Abstract
Background:
Wild-type p53 nuclear phosphoproteins are critical cell cycle regulatory tumor-suppressor gene. Genetic mutation of p53 gene is common in several head–neck cancers, usually associated with smoking and human papillomavirus (HPV) infection. In India, instead of HPV, tobacco/pan masala chewing is more commonly associated with oral cancer.
Aim:
The aim of this study was to investigate p53 codon 72 gene polymorphism and expression of p53 by immunohistochemistry (IHC) in oral lesions as a risk factor for its association with malignancy.
Materials and Methods:
A total of 41 cases of oral lesions comprising 6 cases of leukoplakia and 35 cases of oral squamous cell carcinoma (OSCC), between 30 and 60 years age and tobacco/pan masala chewers were taken. Molecular analysis of p53 codon 72 gene polymorphism was performed by polymerase chain reaction – restriction fragment length polymorphism for Arg/Arg, Arg/Pro, and Pro/Pro. Tissue expression of p53 was done by IHC.
Results:
Genotype frequencies of 35 carcinoma cases of p53 Arg/Arg, Arg/Pro, and Pro/Pro were 23%, 57%, and 20%, respectively, and six leukoplakia cases of p53 Arg/Arg and Arg/Pro genotype were 50% and 50%, respectively. By IHC for expression of p53 out of 35 cases of OSCC biopsies, 17 (48.57%) had weak staining, 14 cases (40%) showed evidence of p53 protein staining, and four cases (11.42%) showed negative staining. Among six cases of leukoplakia, 3 (50%) showed weak staining and 3 (50%) showed negative results.
Conclusion:
The findings of the study indicate that there is no significant association between p53 codon 72 gene polymorphism with OSCC and leukoplakia associated with tobacco/pan masala chewing.
Keywords: Gene polymorphism, oral squamous cell carcinoma, p53 codon
Introduction
Development of squamous cell carcinoma is a multifactorial process associated with various risk factors for oral cancer development, only some of the smokers, alcohol users, and betel quid users develop oral cancer. These environmental exogenous carcinogens play a crucial role in the development of oral cancer either by altering the expression of tumor suppressor gene, apoptosis, or may result in genomic instability by inducing a defective DNA damage response.[1,2] Early detection of oral premalignant lesion can prevent development of invasive cancers, but till date no definitive markers are available for the same.
p53 is the most commonly mutated gene and is altered in over 50% of all cancers, including 25%–70% of oral cancers. The present study was planned with an aim to investigate p53 codon 72 gene polymorphism and expression of p53 by immunohistochemistry (IHC) in oral lesions in India, where genomic differences exist compared to western population. Human papillomavirus (HPV) is not a common risk factor and tobacco chewing; bidi smoking is rampant.
Materials and Methods
This study was carried out in the Department of Pathology, Era's Lucknow Medical College and Hospital. Fifty patients of all age groups attending outpatient department of Era's Lucknow Medical College and Hospital and Surgical Oncology Department of K. G. Medical University, Lucknow, with any complaint related to oral cavity lesions excluding the dental problems, were examined clinically. A detail history was taken. Blood and biopsy from the representative sites of oral lesions were collected with informed consent. All the biopsy sections were studied for expression of p53 was done by IHC, for p53 expression.
For DNA extraction, approximately 3 ml venous blood was collected from each of the aforesaid patients and controls. Blood was mixed slowly with ethylenediaminetetracetic acid for 1 min and stored at −20°C. Genomic DNA was isolated from whole blood using the standard phenol-chloroform extraction method. The DNA concentration was determined by spectrophotometer and stored at −20°C. Polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP) analysis was conducted to identify the p53 polymorphism in codon 72.
PCR-RFLP analysis was conducted to identify the p53 polymorphism in codon 72 with the primers 5'-ATCTACAGTCCCCCTTGCCG-3' and 5'-GCAACTGACCGTGCAAGTCA-3'.[3] The PCR reaction was performed in 25 μl volumes containing 50 ng of genomic DNA template, 12.5 pmol of each primer, 0.1 mM of each deoxynucleoside triphosphate, 1 PCR buffer (50 mM KCl, 10 mM Tris-HCl and 0.1% Triton X-100), 1.5 mM MgCl2, and 1.5 U of Taq polymerase (Promega Corporation, Madison, WI). PCR conditions were as follows: an initial denaturation step at 94°C for 4 min, 35 cycles of 94°C for 40 s, 56°C for 30 s, and 72°C for 30 s and a final extension at 72°C for 10 min. Then, the PCR product (a 296 bp fragment) was digested by BstUI (New England BioLabs, Beverly, MA) overnight at 60°C and then analyzed on 2.5% NuSieve 3:1 agarose gel (FMC BioProducts, Rockland, ME) with ethidium bromide and photographed with polaroid film [Figure 1]. The p53 72 Pro allele, which lacked the BstUI restriction site, had only a single 296 bp band, whereas p53 72 Arg, which had the BstUI restriction site, produced 169 and 127 bp bands. More than 10% of the samples were retested randomly, and the results were 100% concordant.
Figure 1.
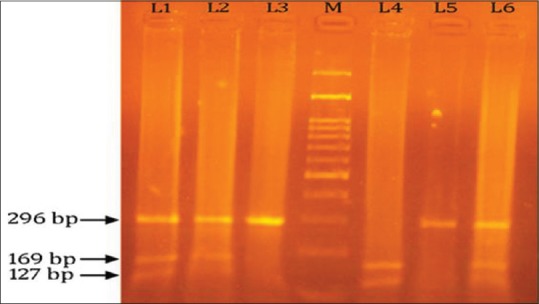
This is 2% agarose gel picture showing polymerase chain reaction-restriction fragment length polymorphism products of p53 gene codon 72 polymorphism; Lane 1, 2, and 6 show heterozygous arginine/proline genotype (296, 169 and 127 bp bands); Lane 3 and 5 show homozyggous proline genotype (296 bp band); Lane 4 show homozygous arginine genotype (169, 127 bp bands); whereas Lane M shows a 100 bp ladder
All the statistical analyses were performed with SPSS version 16 software (IBM). The genotyping data were compared between cases and controls using Chi-square test. P ≤ 0.05 were considered statistically significant.
Results
All the demographic details and clinicopathological characteristics of cases included in the study are shown in Table 1. Of 41 cases included in the present study, 34 (82.92%) were male and 7 (17.07%) were female. All cases were categorized into three subgroups such as 17 cases (41.46%) <40 years, 19 cases (46.34%) between 41 and 55 years, and 5 cases (12.19%) more than 60 years. Highest percentage of oral squamous cell carcinoma (OSCC) patients was identified between 40 and 60 years. Regarding the primary site of lesion, the predominance of buccal mucosa in 26 patients (63.41%) was noted, followed by tongue in 9 patients (21.95%) and low percentage was observed for palate that is in 6 patients (14.63%). The clinical staging of tumor was done (usually numbers I–IV) to know how much tumor had spread. In the present study, Stage III disease showed highest frequency (43.90%) as compared to Stage II (26.82%), Stage I (14.63%), and Stage IV (0%). Histopathologically, of 41 cases of oral lesions, 35 cases (85.36%) were squamous cell carcinoma and 6 cases (14.63%) of leukoplakia. We have also done grading of OSCC out of 35 cases of OSCC, maximum numbers were well-differentiated cases (29, 82.85%) as compared to moderately differentiated cases (6, 17.15%) and no case of poorly differentiated OSCC was found. Of 35 cases of OSCC biopsies, 17 (48.57%) had weak IHC staining for p53, 14 cases (40%) showed evidence of p53 protein staining, and 4 cases (11.42%) showed negative staining [Figures 2 and 3]. Among 6 cases of leukoplakia, 3 (50%) showed weak staining and 3 (50%) showed negative results [Table 1].
Table 1.
Demographical details of site of lesion, histopathology, grading, and staging of cases included in the study
Figure 2.
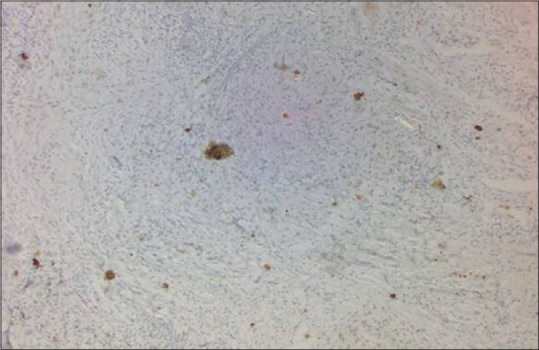
Weak expression of p53 by immunohistochemistry (×10)
Figure 3.
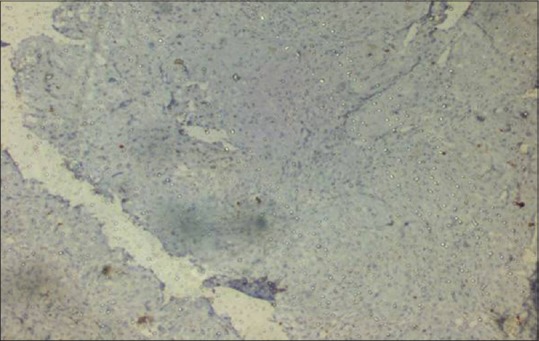
Negative expression of p53 by immunohistochemistry (×10)
In the present study as shown in Table 2, it was observed that frequency of Arg/Arg genotypes was lower in OSCC patients as compared to controls (P < 0.720), whereas frequency of Arg/Pro and Pro/Pro was elevated in OSCC patients as compared to controls (P < 0.809). In leukoplakia patients, frequency of Arg/Arg, Arg/Pro, and Pro/Pro genotype was equal to controls (P < 0.548) [Table 3]. It was observed in that codon 72 polymorphism was not associated significantly neither with OSCC nor with leukoplakia, associated with tobacco/pan masala consumption.
Table 2.
Distribution of the p53 codon 72 alleles and genotypes in oral squamous cell carcinoma samples and controls

Table 3.
Distribution of the p53 codon 72 alleles and genotypes in leukoplakia samples and controls

Discussion
Development of cancer is a multifactorial and multistep process which occurs with an effect of series of progressive genetic alteration. Various genetic studies at molecular level state that a group of protooncogenes and tumor suppressor genes alterations were play an effective role in cancer development. Among group of tumor suppressor genes, p53 is the most important one which plays an important role in various cancers including OSCC. Hence, detection of p53mutations is an important factor for early diagnosis and treatment of OSCC.
Up till now, various studies have provided evidence that p53 polymorphism at codon 72 may be associated with certain cancers such as breast carcinoma,[4] lung cancer,[5] hepatocellular carcinoma,[6] and esophageal carcinoma. Specifically both Arg and Pro alleles have been found to be associated with high risk of the development of cancer. The present study has been planned out to evaluate the gene polymorphism of p53 codon 72 and its correlation with p53 expression on IHC, which form an important tool for the future early diagnostic modalities in the treatment of oral cancers.
Occurrence of a single-nucleotide polymorphism at codon 72 of p53 leads to the presence of either Arg or Pro alleles, which in turn could result in three different genotypes: Arg/Arg, Arg/Pro, and Pro/Pro. The structure of the p53 protein is affected by substitution of the Arg codon with Pro or vice versa although the mechanism by which this might affect function of p53 remains controversial.
The effect of p53 codon 72 polymorphism has been debated in various carcinomas. For example, Storey et al. have proved that overexpression of homozygous Arg 72 p53 protein can increase the susceptibility of cervical cancer which is HPV associated up to seven fold.[7] By contrast, Liu et al. have proved that Pro 72 p53 allele is associated with increased risk of lung squamous cell carcinoma and adenocarcinoma.[8] In other study, Twu et al. have found that the heterozygous Arg/Pro genotype is associated with an increased risk of hypopharyngeal squamous cell carcinoma.[9] These findings have also been supported by some other studies,[10,11,12,13] while they have also opposed by some other investigators.[14,15]
There are limited results on the role of p53 codon 72 polymorphism on OSCC. In a study carried out in Taiwan, Bau et al. reported that the Arg/Arg genotype seems to increase the risk of OSCC by 2.7 fold.[16] By contrast, in the present study, we found no significant association between p53 codon 72 polymorphism genotypes and OSCC. Our observation is similar to the results obtained by Shen et al. on head and neck squamous cell carcinoma[17] as well as Katiyar et al. on HPV-associated oral cancer in India.[18] These variations in results might be possible due to role of various factors such as geographic distribution and racial differences on various predisposing factors involving tobacco consumption, betel quid, bidi smoking, alcohol use, lifestyle, and risk of HPV virus. The major limitation in the present study was small sample size. Thus, additional studies with larger sample size might be required in the near future to further evaluate the role of p53 codon 72 gene polymorphism as a risk factor for the development of OSCC.
To conclude, additional studies with larger sample size are required in the near future to investigate the role of various environment and thenic factors to determine the role of p53 codon 72 gene polymorphism in OSCC.
Financial support and sponsorship
The study was supported by Intramural Grant (Era's Lucknow Medical College and Hospital, Lucknow, Uttar Pradesh, India).
Conflicts of interest
There are no conflicts of interest.
References
- 1.Zain RB, Ikeda N, Gupta PC, Warnakulasuriya S, van Wyk CW, Shrestha P, et al. Oral mucosal lesions associated with betel quid, areca nut and tobacco chewing habits: Consensus from a workshop held in Kuala Lumpur, Malaysia, November 25-27, 1996. J Oral Pathol Med. 1999;28:1–4. doi: 10.1111/j.1600-0714.1999.tb01985.x. [DOI] [PubMed] [Google Scholar]
- 2.Sankaranarayanan R, Ramadas K, Thomas G, Muwonge R, Thara S, Mathew B, et al. Effect of screening on oral cancer mortality in Kerala, India: A cluster-randomised controlled trial. Lancet. 2005;365:1927–33. doi: 10.1016/S0140-6736(05)66658-5. [DOI] [PubMed] [Google Scholar]
- 3.Ji X, Neumann AS, Sturgis EM, Adler-Storthz K, Dahlstrom KR, Schiller JT, et al. p53 codon 72 polymorphism associated with risk of human papillomavirus-associated squamous cell carcinoma of the oropharynx in never-smokers. Carcinogenesis. 2008;29:875–9. doi: 10.1093/carcin/bgn039. [DOI] [PubMed] [Google Scholar]
- 4.Själander A, Birgander R, Hallmans G, Cajander S, Lenner P, Athlin L, et al. p53 polymorphisms and haplotypes in breast cancer. Carcinogenesis. 1996;17:1313–6. doi: 10.1093/carcin/17.6.1313. [DOI] [PubMed] [Google Scholar]
- 5.Fan R, Wu MT, Miller D, Wain JC, Kelsey KT, Wiencke JK, et al. The p53 codon 72 polymorphism and lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2000;9:1037–42. [PubMed] [Google Scholar]
- 6.Yu MW, Yang SY, Chiu YH, Chiang YC, Liaw YF, Chen CJ. A p53 genetic polymorphism as a modulator of hepatocellular carcinoma risk in relation to chronic liver disease, familial tendency, and cigarette smoking in hepatitis B carriers. Hepatology. 1999;29:697–702. doi: 10.1002/hep.510290330. [DOI] [PubMed] [Google Scholar]
- 7.Storey A, Thomas M, Kalita A, Harwood C, Gardiol D, Mantovani F, et al. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature. 1998;393:229–34. doi: 10.1038/30400. [DOI] [PubMed] [Google Scholar]
- 8.Liu G, Miller DP, Zhou W, Thurston SW, Fan R, Xu LL, et al. Differential association of the codon 72 p53 and GSTM1 polymorphisms on histological subtype of non-small cell lung carcinoma. Cancer Res. 2001;61:8718–22. [PubMed] [Google Scholar]
- 9.Twu CW, Jiang RS, Shu CH, Lin JC. Association of p53 codon 72 polymorphism with risk of hypopharyngeal squamous cell carcinoma in Taiwan. J Formos Med Assoc. 2006;105:99–104. doi: 10.1016/S0929-6646(09)60330-2. [DOI] [PubMed] [Google Scholar]
- 10.Dunning AM, Healey CS, Pharoah PD, Teare MD, Ponder BA, Easton DF. A systematic review of genetic polymorphisms and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 1999;8:843–54. [PubMed] [Google Scholar]
- 11.Humbey O, Cairey-Remonnay S, Guérrini JS, Algros MP, Mougin C, Bittard H, et al. Detection of the human papillomavirus and analysis of the TP53 polymorphism of exon 4 at codon 72 in penile squamous cell carcinomas. Eur J Cancer. 2003;39:684–90. doi: 10.1016/s0959-8049(02)00835-3. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki K, Matsui H, Ohtake N, Nakata S, Takei T, Nakazato H, et al. Ap53 codon 72 polymorphism associated with prostate cancer development and progression in Japanese. J Biomed Sci. 2003;10:430–5. doi: 10.1007/BF02256434. [DOI] [PubMed] [Google Scholar]
- 13.Zhang ZW, Newcomb P, Hollowood A, Feakins R, Moorghen M, Storey A, et al. Age-associated increase of codon 72 Arginine p53 frequency in gastric cardia and non-cardia adenocarcinoma. Clin Cancer Res. 2003;9:2151–6. [PubMed] [Google Scholar]
- 14.Hamel N, Black MJ, Ghadirian P, Foulkes WD. No association between P53 codon 72 polymorphism and risk of squamous cell carcinoma of the head and neck. Br J Cancer. 2000;82:757–9. doi: 10.1054/bjoc.1999.0993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Summersgill KF, Smith EM, Kirchner HL, Haugen TH, Turek LP. p53 polymorphism, human papillomavirus infection in the oral cavity, and oral cancer. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2000;90:334–9. doi: 10.1067/moe.2000.107359. [DOI] [PubMed] [Google Scholar]
- 16.Bau DT, Tsai MH, Lo YL, Hsu CM, Tsai Y, Lee CC, et al. Association of p53 and p21(CDKN1A/WAF1/CIP1) polymorphisms with oral cancer in Taiwan patients. Anticancer Res. 2007;27:1559–64. [PubMed] [Google Scholar]
- 17.Shen H, Zheng Y, Sturgis EM, Spitz MR, Wei Q. P53 codon 72 polymorphism and risk of squamous cell carcinoma of the head and neck: A case-control study. Cancer Lett. 2002;183:123–30. doi: 10.1016/s0304-3835(02)00117-9. [DOI] [PubMed] [Google Scholar]
- 18.Katiyar S, Thelma BK, Murthy NS, Hedau S, Jain N, Gopalkrishna V, et al. Polymorphism of the p53 codon 72 Arg/Pro and the risk of HPV type 16/18-associated cervical and oral cancer in India. Mol Cell Biochem. 2003;252:117–24. doi: 10.1023/a:1025546610920. [DOI] [PubMed] [Google Scholar]


