Abstract
Two articles in Cell Research focus on the structure-function relationships in the shelterin complex that binds to telomeres and is essential for their stability and functions. These studies concerning both mammalian and Schizosaccharomyces pombe proteins reveal unexpected structural conservation of a motif called TRFH (Telomeric Repeat Factors Homology) domain between several subunits in these complexes, providing a rationale for further dissection of the role of telomeres in chromosome stability, aging and cancer, and encouraging us to revisit the evolution of telomere proteins.
The somatic erosion of telomeres has emerged as a key driver of aging. It controls cell senescence, tissue function, and the balance between tissue renewal and energy consumption. Thus, the mechanisms behind telomere functions are fundamental questions in biology.
The concept that telomere functionality relies on specific DNA-protein interactions stems from the discovery of the TEBPα/β heterodimer from Oxytrichia nova that binds to and protects the G-rich 3′ overhang that constitutes the end of telomeric DNA across many species1. Since this early discovery, numerous variations in telomere nucleoprotein organization have been found1. For instance, in the budding yeast Saccharomyces cerevisiae, the 3′ overhang is bound by Cdc13, whereas the duplex portion is covered by an array of Rap1 proteins. The telomeres of mammals and Schizosaccharomyces pombe exhibit similar organizations: their 3′ overhangs are bound by orthologs of TEBPα/β, POT1-TPP1 in mammals and Pot1-Tpz1 in S. pombe, whereas the duplex is specifically bound by a family of TRF proteins characterized by the presence of a TRFH domain and by a Myb-like telomeric DNA-binding domain (called a telobox) at the C-terminus. At S. pombe telomeres, there is one known TRF protein, Taz1, whereas two TRF proteins are found at the end of mammalian telomeres: TRF1 and TRF2. The telobox is highly conserved both in overall shape and composition and recognizes the duplex telomeric DNA sequences2. Despite low sequence conservation, TRFHs exhibit similar architectures, including a bundle of nine or ten helices3. The TRFHs of TRF1 and TRF2 mediate their homodimerization and interact with a set of proteins required for telomere protection. They also interact with DNA, forming a complex where DNA is wrapped around them in a right-handed manner to control telomeric DNA topology4. Although the structure of the TRFH in S. pombe Taz1 has been determined5, little is known about its functions and interacting partners.
TRF proteins and 3′-overhang-binding heterodimers are linked by a protein bridge, formed by Rap1 and Poz1 in S. pombe and by TIN2 (an ortholog of Poz1) in mammals. The bridge is closed in S. pombe by Poz1, which contacts Rap1 and Tpz1, and in mammals by TIN2, which interacts simultaneously with TRF1, TRF2 and TPP1. Together with mammalian RAP1, which is recruited to telomeres via TRF2, and S. pombe Ccq1 bound to Tpz1, these bridged telomeric protein complexes have been named shelterin1. The shelterin is involved in most (if not all) telomeric functions: protection against activation of the DNA damage response (DDR), chromosome stability, telomerase regulation, long and short range transcriptional regulation and meiosis. It can exist either as a full complex or as subcomplexes, which perform specific functions during the cell cycle and cellular differentiation6. How shelterin determines cell fate has thus emerged as a major question in telomere research.
A key structure controlling shelterin complex assembly and function is the protein bridge. The bridge is assembled on what can be called the “sensitive” parts of telomeres, i.e., the 3′ overhang, which can be bound by RPA and engage the ATR kinase, and a DNA extremity that can be recognized as an accidental double-stranded break, activating the ATM kinase. Thus, the shelterin bridge is specifically dedicated to the masking of telomeres from unwanted DDR activation. In addition, the bridge is required for the cooperative assembly of shelterin subcomplexes7, heterochromatin formation, and telomerase regulation8. The issues of how the shelterin bridge is structured and regulated are addressed in two elegant studies in Cell Research combining structural, genetic and cell biology approaches.
The results from Xue et al.9 reveal that S. pombe Poz1 is a homodimeric protein with separate interfaces for binding Rap1 and Tpz1. The expression of Poz1 mutants carrying point mutations abolishing these interfaces confirms that the shelterin bridge is required for telomerase regulation and proper sub-telomeric heterochromatin formation.
In the second study, Hu et al.10 analyzed the structure of the N-terminal part of human TIN2, which shows a similar architecture to that of S. pombe Poz1. The expression of various mutants of TIN2 abolishing either its binding to TRF1, TRF2 or TPP1 in human and mouse cells results in telomere deprotection, confirming the importance of the shelterin bridge for telomere functionality. Further analyses also indicate how the different protein interactions forming the bridge repress distinct signaling pathways in the DDR.
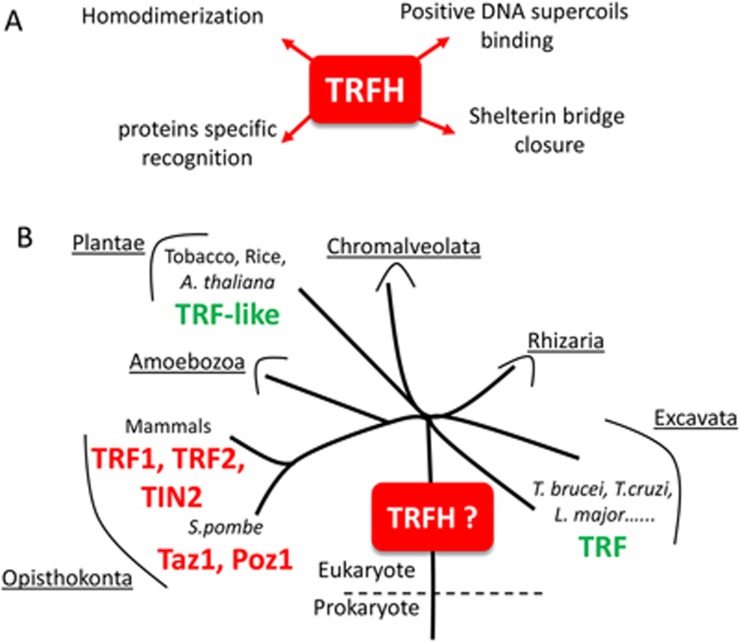
The biggest surprise from these two studies is the similar architecture of the bridge domains in Poz1/TIN2 and the TRFHs harbored by their corresponding double-stranded DNA-binding proteins Taz1/TRF1, TRF2 (Figure 1A). As a consequence of their conserved architecture, the TRFHs create a specific docking site for peptide sequences, thus explaining their function as a hub for various shelterin partners.
Figure 1.
(A) Known properties of the conserved TRFH domains in shelterin complexes. Of note, TRFHs do not exhibit this full set of properties. For instance, TIN2 seems devoid of DNA binding. (B) A schematic tree depicting the phylogenetic relationships of most eukaryotes into six major supergroups (underlined)11. The proteins in red contain a TRFH whose structure was solved, the proteins in green are TRFH protein candidates on the basis of a TRF-like domain organization, i.e., the presence of a telobox at their C-terminus. Structure determination of various putative TRFH in distantly related shelterin proteins should shed light on whether TRFH occurred at the very beginning or even before the expansion of the eukaryotic tree of life.
The discovery of this striking structural conservation between previously known TRFHs and the bridge domains of TIN2 and Poz1 has important implications for telomere biology and evolution. Specifically, it provides a rationale for understanding the hierarchical assembly and stoichiometry of telomere subunits, and their regulation during the cell cycle, cellular differentiation and senescence. It also suggests a common evolutionary origin for these domains despite their low level of sequence identity, which makes their identification by standard homology search algorithms difficult. Various plants and Trypanosoma, organisms that are distantly related to metazoans and fungi, express proteins that contain a telobox at their C-terminus1 and an N-terminal domain that is likely to fold into a TRFH structure (although its structure has not been solved yet). If confirmed, this would suggest that the TRFH and telobox are both ancient telomere motifs that arose at least at the time of prokaryote-eukaryote divergence (Figure 1B). Interestingly, TIN2 can be localized to mitochondria, where it controls cellular metabolism12. Given the bacterial origin of mitochondria, it is tempting to speculate that TRFHs may even have emerged before the divergence of prokaryotes and eukaryotes.
References
- Giraud-Panis MJ, Pisano S, Benarroch-Popivker D, et al. Front Oncol 2013; 3:48. [DOI] [PMC free article] [PubMed]
- Bilaud T, Koering CE, Binet-Brasselet E, et al. Nucleic Acids Res 1996; 24:1294–1303. [DOI] [PMC free article] [PubMed]
- Fairall L, Chapman L, Moss H, et al. Mol Cell 2001; 8:351–361. [DOI] [PubMed]
- Benarroch-Popivker D, Pisano S, Mendez-Bermudez A, et al. Mol Cell 2016; 61:274–286. [DOI] [PMC free article] [PubMed]
- Deng W, Wu J, Wang F, et al. Cell Res 2015; 25:881–884. [DOI] [PMC free article] [PubMed]
- Ye J, Renault VM, Jamet K, et al. Nat Rev Genet 2014; 15:491–503. [DOI] [PubMed]
- O'Connor MS, Safari A, Xin H, et al. Proc Natl Acad Sci USA 2006; 103:11874–11879. [DOI] [PMC free article] [PubMed]
- Pan L, Hildebrand K, Stutz C, et al. Genes Dev 2015; 29:1164–1174. [DOI] [PMC free article] [PubMed]
- Xue J, Chen H, Wu J, et al. Cell Res 2017; 27:1503–1520. [DOI] [PMC free article] [PubMed]
- Hu C, Rai R, Huang C, et al. Cell Res 2017; 27:1485–1502. [DOI] [PMC free article] [PubMed]
- Simpson AG, Roger AJ. Curr Biol 2004; 14:R693–R696. [DOI] [PubMed]
- Chen LY, Zhang Y, Zhang Q, et al. Mol Cell 2012; 47:839–850. [DOI] [PMC free article] [PubMed]