Abstract
Naive hypomethylated embryonic pluripotent stem cells (ESCs) are developmentally closest to the preimplantation epiblast of blastocysts, with the potential to contribute to all embryonic tissues and the germline, excepting the extra-embryonic tissues in chimeric embryos. By contrast, epiblast stem cells (EpiSCs) resembling postimplantation epiblast are relatively more methylated and show a limited potential for chimerism. Here, for the first time, we reveal advanced pluripotent stem cells (ASCs), which are developmentally beyond the pluripotent cells in the inner cell mass but with higher potency than EpiSCs. Accordingly, a single ASC contributes very efficiently to the fetus, germline, yolk sac and the placental labyrinth in chimeras. Since they are developmentally more advanced, ASCs do not contribute to the trophoblast. ASCs were derived from blastocysts in two steps in a chemically defined medium supplemented with Activin A and basic fibroblast growth factor, followed by culturing in ABCL medium containing ActA, BMP4, CHIR99021 and leukemia inhibitory factor. Notably, ASCs exhibit a distinct transcriptome with the expression of both naive pluripotency genes, as well as mesodermal somatic genes; Eomes, Eras, Tdgf1, Evx1, hand1, Wnt5a and distinct repetitive elements. Conversion of established ESCs to ASCs is also achievable. Importantly, ASCs exhibit a stable hypermethylated epigenome and mostly intact imprints as compared to the hypomethylated inner cell mass of blastocysts and naive ESCs. Properties of ASCs suggest that they represent cells at an intermediate cellular state between the naive and primed states of pluripotency.
Keywords: ESCs, pluripotency, blastocysts, chimeras, yolk sac, placenta, hypermethylated epigenome
Introduction
Development of zygotes to blastocysts is accompanied by a sequential change from totipotency to pluripotency, with differentiation of the outer trophectoderm cells and the pluripotent cells in the inner cell mass (ICM). The ICM gives rise to all the somatic tissues, as well as germ cells. Pluripotent embryonic stem cells (ESCs) isolated from the ICM are in vitro counterparts1,2,3, which when introduced into the blastocysts can differentiate into all fetal tissues and germ cells, albeit making an insignificant contribution to the extraembryonic tissues4.
Following blastocyst implantation, epiblast cells in postimplantation embryos retain some characteristics of pluripotency while becoming progressively more hypermethylated before the initiation of somatic cell fates5,6. Epiblast stem cells (EpiSCs) from postimplantation epiblast cells are hypermethylated but retain some features of pluripotency7,8, where female EpiSCs have an inactive X chromosome9. A variety of self-renewing pluripotent stem cells have been reported10,11,12,13,14, with variable potencies, epigenetic states and transcriptional regulators. These cells have been generated in vitro using empirical combinations of cytokines, fetal calf serum and hormones, including mouse embryonic fibroblasts (MEF) as feeders.
Here, ASCs showed unique properties with the expression of naive pluripotency and some mesodermal somatic genes in chemically defined medium without serum. Importantly, these cells were also hypermethylated relative to naive ESCs. Altogether, ASCs represent an early postimplantation epiblast-like state of development. However, when injected into host blastocysts, single ASC contributed to embryonic tissues, germline, yolk sac and placental labyrinth that originate from the postimplantation epiblast, but did not contribute to the trophoblast. ASCs exhibit unique features and provide a new model for investigations on the epigenetic and transcriptional states that regulate cell potency.
Results
Derivation and characterization of AFSCs
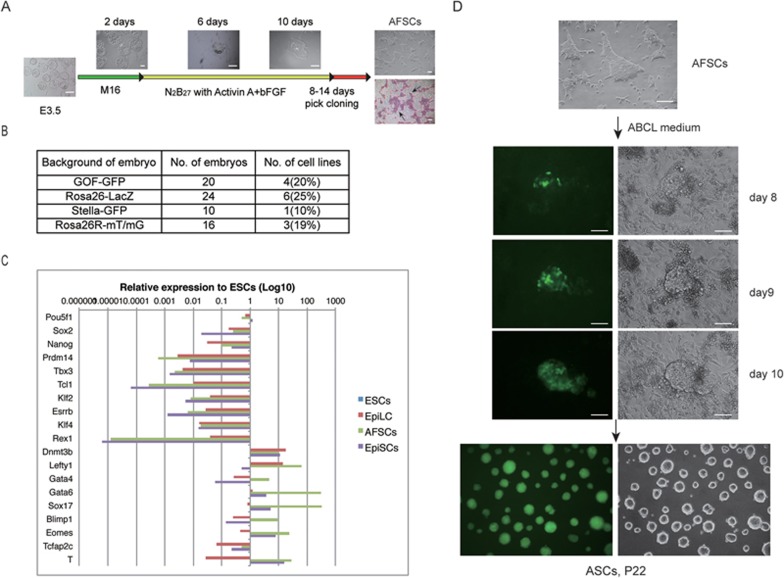
First, we attempted to derive stem cells from blastocysts in a chemically defined medium with N2B27 in the presence of ActA and basic fibroblast growth factor (bFGF) (but excluding KSR and feeder cells). Most of the E3.5 blastocysts attached within 5-6 days, forming ICM outgrowth, of which ∼20% resulted in stable lines from diverse genetic backgrounds; we designated these as ActA/bFGF stem cells (AFSCs) (Figure 1A and 1B). The rate of AFSC derivation was similar to that reported previously for naive ESCs in 2i/LIF15.
Figure 1.
Derivation of AFSCs and ASCs. (A) Schematic overview of AFSC derivation. Note AP-positive cells (arrow indicated) in AFSCs. Scale bars, 100 μm. (B) Derivation rate of AFSCs with diverse genetic backgrounds. (C) Quantitative RT-PCR of key genes in ESCs, EpiLCs, AFSCs and EpiSCs, all derived from 129/sv females mated with GOF-GFP males of mixed background. Note the expression of endoderm and mesoderm genes in AFSCs (at the bottom). (D) Reprogramming AFSC to ASCs. Scale bars, 100 μm.
Immunofluorescence (IF) and RT-qPCR analysis of AFSCs revealed expression of OCT4, SOX2, NANOG, E-cadherin; we also detected H3K27me3 which is a distinctive mark of an inactive X chromosome (Figure 1C; Supplementary information, Figure S1A). AFSCs share similarities with EpiSCs7,8, where expression of some genes of the ICM and ESCs were significantly reduced, including Prdm14, Tbx3, Tcl1, Klf2, Esrrb, Klf4 and Rex1. By contrast, we observed higher expression of early germ layer genes Eomes, Lefty1 and brachyury (T) (Figure 1C), together with the strong expression of Sox17 and Blimp1. AFSCs also uniquely displayed expression of Gata6 and Gata4, which are a part of the primitive endoderm transcriptome (Figure 1C). Overall, AFSCs were clearly at a stage beyond the pluripotent state of epiblast cells in blastocysts but as teratomas, they were still able to differentiate into cells of all the three germ layers (Supplementary information, Figure S1B).
Next, following the injection of 15-20 AFSCs with Rosa26-LacZ reporter into E3.5 blastocysts, we detected their contribution (∼11%-14%) in chimeric embryos at E6.5-E8.5 (Supplementary information, Figure S1C, S1D and Table S1A), which is unlike EpiSCs7,8. More strikingly, their high contribution was found in the posterior primitive streak and the extra-embryonic mesoderm surrounding the epiblast in E6.5-E7.5 chimeric embryos (Supplementary information, Figure S1C and S1D). The latter develops into the yolk sac and contributes in part to the placenta. Thus, AFSCs are distinct and, unlike pluripotent EpiSCs.
Derivation and characterization of ASCs
Next, we explored the impact of Wnt and BMP4 signaling on AFSCs, which can substitute for serum in ESC cultures16,17, and induce differentiation of EpiSCs18,19. Using CHIR99021 (CH) for simulating canonical Wnt signaling20, we started with two established AFSCs cell lines with GOF-GFP or Stella-GFP reporters. AFSCs were dissociated and cultured in N2B27 medium with ActA, BMP4, CH and LIF (henceforth called ABCL medium) (Figure 1D). After 10 days, we detected GOF-GFP-positive clones, which were able to self-renew for more than 30 passages. We designated these cells as advanced pluripotent stem cells (ASCs) and compared them with EpiSCs (Supplementary information, Table S1B).
To determine the key features of ASCs relative to ESCs, we examined expression by IF for cMYC, KLF4, E-cadherin, ESRRB and PRDM14. Excluding the higher expression of cMYC in ASCs than in ESCs, expression of other genes was equivalent to that in ESCs (Figure 2A). Notably, high expression of cMYC in ESCs promotes rapid transition through the G1 phase of the cell cycle, suppresses differentiation and controls pluripotency21.
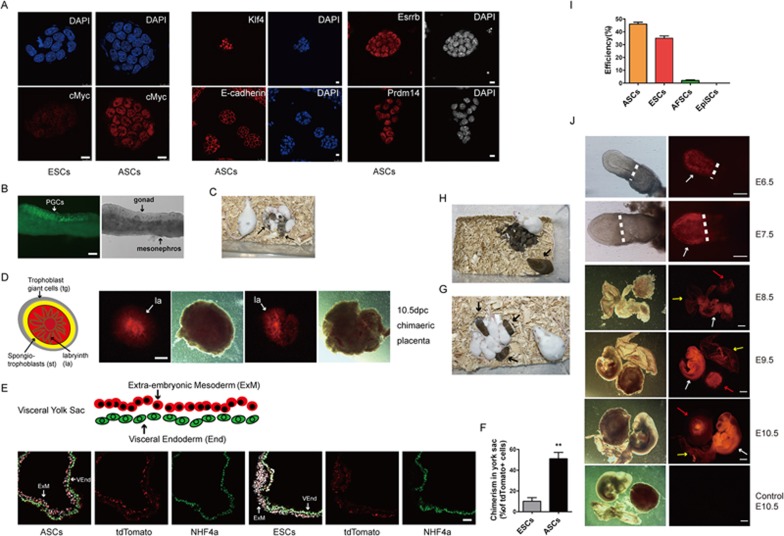
Figure 2.
Characteristics of ASCs. (A) IF staining assays in ESCs and ASCs, including those detect the expression of KLF4, E-CADHERIN, ESSRB and PRDM14 in ASCs. Scale bars, 10 μm. (B) Germline transmission of ASCs in E12.5 chimeras shown by GOF-GFP-positive cells (arrow). Scale bars, 100 μm. (C) Chimeric pups (arrows) generated by injecting ASCs in ICR host blastocysts. (D) A schematic depiction of placenta tissue indicates the contribution of ASCs (tdTomato) to labyrinth of placenta in E10.5 chimeras (white arrows). Scale bars, 1 mm. (E, F) Schematic depiction of the yolk sac, and the contribution of ASCs (tdTomato) to the extraembryonic mesoderm (red) in E10.5 chimeras. Scale bars, 100 μm. (G) Pups (arrows) generated entirely from ASCs (between 129/sv females and GOF-GFP males) in tetraploid ICR host blastocysts. (H) The F1 pups generated by ASCs-tetraploid male (arrows) mated with ICR female. (I) The efficiency of clonal line derived from single ASCs. (J) Chimeras (E6.5-E10.5) generated with single ASC. White arrows indicate embryonic body; yellow arrows mark yolk sac; and red arrows point placenta. Scale bars for E6.5-7.5, 200 μm; scale bars for E8.5-10.5, 2 mm. ExM, extraembryonic mesoderm; la, labyrinth; st, spongetrophectoderm; tg, trophectoderm giant cell; VEnd, visceral endoderm.
Next, we assessed the differentiation potential of ASCs with GOF-GFP reporter in chimeric embryos by injecting 15 cells into individual blastocysts. We found that ASC contributed to the embryo proper and germline of E12.5 embryo, as well as to the full-term pups (Figure 2B and 2C; Supplementary information, Table S1C and S1D). Importantly, ASCs contributed to the placenta in chimeric conceptuses (Supplementary information, Figure S2A), which was also the case with the other ASC line harboring Stella-GFP reporter (Supplementary information, Figure S2B). Furthermore, using ASCs with GOF-GFP and a constitutive tdTomato reporter, we detected these cells in the placental labyrinth (Figure 2D; Supplementary information, Figure S2C), yolk sac (Figure 2E and 2F), embryo proper and the germline at E10.5-E13.5 (Supplementary information, Figure S2D, S2E and Table S1E). Notably, ASCs contributed to more than 50% of the extra-embryonic mesoderm of yolk sac, but there was no detectable contribution to the extra-embryonic endoderm of yolk sac (Figure 2E and 2F). This result indicates that ASCs are in a stable state between pluripotency and the onset of differentiation, with a potential to contribute to embryos, yolk sac and placental labyrinth.
For a further stringent test of the potency of ASCs, we introduced them into tetraploid host blastocysts, where the donor cells contribute predominantly if not exclusively to the embryo22. Using 15 donor ASCs, we obtained live pups with normal fertility at a higher rate than reported previously23,24 (Figure 2G and 2H; Supplementary information, Table S1F). The derivation efficiency of clonal lines from single ASC was equivalent to that of naive ESCs and much higher than that of EpiSCs or AFSCs (Figure 2I). Thus WNT and BMP4 signaling pathways increase the potency of AFSCs in the course of ASC derivation, and this potency exceeds that shown by EpiSCs in vivo.
Finally, we tested the potency of ASCs more stringently by introducing single ASCs into 8-cell stage embryos, which were cultured overnight and then transferred to recipient females. We found that 24%-36% of the embryos at E6.5-E7.5 and 31%-50% of the embryos at E8.5-E10.5 contained derivatives of ASCs with tdTomato reporter (Figure 2J; Supplementary information, Figure S2F and Table S2A). We also observed germline transmission and full-term chimeras from single ASCs (Supplementary information, Table S2B and S2C). Additional analysis of 18 conceptuses at E8.5-E10.5 resulting from single donor ASCs showed 14 chimeras (78%) with contributions to the embryo, yolk sac and the placenta (Figure 2J; Supplementary information, Figure S2F and Table S2A). In contrast, single naive donor ESCs cultured in 2i/LIF resulted in just 2/20 chimeras, with a contribution to the embryo, and no contribution to the extraembryonic tissues. When the number of donor cells was increased to 15, we obtained 18 chimeras, in which donor cell contributed predominantly to the fetus. However, in only one of these chimeras, ESC cells contributed to the fetus, yolk sac and the placenta (Supplementary information, Table S2D). Altogether, ASCs exhibit a unique pluripotent state, with high clonogenicity and potency, and a single ASC contributed to the embryo, placental labyrinth and the germline.
To test for the potential for the conversion of ESCs (2i/LIF) to ASCs, we cultured two ESC cell lines (SQ3.3 and E14). Both cell lines demonstrated normal growth and were passaged over 20 times in ABCL medium; and the resulting cells was designated as ESC-ASCs. We tested the in vivo developmental potential of ESC-ASCs in chimeras using single or multiple ESC-ASCs. We found that both single and multiple ESC-ASCs contributed to the embryo, yolk sac and placenta labyrinth (Supplementary information, Figure S2G; Supplementary information, Table S2E). Thus ESCs can be converted to ASCs by culturing in ABCL, exhibiting increased potency and characteristics resembling ASCs.
Molecular features of ASCs
To investigate the molecular features of ASCs, we profiled their transcriptome by RNA sequencing (RNA-seq), and compared it with naive ESCs (cultured in 2i/LIF), EpiSCs and EpiLCs (epiblast-like cells) (Supplementary information, Table S3A). Unsupervised hierarchical clustering (UHC) showed that ASCs and naive ESCs cluster together, whereas AFSCs are close to EpiSCs (Figure 3A; Supplementary information, Figure S3A). Moreover, the t-distributed stochastic neighbor embedding (t-SNE) analysis indicated a trajectory representing naive to primed cell states from ESCs and ASCs toward EpiLCs, and to more advanced AFSCs and EpiSCs (Figure 3B).
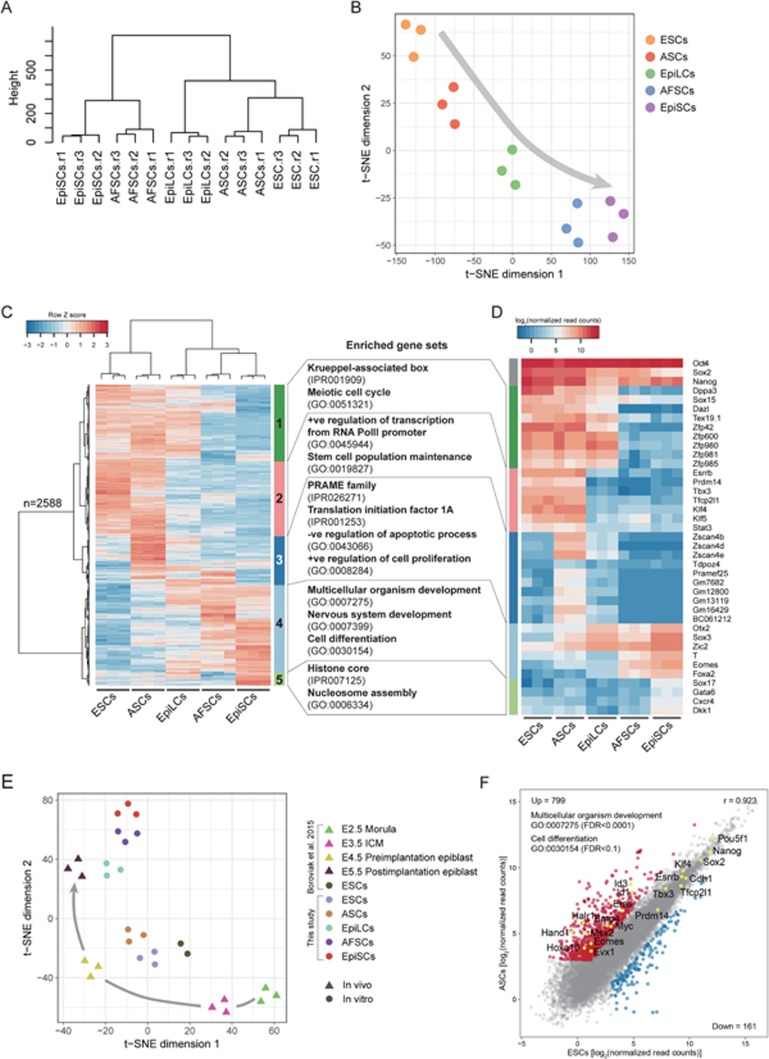
Figure 3.
RNA-seq analysis of AFSCs and ASCs. (A) Unsupervised hierarchical clustering (UHC) of whole-genome transcriptome from three biological replicates. Note that ASCs were clustered close to ESCs but not EpiSCs. (B) t-distributed stochastic neighbor embedding (t-SNE) of whole-genome transcriptome. Arrow represents potential trajectory from naive to primed pluripotent stem cells. (C) Heatmap showing scaled expression values of 2 588 differentially expressed genes (mean log2(normalized read counts) > 3 in any sample, log2(fold change) > 3, adjusted P-value < 0.05) between the five cell types. UHC of five major gene clusters. The top representative GO biological process and InterPro terms for each cluster are indicated on the right. (D) Expression heatmap of representative genes for each cluster in C. (E) t-SNE analysis of gene expression of pluripotent stem cells, and of E2.5-E5.5 embryos25, based on 1 685 dynamically expressed genes25. Arrow indicates developmental progression from E2.5 morula to E5.5 postimplantation epiblast. (F) Pair-wise gene expression comparison between ASCs and ESCs (2i/LIF). Upregulated (red) and downregulated (blue) genes in ASCs were highlighted (mean log2(normalized read counts) > 3 in either samples, log2(fold change) > 2, adjusted P-value < 0.05). Gene ontology analysis (by DAVID) showed ASCs upregulated genes that are involved in multicellular organism development and cell differentiation.
We examined the similarities and differences among the five cell types by analyzing a total of 2 588 differentially expressed genes. These genes were segregated into five distinct classes by UHC (Figure 3C; Supplementary information, Table S3B and S3C). All the five pluripotent cell types expressed core pluripotency factors Oct4, Sox2 and Nanog (Figure 3D), but the expression of naive pluripotency genes (e.g., Esrrb, Tfcp2l1, Klf4, Tbx3 and Prdm14) was detected exclusively in ASCs and ESCs (cluster 2, Figure 3C and 3D), which was confirmed by IF analysis (Figure 2A). On the other hand, AFSCs and EpiSCs showed enrichment for developmental genes, including Otx2, Zic2, T, Eomes and Foxa2 (cluster 4, Figure 3D).
Notably, we found that 421 genes were exclusively expressed in ASCs (cluster 3, Figure 3C), including genes that encode preferentially expressed antigen of melanoma (PRAME) family and translation initiation factor 1A (see below). Principal component analysis (PCA) of transposable element expression also showed a similar trajectory from naive to primed cell states as seen by analyzing the overall gene expression profiles (Supplementary information, Figure S3B). Notably, both ESCs and ASCs showed high expression of RLTR45, RMER16 and LTRIS2 (Supplementary information, Figure S3C), which might distinguish them from EpiSCs.
Next, we tested if ASCs correspond to cells at a particular embryonic developmental stage by comparing the ASC transcriptome with that of E2.5-E5.5 embryos25 (Supplementary information, Table S3D). We performed t-SNE and UHC analyses with 1 685 genes that are dynamically expressed between E2.5 and E5.5 stages25. Naive ESCs and ASCs were clustered with E4.5 preimplantation epiblast, with ASCs being slightly more advanced than ESCs (Figure 3E; Supplementary information, Figure S3D). EpiLCs were clustered with E5.5 postimplantation epiblast, while AFSCs and EpiSCs had progressed to more advanced stages26,27,28 (Figure 3E; Supplementary information, Figure S3D). Consistently, the comparison between ASCs and naive ESCs suggested the expression of the core genes (e.g., Oct4, Nanog and Sox2) and naive pluripotency genes (e.g., Esrrb, Klf4 and Tfcp2l1) in both of them. However, ASCs also showed upregulation of some primitive streak and mesoderm genes, including Eomes, Eras, Evx1, Hand1 and Hoxa10 (Figure 3F; Supplementary information, Figure S3E). Furthermore, ASCs exhibited high gene expression level and protein abundance for cell-adhesion molecule E-cadherin (Cdh1) and c-Myc (Figures 2A and 3F). Although T and Cdx1 IF also showed heterogeneity among ASCs, the consistent contribution of single ASCs to chimeras suggests the presence of a dynamic equilibrium (Supplementary information, Figure S3F). We also noted a slightly higher expression of Hoxa genes in ASCs compared to ESCs, including Hoxa1, Hoxa5, Hoxa9, Hoxa10, Hoxa11, Hoxa11os, and Hoxa13 (Supplementary information, Figure S3G). In this context, the lncRNA Haunt/Halr1 that locates ∼40 kb upstream of the Hoxa cluster29,30,31 also displayed higher expression in ASCs than ESCs (Supplementary information, Figure S3G).
Further examination of ASCs (cluster 3, Figure 3C; Supplementary information, Table S3B) revealed expression of so-called 2C genes: Zscan4b, Zscan4d and Zscan4e (Figure 3D; Supplementary information, Figure S4A and Table S3E), and MERVL transcripts (Supplementary information, Figure S3C)12,32,33,34. However, IF analysis showed that only 8% of the ASCs were MERVL-Gag positive/OCT4 negative, but such cells were rarer in 2i/LIF ESCs (Supplementary information, Figure S4A and S4B). Nonetheless, this does not account for the potency of ASC, since ESC-ASCs showed similar characteristics in the absence of 2C-like gene expression (Supplementary information, Figures S2G, S4C and S4D).
To this end, we injected a single ASC cell with tdTomato and GOF-GFP into an 8-cell stage embryo and examined its chimeric contribution after 48 h of in vitro culturing. Consistently, ASC contributed only to the ICM, and not to the trophoblast in blastocysts (0/42 recovered blastocysts) (Supplementary information, Figure S4E).
ASCs exhibit a hypermethylated stable methylome
Naive ESC pluripotent state displays global DNA hypomethylation, where both X chromosomes are active in female cells15,35,36. Considering the enhanced potency of ASCs described above, we examined the epigenome of ASCs. Our initial IF observations showed bright H3K27me3 foci in female AFSCs, but not in female ASCs, suggesting reactivation of the X-chromosome in ASCs (Supplementary information, Figure S5A).
We further performed DNA methylation analysis of female ASCs, ESCs and EpiSCs by whole-genome bisulfite sequencing (BS-seq) (Supplementary information, Table S3F). Although naive ESCs (2i/LIF) were globally hypomethylated (median CpG methylation of ∼20%), ASCs showed a hypermethylated genome (median ∼70%) (Figure 4A). Indeed, UHC analysis revealed that the methylome of ASCs was more similar to the hypermethylated epigenome of EpiSCs (median 90%) (Figure 4B; Supplementary information, Figure S5B). Most genomic regions, including promoters, enhancers, exons, introns and intergenic regions, were highly methylated in ASCs (Supplementary information, Figure S5C). In line with a previous report35, all genomic imprints were erased in female 2i/LIF ESCs, whereas most imprints were retained in both ASCs and EpiSCs (Figure 4C). The retention of imprints, which are essential for development, would make ASCs more robust in culture, and during subsequent differentiation.
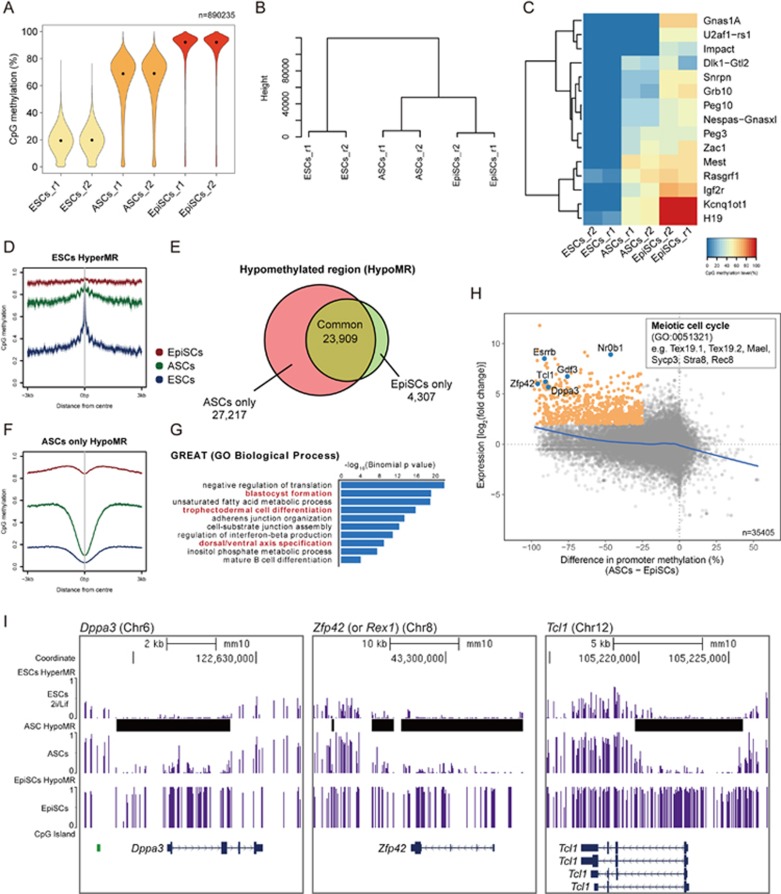
Figure 4.
Methylation analysis of ASCs. (A) Violin plot showing CpG methylation distribution of 2 kilobase (kb) genomic tiles. (B) UHC of methylation levels of 2 kb genomic tiles. (C) Heatmap showing CpG methylation of imprint control regions (ICRs) in ESCs, ASCs and EpiSCs. (D) Methylation profiles of ESCs hypermethylated regions (HyperMR) in ESCs. Note that HyperMR in ESCs were generally hypermethylated in ASCs and EpiSCs. (E) Venn diagram showing overlap of hypomethylated regions (HypoMR) between ASCs and EpiSCs. (F) Methylation profile of ASC-specific HypoMRs in E. (G) GREAT analysis of ASC-specific HypoMRs. (H) Scatterplot of differential gene expression and difference in promoter methylation between ASCs and EpiSCs. Genes with > 20% promoter methylation and log2 (read counts) > 3 in either samples were shown. Genes upregulated in ASCs with promoter demethylation are highlighted in orange. They were enriched for regulating meiotic cell cycle and cell pluripotency. Best fit curve (blue line) was generated by the generalized additive models (GAM). (I) UCSC genome browser snapshots of CpG methylation level at representative naive pluripotency genes.
To characterize the methylome further, we calculated hypermethylated regions (HyperMRs) in ESCs and hypomethylated regions (HypoMRs) in ASCs and EpiSCs using a hidden Markov model37. We found very few HyperMRs in 2i/LIF ESCs (1 910); and most of these regions also showed high methylation levels in ASCs and EpiSCs (Figure 4D). We then focused on comparing the methylomes of ASCs and EpiSCs. Among the 51 126 HypoMRs in ASCs, 27 217 (53%) were not present in EpiSCs (Figure 4E and 4F; Supplementary information, Table S3G). Analysis using Genomic Regions Enrichment of Annotations Tool (GREAT)38 suggested that the ASC-specific HypoMRs (CpG coverage > 40, methylation difference > 40%) were in the vicinity of genes that function in blastocyst formation (e.g., Esrrb and Oct4), trophectodermal cell differentiation (e.g., Tead4, Tfap2c, Eomes) and dorsal/ventral axis specification (e.g., Wnt3, Pax6) (Figure 4G). Promoter methylation and gene expression analysis between ASCs and EpiSCs suggested an overall weak anti-correlation (Figure 4H). However, we found that a small fraction of genes (551) were upregulated in ASCs and exhibited promoter demethylation. They were enriched for genes involved in regulating Meiotic cell cycle (e.g., Mael, Sycp3 and Stra8) (Figure 4H; Supplementary information, Table S3H and S3I). These germline genes are promoter methylation-sensitive and are expressed in naive ESCs, where their promoters are demethylated. In addition, we found that naive pluripotency genes, such as Dppa3, Zfp42 (also known as Rex1), Tcl1, Esrrb, Gdf3 and Fbxo15, were also promoter demethylated and upregulated in ASCs (Figure 4H and 4I; Supplementary information, Figure S5D). Thus, ASCs exhibited an overall hypermethylated genome and maintained imprints, but showed targeted demethylation at loci associated with the preimplantation development and naive pluripotency. This is consistent with their high contribution in chimeras described above.
Discussion
We describe the derivation of two novel cell types, AFSCs and ASCs from mouse blastocysts, which provide new insights on pluripotent stem cells. In particular, the ASCs (advanced pluripotent stem cells) show higher DNA methylation levels than naive ESCs (Figure 4A and 4B), and with the expression of both pluripotent and somatic genes (Figure 3C; Supplementary information, Figure S3E). We also find that genomic imprints are more stable in ASCs relative to naive ESCs (Figure 4C). On the basis of the transcriptome and DNA methylome analysis, ASCs appeared to be at an intermediate state between naive ESCs and primed EpiSCs, and apparently represent the cells with the characteristics of the early postimplantation epiblast. Remarkably, ASC has high developmental potency in chimeras, since a single ASC contributes extensively to the developing embryo, germline and the extraembryonic mesoderm (Figure 2J). Indeed it is possible to obtain a live pup consisting exclusively of ASCs in “tetraploid rescue experiments”. The high potency of individual ASCs may in part be due to the shorter cell cycle time (data not shown) and higher expression of cMYC in ASCs as compared to that in ESCs (Figures 2A and 3F). Accordingly, cMYC might promote rapid cell transition through the G1 phase of the cell cycle while suppressing cell differentiation by activating Dusp2 and Dusp7 phosphatases, which in turn represses FGF/ERK signaling21. The relevance and significance of the cMyc-Dusp2-FGF/ERK pathway in ASCs merit further investigation.
Concerning the extraembryonic tissues, individual ASCs do not contribute to the trophectoderm cells when introduced into 8-cell stage embryos (Supplementary information, Figure S4E), unlike some recently described pluripotent stem cells11,12,13,14. This observation is consistent with ASCs being equivalent to early postimplantation epiblast cells, and therefore developmentally they represent cells at a later stage when we would not expect their contribution to the trophoblast cells, which also accounts for our observation that ASCs did not contribute to the extraembryonic endoderm. Nevertheless, single ASCs contribute significantly to some extraembryonic tissues that originate from the postimplantation epiblast, especially the yolk sac mesoderm and placental labyrinth in chimeric conceptuses as compared to ESCs; this is also a mark of higher potency of ASC39. Altogether, ASC contribute extensively to the tissues that normally originate from postimplantation epiblast cells, which is consistent with their overall molecular state and characteristics.
Another feature of interest concerning ASC involves the Hoxa family genes. We found that the lncRNA Haunt/Halr1 located ∼40 kb upstream of the Hoxa cluster and showed higher expression in ASCs than in ESCs (Supplementary information, Figure S3G). The Huant/Halr1 genomic locus has been shown to be bound by pluripotency regulators, including Oct4, Nanog and Sox2, with enhancer function-related epigenetic marks, such as H3K4me1, DNase I hypersensitivity, H3K4me3 and H3K36me329,30. The Haunt lncRNA and its genomic locus regulate the Hoxa gene cluster during differentiation of pluripotent cells31. Our data suggest that high expression of Haunt/Halr1 in ASCs may promote downregulation of Hoxa cluster to maintain a stable pluripotent state.
How precisely the chemically defined ABCL medium induces and maintains the hypermethylated state in ASC deserves further investigation in the future. The relatively hypermethylated state of ASCs with intact genomic imprints compared to naive ESCs is likely to provide greater stability during self-renewal in vitro, which is consistent with the findings of two recent studies on ESCs40,41.
The AFSCs we describe in this study are quite similar to EpiSCs, which differentiate in the presence of Wnt and BMP418,19. On the other hand, Activin A and LIF support self-renewal of AFSCs and ESC17, respectively. Thus, during the derivation of ASCs, BMP4 and CH would likely promote AFSCs toward differentiation, which would be counteracted by Act A and LIF that supports self-renewal. Under these signaling conditions, a small proportion of surviving cells acquire new phenotypes and we describe these cells as ASCs. Further detailed analyses, including analysis of epigenetic resetting, determining how precisely the ASCs become established in culture, need to be explored in the future studies. Notably, ESCs can also be readily converted to ASCs, referred to here as ESC-ASCs, with similar properties.
In conclusion, ASCs are a novel and distinct self-renewing type of pluripotent stem cells, which exhibit intermediate features between ESCs and EpiSCs. Transcriptionally, ASCs are at a cellular state that beyond the epiblast state of the blastocyst. In this respect, ASCs are like EpiSCs, albeit some loci remain hypomethylated in ASCs, and importantly, female ASCs do not exhibit X-inactivation. Thus, ASC represents a unique pluripotent state with distinct transcriptional and epigenetic features, which confers greater stability and potency.
Materials and Methods
Derivation of AFSC cell lines
Mouse embryos at E3.5 were isolated from 129/sv females mated with Oct4-ΔPE-GFP (GOF-GFP), Stella-GFP, Rosa129/Sv and Rosa26R-mT/mG transgenic males with a mixed background of MF1, 129/sv and C57BL/6J strains42,43,44. The GOF-GFP expression of the reporter that lacked the proximal enhancer and was under the control of the Oct4 promoter and distal enhancer. This GFP transgene showed expression in the ICM of blastocysts, PGC in vivo and in ESC. Rosa129 is a knockin mouse line with a LacZ reporter in the Rosa locus, where LacZ is ubiquitously expressed. Standard AF medium consists of N2B27 medium (Life technology) supplemented with Activin A (20 ng/ml, R&D Systems) and bFGF (12 ng/ml, R&D systems). E3.5 blastocysts were cultured in M16 medium (Sigma-Aldrich) for 2 days. Most of them were zone pellucida free. If blastocysts had zona pellucida, they were removed by Acidic Tyrode's Solution (Sigma-Aldrich). They were then placed in 24-well fibronectin-coated (16.7 μg/ml, Millipore) plate with AF medium. Epiblast cultures grew efficiently and formed “flat” epithelial-like colonies after 5-6 days. The resulting colonies were further cut into smaller pieces by glass needles after 7-9 days of culturing, and then the colonies passaged by Accutase (Life technology) regularly every 2-3 days. These cells, referred to as AFSCs, could be passaged for over 30 passages in AF medium.
Derivation of ASC cell lines
We dissociated GOF-GFP AFSCs using Accutase and placed AFSCs (3 × 105 single cells) in fibronectin-coated 24-well plates with 1 ml ABCL medium. The ABCL medium was N2B27 medium with Activin A (20 ng/ml, R&D systems), BMP4 (50 ng/ml, R&D systems), CHIR9902 (3 μM, Miltenyi Biotech) and leukemia inhibitory factor (1 000 IU/ml, Millipore). Dependent on cell growth, AFSCs were then passaged after 2-4 days. After 6-7 days of ABCL treatment, we noted cell dying. GOF-GFP-positive clones started to appear around day 10-14. When these GFP-positive colonies grew to around 200 μm in diameter, they were cut into smaller pieces using glass needles and then transferred. When these colonies had grown for 6-7 days, they were treated with Accutase, and the resulting cells were cultured to produce GFP-positive colonies, which were capable of self-renewal for over 30 passages. We called these cells ASCs.
Teratoma formation
The AFSCs were disaggregated using Accutase, and 1 × 106 cells were injected under the epithelium of NOD-SCID mice. Three to five weeks after transplantation, tumor(s) were collected and fixed with 4% paraformaldehyde, and processed for paraffin sectioning. Sections were observed following hematoxylin and eosin staining.
Production of chimeras
Approximately, 15-20 cells were injected gently into ICR mice blastocoel cavity of blastocysts (E3.5) using a piezo-assisted micromanipulator attached to an inverted microscope. The injected embryos were cultured in KSOM medium (Millipore) to enable re-expansion of the blastocoel cavity and then transferred to the uteri of pseudopregnant ICR mice at 2.5 days post coitus (dpc). In a similar manner to the injection of blastocysts, 8-cell embryos (E2.5) were injected with a single cell placed carefully into the perivitelline space under the zona pellucida. The embryos were cultured overnight in KSOM medium (Millipore) at 37 °C in a 5% CO2 atmosphere and transferred to the uteri of pseudopregnant ICR mice at 2.5 dpc. The embryos were isolated at embryonic stage E6.5-E13.5 with chimeric contribution checked. Full-term chimeras were confirmed by the coat color pattern of the pups at birth.
Production of full-term pups in tetraploid host blastocysts
Two-cell stage embryos (E1.5) from ICR mice were collected by flushing oviducts; they were subjected to electrofusion to create tetraploid (4N) host blastocysts. Typically 15-20 ASCs were injected into tetraploid host blastocyts, which were transferred to pseudopregnant recipients at 2.5 dpc.
Immunofluorescence
Cells were briefly washed with PBS and fixed in 4% paraformaldehyde in PBS for 15 min at room temperature. Cells were permeabilized for 30 min with 1% BSA and 0.1% Triton X-100 in PBS. Antibody staining was carried out in the same buffer at 4 °C overnight. The slides were subsequently washed three times in PBS with 1% BSA, 0.1% Triton X-100 (5 min each wash), then incubated with secondary antibody for 1 h at room temperature in the dark, washed once for 5 min in 1% BSA, 0.1% Triton X-100 in PBS and twice for 5 min in PBS. The slides were then mounted in Vectashield with DAPI (Vector Laboratories) and imaged using a BioRad Radiance 2100 confocal microscope. Primary antibodies used were: mouse monoclonal Oct4 (BD Biosciences, 1:200), rat monoclonal Nanog (eBioscience, 1:500), goat polyclonal Sox2 (Santa Cruz, 1:200), rabbit polyclonal H3K27me3 (Upstate, 1:500), rat monoclonal E-cadherin (Takara, 1:40), Klf4 (Abcam, 1:300), cMyc (Abcam, 1:200), Esrrb (Abcam, 1:200), Prdm14 (Abcam, 1:200), HNF4a (Santa Cruz, 1:100). All secondary antibodies used were Alexa Fluor highly cross-adsorbed (Molecular Probes).
RNA-seq library preparation
The RNA-seq libraries for this paper were constructed by using NEBNext Ultra Directional RNA Library Prep Kit from Illumina. After isolation of total RNA from bulk amount of mouse stem cells using RNeasy Micro Kit (Qiagen), ∼500 ng total RNA was used to enrich mRNA with polyA tails by using NEBNext Magnetic Oligo d(T)25 beads. After RNA fragmentation and random priming, the first strand cDNAs and second strand cDNAs were synthesized, which was followed by end repair, dA-tailing, adapter ligation and PCR enrichment. The RNA-seq libraries were sequenced on HiSeq4000 platform.
RNA-seq analysis
Paired-end RNA-seq reads were mapped to the mouse reference genome (GRCm38/mm10) with TopHat2 (https://ccb.jhu.edu/software/tophat), and one mismatch was allowed. The alignments were guided by Ensembl gene models (Ensembl release 86). Read counts for either the full transcript or only the first exon were obtained with featureCounts (http://bioinf.wehi.edu.au/featureCounts). Transcript or first exon counts were normalized, and the statistical significance of differential expression between samples was assessed using the R Bioconductor DESeq2 package (https://bioconductor.org/packages/release/bioc/html/DESeq2.html). Transcript and exon counts normalized by DESeq2 size factors were subsequently normalized by their length/1 000. RNA-seq data25 from early mouse embryos were obtained from the European Nucleotide Archive (accession number: ERP007120), and identically processed.
Paired-end reads were further aligned to the mouse reference genome (GRCm38/mm10) with bowtie (options: “-M 1 – v2 – best – strata”). RepeatMasker annotations of transposable elements on the mouse reference genome were obtained from the UCSC genome browser table (GRCm38/mm10). Counts for all TE copies and Ensembl transcripts were obtained using featureCounts, and TE counts were subsequently processed by DESeq2 as described above, with the exception that TE counts were normalized by the total number of transcript counts instead of DESeq2 size factors.
The top 90% expressed genes (log2(normalized counts)) in the whole RNA-seq data set were used for clustering analysis. Pearson correlation analysis was performed by the R cor function with “1-Pearson correlation coefficient” as a distance metric. UHC analysis was performed by the R hclust function with the “ward” method and Euclidean distance metric. t-SNE analysis was performed using the R Rtsne function omitting an initial PCA step. PCA analysis was performed with the R prcomp function on centered and scaled log2(normalized counts) expression values. GSEA analysis was performed by the javaGSEA programme developed by the Broad Institute.
Real-time PCR
The cDNAs from bulk amount (2 μg) of total RNAs were diluted 10 to 40-fold. Then 2 μl of diluted cDNAs was used for each 20 μl real-time PCR reaction (1× Syber Green PCR Master Mix, 0.2 μM of each primer). All reactions were duplicated. The PCR was carried out as follows: first, 95 °C for 10 min to activate the Taq polymerase, followed by 40 cycles at 95 °C for 15 s (for denaturation) and for 1 min at 60 °C for annealing and extension. Finally a dissociation step was run to exclude the possibility of non-specific amplification.
LacZ staining
The embryos were isolated and washed twice in PBS. Then they were transferred to a cold solution (2% formaldehyde, 0.2% glutaraldehyde, 0.02% NP-40, 1 mM MgCl2, 0.01% sodium deoxycholate in PBS) and fixed for 2 h at 4 °C on a rocking platform. The embryos were washed three times in PBS for 2 min each, and stained in lacZ staining solution (0.4 mg/ml Xgal, 4 mM potassium ferrocyanide, 4 mM potassium ferricyanide, 1 mM MgCl2, 0.02% NP-40 in PBS) at 30 °C for 18-36 h in dark. Details of ingredient information are shown below:
Formaldehyde, Sigma-Aldrich; Glutaraldehyde, BioChemika (Fluka); NP-40, Roche;
MgCl2, Sigma-Aldrich; Sodium deoxycholate, Sigma-Aldrich; Xgal, Melford, MB1001;
Potassium ferrocyanide, Sigma-Aldrich; Potassium ferricyanide, Sigma-Aldrich.
Alkaline phosphatase staining
Alkaline phosphatase (AP) staining was carried out using AP Staining Kit from Sigma (86R-1KT) according to manufacturer's instructions. Briefly, the cells were fixed by 4% paraformaldehyde for 10 min, and then were stained overnight by AP staining solution at room temperature in the dark.
BS-seq library preparation
Genomic DNA (two biological replicates per condition) was extracted by QIAamp DNA Micro Kit (Qiagen). Subsequently, 100 ng of genomic DNA was subjected to 3.5 h bisulfite treatment using the Methylcode Bisulfite Conversion Kit (Invitrogen). To monitor the bisulfite conversion efficiency, unmethylated Lambda DNA (0.5 ng, Promega) was spiked-in before conversion. Single-stranded BS-converted DNA was subjected to the post-bisulfite adaptor tagging (PBAT) protocol as described previously45,46. Libraries were subjected to single-end 100 bp sequencing on HiSeq 4000 sequencing system (Illumina). Coverage information was summarized in Supplementary information, Table S3.
BS-seq analysis
PBAT reads were quality-trimmed with Trim Galore (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore), and 4 nt random primer sequences at the 5′ end and 1 nt at the 3′ end of all reads were removed. PBAT reads were then mapped to the computationally bisulfite-converted mouse reference genome (GRCm38/mm10) by using Bismark47 (version: 0.16.3; parameter settings: “bismark --pbat -N 1 -L 40”). Potential PCR duplicates were removed using the deduplicate bismark command.
Post-alignment analysis was performed by using the MethPipe package37. First, aligned sequences were converted to MethPipe format using the to-mr command. Methylation levels and coverage for each symmetric CpG site were calculated by the methcounts and symmetric-cpgs commands. Average CpG methylation levels of annotated genomic regions, i.e., promoters, exons, repeats, imprint control regions, CGIs, enhancers and 2 kb genomic tiles, were calculated with the MethPipe roimethstat program. Only high-confidence genomic regions with at least 40 CpG observations from reads in the region were used in further analyses.
About 2 kb tiles were calculated for all chromosomes with a 1 kb offset. Coordinates of promoters (−1 000 bp and +100 bp from the transcription start site (TSS)), exons, introns and intergenic regions were retrieved from Ensembl (Mouse Release 86). CGI annotations were obtained from the UCSC Table Browser. Imprint control region annotations were obtained from Tomizawa et al48. Tissue-specific enhancer coordinates (± 1 000 bp from peak; mm9 coordinates were converted to mm10 by the UCSC liftOver tool) were retrieved from the Mouse Encode Project49. HyperMRs and hypomethylated regions were calculated by the hmr command from the MethPipe package.
UHC of 2 kb tile methylation was performed using the R hclust command with the “ward” method. Correlation matrix was calculated by the R corrplot package. Profile plot was created by the R seqplots package. Gene ontology analysis was performed by GREAT50 and DAVID Bioinformatics Resource 6.850.
Data accession
All the sequencing data were deposited in the NCBI, Gene Expression Omnibus (GEO) under accession number GSE99494.
Author Contributions
SB, XL and MAS designed the research project, prepared and approved the manuscript; WWCT performed whole-genome bisulfite sequencing experiment and analyzed RNA-seq and BS-seq data; SB, BW and SK derived ASCs and analyzed molecular properties; SB, BW performed in vivo embryo experiment; JL, LL and FT prepared RNA-seq library; SD contributed to RNA-seq and BS-seq analysis; SL performed clonal lines experiments from single cell and helped proof the manuscript; KT, CL, YC and MW provided technical assistance.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We thank G Guo and J Li for technical assistance. This work was supported by grants from the Ministry of Science and Technology project of Inner Mongolia (20130216), the National Natural Science Foundation of China (31560335) and by Wellcome Trust Investigator Award to MAS, and by a core grant from the Wellcome Trust and CRUK to the Gurdon Institute.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Features of AFSCs.
Chimeras produced by ASCs.
RNAseq analysis of AFSCs and ASCs.
2-cell genes expression in ASCs.
Methylation landscape of ASCs.
The contribution of AFSCs and ASCs in chimera and tetraploid.
The summary of contribution of ASCs and ESC-ASCs in chimeras.
References
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981; 292:154–156. [DOI] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA 1981; 78:7634–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H. Molecular mechanism to maintain stem cell renewal of ES cells. Cell Struct Funct 2001; 26:137–148. [DOI] [PubMed] [Google Scholar]
- Beddington RS, Robertson EJ. An assessment of the developmental potential of embryonic stem cells in the midgestation mouse embryo. Development 1989; 105:733–737. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 2008; 133:1106–1117. [DOI] [PubMed] [Google Scholar]
- Oda M, Kumaki Y, Shigeta M, et al. DNA methylation restricts lineage-specific functions of transcription factor Gata4 during embryonic stem cell differentiation. PLoS Genet 2013; 9: e1003574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MWB, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 2007; 448:191–195. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 2007; 448:196–199. [DOI] [PubMed] [Google Scholar]
- Guo G, Yang J, Nichols J, et al. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 2009; 136:1063–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najm FJ, Chenoweth JG, Anderson PD, et al. Isolation of epiblast stem cells from preimplantation mouse embryos. Cell Stem Cell 2011; 8:318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgani SM, Canham MA, Nichols J, et al. Totipotent embryonic stem cells arise in ground-state culture conditions. Cell Rep 2013; 3:1945–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 2012; 487:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YJ, Liu CP, Rissp D, et al. Deficiency of microRNA miR-34a expands cell fate potential in pluripotent stem cells. Science 2017; 355:e1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Liu B, Xu J, et al. Derivation of pluripotent stem cells with in vivo embryonic and extraembryonic potency. Cell 2017; 169:243–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroviak T, Loos R, Bertone P, Smith A, Nichols J. The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat Cell Biol 2014; 16:516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 2003; 115:281–292. [DOI] [PubMed] [Google Scholar]
- Ogawa K, Nishinakamura R, Iwamatsu Y, Shimosato D, Niwa H. Synergistic action of Wnt and LIF in maintaining pluripotency of mouse ES cells. Biochem Biophys Res Commun 2006; 343:159–166. [DOI] [PubMed] [Google Scholar]
- Tsakiridis A, Huang Y, Blin G. et al. Distinct Wnt-driven primitive streak-like populations reflect in vivo lineage precursors. Development 2015; 142: 1209–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurek D, Neagu A, Tastemel M, et al. Endogenous WNT signals mediate BMP-induced and spontaneous differentiation of epiblast stem cells and human embryonic stem cells. Stem Cell Rep 2015; 4:114–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med 2004; 10:55–63. [DOI] [PubMed] [Google Scholar]
- Chappell J, Sun Y, Singh A, Dalton S. MYC/MAX control ERK signaling and pluripotency by regulation of dual-specificity phosphatases 2 and 7. Genes Dev 2013; 27:725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Gocza E, Diaz E, et al. Embryonic stem cells alone are able to support fetal development in the mouse. Development 1990; 110:815–821. [DOI] [PubMed] [Google Scholar]
- Ohta H, Sakaide Y, Yamagata K, Wakayama T. Increasing the cell number of host tetraploid embryos can improve the production of mice derived from embryonic stem cells. Biol Reprod 2008; 79:486–492. [DOI] [PubMed] [Google Scholar]
- Zhao XY, Li W, Lv Z, et al. iPS cells produce viable mice through tetraploid complementation. Nature 2009; 461:86–90. [DOI] [PubMed] [Google Scholar]
- Boroviak T, Loos R, Lombard P, et al. Lineage-specific profiling delineates the emergence and progression of naive pluripotency in mammalian embryogenesis. Dev Cell 2015; 35:366–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Osorno R, Tsakiridis A. Wilson V. In vivo differentiation potential of epiblast stem cells revealed by chimeric embryo formation. Cell Rep 2012; 2:1571–1578. [DOI] [PubMed] [Google Scholar]
- Han DW, Tapia N, Joo JY, et al. Epiblast stem cell subpopulations represent mouse embryos of distinct pregastrulation stages. Cell 2010; 143:617–627. [DOI] [PubMed] [Google Scholar]
- Kojima Y, Kaufman-Francis K, Studdert JB, et al. The transcriptional and functional properties of mouse epiblast stem cells resemble the anterior primitive streak. Cell Stem Cell 2014; 14:107–120. [DOI] [PubMed] [Google Scholar]
- Rivera CM, Ren B. Mapping human epigenomes. Cell 2013; 155:39–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013; 153:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Yan P, Lu J, et al. Opposing roles for the lncRNA Haunt and its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Cell Stem Cell 2015; 16:504–516. [DOI] [PubMed] [Google Scholar]
- Eckersley-Maslin MA, Svensson V, Krueger C, et al. MERVL/Zscan4 network activation results in transient genome-wide DNA demethylation of mESCs. Cell Rep 2016; 17:179–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kigami D, Minami N, Takayama H, Imai H. MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos. Biol Reprod 2003; 68:651–654. [DOI] [PubMed] [Google Scholar]
- Peaston AE, Evsikov AV, Graber JH, et al. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell 2004; 7:597–606. [DOI] [PubMed] [Google Scholar]
- Habibi E, Brinkman AB, Arand J, et al. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell 2013; 13:360–369. [DOI] [PubMed] [Google Scholar]
- Leitch HG, McEwen KR, Turp A, et al. Naive pluripotency is associated with global DNA hypomethylation. Nat Struct Mol Biol 2013; 20:311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q, Decato B, Hong EE, et al. A reference methylome database and analysis pipeline to facilitate integrative and comparative epigenomics. PLoS One 2013; 8:e81148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 2010; 28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Los Angeles A, Ferrari F, Xi R, et al. Hallmarks of pluripotency. Nature 2015; 525:469–478. [DOI] [PubMed] [Google Scholar]
- Choi J, Huebner A, Clement K, et al. Prolonged Mek1/2 suppression impairs the developmental potential of embryonic stem cells. Nature 2017; 548:219–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi M, Kishigami S, Tanaka A, et al. Derivation of ground-state female ES cells maintaining gamete-derived DNA methylation. Nature 2017; 548:224–227. [DOI] [PubMed] [Google Scholar]
- Payer B, Saitou M, Barton SC, et al. Stella is a maternal effect gene required for normal early development in mice. Curr Biol 2003; 13:2110–2117. [DOI] [PubMed] [Google Scholar]
- Yeom Y, Fuhrmann G, Ovitt CE, et al. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development 1996; 122:881–894. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis 2007; 45:593–605. [DOI] [PubMed] [Google Scholar]
- Miura F, Enomoto Y, Dairiki R, et al. Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res 2012; 40:e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Fang F, Miller DF, et al. Global DNA methylation profiling technologies and the ovarian cancer methylome. Methods Mol Biol 2015; 1238:653–675. [DOI] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011; 27:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa S, Kobayashi H, Watanaba T, et al. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development 2011; 138:811–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Wang S, Zhang YJ, et al. Genome-wide DNA methylation profiles in hepatocellular carcinoma. Hepatology 2012; 55:1799–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Features of AFSCs.
Chimeras produced by ASCs.
RNAseq analysis of AFSCs and ASCs.
2-cell genes expression in ASCs.
Methylation landscape of ASCs.
The contribution of AFSCs and ASCs in chimera and tetraploid.
The summary of contribution of ASCs and ESC-ASCs in chimeras.