Dear Editor,
Telomere length homeostasis, dictating cellular proliferative potential, is crucial for proper cellular function. In addition to the well-known telomere shortening during cell division and lengthening by activation of telomerase or an alternative lengthening of telomeres (ALT) mechanism, telomere length is also subject to regulated rapid deletion events referred to as telomere trimming1. This trimming process involves excision of telomeric structures called T-loops, which requires homologous recombination proteins XRCC3 and NBS12,3. Recent studies showed that the balance between telomere trimming and lengthening determines telomere length in germline and stem cells, suggesting that telomere trimming should be under stringent control4,5. However, the regulation of telomere trimming remains largely unknown.
Recently, two groups established human TZAP (ZBTB48, renamed telomeric zinc finger-associated protein) as a factor that directly binds double-stranded telomeric TTAGGG sequence and stimulates telomere trimming6,7,8,9. TZAP is composed of an N-terminal BTB/POZ domain and eleven adjacent C2H2-type zinc fingers (Znf1-11) at its C-terminus (Figure 1A). Li et al.6 mapped TZAP's TTAGGG binding region to Znf9-11, whereas further study refined it specifically to Znf117. This is the first time, to our knowledge, that a C2H2 finger was reported to bind telomeric DNA directly, although the DNA binding specificity and mechanisms of C2H2 finger have been intensely studied. In addition, one canonical C2H2 finger only recognizes three or four base pair sites. This raises the question of how the telomeric hexamer TTAGGG DNA is specifically recognized by TZAP. Notably, TZAP can be classified into a small subset of proteins that directly binds telomeric repeat DNA in mammals, including three members of the shelterin complex (TRF1, TRF2, and single-stranded DNA binding protein POT1) and HOT110,11. TRF1, TRF2 and HOT1 employ homeodomains for double-stranded telomeric DNA recognition and do not recognize subtelomeric DNA, whereas TZAP has preference for distinct types of subtelomeric DNA7,10,11. In contrast to the well-defined DNA binding mechanisms of TRF1, TRF2 and HOT1, the mechanisms by which TZAP specifically interacts with (sub)telomeric DNA requires further investigation.
Figure 1.
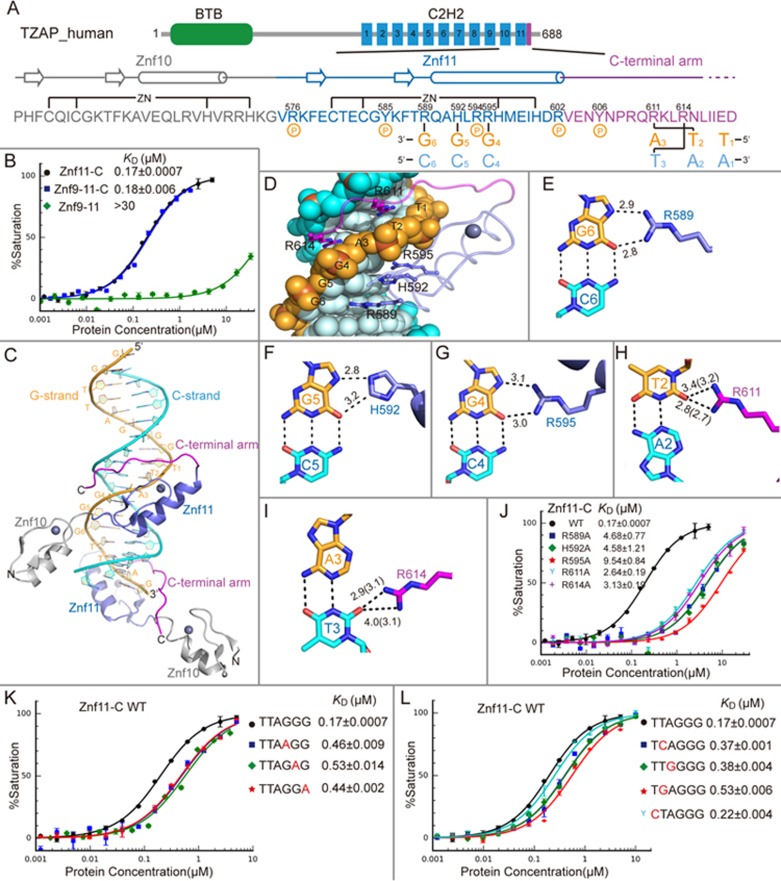
(A) Schematic representation of the domain architecture of human TZAP (top panel). Schematic representation of the Znf10-11-C interactions with TTAGGG sequence (bottom panel). The sequence of Znf10 (Gray), Znf11 (Slate) and C-terminal arm (Purple), together with the secondary structure are shown. Two cysteine and two histidine residues in each finger are responsible for Zn2+ binding (top connecting lines). Three residues in Znf11 and two residues in C-terminal arm interact specifically with the DNA bases (bottom connecting lines). The residues responsible for phosphate backbone interactions are indicated by the circles with a “p”. (B) Comparison of binding affinities of Znf9-11-C, Znf11-C, and Znf9-11 with the TTAGGG probe. (C) Znf10-11-C in complex with telomeric DNA sequence is shown in cartoon representation. Color codes of Znf10, Znf11 and C-terminal arm are defined as in A. The G-strand and C-strand are colored orange and cyan, respectively. Znf10 lies outside the DNA duplex, Znf11 binds in the major groove, and the C-terminal arm crosses the phosphate backbone and binds in the minor groove. (D) Znf11 and C-terminal arm are shown as ribbons, whereas DNA is in sphere representation. The residues responsible for base-specific interactions are shown in stick-ball representation. (E-I) Details of TZAP-DNA base-specific interactions. The hydrogen bonds are depicted as black dashed lines. The hydrogen-bonding distances of base-specific interactions between Chain D and DNA are shown, and values in parentheses are for Chain C. (J) The effects of Znf11-C mutations on TTAGGG binding. (K) Mutating each base pair at positions 4, 5, and 6 to A:T pair reduced the binding by Znf11-C. (L) Comparison of binding affinities of Znf11-C with TTAGGG and subtelomeric DNA sequences.
We first generated a construct of TZAP that included Znf9-11 (residues 516-605). Using FP (fluorescence polarization) assay, we showed that the Znf9-11 construct displayed a relatively low binding affinity (> 30 μM) to a double-stranded oligonucleotide containing TTAGGG sequence (referred to as TTAGGG probe) (Figure 1B and Supplementary information, Table S1). We then noticed that there is an evolutionarily conserved and highly basic region (residues 606-620) located immediately C-terminal to Znf11 (Supplementary information, Figure S1). The construct containing Znf9-11 and the conserved C-terminal region (residues 516-620, referred to as Znf9-11-C) bound the TTAGGG dsDNA probe with a KD of about 0.18 μM in 150 mM NaCl solution, indicating a critical role of the C-terminal region in telomeric DNA binding (Figure 1B).
To elucidate the molecular mechanism of telomeric DNA recognition by TZAP, we crystallized the Znf9-11-C construct bound to an 18 bp double-stranded oligo containing the TTAGGG sequence. The oligo was synthesized with a 5′-overhanging guanine on the G-strand and a 5′-overhanging cytosine on the C-strand (Supplementary information, Table S1). The protein-DNA structure was refined to 2.85 Å in P43212 group (Supplementary information, Table S2). Each asymmetric unit contains two protein molecules bound to a DNA duplex. The DNA molecules in the crystal are coaxially stacked in an end-to-end fashion, and the terminal G and C bases of neighboring DNA molecules pair to form pseudo-continuous TTAGGG duplex (Supplementary information, Figure S2A and S2B). In each asymmetric unit, one protein molecule (Chain D) binds a TTAGGG site in the duplex, whereas the other one (Chain C) binds the end-to-end stacking TTAGGG site (Supplementary information, Figure S2A-S2D). We did not observe electron densities for Znf9 and the last few C-terminus residues (615-620 in Chain D and 619-620 in Chain C). Thus, the refined protein model comprises Znf10-11 and most of the conserved C-terminal region. Znf10 lies outside and does not have direct contacts with the DNA duplex, possessing a higher averaged crystallographic thermal B-factor compared to Znf11-C and the DNA duplex (Figure 1C, Supplementary information, Table S2). Znf11 fits into the DNA major groove where it makes base-specific interactions. The conserved C-terminal region crosses the DNA phosphate backbone and turns back to lie in the DNA minor groove, conferring a shape of an arm, and the entire part of the C-terminal loop is therefore referred to as C-terminal arm (residues 603-618) (Figure 1C). To validate whether Znf11 and the C-terminal region are sufficient for TTAGGG recognition, we carried out FP assay using a Znf11-C construct and found that this construct bound the TTAGGG probe with a similar affinity to Znf9-11-C (Figure 1B). These observations explain why only the mutation of Znf11 prevents the binding of TZAP to the telomeric DNA, as reported earlier7.
Both Znf10 and Znf11 adopt canonical C2H2 zinc-finger fold, consisting of two β-strands and one C-terminal helix (Figure 1A and 1C). The specific recognition of G4G5G6 triplet in T1T2A3G4G5G6 is primarily achieved by H-bonds between the guanines and three conserved residues in the helix of Znf11 and its preceding loop (Figure 1A and 1D). Specifically, the terminal Nη1 and Nη2 groups of Arg595 and Arg589 donate hydrogen bonds to the guanine O6 and N7 atoms of G4 and G6 in a bifurcated hydrogen-bonding pattern, respectively (Figure 1E and 1G). Furthermore, the Nε2 group of His592 donates one hydrogen-bond to the N7 atom of G5, and the adjacent Cε1 atom donates a C-H...O type bond (Figure 1F)12. These hydrogen-bonding interactions confer the specific recognition of G4G5G6 triplet. The guanine-arginine and guanine-histidine recognition patterns are common in other C2H2 fingers. Mutating each of the three residues (Arg589, His592 and Arg595) to alanine significantly reduced the binding affinity to the TTAGGG dsDNA probe (Figure 1J). Accordingly, mutating each base pair at positions 4, 5, and 6 to A:T pair resulted in decrement of binding by Znf11-C (Figure 1K). To validate the importance of G4G5G6 triplet recognition by Znf11 in vivo, we expressed exogenous FLAG-TZAP (wild type) and a triple-point mutant (R589A/H592A/R595A) in U2OS cells. Wild-type FLAG-TZAP showed the expected localization to telomeres as revealed by co-localization with endogenous TRF1, whereas the triple mutant was distributed diffusely throughout the nucleoplasm with no obvious accumulation at telomeres (Supplementary information, Figure S3).
In addition to the recognition of G4G5G6 triplet by Znf11, the C-terminal arm lying in the minor groove provides extra preference and affinity to the telomeric DNA sequence. We observed sequence-specific interactions from residues Arg611 and Arg614 in the arm (Figure 1A and 1D). The Nη1 and Nη2 groups of Arg611 form hydrogen bonds with O2 atom of T2 in the G-strand, and the side chain of Arg614 make hydrogen-bonding interactions with O2 atom of T3 in the C-strand (Figure 1H and 1I). Consistently, mutations of Arg611 and Arg614 to alanine significantly decreased the binding affinity to the TTAGGG dsDNA probe (Figure 1J). Furthermore, a double-point mutant (R611A/R614A) of FLAG-TZAP showed no obvious accumulation at telomeres, emphasizing the importance of the recognition of T2A3 by the C-terminal arm in vivo (Supplementary information, Figure S3). Previous studies showed that full-length TZAP binds subtelomeric variant repeat sequences TCAGGG and TTGGGG more efficiently than TGAGGG using DNA pull-down assays7. Displacement of T2:A2 pair to C:G (TCAGGG) would preserve the hydrogen bonds between O2 atom and Arg611, but the N4 group of guanine would introduce a steric clash to Arg611 (Supplementary information, Figure S4A). Moreover, the TGAGGG sequence (substitution of T2:A2 pair to G:C) would have both unfavorable steric and hydrogen-bonding interactions with the side chain of Arg611 (Supplementary information, Figure S4A). Accordingly, the binding affinity of the TCAGGG probe to Znf11-C was higher than that of TGAGGG and lower than that of TTAGGG (Figure 1L), consistent with previous pull-down results. The interaction between TTGGGG sequence and Znf11-C resembled that between the TCAGGG sequence and Znf11-C, with a comparable binding affinity (Figure 1L, Supplementary information, Figure S4B). Additionally, we noticed that Znf11-C did not have contacts with T1:A1 and Znf11-C bound a CTAGGG probe with a similar affinity to the TTAGGG probe (Figure 1L).
In addition to the base-specific interactions mentioned above, Znf11-C makes extensive contacts with the phosphate backbone as shown in Supplementary information, Figure S5A. Most of the contacts are clustered in two regions flanking the G4G5G6 triplet. Specifically, Arg576, Tyr585, Arg602, and Tyr606 interact with the three phosphates 5′ to G4 on the G-strand, while Arg594 interacts with the two phosphates 5′ to C6 on the C-strand. Besides interacting with the phosphate backbone, the side chains of Arg576 and Tyr585 also form a hydrogen-bond with the backbone carbonyl group and amide group of Arg611, respectively, presumably helping to fix the orientation of the C-terminal arm (Supplementary information, Figure S5B). These interactions must contribute to telomeric DNA recognition. Consistent with this, the R576A, Y585F and Y606F mutants showed significantly reduced affinity towards the TTAGGG probe (Supplementary information, Figure S5C).
The essential role of the C-terminal arm of TZAP in telomeric DNA recognition is reminiscent of the N-terminal arms of TRF1/2 and HOT1 that also confer base-specific recognition in the minor groove10,11. Frequently, base-specific recognition in the minor groove by the N-terminal arm of homeodomain is achieved by the Arginine-Thymine pattern that is also observed for the C-terminal arm of TZAP. Notably, the N-terminal arm usually comprises about six residues, which are apparently shorter than the length of the C-terminal arm of TZAP that is required for its 'turn back' conformation. Moreover, structural superimposition of the DNA moieties of the TRF1, HOT1 and TZAP crystal structures showed major differences in binding. First, the N-terminal arms of TRF1/2 and HOT1 provide sequence-specific recognition towards TTA site of the subsequent telomeric repeat, whereas the C-terminal arm of TZAP recognizes T2A3 in the same telomeric repeat (Supplementary information, Figure S6A-S6C). Second, the helix of TZAP sits in the major groove in a different orientation and makes base-specific contacts in a different manner compared to TRF1/2 and HOT1 (Supplementary information, Figure S6B and S6C).
In summary, our crystal structure provides the molecular basis for the recognition of telomeric DNA by TZAP. We demonstrated that Znf11 is responsible for the recognition of G4G5G6 triplet, and an additional C-terminal arm serves to recognize T2A3. Since TZAP itself does not harbor any obvious enzymatic activity for telomere trimming, it may act as an adaptor protein to recruit other proteins, such as XRCC3 or NBS1, to promote telomere trimming process. Thus, further mechanistic understanding of the function of TZAP requires the knowledge of TZAP protein interaction network. TRF1 and TRF2 each contains an essential N-terminal TRFH domain that mediates protein dimerization for the stable association of these proteins with telomeres and serves as a platform for protein-protein interactions13. Like the TRFH domains of TRF1 and TRF2, the BTB domain of TZAP forms a dimer as shown by an unpublished crystal structure (PDB ID: 3B84) and a static light scattering (SEC-MALS) analysis (Supplementary information, Figure S7A and S7B). In addition, a FLAG-TZAPdelBTB construct localizes to telomeres with a much lesser extent than the wild-type protein, suggesting that the BTB domain of TZAP plays a role in telomere binding in vivo (Supplementary information, Figure S3). Furthermore, it has been reported that BTB domain is capable of mediating protein-protein interactions14. Thus, deciphering the exact role of the BTB domain of TZAP in future studies will provide essential mechanistic insights of the function of TZAP in telomere length homeostasis.
The structure coordinate and structure factor were deposited in the Protein Data Bank (PDB) under accession number 5YJ3.
Acknowledgments
We thank the staffs of BL19U1 beamline at National Center for Protein Science Shanghai and Shanghai Synchrotron Radiation Facility, Shanghai, China, for assistance during data collection. This work was supported by the Ministry of Science and Technology of China (2016YFA0500700), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB08010101), the National Natural Science Foundation of China (31330018, 31500590 and 31400643), and the Fundamental Research Funds for the Central Universities (WK2070000095).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Supplementary information, Tables S1–S2
Supplementary information, Figures S1–S7
References
- Pickett HA, Reddel RR. Cell Cycle 2012; 11:1309–1315. [DOI] [PubMed]
- Wang RC, Smogorzewska A, de Lange T. Cell 2004; 119:355–368. [DOI] [PubMed]
- Compton SA, Choi JH, Cesare AJ, et al. Cancer Res 2007; 67:1513–1519. [DOI] [PubMed]
- Rivera T, Haggblom C, Cosconati S, et al. Nat Struct Mol Biol 2017; 24:30–39. [DOI] [PMC free article] [PubMed]
- Pickett HA, Henson JD, Au AYM, et al. Hum Mol Genet 2011; 20:4684–4692. [DOI] [PubMed]
- Li JS, Miralles Fusté J, Simavorian T, et al. Science 2017; 355:638–641. [DOI] [PMC free article] [PubMed]
- Jahn A, Rane G, Paszkowski-Rogacz M, et al. EMBO Rep 2017; 18:929–946. [DOI] [PMC free article] [PubMed]
- Lossaint G, Lingner J. Science 2017; 355:578–579. [DOI] [PubMed]
- Garcia-Exposito L, O'Sullivan RJ. EMBO Rep 2017; 18:861–863. [DOI] [PMC free article] [PubMed]
- Court R, Chapman L, Fairall L, et al. EMBO Rep 2005; 6:39–45. [DOI] [PMC free article] [PubMed]
- Kappei D, Butter F, Benda C, et al. EMBO J 2013; 32:1681–1701. [DOI] [PMC free article] [PubMed]
- Horowitz S, Trievel RC. J Biol Chem 2012; 287:41576–41582. [DOI] [PMC free article] [PubMed]
- Chen Y, Yang YT, van Overbeek M, et al. Science 2008; 319:1092–1096. [DOI] [PubMed]
- Mathew R, Seiler MP, Scanlon ST, et al. Nature 2012; 491:618–621. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information, Tables S1–S2
Supplementary information, Figures S1–S7