towards the eye by rotating the Z-translator screw counterclockwise.
By rotating and swiveling the cassette and by turning the bite bar and the X-translator screws, pre-set the position of the mouse; the goal is to have the right eye look directly into the lens, and thus to align the optical axes of the eyes and the lens. NOTE: If the mouse is too high or too low, the Y-translator screw should be used first.
- Choose the first scan in the "Exam", click on "Start Aiming", and make any further manual adjustments for retinal imaging.
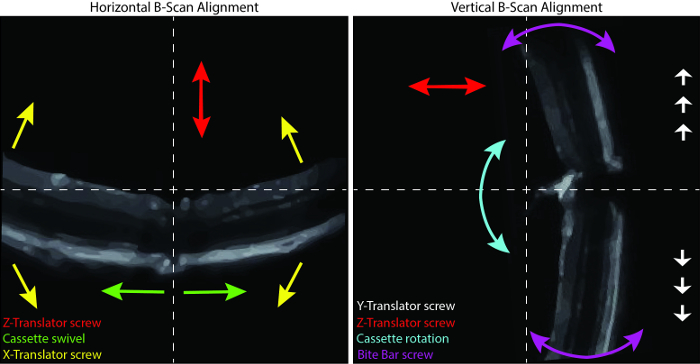
- Rotate the cassette to bring the ONH in the middle of the right panel by moving the ONH up or down. Use the bite bar screw to straighten the retina on the right panel. Swivel the cassette to position the ONH in the middle of the left panel.
- Use the X-translator screw to level the retina on the left panel. Keeping in mind the major function of each modulator, further adjust the position of the retina to centralize the ONH. NOTE: Use the Y-translator screw to move the ONH up and down on the right panel, if necessary. The position may be refined at any time between scans.
4. SD-OCT Imaging of the ONH and Retina
- Once content with the adjustments, click on "Start Snapshot" to begin SD-OCT scanning.
- If a 3D imaging is not required, uncheck the OCU option.
Save the scan and the report.
Proceed with the next scans.
To image the second eye, after retracting the lens, turn the cassette accordingly and repeat steps 3.6 - 4.3.
5. Acquisition Completion
When the acquisition is complete, remove the mouse from the cassette, apply ophthalmic gel with 0.3% hypromellose to each eye, and place the mouse on a heating plate to wake up.
After the last acquisition, close the software and switch off the computer and the OCT machine (power supply button).
Clean the cassette with disinfectant.
6. Analysis
- For the retinal layer thickness measurement, use the automated segmentation software provided by the manufacturer.
- Click on the "Patient", choose the desired "Exam" from then list, and click on the "Review Exam" option.
- Load the desired OCT data onto the OCT software by right-clicking on the folder icon in the desired scan.
- Right click on the B-scan; configure the calipers by enabling up to 10 measuring calipers; and enter their names, angles, and colors.
- Choosing the desired caliper by a right-clicking on the B-scan; place it accordingly onto the retina for measurement. NOTE: For the scans centered on the ONH, set 5 calipers on each side of it, equidistant from each other. For the peripapillary analysis, make sure that the caliper is not placed too far from the ONH. For radial scanning, analyze 10 pictures per scan that were chosen by the OCT software. Their numbers may be found in the "Reports" folder.
- Save the results for analysis in a spreadsheet software program by right-clicking on the B-scan and clicking "Save results". NOTE: The results can be found in the same folder as the scanning data.
- Alternatively, use the homemade caliper macro, "MRI Retina Tool", developed for an open-source image processing program (see the Table of Materials).
- Make sure that the "MRI Retina Tool" and the "Modify Polygon Section Tool" macros are active. Load the image. Click on the m button to start the measurement. NOTE: The macro automatically creates one 0.2 mm-long measuring cassette on both sides of the ONH. Each cassette contains 5 measuring points that function as calipers. Their lateral position is unchangeable and is customized for the peripapilla in the radial scans. The horizontal position of the cassettes is predefined to measure the RNFL/GCL thickness, but it is easily modifiable to measure the GC complex layer instead. If the scan quality is poor, the horizontal position needs to be adjusted, or the picture needs to be excluded from the analysis.
- For horizontal adjustment, click on the e button. In the newly opened "ROI Manager" window, choose the first cassette.
- Click on the blue polygon button and adjust the position of the first cassette by clicking on the borders of the measured layer in the picture.
- Repeat for the second cassette by first choosing it in the "ROI Manager" window and then clicking on the appropriate image. Click on the r button to re-measure. View the results in the "Measurements" window. NOTE: The results may be copied to a spreadsheet software program at any time. They contain the following values for the right cassette (r), left cassette (l), and total: Intden - integrated density within the cassette; Area: area of the measurement, in mm2; Len: length of the caliper within the cassette, in mm; Mean: mean intensity of the signal within the cassette; and Std: standard error of the mean intensity of the signal.
- Proceed with the next image.
- Further analyze the data in a spreadsheet software program. NOTE: To find the mean thickness of the layer, take the ten Len values.
Representative Results
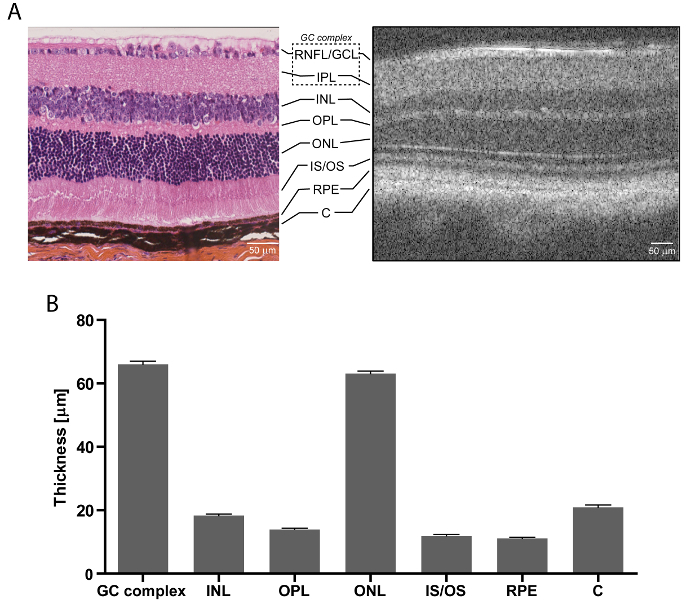
The SD-OCT technology enables retinal imagining and thickness analysis that is comparable to histology, but is faster and more detailed (Figure 3). As presented with wildtype C57Bl/6 mice, even though the quality of an SD-OCT scan (Figure 3A, right) is not as good as that of an image of a retinal cross-section (Figure 3A, left), it visualizes more layers (e.g., OLM). Moreover, it takes only about 40 min, including mouse preparation, versus days or weeks for histological analysis. Finally, it does not require processing and staining, such as haematoxylin, eosin, and saffron, which may damage the tissue and cause the collection of erroneous data. The retinal layers easily measurable in OCT encompass the RNFL/GCL, IPL, INL, OPL, ONL, IS/OS, RPE, and C (Figure 3B), therefore allowing for a complex study of the entire retina. As such, structural retinal changes reflect disease development. In the case of OPNs, this applies to RGCs and the ON, and thus the RNFL/GCL and IPL.
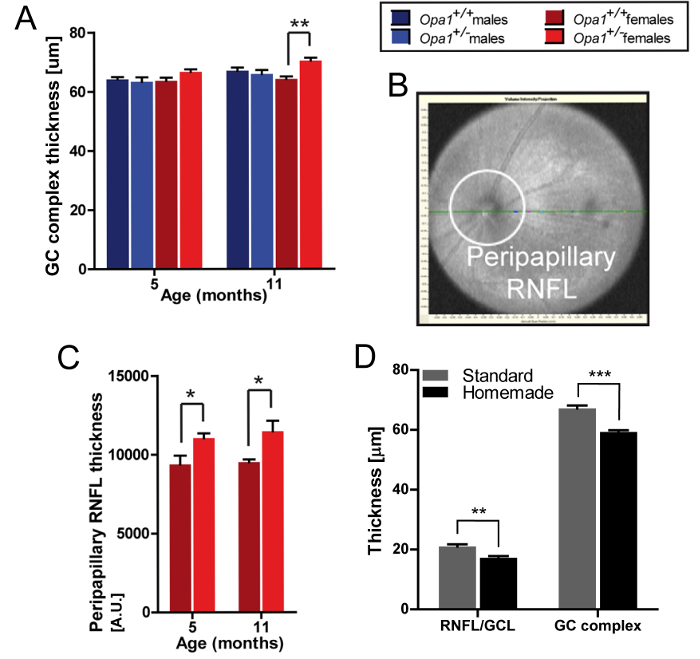
DOA is one of the most common OPNs and is characterized by RGC degeneration and the loss of the RNFL11. Due to mutations in the OPA1 gene12, it leads to visual impairment and blindness. Using the Opa1delTTAG mouse model, which carries the human recurrent c.2708delTTAG mutation, it was discovered that Opa1 haplo-insufficiency hinders vision in a sex-dependent manner8,9. This was determined on the basis of OCT measurements of retinal thickness, which showed a progressive thickening of the GC complex layer (Figure 4A) and the peripapillary RNFL (Figure 4B, 4C) in Opa1+/- females. In these experiments, the calculations were done with the standard calipers for rectangular scans and with an open-source image processing program for the annular scans. For radial scans, which often are of a lower quality and produce a minimum of 10 images per retina for analysis, a homemade macro was developed. A comparison of the standard and homemade calipers (Figure 4D) showed a significantly lower thickness of the RNFL/GCL and GC complex layers measured with the latter. This is because the standard calipers are much thicker and more difficult to place on the border of the layer. Therefore, it is best to avoid using the standard calipers for thin layers, especially on radial scans.
To summarize, the SD-OCT allows for mouse visual phenotyping that can be repeated across several time points. However, the OCT scan type and measuring method must be adapted to the investigated disease, and thus the retinal layer in question. Nevertheless, OCT provides sufficient information to identify defects in retinal structure. However, this must be further analyzed with another method to provide a complete understanding of the underlying mechanisms.
Figure 1: SD-OCT Ophthalmic Imaging System. (A) Overview of the base and AIM-RAS parts of the SD-OCT device. (B) Overview of the AIM-RAS components. A: computer, B: power supply, C: OCT engine reference arm, D: SD-OCT probe, E: mouse-specific lens, F: Z-translator, G: AIM-RAS table, H: Y-translator, I: cassette (rotator), J: bite bar, K – nose band, L: cassette swivel, M: X-translator, and N: aiming tip. This figute is color-coded as in Figure 2, with non-modulators of the retinal position marked in pink. Please click here to view a larger version of this figure.
Figure 2: Positioning the Retina. Horizontal (left) and vertical (right) view of the B-scan alignment. The arrows correspond to movements induced by the color-coded modulators. Please click here to view a larger version of this figure.
Figure 3: Retinal Layers. (A) Wildtype mouse retina histology after haematoxylin, eosin, and saffron staining (left) and SD-OCT (right); scale bar: 50 µm. (B) Retinal thickness measurements for 3 month-old wildtype mice; n = 14, mean ± SEM, scale bar: 50 µm. GC: ganglion cell, RNFL/GCL: retinal nerve fiber layer/ganglion cell layer, IPL: inner plexiform layer, INL: inner nuclear layer, OPL: outer plexiform layer, ONL: outer nuclear layer, IS/OS: photoreceptor inner segments/outer segments, RPE: retinal pigment epithelium, and C: choroid. Please click here to view a larger version of this figure.
Figure 4: Exemplary SD-OCT Measurements. (A) GC complex layer thickness in rectangular scans centered on the ONH, measured with the standard calipers; Opa1delTTAG mouse line, n = 4, mean ± SEM, ** p <0.01 assessed with Student's t-test. (B) Peripapillary RNFL in SD-OCT. (C) Peripapillary RNFL thickness in annular scans, determined as the RNFL area calculation per field; Opa1delTTAG female mice, n = 5-11, mean ± SEM, * p < 0.05 assessed with Student's t-test. (D) RNFL/GCL and GC complex layer thickness in radial scans, measured with the standard or homemade calipers for 3 or 10 scans, respectively; wildtype mice, n = 8, mean ± SEM, ** p <0.01, *** p <0.001 assessed with Student's t-test. RNFL/GCL - retinal nerve fiber layer/ganglion cell layer, GC: ganglion cell. Figures A-C adapted from Sarzi et al.9. Please click here to view a larger version of this figure.
Discussion
The OCT system, a non-invasive in vivo imaging method, provides high-resolution retinal cross-section-like scans. Thus, its main advantage is its potential for detailed analysis, with the wonderful opportunity to transpose protocols routinely applied to humans to mouse models.
In the example of Opa1delTTAG mutant mice, SD-OCT results showed an increase of RNFL and GC complex layer thickness, which allowed for further exploration of DOA pathophysiology9. It would not have been possible solely with histological analysis. In comparison, histology does not provide the possibility to visualize the entire retina, unlike annular or radial OCT scans. Moreover, it is more time-consuming and costly, considering the increased number of animals in studies with several time points. In fact, SD-OCT paved the way to incriminate a new retinal cell type in DOA, the Müller cell9. This was done despite the fact that neither single-cell resolution nor specific cell identifications are possible with the system. On the contrary, the thickness-focused and/or general state-focused analysis of the peripapillary region is largely enough to detect cellular deterioration. Additional investigations with histology can then be conducted with a clear idea of what to look for. Therefore, the same method can also be applied to the evaluation of therapeutic interventions to prevent or brake retinal degeneration.
To further improve the utility of OCT, homemade calipers were developed and had a much higher precision than the standard ones. Even though the standard is thicker than the RNFL/GCL, some teams use it anyway, but for larger layers13. Here, we focused the comparative analysis on RGCs in 10 radial scans per retina, all in the peripapillary region. The RNFL alone was not measurable on the radial scans either way. This layer was too thin and vague; therefore, the RNFL/GCL and the GC complex layer were measured instead. At the same time, we succeeded in measuring the RNFL using annular scans, which proved beneficial to mouse phenotyping. However, the reliability may be controversial. In all these approaches, the critical step was to center the scans on the ONH and to visualize the retina without shadows and opacities. The former can be easily adjusted by following the steps of the protocol in terms of the positioning of the retina. The latter depends upon the transmittance of the cornea and the crystalline. For example, if the ophthalmic gel is spread unevenly, the scan is blurry and/or the retina appears bent. To fix this, it would be enough to properly re-apply the gel. If the crystalline is opaque, the scan is dark or incomplete. The solution here would be to repeat the scan another day, if the transparency of the crystalline returns. Another possible reason for a bad-quality scan is the presence of obstacles, such as whiskers or eyelashes. These can be easily removed by setting aside and applying a little of the ophthalmic gel to hold them in place. Other analytic approaches that differ in terms of equipment type, scan type, angle, and other parameters exist as well and have varying numbers of analyzed images. This must be considered if the result quality is still not satisfactory. For instance, Liu et al. took radial scans at several angles13, in comparison to our radial scans at only one, reporting slightly thicker layers. Nevertheless, the OCT acquisition and analytic approaches proposed in this manuscript are suited for analyzing RGCs in the peripapillary mouse retina.
In conclusion, OCT is a technology with great potential. It enables the detection of subtle changes in the retinal structure—including the RGCs, especially in regards to the OPNs—and proves indispensable to vision science. Therefore, the presented protocol is practical for OPN mouse model phenotyping, as well as for the evaluation of novel therapies.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This work was supported by Inserm, Université Montpellier, Retina France, Union National des Aveugles et Déficients Visuels (UNADEV), Association Syndrome de Wolfram, Fondation pour la Recherche Médicale, Fondation de France, and the Laboratory of Excellence EpiGenMed program.
References
- Drexler W, Fujimoto JG. State-of-the-art retinal optical coherence tomography. Prog Retin Eye Res. 2008;27(1):45–88. doi: 10.1016/j.preteyeres.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Grenier J, et al. WFS1 in Optic Neuropathies: Mutation Findings in Nonsyndromic Optic Atrophy and Assessment of Clinical Severity. Ophthalmology. 2016;123(9):1989–1998. doi: 10.1016/j.ophtha.2016.05.036. [DOI] [PubMed] [Google Scholar]
- Zmyslowska A, et al. Retinal thinning as a marker of disease progression in patients with Wolfram syndrome. Diabetes Care. 2015;38(3):e36–e37. doi: 10.2337/dc14-1898. [DOI] [PubMed] [Google Scholar]
- Fischer MD, et al. Noninvasive, in vivo assessment of mouse retinal structure using optical coherence tomography. PLoS One. 2009;4(10):e7507. doi: 10.1371/journal.pone.0007507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieve K, Thouvenin O, Sengupta A, Borderie VM, Paques M. Appearance of the Retina With Full-Field Optical Coherence Tomography. Invest Ophthalmol Vis Sci. 2016;57(9):OCT96–OCT104. doi: 10.1167/iovs.15-18856. [DOI] [PubMed] [Google Scholar]
- Chang B, et al. Retinal degeneration mutants in the mouse. Vision Res. 2002;42(4):517–525. doi: 10.1016/s0042-6989(01)00146-8. [DOI] [PubMed] [Google Scholar]
- Mustafa S, Pandit L. Approach to diagnosis and management of optic neuropathy. Neurol India. 2014;62(6):599–605. doi: 10.4103/0028-3886.149370. [DOI] [PubMed] [Google Scholar]
- Sarzi E, et al. The human OPA1delTTAG mutation induces premature age-related systemic neurodegeneration in mouse. Brain. 2012;135(Pt 12):3599–3613. doi: 10.1093/brain/aws303. [DOI] [PubMed] [Google Scholar]
- Sarzi E, et al. Increased steroidogenesis promotes early-onset and severe vision loss in females with OPA1 dominant optic atrophy. Hum Mol Genet. 2016;25(12):2539–2551. doi: 10.1093/hmg/ddw117. [DOI] [PubMed] [Google Scholar]
- Delettre-Cribaillet C, Hamel CP, Lenaers G, et al. Optic Atrophy Type 1. In: Pagon RA, editor. Gene Reviews. 2007. Vol. 7. Seattle: University of Washington; 2007. [PubMed] [Google Scholar]
- Lenaers G, et al. Dominant optic atrophy. Orphanet J Rare Dis. 2012;7(46) doi: 10.1186/1750-1172-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delettre C, et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26(2):207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- Liu Y, et al. Monitoring retinal morphologic and functional changes in mice following optic nerve crush. Invest Ophthalmol Vis Sci. 2014;55(6):3766–3774. doi: 10.1167/iovs.14-13895. [DOI] [PubMed] [Google Scholar]


