Abstract
DNA damage is a common phenomenon for each cell during its lifespan, and is defined as an alteration of the chemical structure of genomic DNA. Cancer therapies, such as radio- and chemotherapy, introduce enormous amount of additional DNA damage, leading to cell cycle arrest and apoptosis to limit cancer progression. Quantitative assessment of DNA damage during experimental cancer therapy is a key step to justify the effectiveness of a genotoxic agent. In this study, we focus on a single cell electrophoresis assay, also known as the comet assay, which can quantify single and double-strand DNA breaks in vitro. The comet assay is a DNA damage quantification method that is efficient and easy to perform, and has low time/budget demands and high reproducibility. Here, we highlight the utility of the comet assay for a preclinical study by evaluating the genotoxic effect of olaparib/temozolomide combination therapy to U251 glioma cells.
Keywords: Cellular Biology, Issue 128, Chemo-sensitivity, glioma, DNA damage, comet assay, single cell electrophoresis assay, chemotherapy, genotoxicity, temozolomide, olaparib, PARP
Introduction
The comet assay was first developed by Ostling and Johanson in 1984 by demonstrating the migration of DNA fragments from nuclei under a neutral condition1. The technique was later developed by Singh et al., showing that an alkaline condition substantially increased the specificity and reproducibility of the assay2. Since then, the neutral comet assay is mostly used to detect double-stranded DNA breaks, whereas the alkaline comet assay is more sensitive for smaller amounts of DNA damage, including single and double strand DNA breaks, alkali-labile sites, DNA-DNA or DNA-protein cross-linking, and DNA single-strand breaks associated with incomplete excision repair sites3,4. Both assays allow visualization of fragmented DNA, and provide a straightforward way to quantitatively evaluate DNA damage. The comet assay is regarded as a sensitive method for in vitro and in vivo genetic toxicological studies and is applicable to different research areas, such as early drug-candidate selection, environmental monitoring, human biomonitoring, and fundamental research in DNA damage and repair5.
The principle of the assay is that under an electric field, fragmented DNA migrates out of the nucleoid body (also known as the "comet head") and forms a DNA stain in the agarose gel (also known as the "comet tail"). With nucleotide staining, the extent of DNA damage can be quantified by analyzing "comets" formed by this single cell electrophoresis. Calculation of the tail moment can further help to compare DNA damage among different experimental groups. Compared with traditional methods of DNA damage detection, the comet assay is direct, sensitive, inexpensive, and relatively simple.
Radiotherapy and chemotherapies are common strategies for cancer treatment by generating single strand and double strand DNA breaks in chromosomes6. The recent advancement in DNA repair inhibitors allows a more effective genotoxic effect by combination chemotherapy, and therefore, potentially reduces systemic side effects such as anemia, infection, and bone marrow suppression7,8. In this study, we showed the investigation of a poly (ADP-ribose) polymerase (PARP) inhibitor, olaparib (Ola)9. PARP is an abundant nuclear protein and is responsible for DNA base excision repair by forming a poly (ADP-ribose) polymer10. Temozolomide (TMZ) is an orally available alkylating agent and has been widely used for glioma patient treatment. By using the comet assay to quantify DNA damage, we demonstrate that combining olaparib with temozolomide profoundly enhances the DNA damage in glioma cells, which suggests olaparib/temozolomide combination therapy is an effective strategy to treat glioma, as compared with temozolomide alone11.
Protocol
1. Prepare Reagents
- 1x PBS
- Dilute 100 mL 10x PBS with 900 mL dH2O and adjust the pH to 7.4 using a pH meter. Store at room temperature.
- Lysis solution (LS)
- Prepare 2.5 M NaCl, 100 mM disodium EDTA, 10 mM Tris base, and 200 mM NaOH in 900 mL dH2O; it commonly takes about 20 min to allow the mixture to fully dissolve. Adjust the pH to 10 using a pH meter. Add 1% sodium lauryl sarcosinate and 1% Triton X-100, and adjust the final volume to 1,000 mL. Cool to 4 °C for at least 30 min before use.
- Alkaline electrophoresis solution (AES), pH >13
- Prepare 200 mM NaOH and 1 mM disodium EDTA in 800 mL dH2O. Adjust the pH and make sure that it is pH >13. Adjust the final volume to 1,000 mL. Make fresh before use and cool to 4 °C for at least 30 min before use.
- Neutral electrophoresis sollution (NES)
- Prepare 1,000 mL neutral electrophoresis buffer by mixing 100 mM Tris base and 300 mM sodium acetate to 1,000 mL dH2O. Adjust the pH to 9.0 with glacial acetic acid. Cool to 4 °C for at least 30 min before use.
- DNA precipitation solution (DPS)
- Preparation of 10 mL 7.5 M ammonium acetate stock. For 50 mL of DNA precipitation solution, mix 6.7 mL 7.5 M ammonium acetate with 43.3 mL 95% ethanol. Store at room temperature.
- Staining solution
- Add 1 µL 10,000x green fluorescent nucleic acid stain (e.g., SYBR Green) in 30 mL Tris-EDTA buffer (10 mM Tris-HCl, 1 mM disodium EDTA, pH 7.4) and store at 4 °C. Protect from light.
- 1% low melting agarose
- Melt 1% low melting point agarose (1 g in 100 mL dH2O) in a microwave. Swirl the agarose every 15-20 s to make sure that the agarose is completely molten. Place the agarose in 37 °C water bath for at least 20 min before use.
- Pre-warm pipette tips
- Cut off the narrow ends of P200 pipette tips by 3 mm and warm at 37 °C before pipetting agarose.
2. Prepare Comet Slides
- Slide coating
- Melt 1% agarose (1 g in 100 mL dH2O) in a microwave for 2 - 3 min or until the agarose is completely molten. Dip the glass microscope slides into the agarose and wipe one side of the slide using a lint-free wipe.
- Lay the slides on a flat surface to air-dry or heat at 50 °C for faster drying; a transparent agarose film should be formed after drying. Place the coated slides in 37 °C before use.
- Preparation of single cell suspensions
- Culture and treat the glioma cell
- Culture the U251 MG cells in DMEM-Ham F-12 medium supplemented with 10% FBS, 100 U/mL penicillin, and 10 µg/mL streptomycin at 37 °C with 5% CO2.
- Digest the cells using 1 mL trypsin for 3 min, and neutralize trypsin using DMEM-Ham F-12 medium with FBS. Collect in 15 mL tube, spin at 300 x g for 4 min, aspirate the medium, and suspend cells at 2 x 105 cell/mL in 1x PBS. NOTE: The cell sample should be prepared immediately before starting the assay and all samples should be handled in a dark or dimmed environment to prevent DNA damage from light.
- Combine the cell suspension with 1% molten low melting point agarose (at 37 °C) at a ratio 1:10 (v/v), mix gently by pipetting up and down, and immediately pipette 30 µL onto a slide. Use the side of the the pipette tip to spread the agarose/cell mixture to ensure the formation of a thin layer.
- Place the slide flat at 4 °C in the dark for 10 min. Increasing the gelling time to 30 min improves adherence of samples in high humidity environments.
- Immerse the slide in 4 °C LS in the dark for 1 h to overnight.
3. Single Cell Electrophoresis
Proceed to alkaline (step 3.2) or neutral (step 3.3) comet assay
- For alkaline comet assay
- Gently remove slides from the LS, drain excess buffer, and gently immerse in AES for 1 h at 4 °C to allow DNA unwinding. Keep the slides in the dark.
- Add pre-chilled AES in the electrophoresis slide tray, do not exceed 0.5 cm above the slides (this depends on the size of the electrophoresis units), place the slides inside and cover with a cap. Set the power supply voltage to 1 V/cm (the length between electrodes) and run for 30 min at 4 °C.
- Drain excess electrophoresis solution from slide. Gently immerse slides twice in dH2O for 5 min each at room temperature.
- Gently immerse slides in 70% ethanol for 5 min at room temperature. Proceed to step 4.
- For neutral comet assay
- Gently remove the slides from the LS, drain excess buffer, and gently immerse in NES for 30 min at 4 °C. Keep the slide in the dark.
- Add pre-chilled neutral electrophoresis buffer in the electrophoresis slide tray, do not exceed 0.5 cm above slides (this depends on the size of the electrophoresis units), place the slides inside and cover with a cap. Set the power supply voltage to 1 V/cm (the length between electrodes) and run for 45 min at 4 °C.
- Drain excess buffer from the slides. Gently immerse slides in DPS for 30 min at room temperature.
- Gently immerse the slides in 70% ethanol for 30 min at room temperature. Proceed to step 4.
4. Stain Comet Slides
Dry slides at 37 °C for 10 - 15 min in the dark.
Place 50 - 100 µL green fluorescent nucleic acid staining solution onto each dried agarose and stain for 15 min at room temperature in the dark.
Rinse the slides briefly in dH2O and dry completely at 37 °C in the dark. Proceed to image acquisition and analysis.
5. Image Acquisition and Analysis
NOTE: The visualization and quantification of DNA breaks are based on epifluorescence microscopy and the comet assay software (see Table of Materials)12.
Place the slides on the microscope with a slide holder. Ensure the agarose gel is facing to the objective lens. Randomly capture images from the stained comet slides using a fluorescence microscope with a 10x objective lens. Avoid the edges and the areas around any air bubbles.
Ensure each comet tail is horizontally distributed. Comet heads should originate from the left and the tail from the right.
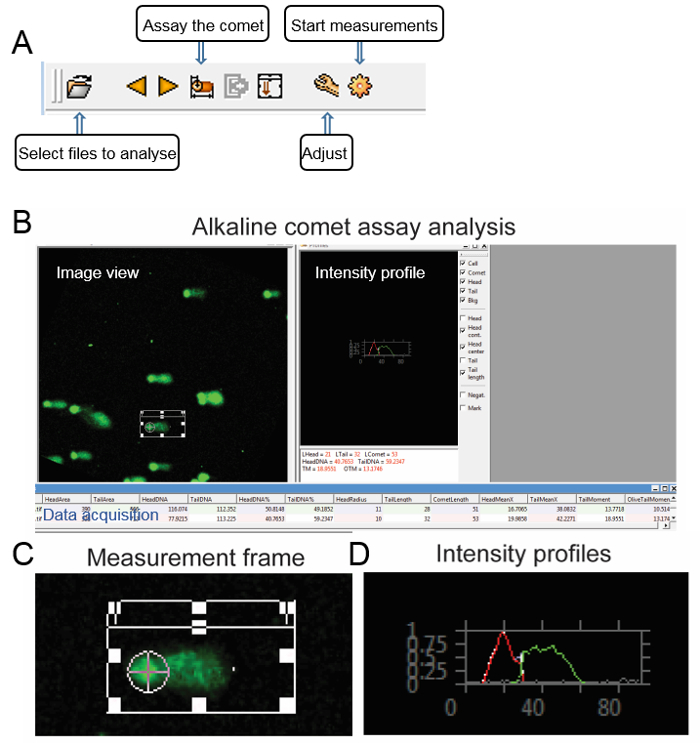
Save each picture in a binary TIF format with bright DNA stain and dark background. Load images to the software using the "Select files to analyze" button, which is located on the left of the toolbar. An image view window should appear (Figure 1).
Draw a measurement frame on the screen and adjust its size in accordance with the comet of the cell. Click the "Adjust" button to set up the threshold of the head, comet, and tail according to the image, then click the "Start measurements" button (Figure 1).
Select a cell using the frame and activate the measurement by clicking with the mouse on the "Assay the comet" button; an intensity image shows up on the "profiles" window with the selected measurement parameters. The results can be saved by clicking the "store result" button (Figure 1). NOTE: The software calculates the parameters including the length of the comet tail, the percentage of the tailed DNA, the tail moment (TM) and the Olive tail moment (OTM). The tail moments are calculated by the formulas as follow:
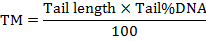
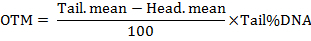
Analyze at least 50 cells per treatment.
Representative Results
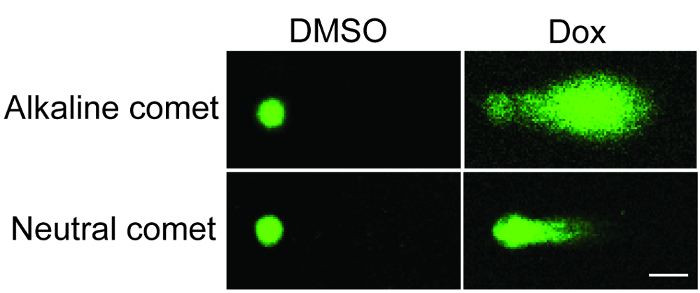
The present protocol describes a step-by-step workflow for the comet assay execution and data analysis (Figure 1). Results from the alkaline and neutral comet assays showed that the comet tail of doxorubicin-treated U251 cells (1 µM, 20 h) was longer and had higher DNA intensity, suggesting a substantial accumulation of fragmented DNA due to chemotherapy (Figure 2).
Quantitative analysis for either the alkaline or neutral comet assay showed significantly increased comet tail formation after doxorubicin treatment (Figure 3A) indicating DNA damage. Results are presented as mean ± SEM, and t-tests were used for statistical comparisons. Quantification of the tail moment from both assays confirmed the increase of average tail moment: For the alkaline comet assay, 0.134 ± 0.0448 (n = 52) vs. 13.84 ± 1.325 (n = 51), p <0.01; For the neutral comet assay, 0.596 ± 0.06668 (n = 78) vs. 9.979 ± 0.658 (n = 51), p <0.01. These data provide a quantifiable index to evaluate DNA damage. Moreover, combination treatment of PARP inhibitor olaparib and temozolomide significantly enhanced DNA damage in glioma cells (Figure 3C), with an increased length and intensity of the comet tail signal. Olaparib alone did not introduce DNA damage, whereas it potentiated the genotoxic effect of the chemo agent temozolomide (DMSO, 0.9836 ± 0.3377 (n = 64); Olaparib, 0.6663 ± 0.2325 (n = 51); TMZ, 6.197 ± 0.572 (n = 51); Olaparib + TMZ, 44.04 ± 1.269(n = 115), p <0.01).
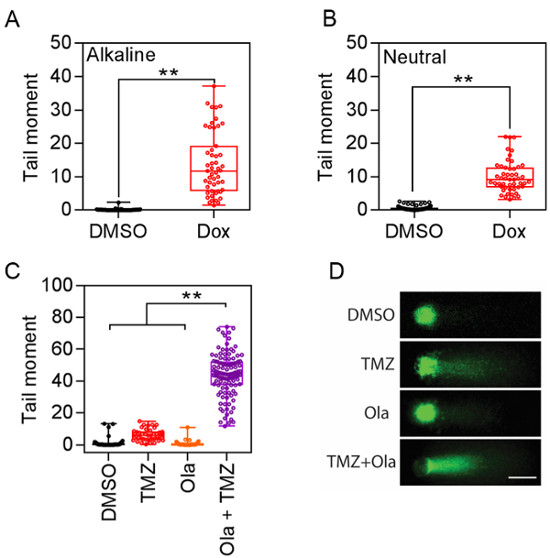
Figure 1: Step-by-step guide of image analysis using comet assay software. (A) The toolbar of the comet assay software. (B) Alkaline comet assay as an example: the user interface of comet assay software consists of an image view, intensity profile, and data acquisition window. (C) The image background, comet region, and comet head were defined in the software. (D) The intensity profiles of the comet head (red) and tail (green) are plotted, respectively. Please click here to view a larger version of this figure.
Figure 2: Representative images of the alkaline and neutral comet assays. U251 glioma cells were treated with doxorubicin and investigated by alkaline (upper panels) and neutral (lower panels) comet assays (Scale bar = 10 µm). Please click here to view a larger version of this figure.
Figure 3: Representative data of the comet tail moment. (A) Alkaline comet assay data plot. **p <0.01. (B) Neutral comet assay data plot. **p <0.01. (C) Comet assay analysis revealed the synergistic effect of combination therapy including temozolomide (TMZ) and olaparib (Ola). **p <0.01. (D) Representative images of glioma cells under combination treatment of temozolomide and olaparib (Scale bar = 10 µm). Please click here to view a larger version of this figure.
Discussion
The comet assay is an efficient tool to measure single and double-strand DNA breaks at the cellular level. The assay has been widely applied as a "golden standard" in studies regarding genotoxicity and biomonitoring13, ranging from base lesions, DNA crosslinks, drug development, and alkali sensitive sites. In the present study, we showed two distinct step-by-step protocols for alkaline and neutral comet assays, respectively. Combining single cell electrophoresis, fluorescent microscopy, and image interpretation, both the alkaline and neutral comet assays provide quantitative approaches to evaluate the extent of DNA damage in vitro.
Several key steps are essential for the successful execution of the comet assay. For example, care should be taken in the steps of reagent preparation. Each solution should be freshly prepared and kept at the appropriate temperature for at least 30 min before use, to prevent endogenous DNA damage or repair during sample preparation. Another vital step is the preparation of viable single-cell suspension. It is important to keep the agarose at 37 °C before use, to maximize cell survival during preparation of the comet slides. The cell/agarose suspension should be prepared immediately before starting the assay. Using the side of pipette tips to spread the cell/agarose suspension helps to achieve a thin layer of agarose on the slides, so that most of the cells can be positioned in the same focal plane during microscopy. Optionally, scratch the edges of the slides using a diamond-tipped pen to improve the agarose attachment.
Despite the advantages of the comet assay, there are several limitations of this traditional method14,15. For example, the throughput of the comet assay is limited by the process of the glass slides preparation. In some occasions, an eight-welled glass slide provides a solution to improve assay throughput. Another limitation is that the comet assay simply represents the ratio of the fragmented DNA in cells. Additional supportive information is required to thoroughly depict the cellular changes besides DNA damage. In that case, apoptosis analysis, γH2A.X staining, and immunoblotting could be very helpful to provide a thorough analysis of different aspects to reveal the effect of DNA damaging agents.
There are numerous software for comet image analysis and the output includes a variety of different parameters. The most commonly used parameters are the tail length, the percentage of DNA in the tail, and the tail moment. Tail length only can be used at low levels of DNA damage, since it does not tend to change once the tail is established14. Subsequently, the intensity of the tail increases as the damage is enhanced. The percentage of DNA in the comet tail is another useful parameter, as it is linearly related to the breaking frequency. The tail moment combines tail length and tail intensity in one single value, thus, it is the most useful and frequently used parameter.
The comet assay has been widely applied in genotoxicity testing and human biomonitoring. With the development of DNA repair enzyme inhibitors, the comet assay could a valuable tool for testing combination therapies that improve the outcome of traditional chemotherapy.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, NCI, and CCR. All authors received Intramural Research Grant from NIH, NCI, and CCR.
References
- Ostling O, Johanson KJ. Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochem Biophys Res Commun. 1984;123(1):291–298. doi: 10.1016/0006-291x(84)90411-x. [DOI] [PubMed] [Google Scholar]
- Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175(1):184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- Tice RR, et al. Single cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testing. Environ Mol Mutagen. 2000;35(3):206–221. doi: 10.1002/(sici)1098-2280(2000)35:3<206::aid-em8>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Shah AJ, Lakkad BC, Rao MV. Genotoxicity in lead treated human lymphocytes evaluated by micronucleus and comet assays. Indian J Exp Biol. 2016;54(8):502–508. [PubMed] [Google Scholar]
- Azqueta A, Collins AR. The essential comet assay: a comprehensive guide to measuring DNA damage and repair. Arch Toxicol. 2013;87(6):949–968. doi: 10.1007/s00204-013-1070-0. [DOI] [PubMed] [Google Scholar]
- Goldstein M, Kastan MB. The DNA damage response: implications for tumor responses to radiation and chemotherapy. Annu Rev Med. 2015;66:129–143. doi: 10.1146/annurev-med-081313-121208. [DOI] [PubMed] [Google Scholar]
- Gavande NS, et al. DNA repair targeted therapy: The past or future of cancer treatment? Pharmacol Ther. 2016;160:65–83. doi: 10.1016/j.pharmthera.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgovnick A, Schumacher B. DNA repair mechanisms in cancer development and therapy. Front Genet. 2015;6:157. doi: 10.3389/fgene.2015.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston VJ, et al. The PARP inhibitor olaparib induces significant killing of ATM-deficient lymphoid tumor cells in vitro and in vivo. Blood. 2010;116(22):4578–4587. doi: 10.1182/blood-2010-01-265769. [DOI] [PubMed] [Google Scholar]
- Brown JS, O'Carrigan B, Jackson SP, Yap TA. Targeting DNA Repair in Cancer: Beyond PARP Inhibitors. Cancer Discov. 2017;7(1):20–37. doi: 10.1158/2159-8290.CD-16-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, et al. Chemosensitivity of IDH1-Mutated Gliomas Due to an Impairment in PARP1-Mediated DNA Repair. Cancer Res. 2017;77(7):1709–1718. doi: 10.1158/0008-5472.CAN-16-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konca K, et al. A cross-platform public domain PC image-analysis program for the comet assay. Mutat Res. 2003;534(1-2):15–20. doi: 10.1016/s1383-5718(02)00251-6. [DOI] [PubMed] [Google Scholar]
- Valverde M, Rojas E. Environmental and occupational biomonitoring using the Comet assay. Mutat Res. 2009;681(1):93–109. doi: 10.1016/j.mrrev.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Collins AR. The comet assay for DNA damage and repair: principles, applications, and limitations. Mol Biotechnol. 2004;26(3):249–261. doi: 10.1385/MB:26:3:249. [DOI] [PubMed] [Google Scholar]
- Karbaschi M, Cooke MS. Novel method for the high-throughput processing of slides for the comet assay. Sci Rep. 2014;4:7200. doi: 10.1038/srep07200. [DOI] [PMC free article] [PubMed] [Google Scholar]


