ABSTRACT
The haloalkane dehalogenase enzyme DmmA was identified by marine metagenomic screening. Determination of its crystal structure revealed an unusually large active site compared to those of previously characterized haloalkane dehalogenases. Here we present a biochemical characterization of this interesting enzyme with emphasis on its structure-function relationships. DmmA exhibited an exceptionally broad substrate specificity and degraded several halogenated environmental pollutants that are resistant to other members of this enzyme family. In addition to having this unique substrate specificity, the enzyme was highly tolerant to organic cosolvents such as dimethyl sulfoxide, methanol, and acetone. Its broad substrate specificity, high overexpression yield (200 mg of protein per liter of cultivation medium; 50% of total protein), good tolerance to organic cosolvents, and a broad pH range make DmmA an attractive biocatalyst for various biotechnological applications.
IMPORTANCE We present a thorough biochemical characterization of the haloalkane dehalogenase DmmA from a marine metagenome. This enzyme with an unusually large active site shows remarkably broad substrate specificity, high overexpression, significant tolerance to organic cosolvents, and activity under a broad range of pH conditions. DmmA is an attractive catalyst for sustainable biotechnology applications, e.g., biocatalysis, biosensing, and biodegradation of halogenated pollutants. We also report its ability to convert multiple halogenated compounds to corresponding polyalcohols.
KEYWORDS: biotechnology, cosolvents, enzyme, haloalkane dehalogenase, marine, microbial, stability, substrate specificity
INTRODUCTION
The haloalkane dehalogenases (HLDs) are a large family of hydrolytic enzymes that have been studied extensively over the last 25 years. In structural terms, they belong to the α/β-hydrolase superfamily (1–3), and they have a common mechanism of action involving the hydrolytic cleavage of carbon-halogen bonds in halogenated aliphatic compounds to produce the corresponding alcohol, a halide ion, and a proton as the reaction products (EC 3.5.1.8). They catalyze the hydrolysis of diverse substrates, including chlorinated, brominated, and iodinated alkanes, alkenes, cycloalkanes, alcohols, epoxides, carboxylic acids, esters, ethers, amides, and nitriles (4, 5). HLDs have many potential applications in biocatalysis, bioremediation, biosensors, and cell imaging (6).
These widespread enzymes originate from various types of organisms. The first HLDs were isolated from bacterial sources such as Xanthobacter autotrophicus GJ10 (2), Sphinghomonas japonicum UT26 (7), and Rhodococcus rhodochrous NCIMB 13064 (8) that had colonized environments highly contaminated with halogenated pollutants such as 1,2-dichloroethane, 1,2,3,4,5,6-hexachlorocyclohexane, or 1-chlorobutane. These bacteria can utilize 1-chloro-n-alkanes, 1-bromo-n-alkanes, and terminal dihalo-n-alkanes as their sole carbon source (1–3, 9). More recently, novel HLDs from diverse sources, including the eukaryotic organism Strongylocentrotus purpuratus (10), symbiotic bacteria such as Bradyrhizobium japonicum USDA 110 (11) and Mesorhizobium loti MAFF 303099 (11), and pathogenic bacteria such as Agrobacterium tumefaciens C58 (12) and Mycobacterium spp. (13), were identified in gene databases on the basis of sequence similarity. Moreover, ongoing genome and metagenome sequencing projects have further enhanced the diversity of known HLDs. Notably, several metagenomics projects have focused on screening marine environments and have the potential to reveal a wide range of proteins with novel functional and structural properties (14, 15).
DmmA is a haloalkane dehalogenase with a known tertiary structure (PDB ID 3U1T) that was identified by a marine metagenomic consortium (16). Inspection of its crystal structure revealed it to have a large active site that can accommodate bulky substrates (17). Here we report the expression, purification, and biochemical characterization of DmmA. The results show a broad pH operational range and extended stability of DmmA in the presence of organic cosolvents. By analyzing a set of diverse halogenated substrates, an exceptionally broad substrate specificity of DmmA has been revealed, including activity toward a number of stable recalcitrant environmental pollutants. All above-mentioned characteristics make DmmA a robust and versatile biocatalyst suitable for biotechnological applications, particularly in the field of environmental protection.
RESULTS
Expression and purification.
DmmA was expressed in Escherichia coli BL21(DE3) and purified by metallo-affinity chromatography. Interestingly, DmmA accounted for around 50% of the cellular protein from this transformed strain (see Fig. S1 in the supplemental material). The yield of DmmA after purification was 20.5 to 22.0 mg per 1 g of wet biomass, corresponding to approximately 200 mg of protein in 1 liter of LB medium culture, calculated from four independent batches. The purity of DmmA after purification was 92 to 95% according to SDS-PAGE.
Secondary structure.
The secondary structure of purified DmmA was studied by circular dichroism (CD) (Fig. S2). Spectroscopic data revealed one maximum at 194 nm and two minima at 208 nm and 220 nm. Obtained CD spectra were loaded into an online tool, BeStSel (27), to determine secondary structure content. The data analysis resulted in 41.3% helical, 2.6% beta, 12.4% turn, and 30.8% random structure content. The spectrum corresponds to an α/β-hydrolase fold known from other characterized HLDs. The prediction of secondary elements obtained by CD spectrum analysis corresponds well with the composition obtained by X-ray crystallography for the helical structure content, with 41% from the prediction versus 42% in the crystal structure. Correlation is lower for beta structures, with 2.6% from the prediction versus 19% in the crystal structure. The difference originates from the usually lower quality of CD spectra in the typical beta structure absorbing area of 190 to 200 nm.
Quaternary structure.
The quaternary structure of DmmA was examined by size exclusion chromatography. The results revealed both monomeric and dimeric states with an equilibrium that favors the dimer under native conditions. The majority of the protein in the samples had a molecular mass of 64.1 ± 2.1 kDa, representing the dimer, while the minority had a lower molecular mass of 33.2 ± 0.7 kDa, corresponding to the monomer (Fig. S3). These values are in good agreement with the molecular mass of 37.1 kDa calculated by UniProtKB for a single DmmA amino acid strand.
Thermostability.
Thermal unfolding experiments with DmmA were conducted over a temperature range of 20 to 80°C, with monitoring of secondary elements by CD spectropolarimetry. The melting temperature (Tm) determined in this way was 49.1 ± 0.2°C (Fig. S4A). This value was confirmed by differential scanning calorimetry, which yielded a Tm of 52.1 ± 0.3°C (Fig. S4B). The Tm of DmmA is comparable to that of similar mesophilic HLDs, whose melting temperatures range from 40°C to 57°C.
Kinetic stability.
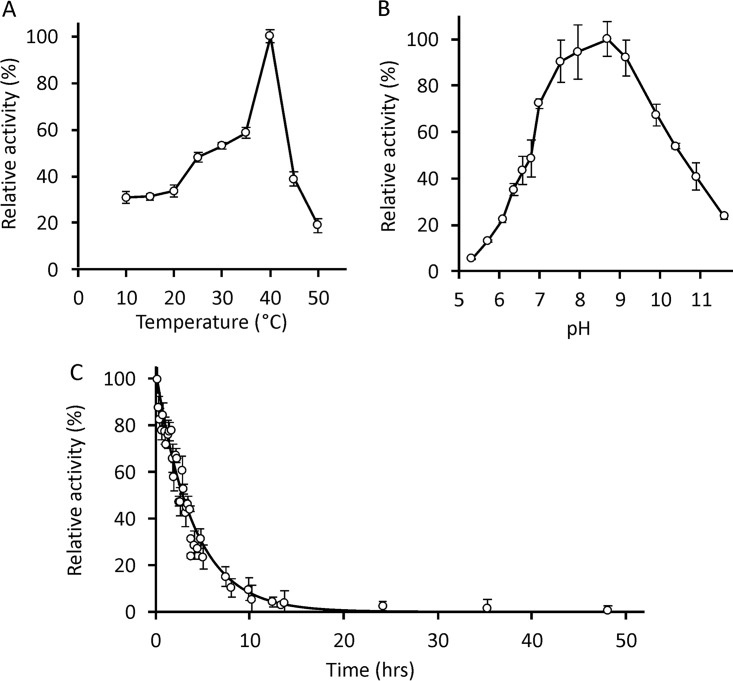
The kinetic stability of DmmA was studied at 45°C with 4-bromobutyronitrile as the best substrate (Fig. 1C). The stability is represented by its half-life of 3.0 h that suggests a faster protein denaturation at the elevated temperature.
FIG 1.
Temperature, pH profile, and kinetic stability of DmmA. (A) A steady increase in the specific activities of DmmA at temperatures ranging from 10 to 35°C with a maximum at 40°C is followed by a rapid loss of activity. Displayed activities are based on an average minimum of three independent reactions with error bars indicating standard deviations of the measurements. (B) Specific activities of DmmA determined at a pH range of 5.3 to 11.6. The displayed profile is based on two independent experiments where the error bars indicate standard deviations. (C) Kinetic stability of DmmA at 45°C at pH 8.0. Each point is an average of the results of three independent experiments. The solid line and error bars represent an exponential fit of the data and the standard deviation, respectively.
Temperature profile.
Significant dehalogenation activity of DmmA was observed at a temperature range of 10 to 50°C, with a narrow optimum at 40°C (Fig. 1A). The activity maximum at 40°C was followed by a rapid decrease and enzyme inactivation above 55°C.
pH profile.
The specific activities of DmmA toward 1,2-dibromoethane at room temperature (21°C) were tested at different pHs ranging from 5.3 to 11.6 by use of a liquid handling robot (Fig. 1B). The pH profile showed an optimum at pH 8.7 and extended pH tolerance. At least 90% of maximal activity was retained within a pH range of 7.5 to 9.2 and more than 20% at pH values of 6.2 to 11.5.
Substrate specificity.
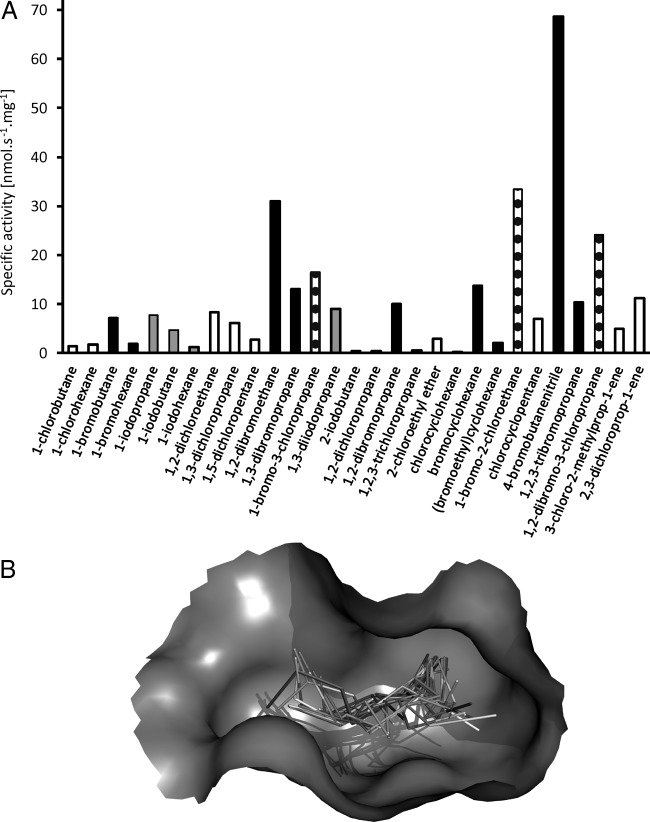
The specific activity (Fig. 2A) of DmmA toward a standard set of 29 representative halogenated substrates was investigated using a liquid handling robot. The activity was measured in the reaction mixture with the substrate concentration at full saturation to establish similar starting conditions for all tested reactions. The highest specific activities were observed with 4-bromobutanenitrile, 1-bromo-2-chloroethane, 1,2-dibromoethane, and 1,2-dibromo-3-chloropropane. DmmA prefers brominated substrates over chlorinated and iodinated substrates. Unusually, the enzyme showed activity toward all tested substrates. A principal-component analysis (Fig. S5) classified DmmA in substrate specificity group 1 (SSG-1) together with other catalytically robust enzymes. Members of SSG-1 such as DbjA, DhaA, DhlA, and LinB are typically active toward at least one of the poorly degradable toxic pollutants, 1,2-dichloroethane, 1,2-dichloropropane, 1,2,3-trichloropropane, and chlorocyclohexane. DmmA was active toward all of these pollutants, making it unique among the known HLDs and potentially applicable in environmental biotechnology (4). Additionally, the enzymatic activity was tested in the presence of 1-bromopentane, 1-bromooctane, 1-iodopropane, 1-iodobutane, 2-iodobutane, and 2-bromo-N-phenylpropionamide (Table S3) as the substrates ubiquitous in seawater and originating in temperate marine macroalgae (19). Positive activity was confirmed for all assayed substrates.
FIG 2.
Substrate specificity profile and visualization of substrates bound to the enzyme active site. (A) Specific activities of DmmA toward the set of 29 representative chlorinated (white), brominated (black), iodinated (gray), and chlorobrominated (dotted) substrates. (B) Graphical visualization of the set of 29 substrates docked into the active site of DmmA. The substrates are represented by sticks, and the active-site cavity is represented by a gray surface (the rest of the protein is omitted for clarity).
Molecular docking and structural analysis.
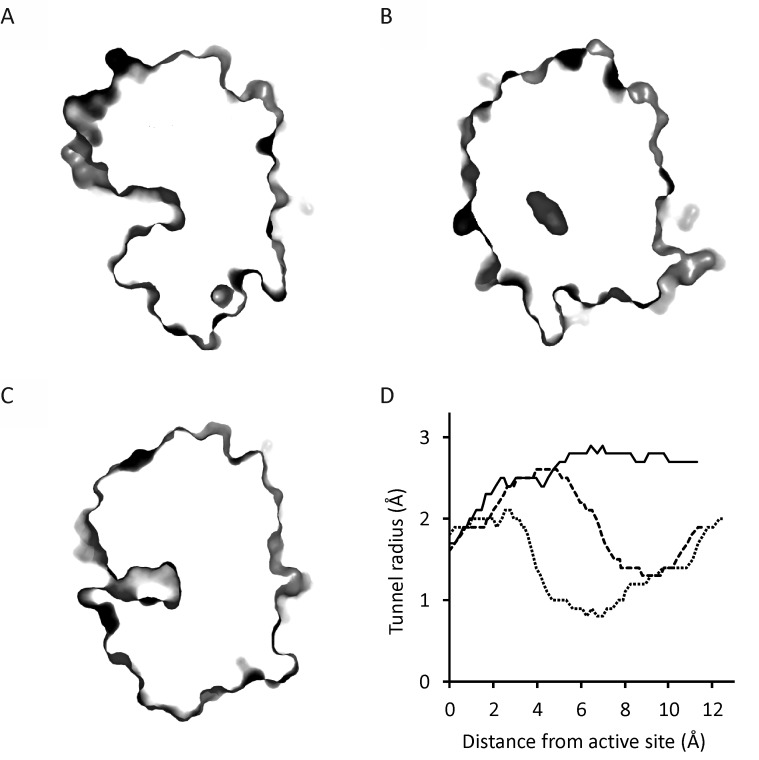
Docking of the 29 substrates to the active site of DmmA showed binding energies between −2.7 and −5.0 kcal/mol (Table S2). These values indicate a significant affinity of DmmA for all tested substrates. In comparison, the calculated binding energies were in the range of −2.6 to −4.1 kcal/mol for DhlA and −2.9 to −5.8 kcal/mol for LinB. The main access tunnels in DmmA, DhlA, and LinB were analyzed by CAVER 3.02 (20) to probe the anatomy of the access pathway connecting the active site with the protein surface. The maximum bottleneck radius of the DmmA main tunnel is 1.32 Å, and its length is 11.53 Å. The tunnel mouth is wide open without any sign of bottleneck (Fig. 3), while access tunnels of two representative dehalogenases, DhlA and LinB, show apparent bottlenecks, forming constrictions in the transport pathway. We expect that such accessibility of the active site to various substrate molecules may contribute to the broad specificity of DmmA.
FIG 3.
Cross-section of the protein molecules and tunnel profiles of DmmA, DhlA, and LinB. (A to C) Cross-sections at main access tunnels of DmmA (A), DhlA (B), and LinB (C) with displayed protein surface. (D) Diameter of the main access tunnels calculated by CAVER for DmmA (solid line), DhlA (dotted line), and LinB (dashed line). No bottleneck is observed for DmmA, as the diameter is increasing toward the tunnel entrance. DhlA and LinB have bottlenecks at distances of 6.5 and 9.8 Å from the active site, respectively.
Steady-state kinetics.
Steady-state kinetics were analyzed for five selected halogenated compounds: 1-chlorobutane, 1-iodopropane, 1-iodobutane, 1,2-dibromoethane, and 4-bromobutyronitrile. The selection represented the diversity of halogenated substrates, including chlorinated, brominated, and iodinated, mono- and dihalogenated analogs of C2 to C4 halocarbons. The catalytic performance of DmmA was closely similar to that of all tested substrates, with the kcat/Km ranging from 0.45 to 1.42 s−1 · mM−1 (Table 1; Fig. S6). Interestingly, 1-iodopropane showed a Hill type sigmoidal trend of the dependence of rate on substrate concentration, while all chlorinated and brominated compounds followed classical hyperbolic kinetics. Although DmmA is present in the dimeric form, the sigmoidal rate dependence was also previously observed in the case of the monomeric HLDs DhaA, LinB, and DpcA (8, 18) and cannot be explained simply by classical Hill-type cooperativity. HLDs are known to be often strongly affected by substrate inhibition, which was not observed for DmmA kinetics with any of the tested substrates.
TABLE 1.
Steady-state kinetic parameters of DmmA toward five selected halogenated substratesa
| Substrate | Km (mM) | kcat (s−1) | kcat/Km (s−1 mM−1) |
|---|---|---|---|
| 1-Chlorobutane | 0.38 ± 0.07 | 0.17 ± 0.02 | 0.45 |
| 1-Iodopropaneb | 0.86 ± 0.03 | 0.43 ± 0.03 | 0.50 |
| 1-Iodobutane | 0.43 ± 0.07 | 0.21 ± 0.03 | 0.49 |
| 1,2-Dibromoethane | 2.3 ± 0.3 | 1.8 ± 0.4 | 0.8 |
| 4-Bromobutyronitrile | 0.9 ± 0.1 | 1.2 ± 0.1 | 1.3 |
Km, Michaelis-Menten constant; kcat, turnover number.
Data were fitted by using the Hill model with an n of 2.2; kinetic parameters were calculated from triplicate measurements.
Effects of cosolvents.
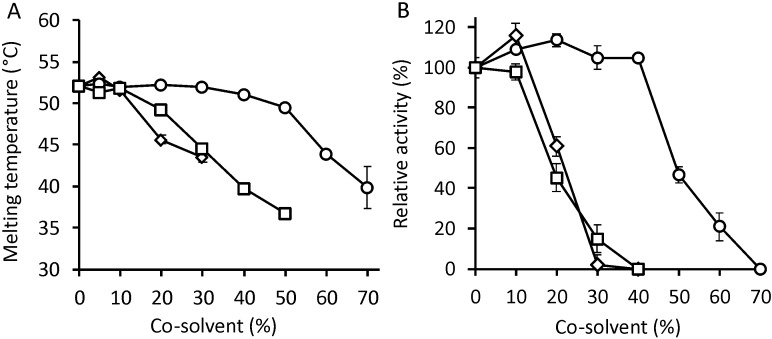
We investigated the effects of cosolvents on DmmA's structure and function. The structural stability of DmmA was determined by differential scanning calorimetry in the presence of 0 to 50% (vol/vol) acetone, 0 to 80% (vol/vol) dimethyl sulfoxide (DMSO), and 0 to 50% (vol/vol) methanol (Fig. 4A). The enzyme exhibits notable structural resistance to all three tested cosolvents: its Tm was unchanged in the presence of up to 10% acetone, 10% methanol, and more than 30% DMSO. Further increases in solvent concentration caused only slight reductions in Tm, with the enzyme's stability occurring at cosolvent concentrations of up to 30% for acetone, 50% for methanol, and 70% for DMSO. In comparison to DbjA, DhaA, and LinB, three dehalogenases that have been studied extensively for stability in the presence of cosolvents (21), DmmA exhibits higher structural stability in acetone and comparable stability in DMSO and methanol.
FIG 4.
Effect of cosolvents on the structure and activity of DmmA. (A) The melting temperature of DmmA was determined in the presence of various concentrations of acetone (diamonds), DMSO (circles), and methanol (squares). (B) The activity of DmmA toward 4-bromobutyronitrile was determined in the presence of various concentrations of acetone (diamonds), DMSO (circles), and methanol (squares). Each point represents an average result of three independent measurements, and the error bar indicates the standard deviation.
The functional stability of DmmA was determined by measuring its specific activity toward 4-bromobutyronitrile in various concentrations of acetone, DMSO, and methanol (Fig. 4B). Its specific activity in buffer solution was comparable to that in 40% DMSO or 10% acetone; further increases in the concentration of either solvent caused significant reductions in activity. Conversely, increasing the concentration of methanol in the solvent caused smooth and proportionate reductions in the activity of DmmA. In keeping with results previously reported for DbjA, DhaA, and LinB, the activity of DmmA also increased its activity in the presence of low concentrations of DMSO. The functional stability of DmmA is not as high as in the case of DbjA but significantly surpasses the performance of LinB and DhaA in acetone and methanol (22).
We also noticed that the decline in the enzyme's activity was more pronounced than the extent of structural denaturation at the same cosolvent concentrations. The effects of cosolvents on steady-state kinetics with 4-bromobutyronitrile were therefore investigated in the presence of 40% DMSO, 10% acetone, and 10% methanol. While these cosolvent concentrations did not affect the enzyme's turnover number (kcat), its Michaelis-Menten constant (Km) increased significantly in the presence of either acetone or DMSO (Table S4; Fig. S7). This result indicates that cosolvent molecules bind the active site of DmmA and compete with the substrate.
Enantioselectivity.
The enantioselectivity of DmmA was studied using four representative chiral substrates: two brominated alkanes (2-bromobutane and 2-bromopentane) and two brominated esters (ethyl-2-bromohexanoate and ethyl-2-bromopropionate). We determined the E values by numerical integration of kinetic resolution experiments measured with the racemic mixture of each substrate. Moderate to high enantioselectivity with a preference for the (R) enantiomer was observed for brominated esters (Table S5; Fig. S8) with the highest E value of 106 achieved with ethyl-2-bromopropionate. Negligible enantioselectivity was observed with either of the tested brominated n-alkanes. The obtained results are in good correspondence with previous studies of HLD enantioselectivity.
Multistep dehalogenation.
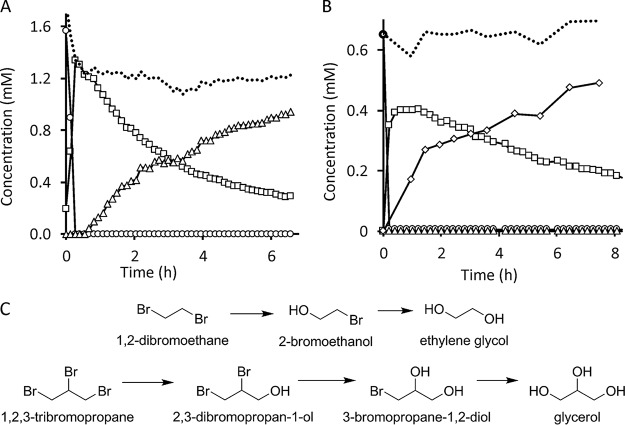
DmmA was tested for its ability to perform a multistep dehalogenation reaction. 1,2-Dibromoethane and 1,2,3-tribromopropane were tested as the model substrates for the sequential conversion leading to ethylene glycol and glycerol, respectively (Fig. 5C). Initial substrate and intermediates were analyzed qualitatively and quantitatively by gas chromatography equipped with mass spectroscopy. The identification of intermediate compounds was confirmed by a combination of three methods: (i) evaluation of the correspondence of retention times with those obtained for analytical standards; (ii) search for similarity of m/z spectra by using NIST 2.0 library (Fig. S9); and (iii) analysis of fragmentation pattern and abundance of isomers by using Mass Hunter software (Fig. S10). Glycerol was detected by using an external kit, since this compound cannot be separated by using gas chromatography. The progress of the 1,2-dibromoethane dehalogenation indicates that it is converted to 2-bromoethanol, which is then turned into ethylene glycol (Fig. 5A). 1,2,3-Tribromopropane is converted to 2,3-dibromopropan-1-ol, followed by formation of 3-bromopropane-1,2-diol and glycerol (Fig. 5B). An accumulation of reaction intermediates indicates a decreasing affinity of the enzyme for a substrate which is converted preferentially to an intermediate that is accumulated and converted only after depletion of the initial substrate.
FIG 5.
Multistep dehalogenation reaction catalyzed by DmmA. (A) Conversion of 1,2-dibromoethane (circles) into the intermediate 2-bromoethanol (squares) and the final product ethylene glycol (triangles). A dotted line indicates the sum of all compounds present in the reaction mixture. (B) Conversion of 1,2,3-tribromopropane (circles) via two intermediates, 2,3-dibromopropan-1-ol (squares) and 3-bromopropane-1,2-diol (triangles), into the final product, glycerol (diamonds). A dotted line indicates the sum of all compounds present in the reaction mixture. (C) Complete multistep dehalogenation scheme for 1,2-dibromoethane and 1,2,3-tribromopropane.
DISCUSSION
The haloalkane dehalogenase DmmA from an unknown marine organism was thoroughly characterized biochemically in this work. The heterologous expression of DmmA was remarkably strong, accounting for 50% of the total soluble protein in the crude extract of E. coli BL21(DE3) (see Fig. S1 in the supplemental material). This unusually high yield will facilitate the biotechnological application of this enzyme by obviating the need for extensive and expensive protein purification (6).
Interestingly, the amino acid sequence of DmmA exhibited high levels of sequence identity (up to 43%) with LinB and DhaA while it exhibited low sequence identity with two other known HLDs of marine origin (Table S1)—21% for DppA from Plesiocystis pacifica SIR-1 (23) and 9% for DadB from Alcanivorax dieselolei B-5 (24). It needs to be stated that while DppA and DadB variants both have a bacterial origin, the biological source of DmmA is unknown. The secondary structure analysis of DmmA by CD spectropolarimetry indicated that DmmA adopts the α/β-hydrolase fold typical for all known HLDs (25). The tertiary structure has previously been resolved at 2.2-Å resolution by X-ray crystallography (16), revealing an unusually large active-site cavity, compared to other structurally characterized HLDs. An analysis of its quaternary structure under native conditions revealed that DmmA exists primarily as a dimeric complex, whereas most HLDs are monomeric (6). These observations are in good agreement with previous findings indicating that DmmA exists in equilibrium between monomeric and dimeric states (16). The temperature optimum of 40°C and the melting temperature are similar to those for other mesophilic HLDs (6). The DmmA pH optimum of 8.7 under slightly alkaline conditions also corresponds with those of other HLDs, but the enzymatic activity is retained in a broader span, with at least 20% of activity from pH 6.2 to pH 11.5 (Fig. 1).
DmmA demonstrated unusual broad substrate specificity, showing activity toward all 29 substrates included in the standard set of representative substrates (4), which has never been observed with previously characterized HLDs (Fig. 2A). Additionally, DmmA is active toward all poorly degradable chlorinated environmental pollutants, namely, 1,2-dichloroethane, 1,2-dichloropropane, 1,2,3-trichloropropane, and chlorocyclohexane, as well as toward newly identified substrates of this enzyme family (17). We speculate that the broad substrate specificity of DmmA may be linked to its large active site and readily accessible active site. Analysis of access tunnels using CAVER identified the widely open mouth without any sign of bottleneck, which is unique to DmmA and has never been observed with other family members. This wide opening provides easy access of a large spectrum of diverse molecules to the enzyme active site (Fig. 2B). Complementary analysis of the two representative HLDs, LinB and DhlA, revealed clear bottlenecks which separate the active site from the surrounding water solvent. The binding energies calculated using AutoDock were as low as −5.8 kcal/mol for all tested enzyme variants and all tested substrates (Table S2). This result suggests that the binding affinity of DmmA is comparable to that of both representative dehalogenases, LinB and DhlA. Nevertheless, DmmA shows only moderate activity with most of the tested substrates in comparison to other HLDs from the same specificity group. This suggests a possible trade-off between reactivity and substrate specificity during evolution. By using protein engineering, we have previously demonstrated that opening the active site can positively increase its accessibility for substrate molecules but at the same time can lower the catalytic performance due to incompatibility of water molecules with the SN2 dehalogenation reaction (25, 26). Broad substrate specificity may be a common characteristic of bacterial HLDs isolated from marine environments. Similarly, marine HLDs DppA and DadB were previously reported to degrade a wide range of diverse halogenated substrates (23, 24). DmmA was tested for activity toward a selected set of halogenated compounds of seawater origin and found to be positive in all cases (Table S3). The ability to sequentially convert di- and trihalogenated compounds was confirmed in the reactions with 1,2-dibromoethane and 1,2,3-tribromopropane, yielding ethylene glycol and glycerol, respectively (Fig. 5). The affinity of the enzyme is significantly lower for haloalcohol intermediates, leading to their accumulation in a reaction medium, before conversion to the final products.
We also investigated the tolerance of DmmA to organic cosolvents. The enzyme was structurally tolerant to DMSO, acetone, and methanol at concentrations of up to 50%, 10%, and 20% (vol/vol), respectively. Compared to the structural changes, DmmA activity declined more sharply with increasing cosolvent concentrations, probably due to competition between the substrate and solvent molecules for the widely accessible active site of this enzyme (Fig. 4). Despite the wide opening of the protein resulting in easy access of solvent molecules to the active site, DmmA still can be rated among the HLDs with the highest cosolvent tolerance (21).
DmmA exhibits moderate to high enantioselectivity toward bromoesters, while it shows negligible enantioselectivity toward bromoalkanes (Fig. S8; Table S5). The higher enantioselectivity with bromoesters is in keeping with the previously reported E values for other HLDs and with the previously reported hypothesis concerning the origin of HLD enantioselectivity (18, 21). The enantiodiscrimination between brominated esters is achieved via the chiral substrate's interactions with halide-stabilizing residues, whereas enantiodiscrimination between n-alkanes is mediated by hydrophobic interactions between the alkyl chain and the walls of the active site. In the same way, the low enantioselectivity of DmmA toward bromoalkanes may be connected to the wide opening of its active site and the low degree of substrate stabilization (22).
In summary, the enzyme DmmA has been subjected to extensive biochemical characterization. The results showed that DmmA possesses a combination of several unique properties attractive for practical applications. In particular, the enzyme is overexpressed in a large quantity and could be obtained in technological grade purity without demanding purification steps, lowering large-scale production costs. A broad substrate specificity, a good tolerance to organic cosolvents, and a wide range of pH values for the operational window makes DmmA an attractive catalyst for applications in biocatalysis as well as for biosensing/biodegradation of halogenated pollutants.
MATERIALS AND METHODS
Materials.
All halogenated compounds were purchased from Fluka or Sigma-Aldrich (Germany), with a purity of 97% or higher. All other chemicals, including buffer components, antibiotics, and SDS gel components, were purchased from Sigma-Aldrich (Germany). Helium and hydrogen used in gas chromatography had 99.995% purity and were purchased from SIAD (Czech Republic). Competent Escherichia coli BL21(DE3) cells were purchased from Thermo Fisher Scientific (USA).
Expression and purification.
The recombinant gene dmmA:His6 with a six-histidine tag at the N terminus was subcloned into the expression vector pET24a with kanamycin resistance. Overexpression in the Escherichia coli BL21(DE3) strain was achieved by IPTG (isopropyl-β-d-thiogalactopyranoside) induction (final concentration, 0.5 mM), and the cells were cultivated in 1 liter of LB medium with kanamycin (final concentration, 50 μg · ml−1) in a New Brunswick Innova 44 orbital incubator (Eppendorf, USA) at 120 rpm and 20°C overnight. Cells were harvested by centrifugation (Sigma, Germany) at 12,400 relative centrifugal force (RCF) at 4°C for 1 h. Harvested cells were then disrupted by pressure homogenization in a One Shot instrument (Constant Systems, United Kingdom) set to 1.5 × 105 kPa. The resulting crude extract was taken up in loading buffer (10 mM imidazole, 50 mM phosphate, 0.5 M sodium chloride, pH 7.5) and loaded onto a 5 ml Ni-nitrilotriacetic acid (Ni-NTA) Superflow column charged with Ni2+ ions (Qiagen, Germany) for purification by metallo-affinity chromatography. Unbound and weakly bound proteins were removed by gradient elution using 0 to 10% solutions of the elution buffer (250 mM imidazole, 50 mM phosphate, 0.5 M sodium chloride, pH 7.5). The recombinant protein was then isolated by progressively increasing the concentration of the elution buffer to 60%. The purified protein fraction was dialyzed against a 50 mM phosphate buffer, pH 7.5. Protein yield was estimated from four independent 1-liter cell culture batches. The centrifuged wet biomass was weighed. The purification yield was determined from the volume of the purified solution, the protein purity, and its concentration. Coomassie brilliant blue-stained and heat-denatured protein samples were run on 12% SDS-PAGE set to 120 V for 80 min. Protein purity was estimated using a calibrated imaging densitometer GS-800 (Bio-Rad, USA). The gels were scanned, and the background was subtracted. Protein concentration was estimated in a DS-11 microdrop spectrophotometer (DeNovix, USA) by measuring sample absorbance at 280 nm.
Secondary structure analysis.
Circular dichroism (CD) spectra were recorded at 20°C using a Chirascan spectropolarimeter (Applied Photophysics, United Kingdom). Data were collected from 185 to 260 nm at 100 nm · min−1 with a 1-s response time and 2-nm bandwidth using a 0.1-cm quartz cuvette containing the studied protein. Each spectrum shown here is the average of 10 individual scans and was corrected for the buffer's absorbance. CD data were expressed in terms of the mean residue ellipticity (θMRE) using the equation:
where θobs is the observed ellipticity in degrees, Mw is the molecular mass of the protein in daltons (Da), n is the number of residues, l is the cell path length, c is the protein concentration in mg · ml−1, and the factor of 100 originates from the conversion of the molecular mass to mg · dmol−1. Secondary structure determination and analysis were carried out on measured ellipticity from 190 to 250 nm using online tool BeStSel with default settings (27).
Quaternary structure analysis.
The quaternary structure of DmmA was investigated by means of size exclusion chromatography using a Zenix SEC column (Sepax Technologies, USA) in conjunction with a Viscotec 305 TDA instrument (Malvern, United Kingdom) equipped with static light scattering, refractive index, UV, and differential viscometer detectors. The device was initially equilibrated with 150 mM phosphate buffer, pH 7.0. The system was calibrated using bovine serum albumin (Sigma-Aldrich, Germany) as a protein standard. A single standard is sufficient for calibration when tetra detection is employed. For analysis, a 1-mg · ml−1 sample solution was injected onto the column and separated at a constant flow rate of 0.5 ml · min−1 using 150 mM phosphate buffer (pH 7.0) as the elution buffer. Retention volumes, molecular weights, hydrodynamic radii, and intrinsic viscosities were calculated using the OmniSec program (Malvern, United Kingdom).
pH profile.
The pH optimum was determined as the enzyme specific activity toward 1,2-dibromoethane using the substrate in a series of Britton-Robinson buffers with a pH range of 5.3 to 11.6 at room temperature (28). The determined activities were normalized to the highest observed activity. The measurement was carried out in two independent experiments with two different enzyme batches.
Thermal stability evaluation.
The thermal unfolding of DmmA was followed by determining its ellipticity at 221 nm between 20 and 80°C at a resolution of 0.1°C and a heating rate of 1°C · min−1. The recorded thermal denaturation curves were roughly normalized to represent signal changes between approximately 1 and 0 and fitted to sigmoidal curves using the Origin 9.0 program (OriginLab, USA). Melting temperatures (Tm) were estimated based on the midpoints of the normalized thermal transitions. The thermal unfolding of DmmA in the presence and absence of various concentrations of dimethyl sulfoxide (DMSO), methanol, and acetone in 100 mM glycine buffer (pH 8.6) was investigated also using a VP-capillary differential scanning calorimeter (MicroCal, USA). The change in the heat capacity of 1 mg · ml−1 DmmA was recorded at temperatures of 20 to 80°C and a heating rate of 1°C · min−1. Data analysis was conducted using Origin 7.0 software (OriginLab, USA) with baseline subtraction, and the Tm was estimated from the inflection point of the first derivative.
Substrate specificity assay.
The catalytic activity of DmmA toward a set of 29 halogenated substrates was measured using a Hamilton Microlab STARlet robot (Hamilton Robotics, Switzerland). The reactions were performed at 37°C in 2-ml vials containing 1 ml of 100 mM glycine buffer (pH 8.6) and 1 μl of each substrate to reach their saturation for all tested reactions. The reactions were initiated by adding 50 μl of enzyme solution (0.1 to 5 mg · ml−1) to the vials. Their progress was monitored by periodically withdrawing 75-μl samples from the reaction mixtures and immediately mixing these samples with 35% nitric acid to terminate the reaction. The release of halide ions in the quenched samples was then analyzed by the colorimetric method of Iwasaki and coworkers (29) whereby the quenched sample is reacted with mercuric thiocyanate and ferric ammonium sulfate and then examined spectrophotometrically at 460 nm using a Sunrise microplate reader (Tecan, Austria). Separate calibration curves were prepared for evaluation formation of chlorides, bromides, and iodides (Fig. S11). Blanks of a buffer, an enzyme, and a substrate were subtracted prior to data evaluation. The dehalogenation activities were quantified as slopes from the linear dependence of the halide evolution over time and divided by the final enzyme concentration in the reaction vessels. The measured specific activities of DmmA were combined with a data set of similar results for other HLDs, namely, LinB, DbjA, DhaA, DmbA, DmbC, DatA, DrbA, DbeA, and DhlA (4). This combined data set was then subjected to multivariate statistical analysis. The untransformed data set was used to construct a t1/t2 score plot, a t1 contribution plot and a p1/p2 loading plot. (t1/t2 and p1/p2 are the highest scores and cosines of loading, respectively.) The transformed data set consisted of log-transformed activities that were weighted according to the following equation:
where vi is the specific activity of a given enzyme toward a particular substrate. A unit value is added to each measured activity in order to avoid obtaining negative numbers after log transformation. The transformed data set was used to construct a t1/t2 score plot and a p1/p2 loading plot.
Molecular docking and structural analysis.
Docking of substrate molecules into the active sites of HLDs was carried out to explore their substrate specificity. The dehalogenases DmmA, DhlA, and LinB (PDB ID 3U1T, 2DHC, and 1MJ5) and a set of 29 halogenated ligands were used. The protein and the substrate docking files were prepared using MGLtools (30). The docking experiments were carried out using AutoDock Vina (31). The exhaustiveness was set to 30; otherwise, default settings were used. The analysis of tunnels in DmmA, DhlA, and LinB was done using the software tool CAVER 3.02 (20). The 0.7-Å probe was used to calculate the tunnels in the crystal structure (PDB ID 3U1T, 2DHC, and 1MJ5). The beginning of the tunnel was set to the OD1 atom of the catalytic nucleophile.
Temperature profile evaluation.
The temperature profile of DmmA was determined in the temperature range of 10 to 55°C with a step of 5°C. Reactions were measured in 10 ml glycine buffer (100 mM, pH 8.6) with 1,2-dibromoethane as a substrate. All reactions were initiated by injection of the enzyme (final concentration, 0.005 to 0.022 mg · ml−1) and monitored by periodically withdrawing 1 ml and mixing it with 35% nitric acid to stop the reaction. The released halide concentration was then determined by the same means as for the substrate specificity.
Kinetic stability.
The kinetic stability of DmmA was determined at 45°C with 4-bromobutyronitrile as the substrate. The enzyme was incubated in aliquots at a concentration of 0.155 to 0.162 mg · ml−1 in a dry bath. The reaction was measured as a fluorescence change at 488-nm excitation and 528-nm emission in 1 mM HEPES buffer, pH 8.0, with 10 μM 8-hydroxypyrene-1,3,6-trisulfonic acid fluorescent as the pH dye, using a Synergy 4 HT reader (BioTek, USA). The fluorescent dye is quenched by the H+ ions formed during the reaction. The reaction was initiated by the injection of 100 μl of the substrate (final concentration, 6.7 mM) into the preincubated 50 μl of the enzyme (final concentration, 0.052 to 0.054 mg · ml−1). The reaction was monitored for 1 min with 6-s intervals to follow the linear decrease of the fluorescence signal. The kinetic half-life was evaluated according to the following equation:
where t1/2 is the half-life and λ is the exponential decay constant.
Enantioselectivity analysis.
The enantioselectivity of DmmA was evaluated by performing kinetic resolution experiments using 2-bromobutane, 2-bromopentane, ethyl-2-bromopropionate, and ethyl-2-bromohexanoate. These reactions were performed in 10 ml of 100 mM glycine buffer (pH 8.6) at 25°C in Erlenmeyer flasks sealed with headspace caps. Both enantiomers were present in the reaction mixture. Enzymatic reactions were initiated by the addition of an enzyme at a final concentration of 0.05 to 0.15 mg · ml−1 (depending on its specific activity). The progress of the reactions was monitored by periodically withdrawing samples from the reaction mixtures. The samples were then extracted with diethyl ether containing 1,2-dichloroethane as an internal standard and analyzed on a 7890A Agilent (Thermo Scientific, USA) gas chromatograph equipped with a flame ionization detector and an Astec Chiraldex B-DM column (Sigma-Aldrich, USA). The enantiomeric ratio was calculated according to the following equation (32):
where kcat and Km are the turnover number and Michaelis constant for the (R) and (S) enantiomers of the substrate in question. Nonspecific degradation of substrates was determined and taken into account during data evaluation. To estimate E values, MicroMath Scientist (Micromath, USA) was used to fit (by numerical integration) the equations describing competitive Michaelis-Menten kinetics to the experimental time courses of the changes in the substrate concentrations obtained from the kinetic resolution experiments.
Steady-state kinetics.
Steady-state kinetic reactions were performed in 1.5-ml gas chromatography glass vials in phosphate buffer (50 mM, pH 7.5) at 25°C. The substrate was incubated in a closed vial for at least 30 min. The reaction was initiated by the addition of the enzyme directly to the vial (final concentration, 522 nM). The mass spectra were obtained on a model 5975C MSD mass spectrometer coupled with a model 7890A gas chromatograph (Agilent, USA) using a ZB-5 column (30 m by 0.25 mm; film thickness, 0.25 μm; Phenomenex, USA) with the following settings: injector temperature, 250°C; carrier gas helium, flow rate, 1.2 ml · min−1; oven temperature programmed from 40 to 180°C at a rate of 20°C min−1; detector temperature, 250°C; quadrupole mass analyzer, electron impact, 70.3 eV; ion source temperature, 230°C; atomic mass unit (amu) range, 25 to 300). Substrate concentrations were evaluated in MassHunter software (Agilent, USA). The slope corresponding to a particular substrate concentration was then fitted using nonlinear regression to kinetic models in the Origin 8.0 software (OriginLab, USA). We used the following equations for evaluation of steady-state kinetics data:
where vobs and vlim are the observed and maximum reaction rates, respectively, S is a substrate concentration, Km is a Michaelis-Menten constant, K0.5 is a substrate concentration where the reaction rate is half of the maximum, and n is the Hill coefficient.
Analysis of multistep dehalogenation.
Dehalogenation reactions were run in 1.5-ml gas chromatography glass vials in phosphate buffer (50 mM, pH 7.5) at 25°C. The substrate was incubated for at least 30 min in a closed vial. The reaction was initiated by the addition of the enzyme (final concentration, 1.3 μM) directly to the vial. The course of the reaction was monitored by using a model 7890A gas chromatograph (Agilent, USA) equipped with a ZB-5 column (30 m by 0.25 mm; film thickness, 0.25 μm; Phenomenex, USA) and coupled with a model 5975C MSD mass spectrometer (Agilent, USA). The settings of the instruments were as follows: injector temperature, 250°C; carrier gas helium, flow rate, 1.2 ml · min−1; oven temperature programmed from 40 to 180°C at a rate of 20°C min−1; detector temperature, 250°C; quadrupole mass analyzer, electron impact, 70.3 eV; ion source temperature, 230°C; amu range, 25 to 300. Retention times were 3.85 min for 2-bromoethanol, 4.14 min for ethane-1,2-diol, 4.44 min for 1,2-dibromoethane, 3.01 min for the internal standard, 6.36 for 3-bromopropane-1,2-diol, 7.02 min for 2,3-dibromopropan-1-ol, and 7.71 min for 1,2,3-tribromopropane. Compounds were also identified by an m/z spectra search in NIST MS Search 2.0 library (NIST, USA). The obtained spectra were compared with predicted fragmentation patterns obtained by using MassHunter software (Agilent, USA). The glycerol concentration was determined using a Free Glycerol assay kit (Biovision, USA) on a microtiter plate reader with absorbance at 570 nm in accordance with the manufacturer's protocol.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by the National Sustainability Programme of the Czech Ministry of Education, Youth and Sports (LO1214), by the RECETOX research infrastructure (LM2011028), and by the Grant Agency of the Czech Republic (GA16-07965S).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01684-17.
REFERENCES
- 1.Janssen DB, Gerritse J, Brackman J, Kalk C, Jager D, Witholt B. 1988. Purification and characterization of a bacterial dehalogenase with activity toward halogenated alkanes, alcohols and ethers. Eur J Biochem 171:67–72. doi: 10.1111/j.1432-1033.1988.tb13759.x. [DOI] [PubMed] [Google Scholar]
- 2.Keuning S, Janssen DB, Witholt B. 1985. Purification and characterization of hydrolytic haloalkane dehalogenase from Xanthobacter autotrophicus GJ10. J Bacteriol 163:635–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagata Y, Miyauchi K, Damborsky J, Manova K, Ansorgova A, Takagi M. 1997. Purification and characterization of haloalkane dehalogenase of a new substrate class from a gamma-hexachlorocyclohexane-degrading bacterium, Sphingomonas paucimobilis UT26. Appl Environ Microbiol 63:3707–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koudelakova T, Chovancova E, Brezovsky J, Monincova M, Fortova A, Jarkovsky J, Damborsky J. 2011. Substrate specificity of haloalkane dehalogenases. Biochem J 2410:345–354. doi: 10.1042/BJ20101405. [DOI] [PubMed] [Google Scholar]
- 5.Schanstra JP, Kingma J, Janssen DB. 1996. Specificity and kinetics of haloalkane dehalogenase. J Biol Chem 271:14747–14753. doi: 10.1074/jbc.271.25.14747. [DOI] [PubMed] [Google Scholar]
- 6.Koudelakova T, Bidmanova S, Dvorak P, Pavelka A, Chaloupkova R, Prokop Z, Damborsky J. 2013. Haloalkane dehalogenases: biotechnological applications. Biotechnol J 8:32–45. doi: 10.1002/biot.201100486. [DOI] [PubMed] [Google Scholar]
- 7.Nagata Y, Nariya T, Ohtomo R, Fukuda M, Yano K, Takagi M. 1993. Cloning and sequencing of a dehalogenase gene encoding an enzyme with hydrolase activity involved in the degradation of gamma-hexachlorocyclohexane in Pseudomonas paucimobilis. J Bacteriol 175:6403–6410. doi: 10.1128/jb.175.20.6403-6410.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curragh H, Flynn O, Larkin MJ, Stafford TM, Hamilton JT, Harper DB. 1994. Haloalkane degradation and assimilation by Rhodococcus rhodochrous NCIMB 13064. Microbiology 140:1433–1442. doi: 10.1099/00221287-140-6-1433. [DOI] [PubMed] [Google Scholar]
- 9.Poelarends GJ, Saunier R, Janssen DB. 2001. trans-3-chloroacrylic acid dehalogenase from Pseudomonas pavonaceae 170 shares structural and mechanistic similarities with 4-oxalocrotonate tautomerase. J Bacteriol 183:4269–4277. doi: 10.1128/JB.183.14.4269-4277.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fortova A, Sebestova E, Stepankova V, Koudelakova T, Palkova L, Damborsky J, Chaloupkova R. 2013. DspA from Strongylocentrotus purpuratus: the first biochemically characterized haloalkane dehalogenase of non-microbial origin. Biochimie 95:2091–2096. doi: 10.1016/j.biochi.2013.07.025. [DOI] [PubMed] [Google Scholar]
- 11.Sato Y, Monincova M, Chaloupkova R, Prokop Z, Ohtsubo Y, Minamisawa K, Tsuda M, Damborsky J, Nagata Y. 2005. Two rhizobial strains, Mesorhizobium loti MAFF303099 and Bradyrhizobium japonicum USDA110, encode haloalkane dehalogenases with novel structures and substrate specificities. Appl Environ Microbiol 71:4372–4379. doi: 10.1128/AEM.71.8.4372-4379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wood DW, Setubal JC, Kaul R, Monks DE, Kitajima JP, Okura VK, Zhou Y, Chen L, Wood GE, Almeida NF Jr, Woo L, Chen Y, Paulsen IT, Eisen JA, Karp PD, Bovee D Sr, Chapman P, Clendenning J, Deatherage G, Gillet W, Grant C, Kutyavin T, Levy R, Li MJ, McClelland E, Palmieri A, Raymond C, Rouse G, Saenphimmachak C, Wu Z, Romero P, Gordon D, Zhang S, Yoo H, Tao Y, Biddle P, Jung M, Krespan W, Perry M, Gordon-Kamm B, Liao L, Kim S, Hendrick C, Zhao ZY, Dolan M, Chumley F, Tingey SV, Tomb JF, Gordon MP, Olson MV, Nester EW. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- 13.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE III, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 14.Lee HS, Kwon KK, Kang SG, Cha S-S, Kim S-J, Lee JH. 2010. Approaches for novel enzyme discovery from marine environments. Curr Opin Biotechnol 21:353–357. doi: 10.1016/j.copbio.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 15.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers Y-H, Smith HO. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 16.Gehret JJ, Gu L, Geders TW, Brown WC, Gerwick L, Gerwick WH, Sherman DH, Smith JL. 2012. Structure and activity of DmmA, a marine haloalkane dehalogenase. Protein Sci 21:239–248. doi: 10.1002/pro.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daniel L, Buryska T, Prokop Z, Damborsky J, Brezovsky J. 2015. Mechanism-based discovery of novel substrates of haloalkane dehalogenases using in silico screening. J Chem Inf Model 55:54–62. doi: 10.1021/ci500486y. [DOI] [PubMed] [Google Scholar]
- 18.Drienovska I, Chovancova E, Koudelakova T, Damborsky J, Chaloupkova R. 2012. Biochemical characterization of a novel haloalkane dehalogenase from a cold-adapted bacterium. Appl Environ Microbiol 78:4995–4998. doi: 10.1128/AEM.00485-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gschwend PM, Macfarlane JK, Newman KA. 1985. Volatile halogenated organic compounds released to seawater from temperate marine macroalgae. Science 227:1033–1035. doi: 10.1126/science.227.4690.1033. [DOI] [PubMed] [Google Scholar]
- 20.Chovancova E, Pavelka A, Benes P, Strnad O, Brezovsky J, Kozlikova B, Gora A, Sustr V, Klvana M, Medek P, Biedermannova L, Sochor J, Damborsky J. 2012. CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput Biol 8:e1002708. doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stepankova V, Khabiri M, Brezovsky J, Pavelka A, Sykora J, Amaro M, Minofar B, Prokop Z, Hof M, Ettrich R, Chaloupkova R, Damborsky J. 2013. Expansion of access tunnels and active-site cavities influence activity of haloalkane dehalogenases in organic cosolvents. Chembiochem 14:890–897. doi: 10.1002/cbic.201200733. [DOI] [PubMed] [Google Scholar]
- 22.Stepankova V, Damborsky J, Chaloupkova R. 2013. Organic co-solvents affect activity, stability and enantioselectivity of haloalkane dehalogenases. Biotechnol J 8:719–729. doi: 10.1002/biot.201200378. [DOI] [PubMed] [Google Scholar]
- 23.Hesseler M, Bogdanović X, Hidalgo A, Berenguer J, Palm GJ, Hinrichs W, Bornscheuer UT. 2011. Cloning, functional expression, biochemical characterization, and structural analysis of a haloalkane dehalogenase from Plesiocystis pacifica SIR-1. Appl Microbiol Biotechnol 91:1049–1060. doi: 10.1007/s00253-011-3328-x. [DOI] [PubMed] [Google Scholar]
- 24.Li A, Shao Z. 2014. Biochemical characterization of a haloalkane dehalogenase DadB from Alcanivorax dieselolei B-5. PLoS One 9:e89144. doi: 10.1371/journal.pone.0089144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagata Y, Prokop Z, Sato Y, Jerabek P, Kumar A, Ohtsubo Y, Tsuda M. 2005. Degradation of β-hexachlorocyclohexane by haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26. Appl Environ Microbiol 71:2183–2185. doi: 10.1128/AEM.71.4.2183-2185.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pavlova M, Klvana M, Prokop Z, Chaloupkova R, Banas P, Otyepka M, Wade RC, Tsuda M, Nagata Y, Damborsky J. 2009. Redesigning dehalogenase access tunnels as a strategy for degrading an anthropogenic substrate. Nat Chem Biol 5:727–733. doi: 10.1038/nchembio.205. [DOI] [PubMed] [Google Scholar]
- 27.Micsonai A, Wien F, Kernya L, Lee Y-H, Goto Y, Réfrégiers M, Kardos J. 2015. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc Natl Acad Sci U S A 112:E3095–E3103. doi: 10.1073/pnas.1500851112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Britton HTS, Robinson RA. 1931. Universal buffer solutions and the dissociation constant of veronal. J Chem Soc 0:1456–1462. doi: 10.1039/JR9310001456. [DOI] [Google Scholar]
- 29.Iwasaki I, Utsumi S, Ozawa T. 1952. New colorimetric determination of chloride using mercuric thiocyanate and ferric ion. Bull Chem Soc Jpn 25:226. doi: 10.1246/bcsj.25.226. [DOI] [Google Scholar]
- 30.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. 2009. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trott O, Olson AJ. 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rakels JLL, Straathof AJJ, Heijnen JJ. 1993. A simple method to determine the enantiomeric ratio in enantioselective biocatalysis. Enzyme Microb Technol 15:1051–1056. doi: 10.1016/0141-0229(93)90053-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.