ABSTRACT
High-pressure processing is a nonthermal method of food preservation that uses pressure to inactivate microorganisms. To ensure the effective validation of process parameters, it is important that the design of challenge protocols consider the potential for resistance in a particular species. Herein, the responses of 99 diverse Salmonella enterica strains to high pressure are reported. Members of this population belonged to 24 serovars and were isolated from various Canadian sources over a period of 26 years. When cells were exposed to 600 MPa for 3 min, the average reduction in cell numbers for this population was 5.6 log10 CFU/ml, with a range of 0.9 log10 CFU/ml to 6 log10 CFU/ml. Eleven strains, from 5 serovars, with variable levels of pressure resistance were selected for further study. The membrane characteristics (propidium iodide uptake during and after pressure treatment, sensitivity to membrane-active agents, and membrane fatty acid composition) and responses to stressors (heat, nutrient deprivation, desiccation, and acid) for this panel suggested potential roles for the cell membrane and the RpoS regulon in mediating pressure resistance in S. enterica. The data indicate heterogeneous and multifactorial responses to high pressure that cannot be predicted for individual S. enterica strains.
IMPORTANCE The responses of foodborne pathogens to increasingly popular minimal food decontamination methods are not understood and therefore are difficult to predict. This report shows that the responses of Salmonella enterica strains to high-pressure processing are diverse. The magnitude of inactivation does not depend on how closely related the strains are or where they were isolated. Moreover, strains that are resistant to high pressure do not behave similarly to other stresses, suggesting that more than one mechanism might be responsible for resistance to high pressure and the mechanisms used may vary from one strain to another.
KEYWORDS: RpoS, Salmonella enterica, serovars, high-pressure processing, outer membrane
INTRODUCTION
High-pressure processing (HPP) is a method of nonthermal food processing in which microorganisms are inactivated by pressures exceeding 300 MPa (2,961 atm). Pressure is transmitted by a fluid medium surrounding the food and hence is applied evenly and instantaneously throughout the food matrix. Because the process does not require added heat or chemicals, it is an attractive method to control the levels of spoilage organisms in minimally processed foods. Examples of HPP-treated foods that are commercially available include fruit juices and jams, guacamole, oysters, and ready-to-eat sliced meats such as chicken, turkey, and ham (1).
The kinetics of microbial inactivation by high pressure are complex (2). Cellular inactivation is not proportional to the magnitude or holding time of the applied pressure (3, 4). Increased holding times, especially at sublethal pressures, may contribute to tailing effects, in which a subpopulation of cells survive the inactivation process (5). The effects of the food matrix on pressure inactivation of cells are variable. Some physiochemical properties of food (e.g., low water activity) may exert protective effects, whereas others (e.g., low pH) may increase sensitivity to high pressure (1, 5).
High pressure exerts its effects through the disruption of noncovalent bonds. Hence, its effects on cells are manifold. The cell membrane is the most pressure-sensitive cellular structure (5, 6). Physical damage to the membrane upon exposure to pressure has been examined microscopically, and the loss of barrier function has been inferred through the loss of membrane proteins, the leakage of cellular materials, and the increased uptake of membrane-impermeable dyes such as propidium iodide (PI) (7–9). Secondary pressure targets include multimeric protein and nucleic acid structures, which can affect diverse cellular processes, including replication and metabolism (6). Consequently, the responses of cells to high pressure vary considerably among and within bacterial species (6, 10–12).
Studies of pressure-resistant bacterial strains have identified a number of adaptations that may be involved in pressure resistance. Piezophilic and pressure-tolerant organisms isolated from high-pressure environments exhibit structural adaptations, such as increased levels of membrane fatty acid unsaturation, that enable life in those environments (13). Foodborne bacteria are exposed to high-pressure environments intermittently, if at all, and demonstrate more dynamic responses to high pressure. A number of studies have implicated members of the rpoS regulon in coordinating the response of Escherichia coli to high pressure (14–16). In particular, mutants that are defective in the synthesis of cyclopropane fatty acids demonstrate increased sensitivity to pressure (7, 17). It has also been suggested that genes involved in heat shock and cold shock responses are involved in pressure tolerance, despite the observation of little to no correlation between temperature and pressure stress (18–20).
Salmonella enterica is an important foodborne pathogen (21). The strains within this species are taxonomically diverse and can belong to one of >2,500 serovars, based on their antigenic properties (22). Members within a serovar may be closely related or genetically diverse (23–25). The response of Salmonella enterica to high-pressure processing has been studied in a variety of experimental systems and food matrices, including milk, juice, almonds, seeds, meat, and peanut butter (10, 12, 26–30). The reported responses are variable and must be examined in the context of process parameters, the food matrix, and strain physiology. Studies investigating the responses of multiple strains of S. enterica to high pressure within the same experimental system demonstrate the inherent variability within the species. Sherry et al. (29) reported an average reduction of 3.3 log10 CFU/ml for 40 S. enterica strains from 33 serovars (1 strain each for 30 serovars, 2 strains each for 2 serovars, and 6 strains for 1 serovar) in spent culture medium exposed to a pressure of 350 MPa for 10 min at 20°C. The reported range was 2.5 log10 CFU/ml, with many strains exhibiting a difference from the most resistant strain of <1 log10 CFU/ml. Alpas et al. (10) investigated the responses of 6 strains (2 strains each for 3 serovars), in culture broth diluted with 1% peptone water, to exposure to 345 MPa for 5 min at 25°C; this population was much more sensitive to pressure, with an average reduction in cell number of 7 log10 CFU/ml and a difference of at least 2.9 log10 CFU/ml between the most pressure-tolerant strains and the most pressure-sensitive strains. Whitney et al. (12) also observed variability among 5 strains (1 strain each for 5 serovars) associated with foodborne outbreaks. When exposed to a pressure of 300 MPa for 2 min at 6°C in tryptic soy broth (TSB), the 5 strains exhibited an average decrease of 2.4 log10 CFU/ml, ranging from 0.53 log10 CFU/ml for the most pressure-resistant isolate to 3.0 log10 CFU/ml for the most pressure-sensitive isolate. These differences were lessened at 550 MPa, at which the strains exhibited an average cell decrease of 4.7 log10 CFU/ml, with a range of 3.8 to 5.4 log10 CFU/ml.
Continued testing of S. enterica strains will establish baseline levels of pressure resistance within the species and allow comparisons to be made between and within serovars; the strains with the greatest potential for resistance could be used as appropriate strains for challenge studies and simulation of worst-case scenarios for risk assessment. To achieve this goal, we examined the responses of a diverse population of 99 strains (3 to 6 strains each for 24 serovars) of S. enterica to a pressure of 600 MPa, and 11 strains with low or high tolerance to pressure were further characterized to elucidate the physiological basis of pressure tolerance in S. enterica.
RESULTS
S. enterica strains demonstrate a range of levels of tolerance to pressure.
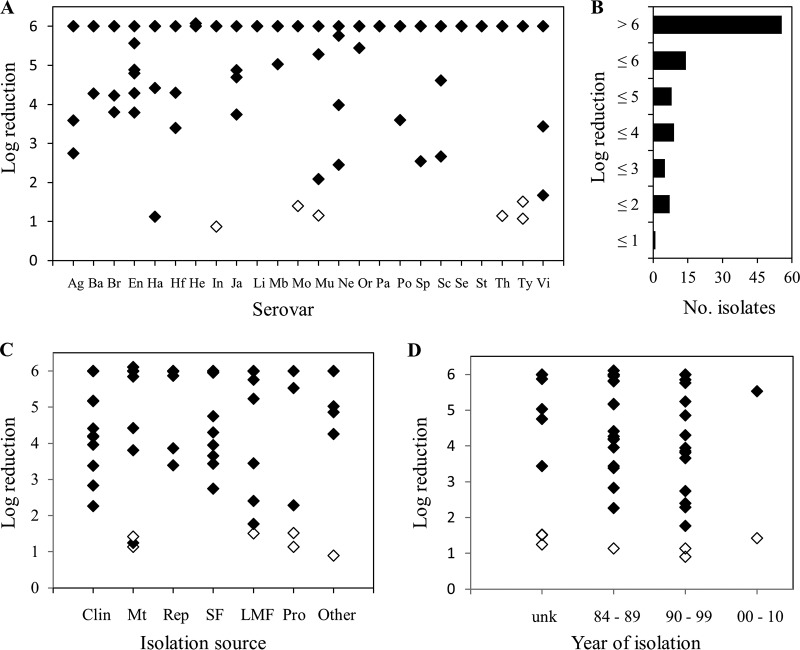
The responses of 99 S. enterica strains to exposure to a pressure of 600 MPa for 3 min involved reductions ranging from 0.9 to 6 log10 CFU/ml (Fig. 1A; also see Table S1 in the supplemental material). Approximately one-half of the strains (55/99 strains) were completely eliminated by the treatment, resulting in a median reduction of 6 log10 CFU/ml and an average reduction of 5.1 log10 CFU/ml for this population of S. enterica strains. The numbers of strains demonstrating reductions of ≤4, ≤3, ≤2, and ≤1 log10 CFU/ml were 22, 13, 7, and 1, respectively (Fig. 1B). The most pressure-tolerant strain was a S. enterica serovar Infantis isolate originally cultured from a noodle salad in 1995; this strain demonstrated a reduction of 0.9 log10 CFU/ml under the conditions of the experiment.
FIG 1.
Decreases in cell numbers for 99 S. enterica strains exposed to 600 MPa for 3 min. (A, C, and D) Decreases in cell numbers according to serovar (A), isolation source (C), and date of isolation (D). (B) Frequency of decreases observed for this population of strains. Open symbols indicate the 6 pressure-tolerant strains selected for further characterization. The limit of detection was 1 log unit below the initial cell concentration. Ag, Agona; Ba, Bareilly; Br, Braenderup; En, Enteritidis; Ha, Hadar; Hf, Hartford; He, Heidelberg; In, Infantis; Ja, Javiana; Li, Litchfield; Mb, Mbandaka; Mo, Montevideo; Mu, Muenchen; Ne, Newport; Or, Oranienburg; Pa, Panama; Po, Poona; Sp, Saintpaul; Sc, Schwarzengrund; Se, Senftenberg; St, Stanley; Th, Thompson; Ty, Typhimurium; Vi, Virchow; Clin, clinical; Mt, meat; Rep, reptile; SF, seafood; LMF, low-moisture food; Pro, produce; unk, unknown.
The responses of the isolates within 20 of the 24 serovars were heterogeneous, with no one serovar exhibiting exclusively pressure-tolerant strains. Only members of S. enterica serovars Litchfield, Panama, Senftenberg, and Stanley demonstrated uniform responses (reductions of >6 log10 CFU/ml) under the test conditions. Similarly, there did not appear to be any relationship between levels of pressure tolerance observed among the strains and either their source or their year of isolation (Fig. 1C and D). The most pressure-resistant strains, demonstrating decreases of <2 log10 CFU/ml, were originally isolated from meat, low-moisture foods, produce, or a Greek noodle salad (Fig. 1C, other) over the span of ≥26 years.
The membrane barrier is affected during and after exposure to high pressure.
To characterize the nature of pressure resistance in Salmonella, 6 pressure-tolerant strains belonging to the S. enterica serovars Infantis, Montevideo, Muenchen, Thompson, and Typhimurium (2 strains) were selected for further characterization. Five pressure-sensitive strains belonging to those serovars were randomly selected for comparison.
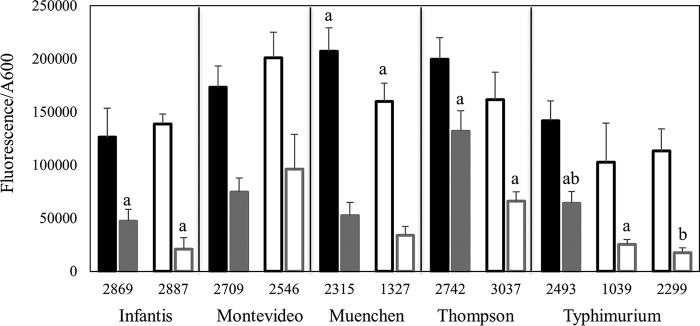
The membrane damage sustained by the 11 strains during exposure to pressure of 600 MPa for 3 min was assessed by exposing cells to the membrane-impermeable fluorescent dye propidium iodide (PI) prior to pressure treatment (Fig. 2). Cells that were not exposed to pressure had low baseline fluorescence values, ranging from 4,130 to 12,500 fluorescence units (average, 7,350 fluorescence units), suggesting that the influx of PI was low or negligible. In contrast, the fluorescence values of cells exposed to high pressure increased by >20-fold, ranging from 82,300 to 236,000 fluorescence units (average, 164,000 fluorescence units). The increase in fluorescence suggested considerable uptake of PI by the cells during pressurization. With the exception of S. enterica serovar Muenchen, the fluorescence values, and hence the permeability of the membranes of the pressure-tolerant strains to PI, were comparable to those of the pressure-sensitive strains. Strain 1327, the pressure-tolerant representative strain from S. enterica serovar Muenchen, demonstrated a fluorescence value of 160,000 fluorescence units, whereas its pressure-sensitive counterpart (strain 2315) demonstrated a modestly but significantly higher fluorescence value of 207,000 fluorescence units.
FIG 2.
Propidium iodide uptake by pressure-sensitive (closed bars) and pressure-tolerant (open bars) strains of S. enterica during (black) and after (gray) exposure to 600 MPa for 3 min. Within each panel, bars that share a lowercase letter are significantly different (P < 0.05).
To examine the restoration of the membrane barrier after pressurization, cells were exposed to pressure and then were exposed to PI for 10 min following pressure removal (Fig. 2). The fluorescence values for the cells under these conditions ranged from 16,500 to 154,000 fluorescence units (average, 64,700 fluorescence units). These values were lower than those for cells exposed to PI prior to pressurization, suggesting that the membrane barrier had been restored to some degree during the interval between the additions of the dye to the two sets of cells. Comparison of the fluorescence values for the pressure-tolerant strains with those for their respective pressure-sensitive counterparts revealed that the members of S. enterica serovars Infantis, Thompson, and Typhimurium demonstrated significantly less PI uptake 10 min after the removal of pressure.
Membrane characteristics of pressure-tolerant S. enterica strains.
The growth of the 11 pressure-tolerant or pressure-sensitive strains in membrane-active detergents was determined, to assess the baseline membrane characteristics of these organisms. In comparison with their pressure-sensitive counterparts, the pressure-tolerant strains belonging to S. enterica serovars Infantis, Montevideo, and Typhimurium demonstrated more robust growth in 1% SDS (Fig. 3). Specifically, these strains exhibited a shorter lag time in the detergent (130 min versus 500 min, 225 min versus 360 min, and 80 min and 130 min versus 710 min for the pressure-tolerant versus pressure-sensitive S. enterica strains belonging to S. enterica serovars Infantis, Montevideo, and Typhimurium, respectively). The S. enterica serovar Thompson pair exhibited similar growth kinetics; however, the turbidity of the pressure-sensitive strain appeared to peak and then to decrease gradually, in comparison with the pressure-tolerant strain. The growth of the 11 strains in other membrane-active agents, including Triton X-100, sodium deoxycholate, EDTA, and MgCl2, was indistinguishable (data not shown). All 11 strains were susceptible to polymyxin B sulfate, gentamicin, and kanamycin.
FIG 3.
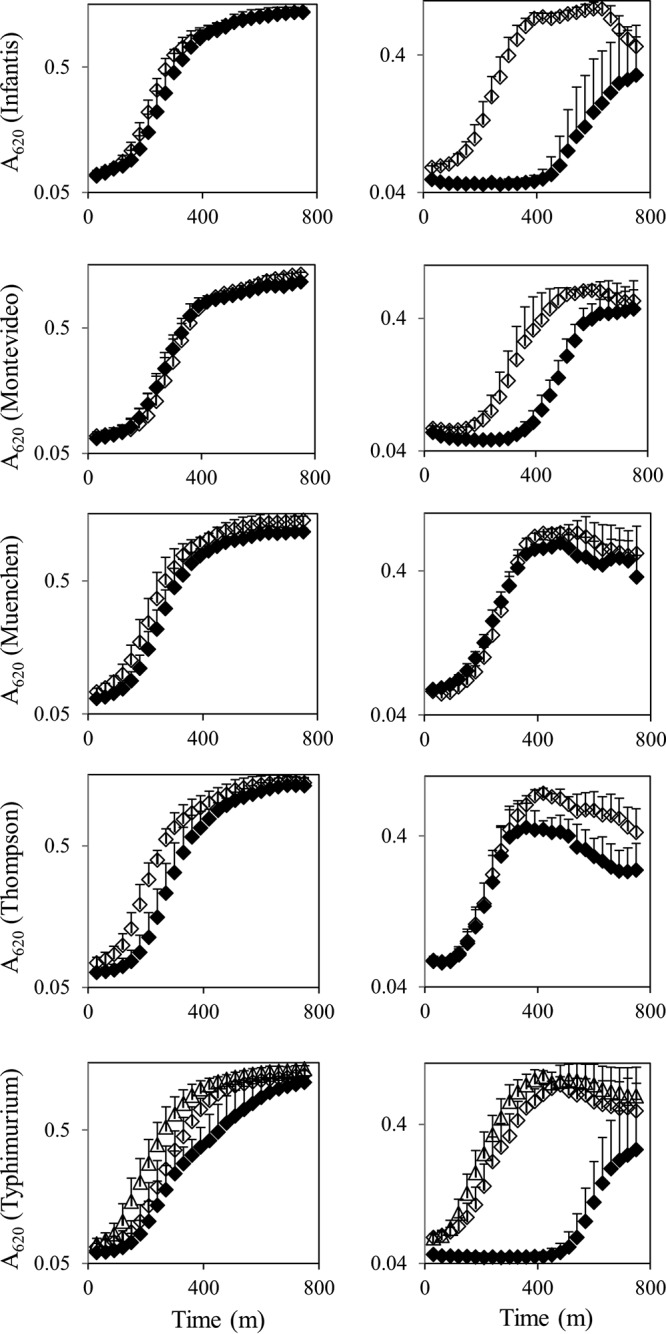
Growth of pressure-tolerant (open symbols) and pressure-sensitive (closed symbols) strains of S. enterica in Luria-Bertani (LB) broth (left) or LB broth with 1% SDS (right). For S. enterica serovar Typhimurium: open triangles, S. enterica serovar Typhimurium strain 1039; open diamonds, S. enterica serovar Typhimurium strain 2299.
The membrane fatty acid compositions of stationary-phase cultures of the 5 pressure-sensitive strains and 6 pressure-resistant strains grown in TSB were determined. For all strains, approximately one-half of the membrane lipids were saturated fatty acid methyl esters, with palmitic aid (C16:0) predominating (Table 1). The second most predominant species were the unsaturated fatty acids palmitoleic acid (9-cis-C16:1) and cis-vaccenic acid (11-cis-C18:1) for S. enterica serovar Montevideo and the cyclopropane fatty acids cis-9,10-methylene-hexadecanoic acid (ΔC17) and lactobacillic acid (ΔC19) for S. enterica serovars Infantis, Muenchen, Thompson, and Typhimurium. Comparison of the fatty acid profiles of the pressure-sensitive strains and the pressure-resistant strains revealed that the resistant counterparts of S. enterica serovars Infantis, Muenchen, Thompson, and Typhimurium had smaller proportions of unsaturated fatty acids than did their pressure-sensitive counterparts (10.9%, 10.1%, 9.48%, and 13.4% and 7.34% for the pressure-tolerant strains versus 12.6%, 11.0%, 11.1%, and 14.6% for the pressure-sensitive strains, respectively). Resistant members of S. enterica serovars Infantis, Muenchen, and Typhimurium were further distinguished from their sensitive counterparts by possessing larger proportions of cyclopropane fatty acids in their membranes (26.9%, 24.8%, and 17.9% and 27.9% for the pressure-tolerant strains versus 25.5%, 21.0%, and 17.0% for the pressure-sensitive strains, respectively). Although the difference in membrane composition was striking for some strains (e.g., S. enterica serovar Typhimurium strains 2493 and 2299), when the findings were viewed collectively there were no significant differences in fatty acid composition between the pressure-resistant and pressure-sensitive strains.
TABLE 1.
Membrane fatty acid composition of S. enterica strains with different levels of pressure tolerance
| Strain | Pressure tolerance | Composition (% of total membrane lipids)a |
|||
|---|---|---|---|---|---|
| SFA | HFA | UFA | CFA | ||
| S. enterica serovar Infantis 2869 | Sensitive | 49.72 | 8.81 | 12.64 | 25.48 |
| S. enterica serovar Infantis 2887 | Resistant | 49.1 | 9.15 | 10.91 | 26.93 |
| S. enterica serovar Montevideo 2709 | Sensitive | 48.75 | 10.59 | 20.11 | 13.42 |
| S. enterica serovar Montevideo 2546 | Resistant | 46.04 | 9.58 | 27.09 | 10.47 |
| S. enterica serovar Muenchen 2315 | Sensitive | 52.99 | 9.71 | 10.95 | 21.04 |
| S. enterica serovar Muenchen 1327 | Resistant | 52.08 | 9.08 | 10.05 | 24.78 |
| S. enterica serovar Thompson 2742 | Sensitive | 50.94 | 8.97 | 11.09 | 25.08 |
| S. enterica serovar Thompson 3037 | Resistant | 53.52 | 9.59 | 9.48 | 21.91 |
| S. enterica serovar Typhimurium 2493 | Sensitive | 53.47 | 9.86 | 14.61 | 17.01 |
| S. enterica serovar Typhimurium 1039 | Resistant | 52.75 | 10.09 | 13.38 | 17.88 |
| S. enterica serovar Typhimurium 2299 | Resistant | 51.97 | 9.16 | 7.34 | 27.89 |
SFA, saturated fatty acids, i.e., lauric acid (C12:0), myristic acid (C14:0), palmitic acid (C16:0), and steric acid (C18:0); HFA, hydroxylated fatty acids, i.e., 3-hydroxy-myrisitc acid (3-OH-C14); UFA, unsaturated fatty acids, i.e., palmitoleic acid (9-cis-C16:1) and cis-vaccenic acid (11-cis-C18:1); CFA, cyclopropane fatty acids, i.e., cis-9,10-methylene-hexadecanoic acid (ΔC17) and lactobacillic acid (ΔC19).
General stress resistance of pressure-tolerant S. enterica strains.
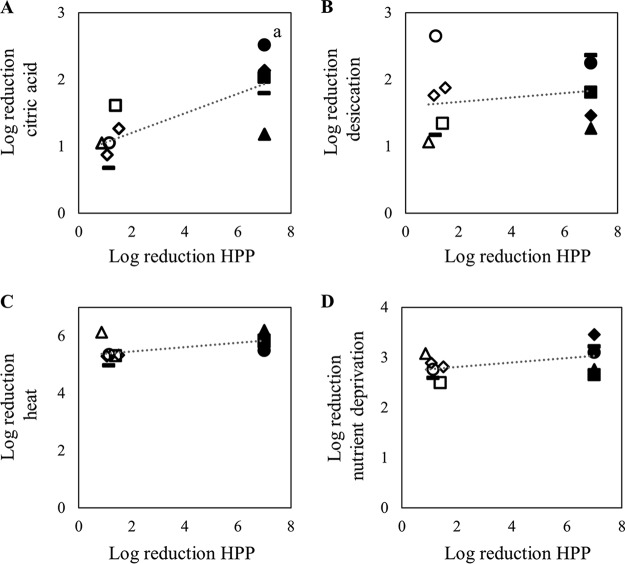
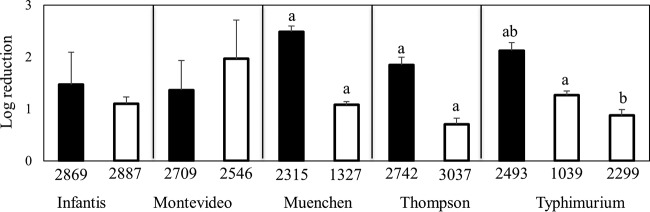
To determine whether pressure resistance in S. enterica could be generalized to resistance to other forms of stress, the 11 pressure-tolerant or pressure-sensitive strains were exposed to stress-inducing conditions, including heat, nutrient deprivation, desiccation, and citric acid. When the cell numbers of the pressure-tolerant strains were compared with those of their pressure-sensitive counterparts, significant differences were not observed with respect to survival at 60°C, with nutrient deprivation, or with desiccation (Fig. 4). Significant differences were observed, however, when the 11 strains were exposed to 50 mM citric acid (pH 2.2) for 2 h (Fig. 4A and 5). Under those conditions, the pressure-tolerant strains from S. enterica serovars Muenchen, Thompson, and Typhimurium had greater numbers of survivors than did the pressure-sensitive strains from the same serovars. These differences were not observed for the members of S. enterica serovars Infantis and Montevideo.
FIG 4.
Correlations between inactivation by high-pressure processing (HPP) (600 MPa for 3 min) and exposure to 50 mM citric acid (pH 2.2) for 2 h (A), desiccation (B), heat (60°C for 5 min) (C), or nutrient deprivation (D) for pressure-tolerant (open symbols) and pressure-sensitive (closed symbols) S. enterica strains. Triangles, S. enterica serovar Infantis; squares, S. enterica serovar Montevideo; circles, S. enterica serovar Muenchen; horizontal bars, S. enterica serovar Thompson; diamonds, S. enterica serovar Typhimurium. a, significant difference between the two groups of bacteria (P < 0.05).
FIG 5.
Average reductions for pressure-sensitive (closed bars) and pressure-tolerant (open bars) strains of S. enterica after exposure to 50 mM citric acid (pH 2.2) for 2 h. Within each panel, bars that share a lowercase letter are significantly different (P < 0.05).
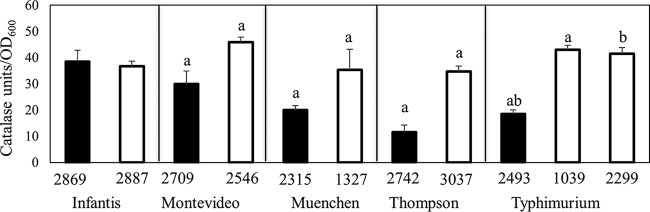
Catalase activity of pressure-tolerant isolates.
The phenotypic differences for some of the pressure-tolerant strains versus the pressure-sensitive strains in response to citric acid suggested the involvement of RpoS. To gauge the levels of RpoS activity in the cells, a catalase assay was performed (Fig. 6). All 11 strains were positive for catalase activity. No differences in catalase activity were observed between the 2 S. enterica serovar Infantis strains. However, the pressure-tolerant strains from S. enterica serovars Montevideo, Muenchen, Thompson, and Typhimurium had significantly higher levels of catalase activity than did their pressure-sensitive counterparts.
FIG 6.
Average catalase activities of pressure-sensitive (closed bars) and pressure-tolerant (open bars) strains of S. enterica. Within each panel, bars that share a lowercase letter are significantly different (P < 0.05).
DISCUSSION
The responses of individual S. enterica strains to high hydrostatic pressure depend on a number of factors related to both the process and the organism (1, 4, 5). In order to define the basic levels of pressure resistance in Salmonella, 99 S. enterica strains from 24 serovars, isolated from diverse sources over a period of ≥26 years, were subjected to high pressure under identical experimental conditions. Upon exposure to 600 MPa for 3 min, a difference of at least 5 log10 CFU/ml between the most pressure-tolerant strain and the most pressure-sensitive strains was observed. Despite the close genetic relationships among strains in some serovars, there did not appear to be any relationship between the degree of pressure resistance and the serovar classification; the isolation source and the date of isolation also did not correlate with pressure resistance. Similar trends were observed in a study investigating the pressure resistance of 100 E. coli strains (11). However, the distributions of the resistance patterns differed between the bacterial species. The E. coli strains demonstrated reductions that appeared to be evenly distributed between 3 log10 CFU/ml and 5.5 log10 CFU/ml, with a median of 3.9 log10 CFU/ml. In contrast, the median reduction observed for the S. enterica strains was 6 log10 CFU/ml, largely due to the complete inactivation of 56% of the strains upon exposure to 600 MPa. The S. enterica population harbored more strains that demonstrated reductions of ≤2 log10 CFU/ml, compared to the E. coli population (8 strains and 3 strains, respectively). Therefore, although the majority of S. enterica strains may be highly sensitive to high-pressure processing, significant proportions may exhibit unusual levels of pressure tolerance. This conclusion contrasts with previous reports that suggested more uniform responses to high pressure (10, 29). These differences may be a result of differences in process parameters and experimental conditions used in the studies or may reflect inherent differences between the tested strains.
The membrane characteristics of 5 S. enterica serovars were investigated to determine the nature of pressure resistance in Salmonella. The pressure-tolerant strains of some serovars demonstrated reduced levels of propidium iodide uptake (S. enterica serovars Infantis, Muenchen, Thompson, and Typhimurium) and higher levels of cyclopropane fatty acids (S. enterica serovars Infantis, Muenchen, and Typhimurium), as reported for some E. coli strains (Table 2) (7, 17). Two exceptions to this trend were the pressure-tolerant strains belonging to S. enterica serovars Thompson and Montevideo. It may be that, in the case of the pressure-tolerant S. enterica serovar Montevideo strain, 10 min was not a suitable time point at which to observe differences in propidium iodide uptake. However, neither strain demonstrated elevated levels of cyclopropane fatty acids, in comparison to their pressure-sensitive counterparts. The S. enterica serovar Thompson strain exhibited reduced propidium iodide uptake, and the S. enterica serovar Montevideo strain showed increased growth in 1% SDS. These results suggest that, at least in these 2 S. enterica serovars, the membrane bilayer may be involved but may not play as critical a role in pressure resistance as has been observed for other S. enterica serovars and E. coli. For example, membrane repair after the removal of pressure is an energy-dependent process requiring the synthesis of both RNA and protein (31). Thus, it may be that those systems were differentially affected in the pressure-sensitive strains upon the application of pressure. Alternatively, other features of the membrane that were not investigated in this study, such as membrane proteins, may play a more critical role in pressure resistance, as has been observed for some strains of S. enterica (8, 9).
TABLE 2.
Summary of phenotypic properties associated with pressure-tolerant strains in five S. enterica serovars
| S. enterica serovar | Membrane propertya |
RpoS regulon |
|||
|---|---|---|---|---|---|
| PI uptake | CFA | Growth in SDS | Acid tolerance | Catalase activity | |
| Infantis | + | + | + | − | − |
| Montevideo | − | − | + | − | + |
| Muenchen | + | + | − | + | + |
| Thompson | + | − | − | + | + |
| Typhimurium | + | + | + | + | + |
+, a difference between the pressure-tolerant and pressure-sensitive strains within the serovar was observed; −, a difference between the pressure-tolerant and pressure-sensitive strains within the serovar was not observed; CFA, cyclopropane fatty acids.
Resistance to pressure in E. coli is correlated with RpoS activity, and exposure to high pressure selects for variants with increased RpoS activity (15, 16). The role of RpoS in maintaining the integrity of the E. coli cell membrane during exposure to stressors such as pressure and SDS has been established, and there are a number of downstream targets that may also mediate pressure resistance independent of the cell membrane (7, 17, 32–35). The differences observed among the 11 S. enterica strains with respect to acid resistance and catalase activity suggested a role for RpoS in coordinating the response to pressure in this organism as well. The overall responses of the 11-strain panel were heterogeneous. Compared to their pressure-sensitive counterparts, the pressure-tolerant strains of some serovars (S. enterica serovars Muenchen, Thompson, and Typhimurium) exhibited phenotypes associated with enhanced RpoS activity (Table 2). In contrast, the pressure-tolerant S. enterica serovar Montevideo strain exhibited only enhanced catalase activity, and the pressure-tolerant S. enterica serovar Infantis strain did not exhibit higher levels of any activity associated with RpoS. These results suggest that the role of RpoS in mediating pressure resistance may be more critical in some S. enterica serovars than in others. At present, there are few data on the contributions of RpoS to stress resistance in different serovars of S. enterica, and this is an area that warrants further study.
The 5 serovars investigated in this study demonstrated unique patterns of membrane properties and stress responses that are correlated with pressure resistance in E. coli (Table 2). There did not seem to be a uniform coordinated response to high pressure among the strains investigated in this study. The strains of some serovars, such as S. enterica serovar Infantis, seemed to rely solely on membrane phenotypes to mediate pressure resistance, whereas other serovars, such as S. enterica serovars Muenchen and Typhimurium, might have also used RpoS to coordinate responses to pressure. Therefore, based on this data set, it appears that there may be multiple paths to pressure resistance in S. enterica.
The responses of S. enterica to high pressure are heterogeneous with respect to both the degree of inactivation and the mechanisms used to overcome it. The unpredictable nature of these responses should be considered during the design of challenge studies and the selection of surrogate strains. This data set will be useful for the identification of the most pressure-resistant populations of S. enterica and the simulation of worst-case scenarios for risk assessment.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The 99 strains used in this study were from the Bureau of Microbial Hazards Salmonella culture collection and comprised 24 serovars. Three to 6 strains from each serovar were tested; the strains are listed in Table S1 in the supplemental material. Prior to each set of experiments, strains were freshly cultured, from storage at −80°C, on tryptic soy agar (TSA) at 35°C for approximately 24 h. Two or 3 colonies from TSA were inoculated into the specified culture medium and incubated for 18 h at 37°C, with shaking at 250 rpm, on the day preceding each experiment. All procedures were carried out at ambient temperatures unless indicated otherwise.
High-pressure treatment of bacterial cells.
Cells were cultured in TSB, collected by centrifugation, washed twice in phosphate-buffered saline (PBS) (10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl), and resuspended in PBS to a cell density of 1 × 107 CFU/ml. A portion of the cell suspension (4 ml) was used to fill sterile sampling bags (7.5 by 18.5 cm; Fisher Scientific, Ottawa, Canada). The bags were sealed without any headspace by using a Midwest Pacific Impulse heat sealer and were trimmed of excess plastic to form pouches (approximately 2.5 by 6 cm). The pouches were placed and sealed in a larger sampling bag (11.5 by 23 cm; Fisher) containing 1% bleach, to inactivate pathogens in the event of a leak during pressure treatment. Pressure treatment was administered using a 1-liter-capacity pressure vessel (Dustec Hochdrucktechnik GmbH, Wismar, Germany), with water as the transmission fluid. A pressure of 600 MPa was applied for 3 min, with a 1-min pressurization/30-s depressurization cycle. The pressure treatment was conducted at 24°C, and adiabatic heating of the unit during pressurization increased the temperature of the water in the pressure cell to a maximum of 39°C. Cells were immediately enumerated by direct TSA plate counting. Plates were incubated at 35°C for 72 h. The limit of detection for the surviving cells was 10 CFU/ml, or a reduction of 1 log unit below the starting concentration. If no surviving cells were recovered, then the reduction was recorded as >6 log units.
To assess membrane permeability during the pressurization/depressurization cycle, bacterial suspensions were prepared to an optical density at 600 nm (OD600) of 0.2 and propidium iodide (Molecular Probes, Eugene, OR) was added to a final concentration of 3 μM prior to pressure treatment. Cells were collected immediately after exposure to pressure, washed twice, and resuspended in PBS.
The degree of membrane permeabilization following pressure treatment was assessed by incubating pressure-treated cells with 3 μM propidium iodide for 10 min at room temperature and processing the cells as described above. Cellular fluorescence was measured in a fluorescence microplate reader (BioTek, Winooski, VT), with an excitation wavelength of 495 nm and an emission wavelength of 615 nm. Fluorescence units were calculated by normalizing the fluorescence readings to the absorbance of the cell suspensions. The background fluorescence of the untreated cells has been subtracted from the reported fluorescence values. Differences between means were calculated using Student's t test, with a significance of 0.05.
Analysis of tolerance to stress.
Stationary-phase cells cultured in TSB were harvested by centrifugation, washed twice in PBS, and resuspended to a final cell concentration of 2 × 109 CFU/ml PBS. Heat tolerance was determined by incubating cells in a heated circulating water bath (VWR, Mississauga, Canada) set to 60°C. After 5 min, the cells were immediately placed in an ice water bath. Surviving cells were enumerated by direct plating on TSA and incubation at 35°C for 24 h. Acid tolerance was determined by incubating cells in an equal volume of 100 mM citric acid (resulting in a final pH of 2.2) for 2 h at room temperature prior to enumeration. Tolerance to nutrient deprivation and desiccation was determined as described previously (36, 37). Reported results are the average and standard deviation of three independent experiments. Differences between means were calculated using Student's t test, with a significance of 0.05.
Catalase assays.
The catalase activity of cells was determined using the method described by Iwase et al. (38). Briefly, stationary-phase cells were collected from TSB, washed twice, and resuspended in PBS. A 100-μl aliquot of this bacterial suspension was added to a borosilicate glass tube (13 mm by 100 mm) containing 100 μl of 1% Triton X-100 (Sigma-Aldrich, Oakville, Canada) and 100 μl of 30% hydrogen peroxide (Sigma-Aldrich). After 15 min of incubation at room temperature, the height of the foam was measured using a metric ruler. Catalase activity was estimated using a standard curve prepared by using catalase from bovine liver (product no. C1345; Sigma), and findings were normalized to the optical density of the cell preparation. Results are reported as the average and standard deviation of three independent experiments. Statistical differences between sample means were calculated using Student's t test, with a significance level of 0.05.
Growth assays.
Overnight cultures of cells grown in Luria-Bertani (LB) broth (1% tryptone peptone, 0.5% yeast extract [wt/vol]) were diluted 100-fold in either LB broth or LB broth containing supplements. Supplements included the detergents SDS, Triton X-100, and sodium deoxycholate, used at concentrations of 1%. EDTA was used at a concentration of 1 mM. For some experiments, 10 mM MgCl2 was added to the detergent solution. Cells were aliquoted in duplicate in a 96-well flat-bottomed polystyrene plate and incubated in a microplate reader (BioTek) set at 37°C, with continuous shaking. The instrument measured and recorded the absorbance at 620 nm every 30 min. Growth parameters (lag time and doubling time) were calculated according to the procedures described by Hall et al. (39). Results reported are the average and standard deviation of six trials conducted with three independent cultures.
Antibiotic sensitivity assays.
The sensitivity of selected strains to antibiotics was assessed with the disk diffusion assay, according to the procedures described by the Clinical and Laboratory Standards Institute (40).
Fatty acid methyl ester analysis.
Strains were grown in TSB to stationary phase and washed with PBS as described previously. Cell pellets were shipped on dry ice to a certified testing laboratory, where they were analyzed by gas chromatography (41).
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by Health Canada (A-base) to support Canada's food safety programs. I thank Alex Gill and Franco Pagotto for critical reading of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01673-17.
REFERENCES
- 1.Daryaei H, Balasubramaniam VM. 2012. Microbial decontamination of food by high pressure processing, p 370–406. In Demirci A, Ngudi MO (ed), Microbial decontamination in the food industry: novel methods and applications. Woodhead Publishing Ltd., Philadelphia, PA. [Google Scholar]
- 2.Serment-Moreno V, Fuentes C, Barbosa-Cánovas G, Torres JA, Welti-Chanes J. 2015. Evaluation of high pressure processing kinetic models for microbial inactivation using standard statistical tools and information theory criteria, and the development of generic time-pressure functions for process design. Food Bioprocess Technol 8:1244–1257. doi: 10.1007/s11947-015-1488-x. [DOI] [Google Scholar]
- 3.Donsi G, Ferrari G, Maresca P. 2010. Pasteurization of fruit juices by means of a pulsed high pressure process. J Food Sci 75:E169–E177. doi: 10.1111/j.1750-3841.2010.01535.x. [DOI] [PubMed] [Google Scholar]
- 4.Earnshaw RG, Appleyard J, Hurst RM. 1995. Understanding physical inactivation processes: combined preservation opportunities using heat, ultrasound and pressure. Int J Food Microbiol 28:197–219. doi: 10.1016/0168-1605(95)00057-7. [DOI] [PubMed] [Google Scholar]
- 5.Patterson MF. 2005. Microbiology of pressure-treated foods. J Appl Microbiol 98:1400–1409. doi: 10.1111/j.1365-2672.2005.02564.x. [DOI] [PubMed] [Google Scholar]
- 6.Ganzle M, Liu Y. 2015. Mechanisms of pressure-mediated cell death and injury in Escherichia coli: from fundamentals to food applications. Front Microbiol 6:599. doi: 10.3389/fmicb.2015.00599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charoenwong D, Andrews S, Mackey B. 2011. Role of rpoS in the development of cell envelope resilience and pressure resistance in stationary-phase Escherichia coli. Appl Environ Microbiol 77:5220–5229. doi: 10.1128/AEM.00648-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ritz M, Freulet M, Orange N, Federighi M. 2000. Effects of high hydrostatic pressure on membrane proteins of Salmonella typhimurium. Int J Food Microbiol 55:115–119. doi: 10.1016/S0168-1605(00)00165-3. [DOI] [PubMed] [Google Scholar]
- 9.Wang CY, Hsu CP, Huang HW, Yang BB. 2013. The relationship between inactivation and morphological damage of Salmonella enterica treated by high hydrostatic pressure. Food Res Int 54:1482–1487. doi: 10.1016/j.foodres.2013.08.004. [DOI] [Google Scholar]
- 10.Alpas H, Kalchayanand N, Bozoglu F, Sikes A, Dunne CP, Ray B. 1999. Variation in resistance to hydrostatic pressure among strains of food-borne pathogens. Appl Environ Microbiol 65:4248–4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Gill A, McMullen L, Ganzle MG. 2015. Variation in heat and pressure resistance of verotoxigenic and nontoxigenic Escherichia coli. J Food Prot 78:111–120. doi: 10.4315/0362-028X.JFP-14-267. [DOI] [PubMed] [Google Scholar]
- 12.Whitney BM, Williams RC, Eifert J, Marcy J. 2007. High-pressure resistance variation of Escherichia coli O157:H7 strains and Salmonella serovars in tryptic soy broth, distilled water, and fruit juice. J Food Prot 70:2078–2083. doi: 10.4315/0362-028X-70.9.2078. [DOI] [PubMed] [Google Scholar]
- 13.Kato C, Bartlett DH. 1997. The molecular biology of barophilic bacteria. Extremophiles 1:111–116. doi: 10.1007/s007920050023. [DOI] [PubMed] [Google Scholar]
- 14.Malone AS, Chung YK, Yousef AE. 2006. Genes of Escherichia coli O157:H7 that are involved in high-pressure resistance. Appl Environ Microbiol 72:2661–2671. doi: 10.1128/AEM.72.4.2661-2671.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robey M, Benito A, Hutson RH, Pascual C, Park SF, Mackey BM. 2001. Variation in resistance to high hydrostatic pressure and rpoS heterogeneity in natural isolates of Escherichia coli O157:H7. Appl Environ Microbiol 67:4901–4907. doi: 10.1128/AEM.67.10.4901-4907.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanlint D, Rutten N, Govers SK, Michiels CW, Aertsen A. 2013. Exposure to high hydrostatic pressure rapidly selects for increased RpoS activity and general stress-resistance in Escherichia coli O157:H7. Int J Food Microbiol 163:28–33. doi: 10.1016/j.ijfoodmicro.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Chen YY, Ganzle MG. 2016. Influence of cyclopropane fatty acids on heat, high pressure, acid and oxidative resistance in Escherichia coli. Int J Food Microbiol 222:16–22. doi: 10.1016/j.ijfoodmicro.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 18.Benito A, Ventoura G, Casadei M, Robinson T, Mackey B. 1999. Variation in resistance of natural isolates of Escherichia coli O157 to high hydrostatic pressure, mild heat, and other stresses. Appl Environ Microbiol 65:1564–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanlint D, Rutten N, Michiels CW, Aertsen A. 2012. Emergence and stability of high-pressure resistance in different food-borne pathogens. Appl Environ Microbiol 78:3234–3241. doi: 10.1128/AEM.00030-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Welch TJ, Farewell A, Neidhardt FC, Bartlett DH. 1993. Stress response of Escherichia coli to elevated hydrostatic pressure. J Bacteriol 175:7170–7177. doi: 10.1128/jb.175.22.7170-7177.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, Dopfer D, Fazil A, Fischer-Walker CL, Hald T, Hall AJ, Keddy KH, Lake RJ, Lanata CF, Torgerson PR, Havelaar AH, Angulo FJ. 2015. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med 12:e1001921. doi: 10.1371/journal.pmed.1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guibourdenche M, Roggentin P, Mikoleit M, Fields PI, Bockemuhl J, Grimont PA, Weill FX. 2010. Supplement 2003–2007 (no. 47) to the White-Kauffmann-Le Minor scheme. Res Microbiol 161:26–29. doi: 10.1016/j.resmic.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Achtman M, Wain J, Weill FX, Nair S, Zhou Z, Sangal V, Krauland MG, Hale JL, Harbottle H, Uesbeck A, Dougan G, Harrison LH, Brisse S. 2012. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog 8:e1002776. doi: 10.1371/journal.ppat.1002776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beltran P, Musser JM, Helmuth R, Farmer JJ III, Frerichs WM, Wachsmuth IK, Ferris K, McWhorter AC, Wells JG, Cravioto A. 1988. Toward a population genetic analysis of Salmonella: genetic diversity and relationships among strains of serotypes S. choleraesuis, S. derby, S. dublin, S. enteritidis, S. heidelberg, S. infantis, S. newport, and S. typhimurium. Proc Natl Acad Sci U S A 85:7753–7757. doi: 10.1073/pnas.85.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobsen A, Hendriksen RS, Aaresturp FM, Ussery DW, Friis C. 2011. The Salmonella enterica pan-genome. Microb Ecol 62:487–504. doi: 10.1007/s00248-011-9880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen H. 2007. Use of linear, Weibull, and log-logistic functions to model pressure inactivation of seven foodborne pathogens in milk. Food Microbiol 24:197–204. doi: 10.1016/j.fm.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 27.D'Souza T, Karwe M, Schaffner DW. 2014. Effect of high hydrostatic pressure on Salmonella inoculated into creamy peanut butter with modified composition. J Food Prot 77:1664–1668. doi: 10.4315/0362-028X.JFP-14-062. [DOI] [PubMed] [Google Scholar]
- 28.Neetoo H, Chen H. 2010. Pre-soaking of seeds enhances pressure inactivation of E. coli O157:H7 and Salmonella spp. on crimson clover, red clover, radish and broccoli seeds. Int J Food Microbiol 137:274–280. doi: 10.1016/j.ijfoodmicro.2009.11.026. [DOI] [PubMed] [Google Scholar]
- 29.Sherry AE, Patterson MF, Madden RH. 2004. Comparison of 40 Salmonella enterica serovars injured by thermal, high-pressure and irradiation stress. J Appl Microbiol 96:887–893. doi: 10.1111/j.1365-2672.2004.02211.x. [DOI] [PubMed] [Google Scholar]
- 30.Tananuwong K, Chitsakun T, Tattiyakul J. 2012. Effects of high-pressure processing on inactivation of Salmonella Typhimurium, eating quality, and microstructure of raw chicken breast fillets. J Food Sci 77:E321–E327. doi: 10.1111/j.1750-3841.2012.02941.x. [DOI] [PubMed] [Google Scholar]
- 31.Chilton P, Isaacs NS, Manias P, Mackey BM. 2001. Biosynthetic requirements for the repair of membrane damage in pressure-treated Escherichia coli. Int J Food Microbiol 71:101–104. doi: 10.1016/S0168-1605(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 32.Mitchell AM, Wang W, Silhavy TJ. 2017. Novel RpoS-dependent mechanisms strengthen the envelope permeability barrier during stationary phase. J Bacteriol 199:e00708-16. doi: 10.1128/JB.00708-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grogan DW, Cronan JE Jr. 1997. Cyclopropane ring formation in membrane lipids of bacteria. Microbiol Mol Biol Rev 61:429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ibanez-Ruiz M, Robbe-Saule V, Hermant D, Labrude S, Norel F. 2000. Identification of RpoS (σS)-regulated genes in Salmonella enterica serovar Typhimurium. J Bacteriol 182:5749–5756. doi: 10.1128/JB.182.20.5749-5756.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schellhorn HE, Audia JP, Wei LI, Chang L. 1998. Identification of conserved, RpoS-dependent stationary-phase genes of Escherichia coli. J Bacteriol 180:6283–6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang FC, Libby SJ, Buchmeier NA, Loewen PC, Switala J, Harwood J, Guiney DG. 1992. The alternative sigma factor KatF (RpoS) regulates Salmonella virulence. Proc Natl Acad Sci U S A 89:11978–11982. doi: 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gruzdev N, Pinto R, Sela S. 2011. Effect of desiccation on tolerance of Salmonella enterica to multiple stresses. Appl Environ Microbiol 77:1667–1673. doi: 10.1128/AEM.02156-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iwase T, Tajima A, Sugimoto S, Okuda K, Hironaka I, Kamata Y, Takada K, Mizunoe Y. 2013. A simple assay for measuring catalase activity: a visual approach. Sci Rep 3:3081. doi: 10.1038/srep03081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hall BG, Acar H, Nandipati A, Barlow M. 2014. Growth rates made easy. Mol Biol Evol 31:232–238. doi: 10.1093/molbev/mst187. [DOI] [PubMed] [Google Scholar]
- 40.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. CLSI document M100-25. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 41.Sasser M. 1990. Identification of bacteria by gas chromatography of cellular fatty acids. MIDI, Newark, DE. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.