Abstract
Clear cell renal cell carcinoma (ccRCC) is one of the leading causes of genitourinary cancer-related death, largely due to the metastasis of ccRCC. Previous profiling study showed that lncRNAs are critical regulators in lots of cancers. However, the roles of specific lncRNAs in ccRCC migration and invasion are still unknown. In this study, we utilized the high-throughput genome sequencing to identify the potential differentially expressed lncRNAs in ccRCC and further determined the underlying regulatory mechanism. We found that lncRNA SNHG14 was significantly up-regulated in ccRCC cell lines in contrast to normal renal epithelial cells. By performing bioinformatics analysis and luciferase reporter assays, we revealed that the transcription factor SP1 can bind to the promoter region of SNHG14, resulting in the overexpression of SNHG14 in ccRCC. Functionally, enhanced expression of lncRNA SNHG14 promoted cell migration and invasion through promoting N-WASP protein level. More importantly, RT-qPCR and in situ RNA FISH analysis showed that SNHG14 was predominantly abundant in the cytoplasm of ccRCC cells. The subsequent RNA immunoprecipitation assay, and gain or loss-function assays showed that SNHG14 functioned as ceRNA to regulate N-WASP expression and cell motility ability via a miR-203-dependent manner. Our results imply that SNHG14 is a critical lncRNA that promotes ccRCC migration and invasion via sponging miR-203 and elevating N-WASP. Therefore, SNHG14 could serve as a promising therapeutic target for ccRCC.
Keywords: Clear cell renal cell carcinoma, LncRNA SNHG14, SP1, N-WASP, miR-203
Introduction
Renal cell carcinoma (RCC) is the most common malignancy of the kidney, and its incidence is increasing [1]. It is estimated that approximately 37.7 men and 16.6 women per 100,000 Chinese people are diagnosed with RCC every year [2]. Approximately 80-90% of RCCs are clear cell renal cell carcinoma (ccRCC), with a characteristic of high metastasis and relapse rate compared with other RCC subtypes [3]. Localized ccRCC remains a surgical disease and about 20%-30% patients who present with limited disease at the time of nephrectomy develop metastasis. Compared with other cancers, there are very few tumor and biomarkers for ccRCC [4]; thus, early detection and treatment are very important for patients with ccRCC.
With the advanced development of whole genome and transcriptome sequencing technologies and the ENCODE project, it is more and more clear that most of the genome DNA is represented in processed transcripts without or lacking of protein-coding capacity [5]. Long non-coding RNAs (lncRNAs), extensively transcribed from the mammalian genome, have gained widespread attention in recent years. They serve as important and powerful regulators of various biological activities and play critical roles in the progression of a variety of diseases, including cancer [6,7]. In the nucleus, lncRNAs can act as scaffolds to bind to specific proteins and to hire gene-modifying body to silence or activate targeted genes [8]. In the cytoplasm, lncRNAs may function as competing endogenous RNAs (ceRNAs), thus inducing the suppression of genes that targeted by specific miRNAs [9]. To date, elucidating the deregulated lncRNAs and identifying their functions in RCC are still an ongoing process in cancer investigation.
Neural Wiskott-Aldrich syndrome protein (N-WASP) is a key regulator of actin polymerization and cytoskeletal remodeling [10]. WASP family proteins are key regulators of actin polymerization during cell adhesion and invasion through activating Arp2/3 complex-mediated actin polymerization [11]. Recent evidences also indicate that N-WASP may be essential for tumor metastasis [12]. However, the functional role of N-WASP in ccRCC metastasis and the interaction between N-WASP and lncRNAs are still not well known.
In this research, we utilized the second-generation Hiseq sequencing followed by RT-qPCR assays to validate the imaginary that some lncRNAs may be promising therapeutic targets in ccRCC. Furthermore, the identified lncRNAs may improve and perfect the clinical treatment strategy. Eventually, we identified the up-regulation of lncRNA SNHG14 in ccRCC cells, and the increased expression of lncRNA SNHG14 promoted ccRCC cell migration and invasion through acting as a ceRNA to promote N-WASP expression.
Materials and methods
Cell culture
Human ccRCC cell lines A-498, 786-O, Caki-2, Caki-1 and human normal renal epithelial cell line HK-2 were purchased from the Chinese of Sciences in Shanghai. All ccRCC cell lines were maintained in RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS) (Sigma-Aldrich, St. Louis, MO, USA) at 37°C in a humidified chamber supplemented with 5% CO2. Human normal renal epithelial cell line HK-2 was grown in keratinocyte-SFM (Gibco, Invitrogen).
cDNA library construction and HiSeq sequencing analysis
Total RNA from four ccRCC cell lines and one normal renal epithelial cell line was extracted by one-step extraction using a Trizol kit (Life Technologies, USA), and the purity and quantity of RNA were determined by UV spectrophotometry. cDNA library construction and sequencing were performed according to previously described methods [13].
RNA oligoribonucleotides and cell transfection
The small interfering RNAs (siRNAs) that target lncRNA SNHG14 were synthesized and named as si-SNHG14 (Ribo Bio Corporation, Guangzhou, China) and embedded with GFP fluorescence for transfection quality control. The si-Negative Control (si-NC) was also provided by Genechem Corporation. The lncRNA SNHG14 overexpression plasmid (p-SNHG14) and control vector (p-Vector) were also synthesized by Ribo Bio (Guangzhou, China). The miR-203 mimics and siRNAs that specifically target N-WASP (si-N-WASP) and SP1 (si-SP1) were synthesized by GenePharma (Shanghai, China). Forty-eight h after planting ccRCC cells into 24-well plate, 100 nM of specific RNA oligoribonucleotides as well as negative controls were transfected into the cells with Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. The sequences of siRNAs are shown in Table 1.
Table 1.
Information of the RT-qPCR primer sequences and siRNA sequences
| RT-qPCR primer name | Primer sequence (5’-3’) |
|
| |
| SNHG14 (Forward) | GGGTGTTTACGTAGACCAGAACC |
| SNHG14 (Reverse) | CTTCCAAAAGCCTTCTGCCTTAG |
| N-WASP (Forward) | GAACGAGTCCCTCTTCACTTTC |
| N-WASP (Reverse) | GTTCCGATCTGCTGCATATAACT |
| miR-203 (Forward) | ACACTCCAGCTGGGGT |
| miR-203 (Reverse) | TGGTGTCGTGGAGTCG |
| SP1 (Forward) | TGGCAGCAGTACCAATGGC |
| SP1 (Reverse) | |
| GAPDH (Forward) | GCACCGTCAAGGCTGAGAAC |
| GAPDH (Reverse) | ATGGTGGTGAAGACGCCAGT |
| U6 (Forward) | CTCGCTTCGGCAGCACA |
| U6 (Reverse) | AACGCTTCACGAATTTGCGT |
| U1 (Forward) | GGGAGATACCATGATCACGAAGGT |
| U1 (Reverse) | CCACAAATTATGCAGTCGAGTTTCCC |
|
| |
| SiRNA name | SiRNA sequence (5’-3’) |
|
| |
| si-SNHG14-(1) sense | GCAAAUGAAAGCUACCAAU |
| si-SNHG14-(1) antisense | AUUGGUAGCUUUCAUUUGC |
| si-SNHG14-(2) sense | GCACAAUAUCUUUGAACUA |
| si-SNHG14-(2) antisense | UAGUUCAAAGAUAUUGUGC |
| si-SNHG14-(3) sense | CUAGAAUCCUAAAGGCAAA |
| si-SNHG14-(3) antisense | UUUGCCUUUAGGAUUCUAG |
| si-SP1 sense | CCAACAGAUUAUCACAAAU |
| si-SP1 antisense | GGUUGUCUAAUAGUGUUUA |
| si-N-WASP sense | ACAACTTAAAGACAGAGAA |
| si-N-WASP antisense | GCAAGAAATGTGTGACTAT |
| NC siRNA sense | UUCUCCGAACGUGUCACGUTT |
| NC siRNA antisense | ACGUGACACGUUCGGAGAATT |
Quantitative real-time PCR (RT-qPCR)
Total RNA from cells was isolated with TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. RNA was reverse transcribed using the SuperScript III® (Invitrogen) and then amplified by RT-qPCR with an SYBR Green Kit (Takara Bio Company, Dalian, China) on an ABI PRISM 7500 Sequence Detection System (Life Technologies, Grand Island, NY, USA). The gene expression levels were normalized by GAPDH/U6 expression. RT-qPCR results were analysed and expressed relative to CT (threshold cycle) values, and then converted to fold changes. All the premier sequences are shown in Table 1.
Luciferase reporter assay
The lncRNA SNHG14 promoter region construct was amplified from genomic DNA of A498 cells. The WT and mutated SNHG14 promoter constructs were cloned into the pGL4-basic reporter gene vector and verified by sequencing. For the identification of interaction between SNHG14 and miR-203, the N-WASP 3’ UTR cDNA was amplified and cloned to pGL4 luciferase expression vector. Renilla was used as the internal control for transfection efficiency.
RNA immunoprecipitation
Magna RIPTM RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) were used for RIP. ccRCC cells were lysed in complete RNA lysis buffer, then cell lysates were incubated with RIP immunoprecipitation buffer containing magnetic beads conjugated with human anti-Argonaute 2 (AGO2) antibody (Millipore) or negative control mouse IgG (Millipore).
Fluorescence in situ hybridization analysis
A-498 and 786-O cells were used for RNA FISH analysis. Nuclear and cytosolic fraction separation was performed using a PARIS kit (Life Technologies), and RNA FISH probes were designed and synthesized by Bogu according to the manufacturer’s instructions. Briefly, cells were fixed in 4% formaldehyde for 15 min and then washed with PBS. The fixed cells were treated with pepsin and dehydrated through ethanol. The air-dried cells were incubated further with 40 nM of the FISH probe in hybridization buffer. After hybridization, the slide was washed, dehydrated and mounted with Prolong Gold Antifade Reagent with DAPI for detection. The slides were visualized for immunofluorescence with an Olympus microscope.
Cell migration and invasion assays
For migration wound healing assay, ccRCC cells were seeded in six well plates and cultured until they reached confluence. Wounds were scratched on the monolayer of cells using 20 μL pipette tips. Plates were washed once with fresh medium to remove non-adherent cells after the cells had been cultured for 48 h, and then photographed. For transwell invasion assay, 100 μl matrigel (BD, USA) was firstly added onto the bottom of the transwell chamber (24-well insert; 8-mm pore size, Corning Costar Corp), then 1×105 ccRCC cells in reduced serum medium (Opti-MEM, Gibco) were placed on the coated membrane in the chamber. RPMI 1640 plus 10% FBS, was placed in the bottom wells as chemoattractants. After 24 h, cells that did not migrate were removed from the top side of the inserts with a cotton swab. Cells that migrated through the permeable membrane were fixed in methanol, stained with crystal violet, and counted under a microscope at 20× magnification in random fields in each well.
Western blot and antibodies
ccRCC cells treated with ESMC for 48 h were lysed with RIPA lysis buffer containing protease inhibitor cocktail and phosphates inhibitor cocktail on ice for 30 min, then lysis buffer was collected, and centrifuged at 12000 g, 4°C for 10 min. The protein lysates were resolved by SDS-PAGE, and separated proteins were transferred to PVDF membranes and blocked with 5% skimmed milk for 2 h. The primary antibodies used for western blotting were rabbit anti-human N-WASP antibody (1:1000; Cell Signaling Technology), rabbit anti-human SP1 antibody (1:1000, Cell Signaling Technology), and rabbit anti-human β-actin antibody (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA, USA). Horseradish peroxidase-conjugated (HRP) anti-rabbit antibodies (1:5000; Santa Cruz Biotechnology) were used as the secondary antibodies. The blots were incubated with the respective antibody overnight at 4°C under gently shaking. Finally, the proteins were detected by using horseradish peroxidase labeled secondary antibodies and an enhanced chemiluminescence detection system.
Statistical analysis
Kolmogorov-Smirnov test was used to determine the normality of the distribution of data in each group. Date differences were shown in median expression and were determined using the Mann-Whitney U test or Kruskal-Wallis test. Count dates were described as frequency and examined using Fisher’s exact test. The results were considered statistically significant at P<0.05. All statistical analyses were performed with SPSS 17.0 software (SPSS Incorporation, Chicago, IL). Error bars in figures represent SD (Standard Deviation). *P<0.05; **P<0.01; ***P<0.001.
Results
LncRNAs expression profile analysis
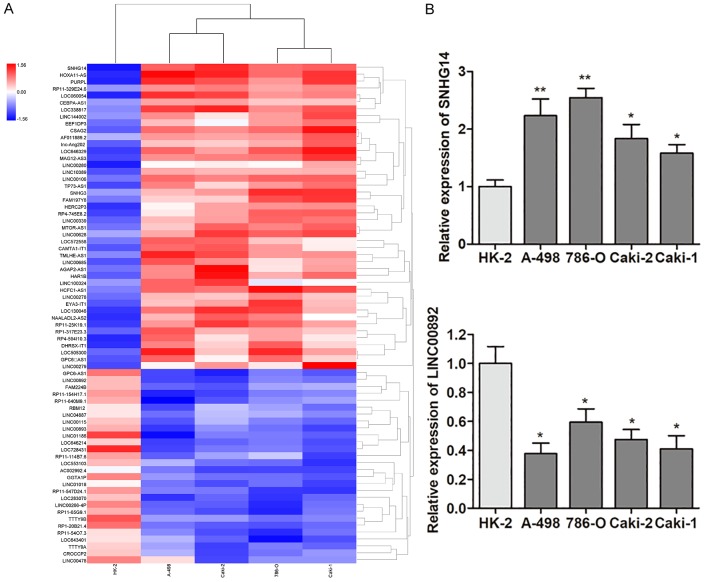
We firstly sought to define the lncRNAs that were differently expressed between ccRCC cells and normal renal epithelial cells via high-throughput Hiseq array. We identified 476 lncRNAs that were differentially expressed between the two groups. Among these, the expression of 72 lncRNAs showed more than 2-fold difference, including 44 lncRNAs that were up-regulated and 28 lncRNAs that were down-regulated (Figure 1A). Subsequently, we focused on the six most dysregulated lncRNAs (Table 2), and RT-qPCR experiment was performed to verify the potential differentially expressed lncRNAs. As shown in Figure 1B, lncRNA SNHG14 was significantly up-regulated while LINC00892 expression was significantly suppressed in ccRCC cell lines in contrast to the normal renal epithelial cells. However, our preliminary results showed that ectopic expression of LINC00892 had no significant effect on ccRCC cell viability and motility (data not shown), which inspired us to focus on the biological significance of lncRNA SNHG14 in ccRCC progression.
Figure 1.
LncRNAs expression profile analysis. A. The heat map showed the top 44 most increased and 28 most decreased lncRNAs in ccRCC cells in contrast to normal renal epithelial cells analyzed by Hiseq sequencing. B. Relative expression of lncRNA SNHG14 and LINC00892 in ccRCC cell lines via RT-qPCR.
Table 2.
Candidate lncRNAs selected on a basis of the Hiseq analysis
| Seq-name | Location | Regulation (C. vs N.) | Fold change | P value |
|---|---|---|---|---|
| SNHG14 | Chr15q11.2 | Up | 42.2689 | 0.00005627 |
| HOXA11-AS | Chr7p15.2 | Up | 23.2731 | 0.00017935 |
| PURPL | Chr5p14.1 | Up | 17.3405 | 0.00065962 |
| GPC6-AS1 | Chr13q31.3 | Down | 30.7465 | 0.00009357 |
| LINC00892 | ChrXq26.3 | Down | 24.8801 | 0.00013587 |
| FAM224B | ChrYq11.222 | Down | 15.4768 | 0.00095764 |
C.: ccRCC cell; N.: Normal cell.
LncRNA SNHG14 is activated by SP1 in ccRCC
Although a lot of lncRNA dysregulation has been reported in cancers, the regulators involved in misregulation of these molecules are not properly understood. Increasing evidence has revealed that several key transcription factors contribute to lncRNA dysregulation in the human cancer cells. Herein, we focused on transcription factors binding to the SNHG14 promoter. Using the online transcription factor prediction software JASPAR, we found that there are several SP1 binding sites in the SNHG14 promoter regions with high score (Figure 2A). In addition, SP1 was up-regulated in ccRCC cell lines in contrast to normal cells at both transcript and protein level (Figure 2B). Then, RT-qPCR showed that SP1 was inhibited in ccRCC cells by transfection of specific si-SP1 vector in A-498 and 786-O cells (Figure 2C), and knockdown of SP1 suppressed the expression of lncRNA SNHG14 in A-498 and 786-O cells (Figure 2D). In addition, the SNHG14 promoter region including 3 potential binding sites of SP1 was inserted into a PGL4 luciferase reporter vector (Figure 2E), and dual-luciferase reporter analysis showed that knockdown of SP1 could inhibit the luciferase activity (Figure 2F). These results indicated that the up-regulation of SNHG14 in ccRCC cells may be induced by SP1.
Figure 2.
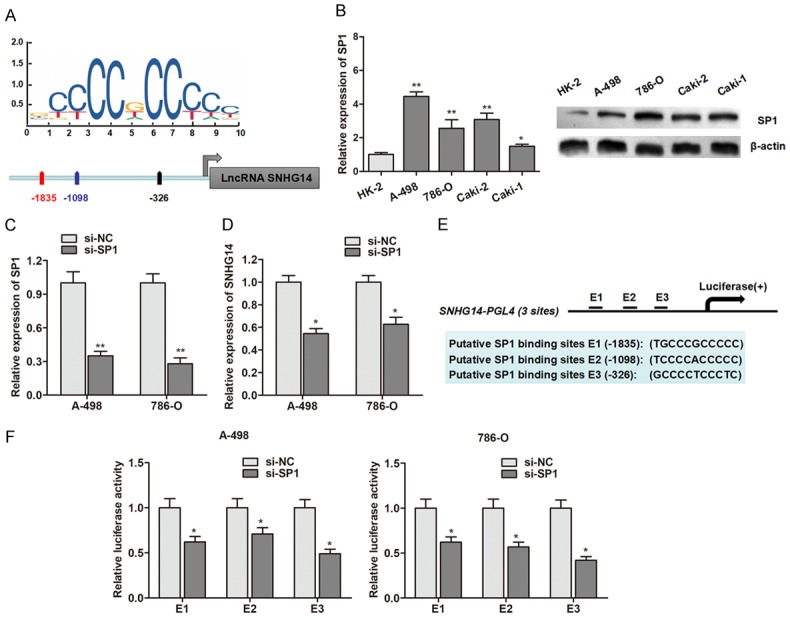
LncRNA SNHG14 is activated by SP1 in ccRCC. A. SP1 binding site prediction in the SNHG14 promoter region using JASPAR. B. The expression of SP1 in ccRCC cell lines and normal renal epithelial cells at transcript (left panel) and protein (right panel) levels. C. SP1 was knocked down by the transfection of specific siRNAs. D. LncRNA SNHG14 was down-regulated by knockdown of transcript factor SP1. E. Potential SP1 binding sites in the promoter region of SNHG14 used for construction of luciferase vector containing the binding region. F. Luciferase activity was significantly decreased in si-SP1-transfected cells compared with control vector in three binding sites.
Knockdown of SNHG14 suppresses ccRCC cell migration and invasion
We chose A-498 and 786-O cells for further investigation, because these two cells had relatively higher SNHG14 expression level than that in other ccRCC cell lines. To determine the functional role of SNHG14 in ccRCC, we designed three different siRNAs and transfected these three siRNAs into ccRCC cells (Figure 3A). As shown in Figure 3B, si-SNHG14-(3) showed the best silencing efficiency, and this siRNA was used for further studies (shortened as si-SNHG14). We evaluated the function of SNHG14 on cell migration and invasion. After transfection of si-SNHG14 for 48 h, a significantly decreased number of ccRCC cells were observed to migrate through the collagen membrane compared with control cells (Figure 3C). Similar effects were also observed that a much smaller numbers of ccRCC invading through the Matrigel-coated membrane (Figure 3D).
Figure 3.
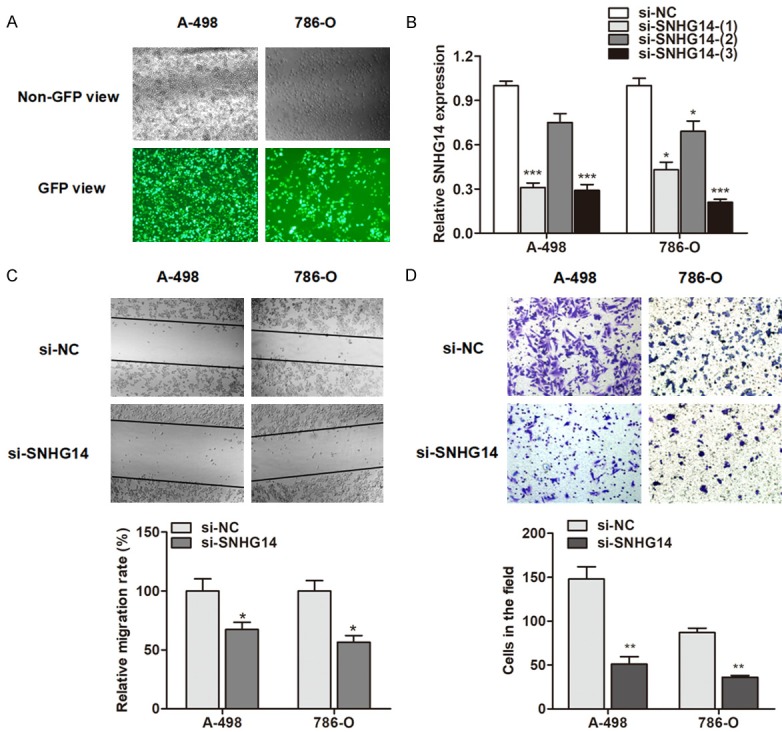
Knockdown of SNHG14 suppresses ccRCC cell migration and invasion. (A) The siRNAs labeled with GFP green fluorescence were transfected as described in Methods. (B) The silencing efficacy was evaluated by transfection of three siRNAs of SNHG14. (C, D) Knockdown of SNHG14 significantly suppressed the migratory (C) and invasive (D) ability of A-498 and 786-O cells.
LncRNA SNHG14 regulates motility of ccRCC cells via targeting N-WASP
To identify the underlying regulatory mechanism of SNHG14 in ccRCC cell migration and invasion, we focused on the metastasis-associated proteins. Our preliminary results suggested that N-WASP was also upregulated in ccRCC cells (Figure 4A). Thus, we wondered if SNHG4 exerted the pro-metastatic function through targeting N-WASP. As expected, N-WASP was dramatically downregulated in ccRCC cells after the reduction of SNHG-14 (Figure 4B). To further identify the functional interaction between SNHG14 and N-WASP, we generated the lncRNA SNHG14 overexpressing vector and N-WASP silencing vector (Figure 4C and 4D). The results showed that overexpression of SNHG14 promoted cell migration and invasion ability, however, this effect was dramatically abrogated by the co-transfection of N-WASP silencing vector in ccRCC cells (Figure 4E). This indicates that SNHG14 regulates cell migration and invasion via elevating N-WASP expression.
Figure 4.
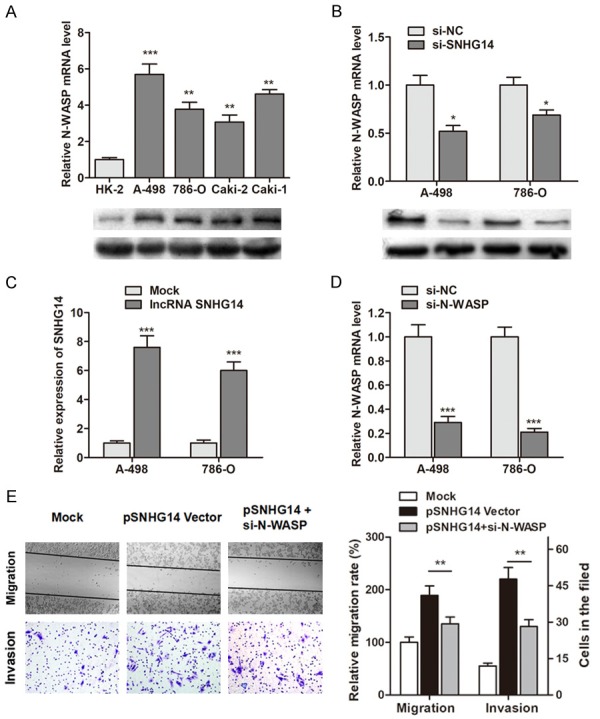
SNHG14 regulates ccRCC cell migration and invasion via targeting N-WASP. A. The expression of N-WASP in ccRCC cells and normal renal epithelial cells were detected. B. N-WASP was down-regulated in ccRCC cells by knockdown of SNHG14 at both mRNA and protein levels. C. SNHG14 was up-regulated by transfection of SNHG14 overexpressing vector. D. N-WASP was knocked down by specific silencing vectors. E. Overexpression of SNHG14 promoted cell migratory and invasive ability, however, this effect was partly reversed by co-transfection of si-N-WASP in A-498 cells.
LncRNA SNHG14 regulates N-WASP expression via a miR-203-dependent manner
It is widely accepted that lncRNAs may function as ceRNA to further regulate the target mRNA of miRNAs. With this hypothesis, we firstly localized the expression of lncRNA SNHG14 in ccRCC cells. As shown in Figure 5A, RT-qPCR analysis of nuclear and cytoplasmic lncRNA showed that SNHG14 was mainly enriched in the cytoplasm section of ccRCC cells. More importantly, our in situ RNA FISH analysis also suggested that SNHG14 was expressed predominately in the cytoplasm part (Figure 5B). Moreover, the RIP assay with AGO2 antibody in ccRCC cells showed an enrichment of both SNHG14 and N-WASP (Figure 5C), indicating that SNHG14 and N-WASP were both recruited to RNA-induced mediating complexes based on AGO2 and may sponge with miRNAs. Subsequently, we sought to define the specific miRNAs. Based on miRanda and miRcode, we identified that miR-203 targeted the transcript of SNHG14 and N-WASP with high score (Figure 5D and 5E). To verify that SNHG14 and N-WASP were both targeted by miR-203, we performed luciferase reporter assay. We found that the luciferase activity were significantly suppressed by overexpression of miR-203 at both SNHG14 and N-WASP wild type reporter, however, miR-203 did not inhibit the luciferase activity of reporter vector containing the mutant sequences lacking miR-203 binding sites (Figure 5F). Moreover, RIP assay showed that SNHG14 overexpressing vector in A-498 cells caused inhibited enrichment of N-WASP by miR-203; while knockdown of SNHG14 in induced a significant increase enrichment of N-WASP by miR-203 (Figure 5G).
Figure 5.
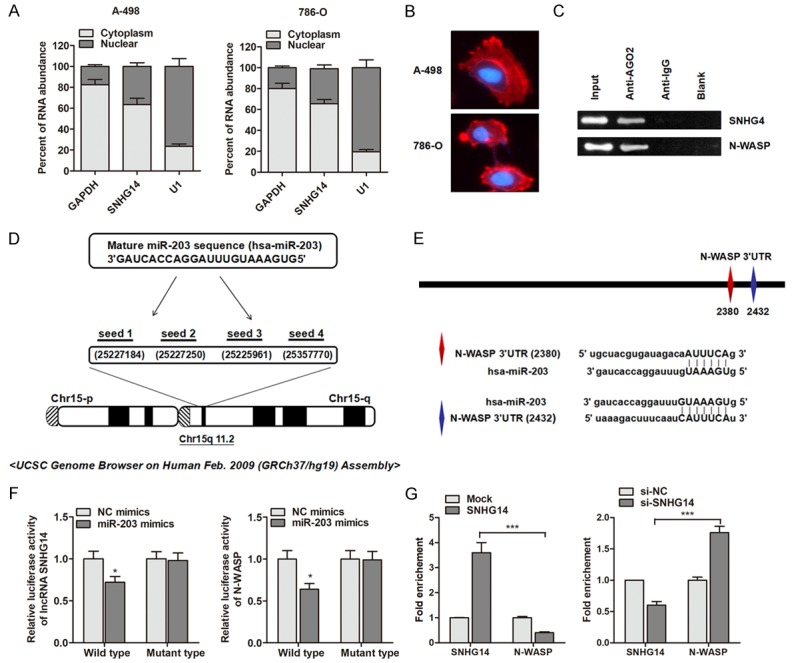
LncRNA SNHG14 regulates N-WASP expression via a miR-203-dependent manner. A. The expression levels of SNHG14 in nuclear and cytoplasm of ccRCC cells. U1 (nuclear retained) and GAPDH (exported to cytoplasm) were used as controls. B. Fluorescence in situ hybridization analysis of the subcellular location of SNHG14 in ccRCC cells. C. RIP experiments were performed using the AGO2 antibody, and specific primers were used to detect the enrichment of SNHG14 and N-WASP in A-498 cells. D. Representation of the miR-203 binding site in SNHG14 based on miRcode (http://www.mircode.org/mircode/). E. Illustration of the the putative predicted miR-203 binding sites in the N-WASP 3’UTR region. F. Firefly luciferase activity normalized to Renilla luciferase activity in A-498 cells co-transfected with luciferase reporters with wild type or mutant transcripts of SNHG14 or N-WASP along with miR-203 mimics or negative control (NC mimics). G. Overexpression of SNHG14 in A-498 cells led to the decreased enrichment of N-WASP transcripts on miR-203. However, SNHG14 knockdown caused a significant increase in the recruitment of N-WASP to miR-203.
MiR-203 is essential for the function of SNHG14 in ccRCC cell migration and invasion
After having validated the functional correlation between SNHG14 and N-WASP, and the regulatory interactions of SNHG14, miR-203 and N-WASP, we further investigated the role of miR-203 in the function of SNHG14. As shown in Figure 6, the enhanced migratory ability induced by overexpression of SNHG14 was reversed by co-transfection of miR-203 mimics. In addition, the increased cells that invading through the Matrigel-coated membrane caused by SNHG14 was also abrogated by co-expression of miR-203 mimics. Combining with the above observations, we finally developed that lncRNA SNHG14 promoted ccRCC cell motility by inhibiting miR-203 and activating N-WASP.
Figure 6.
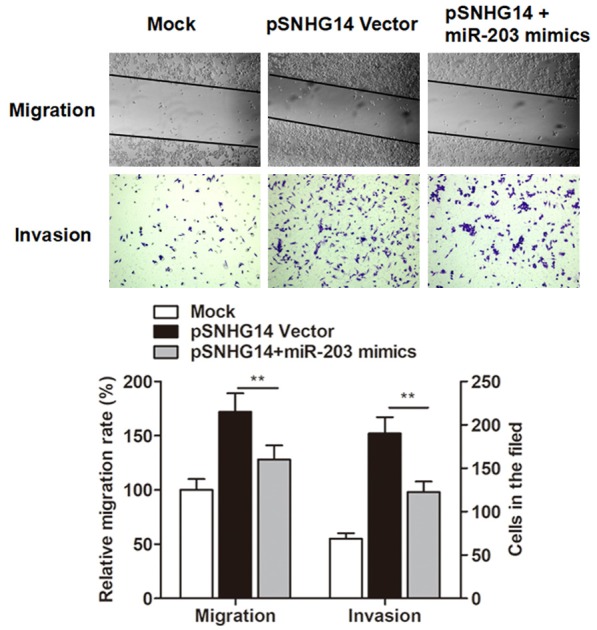
Overexpression of SNHG14 promoted the cell migratory and invasive ability of A-498 cells, however, this effects were reversed by co-transfection of miR-203 mimics.
Discussion
Early-stage ccRCC is curable by surgery; however, locally advanced or metastatic cancer is chemotherapy resistant. To develop a novel therapeutic drug for ccRCC, it is crucial to clarify the underlying molecular mechanisms involved in the development and progression of ccRCC. In the present study, we identified and validated the up-regulation of lncRNA SNHG14 in ccRCC cells based on high-throughput Hiseq sequencing. Furthermore, we also uncovered that the increased expression of SNHG14 was due to the activation by the transcription factor SP1. Mechanistic analysis indicated that SNHG14 promoted ccRCC cell migration and invasion through specifically sponging miR-203 and releasing N-WASP protein.
LncRNAs have gained huge attention in recent years due to its aberrant expression in a large range of cancers and multiple roles in cancer initiation, progression and metastasis. LncRNAs could act as activators or inhibitors to participate in a variety of biological processes via interacting with DNAs, mRNAs, microRNAs or proteins [14]. During recent years, more and more new lncRNAs were identified and functionally characterized. Based on the second-generation sequencing method and subsequent RT-qPCR validation, we found that lncRNA SNHG14 was significantly up-regulated in ccRCC cells. LncRNA SNHG14, alternatively named UBE3A-ATS, is located on chromosome 15q11.2, is rarely reported. Previously, Qi et al. demonstrated that SNHG14 may increase the expression of PLA2G4A by inhibition of miR-145-5p, which resulted in the activation of microglia cells in cerebral infarction [15]. However, the expression and functional role of SNHG14 in cancer progression were not well known. Therefore, this study that identified the up-regulation of SNHG14 in cancer may open a new avenue for the cancer research.
We sought to define the underlying regulatory mechanism of lncRNA SNHG14. Typically, genes were transcribed and activated by the regulation of various factors. The activation or silence of gene promoter area by specific promoters is dramatically important for the gene expression. Currently, the transcription of SNHG14 is regulated by many promoters. However, no study has reported the promoter regulation of SNHG14 gene, therefore, it is still unknown why SNHG14 is up-regulated in cancers, which of these promoters is predominantly used, and how these promoters activate SNHG14 by binding to the specific promoter region. By using bioinformatics databases including Jaspar database, we screened for potential proto-oncogenic transcription factor. We then focused on SP1 transcription factor, because SP1 presented relatively higher score than other regulators. A few studies have revealed that lncRNA transcription can also be regulated by SP1. For example, Xu et al. demonstrated that SP1 binds to the promoter region of lncRNA TINCR and promote its transcription, inducing the enhancement of cell proliferation [16]; Huang et al. also found that LINC00673 is activated by SP1 and exerts oncogenic properties by interacting with LSD1 and EZH2 in gastric cancer [17]; Cai et al. reported that lncRNA XIAP-AS1 promotes XIAP transcription by XIAP-AS1 interacting with SP1 [18]. In our study, we performed luciferase reporter assays, and determined that SP1 could bind to the SNHG14 promoter region and induce its transcription. Our findings combined with previous studies suggest that the abnormal activation of transcription factors may play an important role in lncRNA overexpression in the human cancer cells.
We then determined the functional role of lncRNA SNHG14, and found that knockdown of SNHG14 suppressed ccRCC cell migration and invasion. More importantly, the metastatic protein N-WASP was verified as the downstream target of SNHG14. A large amount of evidence indicated that N-WASP could promote cancer metastasis in various cancer types. Liu et al. clearly reported that a higher level of N-WASP expression in the tumor was associated with advanced stage and relatively poor survival in ccRCC patients [19]. Both Bettencourt et al. and Schwickert et al. demonstrated that N-WASP was regulated by miR-142-3p and promoted cancer cell migration and invasion [20,21]. Our gain and loss functional assay also validated that SNHG14 promoted ccRCC cell invasive ability by activating N-WASP.
It is widely accepted that lncRNAs may function as ceRNA to further regulate the target mRNA of miRNAs. We then focused on the target miRNA of SNHG14 and N-WASP. We used the bioinformatics tool miRanda and miRcode and identified that miR-203 targeted both SNHG14 and N-WASP sequences with high score. MiR-203, located at the chromosome 14q32.33, is one of the most frequently mentioned miRNAs. Previous studies have showed that miR-203 exhibited aberrant expression in multiple malignancies compared with their normal tissues [22]. Although the role of miR-203 in malignancies is conflicting, miR-203 may acts as a tumor suppressor in ccRCC. Xu et al. demonstrated that miR-203 inhibits renal cancer cell proliferation, migration and invasion by targeting of FGF2 [23]. Consistent with this study, our luciferase reporter assay and ChIP assay clearly showed that SNHG14 sponged miR-203 and subsequently induced the up-regulation of N-WASP, a downstream targeted mRNA of miR-203.
Finally, we evaluated the functional role of the SNHG14/miR-203/N-WASP pathway in ccRCC metastasis. The gain and loss functional assays suggested that SNHG14 promoted ccRCC migration and invasion through sponging miR-203 and promoting N-WASP protein. In conclusion, our integrated approach demonstrated that lncRNA SNHG14 was up-regulated and activated by SP1 regulator in ccRCC cells. SNHG14 could promote ccRCC cell migration and invasion as a ceRNA through sponging miR-203 and releasing N-WASP. Therefore, lncRNA SNHG14 can serve as a promising prognostic marker or therapeutic target of patients with ccRCC.
Disclosure of conflict of interest
None.
References
- 1.Qin C, Sun LJ, Cui L, Cao Q, Zhu J, Li P, Zhang GM, Mao X, Shao PF, Wang ML, Zhang ZD, Gu M, Zhang W, Yin CJ. Application of the revised Tumour Node Metastasis (TNM) staging system of clear cell renal cell carcinoma in eastern China: advantages and limitations. Asian J Androl. 2013;15:550–557. doi: 10.1038/aja.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang L, Parkin DM, Ferlay J, Li L, Chen Y. Estimates of cancer incidence in China for 2000 and projections for 2005. Cancer Epidemiol Biomarkers Prev. 2005;14:243–250. [PubMed] [Google Scholar]
- 3.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 4.Linehan WM. Genetic basis of kidney cancer: role of genomics for the development of disease-based therapeutics. Genome Res. 2012;22:2089–2100. doi: 10.1101/gr.131110.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tukiainen T, Villani AC, Yen A, Rivas MA, Marshall JL, Satija R, Aguirre M, Gauthier L, Fleharty M, Kirby A, Cummings BB, Castel SE, Karczewski KJ, Aguet F, Byrnes A, Lappalainen T, Regev A, Ardlie KG, Hacohen N, MacArthur DG. Landscape of X chromosome inactivation across human tissues. Nature. 2017;550:244–248. doi: 10.1038/nature24265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiao F, Hu H, Yuan C, Wang L, Jiang W, Jin Z, Guo Z, Wang L. Elevated expression level of long noncoding RNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer. Oncol Rep. 2014;32:2485–2492. doi: 10.3892/or.2014.3518. [DOI] [PubMed] [Google Scholar]
- 7.Li J, Huang H, Li Y, Li L, Hou W, You Z. Decreased expression of long non-coding RNA GAS5 promotes cell proliferation, migration and invasion, and indicates a poor prognosis in ovarian cancer. Oncol Rep. 2016;36:3241–3250. doi: 10.3892/or.2016.5200. [DOI] [PubMed] [Google Scholar]
- 8.Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, Plath K, Guttman M. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stradal TE, Rottner K, Disanza A, Confalonieri S, Innocenti M, Scita G. Regulation of actin dynamics by WASP and WAVE family proteins. Trends Cell Biol. 2004;14:303–311. doi: 10.1016/j.tcb.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 11.Prehoda KE, Scott JA, Mullins RD, Lim WA. Integration of multiple signals through cooperative regulation of the N-WASP-Arp2/3 complex. Science. 2000;290:801–806. doi: 10.1126/science.290.5492.801. [DOI] [PubMed] [Google Scholar]
- 12.Hou J, Yang H, Huang X, Leng X, Zhou F, Xie C, Zhou Y, Xu Y. N-WASP promotes invasion and migration of cervical cancer cells through regulating p38 MAPKs signaling pathway. Am J Transl Res. 2017;9:403–415. [PMC free article] [PubMed] [Google Scholar]
- 13.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao X, Dhanasekaran SM, Wu YM, Robinson DR, Beer DG, Feng FY, Iyer HK, Chinnaiyan AM. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li T, Mo X, Fu L, Xiao B, Guo J. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget. 2016;7:8601–8612. doi: 10.18632/oncotarget.6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi X, Shao M, Sun H, Shen Y, Meng D, Huo W. Long non-coding RNA SNHG14 promotes microglia activation by regulating miR-145-5p/PLA2G4A in cerebral infarction. Neuroscience. 2017;348:98–106. doi: 10.1016/j.neuroscience.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 16.Xu TP, Liu XX, Xia R, Yin L, Kong R, Chen WM, Huang MD, Shu YQ. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34:5648–5661. doi: 10.1038/onc.2015.18. [DOI] [PubMed] [Google Scholar]
- 17.Huang M, Hou J, Wang Y, Xie M, Wei C, Nie F, Wang Z, Sun M. Long noncoding RNA LINC00673 is activated by SP1 and exerts oncogenic properties by interacting with LSD1 and EZH2 in gastric cancer. Mol Ther. 2017;25:1014–1026. doi: 10.1016/j.ymthe.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Cai J, Wang D, Bai ZG, Yin J, Zhang J, Zhang ZT. The long noncoding RNA XIAP-AS1 promotes XIAP transcription by XIAP-AS1 interacting with Sp1 in gastric cancer cells. PLoS One. 2017;12:e0182433. doi: 10.1371/journal.pone.0182433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu GH, Chen J, Ji ZG, Zhou L. Expression of neural wiskott-aldrich syndrome protein in clear cell renal cell carcinoma and its correlation with clinicopathological features. Urol Int. 2015;95:79–85. doi: 10.1159/000365595. [DOI] [PubMed] [Google Scholar]
- 20.Schwickert A, Weghake E, Bruggemann K, Engbers A, Brinkmann BF, Kemper B, Seggewiss J, Stock C, Ebnet K, Kiesel L, Riethmuller C, Gotte M. microRNA miR-142-3p inhibits breast cancer cell invasiveness by synchronous targeting of WASL, integrin alpha V, and additional cytoskeletal elements. PLoS One. 2015;10:e0143993. doi: 10.1371/journal.pone.0143993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bettencourt P, Marion S, Pires D, Santos LF, Lastrucci C, Carmo N, Blake J, Benes V, Griffiths G, Neyrolles O, Lugo-Villarino G, Anes E. Actin-binding protein regulation by microRNAs as a novel microbial strategy to modulate phagocytosis by host cells: the case of N-Wasp and miR-142-3p. Front Cell Infect Microbiol. 2013;3:19. doi: 10.3389/fcimb.2013.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu J, Wang Y, Wu X. MicroRNA in the pathogenesis and prognosis of esophageal cancer. Curr Pharm Des. 2013;19:1292–1300. doi: 10.2174/138161213804805775. [DOI] [PubMed] [Google Scholar]
- 23.Xu M, Gu M, Zhang K, Zhou J, Wang Z, Da J. miR-203 inhibition of renal cancer cell proliferation, migration and invasion by targeting of FGF2. Diagn Pathol. 2015;10:24. doi: 10.1186/s13000-015-0255-7. [DOI] [PMC free article] [PubMed] [Google Scholar]