Abstract
The incidence of human papillomavirus (HPV)-related oropharynx cancer has steadily increased over the past two decades, and now represents a majority of oropharyngeal cancer cases. Integration of the HPV genome into the host genome is a common event during carcinogenesis that has clinically-relevant effects if the viral early genes are transcribed. Understanding the impact of HPV integration on clinical outcomes of head and neck squamous cell carcinoma (HNSCC) is critical for implementing de-escalated treatment approaches for HPV-positive HNSCC patients. RNA sequencing (RNA-seq) data from HNSCC tumors (n=84), was used to identify and characterize expressed integration events, which were over-represented near known head and neck, lung, and urogenital cancer genes. Five genes were recurrent, including CD274 (PD-L1). A significant number of genes detected to have integration events were found to interact with Tp63, ETS, and/or FOX1A. Patients with no detected integration had better survival than integration-positive and HPV-negative patients. Furthermore, integration-negative tumors were characterized by strongly heightened signatures for immune cells, including CD4+, CD3+, regulatory, CD8+ T cells, NK cells, and B cells, compared to integration-positive tumors. Finally, genes with elevated expression in integration-negative specimens were strongly enriched with immune related gene ontology (GO) terms while up-regulated genes in integration-positive tumors were enriched for keratinization, RNA metabolism and translation.
Keywords: Head and neck cancer, human papillomavirus, HPV-integration (or HPV-host fusional transcripts)
Introduction
Head and neck cancers together represent the sixth most common cancer worldwide. In 2015 the incidence of this type of cancer was estimated at > 742,000 new cases (> 400,000 deaths) [1]. While the most common risk factors associated with head and neck squamous cell carcinomas (HNSCCs) are tobacco use and alcohol consumption, the past few decades reveal a steadily increasing subset of HNSCCs associated with high-risk human papillomavirus (HPV) infection. HPV-associated HNSCC patients tend to be slightly younger, male (75%), more often non-smokers [2, 3] and characteristically demonstrate improved survival in the majority of patients compared to patients with HPV negative cancers [4]. Better understanding the role of HPV integration in oropharyngeal cancer biology is fundamental to the design of new therapeutic strategies and selection of patients for aggressive therapy.
HPV is the most common sexually transmitted infection in the US, with at least fifteen high-risk HPV types classified as carcinogenic (HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, 73, and 82) [5]. The vast majority of persons exposed to HPV successfully clear the infection; however failure of the immune response can result in persistent infection with an increased risk of progression to cancer [6]. The genome organization of HPV comprises a long control region and eight genes necessary at different stages of the viral life cycle. The E1 HPV protein is essential for replication of the viral episome (circular, extrachromosomal HPV DNA) [7], while E2 functions in DNA replication [8], and suppresses expression of oncogenes E6 and E7; E1 and E2 are often lost upon integration into the host genome. E4 and E5 contribute indirectly to genome amplification by modifying the cellular environment, and E5 also possesses pore forming capability and interferes with apoptosis [9]. Oncogenic E6 and E7 proteins are most important for HPV-associated tumorigenesis. High-risk E6 promotes p53 degradation, upregulates telomerase activity and maintains telomere integrity during repeated cell divisions [9], while E7 binds to pRb (retinoblastoma protein) allowing unchecked cell division. E7 can bind and degrade proteins that control cell cycle entry in the basal and upper epithelial layers, and thus is able to stimulate host genome instability through deregulation of the centrosome cycle [9]. Increased E6 and E7 expression predisposes infected cells to an accumulation of genetic changes, which may increasingly contribute to cancer progression, and correlate with the severity of neoplasia [9, 10]. E6 and E7 are maintained in successful HPV integration events into the host genome.
Integration of high-risk HPV genomes into the host genome is observed in most invasive cervical cancers and a majority of HPV(+) HNSCCs, although accurate percentages by tumor site are unknown. It is still not clear whether HPV integration precedes E6/E7 induced genetic instability or rather is a consequence of instability [11]. It is thought that integration occurs relatively late in progression in high-grade lesions such as CIN2 and CIN3 (cervical intraepithelial neoplasia). The evidence is also mixed on whether expression of E6 and E7 is higher with integration [10] or if it is constitutive expression of E6/E7 upon integration rather than an increase in oncogene expression that is relevant to the malignant phenotype [11]. Regardless, both of the above studies were performed for cervical SCCs and thus their results may not translate directly to HNSCC.
The level of HPV integration has been proposed as a marker of disease progression in cervical cancer [12, 13]: during the progression of cervical lesions, the rate of HPV integration was observed to rise from 53.8% of CINs to 81.7% of cervical carcinomas [14]. The longer half-life of integrated viral transcripts compared with episomal transcripts further promotes immortalization and transformation of cancer cells and provides a selective growth advantage [15].
One of the largest collections of characterized HPV-positive HNSCC samples is The Cancer Genome Atlas (TCGA), with 66 collected as of August 2015. From whole-genome and transcriptome sequencing of the first 36 of these tumors, most HPV(+) tumors demonstrated clear evidence of host genome integration (25 HPV(+)/integration-positive tumors), and often in association with amplifications of the genomic region. This TCGA analysis did not identify any genes with recurrent integrations, or any common driver mechanism related to HPV integration [16].
We have analyzed HPV integration sites in the human transcriptome in 84 primary HPV(+) HNSC neoplasms collected at the University of Michigan (UM) (18 tumors) and TCGA. We have expanded the sample size of TCGA tumors analyzed for HPV integration events from 36 [16] to 66 cases, with 47 oropharyngeal and 16 oral cavity tumors. We find five genes with recurrent integration events, including CD274 (PD-L1). We also show strong biologic selection for HPV integration into genes known to play important roles in head and neck cancer. A significant number of genes detected to have integration events were found to interact with Tp63, ETS, and/or FOX1A. As opposed to findings in cervical cancers, we do not find statistically significant evidence for an enrichment of integration events in genomic common fragile regions. However, we do find strong enrichment in specific types of repetitive regions. Our survival analysis shows the clinical relevancy of HPV-integration, which may be partly due to a change in immune response upon integration. If confirmed, these findings have important implications for identifying specific patients for more or less aggressive treatment approaches.
Materials and Methods
Tumor tissue acquisition, RNA extraction and RNA-seq protocol
Eighteen HPV(+) tumor samples were collected at the University of Michigan hospital from patients with untreated oropharynx or oral cavity squamous cell carcinoma. Written informed consents were obtained and the study was approved by the University of Michigan Institutional Review Board. Tumor tissues were collected into a cryogenic storage tube, flash frozen in liquid nitrogen and stored in −80C until prepared for histology. H&E slides were assessed for degree of cellularity (minimum 70%) and necrosis (less than 10%). Frozen scrapings were processed using the Qiagen AllPrep DNA/RNA/Protein Mini Kit (Valencia, CA, USA) as per manufacturer protocol. RNA library construction and sequencing on Illumina HiSeq using 100 nt paired-end reads were performed by the University of Michigan DNA Sequencing Core Facility, as described in [17]. Raw and processed RNA-seq data can be accessed from GEO with the accession number GSE74927.
RNA-seq analysis
The RNA-seq libraries were aligned to human and HPV genomes to quantify the host and viral gene expression and determine HPV status. Samples were classified as HPV(+) if they had more than 1000 read pairs aligned to any HPV genome. We aligned raw reads to following high-risk type HPV genomes: 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68, downloaded from NCBI [17]. RNA-seq fastq files of 66 TCGA HPV(+) tumor samples were downloaded from cghub [18]. The data were re-aligned and analyzed in the same way as UM RNA-seq data (see Supplementary Methods for details). Measuring HPV gene expression levels was also performed as described in [17]. See Supplementary Methods for additional RNA-seq analysis details.
Detection of HPV integration sites
Detection of known viruses, reconfirmation of positive HPV status, and identification of the integration sites into the human genome was performed for HPV-positive UM and TCGA samples using VirusSeq [19]. A positive integration event was defined as having at least four supporting discordant read pairs and at least one junction spanning read. A tumor sample was called integration positive if it contained at least one identified integration event. See Supplementary Methods for additional details.
Gene network construction
To understand the biological relevance and relatedness of the genes harboring HPV-host fusion transcripts, MetaCore software by Thomson Reuters (https://portal.genego.com/) was used to model interactions among these genes. The set of parameters used for network construction were high-confidence, functional, binding and low-trust direct interactions between genes with the shortest path building algorithm. In MetaCore, gene IDs of interest were mapped onto gene IDs of entities (for example Diseases), and ranked based on “relevance” to the analyzed gene set. MetaCore using the hypergeometric distribution for calculating disease enrichment p-values, given the size of the ontology, the dataset and the particular entity. In MetaCore, p-value means the probability of a random intersection of two different gene/protein/compound sets. (MetaCore Glossary).
Additional analyses
Analysis details for (1) associating HPV integration sites with repetitive and fragile regions of the genome; (2) comparing integration results from whole genome sequencing and RNA-seq data; (3) defining the cell type specific signatures; and (4) identifying the CD274 enhancer regions are available in the Supplementary Methods.
Survival analysis
Overall survival for TCGA cases was analyzed for three groups: integration-positive, integration-negative, and HPV(−) samples using the survival and survminer R packages. Kaplan-Meier estimates of survival were determined, and a p-value was calculated using a univariate log-rank test. Cox proportional hazards regression models were used for adjustment of clinical covariate variables (age, clinical stage, tumor site and smoking status.)
Functional enrichment testing
Enriched Gene Ontology (GO) terms and KEGG pathways were tested using RNA-Enrich [20] (http://lrpath.ncibi.org/) which tests for gene sets that have higher significance values (e.g. for differential expression) than expected at random, and takes into account any relationship between gene read count and significance level. RNA-Enrich is able to detect both pathways with a few very significant genes and pathways with many only moderate differentially expressed genes without requiring a cutoff for significance. We implemented the directional RNA-Enrich test (which tests for significantly up-versus down-regulated gene sets) and only terms with less than 500 genes were considered for analysis. Custom code was implemented to reduce redundancy (remove less significant, closely related GO terms) for presenting the top enriched terms by integration status.
Results
Samples description
RNA sequencing data from 84 HPV(+) HNSC malignant neoplasms were interrogated for the presence of HPV integration sites into the cancer transcriptomes. Eighteen tumors were collected at UM and tested for HPV status as previously described in [17]; the other 66 were collected as part of TCGA. Detection of expressed viral integration events and their insertional breakpoints was performed using VirusSeq [19] (see Methods and Supplementary Methods for details). HPV integrations into the host genome (detected as HPV-host fusion transcripts) were detected in 51 (60.7%) of the 84 samples. Among the 18 HPV(+) UM tumors, viral-host fusion transcripts were found in nine (50%) of the samples. In the TCGA cohort we found 42 of the 66 tumors (63.6%) were integration-positive. Among the integration-positive tumors there were 41 HPV16 tumors, one HPV18, six HPV33, and three HPV35. Among the HPV(+) neoplasms investigated in this study, we did not find differences between integration-positive and integration-negative samples by demographic or clinicopathological parameters except by anatomical site (Table 1). We tested only oropharynx versus oral cavity tumors for anatomical site differences, due to insufficient samples from other sites, and found that oral cavity tumors have a higher rate of HPV-host fusion events than oropharyngeal tumors (p = 0.011). However, we note that some of the HPV(+) tumors may have been posterior tongue cancers misclassified as oral cavity.
Table 1.
Demographic and clinicopathologic characteristics of HPV-positive patients by virus integration status.
| Parameter | HPV(+) UM tumors | HPV(+)TCGA tumors | p-value | ||||
|---|---|---|---|---|---|---|---|
| Total | Integration-positive | Integration-negative | Total | Integration-positive | Integration-negative | ||
|
| |||||||
| 18 | 9 | 9 | 66 | 42 | 24 | ||
|
| |||||||
| Age at diagnosis | |||||||
| Median (std) | 58 (7.3) | 54 (6.2) | 59 (7.3) | 57 (9.2) | 59 (9.5) | 56 (8.2) | 0.463 |
| Gender | |||||||
| Male | 17 | 9 (100%) | 8 (89%) | 60 | 38 (90%) | 22 (92%) | 1 |
| Female | 1 | 0 | 1 (11%) | 6 | 4 (10%) | 2 (8%) | |
| HPV type | |||||||
| HPV16 | 14 | 7 (78%) | 7 (78%) | 55 | 34 (81%) | 21 (88%) | 0.772 |
| HPV18 | 1 | 1 (11%) | 0 | 0 | 0 | 0 | |
| HPV33 | 1 | 1 (11%) | 0 | 8 | 5 (12%) | 3 (13%) | |
| HPV35 | 2 | 0 | 2 | 3 | 3 (7%) | 0 | |
| Anatomical Site | |||||||
| Oropharynx | 17 | 8 (89%) | 9 (100%) | 47 | 26 (62%) | 21 (88%) | 0.011 |
| Oral Cavity | 1 | 1 (11%) | 0 | 16 | 14 (33%) | 2 (8%) | |
| Larynx | 0 | 0 | 0 | 1 | 1 (2%) | 0 | |
| Hypopharynx | 0 | 0 | 0 | 2 | 1 (2%) | 1 (4%) | |
| Tumor Stage | |||||||
| I–II | 1 | 0 | 1 (11%) | 12 | 6 (14%) | 6 (25%) | 0.439 |
| III | 2 | 0 | 2 (22%) | 7 | 5 (12%) | 2 (8%) | |
| IV | 15 | 9 (100%) | 6 (67%) | 47 | 31 (74%) | 16 (67%) | |
| T stage | |||||||
| T1–T2 | 8 | 3 (33%) | 5 (56%) | 39 | 22 (52%) | 17 (71%) | 0.176 |
| T3–T4 | 10 | 6 (67%) | 4 (44%) | 26 | 19 (45%) | 7 (29%) | |
| N stage | |||||||
| N0 | 1 | 0 | 1 (11%) | 18 | 12 (29%) | 6 (25%) | 0.674 |
| N1 | 2 | 0 | 2 (22%) | 6 | 4 (10%) | 2 (8%) | |
| N2 | 11 | 6 (67%) | 5 (56%) | 39 | 24 (57%) | 15 (63%) | |
| N3 | 4 | 3 (33%) | 1 (11%) | 2 | 2 (2%) | 0 | |
| Smoking Status | |||||||
| Current | 3 | 1 (11%) | 2 (22%) | 13 | 8 (19%) | 5 (21%) | 0.439 |
| Former | 11 | 6 (67%) | 5 (56%) | 30 | 22 (52%) | 8 (33%) | |
| Never | 4 | 2 (22%) | 2 (22%) | 22 | 12 (29%) | 10 (42%) | |
Tests were performed between integration-positive and integration-negative samples for both combined cohorts. For age, Wilcoxon rank sum test was used for age; Fisher’s exact test was used for other variables. For anatomical sites we tested only oropharynx versus oral cavity tumors due to insufficient samples from other sites.
Analysis of integration at breakpoints
We found 320 virus-host fusion breakpoints, which were broadly distributed across the human and viral genomes (Supplemental Table S1). Breakpoints were localized on all chromosomes except chromosomes 20, 22, and Y, and occurred within or near 89 human genes (Figure 1). All 320 breakpoints were annotated to one of the 89 human genes. Overall, 116 (36.25%) breakpoints were located in exons, 124 (38.75%) were in introns, 41 (12.81%) were downstream of the closest gene, and 39 (12.19%) were upstream of the closest gene (Supplemental Table S1). We use the term “within” a human gene when integration occurred in an exonic or intronic region, and “near” when integration occurred up- or down-stream of the closest gene. Some genes have only 1 breakpoint while others up to 23 (RAD51B); the average number of breakpoints per gene was 3.6. The average number of breakpoints per sample was 6.3 (range = 1 to 19) (Supplemental Table S1).
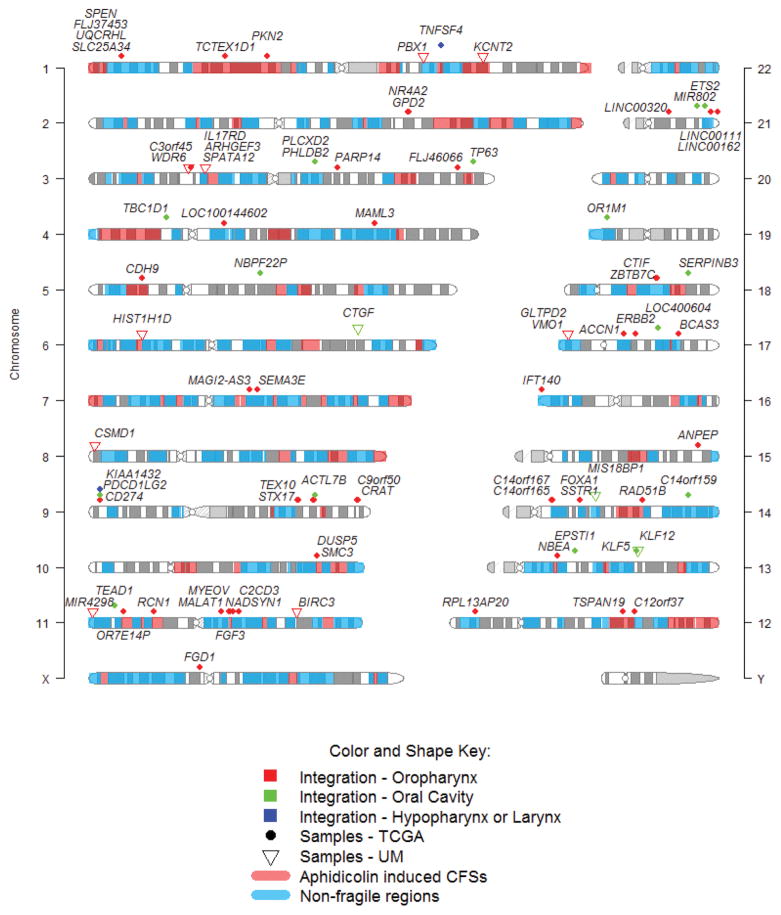
Figure 1. Identified HPV integration sites in the human genome.
Viral-host fusional breakpoints found in HPV-positive head and neck cancer samples from TCGA (42 out of 66 samples are integration-positive) and University of Michigan (UM; 9 out of 18 samples are integration-positive). The integration sites are broadly distributed across the genome and are annotated within or near 89 unique human genes. CFSs = common fragile sites.
Within the viral genome, breakpoints in oncogenes E6 and E7 were more common – 202 (63.13%) compared to breakpoints into other viral genes: E1 and E2 – 99 (30.94%), E4 and E5 – 44 (13.75%), or L1 and L2 – 15 (4.69%) (Table S5). This may be explained by preservation and expression of oncogenes E6 and E7 in all analyzed samples, while expression of genes E1 and E2 were lost in more than half of the integration-positive tumors. For the 36 TCGA tumors previously analyzed for HPV integration sites based on both DNA and RNA[16, 21], we had good agreement using RNA only, with only five samples reclassified based on RNA-seq (See Supplemental Methods and Discussion), suggesting that most tumors with HPV integration events detected with DNA have expressed transcripts from at least one of those integrations.
Comparison of viral gene expression by integration status showed significantly less expression in genes E2, E4, and E5 in integration-positive tumors compared to integration-negative (p-values vary from 4.14 × 10−08 to 3.8 × 10−07), while there was no difference for E6 and E7 oncogenes (p = 0.97 and 0.35 correspondingly; Table S5). Despite the reduced expression of E2 in approximately 2/3 of integration-positive samples, some tumors showed extremely high expression of this viral gene-regulator. Eleven integration-positive samples with ≥1 breakpoints, had elevated expression of E2 (> 100 counts per million (CPM)) and ten had E2/E6 expression ratio > 2 (Table S2). For example, sample UM-P03 had multiple host-fusion breakpoints in BIRC3 and in all HPV genes. Expression of E2 in this sample was the highest among all tumors and expression of BIRC3 was significantly elevated. We note, however, that protein levels of the HPV genes may not be highly correlated with the RNA levels.
We analyzed the distribution of breakpoints throughout the HPV16 genome, and observed the integration locations into E6 and E7 by sample (Figure S2). A significant part of E6 had no integration event across all analyzed samples, which could indicate selective pressure to maintain this region.
HPV integration sites associate with LINE, MIR SINE and LTR repetitive elements but not associated with common fragile sites
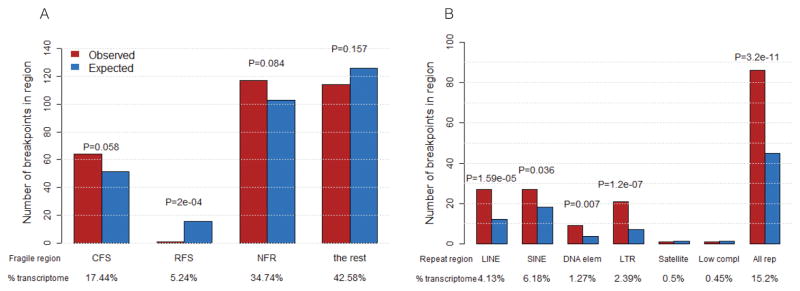
We investigated potential associations of integration sites with fragile and repetitive regions of the human genome, accounting for the regions of the genome covered by the RNA-seq data from all analyzed HPV(+) samples. We found significantly more insertional breakpoints in the following classes of repeats: LINE (Long interspersed nuclear elements), SINE (Short interspersed nuclear elements, including ALUs), DNA (DNA repeat elements), LTR (Long terminal repeat elements, including retroposons) and “All repeats” than expected by chance (Figure 2B and Table S6). When breaking down LINEs and SINEs by subtype, we found that mammalian-wide interspersed repeat (MIR) SINEs were significantly enriched with integration events, but not Alu elements (Table S6). There was no significant enrichment of HPV-host fusion breakpoints within common fragile sites (CFSs) although the p-value suggests a trend (p = 0.058). We did not find an enrichment of host-fusion breakpoints in non-fragile regions (NFR) (p = 0.084) or rare fragile sites (RFS), where fewer than expected by chance were identified (Figure 2A and Table S7).
Figure 2. Assessment of HPV-host fusional breakpoints in fragile and repetitive regions of the human genome.
A. Number of breakpoints in common fragile sites (CFS), rare fragile sites (RFS) and non-fragile regions (NFR), compared to what is expected by chance. HPV is not more prone to integrate into fragile sites in the human genome in HNSCC tumors. B. Number of breakpoints in repetitive regions compared to what is expected by chance. Several repetitive element types (LINE, DNA, LTR and all repeats combined) are determined to be significantly enriched for HPV integrations (Chi-squared test p-values with FDR adjustment = 7.42E-05, 0.015, 8.40E-07, and 4.41E-10 correspondingly). (See also Supplemental Table S6 for a further breakdown by repeat family).
Analysis of integration sites at the gene level – recurrent integrations
Recurrent HPV integration events may signify the natural selection for tumor cells with breakpoints in specific genomic regions, and can suggest novel cancer driver genes. Of the 89 human genes with or near at least one identified integration event (Figure 1, Supplemental Table S1), five were associated with more than one tumor sample (i.e. recurrent integration). These genes were CD274, FLJ37453, KLF12, RAD51B, and TTC6 (Table 2). The genomic distances between integration sites mapped within or near the same gene in two different samples varied between 15 – 440 Kb. In one case, three samples harbored breakpoints within the same locus, but not all annotated to the same gene. Samples TCGA-CV-5443 (Larynx) and TCGA-T2-A6X0 (oropharynx) had HPV16 virus-host fusion breakpoints at intron4 of CD274 and ~100 bp upstream of the same gene, respectively, and CD274 expression was higher in these samples than the average of others. The third sample, TCGA-HL-7533 (oral cavity), harbored breakpoints all within a 207 Kb region in the same cytoband chr9p24.1, just upstream from CD274, and were annotated to exon4 of gene PDCD1LG2 and in exon2 and upstream of gene KIAA1432. CD274 expression was up-regulated in this sample compared to others without integration in that locus (Supplemental Figure S3). Evidence for the identification of two enhancer sites for CD274 in this region (see Supplementary Methods, Identification of the enhancer regions for CD274) suggests this region may regulate CD274 expression. HPV16 breakpoints were also found clustered within or near genes KLF5 and KLF12 (cytoband chr13 q22.1) in samples UM-P17, TCGA-CR-7369, and TCGA-CN-4741, spanning a distance of ~700 Kb.
Table 2. Genes with recurrent integrations in HNSCC tumors.
The integration events occurred either within or near the gene, as detailed in Supplemental Table S1.
| Sample IDs | Gene/cytoband | Summary | Genomic distances between viral integrants in two samples |
|---|---|---|---|
| TCGA-CV-5443 - Larynx TCGA-T2-A6X0 - Oropharynx |
CD274 chr9p24.1 |
CD274 molecule gene encodes an immune inhibitory receptor ligand that is expressed by T cells, B cells and various types of tumor cells. Interaction of this ligand with its receptor inhibits T-cell activation and cytokine production. In tumor microenvironments, this interaction provides an immune escape for tumor cells through cytotoxic T-cell inactivation. | 15Kb |
| TCGA-CN-A49C - Oropharynx TCGA-KU-A6H7 - Oropharynx |
FLJ37453 chr1p36.21 |
Uncharacterized LOC729614 is an RNA gene, and is affiliated with the ncRNA class. | 25 Kb |
| TCGA-CR-7369 - Oral Cavity UM-P17 - Oral Cavity |
KLF12 chr13q22.1 |
Kruppel-like factor 12 - Developmentally-regulated transcription factor and important regulator of gene expression during vertebrate development and carcinogenesis | 440 Kb |
| TCGA-BA-4077 - Oropharynx TCGA-CN-A6V7 - Oropharynx |
RAD51B chr14q24.1 |
RAD51 paralog B - member of the RAD51 protein family which is essential for DNA repair by homologous recombination. Overexpression of this gene was found to cause cell cycle G1 delay and cell apoptosis, which suggested a role of this protein in sensing DNA damage. | 280 Kb |
| TCGA-BA-A4IG - Oropharynx TCGA-MZ-A6I9 - Oropharynx |
TTC6 chr14q21.1 |
Tetratricopeptide repeat domain 6 is a protein coding gene. | 28 Kb |
Genes harboring integrations are strongly enriched with head and neck, lung and urogenital cancer related genes
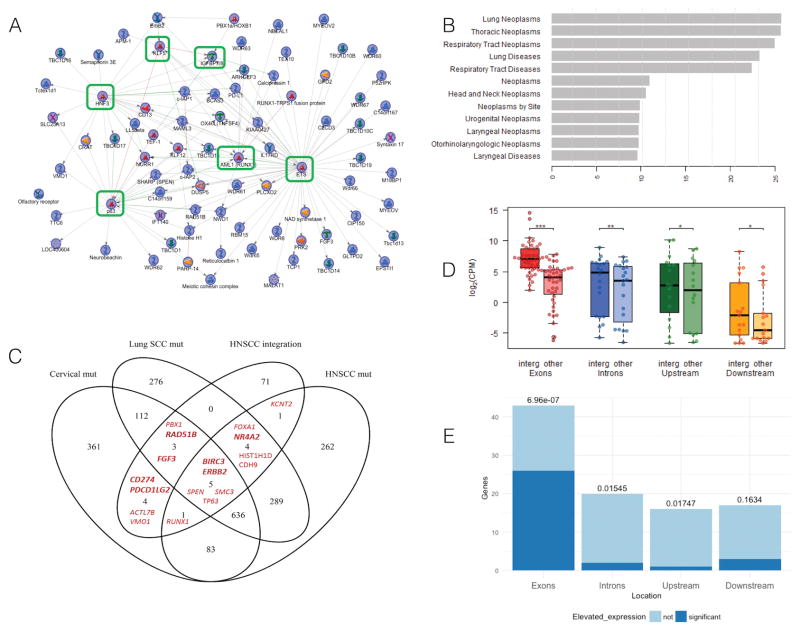
To further understand the biological context of genes associated with one or more insertional HPV breakpoints, we generated a protein interaction network directly connecting 65 of the 89 total genes (MetaCore software by Thomson Reuters; see Methods). Within this resulting subnetwork, there were several hubs (genes with more than five interactions) (Figure 3A). These genes, in order from most to fewest interactions, were: ETS2 (ETS), TP63, FOXA1 (HNF3), RUNX1 (AML1), KLF5, and CTGF (IGFBP7/8). The 89 genes forming the network was highly statistically enriched for genes known to be important specifically in lung neoplasms (p=1.69× 10−26; rank=1), head and neck neoplasms (p= 2.66 × 10−11; rank=7), and urogenital neoplasms (p=1.52× 10−10; rank=9) (Figure 3B; Supplemental Table S3 spreadsheet “Diseases”), suggesting selection for cells with integration events in key carcinogenic genes. Genes associated with head and neck neoplasms included CD274, BIRC3, KCNT2, ERBB2, CDH9, HIST1H1D, SMC3, and TP63.
Figure 3. Genes associated with a detected integration are enriched with HNSCC, urogenital and lung neoplasm related genes, and are often up-regulated.
A. Protein interaction network constructed from the 89 human genes associated with integration event(s); displayed is the highly-connected subnetwork consisting of the 65 (of the 89) host genes that had direct interactions. Genes ETS, TP63, RUNX1, HNF3, KLF5 and CTGF are hubs, indicated with green rectangles, in this network. A legend for the shapes used for the nodes is provided in MetaCore Quick Refernce Guide https://ftp.genego.com/files/MC_legend.pdf. B. Genes in the network are statistically enriched for relevant human diseases. The –log10(p-values) for enrichment were calculated using MetaCore GeneGO with the hypergeometric distribution. The enrichment was tested for the 89 genes and shows the unadjusted significance levels of enrichment. C. Venn diagram of the genes harboring virus-host fusional breakpoints (HNSCC integration) with genes having mutations for: head and neck SCC (HNSCC mut) [16], lung SCC (Lung SCC mut) [36], and cervical SCC and endocervical adenocarcinoma (Cervical mut) (TCGA, Provisional [23, 24]). Gene symbols in bold were significantly up-regulated in the samples where integration occurred compared to all other samples. No genes were significantly down-regulated. D. Distribution of human gene expression levels categorized by the genic or intergenic region where the integration occurred (exons, introns, upstream of the TSS, or downstream of the TES). The left box for each region represents expression in the samples where the integrations occurred (“integr”); the right boxes represent the average expression of the same genes in all other integration-positive samples (“other”). *** Significant difference (paired t-test p-value = 1.60E-09) in expression when insertional breakpoints occur in a gene exons, ** p-value <0.01 and * p-value <0.05. E. The bars represent the number of genes harboring the insertional breakpoints in each type of region (exons, introns, upstream of TSS, or downstream of the TES). Dark blue bars represent number of genes harboring the insertional breakpoints with significantly elevated expression in samples with integration. Numbers above the bars represent the two-sided Fisher’s Exact Test p-values (unadjusted) calculated from testing whether there are more or fewer samples with elevated expression of the gene harboring the integration (in sample with integration) than expected by chance. Integrations into exons of the genes tend to result in upregulation of these genes in the samples with the integration. Conversely, introns and upstream regions have a trend toward fewer than expected by chance.
We next sought to determine which genes harboring virus-host fusion breakpoints were also known to have mutations in lung, head and neck, or cervical SCC. Comparisons were made between the 89 genes from above and mutated genes from TCGA: HNSCC [16], lung SCC [22], and cervical SCC and endocervical adenocarcinoma (TCGA, Provisional [23, 24]) (Figure 3C). Statistical testing for significance of overlapping of the 89 HNSCC-integration genes with mutated gene-sets of diseases of interest was performed using Fisher’s Exact Test with Bonferroni correction (Supplemental Table S9). Overlapping the 89 genes with HNSCC mutated genes was significant (p-value after Bonferroni correction = 0.0116), as was overlap with lung SCC mutated genes (p-value = 0.0006) and cervical mutated genes (p-value = 0.0049) (Supplemental Table S9). Five genes (BIRC3, ERBB2, SPEN, SMC3, and TP63) overlapped between all four data sets. Others that overlapped with lung or cervical SCC mutations (PBX1, RAD51B, FGF3, CD274, PDCD1LG2, ACTL7B, and VMO1) suggest additional novel drivers for HPV(+) HNSCC.
Genes with integration sites into exonic regions show elevated expression
To investigate the impact of HPV integration on the expression of the corresponding host gene, we tested for a significant difference in expression between the gene in the sample harboring the insertional breakpoint versus the same gene in all other integration-positive samples. We then tested whether the set of genes with integration overall had elevated (or decreased) expression. Since the effect may depend on which part of the host gene contains the insertion, we tested for a significant difference in expression for each of the following genic/intergenic regions separately: upstream of the TSS, exon, intron, and downstream of the transcription end site (TES) (Figure 3D and E). Taking into account recurrent integrations, we analyzed 96 gene-sample pairs. Expression of genes at/near an integration site were higher in the tumor with the integration compared to tumors without viral integration near the same gene (79 of the 96 cases), and significantly higher in 32 cases (t-test p-value < 0.05) When integration occurred in a gene exon, the expression of the gene was significantly higher in the sample with the integration, compared with the expression of the same gene in other samples (paired t-test p-value = 1.60E-09) (Figure 3D). Fisher’s exact test also demonstrates that a significant number of the genes with an integration event in an exon had elevated expression (OR = 11.6, p-value = 6.96 × 10−07) (Figure 3E). For HPV-host fusional breakpoints in intronic regions or upstream of the genes, there were actually fewer genes significantly up-regulated than expected by chance (OR = 0.17 and 0.11, Fisher’s exact test p-values = 0.015 and 0.017 correspondingly). When considering genes with increased expression in the integrated sample that were also identified as mutated in HNSCC, lung, or cervical cancer, we found seven genes (NR4A2, RAD51B, FGF3, CD274, PDCD1LG2, BIRC3, and ERBB2) (bold in Figure 3C).
Integration-negative patients have better survival than integration-positive
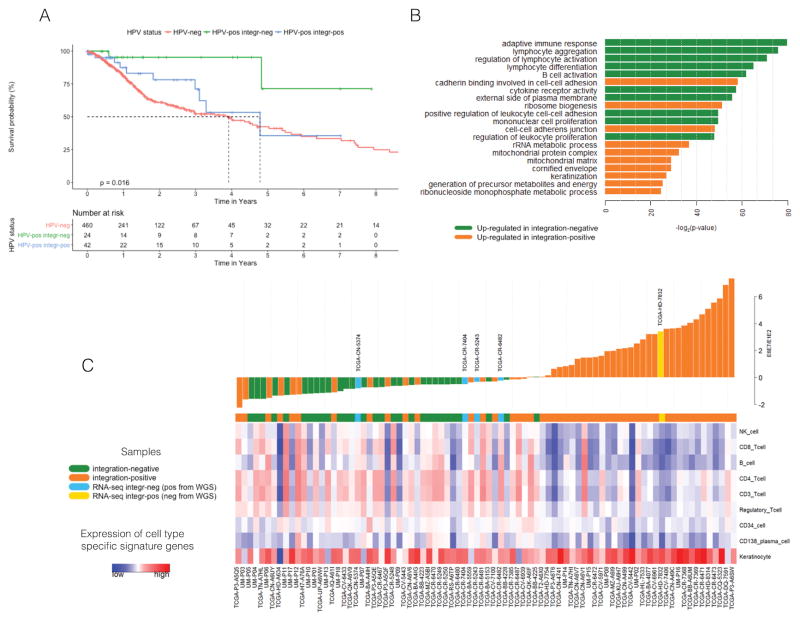
Using HNSCC overall survival data from TCGA, we found that patients with integration-negative tumors had better survival compared to those with integration-positive tumors (for two group comparison log-rank p-value = 0.0436 (Figure S1)), which had a survival rate similar to patients with HPV(−) tumors (log-rank p-value = 0.0158 for three group comparison) (Figure 4A). Univariate and multivariate Cox regression models were performed including clinical covariates: site, sex, clinical stage, smoking status and age for comparison between three groups: integration-negative, integration-positive and HPV(−) (Table S10 and Table S12) and two groups: integrated versus not integrated (Tables S11 and S13). Number of events in groups of integration-negative, integration-positive and HPV(−) are 2, 10 and 158 respectively. Univariate analysis demonstrates that HPV integration was associated with overall survival, and remained significant in multivariate analysis (Tables S10, S11, S12 and S13). There was no difference between integration-positive and HPV(−) samples (p-value = 0.2065). Older age was significantly associated with worse survival. Stage and disease anatomical site were not detected to have a significant effect on survival, but the lack of significance may be due to small sample size. Former smokers had reduced hazard of death compared to current smokers. These results suggest that the variability in survival observed in patients with HPV(+) tumors could be attributed to the better survival of patients with integration-negative tumors. However larger sample sizes are needed for confirmation.
Figure 4. Association of HPV integration events with survival and immune response.
A. Overall survival for TCGA patients with HPV(+) tumors by integration status and HPV(−) patients; p-value was calculated using a univariate Kaplan–Meier log-rank test. B. Gene Ontology terms for genes differentially expressed by HPV integration status. Shown are the top 10 enriched gene sets by integration status after filtering out functional redundancies in the list of gene sets. Genes with elevated expression in integration-negative samples were most strongly enriched for immune related terms; up-regulated genes in integration-positive tumors were enriched for cell-cell adhesion terms related to RNA metabolism and keratinization. C. HPV gene expression (the log2(E6E7/E1E2) ratio) and immune cell type specific signatures of analyzed HNSCC tumors. Waterfall plot of E6E7/E1E2 ratio values demonstrate properties of samples whose integration-status was reclassified using RNA-seq versus WGS data (in blue and yellow). The lower panel shows cell type specific expression signatures (only cell types significantly discriminating integration-positive and integration-negative tumors are shown). Samples that were defined as integration-positive from WGS by TCGA and reclassified as integration-negative from RNA-seq (light blue in waterfall plot) are closer to other integration-negative tumors (green) by their E6E7/E1E2 ratio and by expression of cell type specific signatures. These patients (with ID TCGA-CN-5374, TCGA-CR-7404, TCGA-CR-5243, and TCGA-6482) have survival status alive with follow-up of 9.5, 48.4, 84.2, and 11.3 months respectively. The sample defined as integration-negative from WGS by TCGA and reclassified as integration-positive from RNA-seq (yellow in waterfall plot) has characteristics closer to other integration-positive samples by its E6E7/E1E2 ratio and cell type signatures. This patient (TCGA-HD-7832) did not have any follow-up.
To investigate whether the difference in survival between integration-positive and integration-negative patients could be explained by differences in biological processes, we performed enrichment analysis on the differentially expressed genes (DEGs) between the two groups. Differential expression analysis on all 84 HPV(+) samples using integration status as the group variable revealed 832 significantly DEGs (346 up in integration-positive and 486 up in integration-negative; FDR < 0.05 and |log2(FC)| > 1). Genes with elevated expression in integration-negative samples were most strongly enriched for immune related terms (“T cell activation”, lymphocyte differentiation”, “B cell activation” etc.) (Figure 4B, Table S4); up-regulated genes in integration-positive tumors were enriched for keratinization and terms related to RNA metabolism and translation.
Integration-negative samples are enriched for T-cell and B-cell signatures
We hypothesized that enrichment of integration-negative samples for immune related genes could be explained by increased abundance of inflammatory cell types within these tumors. To test this hypothesis we used a cell-type-specific deconvolution technique to determine how the expression signatures of epithelial-relevant cell types differentiate the two groups. We used cell type specific signatures developed from a microarray database containing 723 samples associated with 25 epithelial-relevant cell types [25], and calculated a signature score across these 25 cell types for each of the 84 HPV(+) tumors (see Supplemental Methods).
We found that integration-negative tumors had stronger immune signatures, characterized by heightened signatures for CD4+ T-cells, CD8+ T-cells, CD3+ T-cells, NK cells, Regulatory T-cells, B cells, NK T-cells and CD34+ cells (Mann-Whitney U test; all FDR < 0.10) (Figure 4C, Table S8), which suggests that these tumors have higher levels of infiltrating immune cells. To confirm this, we performed assessment of lymphocyte infiltration in our 18 HPV-positive UM samples. We validated the trend of higher lymphocyte infiltration in HPV integration-negative tumors (average value degree of infiltration 2.11) compared to integration-positive tumors (average value 1.67), although due to the small sample size (9 vs 9), it did not reach statistical significance (p=0.1428 by Mann-Whitney U non-parametric test). We used H&E (hematoxylin and eosin stained) slides and graded lymphocyte infiltration on a scale of 0–4+ which corresponds to the number of lymphocytes inside tumor area versus outside [26] (Supplemental Table S14). The integration-negative did not have significantly higher signatures for macrophages, gamma-delta T-cells, or neutrophils. The strongest cell type for integration-positive samples was keratinocytes (FDR = 0.0617) (Table S8).
Discussion
Human papillomavirus (HPV)-related oropharyngeal cancer has been rising in prevalence in the United States, and is expected to soon overtake cervical cancer in incidence rate [2–4]. Conventional treatment for patients with advanced cancers generally involves radical surgical resection and/or intensive high dose radiation. Both modalities are associated with significant functional morbidity. Although as a group, survival of HPV(+) oropharyngeal cancer patients is generally better than their HPV(−) counterparts, biomarkers to predict which patients would benefit from a de-escalated therapy regimen versus a more aggressive treatment plan similar to that standard for HPV(−) patients are not clear. Integration of the HPV genome into the host’s genome is one viral-related event that has remarkable downstream consequences affecting viral expression, the host immune response, cellular differentiation and more. Thus, patients harboring one or more viral integrations may have heterogeneous prognoses or responses to treatment, as has been observed in analyses considering the number of detected viral integrations in cervical cancers [13].
Most studies of viral integration have focused on the DNA, however not all DNA integration events are transcribed [27]. There is strong evidence that tumors with presence of viral DNA and RNA (HPV16 DNA+ RNA+) are very different from those which have viral DNA but not RNA (HPV16 DNA+ RNA−) and viral DNA-negative tumors, which were found to be similar and clinically indistinguishable [27]. We hypothesized that since expression is key, identification of viral integration using RNA may be a more clinically relevant marker, especially in a genic region where the tumor could exploit the cell’s promoter region to induce viral gene transcription, knock out the relevant gene’s function, or use viral transcriptional regulation to over-express an oncogene. Two data sources provide support for this hypothesis. Zhang et al, 2016, characterized two subtypes of HPV(+) oral cancers that were correlated with HPV-host fusion transcript status, and this correlated with several other variables known to affect survival (chr16q loss, E2/E5 expression, immune response, and BCL2 expression) [17]. Second, the TCGA HPV(+) tumors that were found to have an HPV integration event from the WGS data, but not from RNA-seq, have properties more consistent with the other integration-negative patients. That is, they had higher immune response signature and higher expression of E2 (Figure 4C). It’s possible that samples with integration and elevated expression of E2 could bear both integrated and episomal forms. However, a limitation of our analysis is that with RNA-seq data we could not confidently distinguish samples with mixed versus integrated-only forms of the HPV oncogenes.
HPV integration events in cervical cancer map broadly across the human genome but with frequent breakpoints in genic regions [14]. In the study of HPV integration in 35 HNSCC TCGA tumors, integration into at least one host gene was identified in 54% of cases and it was found within 20 kb of a gene in another 17% cases, suggesting a selective pressure for viral integration in or near genic regions [21]. Similarly, Akagi and colleagues (2014) observed enrichment of HPV integration sites detected using WGS data within 50 kb of RefSeq genes in a panel of 10 cervical and head and neck cancer cell lines, and also modest enrichment within common fragile sites or DNase I hypersensitivity sites [28]. However, after adjustment for the over-representation of breakpoint clusters, the enrichment of integration in the genomic fragile regions was not significant. Different conclusions were made regarding the association of integration sites with fragile regions. HPV integrations were previously detected within or close to fragile sites in cervical tumors [29–32] and head and neck cell lines [33]. However, other studies, including a comprehensive analysis of 135 cervical cancers and cell lines, did not find statistically significant evidence for this [14]. Doolittle-Hall with colleagues performed meta-analysis of DNA tumor-viral integration sites and showed no evidence for preferential HPV integration in CFSs [34]. Our results also did not show statistically significant association between CFSs and sites of integration in transcribed regions of cancer genomes at the α=0.05 level. Inconsistent outcomes from different studies may be due to a small effect size, confounding variables, and/or ill-defined fragile site boundaries. We did find significant associations between HPV integration sites and repetitive regions of the human genome (LINEs, MIR SINEs, DNA, and LTR). Other studies also demonstrated enrichment of integration sites within repeats [14, 29, 34].
Viral integration is detected in almost 90% of cervical carcinomas [35] and presents a crucial step in carcinogenesis: its appearance correlates with the progression of precancerous lesions (CIN2/3) to invasive carcinoma. In our study, 61% of samples were defined as integration-positive based on virus-host fusion transcripts. Integration is not a normal part of the HPV life cycle and is characterized by loss of E2, which regulates transcription of E6 and E7. In our analysis, E1, E2, E4, and E5 gene expression were significantly reduced in integration-positive samples compared to integration-negative. There was no evidence for a difference in expression of oncogenes E6 or E7 by integration status. These findings are in agreement with Hafner et al (2008), who observed highly variable levels of viral oncogene expression in CIN and cervical cancers, but these levels were independent of the physical state of the viral genome (episomal, integrated or mixed forms) [11]. Thus, our data also support the hypothesis that HPV integration ensures an essential level of expression of the viral oncogenes instead of an elevated level of oncogene expression. The presence of breakpoints in E6 and E7 concurrently with positive expression of these viral oncogenes could be due to (1) another integration event not in the same oncogene, (2) additional episomal expression of the gene, or (3) the cells with the breakpoint could still transcribe an isoform of E6 or E7 that did not violate its carcinogenic function (see Supplemental Figure S2).
Our results show striking overrepresentation of integration events in or near genes known to be important to head and neck cancers, lung cancers, and urogenital cancers. Lung cancers are known to have several molecular similarities to head and neck cancers [36], while genital cancers are also dominated by an HPV-related etiology. These results suggest strong natural selection in HPV(+) tumors for cells either with an integration event that enhances activation of an oncogene or damages the function of a tumor suppressor gene. If true, we would expect to see increased expression of oncogenes with an integration event in the sample with the integration. And for tumor suppressor genes, we would expect an enrichment of integration events in exonic regions, which would functionally knock out the protein. While we did find significant overall increased expression of genes with an integration, especially in exons, this increase may be partially due to increased copy number caused by HPV-driven amplification events. We did not reach statistical significance for whether tumor suppressor genes were more likely to have integration in an exon. However, the study was not powered to detect these differences. Our results are consistent with recent findings from an analysis of 10 patients who differed in tumor response after therapy, which demonstrated that almost all HPV integration events identified in responsive tumors were detected in the intergenic chromosome regions, whereas the majority of integrations in the recurrent tumors were detected in cellular genes [37]. Moreover, they demonstrated that the genes disrupted by viral integration in nonresponsive tumors were related to cancer or differentially expressed in cancers. Our results are also consistent with those found in a recent meta-analysis of integration in cervical cancers which demonstrates that genes targeted by HPV integration are concentrated in transcriptionally active regions and enriched in cancer-related functional terms and pathways [38].
Our analysis revealed that some genes harboring integration events are characterized by carcinogenic functions in a variety of squamous cell neoplasms, and some of these genes were also recurrent and/or were hubs in our protein interaction network. Several lines of evidence point to the importance of CD274. PD-L1 (CD274) is one of two ligands specific to PD-1, and members of the promising immune checkpoint pathways currently investigated in HNSCCs [39]. PD-1 mediated T-cell signaling is characterized by altered cytotoxic killing, cytokine production and T cell proliferation [40]. Ligands PD-L1 (CD274) or PD-L2 (CD273) are upregulated in many human tumors, including HNSCCs [40] and tumor immune evasion can occur by high tumor expression of PD-L1 [41]. Blockade of PD-1 or PD-L1 by specific monoclonal antibodies can reverse the anergic state of tumor-specific T cells and thereby enhance antitumor immunity [40]. Recent clinical trials have demonstrated significant tumor responses and improved survival with anti-PD-L1 and anti-PD-1 therapy in advanced HNSCC, melanoma, lung, and renal cell cancers [42–44]. A recent study of patients with cervical and vulvar SCCs [45] revealed that increased PD-L1 protein expression was caused by co-gain or co-amplification of CD274 and PDCD1LG2 genes in a significant portion of patients, and therefore these patients also were candidates for clinical trials of PD-1 blockade.
Our subsequent analysis of differentially expressed genes by integration status confirmed the importance of HPV integration for clinical outcomes. Immune-related genes were the most highly overrepresented among the genes with significantly elevated expression in integration-negative samples, which may explain the better survival rate for this group of patients. Our cell-type specific signatures showed elevated expression of genes specifically expressed in T-cells (CD4+, Regulatory, CD3+, and CD8+), NK cells, and B cells in integration-negative tumors. High tumor infiltrates of T-cells has been associated with improved survival [46, 47]. These immune cells and genes are important in establishing a tumor immune response, and may be a result of these tumors being in or near a lymph node and the immunogenicity of the non-integrated (episomal) form of HPV. Further, high expression of PD-L1 has been associated with decreased T-cell tumor infiltration [48]. Despite the clear evidence of enrichment for immune-related genes in integration-negative tumors, which is in agreement with better survival for this group of patients, a larger cohort of patients is needed to better estimate the influence of viral integration on the immune response and patient’s survival.
The gene RAD51B (RAD51 paralog B) is essential for DNA repair by homologous recombination. Overexpression of this gene was found to cause cell cycle G1 delay and cell apoptosis. Therefore, disruption of RAD51B by viral integration may facilitate tumor development. We observed recurrent integrations of HPV16 into intronic and exonic regions of this gene in two samples, with slightly to significantly elevated expression in them. Recurrent integrations into the RAD51B gene were also observed in the intronic regions in three cervical tumors [31] suggesting the importance of the homologous recombination repair pathway in multiple HPV(+) SCC types.
Among the genes identified as interaction hubs in our network analysis (TP63, ETS2, RUNX1, and FOXA1), all have important roles in cancer development. ETS2 (ETS proto-oncogene 2) is a tumor suppressor gene that regulates development and apoptosis and is important in cancer-specific epigenetic networks. Over-activation of ETS2 induces hyper-proliferation of epidermal stem cells accompanied by upregulation of SCC super-enhancer-associated genes FOS, JUNB and KLF5 [49]. TP63 is a member of the p53 family of transcription factors (p53, p63, p73), which share a high degree of homology and are important to cell homeostasis. p63 regulates many p53 target genes and can compensate for the loss of p53. In one study, the genomic sequence of p63 was amplified in 88% of squamous carcinomas [50]. Also, in Zhang et al 2016, our group found that one subtype of HPV(+) tumors (which were enriched with “keratinocyte differentiation” and included mostly integration-positive samples) had more amplifications on all or a significant portion of chr3q, where TP63 is located. Mutations in TP63 were found in HNSCC, lung SCC and cervical cancers from TCGA [16, 23, 24, 36]. Another gene affected by HPV integration in our study, and mutated in cervical cancer and HNSCC [16], is RUNX1. RUNX1 (runt related transcription factor 1) is a known hematopoietic stem cell and leukemia factor, and is overexpressed and essential for some human epithelial cancers: skin SCC, oral SCC, and ovarian cancer [51].
We also found the first HPV16 integration in ERBB2 (erb-b2 receptor tyrosine kinase 2) or HER2 gene, which had elevated expression in the sample with the integration compared to other analyzed samples. ERBB2 regulates cell growth, survival, and differentiation via multiple signal transduction pathways and participates in cellular proliferation and differentiation. ERBB2 can form heterodimers with other EGF receptor family members and enhance kinase-mediated activation of downstream signaling pathways, such as MAPK, PI3K-Akt, and protein kinase C (PKC) [52]. Overexpression of ERBB2 occurs in many cancer types, and HER2 aberrations were recently identified in a subset of HNSCCs [53], suggesting HER2 positive HNC patients could benefit from the targeted anti-HER2 therapy. Mutations in this gene also were found in HNSCC, lung SCC and cervical cancers from TCGA [16, 23, 24, 36].
Our survival analysis revealed that HPV(+) patients with integration-negative tumors (defined by absence of expression of viral-host fusion RNA transcripts) have better overall survival compared to those with integration-positive tumors. Moreover, the survival rate for integration-positive patients was similar to that of HPV(−) patients. We speculate that the well-known better survival rate for HPV(+) patients could be attributed mostly to the better survival of patients with integration-negative tumors, which may be related to the enhanced immunogenicity of episomal HPV. The impact integration status had on clinical outcome is in agreement with cervical cancer studies [13, 54]. Das and colleagues (2012) demonstrated that cervical cancer patients with the episomal form of HPV had better disease free survival (after radical radiotherapy) than patients with integrated HPV [54]; this study of Indian women used the APOT assay for identification of integration sites. Also, Shin with colleagues (2014) reported that HPV integration was a significant prognostic factor for poor disease-free survival in patients with cervical cancer [13]. Other studies did not report significant association of integration status with clinical outcomes, but demonstrated a trend toward worse survival for tumors with viral integration [55, 56]. The above-mentioned studies compared tumors with integrated or mixed viral forms versus the episomal form of the virus. There may be inconsistencies among the studies due to different approaches in identification of integration.
A difference between our study and those mentioned above is that we analyzed integration events based on virus-host fusion transcripts, and stratified the samples on this basis. Thus, integration-positive tumors all had a transcribed integration event, but also could carry episomal forms. Integration-negative tumors in our study did not show actively expressed virus-host fusions, but may have low level expression of integrated viral DNA. Thereby, one of the limitations of our study is that we could not directly estimate expression levels of integrated versus episomal forms of the virus in each tumor. We believe the gold standard for detection of HPV integration events should be to use both WGS and RNA-seq data. WGS alone will result in false positive cases in the sense that some patients will have a DNA integration event that is not expressed or clinically relevant. RNA-seq alone will likely result in false negative cases where an integration event is expressed but without transcription of any of the surrounding host genomic sequence. How common these false positives and false negatives are is unknown. Therefore, the relative performance of RNA-seq versus WGS for integration detection is not yet known. In this study, we chose to use RNA-seq, due to the lower cost, the ability to confirm expression of the integrated form, and to contrast with the previous experiments performed with DNA sequencing.
The treatment of oropharyngeal cancer is actively evolving to better reflect a recent appreciation of differences in epidemiology, marked by increased incidence and survivorship associated with HPV-related HNSCC. Biomarkers useful in personalizing therapy for patients with oropharyngeal cancer are urgently needed, particularly with the introduction of new immunotherapeutic strategies. The current findings suggest that HPV viral integration status is an important and potentially useful clinical biomarker that will need confirmation in larger, prospective validation studies.
Supplementary Material
Implications.
These findings demonstrate the clinical relevancy of expressed HPV-integration, which is characterized by a change in immune response and/or aberrant expression of the integration-harboring cancer-related genes, and suggest strong natural selection for tumor cells with expressed integration events in key carcinogenic genes.
Acknowledgments
Funding
This work was funded by National Institutes of Health grants R01 CA158286, the University of Michigan Specialized Programs of Research Excellence (SPORE) grant (P50 CA097248), and the National Human Genome Research Institute (NHGRI) training grant (T32 HG00040).
We would like to acknowledge Emily L. Bellile for help with the survival analysis, William R. Swindell for helping with the cell type specific gene signature analysis, Heather M. Walline for identifying the U of M tumor samples with HPV-MultiPlex PCR MassArray and Heming Yao for providing the enhancer regions that were linked to CD274.
Footnotes
Author contributions
L.A.K. performed the bioinformatics and statistical analyses, and contributed to the interpretation of the data, and writing of the manuscript; Y.Z. and P.B.H. contributed specific bioinformatics analyses and interpretation of the data; S.V. processed samples and helped interpret the data; J.B.M. performed a histopathological assessment of the UM tumor samples; D.B.C contributed in acquisition of the UM samples; G.T.W and T.E.C contributed conceptually to the study, data interpretation, and revision of the manuscript; L.S.R. contributed in sample collection, concept and design of the study, interpretation of the data and reviewing of the manuscript; MAS contributed in concept and design of the study, determined and spearheaded the bioinformatics and statistical analyses, and participated in the interpretation of data and writing of the manuscript. All authors read and approved of the manuscript.
References
- 1.Ferlay J, et al. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet] 2013 Available from: http://globocan.iarc.fr/Pages/burden_sel.aspx.
- 2.Moore KA, 2nd, Mehta V. The Growing Epidemic of HPV-Positive Oropharyngeal Carcinoma: A Clinical Review for Primary Care Providers. J Am Board Fam Med. 2015;28(4):498–503. doi: 10.3122/jabfm.2015.04.140301. [DOI] [PubMed] [Google Scholar]
- 3.Chaturvedi AK, et al. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J Clin Oncol. 2013;31(36):4550–9. doi: 10.1200/JCO.2013.50.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaturvedi AK, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29(32):4294–301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munoz N, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348(6):518–27. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 6.Stanley M. HPV - immune response to infection and vaccination. Infect Agent Cancer. 2010;5:19. doi: 10.1186/1750-9378-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergvall M, Melendy T, Archambault J. The E1 proteins. Virology. 2013;445(1–2):35–56. doi: 10.1016/j.virol.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McBride AA. The papillomavirus E2 proteins. Virology. 2013;445(1–2):57–79. doi: 10.1016/j.virol.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doorbar J, et al. The biology and life-cycle of human papillomaviruses. Vaccine. 2012;30(Suppl 5):F55–70. doi: 10.1016/j.vaccine.2012.06.083. [DOI] [PubMed] [Google Scholar]
- 10.Melsheimer P, et al. DNA aneuploidy and integration of human papillomavirus type 16 e6/e7 oncogenes in intraepithelial neoplasia and invasive squamous cell carcinoma of the cervix uteri. Clin Cancer Res. 2004;10(9):3059–63. doi: 10.1158/1078-0432.ccr-03-0565. [DOI] [PubMed] [Google Scholar]
- 11.Hafner N, et al. Integration of the HPV16 genome does not invariably result in high levels of viral oncogene transcripts. Oncogene. 2008;27(11):1610–7. doi: 10.1038/sj.onc.1210791. [DOI] [PubMed] [Google Scholar]
- 12.Hudelist G, et al. Physical state and expression of HPV DNA in benign and dysplastic cervical tissue: different levels of viral integration are correlated with lesion grade. Gynecol Oncol. 2004;92(3):873–80. doi: 10.1016/j.ygyno.2003.11.035. [DOI] [PubMed] [Google Scholar]
- 13.Shin HJ, et al. Physical status of human papillomavirus integration in cervical cancer is associated with treatment outcome of the patients treated with radiotherapy. PLoS One. 2014;9(1):e78995. doi: 10.1371/journal.pone.0078995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Z, et al. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat Genet. 2015;47(2):158–63. doi: 10.1038/ng.3178. [DOI] [PubMed] [Google Scholar]
- 15.Rusan M, Li YY, Hammerman PS. Genomic landscape of human papillomavirus-associated cancers. Clin Cancer Res. 2015;21(9):2009–19. doi: 10.1158/1078-0432.CCR-14-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas, N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–82. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, et al. Subtypes of HPV-positive head and neck cancers are associated with HPV characteristics, copy number alterations, PIK3CA mutation, and pathway signatures. Clin Cancer Res. 2016 doi: 10.1158/1078-0432.CCR-16-0323. [DOI] [PMC free article] [PubMed]
- 18.Wilks C, et al. The Cancer Genomics Hub (CGHub): overcoming cancer through the power of torrential data. Database (Oxford) 2014 doi: 10.1093/database/bau093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, et al. VirusSeq: software to identify viruses and their integration sites using next-generation sequencing of human cancer tissue. Bioinformatics. 2013;29(2):266–7. doi: 10.1093/bioinformatics/bts665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee C, Patil S, Sartor MA. RNA-Enrich: a cut-off free functional enrichment testing method for RNA-seq with improved detection power. Bioinformatics. 2016;32(7):1100–2. doi: 10.1093/bioinformatics/btv694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parfenov M, et al. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc Natl Acad Sci U S A. 2014;111(43):15544–9. doi: 10.1073/pnas.1416074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanley M. Immunobiology of HPV and HPV vaccines. Gynecol Oncol. 2008;109(2 Suppl):S15–21. doi: 10.1016/j.ygyno.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swindell WR, et al. Dissecting the psoriasis transcriptome: inflammatory- and cytokine-driven gene expression in lesions from 163 patients. BMC Genomics. 2013;14:527. doi: 10.1186/1471-2164-14-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berinstein NL, et al. Increased lymphocyte infiltration in patients with head and neck cancer treated with the IRX-2 immunotherapy regimen. Cancer Immunol Immunother. 2012;61(6):771–82. doi: 10.1007/s00262-011-1134-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wichmann G, et al. The role of HPV RNA transcription, immune response-related gene expression and disruptive TP53 mutations in diagnostic and prognostic profiling of head and neck cancer. Int J Cancer. 2015;137(12):2846–57. doi: 10.1002/ijc.29649. [DOI] [PubMed] [Google Scholar]
- 28.Akagi K, et al. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res. 2014;24(2):185–99. doi: 10.1101/gr.164806.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thorland EC, et al. Human papillomavirus type 16 integrations in cervical tumors frequently occur in common fragile sites. Cancer Res. 2000;60(21):5916–21. [PubMed] [Google Scholar]
- 30.Wentzensen N, Vinokurova S, von Knebel Doeberitz M. Systematic review of genomic integration sites of human papillomavirus genomes in epithelial dysplasia and invasive cancer of the female lower genital tract. Cancer Res. 2004;64(11):3878–84. doi: 10.1158/0008-5472.CAN-04-0009. [DOI] [PubMed] [Google Scholar]
- 31.Ojesina AI, et al. Landscape of genomic alterations in cervical carcinomas. Nature. 2014;506(7488):371–5. doi: 10.1038/nature12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang WS, et al. Simultaneous characterization of somatic events and HPV-18 integration in a metastatic cervical carcinoma patient using DNA and RNA sequencing. Int J Gynecol Cancer. 2014;24(2):329–38. doi: 10.1097/IGC.0000000000000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olthof NC, et al. Viral load, gene expression and mapping of viral integration sites in HPV16-associated HNSCC cell lines. Int J Cancer. 2015;136(5):E207–18. doi: 10.1002/ijc.29112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doolittle-Hall JM, et al. Meta-Analysis of DNA Tumor-Viral Integration Site Selection Indicates a Role for Repeats, Gene Expression and Epigenetics. Cancers (Basel) 2015;7(4):2217–35. doi: 10.3390/cancers7040887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pett M, Coleman N. Integration of high-risk human papillomavirus: a key event in cervical carcinogenesis? J Pathol. 2007;212(4):356–67. doi: 10.1002/path.2192. [DOI] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas Research, N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walline HM, et al. Genomic Integration of High-Risk HPV Alters Gene Expression in Oropharyngeal Squamous Cell Carcinoma. Mol Cancer Res. 2016;14(10):941–952. doi: 10.1158/1541-7786.MCR-16-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang R, et al. Dysregulation of host cellular genes targeted by human papillomavirus (HPV) integration contributes to HPV-related cervical carcinogenesis. Int J Cancer. 2016;138(5):1163–74. doi: 10.1002/ijc.29872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zandberg DP, Strome SE. The role of the PD-L1:PD-1 pathway in squamous cell carcinoma of the head and neck. Oral Oncol. 2014;50(7):627–32. doi: 10.1016/j.oraloncology.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 40.Pai SI, Zandberg DP, Strome SE. The role of antagonists of the PD-1:PD-L1/PD-L2 axis in head and neck cancer treatment. Oral Oncol. 2016;61:152–8. doi: 10.1016/j.oraloncology.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferris RL. Immunology and Immunotherapy of Head and Neck Cancer. J Clin Oncol. 2015;33(29):3293–304. doi: 10.1200/JCO.2015.61.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chow LQ, et al. Antitumor Activity of Pembrolizumab in Biomarker-Unselected Patients With Recurrent and/or Metastatic Head and Neck Squamous Cell Carcinoma: Results From the Phase Ib KEYNOTE-012 Expansion Cohort. J Clin Oncol. 2016 doi: 10.1200/JCO.2016.68.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reck M, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med. 2016 doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 44.Menon S, Shin S, Dy G. Advances in Cancer Immunotherapy in Solid Tumors. Cancers (Basel) 2016;8(12) doi: 10.3390/cancers8120106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howitt BE, et al. Genetic Basis for PD-L1 Expression in Squamous Cell Carcinomas of the Cervix and Vulva. JAMA Oncol. 2016;2(4):518–22. doi: 10.1001/jamaoncol.2015.6326. [DOI] [PubMed] [Google Scholar]
- 46.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489(7414):75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen N, et al. Tumor infiltrating lymphocytes and survival in patients with head and neck squamous cell carcinoma. Head Neck. 2016;38(7):1074–84. doi: 10.1002/hed.24406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cho YA, et al. Relationship between the expressions of PD-L1 and tumor-infiltrating lymphocytes in oral squamous cell carcinoma. Oral Oncol. 2011;47(12):1148–53. doi: 10.1016/j.oraloncology.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 49.Yang H, et al. ETS family transcriptional regulators drive chromatin dynamics and malignancy in squamous cell carcinomas. Elife. 2015;4:e10870. doi: 10.7554/eLife.10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Massion PP, et al. Significance of p63 amplification and overexpression in lung cancer development and prognosis. Cancer Res. 2003;63(21):7113–21. [PubMed] [Google Scholar]
- 51.Scheitz CJ, et al. Defining a tissue stem cell-driven Runx1/Stat3 signalling axis in epithelial cancer. EMBO J. 2012;31(21):4124–39. doi: 10.1038/emboj.2012.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iqbal N, Iqbal N. Human Epidermal Growth Factor Receptor 2 (HER2) in Cancers: Overexpression and Therapeutic Implications. Mol Biol Int. 2014;2014:852748. doi: 10.1155/2014/852748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Birkeland AC, et al. Identification of Targetable ERBB2 Aberrations in Head and Neck Squamous Cell Carcinoma. JAMA Otolaryngol Head Neck Surg. 2016;142(6):559–67. doi: 10.1001/jamaoto.2016.0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Das P, et al. HPV genotyping and site of viral integration in cervical cancers in Indian women. PLoS One. 2012;7(7):e41012. doi: 10.1371/journal.pone.0041012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vojtechova Z, et al. Analysis of the integration of human papillomaviruses in head and neck tumours in relation to patients’ prognosis. Int J Cancer. 2016;138(2):386–95. doi: 10.1002/ijc.29712. [DOI] [PubMed] [Google Scholar]
- 56.Lim MY, et al. Human papillomavirus integration pattern and demographic, clinical, and survival characteristics of patients with oropharyngeal squamous cell carcinoma. Head Neck. 2016;38(8):1139–44. doi: 10.1002/hed.24429. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.