Abstract
Hepatocellular carcinoma (HCC) accounts for ~85% of malignant liver tumors and results in 600,000 deaths each year, emphasizing the need for new therapies. Upregulation of menin was reported in HCC patients and high levels of menin correlate with poor patient prognosis. The protein-protein interaction between menin and histone methyltransferase Mixed Lineage Leukemia 1 (MLL1) plays an important role in the development of HCC, implying that pharmacologic inhibition of this interaction could lead to new therapeutic strategy for the HCC patients. Here, we demonstrate that the menin-MLL inhibitor MI-503 shows anti-tumor activity in in vitro and in vivo models of HCC and reveal the potential mechanism of menin contribution to HCC. Treatment with MI-503 selectively kills various HCC cell lines and this effect is significantly enhanced by a combination of MI-503 with sorafenib, the standard of care therapy for HCC. Furthermore, MI-503 reduces sphere formation and cell migration in in vitro HCC models. When applied in vivo, MI-503 gives a strong anti-tumor effect both as a single agent and in combination with sorafenib in mice xenograft models of HCC. Mechanistically, treatment with MI-503 downregulates expression of several genes known to play a critical role in proliferation and migration of HCC cells, including PEG10, and displaces the menin-MLL1 complex from the PEG10 promoter, resulting in reduced H3K4 methylation and transcriptional repression. Overall, our studies reveal a mechanistic link between menin and genes involved in HCC and demonstrate that pharmacologic inhibition of the menin-MLL interaction might represent a promising therapeutic approach for HCC.
Keywords: menin-MLL inhibitor, liver cancer, targeted therapy, protein-protein interaction inhibitor, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) accounts for ~85% of primary malignant liver tumors and is the fifth most common cancer in the world, leading to over 600,000 deaths each year (1,2). The majority of HCC patients are diagnosed at a late stage of the disease, when curative therapies are no longer effective (3), and most patients die about 6 months after diagnosis (1,4,5). Furthermore, patients who initially respond to curative therapies frequently experience disease recurrence, resulting in an overall 5-year survival of only 7% (6), while patients with surgically resectable tumors have survival rates still below 40% (7). All these findings strongly support the need for new and more effective therapies for HCC. One recent new treatment strategy for advanced HCC patients is the use of the multitargeted kinase inhibitor sorafenib (3), which blocks the activity of Raf serine/threonine kinases and various receptor tyrosine kinases (8). Sorafenib inhibits the proliferation of HCC cells and blocks tumor angiogenesis, resulting in an increase of about three months in the median survival of HCC patients (8). However, sorafenib has only limited clinical efficacy with undesired side effects (9), strongly implying the need for more efficacious therapies for HCC.
Recently it has been demonstrated that menin, a scaffold protein encoded by the multiple endocrine neoplasia type 1 (MEN1) gene, is involved in promoting hepatocellular carcinoma (10). Menin is a highly specific binding partner of Mixed Lineage Leukemia 1 (MLL1, also known as KMT2A) (11,12), a histone methyltransferase that catalyzes the trimethylation of H3K4 (H3K4me3) (13), and is required for the recruitment of the MLL1 complex to the target genes (14). Menin upregulation was reported in HCC patients, and high levels of menin correlate with poor patient prognosis (10). Indeed, menin knockdown substantially represses HCC cell proliferation, blocks colony formation and inhibits tumor growth in the xenograft model of HCC (10). Likewise, heterozygous loss of Men1 reduces development of HCC in vivo (10).
The function of menin in liver tumorigenesis has been linked to the transcriptional regulation of the Yes-associated protein (Yap1), an important oncogene in HCC (15) and a downstream target of the Hippo pathway (16,17). Menin occupancy at the promoter region of Yap1 has been noted to coincide with H3K4me3, a histone mark regulated by MLL1 (10), implying a potential involvement of the menin-MLL1 interaction in HCC development. We, and others, have previously shown a critical role of the menin interaction with MLL1 and/or MLL fusion proteins in acute leukemias with translocations of the MLL gene (12,18), as well as in solid tumors, including metastatic prostate cancer (19). All these studies imply that the menin-MLL1 interaction might play a more general role in cancer, including liver tumorigenesis; therefore inhibition of this interaction with small molecules might represent a novel therapeutic approach for HCC treatment. On the other hand very limited effect observed in HCC cells following treatment with a weak menin-MLL inhibitor MI-1 (no significant effect on cell viability in liquid culture and <25% inhibition of colony formation and cell migration when applied as a single agent at 4 μM) (20) raises questions regarding the potential therapeutic value of blocking the menin-MLL1 interaction as a treatment for liver cancer.
We have recently reported very potent small molecule inhibitors of the menin-MLL1 interaction with optimized drug-like properties, including MI-503 (IC50 = 14 nM, Kd = 9 nM), which demonstrated very potent activity in both in vitro and in vivo models of MLL leukemia (18,21–23) as well as in castration resistant prostate cancer (19). Since the in vitro potency of MI-503 is >140-fold better than MI-1 and it demonstrates strong and selective activity in cancer cells, we selected MI-503 to study the effect of pharmacologic inhibition of the menin-MLL interaction in liver cancer. Here, we performed a systematic evaluation of the MI-503 in HCC models to assess whether pharmacologic inhibition of the menin-MLL1 interaction might represent a new therapeutic strategy for liver cancer. Treatment with MI-503 had a very pronounced effect in various models of HCC, both in vitro and in vivo, and this effect was enhanced when MI-503 was combined with sorafenib, the current standard of care therapy in patients with advanced HCC. Pharmacologic inhibition of menin-MLL1 by MI-503 strongly downregulated the expression level of genes previously linked to HCC, including PEG10 (24,25), thereby providing a new mechanistic insight into the role of menin-MLL1 and H3K4me3 in HCC. Overall, our findings demonstrate that pharmacologic inhibition of the menin-MLL1 interaction can block progression of HCC, thus validating this protein-protein interaction as an attractive target for therapeutic intervention in liver cancer.
Materials and Methods
Chemistry
Chemical synthesis and chemical characterization of MI-503 and MI-372 compounds have been described previously (18).
Cell culture
The HepG2 (low-metastatic) (26) and Hep3B (low-metastatic) (27) human HCC cell lines were obtained from ATCC in 2014 while the remaining cell lines (MHCC97, PLC/PRF/5, SNU449, and SNU423) were received from Dr. Ilona Kryczek, University of Michigan. HepG2, Hep3B and PLC/PRF/5 cells were maintained in Eagles Minimum Essential Medium (EMEM) (ATCC) with 10% fetal bovine serum (FBS) and 1% penicilin/streptomycin (Pen Strep, Gibco) antibiotics. SNU449, SNU423 and MHCC97 were cultured in RPMI-1640 media with 10% FBS and 1% penicillin/streptomycin (Pen Strep, Gibco). ASC52 cell line was received from Dr. Elizabeth Lawlor and cultured in Mesenchymal Stem Cell (MSC) Basal Medium with Mesenchymal Stem Cell Growth Kit for Adipose and Umbilical-derived MSCs Low Serum Components and G418 (ATCC). All cell lines were used in the described experiments before reaching the 10th passage after thawing the cells out and were negative for mycoplasma prior performing these studies as assessed by the TOKU-E PCR Mycoplasma Detection Kit.
Cell viability assay
Cell viability assays are described in the Supplementary Information.
Cellular thermal shift assay
Cellular thermal shift assay (CETSA) was performed as described before (19,28). Experimental details are provided in the Supplementary Information.
Real-time qPCR
Cells were plated at a concentration of 0.25×106 cells/ml in the 12-well plate in duplicates and treated with MI-503 or DMSO for 6 and/or 13 days followed by RNA isolation according to the manufacturer’s protocol (RNeasy kit Qiagen), RNA reverse transcription to cDNA and quantitative PCR (see Supplementary Information for details).
RNAseq studies
HepG2 cells were plated in the 12-well plates at the initial concentration of 0.4×106 cells/ml and treated with 3 μM MI-503 or DMSO (0.25%) in triplicates. After 3 days of treatment cells were trypsinized, counted and viable cell number was adjusted to the original concentration. Media was changed and compound or DMSO was re-supplied at that time. Cells were harvested after 3 more days of incubation. Total RNA was isolated from cells using RNeasy kit (Qiagen), amplified, and quality was assessed using the TapeStation (Agilent). Sequencing was performed at the University of Michigan DNA Sequencing Core. Samples with RINs (RNA Integrity Numbers) of 8 or greater were prepped using the Illumina TruSeq mRNA kit (Illumina). RNA was converted to mRNA using a polyA purification. cDNA library was created using reverse transcriptase, barcoded and sequenced using 4 samples per lane on a HiSeq 2000 (Illumina) in High Output mode. Sequenced reads were aligned to human reference genome using Bowtie and Tophat (version 2.0.3). Differential gene expression analysis was done using program Cuffdiff. Genes with p < 0.05 and a fold change greater that 2-fold were considered significant. Gene Set Enrichment Analysis (http://www.broadinstitute.org/gsea/index.jsp) was performed on gene expression of MI-503 treated versus DMSO treated cells.
Sphere formation and cell migration assays
Sphere formation and cell migration assays are described in Supplementary Information.
In vitro combination studies
HepG2 and Hep3B cells were plated at a concentration of 0.45×105cells/ml in a 24-well plate and treated with 0.25% DMSO, sorafenib, MI-503 or both compounds. After 4 days (for HepG2) or 5 days (for Hep3B) of incubation cells were transferred to the 96-well plate in quadruplicates. The MTT cell proliferation assay kit (Roche) was then applied. Plates were read for absorbance at 570 nm using a PHERAstar BMG microplate reader. Drug interactions were analyzed by CalcuSyn program (Biosoft, Cambridge, UK) based on the analytical method of Chou and Talalay (29).
Chromatin Immunoprecipitation
1.5×106 HepG2 cells plated in 10 cm dish were treated with DMSO (0.25%) or MI-503 at 4 μM, 2 μM and 1 μM concentrations for 6 days. After 3 days of incubation cell number was adjusted to the starting concentration, media was changed and fresh compound was added. Following 6 days of incubation, cells were fixed with 1% paraformaldehyde for 10 min at room temperature. Cells were then quenched with glycine for 5 min at room temperature. ChIP was performed using Magna ChIP A/G kit (Millipore) according to the manufacturer’s protocol. The cell lysate was prepared and equal amounts of DNA isolated from either DMSO or MI-503 treated samples was immunoprecipitated using antibodies against Menin (Bethyl), MLL1 (Millipore), H3 (Abcam), H3K4me3 (Abcam) and IgG (Millipore) overnight at 4°C. The DNA was eluted and subjected to quantitative PCR using SYBR Green primers for PEG10 (see Supplementary Information for primer sequences and corresponding genomic reference coordinates).
Animal studies
All animal studies were performed in compliance with the guidelines of the University of Michigan Committee for Use and Care of Animals and Unit for Laboratory Animal Medicine (ULAM) under the protocol PRO00005043. Freshly thawed HepG2 or Hep3B cells (ATCC) were cultured in EMEM media with 10% fetal bovine serum (FBS) and 1% Pen Strep antibiotics. On the day of injection, cells were re-suspended in serum free medium at 1×107 cells/mL and mixed 1:1 with Matrigel. 0.5×107 cells/mL were injected to the flank of the 6 week old female athymic nude mice. When tumor size has reached ~100 mm3 mice were randomized into four groups with each group containing 8–9 mice to provide over 80% power to detect 50% effect. Mice were then treated with vehicle (25% DMSO, 25% PEG400, 50% PBS), MI-503 (dissolved in vehicle), sorafenib (dissolved in 12.5% Cremophor, 12.5% Ethanol and 75% water), or both compounds once daily at the designated doses using intraperitoneal, i.p. (vehicle and MI-503) or oral, p.o. (sorafenib) administration. Body weight and tumor sizes were monitored 3 times per week. The tumor volume was calculated according to the formula: tumor volume = a x b2/2 (a, long diameter; b, short diameter). Treatment continued until the mean tumor size in the vehicle group reached ~1200 mm3. The experimenters were not blind to group assignment and outcome assessment.
Statistical analysis
Student’s t-test (unpaired, two-tailed) was used to calculate significance level between treatment groups to calculate p values. P values of less than 0.05 were considered significant. Graph generation and statistical analysis were performed using GraphPad Prism version 6.02 software (GraphPad, La Jolla, CA).
Results
Menin is overexpressed in liver cancer cells
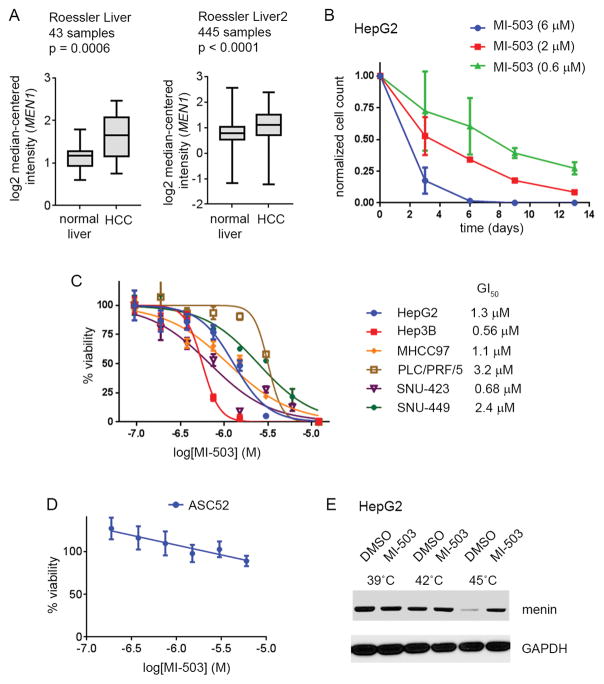
To understand the tumorigenic function of menin in hepatocellular carcinoma we compared the expression level of MEN1, which encodes menin, in normal liver and liver tumor samples by analyzing the Oncomine datasets (30). Comparison of the MEN1 expression level between the two sets of normal liver and HCC patient samples (31) revealed a significant upregulation of MEN1 in the liver tumor samples, Fig. 1A, consistent with the previous studies (10). We further investigated the role of menin in liver cancer by assessing the level of menin in several human HCC cell lines: HepG2, Hep3B, SNU449, SNU423, MHCC97, PLC/PRF/5. Western Blot analysis confirmed that menin is expressed and detectable in each of these HCC cell lines, although some variability in the menin expression was observed, Supplementary Fig. S1A.
Figure 1. Effect of MI-503 menin-MLL inhibitor on HCC cell lines growth.
A. MEN1 expression in normal versus tumor liver cells based on the analysis of the Oncomine datasets (30). B. Growth curves demonstrating the effect of MI-503 on cell growth in HepG2 cells. Viable cells were counted at days: 3, 6, 9, 13. Data are normalized to DMSO treated cells and represent mean of duplicates ± SD. C. MTT cell viability assay performed in HCC cell lines after 12 days of treatment with MI-503. Data are normalized to the DMSO treated cells and represent mean of quadruplicates ± SD. D. MTT cell viability assay performed for MI-503 in normal adipose-derived mesenchymal stem cells, ASC52. Data are normalized to the DMSO treated cells and represent mean of quadruplicates ± SD. E. Results from the Cellular Thermal Shift Assay (CETSA) performed in HepG2 cells. Detection of menin and GAPDH is shown on the SDS-PAGE gel. MI-503 was used at 2 μM.
Studies by Xu et al (10) support the importance of the menin interaction with MLL in hepatocellular carcinoma. Therefore, we used the panel of HCC cell lines to evaluate the expression level of two MLL family members, MLL1 (KMT2A) and MLL2 (KMT2B), which are both capable of strong interaction with menin (11). Interestingly, we found much higher expression level of MLL1 than MLL2 in all HCC cell lines tested, Supplementary Fig. S1B, which suggests that MLL1, rather than MLL2, plays an important role in liver cancer through a direct interaction with menin.
Menin-MLL inhibitor blocks proliferation of HCC cells
Knock down of menin substantially represses proliferation and reduces colony formation in HCC cells (10). To determine whether pharmacologic inhibition of the menin-MLL1 interaction affects growth of liver cancer cells we used our recently reported and very potent menin-MLL1 inhibitor MI-503 (Supplementary Fig. S1C), which has demonstrated strong activity in MLL leukemia (18) and prostate cancer (19) models. Treatment with MI-503 inhibited the proliferation of HepG2 liver cancer cells in a dose- and time-dependent manner, with a more pronounced effect observed upon prolonged treatment (over 7 days), Fig. 1B. The need for a prolonged treatment in the HCC cells is consistent with the effects we observed in MLL leukemia (18) and prostate cancer cells (19). Further examination of MI-503 in a panel of HCC cell lines revealed a strongly decreased viability of these cells after 7 and 12 days of treatment, with a half-maximal growth inhibitory (GI50) concentration ranging from 0.5 – 3.2 μM after 12 days of treatment, Fig. 1C, Supplementary Table 1. Importantly, treatment with a structurally similar but much weaker menin-MLL inhibitor MI-372 (Supplementary Fig. S1C) did not show an effect on HCC cell proliferation, Supplementary Fig. S1D, Supplementary Table 1. These results validate that targeted inhibition of the menin-MLL1 by MI-503 inhibits the growth of HCC cells. We have also tested the effects of MI-503 treatment in normal adipose-derived ASC52 mesenchymal stem cells, which served as a negative control cell line, and observed no substantial effect on cell growth up to 6 μM of MI-503, Fig. 1D, Supplementary Table 1. These data are consistent with no or limited effect of MI-503 in other negative control cell lines as we reported previously (18,19) and demonstrate specificity of MI-503 on proliferation of liver cancer or other menin-MLL dependent cells (18). To assess whether cell growth inhibition correlates with menin engagement by MI-503 we performed the cellular thermal shift assay (CETSA). MI-503 treatment of HepG2 cells at 45°C induced a thermal stabilization of menin when compared to the DMSO treated cells, Fig. 1E, Supplementary Fig. S1E, confirming that MI-503 binds to menin in HCC cells.
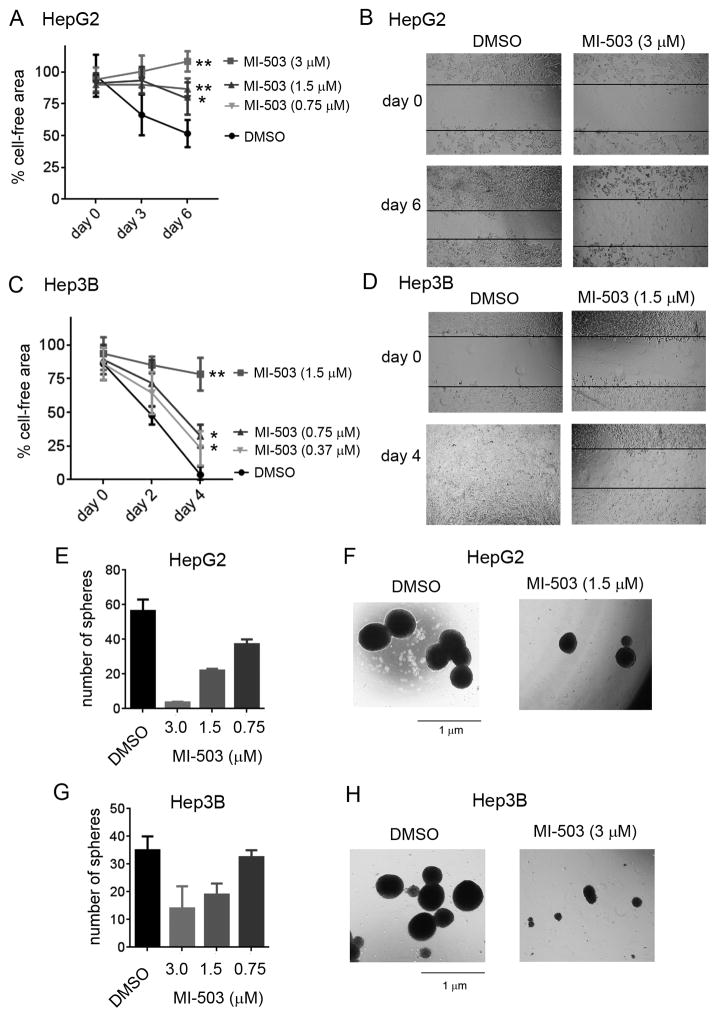
MI-503 blocks cell migration and sphere formation in HCC cells
The menin-MLL1 interaction was shown to contribute to the aggressive nature of HCC (10). As cell migration is a property of cancer cells associated with cancer metastasis, we assess the effect of menin inhibition on HCC cell migration using the cell culture wound closure assay (32) by measuring the rate of wound closure after treatment with MI-503. The migration of both HepG2 and Hep3B cells was markedly reduced upon treatment with MI-503 when compared to the DMSO treated cells, Fig. 2A–D and Supplementary Fig. S2A,B. Furthermore, we have also evaluated the effect of MI-503 on sphere formation in HepG2 and Hep3B cell lines, both of which were previously shown to retain the capacity of sphere formation (33,34), a propensity of cancer stem cells (35). Treatment with MI-503 strongly inhibited sphere formation in both cell lines as indicated by markedly reduced sphere numbers (Fig. 2E,G) and substantially smaller sphere sizes when compared to DMSO-treated cells, Fig. 2F,H. Overall, all these data confirm strong effect of the MI-503 menin-MLL inhibitor in HCC cell lines, supporting a potential therapeutic value of menin-MLL inhibitors in HCC.
Figure 2. MI-503 blocks sphere formation and cell migration in HCC cell lines.
A–D. Inhibition of cell migration in HepG2 (A, B) and Hep3B (C, D) cells upon treatment with MI-503 measured by the wound closure assay. Data in panels A and C represent mean of duplicates ± SD. * indicates p < 0.05, ** indicates p < 0.01. E, G. Quantification of spheres in the sphere formation assay performed in HepG2 (E) and Hep3B (G) cells upon 14 days of treatment with MI-503 and DMSO. Data represent mean of duplicates ± SD. F, H. Pictures of spheres upon treatment of HepG2 (F) and Hep3B (H) cells with MI-503 or DMSO.
MI-503 affects expression of genes linked to HCC
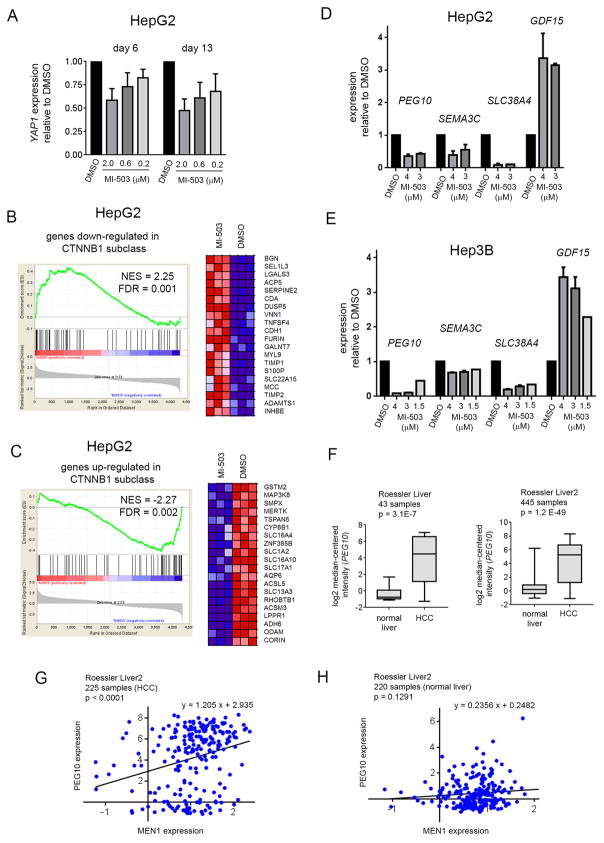
Previous studies have suggested that menin promotes liver tumorigenesis by regulating the expression and function of YAP1, an important “driver gene” in HCC, through an epigenetic mechanism involving the H3K4me3 histone mark (10). To investigate whether pharmacologic inhibition of the menin-MLL1 interaction affects YAP1 expression we have treated HepG2 cells with MI-503 and assessed YAP1 expression by qRT-PCR. We found a dose dependent reduction in YAP1 expression after 6 days of treatment with MI-503, but this effect was relatively modest, with only ~40% downregulation of YAP1 observed following treatment with 2 μM MI-503, Fig. 3A. Furthermore, downregulation of YAP1 expression did not become more pronounced after prolonged (13 days) treatment of HepG2 cells with MI-503, Fig. 3A.
Figure 3. Menin-MLL inhibitor inhibits genes involved in proliferation and migration of HCC cells.
A. Quantitative RT-PCR analysis of YAP1 expression performed in HepG2 cells after 6 and 13 days of treatment with MI-503 or DMSO. Data represent mean of duplicates ± SD. B,C. Gene Set Enrichment Analysis (GSEA) of genes downregulated (B) or upregulated (C) upon treatment with MI-503 in HepG2 cells as compared with genes from the Wnt signaling overexpressed in patient samples. The heat maps show genes comprising the leading edge of the GSEA plots. Red indicates high expression, blue indicates low expression. Triplicate samples were used for global gene expression studies. NES: Normalized Enrichment Score, FDR: False Discovery Rate. D, E. Quantitative RT-PCR analysis of PEG10, SEMA3C, SLC38A4 and GDF15 expression performed in HepG2 (D) or Hep3B (E) cells after 6 days of treatment with MI-503 or DMSO. Data represent mean of triplicates ± SD. Expression of genes in A, D, E was normalized to GAPDH and referenced to DMSO-treated cells. F. Expression level of PEG10 in normal liver samples compared to hepatocellular carcinoma as obtained from the analysis of two datasets (Roessler liver, Roessler liver 2) in the Oncomine database. G, H. Correlation of PEG10 with MEN1 expression in HCC (G) or normal liver samples (H). Gene expression is shown as log2 median-centered intensity.
To further investigate the effect of menin-MLL1 inhibition on gene expression changes in HCC cells we performed the RNA sequencing (RNA-seq) studies in HepG2 cells following treatment with MI-503. We found 291 and 552 genes down- and upregulated, respectively, that showed at least 2-fold change in the expression level following treatment with MI-503, Supplementary Table 2. We then have performed the GSEA analysis and found significant enrichment for genes up and down-regulated in a subclass of HCC with activated CTNNB1 gene encoding β-catenin (36). A set of genes up- and down-regulated upon treatment with MI-503 is, respectively, down and up-regulated in this subtype of HCC, Fig. 3B,C. The CTNNB1 gene encodes β-catenin and its mutations lead to the activation of Wnt signaling, which plays an essential role in development of HCC by controlling cell proliferation, apoptosis, cell cycle and motility (37–39). Mutations in CTNNB1 are highly prevalent in HCC, representing the second most frequently mutated gene in HCC (40). Indeed, the HepG2 cell line represents the liver cancer cell line characterized by activated Wnt signaling and expresses a truncated β-catenin lacking the Ser/Thr domain that regulates its degradation (41), Supplementary Table 3. The observation that MI-503 reverses gene expression signature in HCC with activating CTNNB1 mutations supports the potential application of menin-MLL1 inhibitors in HCC with activated Wnt-signaling pathway.
The RNA-seq data for HepG2 cells treated with MI-503 showed only a minor reduction (~20%) in the YAP1 expression, which did not reach statistical significance, Supplementary Table 2; this was likely due to a lower sensitivity of this method compared to qRT-PCR (see above). Interestingly, within the top genes strongly downregulated following treatment with MI-503 we found several genes known to promote proliferation or migration of HCC cells, including PEG10, SLC38A4 and SEMA3C (Supplementary Table 2). These results were further validated by the qRT-PCR studies in both HepG2 and Hep3B cells, demonstrating a strong reduction in the expression level of these genes following treatment with MI-503, Fig. 3D,E.
The PEG10 (Paternally Expressed Gene 10) gene is not expressed in normal adult liver (24) but is highly overexpressed in HCC (24,25). Indeed, the Oncomine (30) analysis has confirmed the high overexpression of PEG10 in HCC in different sets of patient samples when compared to normal liver samples, Fig. 3F. Furthermore, significant correlation of PEG10 and MEN1 expression was found in the HCC patient samples but not in normal liver samples (Fig. 3G,H), suggesting a positive regulation of PEG10 by menin. Importantly, PEG10 promotes cancer cell growth in HCC and is associated with poor patient survival and with tumor recurrence (24,25). Therefore, inhibition of HCC cell growth induced by MI-503, Fig. 1C, could, at least in part, result from the downregulation of PEG10. As mentioned above, treatment of HCC cells with MI-503 also leads to downregulation of both SLC38A4, a gene activated in HCC that confers growth and survival advantages in the pre-malignant and malignant lesions (42), and SEMA3C (semaphorin 3C), which plays an important role in migration of HCC cells (43), Fig. 3D,E and Supplementary Table 2. These results suggest that pharmacologic inhibition of the menin-MLL interaction can block hepatocellular carcinoma by its effects on pathways associated with the proliferation and migration of HCC cells.
Interestingly, among genes markedly upregulated upon treatment of HepG2 cells with MI-503 we found GDF15 (Growth Differentiation Factor 15), Supplementary Table 2. The qRT-PCR studies performed in HepG2 and Hep3B cells demonstrated ~3-fold increase in GDF15 expression following treatment with MI-503, and this effect was dose dependent, Fig. 3D,E. Downregulation of GDF15 was previously found to be associated with HCC development, while restoration of GDF15 level attenuated the progression of HCC (44); however the role of this gene in HCC seems to be quite complex (45). Nevertheless, we selected this gene as a positive control in the gene expression studies as its level increases following menin-MLL1 inhibition, but the functional consequences of GDF15 changes due to MI-503 treatment of HCC cells still need further investigation.
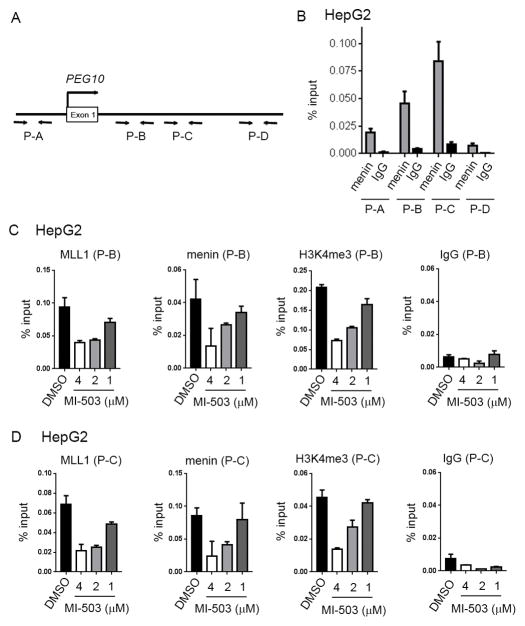
PEG10 is a direct target of menin-MLL1 complex
The finding that treatment of HCC cells with MI-503 resulted in downregulation of PEG10 prompted us to perform chromatin co-immunoprecipitation (ChiP) experiments in HepG2 cells to determine whether the menin-MLL1 complex regulates PEG10 expression through a direct binding to this gene. We first assessed menin binding to PEG10 and found strong enrichment of menin at two sites (P-B and P-C), Fig. 4A,B. We then assessed the effect of MI-503 on the binding of the menin-MLL1 complex to PEG10 at the sites with highest menin occupancy, Fig. 4A,B. We observed a strong, dose-dependent reduction in both menin and MLL1 binding to the PEG10 promoter after 6 days of treatment with MI-503, Fig. 4C,D. MLL1 is a histone methyltransferase that catalyzes H3K4 tri-methylation, representing a chromatin activation mark. Therefore, we assessed whether reduced binding of the menin-MLL1 complex to PEG10 induced by MI-503 affects the H3K4 tri-methylation mark at those sites. We found that MI-503 treatment resulted in marked reduction in H3K4me3 at both sites on PEG10, Fig. 4C,D, consistent with the reduced expression of PEG10, Fig. 3D. These results support an epigenetic regulation of PEG10 by the menin-MLL1 complex and demonstrate that pharmacological inhibition of the menin-MLL1 interaction releases this complex from PEG10 and leads to a reduction in H3K4 tri-methylation and downregulation of PEG10 expression.
Figure 4. Epigenetic regulation of PEG10 by menin-MLL1 in HCC.
A. Diagram showing the location of primers at the promoter and downstream regions of PEG10. Genomic coordinates are provided in Supplementary Methods. B. Menin binding to different regions (P-A – P-D) of PEG10 obtained from the chromatin immunoprecipitation (ChiP) experiment performed in HepG2 cells. C, D. ChiP experiments performed in HepG2 cells upon 6 days of treatment with various concentrations of MI-503 or DMSO to detect menin and MLL1 binding as well as H3K4me3 on PEG10. Real-time PCR was performed on the precipitated DNAs with P-B (C) and P-C (D) primers for PEG10 genomic regions. IgG was used as a control. Data represent mean of triplicates ± SD.
Sorafenib sensitizes HCC cells to MI-503 treatment
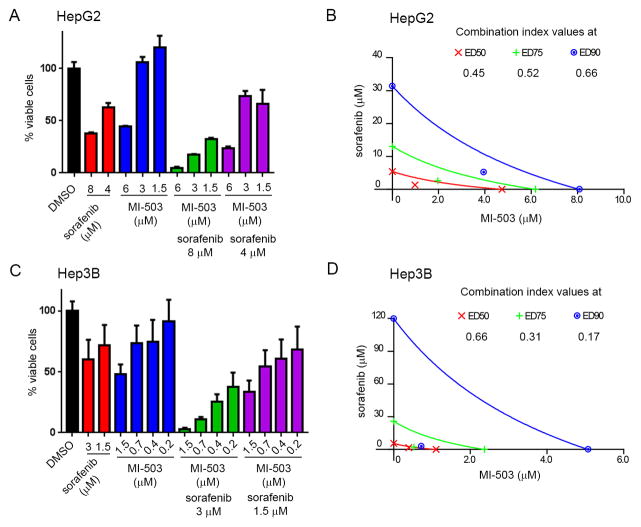
Sorafenib, a multikinase inhibitor of tyrosine protein kinases with a validated therapeutic activity against HCC, has been approved for clinical use in patients with unresectable hepatocellular carcinoma (1). We explored whether sorafenib would sensitize HCC cells to pharmacologic inhibition of the menin-MLL1 interaction by testing a combination of sorafenib and MI-503. Both sorafenib and MI-503 were used at concentrations required for effective growth inhibition in HCC cells (4), Fig. 5A,C. The inhibitory effect of MI-503 on HepG2 cell growth was strongly enhanced by a combination with sorafenib, Fig. 5A. The combination indices (CIs) calculated for the simultaneous treatment of HepG2 cells with MI-503 and sorafenib ranged from 0.45 – 0.66, suggesting strong synergistic effect, Fig, 5A,B. Similarly, the combination of MI-503 and sorafenib resulted in a more pronounced inhibition of cell growth in Hep3B cells when compared to treatment with either compound individually, Fig. 5C. The calculated CI values (<0.66) support synergistic effect of MI-503 and sorafenib, Fig. 5C,D. Overall, these results demonstrate that sorafenib sensitizes HCC cells to the pharmacologic inhibition of the menin-MLL1 interaction.
Figure 5. Sorafenib sensitized HCC cells to the treatment with MI-503.
A. MTT cell viability assay performed after 4 days of treatment of HepG2 cells with MI-503, sorafenib and both agents tested simultaneously. Data represent mean of quadruplicates ± SD. B. Isobologram obtained from analysis of the combination of MI-503 and sorafenib in HepG2 cells after 4 days of treatment. Combinatorial indices (CI < 1) indicate synergistic effect between these two agents in HepG2 cells. C. MTT cell viability assay performed after 5 days of treatment of Hep3B cells with MI-503, sorafenib or both agents tested simultaneously. Data represent mean of quadruplicates ± SD. D. Isobologram obtained from analysis of the combination of MI-503 and sorafenib in Hep3B cells after 5 days of treatment. Combinatorial indices (CI < 1) indicate synergistic effect between these two agents in Hep3B cells.
MI-503 inhibits tumor growth in vivo in HCC xenograft models
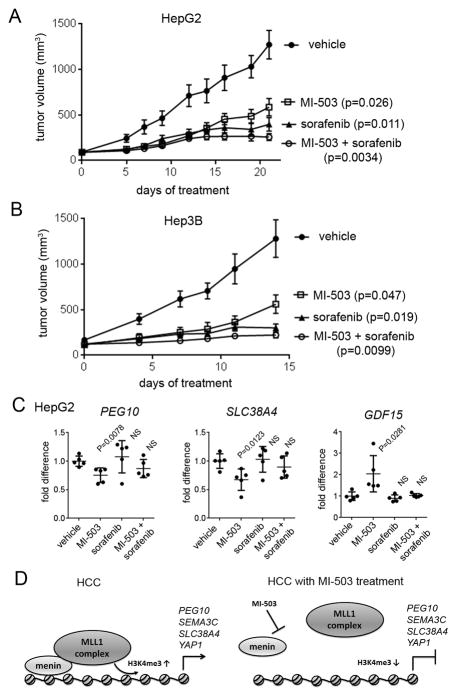
The pronounced effects of MI-503 observed in HCC cell lines prompted us to assess the therapeutic potential of the menin-MLL inhibitor in in vivo models of HCC. We have recently shown that MI-503 has a favorable pharmacokinetic (PK) profile in mice and have demonstrated strong in vivo efficacy in mice models of MLL leukemia (18) and in castration resistant prostate cancer (19), which emphasizes its suitability as a candidate for in vivo studies. Here, we used MI-503 both as a single agent and in combination with sorafenib to assess the in vivo efficacy of the menin-MLL1 inhibition in two mouse models of HCC: HepG2 and Hep3B xenograft models. The HepG2 or Hep3B cells were implanted into the flank of athymic nude mice. Treatment with MI-503 and/or sorafenib was then initiated when the tumors reached ~100 mm3 volume and was continued until the volume of the tumor in the vehicle treated mice reached ~1,200 mm3 (2–3 weeks). Once daily intraperitoneal (i.p.) administration of MI-503 at a 35 mg/kg dose caused a strong reduction (>50%) in the tumor growth in both HepG2 and Hep3B xenograft models, Fig. 6A,B. Furthermore, an 85% inhibition of the tumor growth was observed in both xenograft models when MI-503 was combined with sorafenib, Fig. 6A,B. Importantly, no substantial signs of toxicity (e.g. no significant changes in mouse body weight or liver enzymes level), were observed in the mice during the treatment, Supplementary Fig. S3. These results support a potential therapeutic value of menin-MLL inhibitors in HCC. Since sorafenib alone leads to >50% tumor growth inhibition in both xenograft models, Fig. 6A,B, it remains to be explored whether lower doses of both compounds could result in even stronger combinatorial effect in vivo.
Figure 6. In vivo efficacy of MI-503 in HCC models.
A, B. In vivo inhibition of the tumor growth by MI-503 (35 mg/kg, i.p.), sorafenib (20 mg/kg or 40 mg/kg in HepG2 and Hep3B xenografts, respectively, p.o.) or combination of both agents relative to vehicle control in HepG2 (A) and Hep3B (B) mice xenograft models, n = 8–9 mice per group. Mice were treated once daily at the doses indicated above. Error bars represent SEM. C. Expression of PEG10, SLC38A4 and GDF15 measured by qRT-PCR of RNA extracted from tumor samples harvested at the end point of treatment with MI-503, sorafenib, combination of both agents or vehicle in HepG2 xenograft model (panel A). PEG10, SLC38A4 and GDF15 transcripts levels are normalized to the mean transcript level in tumors from vehicle treated mice; n = 5 per group. p values < 0.05 are considered significant. NS – not significant. D. Menin interaction with the MLL1 histone methyltransferase complex leads to increase in the H3K4 tri-methyl mark on PEG10 and possibly other genes relevant to HCC resulting in increased expression of these genes (left panel). Inhibition of the menin-MLL1 interaction by MI-503 results in reduced H3K4me3 and repression of PEG10 and possibly other genes relevant to HCC (right panel).
We next examined gene expression in the tumor samples collected from the HepG2 xenograft mice treated with MI-503, sorafenib, combination of both agents or vehicle. The tumor samples collected from mice treated with MI-503 as a single agent showed a marked reduction in the expression level of PEG10 and SLC38A4, which are both growth promoting genes in HCC cells, Fig. 6C. Upregulation of GDF15 was also observed in MI-503-treated mice when compared to vehicle-treated control mice, Fig. 6C. These results were consistent with the on target activity of MI-503 and gene expression studies performed in vitro in the HepG2 and Hep3B cells, Fig. 3D,E. In contrast, we did not observe statistically significant effect on the expression level of PEG10, SLC38A4 or GDF15 in the tumor samples isolated from mice treated with sorafenib alone, Fig. 6C. These results indicate a different mechanism of action for sorafenib and MI-503 in HCC cells. Interestingly, combinatorial treatment of MI-503 and sorafenib in mice resulted in a similar trend in the gene expression changes as observed for MI-503 alone (especially for the downregulated genes), but the effects are less pronounced and did not reach statistical significance, Fig. 6C. Taken together, the reduction in the tumor growth in the in vivo models of HCC induced by MI-503 correlated with the decrease in the expression level of genes linked to HCC, thereby validating the on target mechanism of action of MI-503. These results demonstrate that pharmacologic inhibition of the menin-MLL interaction and/or its combination with sorafenib could represent an effective approach for HCC treatment.
Discussion
Hepatocellular carcinoma represents an aggressive and incurable cancer that requires new therapies for effective patient treatment. Menin is upregulated in HCC, Fig. 1A, and a high level of menin expression correlates with poor patient survival, emphasizing an important role of menin in HCC (10). Since menin interaction with MLL1 was indicated to play an important role in the development of hepatocellular carcinoma, we hypothesized that pharmacologic inhibition of this interaction could represent a new therapeutic approach in HCC. Here, we established that MI-503, a potent small molecule inhibitor of the menin-MLL1 interaction, has a pronounced anti-tumor effect in both in vitro and in vivo models of HCC. Treatment with MI-503 results in effective killing of HCC cells and reduces sphere formation and cell migration. The combination of MI-503 with sorafenib, multi-kinase inhibitor currently used in the clinic for HCC treatment, synergistically enhanced the effects of MI-503 on proliferation of HCC cells. Furthermore, in vivo application of MI-503 resulted in marked reduction in tumor growth in HepG2 and Hep3B xenograft models, and these effects were enhanced by combination of MI-503 with sorafenib. Overall, our studies demonstrate that pharmacologic inhibition of the menin-MLL interaction has a pronounced effect in HCC models, both in vitro and in vivo, and this approach might be promising as therapy for HCC patients.
Previous studies have suggested that menin function in liver tumorigenesis is associated with transcriptional activation of YAP1, an important oncogene in HCC (10,15). However, here we found that pharmacologic inhibition of the menin-MLL1 interaction had only a modest effect on the YAP1 expression in HepG2 cells. Instead, we found strong downregulation of other genes involved in HCC cell growth or migration, including PEG10, SLC38A4 and SEMA3C, following treatment with the MI-503 menin-MLL inhibitor. We further validated that MI-503 reduces the binding of menin and MLL1 to PEG10 and decreases the H3K4me3 level at PEG10, providing an important mechanistic link between PEG10 and the menin-MLL1 axis in HCC, Fig. 6D. Furthermore, global gene expression studies have identified that MI-503 affects expression of genes associated with a sub-class of HCC with mutations in CTNNB1, which encodes β-catenin, thereby providing a link to the Wnt signaling pathway that is known to play a crucial role in a subset of HCC (37–39). Overall, our studies demonstrate that the role of menin in HCC is much more complex than the transcriptional regulation of YAP1 and Hippo pathway, as previously suggested (10). The menin-MLL1 complex participates in epigenetic activation of several genes involved in HCC pathogenesis, rather than regulating a specific pathway in HCC.
We have previously demonstrated strong anti-tumor effects of the MI-503 menin-MLL1 inhibitor in in vitro and in vivo models of MLL leukemia (18), prostate cancer (19) and Ewing sarcoma (46). In addition, our earlier menin-MLL1 inhibitor, MI-2, also strongly inhibited tumor cell growth in pediatric gliomas with H3.3K27M mutations (47) and downregulated genes involved in the estrogen receptor positive breast cancer (48). In the study presented here we demonstrate that pharmacologic inhibition of the menin-MLL1 interaction by MI-503 leads to the effective killing of HCC cells and pronounced in vivo tumor growth inhibition, providing an another example of the anti-tumor effect resulting from blocking the menin-MLL1 interaction. The results of these studies might suggest a novel and promising treatment for HCC patients by using the menin-MLL1 inhibitors either as single agents or in combination with known drugs, including sorafenib. While menin-MLL1 inhibitors may represent valuable anti-cancer agents, further studies are needed to assess inhibition of the menin-MLL1 interaction in the context of tumor suppressor function of menin in endocrine tissues (49). Although loss of menin results in MEN1 tumors, many MEN1 missense mutations are outside of the MLL binding site on menin (50), suggesting that other menin interactions may be required for the tumor suppressor activity of menin. Clinical studies are needed to address the utility of menin-MLL1 inhibitors in HCC and other cancers.
Supplementary Material
Acknowledgments
Grant Support. This work was funded by the National Institute of Health (NIH) grants R01 (1R01CA160467) to J.G. and R01 (1R01CA200660) to J.G., LLS Scholar (1215-14) to J.G. and American Cancer Society grant (RSG-13-130-01-CDD to J.G.).
We thank Dr. Ilona Kryczek, University of Michigan, for providing MHCC97, PLC/PRF/5, SNU449 and SNU423 human HCC cell lines for this project and Dr. Elizabeth Lawlor, University of Michigan, for providing the ASC52 cell line. The mouse work was performed under oversight of UCUCA at the University of Michigan.
Footnotes
Accession numbers
RNA-seq data were deposited at the NCBI Gene Expression Omnibus under the accession number GSE103327.
References
- 1.Llovet JM, Bruix J. Molecular targeted therapies in hepatocellular carcinoma. Hepatology. 2008;48:1312–27. doi: 10.1002/hep.22506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907–17. doi: 10.1016/S0140-6736(03)14964-1. [DOI] [PubMed] [Google Scholar]
- 3.Balogh J, Victor D, 3rd, Asham EH, Burroughs SG, Boktour M, Saharia A, et al. Hepatocellular carcinoma: a review. J Hepatocell Carcinoma. 2016;3:41–53. doi: 10.2147/JHC.S61146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu L, Cao Y, Chen C, Zhang X, McNabola A, Wilkie D, et al. Sorafenib blocks the RAF/MEK/ERK pathway, inhibits tumor angiogenesis, and induces tumor cell apoptosis in hepatocellular carcinoma model PLC/PRF/5. Cancer Res. 2006;66:11851–8. doi: 10.1158/0008-5472.CAN-06-1377. [DOI] [PubMed] [Google Scholar]
- 5.Feo F, Pascale RM, Simile MM, De Miglio MR, Muroni MR, Calvisi D. Genetic alterations in liver carcinogenesis: implications for new preventive and therapeutic strategies. Crit Rev Oncog. 2000;11:19–62. [PubMed] [Google Scholar]
- 6.Bosch FX, Ribes J, Diaz M, Cleries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127:S5–S16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 7.Takenaka K, Kawahara N, Yamamoto K, Kajiyama K, Maeda T, Itasaka H, et al. Results of 280 liver resections for hepatocellular carcinoma. Arch Surg. 1996;131:71–6. doi: 10.1001/archsurg.1996.01430130073014. [DOI] [PubMed] [Google Scholar]
- 8.Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S, Kim JS, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10:25–34. doi: 10.1016/S1470-2045(08)70285-7. [DOI] [PubMed] [Google Scholar]
- 9.Bertot LC, Sato M, Tateishi R, Yoshida H, Koike K. Mortality and complication rates of percutaneous ablative techniques for the treatment of liver tumors: a systematic review. Eur Radiol. 2011;21:2584–96. doi: 10.1007/s00330-011-2222-3. [DOI] [PubMed] [Google Scholar]
- 10.Xu B, Li SH, Zheng R, Gao SB, Ding LH, Yin ZY, et al. Menin promotes hepatocellular carcinogenesis and epigenetically up-regulates Yap1 transcription. Proc Natl Acad Sci U S A. 2013;110:17480–5. doi: 10.1073/pnas.1312022110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grembecka J, Belcher AM, Hartley T, Cierpicki T. Molecular basis of the mixed lineage leukemia-menin interaction: implications for targeting mixed lineage leukemias. J Biol Chem. 2010;285:40690–8. doi: 10.1074/jbc.M110.172783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–18. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 13.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–45. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 14.Milne TA, Hughes CM, Lloyd R, Yang Z, Rozenblatt-Rosen O, Dou Y, et al. Menin and MLL cooperatively regulate expression of cyclin-dependent kinase inhibitors. Proc Natl Acad Sci U S A. 2005;102:749–54. doi: 10.1073/pnas.0408836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–67. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16:425–38. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–74. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borkin D, He S, Miao H, Kempinska K, Pollock J, Chase J, et al. Pharmacologic inhibition of the Menin-MLL interaction blocks progression of MLL leukemia in vivo. Cancer Cell. 2015;27:589–602. doi: 10.1016/j.ccell.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malik R, Khan AP, Asangani IA, Cieslik M, Prensner JR, Wang X, et al. Targeting the MLL complex in castration-resistant prostate cancer. Nat Med. 2015;21:344–52. doi: 10.1038/nm.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao SB, Xu B, Ding LH, Zheng QL, Zhang L, Zheng QF, et al. The functional and mechanistic relatedness of EZH2 and menin in hepatocellular carcinoma. J Hepatol. 2014;61:832–9. doi: 10.1016/j.jhep.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 21.He S, Malik B, Borkin D, Miao H, Shukla S, Kempinska K, et al. Menin-MLL inhibitors block oncogenic transformation by MLL-fusion proteins in a fusion partner-independent manner. Leukemia. 2015 doi: 10.1038/leu.2015.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borkin D, Pollock J, Kempinska K, Purohit T, Li X, Wen B, et al. Property Focused Structure-Based Optimization of Small Molecule Inhibitors of the Protein-Protein Interaction between Menin and Mixed Lineage Leukemia (MLL) J Med Chem. 2016;59:892–913. doi: 10.1021/acs.jmedchem.5b01305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grembecka J, He S, Shi A, Purohit T, Muntean AG, Sorenson RJ, et al. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat Chem Biol. 2012;8:277–84. doi: 10.1038/nchembio.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bang H, Ha SY, Hwang SH, Park CK. Expression of PEG10 Is Associated with Poor Survival and Tumor Recurrence in Hepatocellular Carcinoma. Cancer Res Treat. 2015;47:844–52. doi: 10.4143/crt.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ip WK, Lai PB, Wong NL, Sy SM, Beheshti B, Squire JA, et al. Identification of PEG10 as a progression related biomarker for hepatocellular carcinoma. Cancer Lett. 2007;250:284–91. doi: 10.1016/j.canlet.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 26.Liu L, Ren ZG, Shen Y, Zhu XD, Zhang W, Xiong W, et al. Influence of hepatic artery occlusion on tumor growth and metastatic potential in a human orthotopic hepatoma nude mouse model: relevance of epithelial-mesenchymal transition. Cancer Sci. 2010;101:120–8. doi: 10.1111/j.1349-7006.2009.01363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen R, Dong Y, Xie X, Chen J, Gao D, Liu Y, et al. Screening candidate metastasis-associated genes in three-dimensional HCC spheroids with different metastasis potential. Int J Clin Exp Pathol. 2014;7:2527–35. [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez Molina D, Jafari R, Ignatushchenko M, Seki T, Larsson EA, Dan C, et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341:84–7. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 29.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 30.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202–12. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang CC, Park AY, Guan JL. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007;2:329–33. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 33.Cao L, Zhou Y, Zhai B, Liao J, Xu W, Zhang R, et al. Sphere-forming cell subpopulations with cancer stem cell properties in human hepatoma cell lines. BMC Gastroenterol. 2011;11:71. doi: 10.1186/1471-230X-11-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uchida Y, Tanaka S, Aihara A, Adikrisna R, Yoshitake K, Matsumura S, et al. Analogy between sphere forming ability and stemness of human hepatoma cells. Oncol Rep. 2010;24:1147–51. doi: 10.3892/or_00000966. [DOI] [PubMed] [Google Scholar]
- 35.Weiswald LB, Bellet D, Dangles-Marie V. Spherical cancer models in tumor biology. Neoplasia. 2015;17:1–15. doi: 10.1016/j.neo.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiang DY, Villanueva A, Hoshida Y, Peix J, Newell P, Minguez B, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68:6779–88. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laurent-Puig P, Legoix P, Bluteau O, Belghiti J, Franco D, Binot F, et al. Genetic alterations associated with hepatocellular carcinomas define distinct pathways of hepatocarcinogenesis. Gastroenterology. 2001;120:1763–73. doi: 10.1053/gast.2001.24798. [DOI] [PubMed] [Google Scholar]
- 38.Liu LJ, Xie SX, Chen YT, Xue JL, Zhang CJ, Zhu F. Aberrant regulation of Wnt signaling in hepatocellular carcinoma. World J Gastroenterol. 2016;22:7486–99. doi: 10.3748/wjg.v22.i33.7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waisberg J, Saba GT. Wnt-/-beta-catenin pathway signaling in human hepatocellular carcinoma. World J Hepatol. 2015;7:2631–5. doi: 10.4254/wjh.v7.i26.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boyault S, Rickman DS, de Reynies A, Balabaud C, Rebouissou S, Jeannot E, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 41.Lachenmayer A, Alsinet C, Savic R, Cabellos L, Toffanin S, Hoshida Y, et al. Wnt-pathway activation in two molecular classes of hepatocellular carcinoma and experimental modulation by sorafenib. Clin Cancer Res. 2012;18:4997–5007. doi: 10.1158/1078-0432.CCR-11-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kondoh N, Imazeki N, Arai M, Hada A, Hatsuse K, Matsuo H, et al. Activation of a system A amino acid transporter, ATA1/SLC38A1, in human hepatocellular carcinoma and preneoplastic liver tissues. Int J Oncol. 2007;31:81–7. [PubMed] [Google Scholar]
- 43.Liao YL, Sun YM, Chau GY, Chau YP, Lai TC, Wang JL, et al. Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene. 2008;27:5578–89. doi: 10.1038/onc.2008.168. [DOI] [PubMed] [Google Scholar]
- 44.Yu J, Shen B, Chu ES, Teoh N, Cheung KF, Wu CW, et al. Inhibitory role of peroxisome proliferator-activated receptor gamma in hepatocarcinogenesis in mice and in vitro. Hepatology. 2010;51:2008–19. doi: 10.1002/hep.23550. [DOI] [PubMed] [Google Scholar]
- 45.Zimmers TA, Jin X, Gutierrez JC, Acosta C, McKillop IH, Pierce RH, et al. Effect of in vivo loss of GDF-15 on hepatocellular carcinogenesis. J Cancer Res Clin Oncol. 2008;134:753–9. doi: 10.1007/s00432-007-0336-4. [DOI] [PubMed] [Google Scholar]
- 46.Svoboda LK, Bailey N, Van Noord RA, Krook MA, Harris A, Cramer C, et al. Tumorigenicity of Ewing sarcoma is critically dependent on the trithorax proteins MLL1 and menin. Oncotarget. 2017;8:458–71. doi: 10.18632/oncotarget.13444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Funato K, Major T, Lewis PW, Allis CD, Tabar V. Use of human embryonic stem cells to model pediatric gliomas with H3. 3K27M histone mutation. Science. 2014;346:1529–33. doi: 10.1126/science.1253799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dreijerink KM, Groner AC, Vos ES, Font-Tello A, Gu L, Chi D, et al. Enhancer-Mediated Oncogenic Function of the Menin Tumor Suppressor in Breast Cancer. Cell Rep. 2017;18:2359–72. doi: 10.1016/j.celrep.2017.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chandrasekharappa SC, Teh BT. Functional studies of the MEN1 gene. J Intern Med. 2003;253:606–15. doi: 10.1046/j.1365-2796.2003.01165.x. [DOI] [PubMed] [Google Scholar]
- 50.Murai MJ, Chruszcz M, Reddy G, Grembecka J, Cierpicki T. Crystal structure of menin reveals binding site for mixed lineage leukemia (MLL) protein. J Biol Chem. 2011;286:31742–8. doi: 10.1074/jbc.M111.258186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.