ABSTRACT
Many pathogenic bacteria use sophisticated survival strategies to overcome harsh environmental conditions. One strategy is the formation of slow-growing subpopulations termed small colony variants (SCVs). Here we characterize an SCV that spontaneously emerged from an axenic Salmonella enterica serovar Typhimurium water culture. We found that the SCV harbored a frameshift mutation in the glutamine synthetase gene glnA, leading to an ∼90% truncation of the corresponding protein. Glutamine synthetase, a central enzyme in nitrogen assimilation, converts glutamate and ammonia to glutamine. Glutamine is an important nitrogen donor that is required for the synthesis of cellular compounds. The internal glutamine pool serves as an indicator of nitrogen availability in Salmonella. In our study, the SCV and a constructed glnA knockout mutant showed reduced growth rates, compared to the wild type. Moreover, the SCV and the glnA mutant displayed attenuated entry into host cells and severely reduced levels of exoproteins, including flagellin and several Salmonella pathogenicity island 1 (SPI-1)-dependent secreted virulence factors. We found that these proteins were also depleted in cell lysates, indicating their diminished synthesis. Accordingly, the SCV and the glnA mutant had severely decreased expression of flagellin genes, several SPI-1 effector genes, and a class 2 motility gene (flgB). However, the expression of a class 1 motility gene (flhD) was not affected. Supplementation with glutamine or genetic reversion of the glnA truncation restored growth, cell entry, gene expression, and protein abundance. In summary, our data show that glnA is essential for the growth of S. enterica and controls important motility- and virulence-related traits in response to glutamine availability.
IMPORTANCE Salmonella enterica serovar Typhimurium is a significant pathogen causing foodborne infections. Here we describe an S. Typhimurium small colony variant (SCV) that spontaneously emerged from a long-term starvation experiment in water. It is important to study SCVs because (i) SCVs may arise spontaneously upon exposure to stresses, including environmental and host defense stresses, (ii) SCVs are slow growing and difficult to eradicate, and (iii) only a few descriptions of S. enterica SCVs are available. We clarify the genetic basis of the SCV described here as a frameshift mutation in the glutamine synthetase gene glnA, leading to glutamine auxotrophy. In Salmonella, internal glutamine limitation serves as a sign of external nitrogen deficiency and is thought to regulate cell growth. In addition to exhibiting impaired growth, the SCV showed reduced host cell entry and reduced expression of SPI-1 virulence and flagellin genes.
KEYWORDS: Salmonella enterica, small colony variant, glutamine synthetase, motility, SPI-1
INTRODUCTION
Nontyphoidal Salmonella strains, such as Salmonella enterica serovar Typhimurium, are a leading cause of bacterial diarrhea worldwide, and the pathogens are isolated from a wide range of hosts, including humans. Infections in humans exhibit manifestations ranging from enteric symptoms to systemic disease, and they often occur as outbreaks. S. enterica invades host cells and proliferates intracellularly in epithelial and phagocytic cells. Among others, important virulence factors are two needle-like type III secretion systems, encoded on Salmonella pathogenicity island 1 (SPI-1) and SPI-2, and their individual sets of effector proteins (1–3). Furthermore, S. enterica possesses an extraordinary capacity to survive and to persist outside infected hosts in soil, sediments, and water, which poses a major public health concern. Water is thought to be a main reservoir, facilitating bacterial dissemination into the environment and transmission between hosts (4–6).
Natural water reservoirs often provide challenging living conditions, in terms of temperature, salinity, or nutrient availability. Such conditions lead to stress and trigger microbial phenotypic diversity (7–9). A well-known result of such diversification is the occurrence of small colony variants (SCVs), which constitute slow-growing subpopulations with unusual phenotypic traits. SCVs have been described for a range of pathogens, including staphylococci, Escherichia coli, pseudomonads, and Salmonella enterica (10). In addition to the fact that stress, including stress conferred by the immune systems of hosts, stimulates SCV formation, SCVs emerge randomly within fast-growing populations, as shown for Staphylococcus aureus (11). SCV formation is often caused by genetic changes that lead to growth rate reductions and auxotrophy, which can result in pleiotropic phenotypic manifestations. There is no defined core set of SCV genes. Most SCVs studied carry mutations in genes required for hemin, menadione, or thymidine biosynthesis, resulting in diminished electron transport and reduced synthesis of ATP. Importantly, specific phenotypes of auxotrophic SCVs are typically reversed by supplementation with the required metabolite (10).
Knowledge of persistence mechanisms and their reversion is of major importance for developing strategies for pathogen control. Currently, only a few descriptions of S. enterica SCVs are available (12–15). Cano et al. (12) isolated SCVs when fibroblasts restricting S. Typhimurium intracellular growth were infected with the bacteria for long periods. Several SCVs turned out to be nutrient auxotrophs, and some of them revealed a mutation in aroD, a gene involved in the synthesis of aromatic amino acids. The aroD mutants showed increased intracellular persistence rates in fibroblasts and reduced susceptibility to aminoglycoside antibiotics. Pränting and Andersson (13) described the spontaneous appearance of S. Typhimurium SCVs when the parental strain was treated with protamine, a cationic peptide with antimicrobial activity. The SCVs revealed mutations in genes involved in heme biosynthesis and respiration, resulting in reduced bacterial fitness but lower susceptibility to several antimicrobials. Recently, Li and colleagues (15) characterized two S. Typhimurium SCVs obtained with streptomycin treatment. Using a whole-genome sequencing approach, they demonstrated that frameshift mutations in the ubiE gene were responsible for the phenotypic switch from the wild type to the small colony variant. The ubiE gene product is required for coenzyme Q8 and menaquinone biosynthesis; therefore, the resulting mutant strains are expected to have defects in the electron transport chain. Moreover, the authors uncovered two modes of genetic alterations for reversion to a fast-growing phenotype, i.e., (i) acquisition of a secondary mutation in ubiE or (ii) acquisition of a compensatory mutation in pfrB, encoding peptide chain release factor 2.
The aim of this study was to characterize a novel SCV that spontaneously emerged from a water culture inoculated with S. Typhimurium. Our data show that the variant harbors a loss-of-function mutation in the gene for glutamine synthetase, glnA. Defects in glnA attenuated bacterial growth and also affected motility- and virulence-related traits in S. Typhimurium.
RESULTS
Isolation of an S. Typhimurium SCV.
The SCV described in this study was isolated from an S. Typhimurium starvation experiment. A microcosm was set up in autoclaved distilled water with the aim of generating viable but unculturable bacteria and was stored at 23°C in the dark. The microcosm was repeatedly assayed for bacterial culturability as determined by plating aliquots on Luria-Bertani (LB) agar. Shortly after no CFU were detectable (after 913 days), a population of tiny uniform colonies emerged when the culture was plated on LB agar (day 941). To exclude contamination, we confirmed those colonies as Salmonella by using selective media (Oxoid Salmonella chromogenic medium [OSCM] and Rambach agars) and invA PCR (16). We colony purified one clone, termed SCV WPASI01, for further characterization.
Attenuated growth of the S. Typhimurium SCV, compared to the growth of the wild type and revertants.
First, we examined growth of the SCV on LB agar, in LB broth, and on M9-glucose (M9G) minimal agar; the last contains glucose as the sole carbon source. As expected, the SCV showed a reduced growth rate or no growth at all, compared to the wild type (Fig. 1A to C). The SCV specifically exhibited slowed exponential growth (SCV doubling time in the exponential phase of 93 min, compared with 30 min for the wild type). Moreover, we subjected the SCV to repeated passages in rich medium in order to check whether fast-growing revertants appeared. Indeed, after two cycles, revertants showing wild-type-like colony morphology arose spontaneously (Fig. 1A). A phenotypically reverted colony, named WPASI01r (in short, Rev), was included in the growth experiments and behaved like the wild type (doubling time in the exponential phase of 31 min) (Fig. 1A to C).
FIG 1.
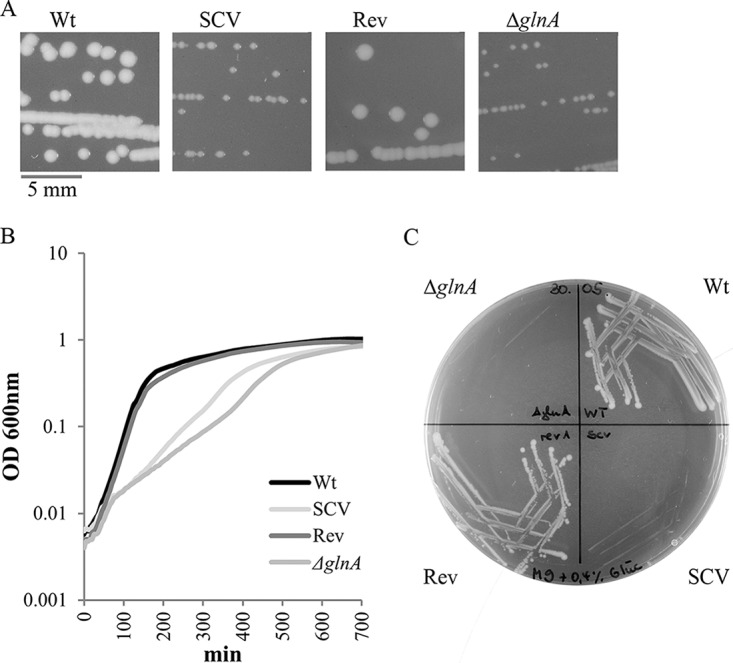
Attenuated growth of the S. Typhimurium SCV and a glnA knockout mutant, compared to the wild type and a revertant. (A) Colony morphology on LB agar. (B) Growth curves for growth in LB broth. (C) Growth behavior on M9G minimal medium. Data shown in panel B are means from quadruplicate cultures for which OD600 values were read at 15-min intervals, and all data are representative of three independent experiments. Wt, wild type; SCV, small colony variant; Rev, revertant; ΔglnA, glnA knockout mutant.
Reading frame truncation in the glutamine synthetase gene glnA determining the SCV phenotype, which can be mimicked by glnA knockout.
In order to clarify the genetic background of the SCV phenotype, we performed shotgun whole-genome sequencing by means of Illumina MiSeq technology. Within the genome, we identified three differences from the parental strain. First, the SCV showed an adenosine insertion at position 120 in the glutamine synthetase gene glnA, resulting in a frame shift and therefore a premature stop codon in glnA at positions 133 to 135. This translated into a truncated GlnA protein (from originally 469 amino acids long to 44 amino acids), presumably with a loss of function (Fig. 2A). Second, the SCV harbored the point mutation G85T in the hfq gene, leading to the amino acid substitution G29C in the RNA-binding protein Hfq (17). Third, the SCV revealed the lack of an 18.9-kb genomic stretch from position 1685885 (in srfC) to position 1704752 (in STM1615) of the S. Typhimurium LT2 genome (GenBank accession no. NC_003197.1) (18).
FIG 2.
Reading frame truncation in the glutamine synthetase gene glnA, triggering the S. Typhimurium SCV phenotype. The addition of glutamine restores SCV and glnA mutant growth. (A) Genetic characteristics of the wild-type, SCV, and revertant strains with regard to the glutamine synthetase gene glnA. The numbering of nucleotides is relative to the translational start site. Insertion of adenosine 120 in the glnA gene of S. Typhimurium SCV (boxed in black) causes a frameshift mutation, resulting in a premature stop codon (marked by an asterisk). The revertant shows a compensatory deletion of cytosine 123 (boxed in gray), leading to restoration of the glnA open reading frame. (B) Growth behavior of the S. Typhimurium strains in liquid LB medium supplemented with 2 mM l-glutamine (+ Gln). Growth of the SCV and ΔglnA mutant was restored to wild-type and revertant levels, respectively, when glutamine was added to the LB broth. The results were derived from the same experiment as shown in Fig. 1B, with the difference that l-glutamine was added to the growth medium. Data shown are means from quadruplicate cultures for which OD600 values were read at 15-min intervals, and data are representative of three independent experiments. (C) Growth of the SCV and ΔglnA mutant, compared to the wild type and revertant, respectively, on 2 mM l-glutamine-supplemented M9G minimal medium (+ Gln). Wt, wild type; SCV, small colony variant; Rev, revertant; ΔglnA, glnA knockout mutant.
The corresponding regions were analyzed in the revertant strain WPASI01r by means of PCR amplification and, in case of the hfq and glnA loci, PCR product sequencing. We found that, in the revertant, the mutated hfqG58T allele was unaffected. However, the glnA allele harbored a secondary mutation, which by deletion of cytosine 123 restored the original glnA reading frame and resulted in two amino acid substitutions, from 40N41A in wild-type GlnA to 40K41C in the revertant (Fig. 2A). Furthermore, we confirmed by means of PCR that the 18.9-kb stretch was still absent in the revertant, which indicates that deficiency of functional GlnA in the SCV determines its small colony phenotype. To test this hypothesis, we constructed an S. Typhimurium glnA knockout mutant and examined its growth behavior. Like the SCV, the glnA mutant formed tiny colonies on LB agar, showed a reduced exponential growth rate (130 min versus 30 min for the wild type) in LB broth, and was not able to grow on M9G minimal agar (Fig. 1A to C).
Restoration of SCV and glnA mutant growth with the addition of glutamine.
In enteric bacteria, glutamine synthetase is the only enzyme capable of synthesizing glutamine (19). In order to test whether possible glutamine auxotrophy caused by GlnA truncation was responsible for the slow growth of the SCV, we analyzed the growth of wild-type, SCV, glnA mutant, and revertant colonies with glutamine supplementation in LB broth and M9G minimal medium. Indeed, the SCV and the glnA mutant grew comparably to the wild type and the revertant in glutamine-supplemented LB broth and M9G agar (Fig. 2B and C). These findings show that limitation of glutamine availability, triggered by a lack of functional GlnA, accounts for the SCV phenotype.
Increased Salmonella O1 phage resistance of the SCV.
For further characterization of the SCV, we performed phage typing according to the scheme described by Anderson et al. (20). Mainly used for epidemiological purposes, the method uses a panel of lytic bacteriophages to subtype S. Typhimurium (20, 21). Despite slower growth, the phage type, determined as definitive type 4 (DT4), was not altered in the SCV or the glnA mutant, compared to that of the parental strain or the revertant, respectively. However, we observed a difference when susceptibility to Salmonella phage Felix O1 was tested (22–24). Specifically, the SCV and the glnA mutant showed resistance to Felix O1, whereas the wild type and the revertant were lysed. Addition of glutamine to the growth medium caused reversion of the phenotype of the SCV and the glnA knockout mutant to that of the wild type (Fig. 3).
FIG 3.
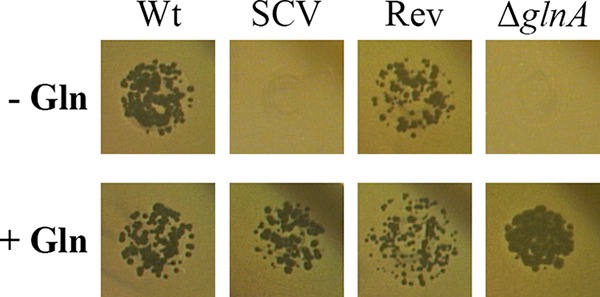
Felix O1 phage resistance phenotype of S. Typhimurium SCV and a glnA knockout mutant, compared to the wild type and a revertant. (Upper) In the absence of glutamine (− Gln), the wild type and revertant were susceptible to Felix O1 phage, whereas the SCV and a glnA knockout mutant were resistant. (Lower) In the presence of 2 mM l-glutamine (+ Gln), all strains were susceptible to Felix O1 phage. Data are representative of at least three independent experiments. Wt, wild type; SCV, small colony variant; Rev, revertant; ΔglnA, glnA knockout mutant.
Reduced host cell entry of the SCV and a glnA knockout mutant.
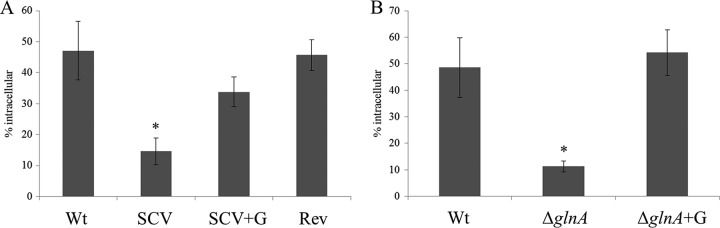
Glutamine is one of only two central intermediates in nitrogen assimilation and functions as an essential amide group donor in many biosynthetic reactions (19). Thus, the defect in glutamine biosynthesis, and potential associated impairment of nitrogen metabolism, may affect the bacterium-host interplay (25). Therefore, we performed infection experiments with differentiated U937 macrophage-like cells permissive for Salmonella intracellular growth (26). We did not observe a difference in intracellular growth, but we noticed that SCV host cell entry was only ∼30% of the wild-type level (Fig. 4A). When the SCV was grown in glutamine-supplemented broth, entry was restored to 70% of the wild-type level. The revertant showed wild-type-like behavior (Fig. 4A). As with the SCV, the number of intracellular ΔglnA bacteria was reduced to ∼25% of the wild-type level, but the ΔglnA strain grown with glutamine behaved like the wild type (Fig. 4B). Our finding suggests that establishment of functional traits facilitating host cell entry in S. Typhimurium is linked to the glutamine pool.
FIG 4.
Reduced entry of the S. Typhimurium SCV and a glnA knockout mutant in U937 cells. Entry is restored in the revertant strain or when strains are grown in culture medium with glutamine before infection. CFU were determined after a 30-min entry period and 90 min of gentamicin killing of extracellular bacteria. (A) Entry of the wild type, SCV, and revertant grown without glutamine and the SCV grown with 2 mM l-glutamine (+G) in U937 cells. (B) Entry of the S. Typhimurium wild type, a glnA knockout mutant grown without glutamine, and a glnA knockout mutant grown in the presence of 2 mM l-glutamine (+G) in U937 cells. Data are representative of at least three independent experiments. *, significantly reduced number of intracellular bacteria, compared to the wild type (P < 0.02, Student's two-tailed t test, type 2). Wt, wild type; SCV, small colony variant; Rev, revertant; ΔglnA, glnA knockout mutant.
Severely reduced exoprotein levels in the SCV and a glnA knockout mutant.
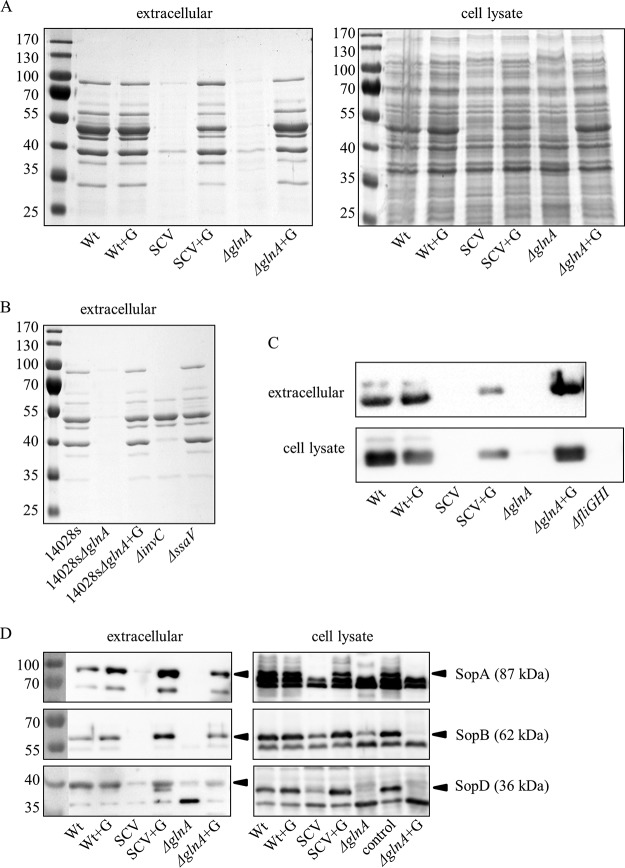
Since S. Typhimurium invasion is driven by the SPI-1-associated type III secretion system and its effector proteins (27, 28), we compared the exoprotein pattern of early-stationary-phase SCV bacteria to that of the wild type. Indeed, we found a dramatic reduction in protein quantity in the SCV (Fig. 5A, left). However, when the SCV was grown in the presence of glutamine, the exoproteome was like that of the wild type (Fig. 5A). Moreover, glnA mutants derived from wild-type strains LT2 and ATCC 14028s showed SCV-like phenotypes with respect to their exoproteins (Fig. 5A and B). The exoprotein levels of a mutant with a nonfunctional SPI-1-associated secretion system (ΔinvC mutant) were less severely reduced than in the SCV or the glnA mutant, suggesting a more general effect of glutamine deficiency on protein export or synthesis than a sole impact on SPI-1-dependent secretion (Fig. 5B). A mutant with a defect in the SPI-2-associated secretion system (ΔssaV mutant) did not show visible exoprotein changes under the conditions tested (Fig. 5B). Interestingly, the whole-cell fractions of the wild-type, SCV, and glnA mutant strains did not show major differences in the total protein amounts, as judged by SDS-PAGE (Fig. 5A, right), implying that glutamine might specifically trigger protein export or production of exported proteins in S. Typhimurium. Nevertheless, some differences in the whole-cell protein patterns of the SCV and glnA mutant were observed; a prominent band of ∼50 kDa was absent and several band size shifts of >100 kDa occurred (Fig. 5A, right). All differences reverted to the wild-type pattern with the addition of glutamine (Fig. 5A, right).
FIG 5.
Severely reduced levels of flagellin and SPI-1-associated effectors in the SCV and a glnA knockout mutant. Protein levels are restored when strains are grown in culture medium with glutamine. (A and B) Coomassie blue-stained SDS-PAGE gels of early-stationary-phase bacteria grown in LB broth. (A) Extracellular (left) and whole-cell (right) proteins of wild-type, SCV, and glnA knockout mutant strains grown without or with (+G) l-glutamine supplementation. (B) S. Typhimurium ATCC 14028s wild-type and isogenic glnA, invC (nonfunctional SPI-1-dependent secretion system), and ssaV (nonfunctional SPI-2-dependent secretion system) knockout mutant strains grown without glutamine and a glnA knockout mutant strain grown with the addition of 2 mM l-glutamine (+G). (C and D) Western blot analysis of the extracellular and whole-cell protein fractions, as described above, with flagellin-specific antibodies (C) and SopA-, SopB-, and SopD-specific antibodies (D). Negative controls were the ΔfliGHI strain for antiflagellin blots, strain M712 for anti-SopA/anti-SopB blots, and strain MvP1895 for anti-SopD blots. Lanes on the left show prestained protein ladders; the values at the left indicate kilodaltons. Wt, wild type; SCV, small colony variant; ΔglnA, glnA knockout mutant; 14028s, ATCC 14028s; control, negative control.
Reduced amounts of flagellin and SPI-1-associated effectors in the SCV and a glnA knockout mutant.
Flagellin is a highly abundant protein in flagellated bacteria. Therefore, the lack of a prominent ∼50-kDa protein band in the SCV cell lysates suggested reduced amounts of the flagellin proteins FliC (51.6 kDa) and/or FljB (52.5 kDa). By means of Western blotting using a Salmonella group flagellar antibody, we established the absence of flagellin in SCV or glnA mutant cell lysates and extracellular fractions. The phenotype was reversed when strains were grown with glutamine supplementation (Fig. 5C). To test whether other exported proteins were absent under low-glutamine conditions, we analyzed the known SPI-1-associated effectors SopA (87 kDa), SopB (62 kDa), and SopD (36 kDa) (29–31). We tested these effectors in particular due to missing protein bands in the extracellular fractions of the SCV, glnA mutant, and invC mutant strains that corresponded to their respective molecular sizes (3). SopA, SopB, and SopD were absent or were only weakly present in cell lysates of the SCV and glnA mutant strains, and levels were reduced in the corresponding extracellular fractions, suggesting decreased production, similar to findings for flagellin (Fig. 5D). The presence of effectors was restored to wild-type levels when glutamine was added for bacterial growth (Fig. 5D). These findings indicate reduced production of flagellin and the SPI-1-associated effectors in the SCV and the glnA mutant when glutamine levels are low; their production is again induced when glutamine levels are replenished.
Reduced expression of flagellin genes, the class 2 motility gene flgB, and several SPI-1-associated effector genes in the SCV and a glnA mutant.
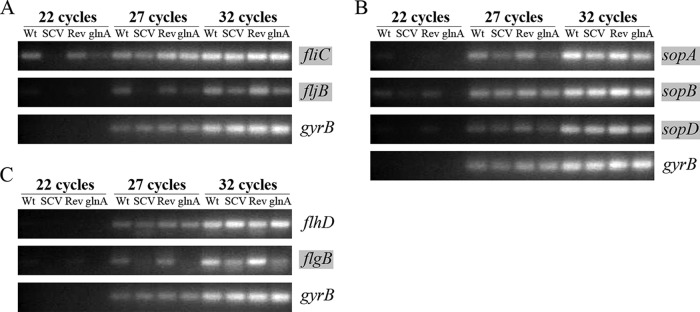
Next, we used semiquantitative reverse transcription (RT)-PCR to test whether decreased quantities of flagellin proteins and SPI-1-associated effectors were due to decreased mRNA levels of the respective genes, implying their reduced expression. Indeed, as shown in Fig. 6A and B, expression of the flagellin genes fliC and fljB and the SPI-1-associated effector genes sopA, sopB, and (to a lesser extent) sopD was reduced in the SCV or the glnA mutant, compared to the wild type or the revertant (Fig. 6A and B). This shows that both flagellin and SPI-1 effector gene transcription is linked to intact glnA and presumably glutamine availability. Next, we asked the question of which stage of the regulatory network controlling flagellum biosynthesis is affected. In E. coli and S. enterica, flagellar genes are organized into a transcriptional hierarchy of three promoter classes. Class 3 promoters drive expression of late motility genes, such as fliC and fljB, and depend on the flagellum-specific alternative sigma factor σ28, coded by fliA. The fliA gene belongs to one of several class 2 operons producing proteins required for the structure and assembly of the flagellar hook basal body. Transcription of class 2 promoters depends, in turn, on the transcriptional activator complex FlhD4C2, which is encoded by the class 1 flhDC operon (32, 33). Therefore, we analyzed expression of representative class 2 and class 1 promoter genes, i.e., flgB, encoding a component of the flagellar basal body, and flhD, respectively. We observed that flgB but not flhD expression was reduced in the SCV and the glnA mutant, compared to the wild type and the revertant (Fig. 6C). This finding reveals that reduced expression of class 2 and class 3 motility genes is not linked to decreased expression of the class 1 flagellar master operon flhDC. The regulation circuit hindered upstream of class 2 flagellar genes remains to be determined.
FIG 6.
Reduced expression of flagellin genes, the class 2 motility gene flgB, and several SPI-1-associated effector genes in the SCV and the glnA mutant, compared to the wild type and a genetic revertant. Analysis of transcript levels in different S. Typhimurium strain backgrounds was performed by semiquantitative RT-PCR, using gene-specific primers for the flagellin-encoding genes fliC and fljB (class 3 motility genes) (A), the SPI-1 effector genes sopA, sopB, and sopD (B), and the class 2 motility gene flgB and the class 1 motility gene flhD (C). Transcript levels of S. Typhimurium gyrB, the constitutively expressed gene encoding gyrase subunit B, were used as the control. Images represent agarose gels of ethidium bromide-stained PCR products after different rounds of amplification. Genes whose transcript levels differed markedly in the different strain backgrounds are marked in gray. Data are representative of three independent experiments. Wt, wild type; SCV, small colony variant; Rev, revertant; glnA, glnA knockout mutant.
DISCUSSION
In this study, we analyzed the genetic basis and the phenotype of an S. Typhimurium SCV that emerged under nutrient limitation. It is important to elucidate the genetic alterations leading to SCV formation because (i) SCVs may arise spontaneously upon stress exposure, (ii) SCVs are slow growing and difficult to eradicate, and (iii) only a few descriptions of S. enterica SCVs are available (34).
We found that the characterized SCV harbors ∼90% truncation of the glnA gene, encoding glutamine synthetase (Fig. 2A). Glutamine synthetase, a major enzyme in nitrogen assimilation, converts glutamate and ammonia to glutamine in an ATP-driven reaction (35). Glutamine is a central intermediate of nitrogen metabolism and serves as an indicator of nitrogen availability in Salmonella. Specifically, when external nitrogen sources are limited, the internal glutamine pool decreases, whereas the glutamate pool remains stable (36, 37). Thus, S. Typhimurium glnA mutants simulate nitrogen limitation internally even when excess nitrogen is available externally. Furthermore, it is thought that a reduced glutamine pool under nitrogen-limiting conditions is responsible for slow growth (36). This is in accordance with our observation that strains with impaired glutamine synthetase (due to loss-of-function gene truncation or glnA knockout) display small colony morphology and reduced growth rates even if cultivated in rich medium, such as LB medium (Fig. 1). Glutamine supplementation of growth media causes reversion of this phenotype (Fig. 2B and C), indicating that the glutamine transporter encoded by glnHPQ is expressed in the SCV and the glnA knockout mutant at sufficient levels to replenish the internal glutamine pool (38, 39).
The amide group in glutamine is required as a nitrogen source in early steps of the synthesis of monomeric building blocks of most cellular macromolecules, such as proteins, nucleic acids, and surface polymers (25). Therefore, it can be expected that the phenotypes of the SCV and a glnA mutant might strongly differ from the wild-type phenotype. We found that the SCV and glnA mutant were severely attenuated in their entry into macrophages (Fig. 4). Earlier studies addressed the fate of glnA mutants in alfalfa seed exudates or intraperitoneal BALB/c mouse infection models. Slight attenuation of mutant growth in the alfalfa exudate but not in the mouse model was observed (39, 40). However, combined knockout of glnA and the glutamine transporter genes glnH and glnQ or genes of the nitrogen regulatory system (ntr), which is responsible for transcription of the glutamine transporter genes, led to severe growth attenuation in the mouse model and impaired intracellular survival in J774 macrophages, showing that glutamine uptake from the host plays an essential role in infection (39). It is important to note that studies with intraperitoneal instead of oral infection of mice might overlook important entry-associated effects.
Invasion into nonphagocytic cells and phagocytes is promoted by effectors of the SPI-1-encoded type III secretion system and by flagellin (2, 3, 28, 41). Accordingly, we found that both the SCV and a glnA mutant lacked flagellin and SPI-1 effector proteins, due to reduced expression of the corresponding genes. Expression of both virulence and motility regulons is under tight control, as these processes require substantial resources from the bacterial cell (42). More than 60 genes contribute to the synthesis, assembly, and functioning of the flagellar system in Salmonella (43). Flagellar operons are divided into three classes, based on the promoters that drive their transcription (33, 44). The class 1 operon flhDC produces the multimeric master regulator of flagellar synthesis, FlhD4C2, which is essential for the expression of all downstream motility genes (32). FlhD4C2 activates transcription of class 2 operons encoding structural genes of the hook basal body, a flagellum-specific type III secretion system, and the alternative sigma factor σ28, which is required for class 3 promoter transcription. Upon completion of the hook basal body substructure, the σ28 anti-sigma factor FlgM is secreted; hence, free σ28 is able to transcribe class 3 promoters, resulting in synthesis of filament and motor proteins and the chemotaxis machinery (Fig. 7). In S. enterica, flagellum and SPI-1 gene expression is interconnected (45–49). The regulatory interlink occurs via the FliZ protein, which is part of the fliAZ operon that can be transcribed from class 2 and class 3 promoters (Fig. 7). In addition to being a positive regulator of FlhD4C2 activity (50), FliZ posttranslationally activates the regulatory protein HilD, which, in turn, positively regulates the expression of hilA, the gene for a key regulator of the SPI-1 system (45, 51, 52). Furthermore, HilD acts as a direct activator of flhDC expression, strengthening the transcriptional cross talk between the flagellar and SPI-1 regulons in Salmonella (52). When measuring mRNA levels in the SCV and a glnA knockout mutant, in comparison to the wild type, we found reduced transcript levels for SPI-1-associated effector genes as well as class 3 and class 2 motility genes (Fig. 6). Reduced expression of class 2 genes such as fliZ would give a plausible explanation for the observed reductions in both flagellin and SPI-1 effector protein levels. Here, flgB mRNA levels were analyzed as an example of class 2 genes (Fig. 6). Interestingly, we did not observe changes in flhD expression in the SCV or glnA mutant, which implies that impairment of flagellar synthesis takes place at a step subsequent to class 1 transcription (Fig. 7). Examples of possible factors involved include the flagellar protein FliT (an anti-FlhD4C2 factor fine-tuning flagellar expression to the state of flagellar assembly [53, 54]), the molecular chaperone DnaK (55), and the EAL-like protein YdiV (56–58). The last is of special interest because YdiV inhibits FlhD4C2 activity when nutrients are scarce (57, 59).
FIG 7.
Model of the flagellar gene network and SPI-1-associated effector expression in the S. Typhimurium SCV and a glnA mutant. Functional impairment of glnA in the SCV or a glnA knockout mutant results in glutamine limitation and negatively affects flagellum biosynthesis and SPI-1-mediated virulence. Specifically, reduced expression of class 2 (flgB) and class 3 (fliC and fljB) motility genes and SPI-1-associated effector genes was detected, accompanied by reduced abundance of flagellin and effector proteins (depicted by orange arrows). The dashed orange lines outline the different possible ways of influencing flagellar and SPI-1-associated effector expression upstream of the class 2 motility genes. It is established that the flagellar and SPI-1 regulons are interconnected by the class 2 motility gene product FliZ, which activates SPI-1 via the HilD regulatory protein (45, 51). Thus, reduced levels of FliZ are expected to lead to reduced levels of the transcriptional regulators HilA (controlled by HilD) and InvF (controlled by HilA), both of which are known to activate the expression of effector genes located inside and outside (indicated by a dotted box) SPI-1 (77–79). While transcriptional regulation of sopA and sopB by HilA/InvF has been described in the literature (78, 80, 81), sopD regulation is less well understood (indicated by a dashed black arrow). HBB, hook basal body; SCV, small colony variant; SPI-1, Salmonella pathogenicity island 1; T3SS-1, SPI-1-encoded type III secretion system; σ70, housekeeping sigma factor; σ28, flagellum-specific alternative sigma factor (FliA).
Finally, the SCV showed reduced susceptibility to the Salmonella phage Felix O1, whereas the wild-type and revertant strains revealed the lysis pattern observed for the vast majority of Salmonella strains (Fig. 3) (23). This demonstrates that, upon exposure to stress, mutations conferring phage resistance may easily arise in Salmonella. The occurrence of phage-resistant strains is of major importance because phage therapy has been studied as a promising alternative to combat zoonotic pathogens, such as Salmonella, in animals, in the food chain, and in human infections (60–65). Bacteriophage Felix O1 in particular, which has a broad host range within the genus Salmonella and historically was used for the identification of Salmonella (66), proved successful in reducing Salmonella colonizing broiler chickens (63) and on the surface of meat (67). However, Salmonella strains resistant to Felix O1 phage have been reported (68, 69). The resistance phenotype has been linked to alterations in the structure of the lipopolysaccharide core, a site that is important for successful adsorption of Felix O1 to its bacterial host (68, 70, 71).
Interestingly, in additional independent starvation experiments, we sporadically detected and isolated other Salmonella strains showing an SCV phenotype within long-term starvation cultures. None of those SCVs, genetically or in terms of auxotrophy, resembled the SCV described here, suggesting that starvation in distilled water does not by itself select for the specific mutations found in SCV WPASI01.
SCVs of S. enterica or other bacterial pathogens might emerge spontaneously upon stress exposure. As their presence can be correlated with recurrent and persistent infections in a number of diseases, it is important to understand the underlying genetic changes and phenotypic consequences (10, 34). In our case, the SCV under investigation showed a loss-of-function mutation in glnA, leading to slow growth, attenuated virulence, and reduced virulence gene expression.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Unless otherwise stated, all experiments were performed with the S. Typhimurium strain LT2, which is designated the wild type. An overview of strains used in this study is given in Table 1. All strains were grown at 37°C in LB medium supplemented with 50 μg/ml kanamycin or 2 mM l-glutamine (product no. 3772.1; Carl Roth), when appropriate, or on M9 supplemented with 0.4% glucose (M9G) agar plates. CFU of plate-grown bacteria were routinely counted after overnight incubation at 37°C. During stress experiments, plates were inspected after overnight incubation and were reinspected after 2 more days of incubation, taking into account bacteria with a prolonged lag phase. A Bioscreen C analyzer (Oy Growth Curves AB Ltd.) was used to determine bacterial growth rates. Strains were cultivated overnight in LB medium and diluted in fresh LB medium to an optical density at 600 nm (OD600) of 0.05, and 200-μl samples were transferred into four replica wells of a Bioscreen plate. Plates were incubated at 37°C with continuous shaking, and absorbance at 600 nm was measured automatically every 15 min for a period of 24 h.
TABLE 1.
Strains used in this study
| S. Typhimurium strain | Characteristics | Source or reference |
|---|---|---|
| LT2 strains | ||
| Wild type | 18 | |
| SCV WPASI01 | glnA120insA, hfqG85T, del_srfC-STM1615 | This study |
| Rev WPASI01r | glnA120insA123delC, hfqG85T, del_srfC-STM1615 | This study |
| ΔglnA mutant | glnA::KmFRT | This study |
| ATCC 14028s strains | ||
| Wild type | Isogenic to wild-type NCTC 12023 | 82 |
| ATCC 14028s ΔglnA | glnA::KmFRT | This study |
| NCTC 12023 P2D6 | ssaV::mTn5 | 83 |
| NCTC 12023 MvP818 | invC::FRT | 84 |
| NCTC 12023 MvP1895 | ΔsipA ΔsopA ΔsopB ΔsopD ΔsopE2 | Unpublished strain provided by M. Hensel (University of Osnabrück) |
| SL1344 strains | ||
| SL1344 M712 | ΔsipA ΔsopA ΔsopB ΔsopE ΔsopE2 | 85 |
| SL1344 M913 | fliGHI::Tn10 | 86 |
Preparation of axenic water cultures.
For starvation microcosms, bacteria were grown overnight on LB plates and subsequently adjusted in distilled water to an OD600 of 0.3 (corresponding to 2 × 108 CFU/ml). Afterwards, 1 ml was added to 200 ml of autoclaved distilled water, and the suspension was stored in 250-ml Erlenmeyer flasks at 23°C in the dark. Each microcosm was set up in duplicate. At the time of inoculation and then routinely, i.e., on a weekly basis for the first year and subsequently twice a year, the microcosms were assessed for CFU by plating of serial dilutions on LB agar.
Mutant generation.
The ΔglnA mutants were constructed by λ Red-mediated mutagenesis (72). The primers for deleting glnA by introducing the pKD4-derived gene for the kanamycin resistance cassette into the chromosome were ABp0_glnA (5′-GAAGTGAAGTTTGTCGATCTGCGCTTCACCGATACCAAAGTGTGTAGGCTGGAGCTGCTT-3′) and ABp2_glnA (5′-CGAAATTTGTCGGATTTTTAATATACGATTAAACGCTGTACATATGAATATCCTCCTTAGTTCC-3′). Deletion of glnA was confirmed by PCR with the primers glnAFL_f (5′-GCGCGTGAGATCAGATTG-3′) and glnAFL_r (5′-GTGGCTTGCGTTCTGTTG-3′), followed by gel electrophoresis and subsequent sequencing of the PCR product.
Phage typing.
O1 phage experiments were performed on Oxoid no. 3 plates, as described previously (21). Briefly, 10 μl of 10-fold serial dilutions (routine test dilutions of 1:10−4 to 1:10−10) of phage suspension was spotted onto bacterial lawns. The plates were incubated overnight at 37°C, and the lytic activity, expressed as the maximal dilution at which no lysis was observed, was determined by monitoring areas of lysis on the bacterial lawns. The routine phage typing of S. Typhimurium was performed according to the Anderson typing system (20, 21).
Gentamicin protection assay.
For determination of invasion/entry in U937 monocytes (CRL-1593.2; American Type Culture Collection, Manassas, VA), a human cell line that differentiates into macrophage-like cells upon treatment with phorbol esters (80 nm phorbol 12-mystrate 13-acetate [product no. P-8139; Sigma]) and incubation for 36 to 48 h, was used. Cells were routinely cultured in RPMI 1640 medium containing 10% fetal bovine serum (FBS), in a humidified atmosphere (37°C in 5% CO2). Infection experiments were performed as described elsewhere (73, 74). Briefly ∼1 × 106 differentiated U937 cells were added to the wells of 24-well plates and allowed to adhere for 2 to 4 h. Bacteria were grown overnight in LB medium under static conditions at 37°C. Infections were performed at a multiplicity of infection of 10. Bacteria were centrifuged onto the cells for 5 min at 800 × g and subsequently incubated for 25 min. Incubation mixtures were then washed twice with phosphate-buffered saline (PBS), and medium containing 100 μg/ml gentamicin was added for 90 min. Subsequently, cells were washed twice and then lysed with PBS containing 0.1% Triton X-100. After repeated pipetting, serial dilutions of lysates were plated onto LB agar to determine the number of internalized bacteria.
Genome sequencing.
Nextera XT sequencing libraries (Illumina) were prepared from 1 ng bacterial DNA and sequenced on a MiSeq instrument, to generate 250-bp paired-end reads for >30-fold average coverage. Illumina reads of the SCV and its parental LT2 wild-type strain were mapped to the Salmonella enterica serovar Typhimurium LT2 reference genome sequence (18) (GenBank accession no. NC_003197.1) (99.5% coverage) by using the map-to-reference tool of Geneious v7.1 software, with default settings. Contigs of the mapped SCV and wild-type genomes were aligned with the reference genome using the Mauve alignment tool implemented in Geneious. Alignments were manually inspected for differences from the reference sequence.
SDS-PAGE and Western blot analysis of cellular and extracellular proteins.
For comparison of extracellular protein patterns, bacteria were grown at 37°C in LB broth, with shaking at 250 rpm, until early stationary phase was reached. For total cellular proteins, 500 μl culture was pelleted, the supernatant was removed, and the pellet was boiled in reducing SDS-PAGE sample buffer (75). For extracellular proteins, 1 ml cell-free culture supernatant (from centrifugation at 8,000 × g for 5 min) was precipitated with trichloroacetic acid, washed with cold acetone, and finally suspended and boiled in SDS-PAGE sample buffer. All samples were run on 10% SDS-PAGE gels and stained with colloidal Coomassie blue (Carl Roth). Antibodies for detection of flagellin (clone 4H2; Biotrend GmbH) or SopA, SopB, and SopD proteins (76) were used at 1,000-fold or 500-fold dilutions and were detected with horseradish peroxidase conjugates of anti-mouse IgG or anti-rabbit IgG (both Sigma-Aldrich), respectively, which were used at 10,000-fold dilutions.
RNA extraction and semiquantitative RT-PCR.
For total RNA extraction, stationary-phase bacteria were diluted in fresh LB medium to an OD600 of 0.05 and were grown to early stationary phase (OD600 of 1.8 to 2.0). Aliquots of 7.5 × 108 bacteria were treated with RNAprotect (Qiagen) as recommended by the manufacturer, and RNA was isolated using the RNeasy minikit (Qiagen), following the standard protocol. Possible DNA contamination was removed by using the Turbo DNA-free kit from Ambion, and then 1 μg RNA was reverse transcribed to cDNA using the Superscript IV enzyme (Invitrogen) as recommended. Subsequently, 2 μl of cDNA diluted 1:5 was used in a semiquantitative PCR with gene-specific primer pairs, as listed in Table 2, and the Taq polymerase (New England BioLabs GmbH). The following cycle conditions were used for all primer combinations: 95°C for 2 min and then 24 to 33 cycles of 95°C for 30 s, 59°C for 30 s, and 68°C for 15 s. Product formation was analyzed by agarose gel electrophoresis and ethidium bromide staining after different rounds of amplification. All semiquantitative RT-PCRs were performed with at least three independent RNA/cDNA preparations. Primers amplifying a part of S. Typhimurium gyrB, the constitutively expressed gene coding for gyrase subunit B, were used as an internal control. cDNA reactions performed without the addition of Superscript IV served as negative controls, to prove that RNA preparations were free of DNA.
TABLE 2.
Primers used for semiquantitative RT-PCRs
Accession number(s).
Sequencing reads were submitted to the European Nucleotide Archive (ENA) and assigned study accession number PRJEB12856.
ACKNOWLEDGMENTS
We are grateful to Michael Hensel (University of Osnabrück) for providing effector-defective strain MvP1895. We thank Roman Gerlach (Robert Koch Institute) for providing S. Typhimurium flagellin-, type III secretion system-, and effector-defective strains. We further acknowledge Marc Erhard (Helmholtz Centre for Infection Research, Braunschweig, Germany) for discussing results and Dagmar Busse and Susanne Kulbe (Robert Koch Institute) for excellent technical assistance with respect to phage typing. We thank Helmut Tschäpe, Wiebke Streckel, and Angelika Fruth for generating and providing the antisera for SPI-1 effector proteins. We thank Sangeeta Banerji for critical reading of the manuscript.
We declare we have no competing interests.
REFERENCES
- 1.Srikanth CV, Mercado-Lubo R, Hallstrom K, McCormick BA. 2011. Salmonella effector proteins and host-cell responses. Cell Mol Life Sci 68:3687–3697. doi: 10.1007/s00018-011-0841-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valdez Y, Ferreira RB, Finlay BB. 2009. Molecular mechanisms of Salmonella virulence and host resistance. Curr Top Microbiol Immunol 337:93–127. [DOI] [PubMed] [Google Scholar]
- 3.Agbor TA, McCormick BA. 2011. Salmonella effectors: important players modulating host cell function during infection. Cell Microbiol 13:1858–1869. doi: 10.1111/j.1462-5822.2011.01701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winfield MD, Groisman EA. 2003. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl Environ Microbiol 69:3687–3694. doi: 10.1128/AEM.69.7.3687-3694.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorski L, Parker CT, Liang A, Cooley MB, Jay-Russell MT, Gordus AG, Atwill ER, Mandrell RE. 2011. Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Appl Environ Microbiol 77:2734–2748. doi: 10.1128/AEM.02321-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Traore O, Nyholm O, Siitonen A, Bonkoungou IJ, Traore AS, Barro N, Haukka K. 2015. Prevalence and diversity of Salmonella enterica in water, fish and lettuce in Ouagadougou, Burkina Faso. BMC Microbiol 15:151. doi: 10.1186/s12866-015-0484-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nevo E. 2001. Evolution of genome-phenome diversity under environmental stress. Proc Natl Acad Sci U S A 98:6233–6240. doi: 10.1073/pnas.101109298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Painter KL, Strange E, Parkhill J, Bamford KB, Armstrong-James D, Edwards AM. 2015. Staphylococcus aureus adapts to oxidative stress by producing H2O2-resistant small-colony variants via the SOS response. Infect Immun 83:1830–1844. doi: 10.1128/IAI.03016-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuchscherr L, Medina E, Hussain M, Volker W, Heitmann V, Niemann S, Holzinger D, Roth J, Proctor RA, Becker K, Peters G, Loffler B. 2011. Staphylococcus aureus phenotype switching: an effective bacterial strategy to escape host immune response and establish a chronic infection. EMBO Mol Med 3:129–141. doi: 10.1002/emmm.201000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Proctor RA, von Eiff C, Kahl BC, Becker K, McNamara P, Herrmann M, Peters G. 2006. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Microbiol 4:295–305. doi: 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- 11.Edwards AM. 2012. Phenotype switching is a natural consequence of Staphylococcus aureus replication. J Bacteriol 194:5404–5412. doi: 10.1128/JB.00948-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cano DA, Pucciarelli MG, Martinez-Moya M, Casadesus J, Garcia-del Portillo F. 2003. Selection of small-colony variants of Salmonella enterica serovar Typhimurium in nonphagocytic eucaryotic cells. Infect Immun 71:3690–3698. doi: 10.1128/IAI.71.7.3690-3698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pränting M, Andersson DI. 2010. Mechanisms and physiological effects of protamine resistance in Salmonella enterica serovar Typhimurium LT2. J Antimicrob Chemother 65:876–887. doi: 10.1093/jac/dkq059. [DOI] [PubMed] [Google Scholar]
- 14.Koskiniemi S, Pränting M, Gullberg E, Nasvall J, Andersson DI. 2011. Activation of cryptic aminoglycoside resistance in Salmonella enterica. Mol Microbiol 80:1464–1478. doi: 10.1111/j.1365-2958.2011.07657.x. [DOI] [PubMed] [Google Scholar]
- 15.Li W, Li Y, Wu Y, Cui Y, Liu Y, Shi X, Zhang Q, Chen Q, Sun Q, Hu Q. 2016. Phenotypic and genetic changes in the life cycle of small colony variants of Salmonella enterica serotype Typhimurium induced by streptomycin. Ann Clin Microbiol Antimicrob 15:37. doi: 10.1186/s12941-016-0151-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rahn K, De Grandis SA, Clarke RC, McEwen SA, Galan JE, Ginocchio C, Curtiss R III, Gyles CL. 1992. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol Cell Probes 6:271–279. doi: 10.1016/0890-8508(92)90002-F. [DOI] [PubMed] [Google Scholar]
- 17.Sauer E. 2013. Structure and RNA-binding properties of the bacterial LSm protein Hfq. RNA Biol 10:610–618. doi: 10.4161/rna.24201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 19.van Heeswijk WC, Westerhoff HV, Boogerd FC. 2013. Nitrogen assimilation in Escherichia coli: putting molecular data into a systems perspective. Microbiol Mol Biol Rev 77:628–695. doi: 10.1128/MMBR.00025-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson ES, Ward LR, Saxe MJ, de Sa JDH. 1977. Bacteriophage-typing designations of Salmonella typhimurium. J Hyg (Lond) 78:297–300. doi: 10.1017/S0022172400056187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rabsch W. 2007. Salmonella Typhimurium phage typing for pathogens. Methods Mol Biol 394:177–211. doi: 10.1007/978-1-59745-512-1_10. [DOI] [PubMed] [Google Scholar]
- 22.Hirsh DC, Martin LD. 1983. Rapid detection of Salmonella spp. by using Felix-O1 bacteriophage and high-performance liquid chromatography. Appl Environ Microbiol 45:260–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welkos S, Schreiber M, Baer H. 1974. Identification of Salmonella with the O-1 bacteriophage. Appl Microbiol 28:618–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhn J, Suissa M, Chiswell D, Azriel A, Berman B, Shahar D, Reznick S, Sharf R, Wyse J, Bar-On T, Cohen I, Giles R, Weiser I, Lubinsky-Mink S, Ulitzur S. 2002. A bacteriophage reagent for Salmonella: molecular studies on Felix 01. Int J Food Microbiol 74:217–227. doi: 10.1016/S0168-1605(01)00682-1. [DOI] [PubMed] [Google Scholar]
- 25.Wingreen NS, Kustu S. 2001. Coordinated slowing of metabolism in enteric bacteria under nitrogen limitation: a perspective. arXiv arXiv:physics/0110037v1 [physics.bio-ph] http://arxiv.org/abs/physics/0110037v1.
- 26.Chateau MT, Caravano R. 1993. Infection of differentiated U937 cells by Salmonella typhimurium: absence of correlation between oxidative burst and antimicrobial defence. FEMS Immunol Med Microbiol 7:111–118. doi: 10.1111/j.1574-695X.1993.tb00389.x. [DOI] [PubMed] [Google Scholar]
- 27.Galan JE. 1999. Interaction of Salmonella with host cells through the centisome 63 type III secretion system. Curr Opin Microbiol 2:46–50. doi: 10.1016/S1369-5274(99)80008-3. [DOI] [PubMed] [Google Scholar]
- 28.Drecktrah D, Knodler LA, Ireland R, Steele-Mortimer O. 2006. The mechanism of Salmonella entry determines the vacuolar environment and intracellular gene expression. Traffic 7:39–51. doi: 10.1111/j.1600-0854.2005.00360.x. [DOI] [PubMed] [Google Scholar]
- 29.Wood MW, Jones MA, Watson PR, Siber AM, McCormick BA, Hedges S, Rosqvist R, Wallis TS, Galyov EE. 2000. The secreted effector protein of Salmonella dublin, SopA, is translocated into eukaryotic cells and influences the induction of enteritis. Cell Microbiol 2:293–303. doi: 10.1046/j.1462-5822.2000.00054.x. [DOI] [PubMed] [Google Scholar]
- 30.Galyov EE, Wood MW, Rosqvist R, Mullan PB, Watson PR, Hedges S, Wallis TS. 1997. A secreted effector protein of Salmonella dublin is translocated into eukaryotic cells and mediates inflammation and fluid secretion in infected ileal mucosa. Mol Microbiol 25:903–912. doi: 10.1111/j.1365-2958.1997.mmi525.x. [DOI] [PubMed] [Google Scholar]
- 31.Jones MA, Wood MW, Mullan PB, Watson PR, Wallis TS, Galyov EE. 1998. Secreted effector proteins of Salmonella dublin act in concert to induce enteritis. Infect Immun 66:5799–5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kutsukake K, Ohya Y, Iino T. 1990. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J Bacteriol 172:741–747. doi: 10.1128/jb.172.2.741-747.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chilcott GS, Hughes KT. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol Mol Biol Rev 64:694–708. doi: 10.1128/MMBR.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johns BE, Purdy KJ, Tucker NP, Maddocks SE. 2015. Phenotypic and genotypic characteristics of small colony variants and their role in chronic infection. Microbiol Insights 8:15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merrick MJ, Edwards RA. 1995. Nitrogen control in bacteria. Microbiol Rev 59:604–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ikeda TP, Shauger AE, Kustu S. 1996. Salmonella typhimurium apparently perceives external nitrogen limitation as internal glutamine limitation. J Mol Biol 259:589–607. doi: 10.1006/jmbi.1996.0342. [DOI] [PubMed] [Google Scholar]
- 37.Hu P, Leighton T, Ishkhanova G, Kustu S. 1999. Sensing of nitrogen limitation by Bacillus subtilis: comparison to enteric bacteria. J Bacteriol 181:5042–5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Betteridge PR, Ayling PD. 1976. The regulation of glutamine transport and glutamine synthetase in Salmonella typhimurium. J Gen Microbiol 96:324–334. doi: 10.1099/00221287-95-2-324. [DOI] [PubMed] [Google Scholar]
- 39.Klose KE, Mekalanos JJ. 1997. Simultaneous prevention of glutamine synthesis and high-affinity transport attenuates Salmonella typhimurium virulence. Infect Immun 65:587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwan G, Pisithkul T, Amador-Noguez D, Barak J. 2015. De novo amino acid biosynthesis contributes to Salmonella enterica growth in alfalfa seedling exudates. Appl Environ Microbiol 81:861–873. doi: 10.1128/AEM.02985-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anderson JK, Smith TG, Hoover TR. 2010. Sense and sensibility: flagellum-mediated gene regulation. Trends Microbiol 18:30–37. doi: 10.1016/j.tim.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Erhardt M, Dersch P. 2015. Regulatory principles governing Salmonella and Yersinia virulence. Front Microbiol 6:949. doi: 10.3389/fmicb.2015.00949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frye J, Karlinsey JE, Felise HR, Marzolf B, Dowidar N, McClelland M, Hughes KT. 2006. Identification of new flagellar genes of Salmonella enterica serovar Typhimurium. J Bacteriol 188:2233–2243. doi: 10.1128/JB.188.6.2233-2243.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chevance FF, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol 6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iyoda S, Kamidoi T, Hirose K, Kutsukake K, Watanabe H. 2001. A flagellar gene fliZ regulates the expression of invasion genes and virulence phenotype in Salmonella enterica serovar Typhimurium. Microb Pathog 30:81–90. doi: 10.1006/mpat.2000.0409. [DOI] [PubMed] [Google Scholar]
- 46.Ellermeier CD, Slauch JM. 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J Bacteriol 185:5096–5108. doi: 10.1128/JB.185.17.5096-5108.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kage H, Takaya A, Ohya M, Yamamoto T. 2008. Coordinated regulation of expression of Salmonella pathogenicity island 1 and flagellar type III secretion systems by ATP-dependent ClpXP protease. J Bacteriol 190:2470–2478. doi: 10.1128/JB.01385-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin D, Rao CV, Slauch JM. 2008. The Salmonella SPI1 type three secretion system responds to periplasmic disulfide bond status via the flagellar apparatus and the RcsCDB system. J Bacteriol 190:87–97. doi: 10.1128/JB.01323-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saini S, Slauch JM, Aldridge PD, Rao CV. 2010. Role of cross talk in regulating the dynamic expression of the flagellar Salmonella pathogenicity island 1 and type 1 fimbrial genes. J Bacteriol 192:5767–5777. doi: 10.1128/JB.00624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saini S, Brown JD, Aldridge PD, Rao CV. 2008. FliZ is a posttranslational activator of FlhD4C2-dependent flagellar gene expression. J Bacteriol 190:4979–4988. doi: 10.1128/JB.01996-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chubiz JE, Golubeva YA, Lin D, Miller LD, Slauch JM. 2010. FliZ regulates expression of the Salmonella pathogenicity island 1 invasion locus by controlling HilD protein activity in Salmonella enterica serovar Typhimurium. J Bacteriol 192:6261–6270. doi: 10.1128/JB.00635-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singer HM, Kuhne C, Deditius JA, Hughes KT, Erhardt M. 2014. The Salmonella Spi1 virulence regulatory protein HilD directly activates transcription of the flagellar master operon flhDC. J Bacteriol 196:1448–1457. doi: 10.1128/JB.01438-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamamoto S, Kutsukake K. 2006. FliT acts as an anti-FlhD2C2 factor in the transcriptional control of the flagellar regulon in Salmonella enterica serovar Typhimurium. J Bacteriol 188:6703–6708. doi: 10.1128/JB.00799-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aldridge C, Poonchareon K, Saini S, Ewen T, Soloyva A, Rao CV, Imada K, Minamino T, Aldridge PD. 2010. The interaction dynamics of a negative feedback loop regulates flagellar number in Salmonella enterica serovar Typhimurium. Mol Microbiol 78:1416–1430. doi: 10.1111/j.1365-2958.2010.07415.x. [DOI] [PubMed] [Google Scholar]
- 55.Takaya A, Matsui M, Tomoyasu T, Kaya M, Yamamoto T. 2006. The DnaK chaperone machinery converts the native FlhD2C2 hetero-tetramer into a functional transcriptional regulator of flagellar regulon expression in Salmonella. Mol Microbiol 59:1327–1340. doi: 10.1111/j.1365-2958.2005.05016.x. [DOI] [PubMed] [Google Scholar]
- 56.Wozniak CE, Lee C, Hughes KT. 2009. T-POP array identifies EcnR and PefI-SrgD as novel regulators of flagellar gene expression. J Bacteriol 191:1498–1508. doi: 10.1128/JB.01177-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wada T, Morizane T, Abo T, Tominaga A, Inoue-Tanaka K, Kutsukake K. 2011. EAL domain protein YdiV acts as an anti-FlhD4C2 factor responsible for nutritional control of the flagellar regulon in Salmonella enterica serovar Typhimurium. J Bacteriol 193:1600–1611. doi: 10.1128/JB.01494-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takaya A, Erhardt M, Karata K, Winterberg K, Yamamoto T, Hughes KT. 2012. YdiV: a dual function protein that targets FlhDC for ClpXP-dependent degradation by promoting release of DNA-bound FlhDC complex. Mol Microbiol 83:1268–1284. doi: 10.1111/j.1365-2958.2012.08007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koirala S, Mears P, Sim M, Golding I, Chemla YR, Aldridge PD, Rao CV. 2014. A nutrient-tunable bistable switch controls motility in Salmonella enterica serovar Typhimurium. mBio 5:e01611-14. doi: 10.1128/mBio.01611-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Endersen L, O'Mahony J, Hill C, Ross RP, McAuliffe O, Coffey A. 2014. Phage therapy in the food industry. Annu Rev Food Sci Technol 5:327–349. doi: 10.1146/annurev-food-030713-092415. [DOI] [PubMed] [Google Scholar]
- 61.Zhang J, Hong Y, Fealey M, Singh A, Walton K, Martin C, Harman NJ, Mahlie J, Ebner PD. 2015. Physiological and molecular characterization of Salmonella bacteriophages previously used in phage therapy. J Food Prot 78:2143–2149. doi: 10.4315/0362-028X.JFP-14-350. [DOI] [PubMed] [Google Scholar]
- 62.Tiwari R, Dhama K, Kumar A, Rahal A, Kapoor S. 2014. Bacteriophage therapy for safeguarding animal and human health: a review. Pak J Biol Sci 17:301–315. doi: 10.3923/pjbs.2014.301.315. [DOI] [PubMed] [Google Scholar]
- 63.Atterbury RJ, Van Bergen MA, Ortiz F, Lovell MA, Harris JA, De Boer A, Wagenaar JA, Allen VM, Barrow PA. 2007. Bacteriophage therapy to reduce Salmonella colonization of broiler chickens. Appl Environ Microbiol 73:4543–4549. doi: 10.1128/AEM.00049-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Higgins JP, Higgins SE, Guenther KL, Huff W, Donoghue AM, Donoghue DJ, Hargis BM. 2005. Use of a specific bacteriophage treatment to reduce Salmonella in poultry products. Poult Sci 84:1141–1145. doi: 10.1093/ps/84.7.1141. [DOI] [PubMed] [Google Scholar]
- 65.Ma Y, Pacan JC, Wang Q, Xu Y, Huang X, Korenevsky A, Sabour PM. 2008. Microencapsulation of bacteriophage Felix O1 into chitosan-alginate microspheres for oral delivery. Appl Environ Microbiol 74:4799–4805. doi: 10.1128/AEM.00246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Whichard JM, Sriranganathan N, Pierson FW. 2003. Suppression of Salmonella growth by wild-type and large-plaque variants of bacteriophage Felix O1 in liquid culture and on chicken frankfurters. J Food Prot 66:220–225. doi: 10.4315/0362-028X-66.2.220. [DOI] [PubMed] [Google Scholar]
- 67.Goode D, Allen VM, Barrow PA. 2003. Reduction of experimental Salmonella and Campylobacter contamination of chicken skin by application of lytic bacteriophages. Appl Environ Microbiol 69:5032–5036. doi: 10.1128/AEM.69.8.5032-5036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.MacPhee DG, Krishnapillai V, Roantree RJ, Stocker BA. 1975. Mutations in Salmonella typhimurium conferring resistance to Felix O phage without loss of smooth character. J Gen Microbiol 87:1–10. doi: 10.1099/00221287-87-1-1. [DOI] [PubMed] [Google Scholar]
- 69.Tsang RS, Nielsen KH, Henning DM, Poppe C, Khakhria R, Woodward DL, Johnson WM. 1997. Screening for Salmonella with a murine monoclonal antibody M105 detects both Felix O1 bacteriophage sensitive and resistant Salmonella strains. Zentralbl Bakteriol 286:23–32. doi: 10.1016/S0934-8840(97)80071-0. [DOI] [PubMed] [Google Scholar]
- 70.Hudson HP, Lindberg AA, Stocker BA. 1978. Lipopolysaccharide core defects in Salmonella typhimurium mutants which are resistant to Felix O phage but retain smooth character. J Gen Microbiol 109:97–112. doi: 10.1099/00221287-109-1-97. [DOI] [PubMed] [Google Scholar]
- 71.Lindberg AA. 1973. Bacteriophage receptors. Annu Rev Microbiol 27:205–241. doi: 10.1146/annurev.mi.27.100173.001225. [DOI] [PubMed] [Google Scholar]
- 72.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martinez-Moya M, de Pedro MA, Schwarz H, Garcia-del Portillo F. 1998. Inhibition of Salmonella intracellular proliferation by non-phagocytic eucaryotic cells. Res Microbiol 149:309–318. doi: 10.1016/S0923-2508(98)80436-1. [DOI] [PubMed] [Google Scholar]
- 74.Popp J, Noster J, Busch K, Kehl A, Zur Hellen G, Hensel M. 2015. Role of host cell-derived amino acids in nutrition of intracellular Salmonella enterica. Infect Immun 83:4466–4475. doi: 10.1128/IAI.00624-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 76.Streckel W, Wolff AC, Prager R, Tietze E, Tschape H. 2004. Expression profiles of effector proteins SopB, SopD1, SopE1, and AvrA differ with systemic, enteric, and epidemic strains of Salmonella enterica. Mol Nutr Food Res 48:496–503. doi: 10.1002/mnfr.200400035. [DOI] [PubMed] [Google Scholar]
- 77.Bajaj V, Hwang C, Lee CA. 1995. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol Microbiol 18:715–727. doi: 10.1111/j.1365-2958.1995.mmi_18040715.x. [DOI] [PubMed] [Google Scholar]
- 78.Darwin KH, Miller VL. 2001. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. EMBO J 20:1850–1862. doi: 10.1093/emboj/20.8.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ellermeier JR, Slauch JM. 2007. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr Opin Microbiol 10:24–29. doi: 10.1016/j.mib.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 80.Dieye Y, Dyszel JL, Kader R, Ahmer BM. 2007. Systematic analysis of the regulation of type three secreted effectors in Salmonella enterica serovar Typhimurium. BMC Microbiol 7:3. doi: 10.1186/1471-2180-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thijs IM, De Keersmaecker SC, Fadda A, Engelen K, Zhao H, McClelland M, Marchal K, Vanderleyden J. 2007. Delineation of the Salmonella enterica serovar Typhimurium HilA regulon through genome-wide location and transcript analysis. J Bacteriol 189:4587–4596. doi: 10.1128/JB.00178-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fields PI, Swanson RV, Haidaris CG, Heffron F. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci U S A 83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shea JE, Hensel M, Gleeson C, Holden DW. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci U S A 93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gerlach RG, Claudio N, Rohde M, Jackel D, Wagner C, Hensel M. 2008. Cooperation of Salmonella pathogenicity islands 1 and 4 is required to breach epithelial barriers. Cell Microbiol 10:2364–2376. doi: 10.1111/j.1462-5822.2008.01218.x. [DOI] [PubMed] [Google Scholar]
- 85.Ehrbar K, Hapfelmeier S, Stecher B, Hardt WD. 2004. InvB is required for type III-dependent secretion of SopA in Salmonella enterica serovar Typhimurium. J Bacteriol 186:1215–1219. doi: 10.1128/JB.186.4.1215-1219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stecher B, Hapfelmeier S, Muller C, Kremer M, Stallmach T, Hardt WD. 2004. Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar Typhimurium colitis in streptomycin-pretreated mice. Infect Immun 72:4138–4150. doi: 10.1128/IAI.72.7.4138-4150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]