Abstract
Currently, there is a limited ability to interactively study developmental cardiac mechanics and physiology. We therefore combined light-sheet fluorescence microscopy (LSFM) with virtual reality (VR) to provide a hybrid platform for 3D architecture and time-dependent cardiac contractile function characterization. By taking advantage of the rapid acquisition, high axial resolution, low phototoxicity, and high fidelity in 3D and 4D (3D spatial + 1D time or spectra), this VR-LSFM hybrid methodology enables interactive visualization and quantification otherwise not available by conventional methods, such as routine optical microscopes. We hereby demonstrate multiscale applicability of VR-LSFM to (a) interrogate skin fibroblasts interacting with a hyaluronic acid–based hydrogel, (b) navigate through the endocardial trabecular network during zebrafish development, and (c) localize gene therapy-mediated potassium channel expression in adult murine hearts. We further combined our batch intensity normalized segmentation algorithm with deformable image registration to interface a VR environment with imaging computation for the analysis of cardiac contraction. Thus, the VR-LSFM hybrid platform demonstrates an efficient and robust framework for creating a user-directed microenvironment in which we uncovered developmental cardiac mechanics and physiology with high spatiotemporal resolution.
Keywords: Cardiology
Keywords: Diagnostic imaging
A user-directed perspective for studying cardiac mechanics, physiology, and developmental biology at single-cell resolution.
Introduction
Despite the advent of optical imaging techniques, 3D and 4D (3D spatial + 1D time) methodologies for visualization and analysis of developmental cardiac mechanics and physiology lack user-directed platforms (1–3). While emerging efforts are being devoted to integrate advanced imaging techniques with interactive visualization methods (4–8), we further coupled interactive interrogation with quantification of microenvironments to uncover ultrastructure and function at high spatiotemporal resolution.
Virtual reality (VR) is an emerging virtual stereoscopic platform for user-directed interaction in a simulated physical environment (9, 10); thus, this process enables us to integrate with advanced imaging to resolve the limitation of the current volume rendering technique for an immersive and interactive experience. Application of VR to life science research and education requires high spatiotemporal resolution to enable visualization of dynamic organ systems. Unlike VR scenarios that are commonly generated by computer graphics or a 360-degree camera (11, 12), the advent of light-sheet fluorescence microscopy (LSFM) (13–21) allows for capturing of physiological events in the 3D or 4D domain that can be further adapted to VR headsets, such as Google Cardboard or Daydream, for effective VR visualization. In comparison with wide-field or confocal microscopy, LSFM enables multidimensional imaging at the single-cell resolution (22–29), which is critical for integrating high-fidelity physiological research with VR visualization. 4D LSFM further enables in vivo imaging of contracting embryonic hearts in zebrafish (Danio rerio) (30, 31), Caenorhabditis elegans (32, 33), and Drosophila melanogaster (34, 35). LSFM therefore offers high-quality imaging data for life science VR applications, allowing for the exploration of developmental and physiological phenomena with high spatiotemporal resolution and low phototoxicity.
To interface VR with LSFM for user-directed cardiovascular research and education, we have developed an effective and robust workflow to convert bitmap files to 3D editable mesh, bypassing the conversion to vector images, which are technically used in VR. To indicate the 3D immersive and interactive experience of a VR-LSFM hybrid platform, we demonstrated the interaction between human dermal fibroblasts (HDFs) and a biomaterials-based scaffold. We also navigated through the endocardiac trabeculated network in a regenerating zebrafish heart, and we localized spatial distribution of potassium channels in adult murine hearts following gene therapy. To further enhance the 4D capacity of our VR-LSFM hybrid, we developed the unattended batch intensity normalized segmentation (BINS) method to post-process the time-variant images over the course of cardiac contraction, and we applied deformable image registration (DIR) to track the contractile function in the zebrafish embryos. Overall, we interface the VR environment with LSFM to navigate through the microenvironments with a user-directed perspective for interactively studying cardiac mechanics, physiology, and developmental biology at a single-cell resolution. This framework is compatible with a low processing power smartphone equipped with a VR headset, leading to a pipeline for developing an interactive virtual laboratory for life science investigation and learning.
Results
We demonstrated the rapid acquisition-interpretation framework of life science phenomena via the VR-LSFM hybrid platform. However, this workflow is not limited to LSFM imaging data. The video recordings of the user-directed movements in VR mode are provided in the Supplemental Videos 1–7 (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.97180DS1 in the VR mode). The panoramic videos established a 360-degree perspective by panning via the cursor on the online-linked video as the user is navigating through the endocardial cavity, thereby, unraveling the cardiac architecture in both zebrafish and mice.
Elucidating the fibroblast proliferation in the hydrogel scaffolds.
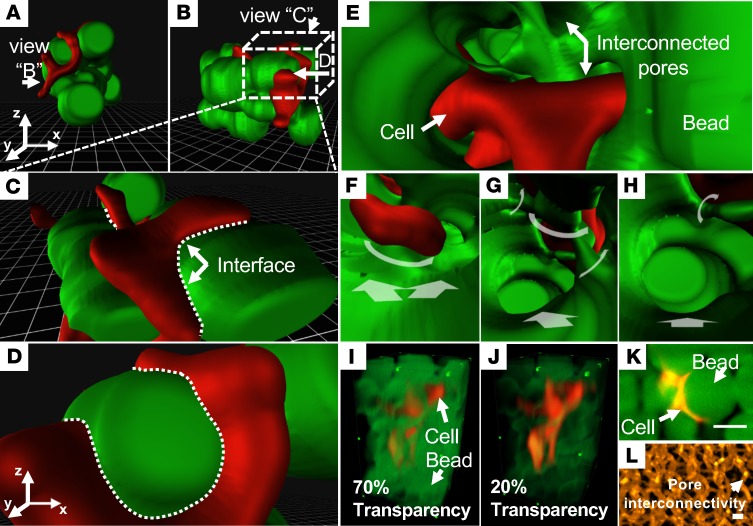
To interrogate cellular interaction with biomaterials, we used HDFs seeded in polymer-based scaffolds, demonstrating proliferating HDFs (Figure 1, red) within porous hyaluronic acid–based (HA-based) beads (Figure 1, green), laying the foundation to further elucidate response to biophysical properties, specifically, degradability and mechanical stiffness (Figure 1). We demonstrated the LSFM-acquired raw images in a large field of view (28), followed by building the 3D VR reconstruction inside the scaffolds. Using an interactive VR, we revealed that HDFs percolated through the hydrogel matrix (Figure 1, A–H). When the user was equipped with a smartphone and a VR viewer, a 3D immersive experience recapitulated the HDFs traversing through the scaffolds. The conventional methods confine the user perception for cellular events with (a) reduced transparency of the 3D microgel (Figure 1, I and J), (b) obstructed display of the beads in the 3D extracellular matrix (Figure 1K), and (c) limited axial resolution in a planar 2D image (Figure 1L). In contrast, the visual perspective of an operator using our VR-LSFM hybrid is unconfined by a specific region or viewpoint, and the user-directed movements are visualized in the recorded VR mode video (Supplemental Video 1).
Figure 1. HDFs embedded in a hydrogel matrix.
(A–D) The physical interface between the HDFs (red) and beads (green) provides a user-initiative perception. (E–H) Applying VR-LSFM accentuates the depth perception and contextual relation. Variable stepping directions are labeled with white arrows. (I and J) Conventional 3D volumetric rendering results are constrained to concurrently depict both beads and HDFs due to the reduced levels of transparency of the beads. (K) The 2D raw data reveal attenuation in spatial resolution of the HDF cells interacting with the beads. (L) The polymer network of the 3D microgel matrix is demonstrated in the conventional mode. HDF, human dermal fibroblast; VR, virtual reality; LSFM, light-sheet fluorescence microscopy. Scale bar: 50 μm. All the images are shown in pseudocolor.
Navigating through the myocardial ridges.
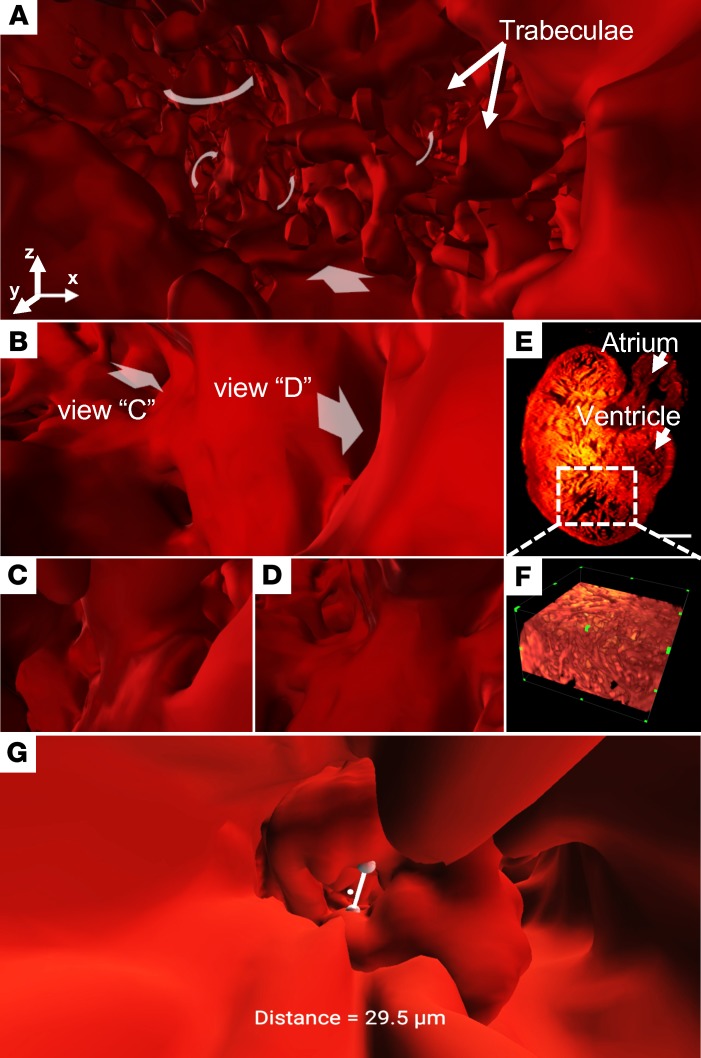
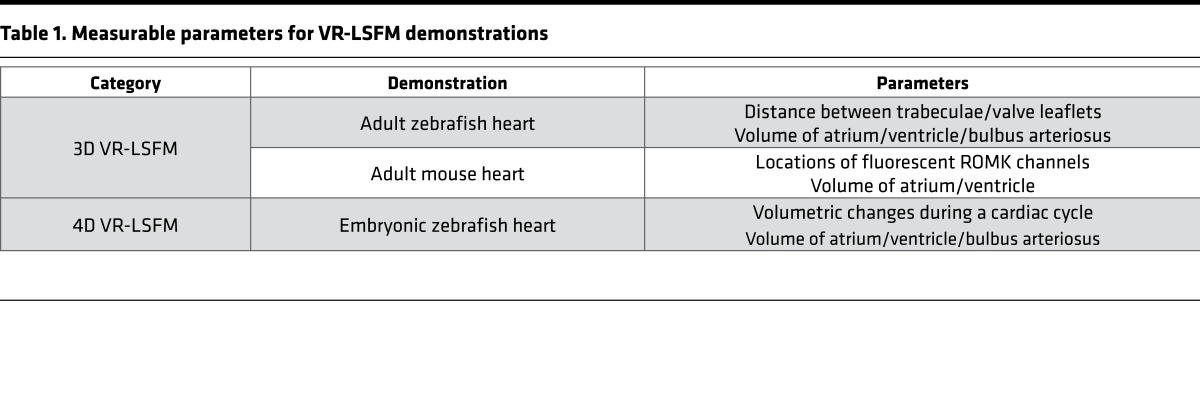
To contrast with conventional methods, we further demonstrated the VR-LSFM hybrid for revealing the endocardiac trabecular network in the cardiac myosin light chain–GFP–transgenic (cmlc2-gfp–transgenic) adult zebrafish (Figure 2). To elucidate user-directed movements navigating through the myocardial ridges, also known as trabeculae, we have shared our video depicting the zebrafish heart in panoramic view (Supplemental Video 2) (36). We further focused on the apical region of the ventricular cavity (Figure 2A) to navigate through the endocardial trabecular network (Figure 2, B–D). These muscular ridges invaginate into the endocardium to participate in contractile function and myocardial oxygen perfusion during development. In contrast to the 2D raw data (Figure 2E) and conventional 3D rendering (Figure 2F), the VR strategy readily allows us to experience muscular ridges from various perspectives. In this context, the VR-LSFM platform provides a biomechanical basis for the user to understand the role of trabeculation in cardiac contractile function (37). Using VR-LSFM, we were also able to measure the distance between the ventriculobulbar valve leaflets (Figure 2G) and the distance between the trabecular ridges in the endocardium of an adult zebrafish (Supplemental Video 3). This demonstration demonstrates the capacity of the VR-LSFM platform to allow for both interactive and quantitative measurements of the 3D digital heart. We listed the parameters for quantitative measurement and interactive visualization in Table 1.
Figure 2. Endocardial trabecular network in a transgenic Tg(cmlc2-gfp) zebrafish ventricle at 60 dpf.
(A) VR accentuates the invaginating muscular ridges in the apical region. (B–D) VR-LSFM enables navigation through various projections into the branching network. Different views are indicated by white arrows (B). (E and F) The conventional (E) 2D raw data and (F) 3D rendering results are limited in revealing the highly trabeculated 2-chambered heart, consisting of an atrium and a ventricle, as the perspective view is predefined. Scale bar: 100 μm. (G) Quantitative measurements of the distance between the ventriculobulbar valve leaflets. dpf, days after fertilization; VR, virtual reality; LSFM, light-sheet fluorescence microscopy. All of these images are shown in pseudocolor.
Table 1. Measurable parameters for VR-LSFM demonstrations.
Elucidating the spatial distribution of exogenous potassium channels.
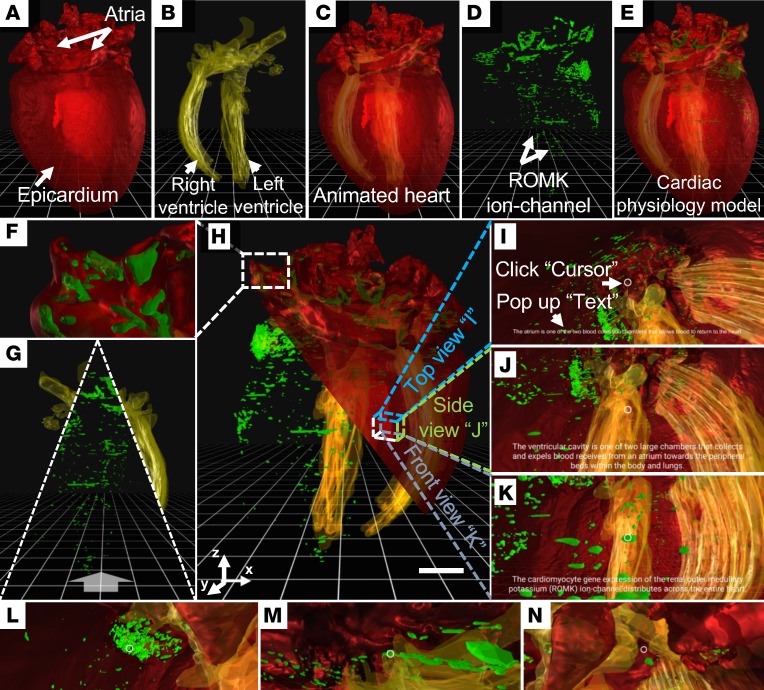
VR-LSFM additionally enabled us to detect the myocardial-specific expression of exogenous renal outer medullary potassium (ROMK) channels in the adult mouse following gene therapy (Figure 3, A–E). A 7.5-month-old male adult mouse (wild-type, C57BL/6) was imaged following intravenous injection of AAV9-ROMK-GFP in the tail at 2 months. LSFM allowed for the localization of the 3D distribution of the specific GFP-labeled ROMK expression in the myocardium, while VR enabled us to explore the distribution of exogenous potassium channels (Figure 3, F–H). We were able to have an interactive experience after segmenting the ventricular cavity (Figure 3, yellow) to contrast with the GFP-labeled potassium channels (Figure 3, green) in the myocardium. We also juxtaposed another half of the epicardium (Figure 3H, red) to visualize the surface topography and to zoom into the atrium and ventricle for the GFP-labeled potassium channels. Furthermore, the 3 images in Figure 3, I–K, interrogate the anatomic architecture from different perceptions. In addition, we were able to explore the individual regions described in the images: in the atrium (Figure 3I), in the ventricular cavity (Figure 3J), and among the ROMK channels (Figure 3K) (Supplemental Video 4). To visualize user-directed navigation through the ventricle to validate the outcome of gene therapy (Figure 3, L–N), we have shared our video depicting the mouse heart in panoramic view (38). We further measured the frame rate of the mouse heart model to test the interactive and immersive experience. In comparison with a uniform rate at 24 frames per second (FPS) for the cinematic films, our 60 FPS VR demonstration (Supplemental Video 5) supports that the current VR-LSFM platform is capable of producing a high FPS on current mobile devices.
Figure 3. 3D perception of the GFP-labeled ROMK channels in a representative adult mouse heart following gene therapy.
The 3D physical presences of (A) epicardium (red) and (B) ventricular cavity (yellow) are superimposed on the (C) animated heart. (D) 3D distribution of ROMK channels (green) is superimposed with C to provide a (E) physiological model. (F) The distribution of ROMK channels in the atrium. (G) The exploration of the animated ventricular cavity or ROMK channels. (H) This illustration integrates the epicardium (red), ventricular cavity (yellow), and ROMK channels (green) for an interactive and immersive experience. Scale bar: 1 mm. (I–K) VR-LSFM allows for reading instructive texts by clicking the 3D anatomic features of (I) the atrium, (J) the ventricular cavity, and (K) ROMK channels. (L–N) VR-LSFM enables zooming into the 3D anatomy to interrogate ROMK channels in relation to the ventricular cavity and myocardium. ROMK, renal outer medullary potassium; VR, virtual reality; LSFM, light-sheet fluorescence microscopy. All of these images are shown in pseudocolor.
Demonstrating 4D VR application for cardiac contractile function.
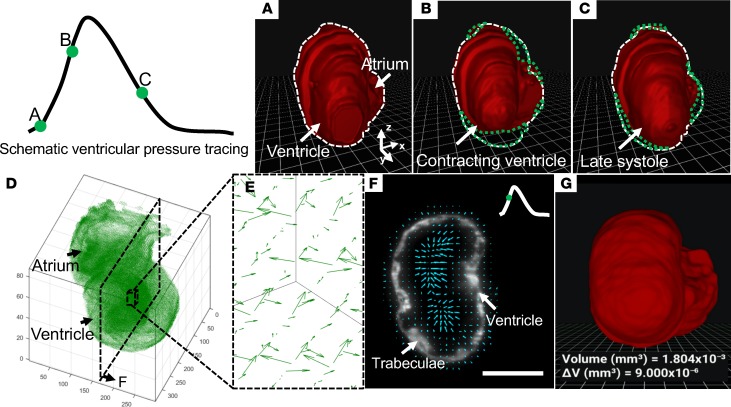
Developing a dynamic VR application based on the 4D scientific data were limited by the changes in anatomical structure that render visualization challenging over time. However, we were able to implement the mobile VR-LSFM platform for the 4D transgenic Tg(cmlc2:gfp) zebrafish line at approximately 120 hours after fertilization (Figure 4). First, we applied the LSFM technique to capture 300 sections from the rostral to caudal end of the zebrafish heart. Each section was captured with 300 xy planes (frames) at 10-ms exposure time per frame. After synchronizing all slices at various time points, we reconstructed approximately 50 digital hearts to demonstrate the entire cardiac cycle (systole + diastole). We established two approaches for generating the 4D segmentation results: (a) applying the BINS method to automatically process all time-variant images and subsequently building the 4D contracting heart or (b) applying the BINS method to segment one static 3D heart as a reference during the entire cardiac cycle and using DIR (39) to generate displacements between the reference heart and the moving hearts at subsequent time points. In reference to the first method, the contracting heart was recapitulated without pauses or foreshortening when operating on the smartphone (Supplemental Video 6). Three representative time points during a cardiac cycle were visualized by demarcating the epicardium with dashed lines at the representative slices (Figure 4, A–C). The second method allowed us to maintain time-dependent cardiac contraction, and the DIR approach enabled us to generate the displacement vectors to quantify the contractile function (Figure 4D), in which the length of the arrows indicated the magnitude and the tip of the arrows indicated the direction of displacement (Figure 4, E and F). Furthermore, we were able to quantify the volumetric changes occurring between consecutive frames (Figure 4G) in the contracting embryonic zebrafish heart (Supplemental Video 7). Thus, coupling quantitative measurements and interactive visualization advances the field of developmental cardiac mechanics and physiology at a microscale that was previously unattainable without the LSFM-acquired imaging data. We listed the parameters for quantitative measurement and interactive visualization in Table 1.
Figure 4. 4D perception of the contracting heart in a transgenic Tg(cmlc2:gfp) zebrafish embryo at 5 dpf.
(A–C) We demonstrate schematic ventricular pressure tracing and representative contractile images at 3 time points during a cardiac cycle. We depict the cardiac volume in white dashed lines in A as the baseline and accentuate the volumetric changes with green dotted lines (B and C) during the cardiac cycle. (D) Displacement vectors reveal the ventricular deformation in terms of direction and magnitude at an instantaneous moment in systole. (E) 3D and (F) 2D displacement vectors. (G) DIR-derived representative 4D scenario. VR-LSFM enables the quantification of the volumetric changes occurring between consecutive frames as the user operates on the 4D contracting heart. VR, virtual reality; LSFM, light-sheet fluorescence microscopy; DIR, deformable image registration. Scale bar: 50 μm. All of these images are shown in pseudocolor.
Discussion
Our VR-LSFM hybrid method establishes an efficient and robust platform to recapitulate cardiac mechanics and physiology. This framework builds on the high spatiotemporal resolution provided by LSFM to offer user-directed acquisition and visualization of life science phenomena, thereby facilitating biomedical research and learning via readily available smartphones and VR headsets. The application of BINS and DIR on LSFM-acquired data generates the input for a VR system, allowing for multiscale interactive use. The terminal platform composed of a smartphone and the Google Cardboard or Daydream viewer effectively reduces barriers to interpret the optical data, thereby paving the way for a virtual laboratory for discovering, learning, and teaching at single-cell resolution.
Our VR-LSFM hybrid platform holds promises for the next generation of microscale interactions with live models at the cellular and material interface. Elucidating the dynamics of cardiac development and repair would accelerate the fields of developmental biology. The VR-LSFM, in collaboration with BINS and DIR, enables us to provide both quantitative and qualitative insights into organ morphogenesis. This potential capacity allows for incorporating endocardial topography and elastography to simulate hemodynamic and myocardial contractile forces in our current applications. We have further recapitulated the 3D intracardiac trabecular network in the adult zebrafish model of injury and regeneration, allowing for the uncovering of hypertrabeculation and ventricular remodeling following ventricular injury and regeneration. In addition, we have localized the spatial distribution of ROMK channels in the adult murine myocardium, localizing the 3D spatial distribution of ion channels critical for contractile function following gene modulation/therapy. Thus, the VR-LSFM platform demonstrates the capacity for improving the understanding of developmental cardiac mechanics and physiology with an interactive and immersive method.
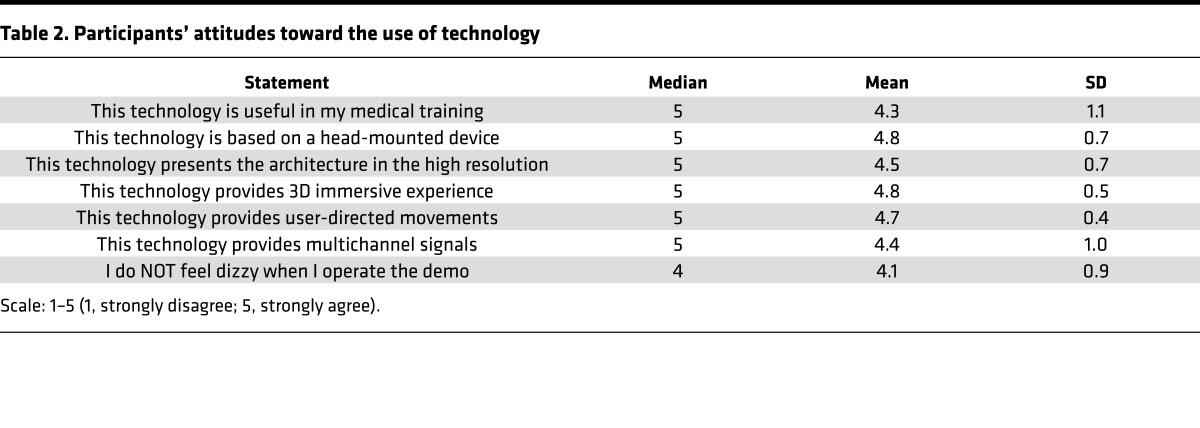
To validate the structures visualized by our VR-LSFM hybrid platform, we performed comparisons (Supplemental Figure 1) of LSFM imaging raw data, LSFM imaging segmented results, and histological reference images. The VR-LSFM–acquired imaging data demonstrate high spatial resolution as compared with the histological data. Furthermore, to evaluate our VR-LSFM platform, we have conducted independent interviews with participants who study or work at the David Geffen School of Medicine at UCLA. Altogether, 20 individuals took part in all interview sessions, with 16 men and 4 women ranging from 22 to 35 years old, with a median age of 30 years. The participants were interested in new technology for the future VR-LSFM imaging modality. To provide an introduction to the VR-LSFM methods to the participants, we asked the participants to test three VR-LSFM demonstrations via Google Cardboard combined with an Android smartphone, followed by responding to a questionnaire about demographic information and their general attitudes toward the visualization and interaction of these demonstrations. In addition, the participants were encouraged to comment on their opinions of the VR-LSFM method. Based on the results in Table 2, most participants underscored the integration of these interactive experiences as part of their medical training. VR-LSFM was unanimously deemed to be instrumental and informative to enhancing learning via the head-mounted device. Specifically, we believe the immersive experience of coupling anatomical structure with cardiac contractile function is novel for 4D user-directed operation with high spatiotemporal resolution. Some participants reported dizziness from their interactive and immersive experience. This type of experience is well-characterized to induce motion sickness in response in response to the VR field (40, 41), and we plan to enhance the frame rate to 90 FPS or even higher in our future exploitation.
Table 2. Participants’ attitudes toward the use of technology.
Furthermore, parallel advances in imaging technologies should lead to even better spatiotemporal resolution, higher image contrast, and less photobleaching as well as phototoxicity. Integration of Bessel beam (33) and 2-photon excitation (34) with our current cardiac LSFM will lead to better spatial resolution. Other imaging modalities (42–46), such as photoacoustic, computed tomography, and magnetic resonance imaging, are compatible with our developing workflow for the future translational VR platform of diagnosis and therapy. The methodology used for developing the VR-LSFM application will also be transferable to other VR head-mounted devices for enhancing user-interactivity features and improving the quantitative analysis. To elucidate cardiovascular architecture and function, we also need to perform precise segmentation of the fluorescently labeled tissues. Developing a novel convolutional or recurrent neural network (47–50) for automatic segmentation may benefit image post-processing procedures that are otherwise limited in accuracy and efficiency by manual segmentation in the settings of large data sets. In addition, optimization of the CLARITY method (51) is also critical for minimizing the risk of diminishing or losing fluorescence with prolonged clearing that is necessary for rodent models. Overall, we believe our framework will be transformative as it advances imaging and visualization, with great value to fundamental and translational research.
Methods
Animal models. An adult 7.5-month-old male mouse (wild-type, C57BL/6 without backcross) was imaged. This mouse was intravenously injected with 8.7 × 1012 viral genomes of adenoassociated virus vector 9 (AAV9; Vector Biolabs) in the tail at 2 months. This vector system employed a cardiac-specific troponin T promoter to drive cardiomyocyte gene expression of a customized construct (UCLA Cardiovascular Research Laboratory) in which the ROMK channel was fused at its C-terminus to one GFP molecule (AAV9-ROMK-GFP). The primary antibodies against GFP (sc-8334, Santa Cruz Biotechnology) and against ROMK (APC-001, Alomone Labs) are commercially available. The secondary antibody was Envision+ System-HRP anti-rabbit polyclonal antibody conjugated to HRP (K4002, Dako). Male zebrafish were bred and maintained at the UCLA Core Facility, the transgenic Tg(cmlc2:gfp) line was used for demonstrating the contracting heart and myocardial ridges.
Sample preparation. A HA backbone functionalized to contain acrylates was prereacted with K-peptide (Ac-FKGGERCG-NH2), Q-peptide (Ac-NQEQVSPLGGERCG-NH2), and RGD (Ac-RGDSPGERCG-NH2) at 500 μM, 500 μM, and 1,000 μM, respectively. The cross-linker solution was prepared by dissolving the dithiol matrix metalloproteinase–sensitive linker peptide (Ac-GCRDGPQGIWGQDRCG-NH2, Genscript) in distilled water at 7.8 mM and then immediately placed in a reaction with 10 μM Alexa Fluor 488–maleimide (Life Technologies) for 5 minutes to fluorescently label the final microgels. These two solutions merged into one channel and were immediately pinched by heavy mineral oil containing 1% span-80 surfactant. Overnight gelation occurred, and then the microgels were purified by repeated washing and centrifugation with buffer. HDFs (Life Technologies) were added to the microgels at a concentration of 1,500 cells/μl along with activated factor XIII at a concentration of 5 units/ml. The microgels were allowed to anneal for 90 minutes at 37°C. Two days after initial cell loading, the cells were fixed with 4% PFA at 4°C overnight and then stained for F-actin using a rhodamine B conjugate of Phalloidin (Life Technologies).
For the imaging of adult mouse and adult zebrafish hearts, the hearts immersed in the refractive index matching solution with 1% agarose were mounted in a Borosilicate glass tubing (Pyrex 7740, Corning) to eliminate refraction and reflection among various interfaces (Supplemental Figure 2 and Supplemental Table 1) (52). For the imaging of zebrafish embryos, the fish immersed in water with 0.5% agarose and 20× tricaine solution were mounted in a fluorinated ethylene propylene (FEP) tube to eliminate refraction and reflection among various interfaces. For the imaging of hydrogel, 20 μl of microgels immersed in 1× PBS with 0.5% agarose solution were injected into a FEP tube to eliminate refraction and reflection among various interfaces.
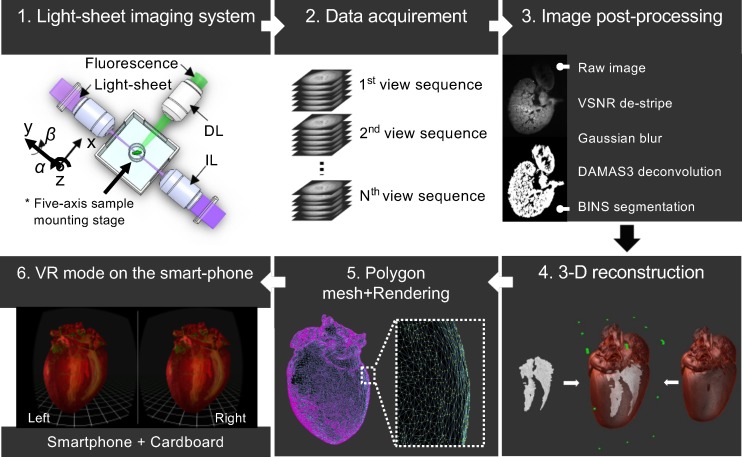
Developing an immersive VR environment. We illustrated the development of a mobile VR-LSFM hybrid platform in 6 steps (Figure 5) and also provide a time chart for representative procedurals (Supplemental Figure 3). All the software used in the following steps are not mandatory, and therefore ImageJ (NIH), Fiji, or ChimeraX (3) are also feasible.
Figure 5. Flow chart highlighting the process of constructing a VR application.
The development of a mobile VR-LSFM hybrid platform in 6 steps. IL, illumination lens; DL, detection lens; VSNR, variational stationary noise remover; DAMAS3, deconvolution approach for mapping the acoustic sources 3D; BINS, batch intensity normalized segmentation; VR, virtual reality.
In step 1, we applied in-house LSFM systems (26, 27, 29, 31) to acquire image stacks of cardiac events. For the large-scale samples, such as the adult mouse hearts and hydrogel scaffolds, dual-sided laser beams were applied to evenly illuminate the samples, followed by orthogonal fluorescent or autofluorescent detection (Supplemental Figure 4). In reference to multidirectional selective plane illumination microscopy (mSPIM) (19), we did not need to use the resonant mirrors to reduce absorption and scattering artifacts. Our systems have the capacity to image the entire cardiac specimen without the need for stitching the image columns in comparison with other systems for large-scale specimens (17, 18, 53, 54). In addition, we replaced the water-dipping objective lenses used in mSPIM with the dry objective lenses (26, 29, 55). Our light-sheet imaging systems enable us to increase the working distance with sufficient spatial resolution needed to track the organ development.
In step 2 (optional), we acquired the contracting hearts at the individual planes over 3~4 cycles (31), followed by a synchronization algorithm to address the varying periodicity.
In step 3, we performed pre- and post-processing of the raw data to construct the volumetric target in MATLAB (The MathWorks Inc.) and ImageJ. In the pre-processing step, we applied a variational stationary noise remover algorithm to remove artificial stripes from photon absorption and scattering (56, 57). Next, we applied the Gaussian blur method to normalize the distribution of brightness and contrast, followed by a deconvolution approach for mapping the acoustic sources 3D (DAMAS3) (58) to enhance the image contrast. In the post-processing step, we applied manual segmentation to label the region of interest (ROI) for the static hearts and hydrogel scaffolds and developed the BINS method (validation in the Supplemental Figure 5) for the contracting hearts in zebrafish embryos. In general, steps 1–3 are essential for obtaining the raw data with high spatiotemporal resolution and specific fluorescence for the final output.
In step 4, we constructed the 3D model for the individual ROI from the corresponding image stack with a priori information indicating the voxel size. Here, the 3D model was transfered from the 2D bitmap images, thereby bridging the current VR developmental engines with the fundamental imaging system. We established this step by Amira to control over the number of vertices, edges, and faces of the mesh so that we were able to validate the reduction of data complexity (Supplemental Figure 6). The data conversion is able to be performed in ImageJ, Fiji, and ChimeraX. For this reason, Amira is not the mandatory software.
In step 5, we edited the polygon mesh of the entire object to generate the scene, while maintaining the original anatomical structure to ensure the applicability of the model for the authentic output. We imported the 3D editable object into the VR developmental engine (a) to generate the motion path for the movement, (b) to retrieve a dynamic process for a living target, (c) to provide texts for manual instruction, and (d) to generate the panorama video for user-directed visualization. We finalized this step in Maya and Unity with educational licenses.
In step 6, we published all of the objects along with the scenes as a single installable application from the VR engine to the Android platform. This approach enabled us to operate in VR mode on the smartphone equipped with the Google VR viewer.
Design and construction of the imaging system. The details of the in-house LSFM systems have been covered by existing publications (26, 27, 29, 31). Our light-sheet imaging systems enable us to increase the working distance with sufficient spatial resolution needed to track the organ development. The light-sheet microscope was built using a continuous-wave laser (Laserglow Technologies) as the illumination source. The detection module was installed perpendicular to the illumination plane, and it was composed of the scientific CMOS (ORCA-Flash4.0) and a set of filters (Semrock). During scanning, the sample holder was oriented by a 5-axis mounting stage. Both illumination and detection modules were controlled by a computer with dedicated solid-state drive redundant array of independent disks level 0 storage for fast data streaming.
Computational algorithm of in vivo image synchronization. Briefly, the computational algorithm is composed of the following steps:
(a) Since the zebrafish heart rates were often not in synchrony with the scanning time, we minimized the difference in the least-square intensity with respect to the periodic hypothesis (T) by estimating the period of each subsequence. Three to four cardiac cycles were acquired in each slice. We adopted the following cost function (Equation 1) to estimate the period and correlate the same instantaneous moment in different periods:
(Equation 1),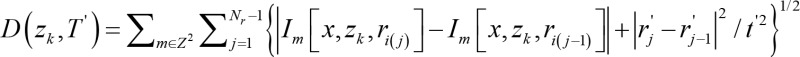
where D denotes the cost function of fitting a period hypothesis, Im denotes the captured image, x represents a vector denoting the pixel index, zk denotes the z-direction index of a slice, ri denotes the phase locked time positions of the acquired image, i(j) denotes a bijective mapping linking the actual acquisition time with the phase locked time position, Z2 is the entire imaging space, j=1 is the initial frame, Nr is the number of acquired frames, r’j denotes the estimate of the phase locked time positions, and t’ denotes a candidate period.
(b) We aligned two image sequences before the 3D structure reconstruction due to the idle time between slices. We formulated the relative shift (Equation 2) in the form of quadratic minimization:
(Equation 2),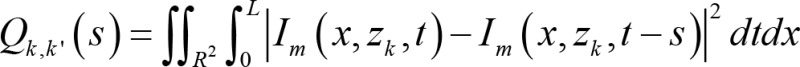
where Qk,k’ denotes a cost function of fitting a relative shift hypothesis, R denotes the possible spatial neighborhood, L represents the total time of image capturing, s represents the relative shift hypothesis, dt is interpreted as temporal infinitesimals, and dx are interpreted as spatial infinitesimals.
(c) We converted the relative shift to the absolute shift with respect to the first image sequence. Although relative shift could be used to align two adjacent frames, all of the frames were aligned to reconstruct the entire heart. Upon obtaining all relative shifts, a relation to align the next slice given the previous slice was built. This recurrent relation was transformed into an absolute relation by solving a linear equation using the pseudoinverse approach.
Post-processing methods and BINS.
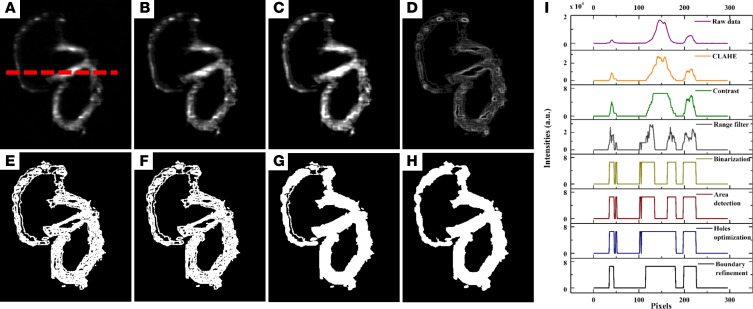
Manual segmentation in ImageJ remained the gold standard to label the ROI for post-processing the adult hearts and hydrogel scaffolds. To improve the efficiency of processing 4D time-variant image stacks, we developed an unattended pipeline known as BINS to segment the pericardium from the zebrafish embryos. The BINS algorithm was developed in MATLAB (The MathWorks Inc.), and all of its individual image enhancing and filtering operations were performed using a mix of functions that were either built-in or provided by the Image Processing and Statistics toolboxes. The BINS segmentation algorithm involved the application of a series of image enhancing and filtering functions on the raw data to yield a primary ROI consisting of a single slice of the zebrafish heart (Figure 6A). First, the contrast and intensity distribution of the image was optimized with contrast-limited adaptive histogram equalization using the adapthisteq function (Figure 6B). The image was processed in 64-row and 64-column tiles, with each tile analyzed by using a 36-bin uniform histogram. Furthermore, a contrast enhancement limit of 0.01 was applied (Figure 6C). Next, the local range of the image was found using rangefilt by determining the range value (maximum–minimum) of the 3-by-3 neighborhood around the corresponding pixels in the image (Figure 6D). The maximum and minimum values were defined by Equations 3 and 4, respectively.
Figure 6. BINS algorithm.
(A) Raw data of an embryonic zebrafish heart at 5 dpf. (B) Adaptive histogram equalization by CLAHE. (C) Contrast adjustment. (D) Range filter. (E) Binary by imbinarize. (F) Filtering the largest area. (G) Filling holes. (H) Subtracting outside boundary by inside boundary after localizing and filling the contour. (I) Pixel intensity values for A–H measured along the red dashed line in A. BINS, batch intensity normalized segmentation; CLAHE, contrast-limited adaptive histogram equalization.
(Equation 3),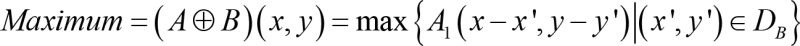
(Equation 4),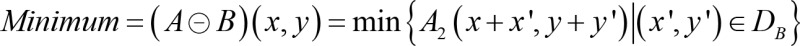
where DB is the domain of structuring element B, B(x, y) = 0; A1(x, y) and A2(x, y) are assumed to be –∞ and +∞ outside of the domain of the image, respectively, and A represents the pixel intensity measured in the grayscale images in Figure 6. Next, the image was binarized with Otsu’s method using the imbinarize function (Figure 6E). All of the regions in the image were detected by using the regionprops function, and all but the one with the largest area were filtered using the bwpropfilt function (Figure 6F). Small holes within this region were then filled using bwareaopen if their areas were smaller than the bottom fifth percentile (Figure 6G). Finally, the outer and inner boundaries of this region were extracted, refined, and used to define an continuous and unfragmented cardiac boundary (Figure 6H). The effectiveness of the BINS algorithm was illustrated in Figure 6, A–H, and as the pixel intensity along the red dashed line in all of the images in Figure 6I. To validate this method, we computed the dice similarity coefficients (59) to demonstrate the outcome of 0.92 ± 0.02, supporting excellent overlap after measuring 60 images (Supplemental Figure 5 and Supplemental Table 2).
Code availability. The computer code generated during and/or analyzed during the current study are available from the corresponding author.
Data availability. The data sets generated during and/or analyzed during the current study are available from the corresponding author.
Statistics. Data are presented as mean ± SD.
Study approval. All animal studies were performed in compliance with the IACUC protocol approved by the UCLA Office of Animal Research.
Author contributions
YD proposed the experiments and built the imaging system. AA developed the BINS method. YD and AA performed experiments and refined the VR applications. SL, JL, CCC, KIB, ES, YL, TPN, RPSP, and TS provided all the samples. JJH conducted the subjective test. AB and PF reviewed the process. YD, AA, PA, and TKH wrote the manuscript, with contributions from all authors. All authors reviewed the manuscript.
Supplementary Material
Acknowledgments
The authors thank Carlos Pedroza for the initial discussion on the VR technique, Rajan P. Kulkarni and Kevin Sung for the tissue clearing, Scott John for invaluable assistance with the preparation of the cardiac reporter gene, and C.-C. Jay Kuo and Hao Xu in the USC Media Communications lab for the computational algorithm. This work was supported by the NIH (5R01HL083015-11, 5R01HL118650-04, 5R01HL129727-03, 2R01HL111437-05A1, U54 EB022002, P41-EB02182), the UCLA Harvey Karp Discovery Award, and the American Heart Association (Scientist Development Grants 13SDG14640095 and 16SDG30910007 and Predoctoral Fellowship 15PRE21400019).
Version 1. 11/16/2017
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2017;2(22): e97180. https://doi.org/10.1172/jci.insight.97180.
Contributor Information
Yichen Ding, Email: ycding@g.ucla.edu.
Arash Abiri, Email: abiria@uci.edu.
Parinaz Abiri, Email: pabiri@ucla.edu.
Shuoran Li, Email: sandylsr89@gmail.com.
Chih-Chiang Chang, Email: Changc4@ucla.edu.
Kyung In Baek, Email: Qorruddls122@gmail.com.
Elias Sideris, Email: esideris@ucla.edu.
Yilei Li, Email: liyilui@gmail.com.
Juhyun Lee, Email: juhyunlee@ucla.edu.
Tatiana Segura, Email: tsegura@ucla.edu.
Peng Fei, Email: feipeng@hust.edu.cn.
References
- 1.Ntziachristos V, Ripoll J, Wang LV, Weissleder R. Looking and listening to light: the evolution of whole-body photonic imaging. Nat Biotechnol. 2005;23(3):313–320. doi: 10.1038/nbt1074. [DOI] [PubMed] [Google Scholar]
- 2.Scherf N, Huisken J. The smart and gentle microscope. Nat Biotechnol. 2015;33(8):815–818. doi: 10.1038/nbt.3310. [DOI] [PubMed] [Google Scholar]
- 3.Goddard TD, et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. doi: 10.1002/pro. [published online ahead of print July 14, 2017]. https://doi.org/10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng H, Ruan Z, Long F, Simpson JH, Myers EW. V3D enables real-time 3D visualization and quantitative analysis of large-scale biological image data sets. Nat Biotechnol. 2010;28(4):348–353. doi: 10.1038/nbt.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peng H, et al. Virtual finger boosts three-dimensional imaging and microsurgery as well as terabyte volume image visualization and analysis. Nat Commun. 2014;5:4342. doi: 10.1038/ncomms5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Theart RP, Loos B, Niesler TR. Virtual reality assisted microscopy data visualization and colocalization analysis. BMC Bioinformatics. 2017;18(Suppl 2):64. doi: 10.1186/s12859-016-1446-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Usher W, et al. A virtual reality visualization tool for neuron tracing. IEEE T Vis Comp Graph. doi: 10.1109/TVCG.2017. [published online ahead of print August 29, 107]. https://doi.org/10.1109/TVCG.2017.2744079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolf S, et al. Sensorimotor computation underlying phototaxis in zebrafish. Nat Commun. 2017;8(1):651. doi: 10.1038/s41467-017-00310-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hale KS, Stanney KM. Handbook of virtual environments: Design, implementation, and applications. Boca Raton, FL: CRC Press; 2014. [Google Scholar]
- 10.Steuer J. Defining virtual reality: Dimensions determining telepresence. J Comm. 1992;42(4):73–93. doi: 10.1111/j.1460-2466.1992.tb00812.x. [DOI] [Google Scholar]
- 11.Greenbaum P. The lawnmower man. Film and Video. 1992;9(3):58–62. [Google Scholar]
- 12.Yang JC, Chen CH, Jeng MC. Integrating video-capture virtual reality technology into a physically interactive learning environment for English learning. Comput Educ. 2010;55(3):1346–1356. doi: 10.1016/j.compedu.2010.06.005. [DOI] [Google Scholar]
- 13.Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EH. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305(5686):1007–1009. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- 14.Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EH. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science. 2008;322(5904):1065–1069. doi: 10.1126/science.1162493. [DOI] [PubMed] [Google Scholar]
- 15.Power RM, Huisken J. A guide to light-sheet fluorescence microscopy for multiscale imaging. Nat Methods. 2017;14(4):360–373. doi: 10.1038/nmeth.4224. [DOI] [PubMed] [Google Scholar]
- 16.Buytaert JA, Dirckx JJ. Design and quantitative resolution measurements of an optical virtual sectioning three-dimensional imaging technique for biomedical specimens, featuring two-micrometer slicing resolution. J Biomed Opt. 2007;12(1):014039. doi: 10.1117/1.2671712. [DOI] [PubMed] [Google Scholar]
- 17.Dodt HU, et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Methods. 2007;4(4):331–336. doi: 10.1038/nmeth1036. [DOI] [PubMed] [Google Scholar]
- 18.Santi PA, Johnson SB, Hillenbrand M, GrandPre PZ, Glass TJ, Leger JR. Thin-sheet laser imaging microscopy for optical sectioning of thick tissues. BioTechniques. 2009;46(4):287–294. doi: 10.2144/000113087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huisken J, Stainier DY. Even fluorescence excitation by multidirectional selective plane illumination microscopy (mSPIM) Opt Lett. 2007;32(17):2608–2610. doi: 10.1364/OL.32.002608. [DOI] [PubMed] [Google Scholar]
- 20.Mertz J, Kim J. Scanning light-sheet microscopy in the whole mouse brain with HiLo background rejection. J Biomed Opt. 2010;15(1):016027. doi: 10.1117/1.3324890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Vos WH, et al. Invited review article: Advanced light microscopy for biological space research. Rev Sci Instrum. 2014;85(10):101101. doi: 10.1063/1.4898123. [DOI] [PubMed] [Google Scholar]
- 22.Huisken J, Stainier DY. Selective plane illumination microscopy techniques in developmental biology. Development. 2009;136(12):1963–1975. doi: 10.1242/dev.022426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amat F, et al. Fast, accurate reconstruction of cell lineages from large-scale fluorescence microscopy data. Nat Methods. 2014;11(9):951–958. doi: 10.1038/nmeth.3036. [DOI] [PubMed] [Google Scholar]
- 24.Chhetri RK, Amat F, Wan Y, Höckendorf B, Lemon WC, Keller PJ. Whole-animal functional and developmental imaging with isotropic spatial resolution. Nat Methods. 2015;12(12):1171–1178. doi: 10.1038/nmeth.3632. [DOI] [PubMed] [Google Scholar]
- 25.Dodt HU, et al. Ultramicroscopy: development and outlook. Neurophotonics. 2015;2(4):041407. doi: 10.1117/1.NPh.2.4.041407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fei P, et al. Cardiac light-sheet fluorescent microscopy for multi-scale and rapid imaging of architecture and function. Sci Rep. 2016;6:22489. doi: 10.1038/srep22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guan Z, et al. Compact plane illumination plugin device to enable light sheet fluorescence imaging of multi-cellular organisms on an inverted wide-field microscope. Biomed Opt Express. 2016;7(1):194–208. doi: 10.1364/BOE.7.000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sideris E, et al. Particle hydrogels based on hyaluronic acid building blocks. ACS Biomater Sci Eng. 2016;2(8):2034–2041. doi: 10.1021/acsbiomaterials.6b00444. [DOI] [PubMed] [Google Scholar]
- 29.Ding Y, et al. Light-sheet fluorescence imaging to localize cardiac lineage and protein distribution. Sci Rep. 2017;7:42209. doi: 10.1038/srep42209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mickoleit M, et al. High-resolution reconstruction of the beating zebrafish heart. Nat Methods. 2014;11(9):919–922. doi: 10.1038/nmeth.3037. [DOI] [PubMed] [Google Scholar]
- 31.Lee J, et al. 4-Dimensional light-sheet microscopy to elucidate shear stress modulation of cardiac trabeculation. J Clin Invest. 2016;126(5):1679–1690. doi: 10.1172/JCI83496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Y, et al. Spatially isotropic four-dimensional imaging with dual-view plane illumination microscopy. Nat Biotechnol. 2013;31(11):1032–1038. doi: 10.1038/nbt.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen BC, et al. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science. 2014;346(6208):1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Truong TV, Supatto W, Koos DS, Choi JM, Fraser SE. Deep and fast live imaging with two-photon scanned light-sheet microscopy. Nat Methods. 2011;8(9):757–760. doi: 10.1038/nmeth.1652. [DOI] [PubMed] [Google Scholar]
- 35.Krzic U, Gunther S, Saunders TE, Streichan SJ, Hufnagel L. Multiview light-sheet microscope for rapid in toto imaging. Nat Methods. 2012;9(7):730–733. doi: 10.1038/nmeth.2064. [DOI] [PubMed] [Google Scholar]
- 36. Hsiai-Lab. Adult Zebrafish Heart. YouTube. https://youtu.be/HwBRnamD4PA Published April 21, 2017. Accessed October 31, 2017.
- 37.Chang CP, Bruneau BG. Epigenetics and cardiovascular development. Annu Rev Physiol. 2012;74:41–68. doi: 10.1146/annurev-physiol-020911-153242. [DOI] [PubMed] [Google Scholar]
- 38. Hsiai-Lab. Mouse Heart. YouTube. https://youtu.be/eoDKijdQokQ Published April 21, 2017. Accessed October 31, 2017.
- 39.Varadhan R, Karangelis G, Krishnan K, Hui S. A framework for deformable image registration validation in radiotherapy clinical applications. J Appl Clin Med Phys. 2013;14(1):4066. doi: 10.1120/jacmp.v14i1.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lawson B. Changes in subjective well-being associated with exposure to virtual environments. Usab Eval Interf Design. 2001;1:1041–1045. [Google Scholar]
- 41.Kennedy RS, Drexler J, Kennedy RC. Research in visually induced motion sickness. Appl Ergon. 2010;41(4):494–503. doi: 10.1016/j.apergo.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 42.Weissleder R, Pittet MJ. Imaging in the era of molecular oncology. Nature. 2008;452(7187):580–589. doi: 10.1038/nature06917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding Y, et al. Laser oblique scanning optical microscopy (LOSOM) for phase relief imaging. Opt Express. 2012;20(13):14100–14108. doi: 10.1364/OE.20.014100. [DOI] [PubMed] [Google Scholar]
- 44.Lu Y, et al. An integrated quad-modality molecular imaging system for small animals. J Nucl Med. 2014;55(8):1375–1379. doi: 10.2967/jnumed.113.134890. [DOI] [PubMed] [Google Scholar]
- 45.Ding Y, et al. In vivo study of endometriosis in mice by photoacoustic microscopy. J Biophotonics. 2015;8(1-2):94–101. doi: 10.1002/jbio.201300189. [DOI] [PubMed] [Google Scholar]
- 46.Wang LV, Hu S. Photoacoustic tomography: in vivo imaging from organelles to organs. Science. 2012;335(6075):1458–1462. doi: 10.1126/science.1216210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lawrence S, Giles CL, Tsoi AC, Back AD. Face recognition: a convolutional neural-network approach. IEEE Trans Neural Netw. 1997;8(1):98–113. doi: 10.1109/72.554195. [DOI] [PubMed] [Google Scholar]
- 48. Krizhevsky A, Sutskever I, and Hinton GE. ImageNet Classification with Deep Convolutional Neural Networks. Adv Neur Inform Proc Systems. 2012:1097–1105. https://papers.nips.cc/paper/4824-imagenet-classification-with-deep-convolutional-neural-networks Accessed November 8, 2017.
- 49. doi: 10.1109/TPAMI.2016.2572683. Long J, Shelhamer E, Darrell T. Fully Convolutional Networks for Semantic Segmentation. Proc IEEE Conf Comp Vis Patt Recog. 2015:3431–3440. [DOI] [PubMed] [Google Scholar]
- 50. Zheng S, et al. Conditional Random Fields as Recurrent Neural Networks. Proc IEEE Intl Conf Comp Vision. 2015:1529–1537. [Google Scholar]
- 51.Murray E, et al. Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. Cell. 2015;163(6):1500–1514. doi: 10.1016/j.cell.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sung K, et al. Simplified three-dimensional tissue clearing and incorporation of colorimetric phenotyping. Sci Rep. 2016;6:30736. doi: 10.1038/srep30736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buytaert JA, Dirckx JJ. Tomographic imaging of macroscopic biomedical objects in high resolution and three dimensions using orthogonal-plane fluorescence optical sectioning. Appl Opt. 2009;48(5):941–948. doi: 10.1364/AO.48.000941. [DOI] [PubMed] [Google Scholar]
- 54.Buytaert JA, Descamps E, Adriaens D, Dirckx JJ. The OPFOS microscopy family: High-resolution optical sectioning of biomedical specimens. Anat Res Int. 2012;2012:206238. doi: 10.1155/2012/206238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomer R, et al. SPED light sheet microscopy: Fast mapping of biological system structure and function. Cell. 2015;163(7):1796–1806. doi: 10.1016/j.cell.2015.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fehrenbach J, Weiss P, Lorenzo C. Variational algorithms to remove stationary noise: applications to microscopy imaging. IEEE Trans Image Process. 2012;21(10):4420–4430. doi: 10.1109/TIP.2012.2206037. [DOI] [PubMed] [Google Scholar]
- 57.Fehrenbach J, Weiss P. Processing stationary noise: Model and parameter selection in variational methods. SIAM J Imag Sci. 2014;7(2):613–640. doi: 10.1137/130929424. [DOI] [Google Scholar]
- 58. Brooks TF, Humphreys WM. Extension of DAMAS phased array processing for spatial coherence determination (DAMAS-C). NTRS: NASA Technical Reports Server. https://ntrs.nasa.gov/search.jsp?R=20060020679 Published January 1, 2006. Accessed October 31, 2017.
- 59.Zou KH, et al. Statistical validation of image segmentation quality based on a spatial overlap index 1: Scientific reports. Acad Radiol. 2004;11(2):178–189. doi: 10.1016/S1076-6332(03)00671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.