Abstract
Arginine methylation of the epidermal growth factor receptor (meEGFR) increases the binding affinity of EGFR ligands and is reported to have a role in predicting response to anti-EGFR agents. This study investigated the predictive impact of meEGFR in metastatic colorectal cancer (mCRC) patients treated with anti-EGFR agents. Two patient cohorts were evaluated. Cohort 1 consisted of mCRC patients with documented disease progression following anti-EGFR treatment. Circulating tumor cells (CTCs) were isolated and distinguished based on CD45- and Epcam+. Cohort 2 consisted of formalin fixed paraffin-embedded (FFPE) blocks from a prospective cohort. meEGFR in both cohorts was identified by positive staining for me-R198/200 EGFR signal. CTCs were identified in 30 out of 47 cases in cohort 1. Of those 30, meEGFR-CTCs were identified in 19 cases. Mean total meEGFR-CTCs counts was 2.3 (range 0-30) cells per 7.5 ml. There was no association between meEGFR-CTCs and clinic-pathological-molecular features. In RAS wt/BRAF wt patients with high levels of meEGFR-CTCs ratio (≥ 0.23) had significantly inferior PFS with anti-EGFR treatment (HR = 3.4, 95% CI 1.5-7.9, P = 0.004). By contrast, high levels of meEGFR in the untreated tumor tissues had no correlation with anti-EGFR treatment duration in cohort 2. Therefore, meEGFR-CTCs may have the potential to serve as a “liquid biopsy” biomarker to predict anti-EGFR treatment efficacy.
Keywords: Liquid biopsy, circulating tumor cells, EGFR, arginine methylation, colorectal cancer, predictive marker
Introduction
Colorectal cancer (CRC) is one of the most commonly diagnosed cancers worldwide. In the Unit-ed States, it is estimated that more than 130,000 new cases will be diagnosed with nearly 50,000 deaths from CRC in 2016. Despite the recent increase in cases with molecular descriptions, treatment advances have not kept pace with the new information, and the 5-year survival rate of advanced-stage CRC is only 15% [1]. Monoclonal antibodies against epidermal growth factor receptor (anti-EGFRab), including cetuximab and panitumumab, are currently the standard treatment for metastatic colorectal cancer (mCRC). The U.S. Food and Drug Administration has recommended the use of anti-EGFRab treatment in colorectal cancers with wild type (WT) RAS (both KRAS/NRAS) as mutant RAS is associated with poor response to cetuximab [2-4]. However, only 40-60% of the RAS WT patient population respond to anti-EGFRab [5], and not all patients harboring mu-tant KRAS show resistance to anti-EGFRab treatment [6]. Therefore, these outcomes suggested there exists some heterogeneity in EGFR signaling and dependency even among RAS WT patients. Similarly those patients who initially respond to anti-EGFRab treatment often develop resistance within a year.
Resistance mechanisms to anti-EGFRab have been wildly studied. Primary resistance mechanisms have been reported, including: 1) Alteration in EGFR and EGFR ligand [7,8]; 2) RAS mutation [9]; 3) Mutation of V-raf murine sarcoma viral oncogene homolog B (BRAF) [10]; 4) Activation of phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA)/phosphatase and tensin homolog (PTEN) pathway [11,12]. Mechanisms underlying acquired resistance to EGFRab have also been proposed: 1) Acquired mutation of RAS and EGFR [13]; 2) Acquired mutation of BRAF [14,15]; 3) Amplification of human epidermal growth factor receptor 2 (HER2) [16,17] or MET [18] signaling; 4) Mutation of PIK3CA [11,12]; 5) Loss of expression of PTEN [11,19,20]. However, accumulating data on these signal transduction pathways currently shows that these mutations serve as prognostic markers only and not predictive markers [21]. Hence, further investigation into the underlying mechanism of both primary and acquired anti-EGFRab resistance and identification of better predictors for anti-EGFRab response are needed.
Aberrant EGFR activation caused by EGFR gene mutation, amplification and/or ligand overexpression is involved in the pathogenesis of multiple cancers [22]. Although EGFR mutations are common in many cancer types, very few occur in CRC. When they do occur, the EGFR mutations are generally localized to the intracellular catalytic domain, resulting in oncogenic activation. There is emerging evidence to suggest that alterations affecting the extracellular domain of EGFR also drive oncogenic activities [23]. Recently, our group reported a post-translational arginine methylation on the extracellular domain of EGFR by protein arginine methyltransferase (PRMT) 1 at R198 and R200 that resulted in increased ligand binding to promote EGFR receptor dimerization and activation, and alters EGFR signaling. Additionally, patients with high methylated EGFR expression in the tumor tissues correlated with shorter duration of cetuximab response [24]. Overall, these results suggested that EGFR R198/200 has the potential to serve as a predictive biomarker for anti-EGFRab treatment response.
Liquid biopsies are innovative types of molecular tumor sampling methods because they can serve as a non-invasive and an easy technique to obtain the gene mutation profiles from either circulating tumor cells (CTCs) or cell-free circulating tumor DNA (cfDNA). Mutation detection in blood can produce results highly similar to those of traditional biopsies [25]. Moreover, liquid biopsies can also identify mutations that are associated with treatment resistance that is not possible to detect in the original tissue biopsy [26]. Several studies reported the unfavorable prognostic impact of high CTCs number on patient survival in CRC [27-30]; however, none of those studies demonstrated the predictive impact on CRC treatment.
Here, we evaluated the possibility of using protein arginine methylation of the EGFR (meEGFR) to predict response to anti-EGFR agents by systemically analyzing two different CRC patient sample cohorts. We analyzed the expression of meEGFR on CTCs in blood samples from patients from first cohort who were previously treated with EGFRab using the ParsortixTM system. We evaluated meEGFR expression in formalin fixed paraffin embedded (FFPE) tumor tissues from patients in the second cohort [tissues]. The association between meEGFR expression and progression-free survival (PFS) were evaluated for both cohorts.
Material and methods
All studies performed were approved by the Institutional Review Board at The MD Anderson Cancer Center.
The CTC sample cohort involved a prospective study. The inclusion criteria were mCRC patients whose histology confirmed colorectal adenocarcinoma, document disease progression after anti-EGFR agents, and age ≥ 18 years and an Eastern Cooperative Oncology group performance status ≤ 2. Patients’ blood was obtained between September 2015 and July 2016. Circulating tumor cell (CTC) isolations involved collecting a maximum 15 mL of blood in Vacutainer tubes containing EDTA (BD Biosciences).
The FFPE cohort involved mCRC patient medical record review and sample identification as part of the Assessment of Targeted Therapies Against Colorectal Cancer (ATTAAC) program. These ATTACC patients were enrolled between February 13, 2009, and November 18, 2015. Last follow up date was January 31, 2017. All patients were provided with a written informed consent for blood collection under IRB protocols 2009-0091 or LAB 10-0963 protocol. The primary objective was to investigate the association of meEGFR expression with progression free survival (PFS) in patients receiving anti-EGFR treatment; the secondary objective was to examine the associations between meEGFR expression and with various clinico-pathological-molecular variables.
Clinical characteristics
Demographic information was collected from a medical record review, including age, gender, primary tumor site, dates of anti-EGFR treatment, lines of anti-EGFR agents used, tumor metastatic sites, previous treatment with irinotecan, date of last follow-up, and date of death. Right-sided colon cancer was defined as cancer in the region from the cecum to the transverse colon, whereas left-sided colon cancer was defined as cancer in the region from the splenic flexor through the rectum. The staging was done per the American Joint Committee on Cancer/Union for International Cancer Control TMN staging system (version 7, 2010) [31]. Progression free survival (PFS) is defined as the interval between the start date for anti-EGFR agents and the stop date of anti-EGFR agents due to disease progression.
Isolation of circulating tumor cells
Tumor cells were isolated from patient blood using the ANGLE Parsorter PR1 system (ParsortixTM). This system uses a microfluidic cassette, which separates CTCs by size differences of blood cells in a micro-flow environment. No antibodies are used in this system. Detailed methods associated with this assay have been previously published [32]. In brief, a CTC separation cassette narrows stepwise to a 10-µm gap and traps larger cells (> 10 µm in diameter). After rinsing the microfluidics cassette with 70% Ethanol and PBS, whole blood containing EDTA is loaded on the ParsorterTM system (Angel, Inc.) and then washed with buffer. Each blood sample was then separated by size, and CTCs were isolated over the course of approximately two hours. CTCs were then spread onto two glass slide by Cytospin 2TM (Shandon Inc.) and fixed with 4% paraformaldehyde (Electron Microscopy Sciences) for 15 min at room temperature, then washed with PBS three times. Sample slides were then stored in -80°C for further analysis.
Identification CTCs and meEGFR-CTCs
CTCs were identified based on the combination of positive Epcam signal and lack of CD45 biomarker expression. In brief, sample slides were first blocked with goat serum at room temperature for 60 min. After blocking, Alexa647 conjugated anti-Epcam antibody (Cell Signaling) and Alexa488 conjugated CD45 antibody (abcam) were applied to the sample with 1:100 and 1:500 dilution, respectively, in antibody dilution buffer (1% BSA, 0.3% Triton X-100). After overnight incubation at 4°C, unbound antibodies were removed by phosphate-buffered saline (PBS) wash and coverslip slides with Prolong Gold Antifade Reagent with DAPI (Cell Signaling). Stained slides were scanned using a high-content imaging system (Molecular Devices), and total CTCs numbers were determined by counting Epcam+ and CD45- cells staining across the entirety of each slide image. A high CTC count was defined as ≥ 3 CTC per 7.5 ml of blood based on the data from a previous study [27]. The number of meEGFR positive cells were determined by immunohistochemistry staining (IHC) using me-R198/200 antibody generated by our lab as previously described [24]. Interpretation of immunohistochemical analysis for meEGFR-CTCs was shown in Supplementary Figure 1.
IHC analysis of meEGFR expression in FFPE
To detect meEGFR in FFPE tumor samples, slides were deparaffinized and rehydrated, and antigen retrieval was performed by the Lab VisionTM PT Module (ThermoFisher Scientific) The sections were treated with 1% hydrogen peroxide in methanol for 30 minutes to block endogenous peroxidase activity. After 1 hour of serum blocking, the samples were incubated with primary antibodies at 4°C overnight. The sections were then treated with biotinylated secondary antibody, followed by incubations with avidinbiotin peroxidase complex solution for 1 hour at room temperature. Color was developed using 3-amino-9-ethylcarbazole solution. Counterstaining was done using Mayer’s hematoxylin. The total protein expression score was calculated as a function of the percentage of immunopositive cells and immunostaining intensity. High meEGFR expression was defined as more than 50% of the immune score activity which was greater than 150. Interpretation of immunohistochemical (IHC) analysis for meEGFR on tissues was shown in Supplementary Figure 2.
Gene mutational analysis
DNA was extracted from FFPE tumor tissue. Samples were evaluated for somatic mutation using a next-generation sequencing platform with 46- or 50-gene panels. Alternately, samples were analyzed for targeted gene mutation of frequently reported point mutations found in human malignancies. Targeted mutation analysis was conducted in a Clinical Laboratory Improvement Amendments (CLIA)-certified molecular diagnostics laboratory. This testing determined the effective lower limit of detection (analytical sensitivity) for single nucleotide variations to be in the range of 5% (one mutant allele in the background of nineteen wild type alleles) to 10% (one mutant allele in the background of nine wild type alleles).
Determination of mismatch repair (MMR) status
MMR status was determined by IHC analysis of MMR protein expression or by polymerase chain reaction (PCR) in the clinical lab. Detailed methods associated with both assays have been previously published [33]. dMMR was defined as the presence of high-level microsatellite instability on PCR and/or the loss of MMR protein expression in IHC. pMMR was defined as the presence of microsatellite stability or low-level microsatellite instability on PCR and/or no loss of MMR protein expression in IHH.
Statistical analysis
Patient characteristics are reported as categorical frequency and percent for each cohort. Correlations between clinical-pathological-molecular variables and meEGFR-CTCs status or meEGFR expression status on tissues were initially tested using Pearson’s χ2 or Fisher exact test. The association between patient and molecular characteristics with PFS was further explored using Kaplan-Meier curves. Cox proportional hazards regression was used to adjust for potential confounders and significant differences were assessed using the log-rank test. Calculations were performed with SPSS-version 23.0 software (IBM Corp., Armonk, NY). P values of less than 0.05 were considered statistically significant.
Results
In CTC cohort
A total of 47 mCRC patients were included in this cohort between September 2015 and July, 2016. The median age of the cohort was 52 years (range 25-71 years), and the ratio of males to females was 1.1. The majority of primary tumors were left-sided colon tumors (29 patients), followed by right-sided colon tumors (13 patients), then rectal tumors (5 patients). Anti-EGFRab were most commonly used in the second line of treatment in 26 patients, followed by third line in 15 patients and first line in 6 patients. Previous irinotecan used in 42.6% of all patients. Patient and tumor characteristics are shown in Table 1A.
Table 1.
Clinical-pathological and molecular characteristic of study populations, n (%)
| A. CTC cohort | ||
|
| ||
| Variable | Value | % |
|
| ||
| No. of patients | 47 | 100 |
| Median age (yr, range) | 52, 25-71 | |
| Age | ||
| < 50 years | 19 | 40.4 |
| ≥ 50 years | 28 | 59.6 |
| Sex | ||
| Female | 22 | 46.8 |
| Male | 25 | 53.2 |
| Primary tumor site | ||
| Ascending | 12 | 25.5 |
| Transverse | 1 | 2.1 |
| Descending | 7 | 14.9 |
| Sigmoid | 22 | 46.8 |
| Rectum | 5 | 10.6 |
| Line of anti-EGFR Rx | ||
| 1st line | 6 | 12.8 |
| 2nd line | 26 | 55.3 |
| 3rd line | 15 | 31.9 |
| Previous treatment | ||
| Irinotecan | 20 | 42.6 |
| Oxaliplatin | 40 | 85.1 |
| Bevacizumab | 36 | 76.6 |
| Chemotherapy regimen | ||
| Anti-EGFRab monotherapy | 8 | 17.0 |
| Irinotecan-based+anti-EGFRab | 35 | 74.5 |
| Oxaliplatin-based+anti-EGFRab | 2 | 4.3 |
| Vemurafenib+Cetuximab+irinitocan | 2 | 4.2 |
| Liver metastasis | ||
| No | 11 | 23.4 |
| Yes | 36 | 76.6 |
| Lung metastasis | ||
| No | 21 | 44.7 |
| Yes | 26 | 55.3 |
| Differentiated | ||
| Moderate | 40 | 85.1 |
| Poorly | 7 | 14.9 |
| NRAS | ||
| wt | 41 | 87.2 |
| mt | 2 | 4.3 |
| No data | 4 | 8.5 |
| BRAF | ||
| wt | 39 | 83 |
| mt | 6 | 12.8 |
| No data | 2 | 4.3 |
| PIK3CA | ||
| wt | 33 | 70.2 |
| mt | 4 | 8.5 |
| Variant | 2 | 4.3 |
| No data | 8 | 17 |
| MSI | ||
| MSS/MSI-L | 35 | 74.5 |
| MSI-H | 3 | 6.4 |
| No data | 9 | 19.1 |
|
| ||
| B. Tissues cohort | ||
|
| ||
| Variable | Value | % |
|
| ||
| No. of patients | 176 | 100 |
| Median age (yr, range) | 55, 20-79 | |
| Age | ||
| < 50 years | 51 | 29 |
| ≥ 50 years | 125 | 71 |
| Sex | ||
| Female | 79 | 44.9 |
| Male | 97 | 55.1 |
| Primary tumor site | ||
| Ascending | 50 | 28.4 |
| Transverse | 14 | 8 |
| Descending | 10 | 5.7 |
| Sigmoid | 66 | 37.5 |
| Rectum | 36 | 20.5 |
| Type of tissue tested | ||
| Primary CRC tissues | 156 | 88.6 |
| Metastatic tissues | 20 | 11.4 |
| Line of anti-EGFR Rx* (n = 74) | ||
| 1st line | 8 | 10.8 |
| 2nd line | 34 | 45.9 |
| 3rd line | 32 | 42.3 |
| Previous treatment* (n = 74) | ||
| Irinotecan | 42 | 56.8 |
| Differentiated | ||
| Moderate | 109 | 61.9 |
| Poorly | 64 | 36.4 |
| Unknown | 3 | 1.7 |
| KRAS | ||
| wt | 127 | 72.2 |
| mt | 48 | 27.3 |
| No data | 1 | 0.6 |
| NRAS | ||
| wt | 140 | 79.5 |
| mt | 4 | 2.3 |
| No data | 32 | 18.2 |
| BRAF | ||
| wt | 136 | 77.3 |
| mt | 24 | 13.6 |
| No data | 16 | 9.1 |
| PIK3CA | ||
| wt | 132 | 75 |
| mt | 19 | 10.8 |
| No data | 25 | 14.2 |
| MSI | ||
| MSS/MSI-L | 98 | 55.7 |
| MSI-H | 7 | 4 |
| No data | 71 | 40.3 |
only the patients that confirmed RASwt and treated with anti-EGFR agents.
wt: wild type, mt: mutation.
Detection of CTCs and meEGFR-CTCs
In this cohort, CTCs were identified in 30 out of 47 cases (63.8%). Of these 30 cases, meEGFR-CTCs were identified in 19 cases (63.3%) (Figure 1). Mean total CTCs and cell counts of CTCs positive for meEGFR were 3.6 cells (range 0-52) and 2.3 cells (range 0-30) per 7.5 ml, respectively. The ratio of meEGFR CTCs per total CTCs is shown in Figure 2. The mean ratio of mEGFR CTCs to total CTCs was 0.23 with the range from 0 to 1. Therefore, we considered cases with a ratio ≥ 0.23 as high meEGFR-CTC cases.
Figure 1.
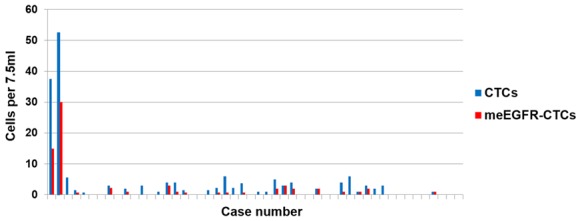
Total CTCs and meEGFR-CTCs detected. CTCs were identified in 30 out of 47 cases (63.8%). Of these 30 cases, meEGFR-CTCs were identified in 19 cases (63.3%).
Figure 2.
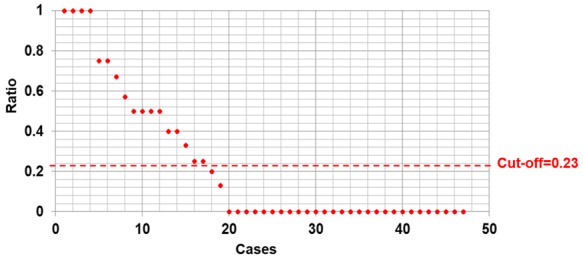
Ratio of meEGFR per total CTCs. The mean ratio of mEGFR CTCs to total CTCs was 0.23 with the range from 0 to 1.
Association between total CTCs or meEGFR-CTCs and clinic-pathologic-molecular characteristic
We compared the clinic-pathological and molecular variables of patients, including age, sex, site of the primary tumor, histologic grade, previous irinotecan used, line of anti-EGFR treatment, and, NRAS, BRAF, PIK3CA, and MSI status, by the status of CTCs and meEGFR-CTCs. No clinic-pathological-molecular features were associated with either detectable/non-detectable meEGFR-CTCs or total CTCs less than/at least 3 cells per 7.5 ml (Supplementary Table 1). Further, there was no significant difference between patients with high vs. low meEGFR-CTCs ratio (Table 2). This suggests that the meEGFR-CTCs are not a surrogate for existing prognosis or predictive features but represents unique molecular feature.
Table 2.
Association between meEGFR-CTCs ratio and clinical-pathological and molecular factors
| Variable | meEGFR-CTCs ratio | P value | |
|---|---|---|---|
|
| |||
| < 0.23 | ≥ 0.23 | ||
| Age | |||
| < 50 years | 15 (50) | 4 (23.5) | 0.08 |
| ≥ 50 years | 15 (50) | 13 (76.5) | |
| Sex | |||
| Female | 14 (46.7) | 8 (47.1) | 0.99 |
| Male | 16 (53.3) | 9 (52.9) | |
| Site | |||
| Right-sided | 8 (26.7) | 5 (29.4) | 0.84 |
| Left-sided | 22 (73.3) | 12 (70.6) | |
| Line of anti-EGFR Rx | |||
| 1st line | 4 (13.3) | 1 (5.9) | 0.38 |
| 2nd line | 15 (50) | 12 (70.6) | |
| 3rd line | 11 (36.7) | 4 (23.5) | |
| Previous irinotecan | |||
| No | 18 (60) | 9 (52.9) | 0.64 |
| Yes | 12 (40) | 8 (47.1) | |
| Liver metastasis | |||
| No | 5 (16.7) | 6 (35.3) | 0.15 |
| Yes | 25 (83.3) | 11 (64.7) | |
| Lung metastasis | |||
| No | 13 (43.3) | 8 (47.1) | 0.81 |
| Yes | 17 (56.7) | 9 (52.9) | |
| Differentiated | |||
| Moderate | 25 (83.3) | 15 (88.2) | 0.65 |
| Poorly | 5 (16.7) | 2 (11.8) | |
| NRAS (n = 43) | |||
| wt | 26 (96.3) | 14 (93.8) | 0.70 |
| mt | 1 (3.7) | 1 (6.3) | |
| BRAF (n = 45) | |||
| wt | 24 (85.7) | 15 (18.2) | 0.81 |
| mt | 4 (14.3) | 2 (11.8) | |
| PIK3CA (n = 37) | |||
| wt | 18 (85.7) | 15 (93.8) | 0.44 |
| mt | 3 (14.3) | 1 (6.3) | |
| MSI (n = 38) | |||
| MSS/MSI-L | 21 (91.3) | 14 (93.3) | 0.82 |
| MSI-H | 2 (8.7) | 1 (6.7) | |
wt: wild type, mt: mutation.
Progression free survival analysis
To test the potential associations between meEGFR-CTCs and progression free survival (PFS), we first performed univariate analyses of PFS by meEGFR-CTC ratio and previously established prognostic factors: sidedness, line of anti-EGFR treatment, and PIK3CA status. The only factor that was significantly associated with worse PFS in this cohort was meEGFR ratio ≥ 0.23. In RASwt BRAFwt mCRC patients with meEGFR ratio ≥ 0.23 had significantly worse PFS for anti-EGFR treatment compared with patients with the ratio < 0.23 (HR 3.4, 95% CI 1.47-7.90. P = 0.004) and remain statistically significant in multivariate analysis (HR = 3.0, 95% CI 1.03-8.5, P = 0.04) (Table 4A; Figure 3A). This ratio had 100% sensitivity and 59% specificity to detect the difference if we used the cut point of 3 months to define the patients as responder and non-responder group.
Table 4.
Univariate and multivariate analysis of prognostic factors influencing PFS on RASwt BRAFwt with anti EGFR treatment
| A. CTC cohort (n = 32) | |||||||
|
| |||||||
| Variables | N | Univariate analysis | Multivariate analysis | ||||
|
| |||||||
| HR | 95% CI | P value | HR | 95% CI | P value | ||
|
| |||||||
| Sidedness | |||||||
| Rt. sided | 7 | Ref | |||||
| Lt. sided | 25 | 0.85 | 0.4-2.0 | 0.71 | |||
| Line of anti-EGFR Rx | |||||||
| 1st line | 3 | Ref | Ref | ||||
| 2nd line | 17 | 2.1 | 0.6-7.4 | 0.26 | 2 | 0.5-8.0 | 0.34 |
| 3rd line | 12 | 1.6 | 0.4-5.9 | 0.47 | 1.5 | 0.4-5.7 | 0.58 |
| PIK3CA | |||||||
| wt | 24 | Ref | Ref | ||||
| mt | 2 | 0.9 | 0.2-3.9 | 0.89 | 1.3 | 0.2-7.3 | 0.77 |
| meEGFR ratio | |||||||
| < 0.23 | 20 | Ref | 0.004 | Ref | |||
| ≥ 0.23 | 12 | 3.4 | 1.5-7.9 | 3.0 | 1.03-8.5 | 0.04 | |
|
| |||||||
| wt: wild type, mt: mutation, Ref: Reference. | |||||||
|
| |||||||
| B. RASwt, BRAFwt in tissue cohort | |||||||
|
| |||||||
| Variables | N | Univariate analysis | |||||
|
| |||||||
| Median PFS (mo) | 95% CI | P value | |||||
|
| |||||||
| Site | |||||||
| Rt. sided | 16 | 9.2 | 4.0-14.4 | 0.72 | |||
| Lt. sided | 54 | 7.8 | 5.3-10.4 | ||||
| Line of anti-EGFR Rx | |||||||
| 1st line | 8 | 11.0 | 8.5-13.5 | 0.02 | |||
| 2nd line | 33 | 8.0 | 3.5-12.6 | ||||
| 3rd line | 29 | 6.0 | 4.1-7.9 | ||||
| PIK3CA | |||||||
| wt | 64 | 7.8 | 5.6-10.1 | 0.37 | |||
| mt | 5 | 11.1 | 2.3-20.0 | ||||
| meEGFR | |||||||
| Low expression | 47 | 7.4 | 5.0-9.7 | 0.46 | |||
| High expression | 20 | 9.5 | 2.9-16.0 | ||||
wt: wild type, mt: mutation.
Figure 3.
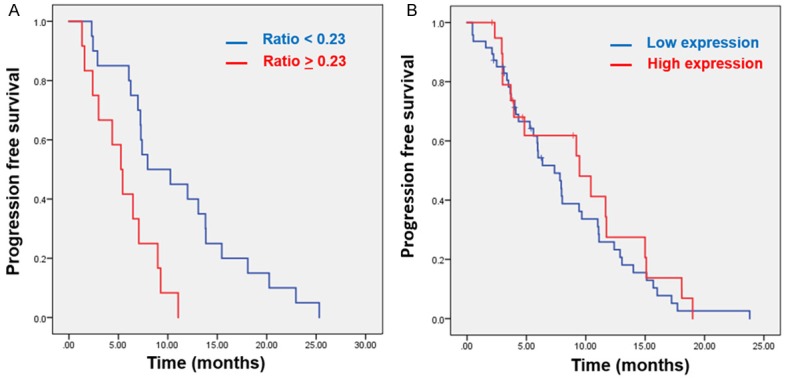
Kaplan-Meier survival curve of RAS wt BRAF wt mCRC. A. RASwt BRAFwt mCRC patients with high meEGFR ratio (≥ 0.23) had significantly worse PFS for anti-EGFR treatment (median PFS 5.3 mo, 95% CI 3.5-7 mo) compared with pts with the ratio < 0.23 (median PFS 8 mo, 95% CI 1.7-14.2 mo, P = 0.002). B. There was no correlation between high or low meEGFR expression in the tumor tissues and PFS for anti-EGFR treatment treatment (median PFS 7.4 mo, 95% CI 5.0-9.7 mo in low expression compared with median PFS 9.5 mo, 95% CI 2.9-16.0 mo in high expression, P = 0.46).
In tissues cohort
A total of 176 mCRC patients were included in the tissue analysis cohort. Of these, we had tumor samples from primary CRC in 156 cases, and metastatic tumor samples from 20 cases. Tissues were collected prior to anti-EGFR treatment. The median age was 55 years (range 20-79 years), and the ratio of males to females was 1.24. Patient and tumor characteristics are shown in Table 1B.
Association between meEGFR expression and clinic-pathologic-molecular characteristic
Out of 176 samples, 164 had data on meEGFR expression. A total of 76 cases (46.3%) exhibited high expression of meEGFR. Further, 63 cases exhibited low expression of EGFR (score > 0-150) and 25 cases showed no expression (score = 0). Comparing the clinical-pathological and molecular variables by meEGFR expression revealed that only KRASmt and NRASmt were significantly associated with meEGFR high expression (P = 0.03 and P = 0.02, respectively) (Table 3).
Table 3.
Association between meEGFR expression and clinical-pathological and molecular factors (N = 164, exclude 12 cases that had no data on meEGFR expression)
| Variable | meEGFR | P value | |
|---|---|---|---|
|
| |||
| Low/No expression | High expression | ||
| Age (n = 164) | |||
| < 50 years | 25 (28.4%) | 21 (27.6%) | 0.91 |
| ≥ 50 years | 63 (71.6%) | 55 (72.4%) | |
| Sex (n = 164) | |||
| Female | 36 (40.9%) | 36 (47.4%) | 0.41 |
| Male | 52 (59.1%) | 40 (52.6%) | |
| Site (n = 164) | |||
| Right-sided | 34 (38.6%) | 27 (35.5%) | 0.68 |
| Left-sided | 54 (61.4%) | 49 (64.5%) | |
| Differentiated (n = 161) | |||
| Moderate | 45 (56.3%) | 52 (64.2%) | 0.30 |
| Poorly | 35 (43.8%) | 29 (35.8%) | |
| KRAS (n = 163) | |||
| wt | 73 (83%) | 51 (68%) | 0.03 |
| mt | 15 (17%) | 24 (32%) | |
| NRAS (n = 136) | |||
| wt | 76 (100%) | 56 (93.3%) | 0.02 |
| mt | 0 (0%) | 4 (6.7%) | |
| BRAF (n = 150) | |||
| wt | 68 (84%) | 58 (84.1%) | 0.97 |
| mt | 13 (16%) | 11 (15.9%) | |
| PIK3CA (n = 142) | |||
| wt | 73 (90.1%) | 51 (83.6%) | 0.25 |
| mt | 8 (9.9%) | 10 (16.4%) | |
| MSI (n = 99) | |||
| MSS/MSI-L | 41 (91.1%) | 51 (94.4%) | 0.52 |
| MSI-H | 4 (8.9%) | 3 (5.6%) | |
wt: wild type, mt: mutation.
Progression free survival analysis
Univariate analysis of PFS was performed using previously established prognostic factors: sidedness, line of anti-EGFR treatment, and PIK3CA status. In 176 cases, there were 107 (60.7%) RAS wt mCRC patients. Of these 107, 67 cases were RASwt BRAFwt mCRC and had available data on outcome with anti-EGFR treatment. Median PFS were 6, 8, 11 mo in 3rd, 2nd, and 1st line, respectively (P = 0.02). There was no correlation between high meEGFR expression in the tumor tissues and PFS in RASwt/RAFwt mCRC in this cohort (HR 0.8, 95% CI 0.45-1.44, P = 0.46) (Table 4B; Figure 3B).
meEGFR expression in CTCs vs. tissues
In this study, there were 10 cases that had data on both meEGFR-CTCs and meEGFR expression from tissues. There was no association between meEGFR-CTC ratio and tumor meEGFR expression status (P = 0.67). Detail of these cases was shown in Supplementary Table 2.
Discussion
In this study, we successfully isolated CTCs from CRC patients’ blood and were able to assess arginine methylated EGFR in the isolated CTCs. We showed for the first time that elevated levels meEGFR-CTCs were associated with a shorter duration of anti-EGFR-based treatment. EGFR arginine-methylation in CTC may serve as a biomarker to stratify the patients that response to anti-EGFR therapy.
EGFR methylation in CRC has been reported at the level of pre-transcriptional and post-translational modification. Arginine methylation represents a common post-translational modification of EGFR [34]. Protein arginine methylatransferases (PRMTs) mediate the methylation of protein substrates of arginine residue and can play an important function in cancer development [35]. The activity of PRMT1, a member of the PRMT family, accounts for more than 90% of the methylarginine residues in mammalian cells [36]. PRMT1 is the major asymmetric arginine methyltransferase and is deregulated in multiple cancers, including breast, prostate, lung, bladder, leukemia, and colon cancer [37-41]. More recently, our group demonstrated that patients with high levels of PRMT1-mediated EGFR methylation had worse PFS with cetuximab treatment and poor OS [24] compared with patients with low levels of PRMT1-mediated EGFR methylation. Although EGFR expression does not appear to be a predictive marker for anti-EGFR treatment [42], methylated EGFR expression may serve as a potential predictive marker in anti-EGFR therapy. However, further validation of this result is needed.
Compared to standard tissue biopsy, liquid biopsy has several unique advantages. First, it is minimally invasive, avoiding the potential complications of biopsies. Second, it provides an opportunity to obtain tumor information when tissue biopsy is difficult or contraindicated. Additionally, the safety and simplicity of such an option allows for serial sampling, which are important for assessing treatment response [43]. Previous studies demonstrated that high CTC numbers in blood correlate with poor prognosis in many cancer types, including colorectal [27], breast [44], and prostate [45] cancers. Data in a prospective multicenter study demonstrated that mCRC patients with at least three CTCs per 7.5 ml at baseline constitutes a strong independent prognostic factor for inferior PFS and OS [27]. Hence, liquid biopsies are growing in popularity as standard tests and have potential for routine cancer patient care. While the prognostic impact on CTCs in CRC has been established [27,46,47], the predictive impact on liquid biopsy in CRC has not been reported.
In this study, CTCs were isolated from patients with Parsortix PR1 system, which isolates CTCs by size and deformation capability. CTCs were identified in 64% of the patients in this cohort, which was higher than the range 28-49% previously reported [27,28,48,49]. However, patients in this cohort were all at stage IV, and when only stage IV disease was considered in other published cohorts, we found similar positive CTCs rate (59.3-60.7%) [29,48]. meEGFR-CTCs was also identified in 63% of all detected CTCs cases, which indicated that meEGFR occurred in the majority of patients with positive CTC detection. As our study is the first report meEGFR-CTCs in mCRC, further studies are warranted to confirm this finding.
We found no correlation between the occurrences of meEGFR-CTCs and PFS of anti-EGFR treated patients when grouping the patients simply based on the amount of detected meEGFR-CTC (≥ 1 or ≥ 3 per 7.5 ml of blood). However, since meEGFR-CTCs and non-meEGFR-CTCs were both simultaneously detected in most of the cases, simply grouping the patients based on the meEGFR-CTC counts may not accurately reflect anti-EGFRab treatment response. Therefore, we hypothesized that tumor with dominant populations of meEGFR positive tumor cells would have poor response to anti-EGFRab treatment, i.e., the ratio of meEGFR-positive tumor cells in tumor may correlate better with anti-EGFRab treated patients’ PFS. Therefore, we used the ratio of meEGFR-CTCs over total CTCs with a cut-off point 0.23 (average ratio = 0.23) to classify the patients into 2 groups. Our study showed that patients with high meEGFR-CTC per total CTCs ratio had significant worse PFS than those who had the ratio < 0.23 (median PFS 5.3 vs. 8 months, HR = 3, 95% CI = 1.03-8.5, P = 0.002). This finding confirms our hypothesis that the ratio of meEGFR-CTCs may help predict treatment response and supports the result from our previous paper [24]. No correlation was found between either meEGFR positive or meEGFR ratio with any clinical-pathological and molecular characteristics implying that this is an independent molecular feature not represented by other known factors. Given the small number samples in the current study, these results will need to be confirmed in lager dataset.
In FFPE tissue staining cohort, this study demonstrated meEGFR-positive staining (either low or high expression) in 127/145 (88%) in primary CRC tissues and 12/19 (63%) in metastatic tissues. However, there was no correlation between meEGFR levels in CTCs and tumor tissues (Supplementary Table 2). This suggests that arginine methylation of EGFR may be a dynamic process influenced by prior chemotherapy and/or clonal drift in a heterogenous tumors. It is also possible that CTCs do not accurately reflect the protein methylation status of the bulk tumor. However, in contrast to our previous report [24] the meEGFR expression on CRC tissue was not correlated with PFS on anti-EGFR treatment. One potential explanation would be the difference in patients’ populations and the difference in the cut-off point to define into high or low meEGFR expression groups. However, our data showed a positive correlation between expression level of meEGFR and PRMT1 (P = 0.03) which confirmed the previous report paper in our group [24] (Supplementary Table 3).
Our group previously reported that meEGFR is a potential for predicting response to anti-EGFR treatment [24]. This occurred only in CTCs but not in the tumor tissues in the present study. Consequently this finding raises the possibility that meEGFR occur during tumor development and increase over time. Therefore, meEGFR-CTCs maybe better predict response than primary tumor tissues. Additional work based on the current finding could refine cut-off point used to define the correlation between high meEGFR-CTCs ratio and high meEGFR expression in FFPE tissues and could be coupled with serial monitoring of meEGFR-CTCs ratio with treatment response. Furthermore, since PRMT1 mediates meEGFR, PRMT1 inhibitors may reduce meEGFR, potentially sensitizing some tumors to anti-EGFRabs. This represents an exciting area for future study, and the appropriate patient identification could potentially increase those that benefit from anti-EGFR therapy.
We recognize the limitations of the current study. First, it was a small, retrospective study with lack of statistical power. Second, we have no data on longitudinal CTC sampling, so we do not know whether meEFGR can develop as a method of acquired resistance.
In summary, this study is the first to indicate that PRMT1 methylated-EGFR detected in CTCs may serve as a potential liquid biopsy biomarker for predicting anti-EGFR response. Further studies are required to identify the patients most likely to benefit from anti-EGFR treatment. Assessment of meEGFR-CTCs may provide a useful “liquid biopsy” biomarker for identifying patients that may exhibit reduced benefit from anti-EGFR treatment.
Acknowledgements
The authors are grateful to Shadarra Crosby for helping with patient appointment. This study was partially funded by Angle Company, CPRIT grant number RP150245 (SK, MCH), and NCI cancer center grant (P30 CA016672).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, Rivera F, Kocákova I, Ruff P, Błasińska-Morawiec M, Šmakal M, Canon JL, Rother M, Williams R, Rong A, Wiezorek J, Sidhu R, Patterson SD. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–34. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 3.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, Penault-Llorca F, Rougier P, Vincenzi B, Santini D, Tonini G, Cappuzzo F, Frattini M, Molinari F, Saletti P, De Dosso S, Martini M, Bardelli A, Siena S, Sartore-Bianchi A, Tabernero J, Macarulla T, Di Fiore F, Gangloff AO, Ciardiello F, Pfeiffer P, Qvortrup C, Hansen TP, Van Cutsem E, Piessevaux H, Lambrechts D, Delorenzi M, Tejpar S. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–62. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 4.Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au HJ, Langer C, Moore MJ, Zalcberg JR. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–65. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 5.Linardou H, Dahabreh IJ, Kanaloupiti D, Siannis F, Bafaloukos D, Kosmidis P, Papadimitriou CA, Murray S. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 2008;9:962–72. doi: 10.1016/S1470-2045(08)70206-7. [DOI] [PubMed] [Google Scholar]
- 6.Mao C, Huang YF, Yang ZY, Zheng DY, Chen JZ, Tang JL. KRAS p. G13D mutation and codon 12 mutations are not created equal in predicting clinical outcomes of cetuximab in metastatic colorectal cancer: a systematic review and meta-analysis. Cancer. 2013;119:714–21. doi: 10.1002/cncr.27804. [DOI] [PubMed] [Google Scholar]
- 7.Moroni M, Veronese S, Benvenuti S, Marrapese G, Sartore-Bianchi A, Di Nicolantonio F, Gambacorta M, Siena S, Bardelli A. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005;6:279–86. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 8.Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbison CT, Wu S, Wong TW, Huang X, Takimoto CH, Godwin AK, Tan BR, Krishnamurthi SS, Burris HA 3rd, Poplin EA, Hidalgo M, Baselga J, Clark EA, Mauro DJ. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J. Clin. Oncol. 2007;25:3230–7. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 9.Bardelli A, Siena S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J. Clin. Oncol. 2010;28:1254–61. doi: 10.1200/JCO.2009.24.6116. [DOI] [PubMed] [Google Scholar]
- 10.Rowland A, Dias MM, Wiese MD, Kichenadasse G, McKinnon RA, Karapetis CS, Sorich MJ. Meta-analysis of BRAF mutation as a predictive biomarker of benefit from anti-EGFR monoclonal antibody therapy for RAS wild-type metastatic colorectal cancer. Br J Cancer. 2015;112:1888–94. doi: 10.1038/bjc.2015.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perrone F, Lampis A, Orsenigo M, Di Bartolomeo M, Gevorgyan A, Losa M, Frattini M, Riva C, Andreola S, Bajetta E, Bertario L, Leo E, Pierotti MA, Pilotti S. PI3KCA/PTEN deregulation contributes to impaired responses to cetuximab in metastatic colorectal cancer patients. Ann Oncol. 2009;20:84–90. doi: 10.1093/annonc/mdn541. [DOI] [PubMed] [Google Scholar]
- 12.Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, Di Nicolantonio F, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69:1851–7. doi: 10.1158/0008-5472.CAN-08-2466. [DOI] [PubMed] [Google Scholar]
- 13.Morelli MP, Overman MJ, Dasari A, Kazmi SM, Mazard T, Vilar E, Morris VK, Lee MS, Herron D, Eng C, Morris J, Kee BK, Janku F, Deaton FL, Garrett C, Maru D, Diehl F, Angenendt P, Kopetz S. Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann Oncol. 2015;26:731–6. doi: 10.1093/annonc/mdv005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sartore-Bianchi A, Di Nicolantonio F, Nichelatti M, Molinari F, De Dosso S, Saletti P, Martini M, Cipani T, Marrapese G, Mazzucchelli L, Lamba S, Veronese S, Frattini M, Bardelli A, Siena S. Multi-determinants analysis of molecular alterations for predicting clinical benefit to EGFR-targeted monoclonal antibodies in colorectal cancer. PLoS One. 2009;4:e7287. doi: 10.1371/journal.pone.0007287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J. Clin. Oncol. 2008;26:5705–12. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 16.Bertotti A, Migliardi G, Galimi F, Sassi F, Torti D, Isella C, Corà D, Di Nicolantonio F, Buscarino M, Petti C, Ribero D, Russolillo N, Muratore A, Massucco P, Pisacane A, Molinaro L, Valtorta E, Sartore-Bianchi A, Risio M, Capussotti L, Gambacorta M, Siena S, Medico E, Sapino A, Marsoni S, Comoglio PM, Bardelli A, Trusolino L. A molecularly annotated platform of patientderived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov. 2011;1:508–23. doi: 10.1158/2159-8290.CD-11-0109. [DOI] [PubMed] [Google Scholar]
- 17.Martin V, Landi L, Molinari F, Fountzilas G, Geva R, Riva A, Saletti P, De Dosso S, Spitale A, Tejpar S, Kalogeras KT, Mazzucchelli L, Frattini M, Cappuzzo F. HER2 gene copy number status may influence clinical efficacy to anti-EGFR monoclonal antibodies in metastatic colorectal cancer patients. Br J Cancer. 2013;108:668–75. doi: 10.1038/bjc.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bardelli A, Corso S, Bertotti A, Hobor S, Valtorta E, Siravegna G, Sartore-Bianchi A, Scala E, Cassingena A, Zecchin D, Apicella M, Migliardi G, Galimi F, Lauricella C, Zanon C, Perera T, Veronese S, Corti G, Amatu A, Gambacorta M, Diaz LA Jr, Sausen M, Velculescu VE, Comoglio P, Trusolino L, Di Nicolantonio F, Giordano S, Siena S. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov. 2013;3:658–73. doi: 10.1158/2159-8290.CD-12-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frattini M, Saletti P, Romagnani E, Martin V, Molinari F, Ghisletta M, Camponovo A, Etienne LL, Cavalli F, Mazzucchelli L. PTEN loss of expression predicts cetuximab efficacy in metastatic colorectal cancer patients. Br J Cancer. 2007;97:1139–45. doi: 10.1038/sj.bjc.6604009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Negri FV, Bozzetti C, Lagrasta CA, Crafa P, Bonasoni MP, Camisa R, Pedrazzi G, Ardizzoni A. PTEN status in advanced colorectal cancer treated with cetuximab. Br J Cancer. 2010;102:162–4. doi: 10.1038/sj.bjc.6605471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sepulveda AR, Hamilton SR, Allegra CJ, Grody W, Cushman-Vokoun AM, Funkhouser WK, Kopetz SE, Lieu C, Lindor NM, Minsky BD, Monzon FA, Sargent DJ, Singh VM, Willis J, Clark J, Colasacco C, Rumble RB, Temple-Smolkin R, Ventura CB, Nowak JA. Molecular biomarkers for the evaluation of colorectal cancer: guideline from the American society for clinical pathology, college of American pathologists, association for molecular pathology, and the american society of clinical oncology. J. Clin. Oncol. 2017;35:1453–1486. doi: 10.1200/JCO.2016.71.9807. [DOI] [PubMed] [Google Scholar]
- 22.Sasaki T, Hiroki K, Yamashita Y. The role of epidermal growth factor receptor in cancer metastasis and microenvironment. Biomed Res Int. 2013;2013:546318. doi: 10.1155/2013/546318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yalak G, Vogel V. Extracellular phosphorylation and phosphorylated proteins: not just curiosities but physiologically important. Sci Signal. 2012;5:re7. doi: 10.1126/scisignal.2003273. [DOI] [PubMed] [Google Scholar]
- 24.Liao HW, Hsu JM, Xia W, Wang HL, Wang YN, Chang WC, Arold ST, Chou CK, Tsou PH, Yamaguchi H, Fang YF, Lee HJ, Lee HH, Tai SK, Yang MH, Morelli MP, Sen M, Ladbury JE, Chen CH, Grandis JR, Kopetz S, Hung MC. PRMT1-mediated methylation of the EGF receptor regulates signaling and cetuximab response. J Clin Invest. 2015;125:4529–43. doi: 10.1172/JCI82826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyberopoulou A, Aravantinos G, Efstathopoulos EP, Nikiteas N, Bouziotis P, Isaakidou A, Papalois A, Marinos E, Gazouli M. Mutational analysis of circulating tumor cells from colorectal cancer patients and correlation with primary tumor tissue. PLoS One. 2015;10:e0123902. doi: 10.1371/journal.pone.0123902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diaz LA, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, Allen B, Bozic I, Reiter JG, Nowak MA, Kinzler KW, Oliner KS, Vogelstein B. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–40. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, Doyle GV, Tissing H, Terstappen LW, Meropol NJ. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008;26:3213–21. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 28.Romiti A, Raffa S, Di Rocco R, Roberto M, Milano A, Zullo A, Leone L, Ranieri D, Mazzetta F, Medda E, Sarcina I, Barucca V, D’Antonio C, Durante V, Ferri M, Torrisi MR, Marchetti P. Circulating tumor cells count predicts survival in colorectal cancer patients. J Gastrointestin Liver Dis. 2014;23:279–84. doi: 10.15403/jgld.2014.1121.233.arom1. [DOI] [PubMed] [Google Scholar]
- 29.Zhang D, Zhao L, Zhou P, Ma H, Huang F, Jin M, Dai X, Zheng X, Huang S, Zhang T. Circulating tumor microemboli (CTM) and vimentin+ circulating tumor cells (CTCs) detected by a sizebased platform predict worse prognosis in advanced colorectal cancer patients during chemotherapy. Cancer Cell Int. 2017;17:6. doi: 10.1186/s12935-016-0373-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsai WS, Chen JS, Shao HJ, Wu JC, Lai JM, Lu SH, Hung TF, Chiu YC, You JF, Hsieh PS, Yeh CY, Hung HY, Chiang SF, Lin GP, Tang R, Chang YC. Circulating tumor cell count correlates with colorectal neoplasm progression and is a prognostic marker for distant metastasis in nonmetastatic patients. Sci Rep. 2016;6:24517. doi: 10.1038/srep24517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–4. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 32.Hvichia GE, Parveen Z, Wagner C, Janning M, Quidde J, Stein A, Müller V, Loges S, Neves RP, Stoecklein NH, Wikman H, Riethdorf S, Pantel K, Gorges TM. A novel microfluidic platform for size and deformability based separation and the subsequent molecular characterization of viable circulating tumor cells. Int J Cancer. 2016;138:2894–904. doi: 10.1002/ijc.30007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korphaisarn K, Morris VK, Overman MJ, Fogelman DR, Kee BK, Raghav KPS, Manuel S, Shureiqi I, Wolff RA, Eng C, Menter D, Hamilton SR, Kopetz S, Dasari A. FBXW7 missense mutation: a novel negative prognostic factor in metastatic colorectal adenocarcinoma. Oncotarget. 2017;8:39268–79. doi: 10.18632/oncotarget.16848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boffa LC, Karn J, Vidali G, Allfrey VG. Distribution of NG, NG,-dimethylarginine in nuclear protein fractions. Biochem Biophys Res Commun. 1977;74:969–76. doi: 10.1016/0006-291x(77)91613-8. [DOI] [PubMed] [Google Scholar]
- 35.Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013;13:37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 36.Tang J, Frankel A, Cook RJ, Kim S, Paik WK, Williams KR, Clarke S, Herschman HR. PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J Biol Chem. 2000;275:7723–30. doi: 10.1074/jbc.275.11.7723. [DOI] [PubMed] [Google Scholar]
- 37.Cheung N, Chan LC, Thompson A, Cleary ML, So CW. Protein arginine-methyltransferase-dependent oncogenesis. Nat Cell Biol. 2007;9:1208–15. doi: 10.1038/ncb1642. [DOI] [PubMed] [Google Scholar]
- 38.Shia WJ, Okumura AJ, Yan M, Sarkeshik A, Lo MC, Matsuura S, Komeno Y, Zhao X, Nimer SD, Yates JR 3rd, Zhang DE. PRMT1 interacts with AML1-ETO to promote its transcriptional activation and progenitor cell proliferative potential. Blood. 2012;119:4953–62. doi: 10.1182/blood-2011-04-347476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Le Romancer M, Treilleux I, Bouchekioua-Bouzaghou K, Sentis S, Corbo L. Methylation, a key step for nongenomic estrogen signaling in breast tumors. Steroids. 2010;75:560–4. doi: 10.1016/j.steroids.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 40.Yoshimatsu M, Toyokawa G, Hayami S, Unoki M, Tsunoda T, Field HI, Kelly JD, Neal DE, Maehara Y, Ponder BA, Nakamura Y, Hamamoto R. Dysregulation of PRMT1 and PRMT6, Type I arginine methyltransferases, is involved in various types of human cancers. Int J Cancer. 2011;128:562–73. doi: 10.1002/ijc.25366. [DOI] [PubMed] [Google Scholar]
- 41.Papadokostopoulou A, Mathioudaki K, Scorilas A, Xynopoulos D, Ardavanis A, Kouroumalis E, Talieri M. Colon cancer and protein arginine methyltransferase 1 gene expression. Anticancer Res. 2009;29:1361–6. [PubMed] [Google Scholar]
- 42.Valentini AM, Pirrelli M, Caruso ML. EGFR-targeted therapy in colorectal cancer: does immunohistochemistry deserve a role in predicting the response to cetuximab? Curr Opin Mol Ther. 2008;10:124–31. [PubMed] [Google Scholar]
- 43.Ilié M, Hofman P. Pros: can tissue biopsy be replaced by liquid biopsy? Transl Lung Cancer Res. 2016;5:420–3. doi: 10.21037/tlcr.2016.08.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–91. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 45.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LW, Pienta KJ, Raghavan D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–9. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 46.Sastre J, Maestro ML, Gómez-España A, Rivera F, Valladares M, Massuti B, Benavides M, Gallén M, Marcuello E, Abad A, Arrivi A, Fernández-Martos C, González E, Tabernero JM, Vidaurreta M, Aranda E, Díaz-Rubio E. Circulating tumor cell count is a prognostic factor in metastatic colorectal cancer patients receiving first-line chemotherapy plus bevacizumab: a Spanish cooperative group for the treatment of digestive tumors study. Oncologist. 2012;17:947–55. doi: 10.1634/theoncologist.2012-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse MA, Mitchell E, Miller MC, Doyle GV, Tissing H, Terstappen LW, Meropol NJ. Prognostic significance of circulating tumor cells in patients with metastatic colorectal cancer. Ann Oncol. 2009;20:1223–9. doi: 10.1093/annonc/mdn786. [DOI] [PubMed] [Google Scholar]
- 48.Sastre J, Maestro ML, Puente J, Veganzones S, Alfonso R, Rafael S, García-Saenz JA, Vidaurreta M, Martín M, Arroyo M, Sanz-Casla MT, Díaz-Rubio E. Circulating tumor cells in colorectal cancer: correlation with clinical and pathological variables. Ann Oncol. 2008;19:935–8. doi: 10.1093/annonc/mdm583. [DOI] [PubMed] [Google Scholar]
- 49.Chen J, Guo F, Shi X, Zhang L, Zhang A, Jin H, He Y. BRAF V600E mutation and KRAS codon 13 mutations predict poor survival in Chinese colorectal cancer patients. BMC Cancer. 2014;14:802. doi: 10.1186/1471-2407-14-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


