Abstract
Growers often use fungicide sprays during bloom to protect crops against disease, which exposes bees to fungicide residues. Although considered "bee-safe," there is mounting evidence that fungicide residues in pollen are associated with bee declines (for both honey and bumble bee species). While the mechanisms remain relatively unknown, researchers have speculated that bee-microbe symbioses are involved. Microbes play a pivotal role in the preservation and/or processing of pollen, which serves as nutrition for larval bees. By altering the microbial community, it is likely that fungicides disrupt these microbe-mediated services, and thereby compromise bee health. This manuscript describes the protocols used to investigate the indirect mechanism(s) by which fungicides may be causing colony decline. Cage experiments exposing bees to fungicide-treated flowers have already provided the first evidence that fungicides cause profound colony losses in a native bumble bee (Bombus impatiens). Using field-relevant doses of fungicides, a series of experiments have been developed to provide a finer description of microbial community dynamics of fungicide-exposed pollen. Shifts in the structural composition of fungal and bacterial assemblages within the pollen microbiome are investigated by next-generation sequencing and metagenomic analysis. Experiments developed herein have been designed to provide a mechanistic understanding of how fungicides affect the microbiome of pollen-provisions. Ultimately, these findings should shed light on the indirect pathway through which fungicides may be causing colony declines.
Keywords: Environmental Sciences, Issue 128, Colony collapse disorder, metagenomics, microbial diversity, microbiome, pollen, yeast
Introduction
Managed and wild bee species are experiencing widespread declines, with major implications for both natural and agricultural systems1. Despite concerted efforts to understand the causes of this problem, the factors driving honey bee declines are still not well understood2,3,4. For certain species of wild, native bees, the situation has become dire5,6. If bee populations cannot be sustained when they intersect with industrial agriculture, their populations will continue to fall, and the crops requiring pollinators (35% of worldwide production7) will endure reduced harvests.
While many potential factors such as pesticide exposure, disease, and habitat loss1,4,8,9,10 have been implicated in honey bee decline, relatively little is known about the interactive effect of these stressors on native bees health, within or near agricultural systems. Many current research efforts continue to focus on insecticides, (e.g., neonicotinoids11,12), although past research indicates that fungicides may also play a role in bee decline by impairing memory formation, olfactory reception13, nest recognition14, enzyme activity and metabolic functions15,16,17. Globally, fungicides continue to be applied to flowering crops during bloom. Recent studies have documented that bees commonly bring fungicide residues back to the hive18, indeed, studies have shown a large proportion of tested hives contained fungicide residues19,20. Further work has revealed that fungicide residue is associated with high rates of honey bee larval mortality21,22,23 and the presence of "entombed pollen" within colonies, which although non-toxic, is devoid of microbial activity and is nutritionally compromised24. Despite the fact that fungicides have long been considered "bee-safe," there is now evidence that exposure to fungicide alone can cause severe colony losses in a native bumble bee species, Bombus impatiens25.
To establish causality between fungicide exposure and colony mortality, the modus operandi of these chemicals need to be determined. As evidenced in soils26, sediments27, and aquatic environments28, by targeting fungi, fungicides most likely alter fungal abundance and diversity within pollen-provisions, thereby invoking a major community shift that may strongly favor bacteria. Without fungal competitors or antagonists, certain pathogenic bacteria can proliferate relatively unchecked, facilitating the spoilage of pollen-provisions. Past research has demonstrated that microorganisms, particularly yeasts and filamentous fungi, serve as nutritional symbionts for bees29,30,31, protect against parasites and pathogens32,33, and provide long-term preservation of pollen stores. Fungicides, therefore, may indirectly harm immature bees by disrupting the microbial community that is needed to provide these services and/or by increasing susceptibility to opportunistic pathogens and parasites12. With increasing demands on food production, crops worldwide are being sprayed each year with fungicides during bloom, underscoring the need to understand the magnitude of such fungicide-induced effects.
To-date, the primary knowledge gaps relating to native bee microbial ecology can be represented by the following questions: To what extent does fungicide change the microbial community within bee pollen-provisions? What are the downstream impacts of consuming pollen with a profoundly altered microbial community? In keeping with these ecologically germane questions, experiments were developed with the primary goals of revealing 1) that fungicide residue alone can cause severe colony decline in a native bee species; 2) the degree to which microbial communities in pollen-provisions are altered by fungicides, and 3) how bee health is affected by a severely altered microbial community. The experimental objectives were defined to address the above questions using a combination of laboratory- and field-based experiments. Using state-of-the-art metagenomic and molecular techniques alongside traditional methods of field observation, this research aims to piece together the potential effects of fungicides on bee health.
The first objective of this study is to demonstrate that fungicide exposure alone can cause significant colony losses among native bee species. A study involving large field cages was used to investigate the effects of fungicide exposure on the colony growth of Bombus impatiens, a ubiquitous, abundant native bee in the US (Figure 1, Figure 2, Figure 3). It was hypothesized that fungicide-treated hives would present lower fitness, and atypical demography compared to non-exposed hives. Data obtained from this experiment supported this hypothesis, demonstrating that fungicide residues within pollen can be the sole cause of profound colony losses in a native bumble bee species25. The second objective of this study is to investigate the response of the pollen microbiome to fungicide exposure. It is hypothesized that the community composition of microbes within pollen-provisions exposed to fungicides will be different from that of untreated pollen. While fungal abundance and diversity are expected to decline significantly, bacteria and/or a single dominant fungal species will likely grow unchecked in the absence of other competing fungi. Through a series of in-vivo trials, these shifts in microbial community composition will be analyzed using metagenomics.



Protocol
1. Examine the Effect of Fungicide Exposure on Bumble Bee Colony Success Using Field Cage Experiments
Set up ten mesh cages in a field planted with oats. Dig a trench around each cage, and dig all four edges of the mesh cage into the ground to ensure that bees cannot escape. Stock the cages with potted, flowering plants that are known to be attractive to bees (e.g. buckwheat, borage, alyssum, cosmos, and sunflowers) (Figure 2).
Supplement the cages with a single tray (36 cm x 42 cm) of in-bloom clover. Cluster floral resources within one corner of the cage, occupying a space approximately 2.5 m x 1 m. Vegetate the remaining cage area by the oats.
Randomly assign the bumble bee colonies, each containing workers and a single queen, a treatment (fungicide present/absent, N = 5 colonies per treatment, 10 colonies total), then place them within a field cage (N = 1/cage) for 29 days (23 June - 21 July 2014).
Orient the colony boxes such that the colony's openings point to the south to provide the bees with optimal navigational conditions. Subsidize the colonies with sugar water bladders, placed inside the hive boxes to supplement nectar availability.
Apply chlorothalonil-based fungicide at a field-relevant level (20 g/L) to flowering plants in the five fungicide treatment cages, using a hand held pesticide sprayer, twice during the study (day 0 and 13). Coat the flowers uniformly such that no further liquid adheres to floral surfaces (Figure 3).
At the conclusion of the field cage study, remove the B. impatiens colonies from the cages by hand, cool the hives by placing in -20 °C freezer for 20 min.
Remove bees using sterile forceps and record the number of larvae, pupae, adult females (i.e. foragers), and adult males. Using an analytical balance record the dry-weight of the mother queen, larvae, pupae, adult females (i.e. foragers), and adult males.
2. Examine the Effects of Fungicide Exposure on Microbial Communities in the Pollen-provisions of Bumble Bee Nests Using Laboratory Based In Vivo Trials
- Pulverize commercially purchased pollen to a fine powder using a standard laboratory ball-mill. Sterilize powdered pollen by soaking in 70% ethanol, allowing it to evaporate overnight under UV light. Verify sterility of pollen by plating ~0.5 mg on general-purpose agar media.
- To the dry, sterilized pollen, using sterilized pipettes, add field relevant dosage of fungicide: propiconazole at 14.3%; azoxystrobin at 22.9% for treatments (0.74 µL and 0.65 µL respectively/ day/ hive). Mix well using sterilized wooden sticks.
- Place 6 experimental hives (n = 3 each for control and treatment) in a clean, hygienic laboratory benchtop maintained at room temperature. Each day weigh 4.27 g of pollen34,35 mixed with fungicides (for treatments) or sterile water (for control) inside a hood using standard aseptic technique.
- Using the trap doors provided by the side of the cardboard box enclosing the hives, introduce the pollen inside the hives. Supplement the hives with sterilized sugar solution each week. Continue feeding regime for four weeks.
At the conclusion of the lab-based study, cool the hives by placing in -20 °C freezer for 20 min. Scrape out the pollen-provisions contained within brood chambers using sterilized forceps and spatulas and place in sterile storage tubes. Store at -80 °C. Count and record the weight of the workers and queen mother at the start and end of the experiment (step 1.7).
- Isolate DNA from pollen-provision sample using commercially available DNA isolation kits (see Materials Table for details).
- Add 0.25 g of pollen-provision to extraction tubes, briefly vortex to mix.
- Make a 200 mg/mL lysozyme solution in deionized distilled water, enough for 50 µL/sample. Shake vigorously to completely form solution.
- Add 50 µL of the lysozyme solution to the extraction tubes with sample in it, and mix well by inverting several times. Incubate the tube for 10 min at 37 °C in a water bath. If precipitate has formed, heat solution to 60 °C until dissolved before use.
- Add 70 µL of aqueous lysis solution to extraction tubes, secure horizontally on a flatbed vortex pad with tape, and vortex for 10 min. Centrifuge tubes at 10,000 x g for 30 s at room temperature.
- Transfer the supernatant to a clean 2 mL collection tube. Add 250 µL of protein precipitation solution, and vortex for 5 s. Incubate at 4 °C for 5 min in an ice bath. NOTE: Expect between 400 µL to 500 µL of supernatant. Supernatant may still contain some particles.
- Centrifuge the tubes at room temperature for 1 min at 10,000 x g, and transfer up to 600 µL of supernatant to a clean 2 mL collection tube. Add 200 µL of aqueous inhibitor removal solution, vortex briefly, and incubate at 4 °C for 5 min. Centrifuge the tubes at room temperature for 1 min at 10,000 x g. Transfer up to 750 µL of supernatant into a clean 2 mL collection tube.
- Add 1200 µL of aqueous bind solution to the supernatant, and vortex for 5 s. Load approximately 675 µL of the supernatant onto a spin filter, and centrifuge at 10,000 x g for 1 min at room temperature. Discard the flow through.
- Repeat 2.4.7. twice. NOTE: A total of three loads for each sample processed are required.
- Add 500 µL ethanol, and centrifuge at room temperature for 30 s at 10,000 x g. Discard the flow through, and centrifuge again at room temperature for 1 min at 10,000 x g. Place spin filter in a clean 2 mL collection tube.
- Add 100 µL of elution buffer to the center of the filter membrane. Centrifuge at room temperature for 30 s at 10,000 x g. Discard the spin filter. Store collected DNA between -20 °C and -80 °C
- Use isolated DNA for sequencing.
- Quantify, and normalize isolated DNA to 2 ng/µL by fluorometric analysis.
- Prepare reactions in triplicate for each extracted sample to compare the relative amounts of 28S (plant), analyze ITS (fungal), and 16S (bacterial) components of each pollen-provision sample36. Ensure that each reaction contains 10 ng of total DNA, 2x of asymmetrical cyanine dye based master mix, and 2.5 µL each of forward and reverse primers pairs 28KJ/28B37 for plant and ITS1/ITS5.8R for fungal DNA38.
- Amplify DNA using the following parameters: 2 min pre-denaturation at 50 °C, 2 min initial denaturation at 95 °C, 40 cycles of (15 s at 98 °C, 15 s at 58 °C, 60 s at 72 °C), followed by a melt curve.
- Prepare a 2-step, nested PCR protocol using next-generation sequencing libraries targeting the 16S rRNA V3/V4 variable region and ITS/5.8s rRNA spacer region.
- Perform initial amplification using the following parameters: 3 min initial denaturation at 95 °C, 25 cycles of (30 s at 95 °C, 30 s at 55 °C, 30 s at 72 °C), 5 min final extension at 72 °C.
- Following initial amplification, verify library size, and quantity by electrophoretic mobility, and clean using a 1x volume solid phase reversible immobilization beads to remove residual primers and reaction reagents. Pool 16S and ITS amplicons quantitatively to create a single amplicon pool for each sample.
- Add sequencer specific adapters and sample specific dual indexes using the following primers (see Table 1). Add 2.5 µL of amplified DNA to 5 pmol of each primer and 2x master mix. Perform library amplification using the following parameters: 3 min initial denaturation at 95 °C, 8 cycles of (30 s at 95 °C, 30 s at 55 °C, 30 s at 72 °C), 5 min final extension at 72 °C.
- Following PCR, clean finished libraries using a 1 x volume of solid phase reversible immobilization beads. Assess quality and quantity of the finished libraries using electrophoretic mobility, and fluorometry, respectively. Standardize libraries to 2 µM and pool prior to sequencing.
- Perform next-generation sequencing on a platform analogous to the adapters added in the secondary PCR, with an appropriate length to cover the entire amplicon41.
- Sequence annotation and microbial composition analysis.
- Combine pair-end sequencing data (R1 and R2) into single contigs for all sequenced libraries. After merging both R1 and R2 files, a single fasta file for each of the libraries is produced. NOTE: This step and the processes described below (unless otherwise indicated) are performed in Mothur version 1.38.038.
- Screen each fasta file to remove ambiguous bases, unexpected long sequences, and long homopolymers. NOTE: The parameters for screening, and sequence removal are maxambig = 0, maxlength = 600, and maxhomop = 8 for both genes. Remove identical sequences, but keep a contingence table separately for all the libraries (a.k.a. count table in Mothur).
- Align unique sequences of the gene 16S library against the SILVA database version 123, and the gene ITS library against the UNITE ITS database42. NOTE: Sequences must be annotated from the kingdom level to the genus level.
- Subsequently, cluster aligned sequences with the function pre.cluster using diff=5, and remove chimeras.
- Classify sequences using the method Wang43 based on the SILVA (for gene 16S) and UNITE ITS (for gene ITS) taxonomy files (cutoff value of 80%).
- Load into R44 the contingence tables (one per gene) generated during the classification process in Mothur. At each taxonomic level, obtain relative abundance per library for further analysis of microbial community changes. NOTE: At each taxonomic level, merge together taxonomic groups when relative abundance is less than 2% for all the experimental repetitions.
Representative Results
Field cage study:
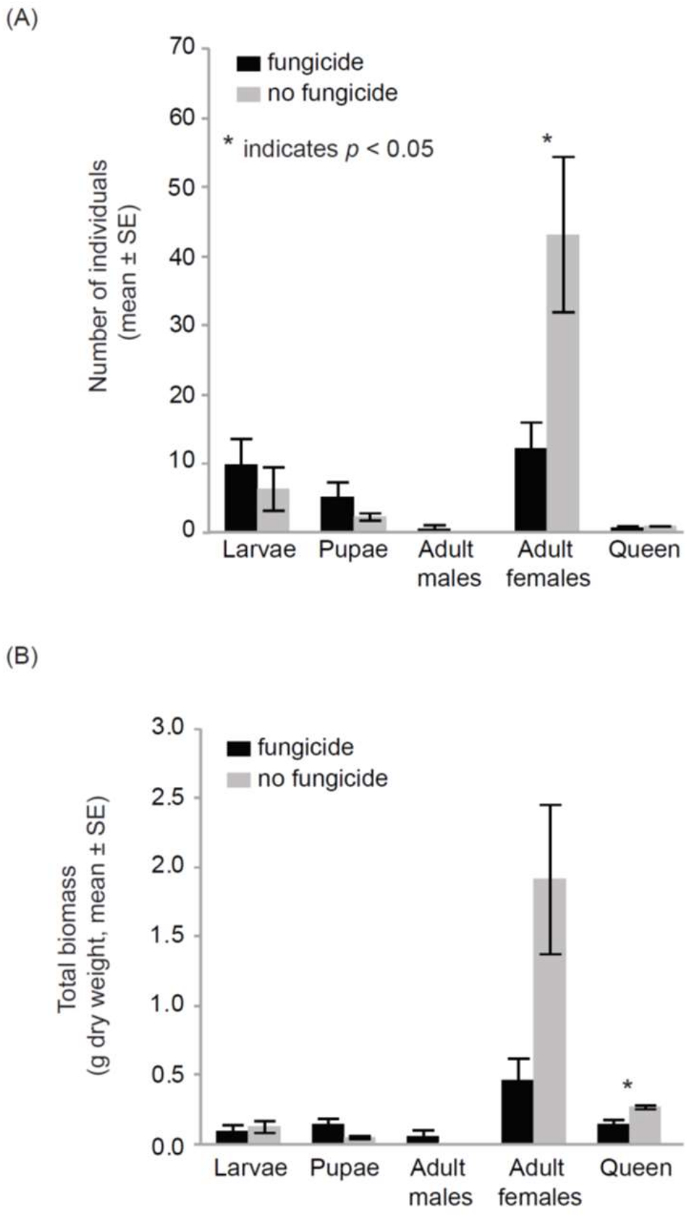
Data obtained from the cage experiments showed that the bumble bee colonies had a significant response to fungicide exposure. The fungicide-treated hives produced significantly fewer workers (12.2 ± 3.8, mean ± SE) than the control hives (43.2 ± 11.2, F1,9= 6.8, p = 0.03) (Figure 4). Additionally, the bee biomass of the fungicide-treated hives (0.91 g ± 0.15) was significantly lower than the control hives (2.36 g ± 0.55; F1,9 = 8.3, p = 0.02). This pattern of lowered biomass in fungicide-treated hives was also observed among the mother queens. The queens of the fungicide-treated hives presented with a significantly lower biomass (0.14 g ± 0.04) than the queens of the control hives (0.27 g ± 0.01; Z = 2.5, p = 0.01). However, fungicide exposure did not affect the number of larvae, pupae, and males across the treatments. The biomass of discrete life stages (larvae, pupae, workers, and adult males) and individual weights for the larvae, pupae, and workers did not show any difference across fungicide-treated and control hives.
Lab-based study:
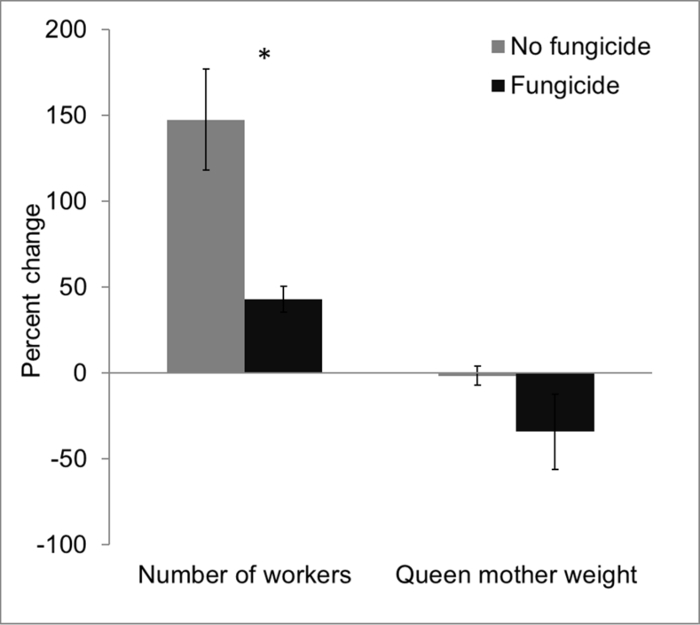
Data obtained from laboratory-based experiments indicated that prior to fungicide exposure, control and fungicide-treated hives had statistically comparable mean worker count (control hives = 28.0 ± 3.1; fungicide-treated hives = 31.67 ± 2.0; n = 3 each) and queen weight (control hives = 0.77 g ± 0.04; fungicide-treated hives = 0.74 g ± 0.01). However, at the end of the study, worker count was significantly higher in control hives (67.67 ± 4.3), compared to fungicide-treated hives (45.33 ± 3.8) (t4 = 3.89, p = 0.01). Worker population increased by ~150% in control hives compared to ~45% in fungicide-treated hives. Similarly, final weight of queen mother remained relatively unchanged in control hives (0.76 g ± 0.02) compared to a ~35% decrease in fungicide-treated hives (0.49 g ± 0.16). Taken together, these results are consistent with previously published results25 and indicate fungicide exposure affected colony fitness as evidenced by fewer worker numbers and reduced queen mother weights (Figure 5).
Metagenomic analysis of pollen-provisions indicated distinct differences between the microbial communities collected from fungicide-treated and control hives (Figure 6). There was a decrease (>95%) in the relative abundance of commonly isolated Streptomycetales, Enterobacteriales in the fungicide-treated hives. Interestingly, both these groups are known for their antifungal activity and role in pollen-preservation30,45 within bumblebee hive environments. Bacterial members of the order Rickettsiales, which includes common pathogens of arthropods46, showed much greater abundance in the fungicide treated hives. Fungicide-treated hives had a lower abundance of fungi belonging to the orders Eurotiales and Sordariales, and a higher abundance of orders Capnodiales and Ascosphaerales compared to control hives (Figure 7). As expected, for both bacteria and fungi, Shannon's diversity index (H) and evenness (E) was lower in fungicide-treated hives compared to controls, although these differences were not statistically significant (bacteria: Hcontrol hives =1.25 ± 0.3, Hfungicide-treated hives = 0.82 ± 0.2, Econtrol hives = 0.46 ± 0.1; Efungicide-treated hives = 0.31 ± 0.1; fungi: Hcontrol hives =0.99 ± 0.3, Hfungicide-treated hives = 0.72 ± 0.4, Econtrol hives = 0.57 ± 0.2; Efungicide-treated hives = 0.42 ± 0.2). Although detailing the metabolic and functional implications of such community shifts was beyond the scope of this study, combined with the colony count, and weight data, these results suggest that fungicide exposure could affect decline in colony health by disrupting the symbiosis between bees and the pollen microbiome. Further investigation into the role of specific microbial groups affecting larval survivorship is recommended to better evaluate the role of the pollen microbiome in sustaining healthy bee populations.
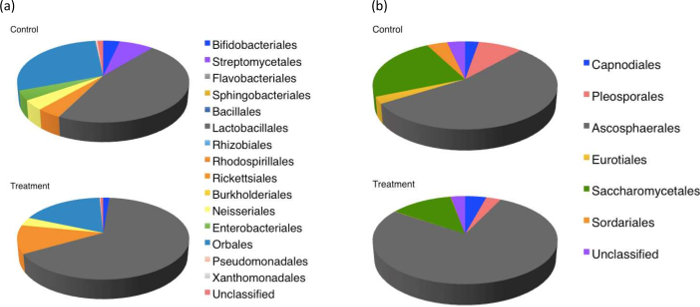
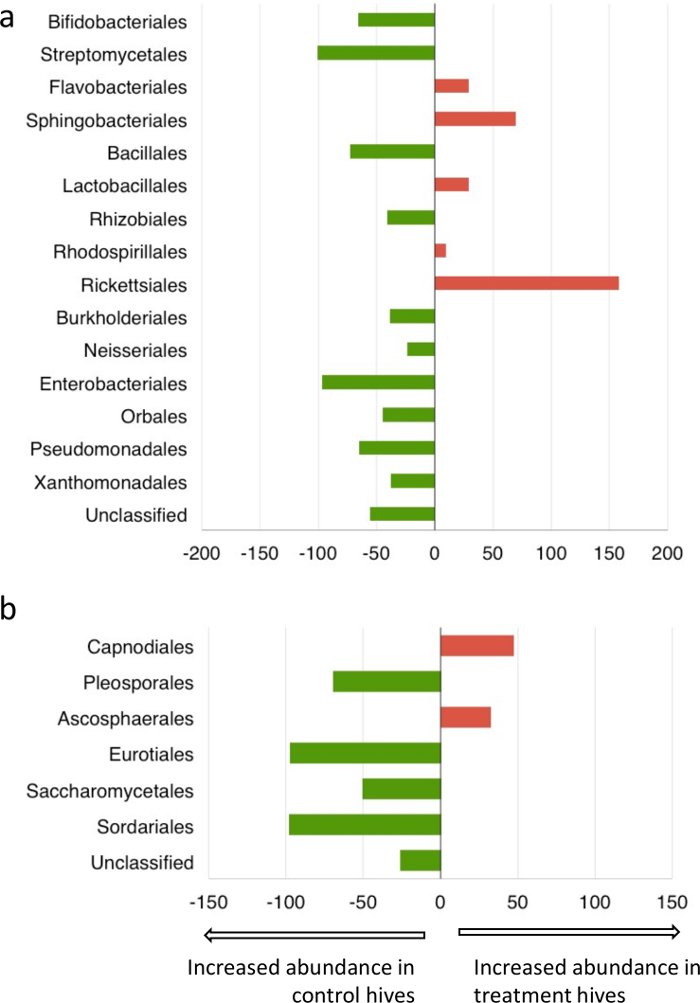
Figure 1: Inside a Bombus impatiens nest. High-resolution image of the workers tending to brood cells. Brood cells can contain several eggs and larvae at any given time. Developing larvae feed on the pollen-provisions periodically introduced into the chamber. Please click here to view a larger version of this figure.
Figure 2: Large field cages erected to house the bee colonies. Each mesh cage (N = 10) was stocked with one commercially purchased Bombus impatiens hive, as well as in-bloom plant species known to attract bees. Flowers were stocked in one corner of the cage and the remaining area was vegetated with oat grass. Please click here to view a larger version of this figure.
Figure 3: Fungicide residue on a recently sprayed sunflower. Field relevant doses of fungicide were sprayed on day 0 and 13 of the experiment. Using a pesticide sprayer, the flowers were evenly coated with a fungicide solution at dusk/evening to avoid direct contact with foraging bees. Please click here to view a larger version of this figure.
Figure 4: Impacts of fungicide residue on bumble bees in cage experiment. Bumble bee colonies that were exposed to fungicide residues on pollen exhibited significant declines in colony size (reductions in adult female abundance) over the course of a single month. Error bars represent ± 1SE; p <0.05. This Figure was modified from Bernauer et al.25. Please click here to view a larger version of this figure.
Figure 5: Impacts of fungicide residue on bumble bees in a laboratory-based experiment. Bumble bee colonies that were exposed to fungicide-treated pollen exhibited significant declines in colony size (reductions in adult worker abundance) and decline in queen mother weight over the course of a single month. Error bars represent ± 1SE; p = 0.03. Please click here to view a larger version of this figure.
Figure 6: Pie chart of metagenomic classification of fungal and bacterial diversity at the order the rank. Analyses based on (a) 16S and (b) ITS based classification of pollen-provision samples collected from control and fungicide-treated hives. Control hives demonstrated higher diversity and evenness in microbial distribution. Please click here to view a larger version of this figure.
Figure 7: Percent change in order rank relative abundance of bacteria and fungi in response to fungicide exposure. Metagenomic classification of microbial communities using (a) 16S and (b) ITS based primers Please click here to view a larger version of this figure.
| Primer pair | Primer type | Primer sequence |
| 28KJ/28B | Plant | GGC GGT AAA TTC CGT CC/ |
| CGT CCG TGT TTC AAG ACG | ||
| ITS1/ ITS5.8 | Fungal | TCC GTA GGT GAA CCT GCG G/ |
| GAG ATC CGT TGT TGA AAG TT | ||
| 16s forward/ reverse | Nested bacterial | 5’- ACACTCTTTCCCTACACGACGCTCTTCCGATCTCCTACGGGNGGCWGCAG-3’/ |
| 5’- GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTGACTACHVGGGTATCTAATCC-3’ | ||
| ITS1F primer/ ITS4 | Nested fungal | 5’- ACACTCTTTCCCTACACGACGCTCTTCCGATCTTTCGTAGGTGAACCTGCGG - 3’ |
| 5’-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTCCTCCGCTTATTGATATGC - 3’ | ||
| Adapter primers | 5’-AATGATACGGCGACCACCGAGATCTACAC[55555555] ACACTCTTTCCCTACACGACGCTCTTCCGATCT-3’ | |
| 5’-CAAGCAGAAGACGGCATACGAGAT[77777777] GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT -3’ |
Table 1: List of primer pairs and primer sequences used in DNA amplification. See text for references.
Discussion
Investigations into the effects of fungicides on bee health have remained an understudied aspect of pest management strategies. Our study aims to bridge this knowledge gap by using a suite of complementary techniques that explicitly isolate the potential factors driving bee declines. The planning, rationale, and rendering of these experiments are detailed below.
It is important to ensure that no bees are allowed to escape the mesh of the cage experiments, since this would compromise demography analysis. It is also important that the artificial nests have sufficient insulation to protect from rain and direct sunlight. Care should be taken so that fungicides are not sprayed directly on the nests. The colonies should be provided with sugar water bladders to supplement nectar gathered and prevent dehydration.
For the laboratory based feeding trials, strict aseptic conditions should be maintained to prevent contamination when preparing the pollen meals. Commercially purchased pollen must be powdered and UV sterilized prior to use. Hive hygiene must be maintained to prevent infestation from extraneous parasites such as pantry moths and roaches. Fluid bladders should be supplemented with sterilized sugar solution to prevent dehydration. For pre-treatment census, care should be taken not to the stress the bees by excessive handling and keeping them outside the hived for prolonged periods.
As DNA yield depends on the age and type of starting material, only fresh or well-frozen samples must be used in DNA extraction and amplification. Sample weight for DNA extraction should not exceed 0.25 gm, as this will lower DNA yield. DNA yields should be quantified using gel electrophoresis. Upfront electrophoretic quantification of isolated DNA is critical before proceeding to qPCR reactions to ensure efficient amplification47.
For the metagenomic analysis, a more comprehensive analysis of microbial dynamic can be obtained by limiting the amount of annotated sequences (by relaxing the fraction of tolerable differences between sequences) and reducing the number of taxonomic groups, as well as merging those for which relative abundance is considerably low (<2%).
The conservative sample size (N = 5 for both fungicide treated and untreated cage study hives) can inflate the variances in data, which will be minimized in future studies by using greater replication. To add greater resolution and statistical power to the results, the bee colonies will be censused and weighed before and after fungicide exposure. Running the cage experiment for longer periods will allow the production of new queens, which can serve as an additional response variable. With these modifications to the current protocol, future studies will provide greater details about hive population dynamics.
The hydrophobic nature of pollen deters its efficient mixing with water. Pulverizing and sifting the pollen granules to a fine powder, aids in the mixing process. The amount of the pollen powder can be adjusted to achieve the desired consistency. Care should be taken to thoroughly mix the fluid (water or fungicide) to achieve a homogenous distribution of active ingredients and even delivery to the foragers.
As excess DNA can inhibit PCR reactions, it is recommended that sample should not weigh more than 0.25 g. Excessive vortexing should be avoided as this shears DNA, lowering yield. Low DNA yield can also be a result of prolonged exposure to the lysis buffer. Bacterial primers should be chosen preferably from the V7-V9 region of 16S rRNA to minimize non-specific amplification of chloroplast DNA48. In addition, sequencing of other genes, such as the 18S ribosomal RNA gene can provide a better understanding of the microbial diversity in pollen, as well as the population changes upon fungicide utilization.
Given the amount of libraries to process with Mothur, computing time can be considerably high. Removing singletons when comparing sequences against each other (if computing resources are limited, remove them before chimera filtration) can considerably decrease the required time for computing operational taxonomic units.
The cage experiments confine the natural flight range of the bees (as opposed to allowing them to forage widely across the landscape), and it is unclear the degree to which such restrictions can influence the response variables. Although the control and treatment cages experience the same range of biotic and abiotic variables, it is logistically impossible to control for the microenvironment within each cage.
The true scope of within-community microbial interactions is far too complex and intricate to replicate through experimental trials. While our data likely document a subset of total microbial diversity within pollen-provisions, it is the first insight into the interactive effects that may prevail between bacteria and fungi in the presence/absence of fungicides.
Using alternative databases for aligning sequences49 or predicting functional metabolic states based on pollen bacterial diversity50 can provide a wider view of the pollen microbiome. Eventually, to better characterize the metabolic and functional dynamic of microbial communities after fungicide application, whole genome sequencing of pollen microbiome is necessary.
Cage experiments using commercially purchased hives provide one of the best frameworks to measure response variables in a semi-controlled setting. Causing minimal alteration to the bees' natural ecology (foraging efficiency, social structure, offspring care), cage experiments minimize the contrivance and stress, introduced by the artificial environment and manual handling during laboratory-based experiments.
To the best of our knowledge, no study has yet been conducted to assess the effects of fungicides on microbial community dynamics within the pollen-provision of bumble bees. Exposure to fungicides may eliminate one or more sensitive species, disrupting the ecological truce between fungi and bacteria. Using field and lab-based trials as a crucible to play out these interactions, this study aims to demonstrate that extermination of resident microbial species can pervert the ecological balance of the pollen microbiome, which may in turn compromise bee health.
Culture-dependent techniques such as dilution and plating on standard media capture only a fraction of the microflora from a given environment. This can lead to a gross underrepresentation of microbial diversity, and unculturable microbes may go undetected51. To study the breadth of microorganisms, scientists must rely on culture-independent techniques, which are more inclusive and comprehensive, for e.g. metagenomic analysis and next-generation sequencing. Drawing heavily from these powerful molecular techniques, this study aims to obtain greater resolution into the pollen microbiome than provided by traditional culture-based techniques alone.
Results from this study have revealed profound losses in the number of workers within bumble bee colonies exposed to fungicide. For a bumble bee colony to be successful, the workers need to provide sufficient resources and care for the mother-queen and particularly for developing larvae. Ultimately, the goal of the colony is to maximize resource capture (pollen and nectar) such that as many daughter-queens as possible can be produced by the end of summer. Healthy daughter-queens are the measure of fitness for a colony. Major reductions in workers, then, effectively undercuts the colony's ability to produce queens for the following year. In this study, not only were worker numbers significantly decreased, but the mother-queens from the fungicide treated hives presented with lower biomass, presumably as a result of insufficient food supply by the foragers. Given the complexity of microbial interactions within pollen-provisions, it is hard to discern the exact interactive effects between fungi and bacteria contributing to these patterns. However, results derived from replicated and repeated experiments as described here, will provide vital insights into the shifting symbioses between these two microbial groups (whether competitive, mutualistic, or commensal) in response to xenobiotic stress, and its downstream effects on the pollen microbiome.
Through the use of powerful molecular tools, the ecological complexity of the pollen microbiome can be better resolved. The preliminary understanding reveals a plethora of naturally occurring bacteria and fungi, contributing collectively to maintain colony fitness. In particular, the yeasts that were isolated from pollen-provisions are known to bear bactericidal and fermentative properties, both of which are crucial for the development of larval bees. Such ecological associations are suggestive of a high degree of mutualism between bees and their pollen microbiome. Pollen-provisions, therefore, represent an emergent diversity effect, brought about by the cultivation of a microbial community comprised of key functional roles. In the absence of these key symbionts, the pollen-provision appears to be compromised to unknown degrees. Continued work in this direction, will likely explain the functional diversity of these microbes and unmask their role as bee symbionts.
Recent research has debunked the notion of fungicides as being harmless to bees19,24,52,53,54. The objective of this research is to isolate the mechanisms driving fungicide impacts on bees, thereby illuminating the cause-effect relationship between fungicide use and bee declines. The hypotheses center around the concept that bee-microbe symbioses are being altered significantly by fungicide residues in pollen. Ultimately, this work will re-frame how fungicides are viewed and used in agriculture and urban environments. In light of the current global pollinator crisis, the proposed research generates new knowledge on native bee ecology, as it relates to agricultural production practices. Fungicides remain the last agrichemical group to be effectively exempted from regulations limiting applications to crops during bloom. As a result, both managed and wild bees are consistently exposed to fungicides. A growing body of literature suggests that, although fungicides are not lethal to bees on contact, at elevated doses, exposure is quite detrimental, leading to higher larval mortality21 and altered forager behavior. It is expected that this work will illuminate the mechanism(s) by which fungicides are harming pollinators. Furthermore, this research will form the basis of wiser pest management policy and inform growers of best management practices. This work will also be instrumental in delivering guidelines for improved pesticide spraying, which may influence pesticide laws and regulations. More broadly, these findings will be relevant to any managed ecosystem in which flowering plants are visited by pollen-collecting insects. With global estimates of bee-specific pollination services being extraordinarily high55,56, if this work can mitigate the decline of bee populations, the potential impacts will be substantial.
Disclosures
The authors have nothing to disclose.
Acknowledgments
The author(s) thank the University of Wisconsin Biotechnology Center DNA Sequencing Facility for providing amplification and sequencing facilities and services, Caitlin Carlson, Jennifer Knack, Jake Otto, and Max Haase for providing technical assistance with molecular analysis. This work was supported by USDA-Agricultural Research Service appropriated funds (Current Research Information System #3655-21220-001). Further support was provided by the National Science Foundation (under Grant No. DEB-1442148), the DOE Great Lakes Bioenergy Research Center (DOE Office of Science BER DE-FC02-07ER64494), and the USDA National Institute of Food and Agriculture (Hatch project 1003258). C.T.H. is a Pew Scholar in the Biomedical Sciences and an Alfred Toepfer Faculty Fellow, supported by the Pew Charitable Trusts and the Alexander von Humboldt Foundation, respectively. The authors thank Hannah Gaines-Day and Olivia Bernauer for their contributions to our earlier, published field work.
References
- Potts SG, Biesmeijer JC, Kremen C, Neumann P, Schweiger O, Kunin WE. Global pollinator declines: Trends, impacts and drivers. Trends Ecol Evolut. 2010;25(6):345–353. doi: 10.1016/j.tree.2010.01.007. [DOI] [PubMed] [Google Scholar]
- Vanengelsdorp D, Meixner MD. A historical review of managed honey bee populations in Europe and the United States and the factors that may affect them. J Invertebr Pathol. 2010;103(Suppl 1):S80–S95. doi: 10.1016/j.jip.2009.06.011. [DOI] [PubMed] [Google Scholar]
- Ellis JD, Evans JD, Pettis J. Colony losses, managed colony population decline, and Colony Collapse Disorder in the United States. J. Apic. Res. 2010;49(1):134–136. [Google Scholar]
- Vanbergen AJ Insect Pollinators Initiative. Threats to an ecosystem service: pressures on pollinators. Front Ecol Environ. 2013;11(5):251–259. [Google Scholar]
- Cameron SA, et al. Patterns of widespread decline in North American bumble bees. Proc. Natl. Acad. Sci. U.S.A. 2011;108(2):662–667. doi: 10.1073/pnas.1014743108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo ND, Colla SR, Wagner DL, Gall LF, Kerr JT. Do pathogen spillover, pesticide use, or habitat loss explain recent North American bumblebee declines? Conser Lett. 2012;5(3):232–239. [Google Scholar]
- Klein A-M, et al. Importance of pollinators in changing landscapes for world crops. Proc R Soc Lond [Biol] 2007;274(1608):303–313. doi: 10.1098/rspb.2006.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Bayo F, Goulson D, Pennacchio F, Nazzi F, Goka K, Desneux N. Are bee diseases linked to pesticides? - A brief review. Environ Int. 2016;89:7–11. doi: 10.1016/j.envint.2016.01.009. [DOI] [PubMed] [Google Scholar]
- Kwong WK, Moran NA. Gut microbial communities of social bees. Nature Rev. Microbiol. 2016;14(6):374–384. doi: 10.1038/nrmicro.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel P, et al. The Bee Microbiome: Impact on Bee Health and Model for Evolution and Ecology of Host-Microbe Interactions. mBio. 2016;7(2):e02164–e02115. doi: 10.1128/mBio.02164-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry M, et al. Science. 6079. Vol. 336. New York, N.Y: 2012. A common pesticide decreases foraging success and survival in honey bees; pp. 348–350. [DOI] [PubMed] [Google Scholar]
- Pettis JS, vanEngelsdorp D, Johnson J, Dively G. Pesticide exposure in honey bees results in increased levels of the gut pathogen Nosema. Die Naturwissenschaften. 2012;99(2):153–158. doi: 10.1007/s00114-011-0881-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson SM, Wright GA. Exposure to multiple cholinergic pesticides impairs olfactory learning and memory in honeybees. J. Exp. Biol. 2013;216(10):1799–1807. doi: 10.1242/jeb.083931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artz DR, Pitts-Singer TL. Effects of fungicide and adjuvant sprays on nesting behavior in two managed solitary bees, Osmia lignaria and Megachile rotundata. PLoS ONE. 2015;10(8) doi: 10.1371/journal.pone.0135688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RM, Wen Z, Schuler MA, Berenbaum MR. Mediation of Pyrethroid Insecticide Toxicity to Honey Bees (Hymenoptera: Apidae) by Cytochrome P450 Monooxygenases. J. Econ. Entomol. 2006;99(994):1046–1050. doi: 10.1603/0022-0493-99.4.1046. [DOI] [PubMed] [Google Scholar]
- Pilling ED, Bromleychallenor KAC, Walker CH, Jepson PC. Mechanism of synergism between the pyrethroid insecticide lambda-cyhalothrin and the imidazole fungicide prochloraz, in the honeybee (Apis mellifera L) Pest Biochem Physiol. 1995;51(1):1–11. [Google Scholar]
- Iwasa T, Motoyama N, Ambrose JT, Roe RM. Mechanism for the Differential Toxicity of Neonicotinoid Insecticides in the Honey Bee Mechanism for the differential toxicity of neonicotinoid insecticides in the honey bee, Apis mellifera. Crop Protection. 2016.
- Mullin CA, et al. High levels of miticides and agrochemicals in North American apiaries: implications for honey bee health. PLoS One. 2010;5(3):e9754. doi: 10.1371/journal.pone.0009754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettis JS, Lichtenberg EM, Andree M, Stitzinger J, Rose R, Vanengelsdorp D. Crop pollination exposes honey bees to pesticides which alters their susceptibility to the gut pathogen Nosema ceranae. PLoS One. 2013;8(7):e70182. doi: 10.1371/journal.pone.0070182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David A, et al. Widespread contamination of wildflower and bee-collected pollen with complex mixtures of neonicotinoids and fungicides commonly applied to crops. Environ Int. 2016;88:169–178. doi: 10.1016/j.envint.2015.12.011. [DOI] [PubMed] [Google Scholar]
- Zhu W, Schmehl DR, Mullin CA, Frazier JL. Four common pesticides, their mixtures and a formulation solvent in the hive environment have high oral toxicity to honey bee larvae. PLoS One. 2014;9(1):e77547. doi: 10.1371/journal.pone.0077547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Delso N, Martin GS, Bruneau E, Minsart LA, Mouret C, Hautier L. Honeybee colony disorder in crop areas: The role of pesticides and viruses. PLoS ONE. 2014;9(7) doi: 10.1371/journal.pone.0103073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MG, Blitzer EJ, Gibbs J, Losey JE, Danforth BN. Negative effects of pesticides on wild bee communities can be buffered by landscape context. Proc R Soc Lond [Biol] 2015;282(1809) doi: 10.1098/rspb.2015.0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Engelsdorp D, et al. "Entombed Pollen": A new condition in honey bee colonies associated with increased risk of colony mortality. J Invertebr Pathol. 2009;101(2):147–149. doi: 10.1016/j.jip.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Bernauer OM, Gaines-Day HR, Steffan SA. Colonies of bumble bees (Bombus impatiens) produce fewer workers, less bee biomass, and have smaller mother queens following fungicide exposure. Insects. 2015;6(2):478–488. doi: 10.3390/insects6020478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu CM. Effect of fungicides, captafol and chlorothalonil, on microbial and enzymatic activities in mineral soil. J Environ Sci Health B. 1993;28(B28):67–80. doi: 10.1080/03601239309372815. [DOI] [PubMed] [Google Scholar]
- Huang C-Y, Ho C-H, Lin C-J, Lo C-C. Exposure effect of fungicide kasugamycin on bacterial community in natural river sediment. J Environ Sci Health B. 2010;45(5):485–491. doi: 10.1080/03601231003800412. [DOI] [PubMed] [Google Scholar]
- Artigas J, et al. Effects of the fungicide tebuconazole on microbial capacities for litter breakdown in streams. Aquat. Toxicol. 2012;122:197–205. doi: 10.1016/j.aquatox.2012.06.011. [DOI] [PubMed] [Google Scholar]
- Goerzen DW. Microflora associated with the alfalfa leafcutting bee, Megachile rotundata (Fab) (Hymenoptera: Megachilidae) in Saskatchewan, Canada. Apidologie. 1991;22(5):553–561. [Google Scholar]
- Anderson KE, Sheehan TH, Eckholm BJ, Mott BM, DeGrandi-Hoffman G. An emerging paradigm of colony health: Microbial balance of the honey bee and hive (Apis mellifera) Insectes Sociaux. 2011;58(4):431–444. [Google Scholar]
- Crotti E, et al. Microbial symbionts of honeybees: a promising tool to improve honeybee health. N. Biotechnol. 2013;30(6):716–722. doi: 10.1016/j.nbt.2013.05.004. [DOI] [PubMed] [Google Scholar]
- Koch H, Schmid-Hempel P. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc. Natl. Acad. Sci. U.S.A. 2011;108(48):19288–19292. doi: 10.1073/pnas.1110474108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KE, et al. Microbial ecology of the hive and pollination landscape: bacterial associates from floral nectar, the alimentary tract and stored food of honey bees (Apis mellifera) PloS One. 2013;8(12):e83125. doi: 10.1371/journal.pone.0083125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans EC, Spivak M. Effects of Honey Bee (Hymenoptera: Apidae) and Bumble Bee (Hymenoptera: Apidae) Presence on Cranberry (Ericales: Ericaceae) Pollination. J Econ Entomol. 2006;99(3):614–620. doi: 10.1603/0022-0493-99.3.614. [DOI] [PubMed] [Google Scholar]
- Goulson D, et al. Can alloethism in workers of the bumblebee, Bombus terrestris, be explained in terms of foraging efficiency? Anim. Behav. 2002;64(1):123–130. [Google Scholar]
- ThermoFisher Scientific. User Guide: Qubit dsDNA HS Assay Kits. 2010. pp. 3–6. Available from: https://tools.thermofisher.com/content/sfs/manuals/Qubit_dsDNA_HS_Assay_UG.pdf.
- Khadempour L, LeMay V, Jack D, Bohlmann J, Breuil C. The Relative Abundance of Mountain Pine Beetle Fungal Associates Through the Beetle Life Cycle in Pine Trees. Microbial Ecol. 2012;64(4):909–917. doi: 10.1007/s00248-012-0077-z. [DOI] [PubMed] [Google Scholar]
- Dorn-In S, Hölzel CS, Janke T, Schwaiger K, Balsliemke J, Bauer J. PCR-SSCP-based reconstruction of the original fungal flora of heat-processed meat products. Int J Food Microbiol. 2013;162(1):71–81. doi: 10.1016/j.ijfoodmicro.2012.12.022. [DOI] [PubMed] [Google Scholar]
- Klindworth A, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41(1) doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications. 1990;18(1):315–322. [Google Scholar]
- Illumina. 16S Metagenomic Sequencing Library Preparation: Preparing 16S Ribosomal RNA Gene Amplicons for the Illumina MiSeq System. 2017. Available from: http://ngs.biodiv.tw/NGSCore/wp-content/uploads/Documents/16s-metagenomic-library-prep-guide-15044223-b.pdf.
- Kõljalg U, et al. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol. 2013;22(21):5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team RC. R: A language and environment for statistical computing [Computer software] Vienna, Austria: R Foundation for Statistical Computing; 2015. [Google Scholar]
- Kaltenpoth M, Engl T. Defensive microbial symbionts in Hymenoptera. Funct Ecol. 2014;28(2):315–327. [Google Scholar]
- Gerth M, Saeed A, White JA, Bleidorn C. Extensive screen for bacterial endosymbionts reveals taxon-specific distribution patterns among bees (Hymenoptera, Anthophila) FEMS Microbiol Ecol. 2015;91(6) doi: 10.1093/femsec/fiv047. [DOI] [PubMed] [Google Scholar]
- Smith CJ, Osborn AM. Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology. FEMS Microbiol Ecol. 2009;67(1):6–20. doi: 10.1111/j.1574-6941.2008.00629.x. [DOI] [PubMed] [Google Scholar]
- Kim M, Morrison M, Yu Z. Evaluation of different partial 16S rRNA gene sequence regions for phylogenetic analysis of microbiomes. J Microbiol Methods. 2011;84(1):81–87. doi: 10.1016/j.mimet.2010.10.020. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, et al. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MGI, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnol. 2013;31(9):814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Beer M, Megharaj M, Naidu R. The use of molecular techniques to characterize the microbial communities in contaminated soil and water. Environ Int. 2008;34(2):265–276. doi: 10.1016/j.envint.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Ladurner E, Bosch J, Kemp WP, Maini S. Assessing delayed and acute toxicity of five formulated fungicides to Osmia lignaria and Apis mellifera. Apidologie. 2005;36(3):449–460. [Google Scholar]
- Huntzinger CI, James RR, Bosch J, Kemp WP. Fungicide Tests on Adult Alfalfa Leafcutting Bees (Hymenoptera: Megachilidae) J Econ Entomol. 2008;101(4):1088–1094. doi: 10.1603/0022-0493(2008)101[1088:ftoaal]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Gradish AE, Scott-Dupree CD, Shipp L, Harris CR, Ferguson G. Effect of reduced risk pesticides for use in greenhouse vegetable production on Bombus impatiens (Hymenoptera: Apidae) Pest Manag. Sci. 2010;66(2):142–146. doi: 10.1002/ps.1846. [DOI] [PubMed] [Google Scholar]
- Calderone NW. Insect pollinated crops, insect pollinators and US agriculture: trend analysis of aggregate data for the period 1992-2009. PloS One. 2012;7(5):e37235. doi: 10.1371/journal.pone.0037235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollerton J, Winfree R, Tarrant S. How many flowering plants are pollinated by animals? Oikos. 2011;120(3):321–326. [Google Scholar]


