Abstract
There is an emerging consensus that genomic researchers should, at a minimum, offer to return to individual participants clinically valid, medically important, and medically actionable genomic findings (e.g., pathogenic variants in BRCA1) identified in the course of research. However, this is not a common practice in psychiatric genetics research. Furthermore, psychiatry researchers often generate findings that do not meet all of these criteria, yet there may be ethically compelling arguments to offer selected results. Here, we review the return of results debate in genomics research and propose that, as for genomic studies of other medical conditions, psychiatric genomics researchers should offer findings that meet the minimum criteria stated above. Additionally, if resources allow, psychiatry researchers could consider offering to return pre-specified “clinically valuable” findings even if not medically actionable – for instance, findings that help corroborate a psychiatric diagnosis, and findings that indicate important health risks. Similarly, we propose offering “likely clinically valuable” findings, specifically, variants of uncertain significance potentially related to a participant’s symptoms. The goal of this Perspective is to initiate a discussion that can help identify optimal ways of managing the return of results from psychiatric genomics research.
INTRODUCTION
Modern genomic analysis (whole genome or exome sequencing and array-based assays) is helping uncover the genetic architecture of psychiatric disorders.1–8 Nevertheless, genomic testing raises complex ethical, scientific, and procedural challenges for psychiatry researchers, including how to manage the increasing amount of clinically relevant information these technologies can generate.9, 10 For example, consider whether researchers should offer to return findings to individual participants in the following scenarios: 1) a genome-wide association study that will generate schizophrenia genetic risks scores for all case and control subjects; 2) a whole genome sequencing study of women with anorexia nervosa that will yield data on proven breast and ovarian cancer risk loci (e.g., BRCA1 and BRCA2; OMIM # 113705; 600185) which are not known to be related to eating disorders, but could generate clinically relevant findings for which there are medical interventions that decrease the risk of poor health outcomes (e.g., bilateral mastectomy, oophorectomy, chemoprevention)11; 3) a genomic study of individuals with treatment-resistant psychosis and some degree of cognitive impairment that will yield data on genetic variation in HTT12 (OMIM # 143100) and PSEN113 (OMIM # 607822)—rare causes of Huntington’s disease and Alzheimer’s disease for which no treatments exist but of clear relevance to the study as well as research participant’s clinical status and prognosis.
Psychiatric research has seen a marked increase in the number of array-based genome-wide association studies (GWAS)2, 14 and to a lesser but growing extent whole genome and exome sequencing (WGS/WES). New genomic testing tools and decreasing costs15 will lead psychiatry researchers to generate a rapidly increasing number of clinically relevant findings (Table 1). For example, for less than $40 per sample, Illumina’s Global Screening Array (GSA) contains approximately 50,000 probes for variants claimed to be clinically relevant in addition to its capacity to identify large CNVs.16 In the next three years, at least 3 million samples are expected be run on the GSA.16 Many of these samples will likely come from psychiatric research studies.14 Numerous other psychiatric genomic studies will employ other arrays and WGS/WES, which could find even more clinically relevant findings, particularly ultra-rare, damaging or disruptive exon variants,17 that are not usually practical to genotype with array-based assays.
Table 1.
Key terms for the return of results debate in genomics research
| Term | Description | Example |
|---|---|---|
| Analytically Valid | the sequencing test or array-based assay reliably measures what it purports to measure | genomic sequencing test generates sequencing data that corresponds to the sample under study |
| Clinically Valid | enough evidence is available to support a strong association between the variant and a severe health outcome | pathogenic variants in LDLR are associated with familial hypercholesterolemia |
| Medically Important | a variant associated with a severe health outcome; higher penetrance increases the medical importance a variant | MLH1, MSH2, MSH6, or PMS2 pathogenic variants (Lynch syndrome) are associated with a high risk of colon cancer |
| Medically Actionable | an intervention is available to minimize the risk or manage poor health outcomes associated with the variant | breast cancer associated with BRCA1 or BRCA2 pathogenic variants may be prevented with bilateral mastectomy |
| Finding Identified in the Course of Research | analysis of the variant is part of the scope of the study; researchers do not have a duty to hunt for clinically relevant findings outside of the scope of their study | depends on the scope of the study (e.g., analysis of 16p11.2 copy number variant would likely be within the scope of an autism spectrum disorder study) |
| Clinically Relevant Finding | a genomic finding that could—immediately or in the future—impact individual medical care by facilitating prevention, diagnosis, treatment selection, or more comprehensive understanding of the pathogenesis of a participant’s symptoms | an actionable finding such as pathogenic variants in BRCA1 or BRCA2; a clinically valuable finding that helps corroborate a diagnosis; a likely clinically valuable findings such as VUS potentially associated to a participant’s symptoms |
| Primary Target Finding | a genomic finding associated with the psychiatric disorder or symptoms under study | finding a deletion of 22q11.2 in a study of the genomics of schizophrenia |
| Secondary Target Finding | a genomic finding identified from variants or genes targeted for analysis by the researchers, but unrelated to the disorder or symptoms under study | finding pathogenic variants in BRCA1, BRCA2, or LDLR when studying the genomics of schizophrenia |
| Incidental Finding | a genomic finding identified in the course of research that was not part of the genes or variants originally intended for analysis in the study | finding that variants in a gene or genomic loci under study for their potential association with schizophrenia are also associated with risk for some type of cancer |
| Clinically Valuable Finding | a genomic finding that is not medically actionable, but it is clinically valid and may facilitate diagnosis, risk prediction, or more comprehensive understanding of the pathogenesis of a participants’ symptoms | genetic diagnosis of Fragile X Syndrome in a study of autism |
| Likely Clinically Valuable Finding | a VUS that lies in loci related to a participant’s symptoms and has characteristics that suggest it could be pathogenic (very rare; nonsense or damaging missense) | very rare, nonsense or damaging missense VUS identified in SETD1A in a participant with symptoms of psychosis |
In this Perspective we examine ethical, scientific, and practical considerations about what findings should be offered to participants in psychiatric genomics research. Finally, we offer a framework for making determinations about the return of results (RoR) to participants.
CLINICALLY RELEVANT FINDINGS IN GENOMICS RESEARCH
Researchers who use brain imaging technologies often identify incidental findings (“incidentalomas.”)18–21 These are unrelated to the reason the brain scan was requested (e.g., to measure the sizes of brain regions) but are detected nonetheless. The clinical relevance varies. Rarely, there might be an unsuspected finding that provides a general medical explanation for a psychiatric presentation – new onset major depressive disorder with brain metastases from a primary lung cancer or findings highly suggestive of multiple sclerosis. One of the more common incidental findings (although still <1%) is the detection of an asymptomatic primary brain neoplasm (e.g., meningioma). The clinical significance of other incidental findings may be uncertain (e.g., an old brain infarct or periventricular hyperintensities).
Genomics research presents a somewhat similar22,23 situation when sequencing or array-based assays are used. In the genomics arena this issue is often referred to as “the incidentalome.”24 The original purpose is to generate generalizable knowledge about a particular disorder based on the study of large groups of cases and controls. Whatever the original intent, these data can contain information unrelated to the purpose for conducting the study, but of clinical relevance to individual research participants. Genomic researchers struggle with how to manage these incidental findings as well as clinically relevant findings generated when examining the primary and secondary target genes (Table 1).20,25,26 However, recent global debate about these issues have outlined some guiding principles.
EMERGING ETHICAL CONSENSUS ABOUT THE RoR
Until recently, many institutional review boards limited the use of genomic data to research purposes only, and explicitly did not allow researchers to return individual findings.27,28 As noted on Table 2, there are strong arguments for and against the RoR in genomics research. Some of the strongest arguments against the RoR are that the principal goal of research is to generate generalizable knowledge not to provide individual benefit, and there are important practical constraints such as the cost and resources necessary for the RoR. There are also technical and interpretative difficulties for determining the pathogenicity of variants, particularly novel ones identified in the course of research. Even the penetrance of variants that are known to be pathogenic may not be well estimated in the general population because most studies have examined the penetrance of these variants in clinical populations.29 Providing erroneous information regarding the pathogenicity and penetrance of a variant could lead to unnecessary treatments and harms for participants, and generate mistrust towards scientific research.
Table 2.
Arguments for and against the return of genomic results to individual research participants
| RoR | Arguments | Description of Arguments |
|---|---|---|
| Arguments Against | Goal of research | to generate generalizable knowledge, not provide individual care; there is no duty to return individual results27,29 |
| Cost | deviating from the goal of research to return individual results drains already limited research resources | |
| Minimize therapeutic misconception | by returning clinically relevant findings researchers may promote the misconception that the research is being conducted for the therapeutic benefit of the participant30,31 | |
| Respect for Autonomy | denying RoR is respectful of participants’ autonomy if researchers make it clear during the informed consent process that there will not be RoR and participants agree to these terms32 | |
| Difficult to obtain meaningful informed consent | Genome-wide testing could generate a plethora of clinically relevant findings and it would be impractical to describe potential findings to obtain meaningful informed consent for RoR33 | |
| Difficult to determine the pathogenicity of variants | it may be difficult for researchers to identify clinically relevant findings, particularly when they are not clinicians34 or if annotations are ambiguous | |
| Non-maleficence | given the complexity of genomic information, participants may overestimate their risk, suffer needless emotional distress, and seek unnecessary treatments; this may be exacerbated in people with severe psychiatric disorders | |
| Lack of genetics training among clinicians | Most internists35 and psychiatrists36–38 report inadequate understanding of genetics and the interpretation and management of genomic tests; this could lead to unnecessary tests and procedures that will increase health care costs, and could potentially harm patients | |
| Unknown impact of RoR on participants | there is lack of research about the impact of RoR on individuals with a history of, currently suffering from, or at risk of developing a psychiatric disorder; it is conceivable that this could exacerbate suicidality or become part of a delusional scheme | |
| Arguments in Favor | Beneficence | genomics research can generate clinically relevant findings that, if known, could improve health outcomes;11,39,40 denying participants access to these findings is inconsistent with beneficence, a basic principle of ethical research advocated by the Belmont Report and others.39,41–44 |
| Respect for Persons | implies not using participants as just means to an end, thus, using participants to generate data but not offer certain clinically relevant findings generated in the course of research may be considered unethical.28, 45 | |
| Respect for Autonomy | if researchers are in possession of clinically relevant findings—and it is feasible to make these available to participants—researchers should not decide for participants whether they should know this information, but allow participants or their legally authorized representative to make this determination39,45,46 | |
| Justice | although this may change in the future, given the cost of genetic testing and usual lack of insurance coverage for these tests, participants may not be able to access this genomic information through other means | |
| Reciprocity | should not withhold clinically relevant findings from participants who contributed and made study possible30,47–49 | |
| Participants want findings | a large majority of individuals would like to know their genomic research results50–52 | |
| Many researchers and other stakeholders support return of certain clinically relevant findings | in 2013, 95% of 234 genomic researchers surveyed in the US believed that highly penetrant and medically actionable findings should be offered;34,53 funding agencies, professional groups, and other stakeholders support the return of certain findings54–60 |
Yet, for more than 15 years there has been growing support for offering some genomic findings to participants. On balance, arguments in favor of offering to return results (Table 2) led US advisory bodies and funding institutes to publish recommendations25,56,57,61 in favor of offering to return findings that are analytically valid, clinically valid, medically important, and medically actionable (Table 1). The US National Human Genome Research Institute (NHGRI), the US All of Us Research Program, the UK Genomes England Project, the Parliaments of Spain, Finland and Estonia, various Japanese government ministries, the Indian Council of Medical Research, and the H3 Africa Consortium, among others, also support participants’ access to findings.58–60, 62–68 Some regulatory bodies or advisory groups in places such as Singapore, Denmark, and Taiwan do not support offering participant access to research data or findings. Overall, there is a clear movement towards favoring RoR across the globe.67,69–71
In 2014, genetic researchers and bioethicists from two US NHGRI-funded consortia – Clinical Sequencing Exploratory Research (CSER) Consortium, and Electronic Medical Records and Genomics (eMERGE) Network – published a consensus statement examining whether genomic researchers should adopt a policy of analyzing and offering findings about a specific set of genes to all participants. The CSER/eMERGE working group did not endorse the analysis and return of pathogenic variants in a specific set of genes, but concluded that analytically and clinically valid, medically important, and medically actionable findings should be the minimum findings offered by genetic researchers (“minimum criteria”; Figure 1).27 The working group also concluded that because “resources for research should be primarily directed at scientific discovery” researchers do not have a duty to hunt for these type of findings if they are not within the scope of the study, and that “participants have a right to decline the receipt of genomic results.”27 Notably, while many guidelines have been published, there are no regulations or laws that specifically address the RoR in the US and most other countries. Thus, determinations about which findings, if any, are offered are generally made by researchers and research ethics committees.
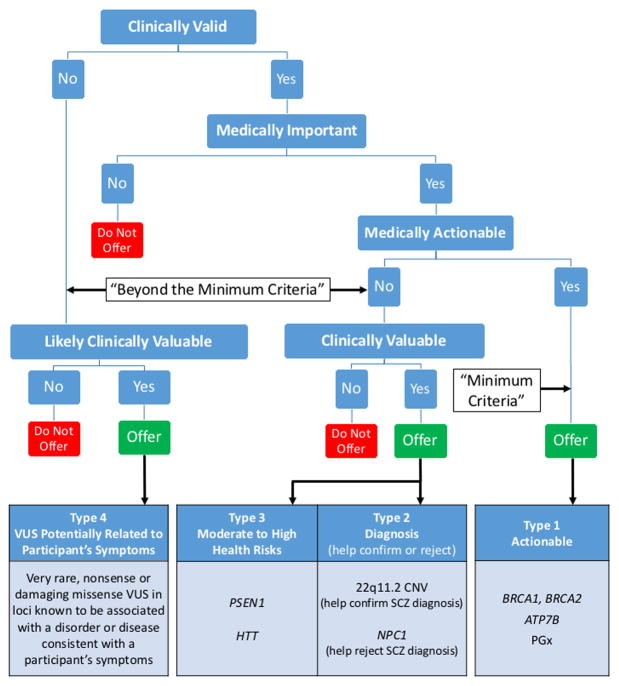
Figure 1. Return of Results Framework.
We propose that psychiatric genomics researchers offer to return findings generated in the course of research that meet the minimum criteria (Type 1): clinically valid, medically important, medically actionable, and identified within the scope of the research study. In our view, if resources allow, researchers should also offer to return clinically valuable findings generated in the course of research even if not medically actionable such as (Type 2) genomic findings that can help confirm or reject a diagnosis. In the example above, 22q11.2 deletion could help confirm a schizophrenia diagnosis, while NPC1 pathogenic variants may help reject such diagnosis. Similarly, we believe researchers should offer clinically valuable (Type 3) findings that suggest moderate to high genomic risks for a severe condition even if not medically actionable. Finally, we propose that researchers should offer (Type 4) likely clinically valuable findings, such as VUS potentially associated with a participant’s known symptoms. These findings will only be identified in a small subset of participants. If the results will be returned, results should ideally be corroborated by a certified clinical laboratory or some other reliable method and returned by a clinician (e.g., genetic counselor) who can explain the results, implications, and alternatives. ATP7B, gene associated with Wilson disease; BRCA1 and BRCA2, genes associated with hereditary breast and ovarian cancer; CNV, copy number variant; HTT, gene associated with Huntington disease; NPC1, gene associated with Niemman-Pick Disease Type C; PGx, pharmacogenetics; PSEN1, gene associated with early-onset Alzheimer disease; SCZ, schizophrenia; VUS, variants of uncertain significance.
WHICH FINDINGS SHOULD BE OFFERED in PSYCHIATRIC GENOMICS RESEARCH?
RoR Type 1: Minimum Criteria-Medically Actionable
Genomic analysis in psychiatric research can generate findings that meet the minimum criteria for RoR (Figure 1; Type 1). Medically actionable findings are expected in approximately 1% of the population.72 As with genomic research for other medical conditions,27 RoR of findings that meet these minimum criteria should ideally be offered in psychiatric research given potential clinical benefits, the universally acknowledged ethical principle of respect for participants,42–44 and several other reasons (Table 2). For example, in a study of the genomics of highly treatment-resistant psychosis, we are using WGS and a SNP array to search for variants that may provide an alternative diagnosis, help explain patients’ symptoms, and offer ideas as to why antipsychotics are ineffective in these patients. We look carefully at exonic variation in ATP7B which encodes a copper transporter that is an autosomal recessive cause of Wilson’s disease (OMIM # 277900; clinically valid and medically important).73 Wilson’s disease is rare but can cause a clinical portrait initially confusable with schizophrenia (therefore, any such finding is within the scope of this study). Early detection is crucial as relatively benign therapies (e.g., chelation and diet) can be highly beneficial, even curative, for psychotic symptoms (medically actionable). Thus, a pathogenic variant suggestive of risk for Wilson’s disease would meet the minimum criteria and we offer RoR for these types of findings.
Some psychiatry researchers may also examine secondary targets and identify findings that meet the minimum criteria for non-brain disorders (e.g., pathogenic variants in BRCA1 or BRCA2). If that is the case, the emerging consensus is RoR should also be offered to participants. For general guidance about specific genes that may generate medically actionable findings, researchers can refer to the list of genes the American College of Medical Genetics and Genomics has deemed appropriate to analyze and offer whenever clinical genomic sequencing is performed (https://www.ncbi.nlm.nih.gov/clinvar/docs/acmg/)29,74–80 and related literature in the research context.9,10,81,82 Many of these genes are for cancer and cardiomyopathies (e.g., APC and adenomatous polyposis coli or MYH7 and familial hypertrophic cardiomyopathy). Several are highly relevant for clinical psychiatry and these include multiple genes for long-QT syndrome (e.g., KCNQ1 or SCN5A; psychiatric medicines worsen long-QT in these individuals) and for single-gene disorders with prominent psychiatric manifestations (e.g., TSC1, TSC2, and tuberous sclerosis along with Wilson’s disease (ATP7B, discussed above).
Researchers may also consider returning genetic risk scores for disorders such as schizophrenia. However, the probability that an individual with a high number of markers will develop schizophrenia is far from being deterministic,83 therefore, this finding would not be considered medically important and thus would not meet the minimum criteria. It would also not meet the criteria for any of the “beyond the minimum” types of findings described in the next section.
“Beyond the Minimum” Findings
Psychiatry researchers can generate findings that do not meet the minimum criteria, but there may be compelling ethical arguments to offer RoR for some of them. At present, there is a lack of clear guidance in genomics research about which “beyond the minimum” findings should be offered. The CSER/eMERGE working group recognized that: “Researchers might be ethically and scientifically justified in returning all genomic information (the “ceiling”) in some format and any level of information in between the “floor” of actionable results identified during the course of research and the “ceiling” of all genomic information.”27 Nevertheless, the report does not offer much direction about how to make these determinations, and there is little in the history of RoR in psychiatric genomics to guide researchers.
Based on ethical and legal analysis of RoR policies, relevant bioethics literature, and our recent experience returning results in psychiatric genomics research, we propose four types of findings that we consider appropriate to offer to participants (Figure 1). In our view, in addition to Type 1 findings (actionable results that meet the minimum criteria), if resources allow, psychiatric genomics researchers should ideally also offer non-medically actionable findings, that are “clinically valuable” (defined in Table 1). Such findings may include: Type 2-clinically valid findings that help corroborate or reject a psychiatric diagnosis; and Type 3-clinically valid findings that provide information about important health risks. We propose that researchers should consider offering Type 4-”likely clinically valuable” findings such as variants of uncertain significance (VUS) potentially related to a participant’s symptoms.
An informed consent process which clearly states that while individuals are participating in a research study; the researchers may generate and return clinically relevant information will be critical. Many potential participants may not be comfortable with having clinically relevant information managed in a research context as opposed to a clinical context where there are different regulatory protections and clinicians who have a fiduciary relationship with the individual.84 A description of the different types of findings that will be analyzed and offered will also be critical to allow participants (or their legally authorized representative) to decide which findings, if any, will have more utility for individual participants and are in their best overall interest to learn.85–87
In the US, new regulations will require studies to specify whether or not clinically relevant data may be returned and under what circumstances.88 Furthermore, researchers will be able to request “broad consent” for future unspecified secondary research using identifiable biospecimens or information. This has important implications for researchers, biobanks, and repositories regulated under the Common Rule. If these groups want their collected identifiable biospecimens or information to be used for future secondary research, they will need to provide “sufficient information to allow a reasonable person to expect that the broad consent would permit the types of [secondary] research conducted.”88 Psychiatry researchers conducting secondary research with these identifiable biospecimens or information should ideally also consider offering to return the clinically relevant results proposed here (Figure 1). To do this, investigators will need to contact their IRBs to evaluate whether RoR is permissible based on what participants were informed during the initial broad consent process.88 Therefore, researchers, biobanks, and repositories will need to address the possibility of RoR in the original consent process to make it more feasible for future secondary research studies to offer RoR.
Why Offer “Beyond the Minimum” Findings?
Some may argue against offering each of the clinically valuable and likely clinically valuable types of findings. Here, we describe these types of findings in more detail and explain why we propose it is appropriate to offer these. The clinically valuable findings (Types 2 and 3) are clinically valid but not medically actionable. For years, medical actionability has perhaps been the main argument for offering the RoR in research.9,10,27,40,56,57,61,82,89–91 One could argue that the research ethics principle of non-maleficence implies that if there is nothing the participant can do to reduce the risk of poor health outcomes associated with the genomic risk, researchers should not burden participants with this information. However, as described below, this genomic risk information may still be clinically valuable, and returning these findings—if the research participant or representative provided consent—would be consistent with the research ethics principles of beneficence, respect for persons and autonomy.
RoR Type 2: Diagnosis
We propose that, if resources allow, genomic findings that help corroborate a psychiatric diagnosis be offered to research participants, even if not medically actionable. For example, a 22q.11.2 deletion92,93 (OMIM #188400) in a participant diagnosed with treatment-resistant schizophrenia would not be medically actionable with regards to the schizophrenia symptoms because it is already known that the participant does not respond to available antipsychotics—although one could argue that given this variant’s pleiotropic effects it could be medically actionable for other purposes (i.e., documented risks of impaired immunity, thrombocytopenia, and hypocalcemia).93–95 Nevertheless, because of the association between 22q.11.2 deletion and risk for schizophrenia,92,93 this finding can help substantiate the schizophrenia diagnosis. This is clinically valuable given the numerous disorders, diseases, injuries, or agents that may cause symptoms that mimic schizophrenia. Similarly, pathogenic variants in NPC1 (OMIM #257220)—which increases the risk for Niemman-Pick disease type C and can present with psychosis96—in a participant diagnosed with schizophrenia would not be medically actionable, but it would suggest that the primary diagnosis is not idiopathic schizophrenia, but rather a single-gene disorder which is clinically valuable information.
Additionally, genomic information that can help corroborate or reject a diagnosis would provide a more complete clinical picture of the participant, including the potential pathogenesis of the symptoms. In a statement on clinical genetic testing, the International Society of Psychiatric Genetics recognized that some genomic information can be valuable even if not medically actionable: “Although there are no effective therapies yet for Fragile X or HD [Huntington’s disease], confirming the diagnosis provides the clinician and the family with useful information about how the patient’s illness is likely to progress and can help anticipate the needs of patients and their caregivers.”97 Furthermore, if treatments that target the pathogenesis of their symptoms are developed, participants informed of these variants will be in a better position to seek and access novel treatments.
RoR Type 3: Significant Health Risks
In our view, clinically valid genomic findings associated with important health risks should ideally also be offered even if not medically actionable such as PSEN1 pathogenic variants associated with early-onset Alzheimer.13,98 Non-medically actionable findings could be emotionally burdensome since there are, at present, no clinical interventions available to help decrease the risk of poor health outcomes. However, as with the return of any finding, informed consent would be paramount to help participants decide if knowing this type of information is in their best overall interest.85 Allowing participants to decide whether they want these findings returned would be consistent with respect for persons and their autonomy. Furthermore, participants could benefit in numerous ways, for example: by not being unnecessarily surprised with the onset of symptoms if the disease is ever expressed; seeking genetic and mental health counseling to learn more about the disease and how to cope with the risk; joining support groups; being attentive to novel therapies or clinical trials; planning certain aspects of their lives such as finances, insurance, and housing arrangements; and informing relatives so they can decide if they want to get tested.
RoR Type 4: VUS Potentially Related to Symptoms
We propose a fourth – and controversial – type of finding for which we believe RoR might be offered: a subset of VUS potentially related to a participant’s known symptoms. Some argue against the return of any VUS on the grounds that by definition these are variants for which not enough evidence has been gathered to determine their pathogenicity, and therefore not clinically valid.83 Participants and their clinicians36–39 may misinterpret the finding and order unnecessary tests or medical interventions that may generate harms with little prospect of benefit.
Nevertheless, we propose offering a small subset of VUS that may help explain a participant’s symptoms because they meet the following criteria: 1) very rare; 2) nonsense or damaging missense variants (particularly if they occur in genes or exons known to be intolerant to variation; 3) occur in genomic loci known to be associated with a psychiatric disorder or related neurological disease; and 4) the participant has known symptoms that are consistent with that disorder or disease.
We believe that RoR for these VUS should be offered to participants for the following reasons. They have characteristics that suggest they may be associated with a participant’s known symptoms. Psychiatric genomics is developing its knowledge base, and there are still high numbers of variant-phenotype associations that may eventually be shown to be clinically valid, but for which the field has simply not collected enough data yet. A number of reports argue that researchers should not have a duty to return any findings beyond their funding period, given the lack of resources to do so, among other practical obstacles.27 However, with the current pace of data collection, it is likely that in the near future the pathogenicity or the role of many of these VUS will be identified. If participants have access to these findings and, for example, any of these VUS are later identified as pathogenic, they could help provide a more complete clinical picture of the participant and information about the pathogenesis of symptoms, which could potentially improve clinical management. If these VUS are not made available by psychiatry researchers, the vast majority of participants are not likely to have access to this genomic information through other means until the cost of genetic testing and analysis decreases significantly more.
Practical challenges for the RoR in Psychiatric Genomics Research
There are two key practical challenges for the RoR in psychiatric genomics research: conflicting RoR policies across countries and cost. Psychiatric genomics research is often conducted through multinational consortia.14 However, countries have different policies regarding the RoR: some prohibit the return, others provide general guidelines, and many do not have any guidelines.67,99 These conflicting policies are problematic because, within an international consortium, some participants may benefit from the RoR and others not. Additionally, projects could decide not to offer the RoR to avoid conflicts with regulators in countries that restrict or prohibit the RoR. Therefore, it is important to begin a dialogue that can help harmonize guidelines regarding the RoR to facilitate research collaborations and maximize the benefits of the research endeavor by directly benefiting participants with clinically relevant information. Developing a consistent informed consent document would be an important step for the RoR Another key step will be to develop a website or software with up-to-date information about clinically relevant variants for psychiatric research that helps standardize the variants offered and allows researchers to sift through data more efficiently in order to identify clinically relevant findings. This could also help minimize the amount of individual resources and time specific projects devote to the analysis portion of the RoR.
The most important challenge for the RoR in psychiatric genomics research is cost. The principal costs of RoR include corroborating research findings in clinical laboratories and having a clinician conduct the RoR. Countries such as the US, require that researchers validate findings in a certified clinical laboratory100 before returning any results to participants, which significantly increases the cost of RoR. Amending regulations to allow less expensive ways of corroborating findings is another way to help decrease cost while still protecting participants from the return of erroneous findings. Given the complex nature of genomic information, it is important that the RoR is performed by a clinician (e.g., a genetic counselor, clinical geneticist, or psychiatrist with genetics training) who can carefully explain the finding, its implications, and suggest specific next steps. Psychiatrists and other clinicians generally report a lack of competence or preparation to manage genomic testing and findings and, thus, might overestimate risk and order unnecessary tests and procedures.37–39,97,101,102 Therefore, ideally, the RoR clinician will also be available to communicate with the research participant’s physician. However, the use of clinicians to conduct RoR significantly raises costs.
Funding agencies in the US and other countries should provide funds for the RoR. This may be difficult because of limited research budgets. However, as we described above, funding agencies and advisory bodies in many countries have recognized the importance of offering to return clinically relevant findings. Thus, funding agencies should make every effort to act in accordance with those statements and the emerging consensus about the importance of offering the RoR of certain clinically relevant findings, by providing funds to allow researchers to offer RoR. One possibility could be to offer supplements for psychiatric genomics studies that are most likely to identify clinically relevant findings. Some studies are using a less expensive “outsourcing” approach to RoR by offering participants their raw genomic data, which allows participants the possibility of getting it interpreted by a third party.86,103 A drawback of this approach is that many participants may not have resources to get their data interpreted.
It is important to note that researchers will not identify clinically relevant findings in the vast majority of participants. Under the proposal presented here, researchers would only offer clinically relevant findings they will generate within the scope of their study. Current estimates are that only about 1% of participants are expected to have medically actionable findings.72 The number of individuals with clinically valuable or likely clinically valuable findings in psychiatric genomics is currently difficult to estimate, may vary between cases and controls, by disorder (e.g., greater with intellectual disability and autism), and, within disorder, might vary with clinical severity and age of onset. There will also be a number of participants who decline the RoR completely and studies suggest that approximately 39% of participants may refuse some types of findings.104 In addition, RoR is one of the main motivations51–53 for participating in research, therefore, by offering findings researchers will likely save significant time and resources in recruitment. The primary goal of research is to generate generalizable knowledge but, if psychiatric genomics researchers have the resources to return results while achieving the scientific goals of their studies, offering these findings can help maximize the societal benefits of psychiatric genomics research.
CONCLUSION
With this Perspective, we hope to spark a discussion about which kinds of findings may be offered in psychiatric genomics research considering the particularities of this field and the potential risks and benefits to participants. We propose that, as in genomics research for other medical conditions, psychiatric researchers should ideally offer to return medically actionable findings identified in the course of research. If resources allow, researchers should consider offering clinically valuable and likely clinically valuable findings. There are obstacles that need to be addressed to facilitate the RoR. However, the RoR from psychiatric genomics research can help maximize the benefits of this research for society and promote the best interest of participants.
Acknowledgments
We would like to acknowledge Maria D. Iglesias de Ussel, Eric T. Ward, Genna Finkel, Jonathan S. Berg, and James P. Evans for helpful work and discussions about the topics addressed in this paper. We would also like to thank the anonymous reviewers for their helpful observations.
Funding
Research for this article was funded by the National Human Genome Research Institute (NHGRI) of the National Institutes of Health (NIH) Grant R00HG008689 (Lázaro-Muñoz, G). The views expressed are those of the authors alone, and do not necessarily reflect views of NIH.
Footnotes
Conflict of Interest
PF Sullivan reports the following potentially competing financial interests: Element Genomics (Scientific Advisory Board member, stock options), Lundbeck (advisory committee), Pfizer (Scientific Advisory Board member), and Roche (grant recipient, speaker reimbursement). The other authors declare no potential conflicts of interest.
References
- 1.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sullivan PF, Daly MJ, O’Donovan M. Genetic architectures of psychiatric disorders: The emerging picture and its implications. Nat Rev Genet. 2012;13:537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Need AC, Goldstein DB. Schizophrenia genetics comes of age. Neuron. 2014;83:760–763. doi: 10.1016/j.neuron.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 6.McCarroll SA, Feng G, Hyman SE. Genome-scale neurogenetics: Methodology and meaning. Nat Neurosci. 2014;17:756–763. doi: 10.1038/nn.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cross-Disorder Group of the Psychiatric Genomics Consortium. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuen RKC, Mericio D, Bookman M, Howe JL, Thiruvahindrapuram B, Patel RV, et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat Neurosci. 2017;20:602–611. doi: 10.1038/nn.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berg JS, Adams M, Nassar N, Bizon C, Lee K, Schmitt CP, et al. An informatics approach to analyzing the incidentalome. Genet Med. 2013;15:36–44. doi: 10.1038/gim.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berg JS, Foreman AK, O’Daniel JM, Booker JK, Boshe L, Carey T, et al. A semiquantitative metric for evaluating clinical actionability of incidental or secondary findings from genome-scale sequencing. Genet Med. 2016;18:467–475. doi: 10.1038/gim.2015.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petrucelli N, Daly MB, Pal T. BRCA1- and BRCA2-Associated Hereditary Breast and Ovarian Cancer. In: Pagon RA, Adam MP, Ardinger HH, et al., editors. GeneReviews® [Internet] University of Washington; Seattle, WA: [accessed May 17, 2017]. Available at https://www.ncbi.nlm.nih.gov/books/NBK1247/ [Google Scholar]
- 12.Warby SC, Graham RK, Hayden MR. Huntington Disease. In: Pagon RA, Adam MP, Ardinger HH, et al., editors. GeneReviews® [Internet] University of Washington; Seattle, WA: [accessed May 17, 2017]. Available at https://www.ncbi.nlm.nih.gov/books/NBK1305/ [Google Scholar]
- 13.Bird TD. Early-Onset Familial Alzheimer Disease. In: Pagon RA, Adam MP, Ardinger HH, et al., editors. GeneReviews® [Internet] University of Washington; Seattle, WA: [accessed May 17, 2017]. Available at https://www.ncbi.nlm.nih.gov/books/NBK1236/ [Google Scholar]
- 14.Sullivan PF, Agrawal A, Bulik CM, Andreassen OA, Borglum A, Breen G, et al. Psychiatric Genomics: An Update and an Agenda. BioRxiv. 2017 Mar 10; doi: 10.1176/appi.ajp.2017.17030283. http://dx.doi.org/10.1101/115600. [DOI] [PMC free article] [PubMed]
- 15.National Human Genome Research Institute. [accessed May 17, 2017];DNA Sequencing Costs: Data. Available at https://www.genome.gov/sequencingcostsdata/
- 16.Petrone J. New Illumina SNP Array Fuels European Consortium Founded to Foment ‘Third Generation GWAS Era’ GenomeWeb. 2016 Jun 15; Available at https://www.genomeweb.com/microarrays-multiplexing/new-illumina-snp-array-fuels-european-consortium-founded-foment-third.
- 17.Genovese G, Fromer M, Stahl EA, Ruderfer DM, Chambert K, Landen M, et al. Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nature Neuroscience. 2016;19:1433–1441. doi: 10.1038/nn.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berland LL, Silverman SG, Gore RM, Mayo-Smith WW, Megibow AJ, Yee J, et al. Managing Incidental Findings on Abdominal CT: White Paper of the ACR Incidental Findings Committee. J Am Coll Radiol. 2010;7:754–773. doi: 10.1016/j.jacr.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 19.Munk MD, Peitzman AB, Hostler DP, Wolfson AB. Frequency and follow-up of incidental findings on trauma computed tomography scans: Experience at a level one trauma center. J Emerg Med. 2010;38:346. doi: 10.1016/j.jemermed.2008.01.021. [DOI] [PubMed] [Google Scholar]
- 20.Wolf SM, Lawrenz FP, Nelson CA, Kahn JP, Cho MK, Clayton EW, et al. Managing Incidental Findings in Human Subjects Research: Analysis and Recommendations. J Law Med Ethics. 2008;36:19–48. doi: 10.1111/j.1748-720X.2008.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Illes J, Kirschen MP, Edwards E, Stanford LR, Bandettini P, Cho MK, et al. Incidental findings in brain imaging research. Science. 2006;311:783–784. doi: 10.1126/science.1124665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ross LF, Rothstein MA, Clayton EW. Mandatory extended searches in all genome sequencing: “incidental findings,” patient autonomy, and shared decision making. JAMA. 2013;310:367–368. doi: 10.1001/jama.2013.41700. [DOI] [PubMed] [Google Scholar]
- 23.Klitzman R, Appelbaum PS, Chung W. Return of secondary genomic findings vs patient autonomy: implications for medical care. JAMA. 2013;310:369–370. doi: 10.1001/jama.2013.41709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohane IS, Masys DR, Altman RB. The incidentalome: A threat to genomic medicine. JAMA. 2006;296:212–215. doi: 10.1001/jama.296.2.212. [DOI] [PubMed] [Google Scholar]
- 25.Presidential Commission for the Study of Bioethical Issues. [accessed May 17, 2017];Anticipate and communicate: ethical management of incidental and secondary findings in the clinical, research, and direct-to-consumer contexts. 2013 doi: 10.1093/aje/kwu217. Available at http://bioethics.gov/sites/default/files/FINALAnticipateCommunicate_PCSBI_0.pdf. [DOI] [PubMed]
- 26.Allyse M, Michie M. Not-so-incidental findings: The ACMG recommendations on the reporting of incidental findings in clinical whole genome and whole exome sequencing. Trends Biotechnol. 2013;31:439–441. doi: 10.1016/j.tibtech.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarvik GP, Amendola LM, Berg JS, Brothers K, Clayton EW, Chung W, et al. Return of genomic results to research participants: the floor, the ceiling, and the choices in between. Am J Hum Genet. 2014;94:818–826. doi: 10.1016/j.ajhg.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shalowitz DI, Miller FG. Disclosing individual results of clinical research: Implications of respect for participants. JAMA. 2005;294:737–740. doi: 10.1001/jama.294.6.737. [DOI] [PubMed] [Google Scholar]
- 29.Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGuire AL, Knoppers BM, Zawati MH, Clayton EW. Can I be sued for that? Liability risk and the disclosure of clinically significant genetic research findings. Genome Res. 2014;24:719–723. doi: 10.1101/gr.170514.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bredenoord AL, Kroes HY, Cuppen E, Parker M, van Delden JJM. Disclosure of individual genetic data to research participants: The debate reconsidered. Trends Genet. 2011;27:41–47. doi: 10.1016/j.tig.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 32.Appelbaum PS, Roth LH, Lidz C. The thereapeutic misconception: Informed consent in psychiatric research. Int J Law Psychiatry. 1982;5:319–329. doi: 10.1016/0160-2527(82)90026-7. [DOI] [PubMed] [Google Scholar]
- 33.Meltzer LA. Undesirable implications of disclosing individual genetics results to research participants. Am J Bioeth. 2006;6:28–30. doi: 10.1080/15265160600935811. [DOI] [PubMed] [Google Scholar]
- 34.Evans J. Finding Common Ground. Genet Med. 2013;15:852–853. doi: 10.1038/gim.2013.150. [DOI] [PubMed] [Google Scholar]
- 35.Klitzman R, Appelbaum PS, Fyer A, Martinez J, Buquez B, Wynn J, et al. Researchers’ views on return of incidental genomic results: qualitative and quantitative findings. Genet Med. 2013;15:888–895. doi: 10.1038/gim.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klitzman R, Chung W, Marder K, Shanmugham A, Chin LJ, Stark M, et al. Attitudes and Practices Among Internists Concerning Genetic Testing. J Genet Couns. 2013;22:99–100. doi: 10.1007/s10897-012-9504-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finn CT, Wilcox MA, Korf BR, Blacker D, Racette SR, Sklar P, et al. Psychiatric genetics: a survey of psychiatrists’ knowledge, opinions, and practice patterns. J Clin Psychiatry. 2005;66:821–830. doi: 10.4088/jcp.v66n0703. [DOI] [PubMed] [Google Scholar]
- 38.Hoop JG, Roberts LW, Hammond KA, Cox NJ. Psychiatrists’ attitudes, knowledge, and experience regarding genetics: a preliminary study. Genet Med. 2008;10:439–449. doi: 10.1097/GIM.0b013e318177014b. [DOI] [PubMed] [Google Scholar]
- 39.Klitzman R, Abbate KJ, Chung WK, Marder K, Ottman R, Taber KJ, et al. Psychiatrists’ views of the genetic bases of mental disorders and behavioral traits and their use of genetic tests. J Nerv Ment Dis. 2014;202:530–538. doi: 10.1097/NMD.0000000000000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ravitsky V, Wilfond BS. Disclosing individual genetic results to research participants. Am J Bioeth. 2006;6:8–17. doi: 10.1080/15265160600934772. [DOI] [PubMed] [Google Scholar]
- 41.Kohlman W, Gruber SB. Lynch Syndrome. In: Pagon RA, Adam MP, Ardinger HH, et al., editors. GeneReviews® [Internet] University of Washington; Seattle, WA: [accessed May 17, 2017]. Available at https://www.ncbi.nlm.nih.gov/books/NBK1211/ [Google Scholar]
- 42.National Commission for the Protection of Human Subjects of Biomedical and Behavioral Research. The Belmont Report: Ethical principles and guidelines for the protection of human subjects of research. Available at: https://www.hhs.gov/ohrp/regulations-and-policy/belmont-report/ [PubMed]
- 43.The Nuremberg Code. Trials of war criminals before the Nuremberg military tribunals under control council law. 10. Vol. 2. Washington, D.C: U.S. Government Printing Office; 1949. pp. 181–182. Available at: https://history.nih.gov/research/downloads/nuremberg.pdf. [Google Scholar]
- 44.World Medical Association. World Medical Association Declaration of Helsinki: Ethical principles for medical research involving subjects. JAMA. 2013;310:2191–2194. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- 45.Beauchamp TL, Childress JF. Principles of Biomedical Ethics. 7. University Press; New York: 2012. [Google Scholar]
- 46.Fernandez CV, Kodish E, Weijer C. Informing study participants of research results: An ethical imperative. IRB. 2003;25:12–19. [PubMed] [Google Scholar]
- 47.Foster MW, Sharp RR. Ethical issues in medical-sequencing research: Implications of genotype-phenotype studies for individuals and populations. Hum Mol Genet. 2006;15:R45–R49. doi: 10.1093/hmg/ddl049. [DOI] [PubMed] [Google Scholar]
- 48.Quaid KA, Jessup NM, Meslin EM. Disclosure of genetic information obtained through research. Genet Test. 2004;8:347–355. doi: 10.1089/gte.2004.8.347. [DOI] [PubMed] [Google Scholar]
- 49.Bredenoord AL, Onland-Moret NC, Van Delden JJM. Feedback of individual genetic results to research participants: In favor of a qualified disclosure policy. Hum Mutat. 2011;32:861–867. doi: 10.1002/humu.21518. [DOI] [PubMed] [Google Scholar]
- 50.Hoeyer K. Donors perceptions of consent to and feedback from biobank research: time to acknowledge diversity? Public Health Genomics. 2010;13:345–352. doi: 10.1159/000262329. [DOI] [PubMed] [Google Scholar]
- 51.Kaufman D, Murphy J, Scott J, Hudson K. Subjects matter: a survey of public opinions about a large genetic cohort study. Genet Med. 2008;10:831–839. doi: 10.1097/GIM.0b013e31818bb3ab. [DOI] [PubMed] [Google Scholar]
- 52.Kaufman DJ, Baker R, Milner LC, Devaney S, Hudson KL. A Survey of U S. Adults’ Opinions about Conduct of a Nationwide Precision Medicine Initiative® Cohort Study of Genes and Environment. PLoS One. 2016;11(18):e0160461. doi: 10.1371/journal.pone.0160461. doi:0160410.0161371/journal.pone.0160461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murphy J, Scott J, Kaufman D, Geller G, LeRoy L, Hudson K. Public expectations for return of results from large-cohort genetic research. Am J Bioeth. 2008;8:36–43. doi: 10.1080/15265160802513093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramoni RB, McGuire AL, Robinson JO, Morley DS, Plon SE, Joffe S. Experiences and attitudes of genome investigators regarding return of individual genetic test results. Genet Med. 2013;15:882–887. doi: 10.1038/gim.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Middleton A, Morley KI, Bragin E, Firth HV, Hurles ME, Wright CF, et al. Attitudes of nearly 7000 health professionals, genomic researchers and publics toward the return of incidental results from sequencing research. Eur J Hum Genet. 2016;24:21–29. doi: 10.1038/ejhg.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.National Bioethics Advisory Committee. Research involving human biological materials: Ethical issues and policy guidance. Vol. 1. Rockville, MD: 1999. [Google Scholar]
- 57.Bookman EB, Langehorne AA, Eckfeldt JH, Glass KC, Jarvik GP, Klag M, et al. Reporting genetic results in research studies: Summary and recommendations of an NHLBI working group. Am J Med Genet A. 2006;140:1033–1040. doi: 10.1002/ajmg.a.31195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.National Human Genome Research Institute. [accessed May 17, 2017];NHGRI Policy Recommendations on Research Privacy Guidelines: Federal Policy Recommendations on Including HIPAA. Available at https://www.genome.gov/11510216/
- 59.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Genomics England. [accessed May 17, 2017];Results. Available at: https://www.genomicsengland.co.uk/taking-part/information-for-participants/results/
- 61.National Heart, Lung, and Blood Institute working group. Fabsitz RR, McGuire A, Sharp RR, Puggal M, Beskow LM, et al. Ethical and practical guidelines for reporting genetic research results to study participants: Updated guidelines from a National Heart, Lung, and Blood Institute working group. Circ Cardiovasc Genet. 2010;3:574–580. doi: 10.1161/CIRCGENETICS.110.958827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parliament of Finland. Biobank Act 688/2012. [accessed June 2, 2017];Finland Ministry of Social Affairs and Health. Avaialble at http://www.finlex.fi/en/laki/kaannokset/2012/en20120688.pdf.
- 63.Parliament of Estonia. [Accessed June 2, 2017];Human Genes Research Act. 2000 Available at https://www.riigiteataja.ee/en/eli/531102013003/consolide.
- 64.Ministry of Education C, Sports, Science and Technology (MEXT), Ministriy of Health, Labour and Welfare (MHLW), and Ministry of Economy Trade and Industry (METI). Ethical guidelines for human genome/gene analysis research (2001). Fully revised in 2004 and 2013, partially revised in 2005 and 2008.
- 65.H3Africa Working Group on Ethics and Retulatory Issues for the Human Heredity and Health (H3Africa) Consortium. [Accessed June 2, 2017];H3Africa Guidelines for Informed Consent. 2013 Avaialble at http://www.health.uct.ac.za/sites/default/files/image_tool/images/116/H3AfricaGuidelines on Informed Consent August 2013.pdf.
- 66.Indian Council of Medical Research. [Accessed June 2, 2017];Ethical guidelines for biomedical research on human participants. 2006 Available at http://icmr.nic.in/ethical_guidelines.pdf.
- 67.Knoppers BM, Zawati MH, Sénécal K. Return of genetic testing results in the era of whole-genome sequencing. Nat Rev Genet. 2015;16:553–559. doi: 10.1038/nrg3960. [DOI] [PubMed] [Google Scholar]
- 68.Parliament of Spain. [accessed June 2, 2017];Law 14/2007, of 3 July, on Biomedical Research. Available at http://www.isciii.es/ISCIII/es/contenidos/fd-investigacion/SpanishLawonBiomedicalResearchEnglish.pdf.
- 69.The Bioethics Advisory Committee Singapore. [accessed June 2, 2017];Ethical, legal and social issues in genetic testing and genetics research. Available at http://www.bioethics-singapore.org/images/uploadfile/21929PMGT CP Final.pdf.
- 70.Danish Council of Ethics. [Accessed June 2, 2017];Genome Testing: Ethical dilemmas in relation to diagnostics, research and direct-to-consumer testing. 2012 Available at http://www.etiskraad.dk/~/media/Etisk-Raad/en/Publications/Genome-testing-report-2012.pdf?la=da.
- 71.Parliament of Taiwan. [accessed June 2, 2017];Human Biobank Management Act. 2012 Available at http://law.moj.gov.tw/Eng/LawClass/LawAll.aspx?PCode=L0020164.
- 72.Evans JP, Berg JS, Olshan AF, Magnuson T, Rimer B. We screen newborns, don’t we?: Realizing the promise of public health genomics. Genet Med. 2013;15:332–334. doi: 10.1038/gim.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weiss KH. Wilson Disease. In: Pagon RA, Adam MP, Ardinger HH, et al., editors. GeneReviews® [Internet] University of Washington; Seattle, WA: [accessed May 17, 2017]. Available at https://www.ncbi.nlm.nih.gov/books/NBK1512/ [Google Scholar]
- 74.American College of Medical Genetics and Genomics. ACMG practice guidelines: Incidental findings in clinical genomics: A clarification. Genet Med. 2013;15:664–666. doi: 10.1038/gim.2013.82. [DOI] [PubMed] [Google Scholar]
- 75.McGuire AL, Joffe S, Koenig BA, Biesecker BB, McCullough LB, Blumenthal-Barby JS, et al. Ethics and genomic incidental findings. Science. 2013;340:1047–1048. doi: 10.1126/science.1240156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wolf SM, Annas GJ, Elias S. Patient autonomy and incidental findings in clinical genomics. Science. 2013;340:1049–1050. doi: 10.1126/science.1239119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Burke W, Antommaria AH, Bennet R, Botkin J, Clayton EW, Henderson GE, et al. Recommendations for returning genomic incidental findings? We need to talk! Genet Med. 2013;15:855–859. doi: 10.1038/gim.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.American College of Medical Genetics and Genomics. ACMG updates recommendation on “opt out” for genome sequencing return of results. Available at https://www.acmg.net/docs/Release_ACMGUpdatesRecommendations_final.pdf.
- 79.Kalia SS, Adelman K, Bale SJ, Chung WK, Eng C, Evans JP, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2. 0): A policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19:249–255. doi: 10.1038/gim.2016.190. [DOI] [PubMed] [Google Scholar]
- 80.Geinsinger Health System. [accessed June 8, 2017];Geisinger Health System list of clinically actionable genes for the Mycode Community Health Initiative. Avaliable at https://www.geisinger.org/-/media/OneGeisinger/Images/GHS/Research/Centers and Institutes/Clinically Actionable Genes.ashx?la=en.
- 81.Berg JS, Khoury MJ, Evans JP. Deploying whole genome sequencing in clinical practice and public health: meeting the challenge one bin at a time. Genet Med. 2011;13:499–504. doi: 10.1097/GIM.0b013e318220aaba. [DOI] [PubMed] [Google Scholar]
- 82.Berg JS, Amendola LM, Eng C, Van Allen E, Gray W, Wagle N, et al. Processes and preliminary outputs for identification of actionable genes as incidental findings in genomic sequence data in the Clinical Sequencing Exploratory Research Consortium. Genet Med. 2013;15:860–867. doi: 10.1038/gim.2013.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lu Y, Pouget JG, Andreassen OA, Djurovic S, Esko T, Hansen T, Hultman CM, Metspalu A, Milani L, Werge T, Sullivan PF. Genetic risk scores and family history as predictors of schizophrenia in Nordic registers. doi: 10.1017/S0033291717002665. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lázaro-Muñoz G. The fiduciary relationship model for managing clinical genomic “incidental” findings. J Law Med Ethics. 2014;42:576–589. doi: 10.1111/jlme.12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lázaro-Muñoz G, Conley JM, Davis AM, Van Riper M, Walker RL, Juengst ET. Looking for Trouble: Preventive genomic sequencing in the general population and the role of patient choice. Am J Bioeth. 2015;15:3–14. doi: 10.1080/15265161.2015.1039721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Appelbaum PS, Parens E, Waldman CR, Klitzman R, Fyer A, Martinez J, et al. Models of consent to return incidental findings in genomic research. Hastings Cent Rep. 2014;44:22–32. doi: 10.1002/hast.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Regier DA, Peacock SJ, Pataky R, et al. Societal preferences for the return of incidental findings from clinical genomic sequenicng: A discrete-choice experiment. CMAJ. 2015;187(6):E190–197. doi: 10.1503/cmaj.140697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Federal Register. 82 Fed Reg 7149, 7221. Jan 19, 2017. Federal Policy for the Protection of Human Subjects. [PubMed] [Google Scholar]
- 89.Fullerton SM, Wolf WA, Brothers KB, Clayton EW, Crawford DC, Denny JC, et al. Return of individual research results from genome-wide association studies: experience of the Electronic Medical Records and Genomics (eMERGE) Network. Genet Med. 2012;14:424–431. doi: 10.1038/gim.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Goddard KA, Whitlock EP, Berg JS, Williams MS, Webber EM, Webster JA, et al. Description and pilot results from a novel method for evaluating return of incidental findings from next generation sequencing technologies. Genet Med. 2013;15:721–728. doi: 10.1038/gim.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lázaro-Muñoz G, Conley JM, Davis AM, Prince AE, Cadigan RJ. Which results to return: Subjective judgments in selecting medically actionable genes. Genet Test Mol Biomarkers. 2017;21:184–194. doi: 10.1089/gtmb.2016.0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vassos E, Collier DA, Holden S. Penetrance for copy number variants associated with schizophrenia. Hum Mol Genet. 2010;19:3477–3481. doi: 10.1093/hmg/ddq259. [DOI] [PubMed] [Google Scholar]
- 93.Kirov G, Rees E, Walters JTR, Escott-Price, Georgieva L, Richards AL, et al. The penetrance of copy number variants for schizophrenia and developmental delay. Biol Psychiatry. 2014;75:378–385. doi: 10.1016/j.biopsych.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kocarnik JM, Fullerton SM. Returning pleiotropic results from genetic testing to patients and research participants. JAMA. 2014;311:795–796. doi: 10.1001/jama.2014.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McDonald-McGinn DM, Emanuel BS, Zackai EH. 22q11.2 Deletion Syndrome. In: Pagon RA, Adam MP, Ardinger HH, et al., editors. GeneReviews® [Internet] University of Washington; Seattle, WA: [accessed May 17, 2017]. Available at https://www.ncbi.nlm.nih.gov/books/NBK1523/ [PubMed] [Google Scholar]
- 96.Patterson M. Niemman-Pick Disease Type C. In: Pagon RA, Adam MP, Ardinger HH, et al., editors. GeneReviews® [Internet] University of Washington; Seattle, WA: [accessed May 17, 2017]. Available at https://www.ncbi.nlm.nih.gov/books/NBK1296/ [Google Scholar]
- 97.International Society for Psychiatric Genomics. [accessed May 17, 2017];Genetic Testing Statement. Available at: https://ispg.net/genetic-testing-statement/
- 98.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Budin-Ljøsne I, Mascalzoni D, Soini S, Machado H, Kaye J, Bentzen HB, et al. Feedback of individual genetic results to research participants: Is it feasible in Europe? Biopreserv Biobank. 2016;14:241–248. doi: 10.1089/bio.2015.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Clinical Laboratory Improvements Act of 1988, Pub. L. No. 100–578, 102 Stat. 2903; see regulations at 42 CFR 493(b)2.
- 101.Finn CT. Increasing genetic education for psychiatric residents. Harv Rev Psychiatry. 2007;15:30–33. doi: 10.1080/10673220601183998. [DOI] [PubMed] [Google Scholar]
- 102.Hoop JG, Savla G, Roberts LW, Zisook S, Dunn LB. The current state of genetics training in psychiatric residency: Views of 235 U.S. Educators and Trainees. Acad Psychiatry. 2010;34:109–114. doi: 10.1176/appi.ap.34.2.109. [DOI] [PubMed] [Google Scholar]
- 103.Genes for Good. [accessed September 25, 2017];Learning about your genome. Available at https://genesforgood.sph.umich.edu/return_of_results/ancestry.
- 104.Jamal L, Robinson JO, Christensen KD, Blumenthal-Barby J, Slashinski MJ, Perry DL, et al. When bins blur: Patient perspectives on categories of results from clinical whole genome sequencing. AJOB Empirical Bioethics. 2017;8:82–88. doi: 10.1080/23294515.2017.1287786. [DOI] [PMC free article] [PubMed] [Google Scholar]