ABSTRACT
Rhizomucor miehei lipase (RML), as a kind of eukaryotic protein catalyst, plays an important role in the food, organic chemical, and biofuel industries. However, RML retains its catalytic activity below 50°C, which limits its industrial applications at higher temperatures. Soluble expression of this eukaryotic protein in Escherichia coli not only helps to screen for thermostable mutants quickly but also provides the opportunity to develop rapid and effective ways to enhance the thermal stability of eukaryotic proteins. Therefore, in this study, RML was engineered using multiple computational design methods, followed by filtration via conservation analysis and functional region assessment. We successfully obtained a limited screening library (only 36 candidates) to validate thermostable single point mutants, among which 24 of the candidates showed higher thermostability and 13 point mutations resulted in an apparent melting temperature () of at least 1°C higher. Furthermore, both of the two disulfide bonds predicted from four rational-design algorithms were further introduced and found to stabilize RML. The most stable mutant, with T18K/T22I/E230I/S56C-N63C/V189C-D238C mutations, exhibited a 14.3°C-higher and a 12.5-fold increase in half-life at 70°C. The catalytic efficiency of the engineered lipase was 39% higher than that of the wild type. The results demonstrate that rationally designed point mutations and disulfide bonds can effectively reduce the number of screened clones to enhance the thermostability of RML.
IMPORTANCE R. miehei lipase, whose structure is well established, can be widely applied in diverse chemical processes. Soluble expression of R. miehei lipase in E. coli provides an opportunity to explore efficient methods for enhancing eukaryotic protein thermostability. This study highlights a strategy that combines computational algorithms to predict single point mutations and disulfide bonds in RML without losing catalytic activity. Through this strategy, an RML variant with greatly enhanced thermostability was obtained. This study provides a competitive alternative for wild-type RML in practical applications and further a rapid and effective strategy for thermostability engineering.
KEYWORDS: Rhizomucor miehei lipase, rational design, thermostability, disulfide bond
INTRODUCTION
Lipases (EC 3.1.1.3), which catalyze the incorporation of new fatty acids into the triglyceride component of a fat or rearrange the existing fatty acids in a triglyceride, have unparalleled advantages in academic and industrial applications, such as mild reaction conditions, extensive adaptability to raw materials, environmental friendliness, and easy downstream processes (1, 2). In order to broaden their applications at higher temperature ranges, many studies have focused on improving the thermal stability of lipases. Most dramatic increases in thermostability (>15°C) have been found in bacterial lipases, such as Bacillus subtilis lipase A, which can be expressed in Escherichia coli and subjected to high-throughput screening of the modified mutants (3, 4). However, E. coli cannot express very large proteins. Moreover, for disulfide (SS) bond-rich proteins and proteins that require posttranslational modifications, E. coli is not a suitable system (5). For hydrophobic proteins, such as lipases from filamentous fungi or yeasts, the formation of cytoplasmic or periplasmic inclusion bodies is common for expression in E. coli. Hence, eukaryotic host cells such as Saccharomyces cerevisiae and Pichia pastoris are usually used to obtain high-level production of soluble proteins with high catalytic activity (6). Enhancing the thermostability of proteins expressed in eukaryotic systems is a particularly exciting but challenging field, since eukaryotic cells have a relatively long culture time and are not suitable for high-throughput screening of the thermostable mutants (7). Therefore, methods that can effectively reduce the size of mutant libraries to obtain thermostable mutants are urgently needed for stabilizing proteins expressed in eukaryotic systems.
Rhizomucor miehei lipase (RML) is highly active under diverse conditions, making it an important industrial biocatalyst in food chemistry, biodiesel production, and organic chemistry (8). It is commercially available in both soluble and immobilized forms (e.g., Lipozyme RM IM). Although RML possesses most advantageous properties as an industrial catalyst, poor thermostability to withstand elevated temperatures confines its wider applications, because thermal denaturation will cause a precipitous drop in the reaction velocity at higher temperatures (9). Noel and Combes (10) investigated the effect of thermal deactivation on the stability of R. miehei lipase in pure water. The results showed that the half-lives (t1/2) of the enzyme at 55°C and 60°C were only 35 min and 5 min, respectively, indicating that RML is unstable at higher temperatures. On the other hand, RML is usually expressed in eukaryotic systems such as P. pastoris to obtain correctly folded, precisely modified, and highly expressed soluble proteins (11). Thus, soluble expression of RML in E. coli allows for high-throughput screening of thermostable mutants, which makes RML a model system to test methods of improving the thermostability of proteins expressed in eukaryotic systems (12).
A variety of methods have been used to improve the thermodynamic and kinetic stability of lipases, such as immobilization (1, 13), medium engineering (14), and protein engineering (15). In protein engineering, irrational design (i.e., directed evolution), semirational design, and rational design are three general methods employed to obtain thermostable mutants of a target enzyme (14). Numerous studies have shown that directed evolution can enhance the performance of diverse enzymes at elevated temperatures based on error-prone PCR and DNA shuffling (3, 16–19). Nimpiboon et al. improved the thermostability of amylomaltase from a mesophilic Corynebacterium glutamicum strain by error-prone PCR and site-directed mutagenesis (20). Mutants A406V and A406L both showed increases in activity and thermostability. However, screening large numbers of colonies (usually >104) is time-consuming and is infeasible for enzymes expressed in slow-growing hosts. As another popular method, the B-factor iterative test (B-FIT), which rigidifies the most flexible residues in a protein, has been employed as a semirational strategy to improve the thermostability of many enzymes (4, 21–24). Roth et al. engineered uronate dehydrogenase from Agrobacterium tumefaciens by semirational design (25). The triple variant A41P/H101Y/H236K showed higher thermodynamic stability, with a value of 62.2°C (a 3.2°C improvement compared to the wild type [WT]). Nevertheless, thousands of clones have to be constructed for screening, and a large number of stabilized mutations are missed if residues with high B factors are chosen for mutagenesis, as they are not located in the most flexible regions (7). As the mechanisms of thermal stability for many enzymes have been clarified, computational design has become feasible as a rational-design method to improve thermostability (26). Compared with irrational and semirational strategies, rational design provides reasonable predictive accuracy and reduces labor-intensive experimental screening. By using computational protein predictions, Mohammadi et al. (27) constructed four mutants of Serratia marcescens lipase. G2P and G59P were more thermostable than the wild-type enzyme. In view of this, many computational approaches have been developed to predict the stabilizing effects of mutations, such as I-mutant (28), FoldX (29), and Rosetta (30). Kim et al. (31) selected appropriate sites of Candida antarctica lipase B (CALB) with higher B-factor values and then mutated them with more-rigid amino acids as determined by RosettaDesign algorithms. The melting temperature of the mutant R249L obtained from this rational approach was increased by 2.3°C compared to that of wild-type CALB.
Disulfide bond engineering, the directed design of novel disulfide bonds into target proteins, is another means to enhance protein thermostability. Results show that each native SS bond increases the stability of a protein by 2.3 to 5.2 kcal/mol (32). Growing interest in disulfide bond research has also led to the development of a variety of computational methods to assist in screening for potential sites. SSBOND (33), MODIP (34), Disulfide by Design 2 (DbD2) (35), and BridgeD (36) are extensively used algorithms to predict candidate disulfide regions. The stability of a variety of proteins has been successfully enhanced with the aid of these computational methods (37, 38). Farnoosh et al. (39) used Disulfide by Design software, homology modeling, and molecular dynamics simulations to select appropriate amino acid pairs for the introduction of a disulfide bridge to improve the thermostability of organophosphorus hydrolase. Data analysis showed that half-lives of the A204C/T234C and T128C/E153C mutants were increased up to 4 and 24 min, respectively.
Recently, several methods were applied to enhance the thermostability of RML, but these did not result in a significant increase. The double mutant N120K/K131F, introduced by B-factor screening, retained 63.1% of its residual activity at 70°C, compared to 51.0% for the wild-type after a 5-h incubation (24). Disulfide bond engineering was also applied to RML. Newly formed C96-C106, predicted by the Disulfide by Design server, added 3°C to the optimal temperature (37).
Therefore, the aim of this study was to obtain thermostable mutants of RML with a limited screening library via rational design. Since the three-dimensional (3D) structure of RML has been determined, potential stabilizing point mutations and disulfide bonds were first predicted via multiple computational methods (point mutations with Rosetta ddg_monomer, FoldX, and I-Mutant, and disulfide bonds with SSBOND, MODIP, Disulfide by Design 2 [DbD2], and BridgeD). Identification of sites with critical functions or in conservative regions further decreased the number of mutants to be screened. The mutants were expressed, purified, and characterized. The enzymatic properties of the most thermostable mutant were compared with those of wild-type RML.
RESULTS
Rational design of stable mutants by computational algorithms.
Rosetta ddg_monomer, FoldX, and I-Mutant were used to predict stable mutants of RML. Rosetta ddg_monomer was applied to calculate all possible mutation sites and identified 317 stable mutations. Subsequently, 537 mutations were obtained by FoldX. Positive results from both predictors were then entered into the I-Mutant 3.0 website manually. After combining the results, 60 mutations were recognized as stable by all three predictors and were further processed to eliminate sites with vital functions.
Analysis of functional sites.
The Consurf Server (40) was used to estimate the evolutionary conservation of amino acids in RML among homologous sequences. Interestingly, none of the 60 stable mutations belonged to conserved amino acid sites. Thermostable mutations may be located in critical functional domains that potentially affect catalytic properties. All residues around 5 Å from the catalytic site were selected as immutable because they probably directly contact the substrate (41). In addition, according to Derewenda et al. (42), mutations disrupting salt bridges, disulfide bonds, and several important functional regions (such as the substrate binding pocket and lid region) were not selected as potential mutable positions. As a result, 36 mutations were retained for further experimental validation. The predicted values of these mutants obtained by each predictor are listed in Table S1 in the supplemental material.
Characterization of single mutants and combined mutants.
Primers used for construction of mutants are listed in Table 1 and in Table S2 in the supplemental material. Mutated sequences were inserted between EcoRI and HindIII restriction sites of the expression vector pET-28a(+). Each single point mutant was expressed and purified to measure its apparent melting temperature () to determine any change in thermostability. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) showed that there was a single band at 37 kDa, which coincided with the molecular mass of RML (see Fig. S1 in the supplemental material).
TABLE 1.
Primers used in this study

a Mutant sites are shown in bold italic.
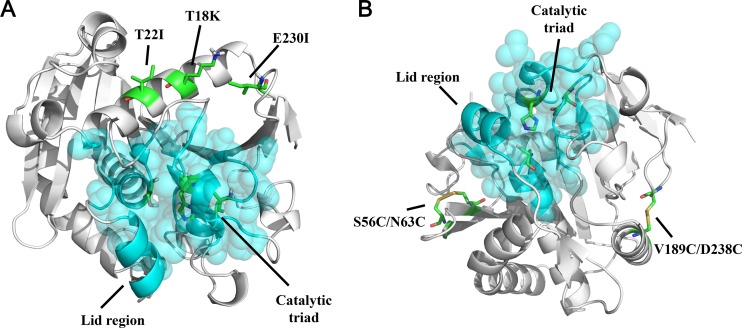
The values of the mutants were examined by differential scanning fluorimetry (43). The results showed that there were 7 unstable, 5 unchanged, and 24 stable candidates (see Table S1 in the supplemental material). Due to the average deviation of the experimental data being lower than 1°C, candidates whose value was at least 1°C higher than that of the wild-type were selected for further experiments. Thr18Lys (T18K) (Δ, +4.5°C), Thr22Ile (T22I) (Δ, +2.4°C), and the best mutant of 11 mutants of Glu230, Glu230Ile (E230I) (Δ, +5.7°C), were selected (Table 2). Double and triple mutants were also designed to observe the synergic effect between the mutants. The values of T18K/T22I, T18K/E230I, T22I/E230I, and T18K/T22I/E230I separately expressed were 5.2, 8.0, 6.9, and 9.4°C higher than that of the original RML, respectively, indicating that these double or triple mutations synergistically increased the thermostability of RML (Table 2). Figure 1A shows the positions of these three mutations in RML.
TABLE 2.
Apparent melting temperatures of single point mutants and disulfide bonds
| Enzyme variant | (°C)a | Δ (°C) |
|---|---|---|
| WT | 58.7 ± 0.7 | 0 |
| E230R | 59.9 ± 0.4 | 1.2 |
| E230M | 60.0 ± 0.3 | 1.3 |
| E230F | 60.3 ± 0.2 | 1.6 |
| E230Y | 60.8 ± 0.6 | 2.1 |
| T22I | 61.1 ± 0.9 | 2.4 |
| E230P | 61.2 ± 0.4 | 2.5 |
| E230W | 61.4 ± 0.2 | 2.7 |
| E230C | 62.0 ± 0.3 | 3.3 |
| E230T | 62.5 ± 0.5 | 3.8 |
| T18K | 63.2 ± 0.0 | 4.5 |
| E230L | 64.1 ± 0.2 | 5.4 |
| E230V | 64.1 ± 0.3 | 5.4 |
| E230I | 64.4 ± 0.2 | 5.7 |
| T18K/T22I | 64.0 ± 0.4 | 5.3 |
| T22I/E230I | 65.6 ± 0.4 | 6.9 |
| T18K/E230I | 66.7 ± 0.0 | 8.0 |
| T18K/T22I/E230I | 68.1 ± 0.4 | 9.4 |
| T18K/T22I/E230I/S56C/N63C | 69.3 ± 0.3 | 10.6 |
| T18K/T22I/E230I/V189C/D238C | 72.3 ± 0.4 | 13.6 |
| T18K/T22I/E230I/S56C/N63C/V189C/D238C (M7) | 73.0 ± 0.3 | 14.3 |
Mean ± standard deviation.
FIG 1.
Locations of different mutations in RML. (A) Distribution of three stabilizing residues (sticks) and functionally important regions of RML (residues within 5 Å of the catalytic triad are shown as cyan spheres). (B) Locations of two stabilizing disulfide bonds (sticks) and functionally important regions of RML (residues within 5 Å of the catalytic triad are shown as cyan spheres).
Comparison of enzyme properties of WT RML and the T18K/T22I/E230I mutant.
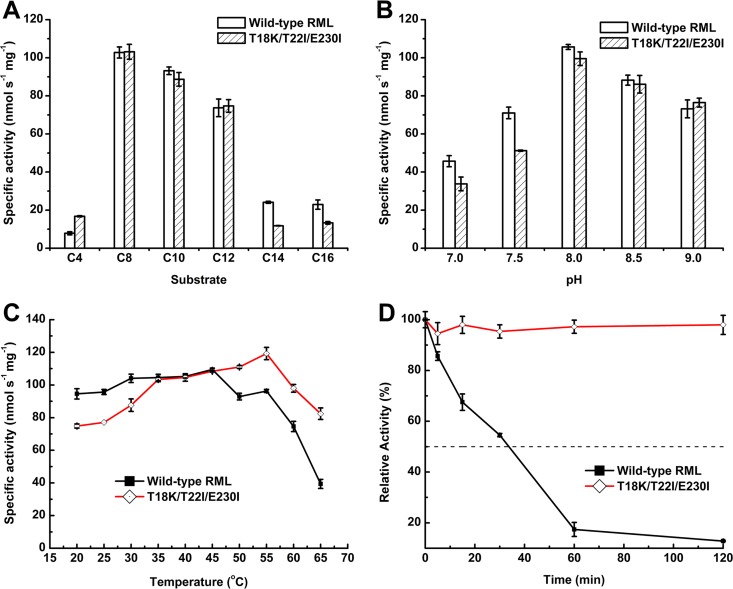
The catalytic properties of the wild-type (WT) enzyme and the triple mutant were further characterized. The WT and the mutant had identical substrate and pH profiles. Their optimal substrate was p-nitrophenyl octanoate (p-NP C8) (Fig. 2A), and the optimal pH for hydrolysis was 8.0 (Fig. 2B). It is worth noting that the optimum temperature for hydrolyzing the optimal substrate was increased from 45°C to 55°C (Fig. 2C), nearly the same as the elevation in . The tolerance of enzymes against high temperatures is closely related to protein stability. The triple mutant retained as much as 95% of its initial activity after a 120-min incubation at 65°C, while the wild-type lipase retained only 13% (Fig. 2D). Under optimum conditions, the kinetic parameters of the two enzymes were assayed using p-NP C8 as the substrate (Table 3). The comparison of the kcat/Km ratio showed that the catalytic efficiencies of the two enzymes were almost the same.
FIG 2.
Enzymatic properties of wild-type RML and the triple mutant (T18K/T22I/E230I). (A) Optimal hydrolytic substrate determined at 40°C. (B) Optimal pH for hydrolytic reaction measured at 40°C. (C) Optimal reaction temperature measured using p-NP C8 as the substrate at pH 8.0. (D) Thermal inactivation by incubation at 65°C.
TABLE 3.
Comparison of kinetic parameters between wild-type RML, the T18K/T22I/E230I mutant, and the M7 mutant
| Enzyme | Topt (°C) | Km (mM)a | kcat (s−1)a | kcat/Km (s−1 · mM−1) | Enzyme activity (U · mg protein−1)a |
|---|---|---|---|---|---|
| WT | 45 | 1.17 ± 0.16 | 24.05 ± 1.96 | 20.56 | 62.35 ± 9.58 |
| T18K/T22I/E230I | 55 | 2.01 ± 0.13 | 42.34 ± 1.51 | 21.06 | 72.35 ± 1.47 |
| M7 | 60 | 1.53 ± 0.07 | 43.71 ± 1.28 | 28.57 | 92.02 ± 4.64 |
Kinetic parameters of all enzymes were determined at their optimal temperatures and are expressed as mean ± standard deviation.
Potential residue pairs predicted to form disulfide bonds.
All residue pairs that may form disulfide bonds were obtained through four web-accessible methods: DbD2, SSBOND, MODIP, and BridgeD. There were 33 residue pairs meeting the default criteria of DbD2, while SSBOND predicted 92 residue pairs where disulfide bonds can be introduced. Eighty-six potential residue pairs were graded from A to D through the MODIP algorithm. By restricting the distance and dihedral angles of newly formed disulfide bonds, 112 residue pairs were suggested by BridgeD as candidates. Based on these results, 155 residue pairs were predicted by at least one method.
Analysis of functional sites.
Residues whose conservation score was above eight and residues within 5 Å of the catalytic triad and other important regions (e.g., the lid region and oxyanion hole) were treated as immutable. After filtering, 56 potential residue pairs were prepared for stability assessment (see Table S3 in the supplemental material).
Stability assessment and visual inspection of designed disulfide bonds.
FoldX was used to predict the stabilizing effects of designed disulfide bonds. As shown in Table S3 in the supplemental material, 47 residue pairs for possible disulfide bonds showed a positive value, indicating that they destabilized the protein. Models of eight residue pairs predicted to stabilize the protein were constructed and energy optimized for visual inspection. In the optimized model, six residue pairs were only free cysteines and could not form disulfide bonds. Finally, S56C/N63C and V189C/D238C potentially formed new disulfide bridges and were chosen as promising candidates for site-directed mutagenesis to improve the thermal stability of RML. Figure 1B shows the positions of the disulfide bridges and functionally important regions in the enzyme. The S56C/N63C bond is located between two beta-strands (positions 50 to 56 and 63 to 69), while V189C/D238C connects an alpha-helix from Pro182 to Ser190 and a free loop from Ser237 to Cys244. Both cysteine pairs were more than 5 Å away from the active site and should not interfere with enzyme activity.
Expression, purification, and determination of disulfide bond mutants.
S56C/N63C and V189C/D238C were constructed in the same manner as the T18K/T22I/E230I mutant of wild-type RML. The two disulfide bond mutants were successfully expressed and purified in E. coli. Figure S1 in the supplemental material shows a single band near 37 kDa by SDS-PAGE analysis, which coincided with the theoretical molecular mass of RML. After purification by nickel affinity chromatography, the solubility of mutant S56C/N63C (50 to 60 mg per 1 liter of culture medium) was comparable to those of wild-type RML and the triple mutant, while the solubility of V189C/D238C was decreased (20 to 30 mg per 1 liter of culture medium). The formation of disulfide bonds in the two mutants was verified by Ellman's method. Under nonreducing conditions, one mole of free thiol was detected in all three protein samples, because there is a free cysteine at position 153. After treatment with dithiothreitol (DTT), 6 mol of free thiol groups was released from T18K/T22I/E230I, while 8 mol of sulfhydryl groups was released from the two mutants that contained designed disulfide bonds (see Table S4 in the supplemental material). According to the study of RML's crystal structure by Derewenda et al., there were three native disulfide bridges, Cys40-Cys43, Cys29-Cys268, and Cys235-Cys244, which stabilized the folding of the polypeptide chain in RML (42). This indicated that T18K/T22I/E230I had three native disulfide bridges and S56C/N63C and V189C/D238C each had four disulfide bonds, indicating successful formation of S56C/N63C and V189C/D238C in the structure of RML.
Effect of introduced disulfide bonds on thermostability.
Enzyme thermodynamic stability is defined as the difference between the Gibbs free energies of the folded and unfolded conformations of a protein. To evaluate the change in thermodynamic stability via mutation, the values of the purified proteins were assessed. The value for T18K/T22I/E230I was 68.1°C. S56C/N63C and V189C/D238C had values of 69.3°C and 72.3°C, respectively, which were 1.2°C and 4.2°C higher than that of the T18K/T22I/E230I mutant. These results validated that S56C/N63C and V189C/D238C had a stabilizing effect on RML (see Fig. S2 in the supplemental material).
Expression of the combined mutants containing stabilizing disulfide bridges.
S56C/N63C and V189C/D238C were combined and introduced into the T18K/T22I/E230I mutant of RML. Due to the low solubility of the disulfide V189C/D238C, 20 to 30 mg of soluble mutant T18K/T22I/E230I/S56C-N63C/V189C-D238C was collected per 1 liter of culture medium. Ellman's test showed that there were five disulfide bridges in the S56C-N63C/V189C-D238C mutant, which meant that these two disulfide bonds were successfully introduced (see Fig. S1 in the supplemental material). We then investigated the enzymatic properties of the combined mutant, T18K/T22I/E230I/S56C-N63C/V189C-D238C, designated M7.
Enzymatic properties of the combined mutants.
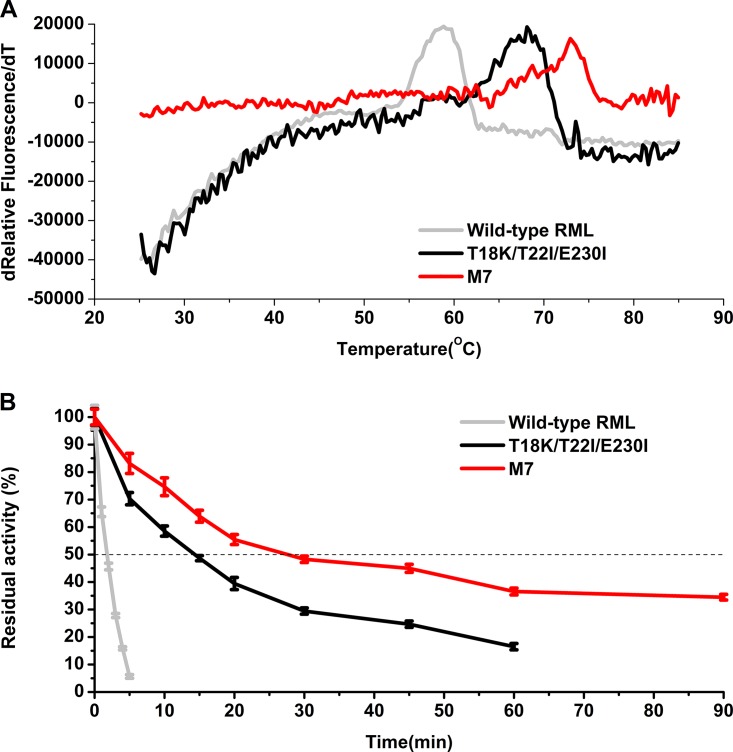
When tested by differential scanning fluorimetry, mutant M7 exhibited a of 73.0°C, which was 4.9°C higher than that of the T18K/T22I/E230I mutant (Fig. 3A). Thus, the effect of beneficial disulfide bridge mutations is synergistic or additive on the thermostability of the target enzyme.
FIG 3.
Thermostability profiles of wild-type RML, T18K/T22I/E230I, and M7. (A) Apparent melting temperatures of wild-type RML, T18K/T22I/E230I, and M7 determined by DSF. (B) Half-lives of wild-type RML and its mutants at 70°C.
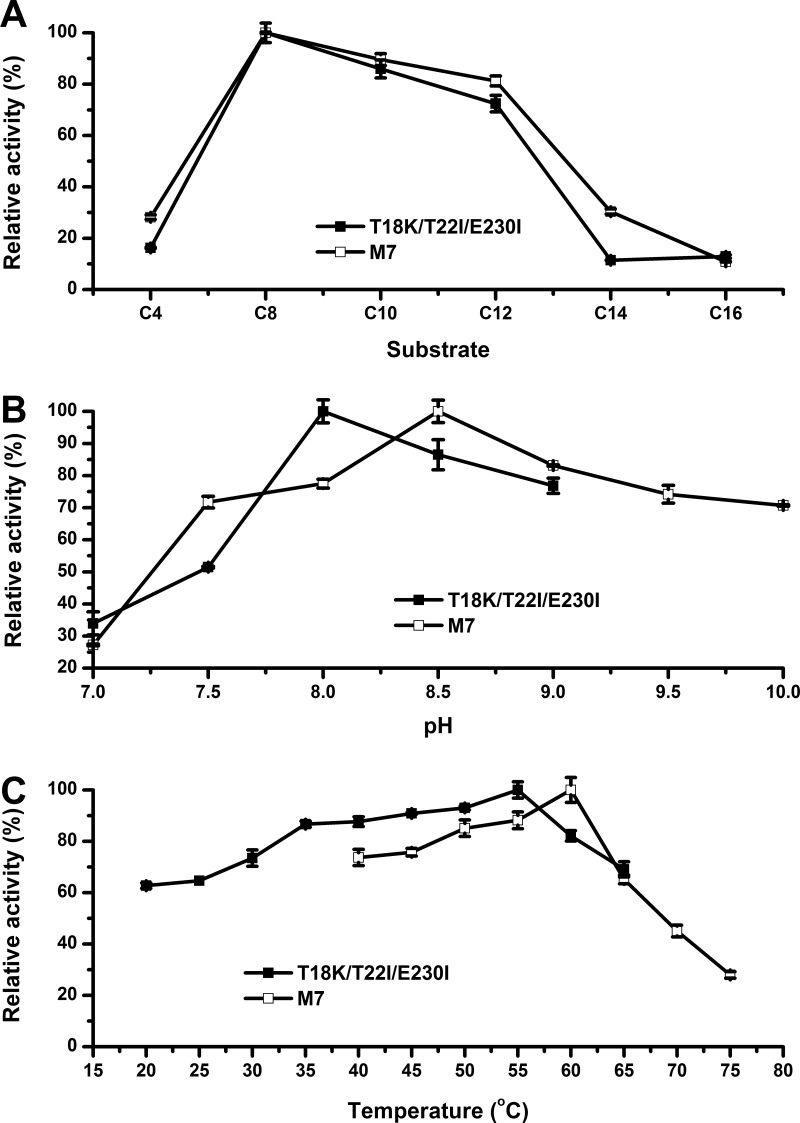
According to the optimum substrate assays (Fig. 4A), we found that for both enzymes, the substrate profile showed no difference. Both enzymes showed the highest catalytic activity toward p-NP C8. This demonstrated that the introduced disulfide bridges had minimal impact on the enzyme's optimum substrate. By utilizing p-NP C8 as the substrate for hydrolysis, the pH preference was determined. The optimum pH of M7 shifted from 8.0 to 8.5 via the introduction of the two disulfide bonds (Fig. 4B). Under alkaline conditions (pH 7.5 to 10.0), M7 maintained more than 70% of its maximal activity and was more stable than T18K/T22I/E230I. The increase of optimum temperature agreed with the obtained . The T18K/T22I/E230I mutant had an optimum temperature of 55°C, whereas mutant M7 was most active at 60°C (Fig. 4C).
FIG 4.
Enzymatic properties of T18K/T22I/E230I and M7. (A) Optimal hydrolytic substrate determined at 40°C. (B) Optimal pH for hydrolytic reaction measured at 40°C. (C) Optimal reaction temperature measured using p-NP C8 as the substrate (at the respective optimal pH).
The kinetic stability of wild-type RML was also compared with those of T18K/T22I/E230I and mutant M7 by determining their half-lives (t1/2) at 70°C. The half-life of wild-type RML at 70°C was only 2.2 min, indicating that the catalytic activity was lost very quickly. The half-life of M7 at 70°C was 27.5 min, which was 93% and 12.5-fold longer than those of T18K/T22I/E230I and the wild type, respectively (Fig. 3B).
The kinetic parameters of both enzymes were determined with p-NP octanoate as the substrate at their own optimum temperatures and pHs (Table 3). The introduced disulfide bridges impacted the kinetics of the enzyme. As shown in Table 3, the Km of mutant M7 decreased by 24%, indicating that the substrate affinity of M7 was higher than that of T18K/T22I/E230I. The catalytic constant (kcat) of mutant M7 increased by 3%. Thus, the catalytic efficiency (kcat/Km) of M7 increased by 36%. The enzyme activities of T18K/T22I/E230I and M7 were measured at their optimum temperatures with p-NP octanoate as the substrate (Table 3). The enzyme activity of M7 was 92.02 ± 4.64 U · mg protein−1, a 27% increase compared to that of T18K/T22I/E230I. This demonstrated that the introduced disulfide bonds improved the thermostability and enzyme activity of RML.
DISCUSSION
R. miehei lipase is a versatile biocatalyst used to produce biodiesel via transesterification or alcoholysis, to synthesize esters from carboxylic acids, to resolve racemic mixtures, to modify polymers, and to catalyze other reactions (8). RML properly retains its catalytic activity only below 50°C, which limits its industrial applications at higher temperatures (10). Soluble expression in E. coli and P. pastoris allows RML to be used as a model system for exploring methods that can enhance thermal stability of proteins expressed in eukaryotic systems. In this study, we aimed to enhance the thermostability of RML by multiple rational-design methods.
Soluble expression of wild-type RML in E. coli makes it rapid and effective to study ways to improve the thermal stability of eukaryotic proteins. However, in our work to engineer RML, different codon usage bias and formation of multiple disulfide bonds made it a great challenge to express eukaryotic proteins in E. coli. RML is expressed in the microenvironment of E. coli, which may differ from that of the original source. All of these factors may lead to protein instability and aggregation (44). Therefore, selecting the appropriate expression system to obtain sufficient soluble protein is also very important for RML thermostability engineering. In this study, the pET-28a expression vector was selected due to the presence of a strong T7 promoter and an effective purification system (45). The Rosetta-gami 2(DE3) strain (Novagen) was used, aiming at enhancing the expression of heterologous proteins by providing codons rarely used in E. coli and improving the formation of disulfide bonds in its cytoplasm (46, 47). Culture conditions may also increase gene expression and protein solubility. The solubility of a recombinant protein is increased by prolonged induction at low temperatures with decreased amounts of IPTG (isopropyl-β-d-thiogalactopyranoside) (48, 49). The optimization of culture conditions in our study resulted in cultivation at 16°C for 16 h after induction with 0.1 mM IPTG.
For protein thermostability enhancement, random mutagenesis and B-factor screening are often conducted at the cost of screening thousands of clones. Computer-assisted design for stability-enhanced mutations is a reliable alternative to screening-based approaches (50), but to improve predictive accuracy, it is necessary to devise reliable methods (51). Rosetta and FoldX have been used in combined strategies such as FRESCO and Fireprot to predict mutants with improved stability when supplemented by molecular modeling, evolutionary conservation analysis, and other screening methods (7, 51). These results showed that it is feasible to combine the prediction results of various algorithms. However, filtering of over 100 RML clones was still needed when using the above-mentioned strategies before conducting wet-lab experiments. Considering this, we assumed that the introduction of new algorithms combined with Rosetta and FoldX would further reduce the screening library. I-Mutant has been successfully used to predict stable point mutations (52) and was one of the most reliable predictors in a systematic analysis of 11 online stability predictors using 1,784 single mutations found in 80 proteins (53). We thus combined the results of Rosetta, FoldX, and I-Mutant to predict stable mutants of RML, followed by an analysis of conserved and functionally important regions, and we obtained 36 mutants with potential stabilizing effects.
Since many mutants are predicted by computational algorithms to enhance thermostability, creating mutants with the best predictive values and characterizing only combined mutants is a common strategy (50, 51, 54). However, this approach inevitably introduces false-positive results and weakly stabilizing mutations (7). Here, we examined the melting temperatures of all single mutants to find the truly stabilizing mutants. Techniques such as differential scanning calorimetry (DSC), differential scanning fluorimetry (DSF), and circular dichroism (CD) spectroscopy are used to determine changes in melting temperature caused by mutations (15). DSF is a rapid, sensitive, and versatile method for measuring the thermal stability of proteins. DSF experiments require only a small amount (10 to 25 μl) of protein with a minimum protein concentration (0.100 to 0.300 mg/ml). A 96-well plate inserted into a real-time PCR machine allows the entire measurement process to be completed in less than 2 h. These features make the DSF method more suitable for rapid measurement of protein thermal stability. Thermostability assays of single mutants of RML using the DSF method revealed that T18K, T22I, and 11 mutants of E230 exhibited a >1°C increase in , which indicated that these mutants had a strong stabilizing effect and that E230 is a “hot spot” for thermostability engineering (Table 2). The most stabilizing mutation of E230, E230I, was rationally combined with the other two mutations. The results showed that T18K, T22I, and E230I synergistically improved the thermostability of RML (Table 2). The more the mutation sites were superposed, the higher was the thermostability. The most stable mutant, T18K/T22I/E230I, which combined the most favorable mutations at each position, showed a 9.4°C-higher than the wild-type (Fig. 3). Enzymatic assays showed that the thermostability profiles of T18K/T22I/E230I were increased compared to that of wild-type RML, while the catalytic properties of the mutated enzyme at higher temperature were almost unchanged.
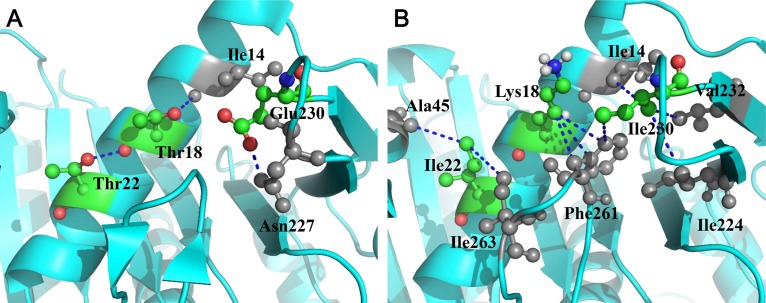
Moreover, in order to explain the possible reasons for the high thermal stability of the T18K/T22I/E230I mutant, the interactions between mutation sites and adjacent amino acids were analyzed. The three-dimensional structure of T18K/T22I/E230I was homology modeled with SWISS-MODEL (55) using RML (PDB ID 3TGL) as the template. The modeled structure was optimized by the GalaxyRefine Server (56) to eliminate steric clashes and disallowed conformations and then assessed by PROCHECK (57) and MolProbity (58). The structure that had the lowest MolProbity score with 95.3% of residues in the favored region of the Ramachandran plot was selected for further analysis. We simulated the stabilizing mechanisms of the three mutants (Fig. 5A and B). The change of interactions via mutation was predicted by iMutants (59), and the overall results are presented in Table S5 in the supplemental material. Mutant T18K might eliminate two side chain-main chain hydrogen bonds and form a new cation-pi interaction between Lys18 and Phe261. The numbers of hydrogen bonds between the main chain and side chain are similar in thermophilic and mesophilic proteins (60); thus, they are not key determinants in stabilizing proteins. The role of cation-pi interactions in protein stability has been analyzed previously, and Lys significantly contributes to the increased stability of thermophiles (61). Mutants T22I and E230I both introduced hydrophobic interactions and formed additional interactions to stabilize the protein. The strength of hydrophobic interactions, such as those between nonpolar amino acid side chains, increases with elevated temperature until it reaches a maximum value (62). In addition, it has been shown that interactions involving hydrophobic residues, such as Ile, are predominant in thermophilic proteins rather than mesophilic ones, thereby contributing to thermal stabilization (60, 63–65).
FIG 5.
Interactions around the mutated positions of WT RML and T18K/T22I/E230I. The interactions around the mutated residues in the WT (A) and the T18K/T22I/E230I mutant (B) are shown. The three mutated residues have colored atoms, while other residues are gray.
A disulfide bond in a protein is a covalent bond formed between two sulfur atoms of sulfhydryl groups in two cysteine residues. Upon oxidation of two thiols, the disulfide bond links two cysteines and their respective peptide chains. The disulfide bridges may increase the stability of enzymes, since it is not energetically conducive to break these covalent physical forces (66). The motion restriction of the random coil or free loop by disulfide bond cross-link also reduces the conformational entropy of the unfolded state, thus contributing the increase in protein stability (67). However, the introduction of disulfide bonds in proteins does not always enhance stability, and it is still difficult to predict appropriate loci and stabilizing effects of disulfide bonds. This is because existing favorable interactions may be lost while unfavorable contacts are formed in the surrounding residues around disulfide bonds (68). MODIP and DbD have been used successfully to increase the thermal stability of certain proteins, demonstrating their usefulness in predicting potential disulfide bonds (38). It has been suggested that the geometrical models used in most current disulfide bond prediction algorithms may overlimit the dynamic movement of the protein chains and their ability to tolerate some modifications without strain (69), meaning that the use of single algorithms such as MODIP and DbD will not predict nearly half of the formable disulfide bonds (34, 70). These unpredictable disulfide bonds are mainly due to violations of distance and angle limits. BridgeD addresses this problem by using several structural models of the target protein to mimic the flexibility of the backbone. The algorithm includes optimizing the residual side chains nearby to adjust the inserted disulfide bonds. Therefore, we summarized all potential pairs that may form disulfide bonds from MODIP, DbD2, SSBOND, and BridgeD to obtain as many disulfide bonds as possible to make the protein more thermostable. However, these tools are concerned only with possible disulfide bond formation, and there is no tool to assess the effect of newly introduced disulfide bonds on protein stability (38). YASARA's FoldX plugin may solve this problem. FoldX minimizes the PDB structure, mutates potential residue pairs to cysteines, and predicts the effects of mutations on protein stability. Whether a disulfide bond is formed or not in the 3D structure can be viewed by YASARA's 3D graphics and intuitive interface. S56C/N63C and V189C/D238C were predicted to stabilize RML and preferentially form in the 3D structure after visual inspection and were thus characterized to determine their thermostability profiles. The presence of the disulfide bridges S56C/N63C and V189C/D238C was confirmed by quickly measuring protein sulfhydryls. Similar to the case for the T18K/T22I/E230I mutant, S56C/N63C and V189C/D238C further contributed 1.2°C- and 4.2°C-higher values, respectively. The newly formed disulfide bonds S56C/N63C and V189C/D238C that increased the thermostability of RML were both located at the protein surface and far from the enzyme catalytic center (Fig. 1B). The disulfide bridge S56C/N63C was situated at the β-sheet of the lipase tertiary structure, while the disulfide bridge V189C/D238C was located between an α-helix and the random coil of the protein. Mutation of amino acid residues to cysteines in flexible surface regions is more likely to generate beneficial mutants due to avoiding formation of strained linkages, minimizing unfavorable contacts with inner residues, and maximizing entropy effects (68).
M7, the mutant that combined the three single point mutations with two introduced disulfide bonds, had a 14.3°C-higher , a 12.5-fold longer half-life at 70°C, a slight shift in optimal pH, and 47% higher catalytic activity than wild-type RML (Fig. 3 and 4 and Table 3). Moreover, according to kinetic experiments, the catalytic efficiency of M7 was 1.39-fold higher than that of the wild-type enzyme mainly due to the increase of the apparent kcat value. A generally accepted opinion is that there is a trade-off between thermal stability and catalytic activity in the natural evolution process of enzymes to accommodate different temperatures in the environment (71). Enzymes with high catalytic efficiency at low temperatures often lose their catalytic activity at high temperatures due to the loss of natural conformation. However, the thermostability of the protein is not always at the expense of the enzyme's affinity and activity. To some extent, it is not contradictory to increase rigidity while increasing affinity and improving catalytic efficiency (72). Several reports have shown that enzymes with both high thermal stability and high catalytic activity were obtained by means of mutagenesis (37, 73).
Finally, recent studies on improvement of lipase thermostability by protein engineering were compared with this study (Table 4). We found that by means of efficient bacterial expression systems, most bacterial lipases, such as Bacillus subtilis lipases, can undergo large-scale screening of mutants, thereby greatly improving their thermostability. However, for most fungal lipases, an over 10°C increase in operating temperature is based on the screening of more than 1,000 mutants, which represents a huge workload and long experimental period for eukaryotic expression systems such as yeast. Previous rational-design methods significantly reduced the experimental screening process for fungal lipases but rarely obtained a substantial increase in the thermal stability of mutants. In contrast to previous studies, a reduced screening library and a highly stable mutant were simultaneously obtained by using the method in this study.
TABLE 4.
Studies on improvement of lipase thermostability
| Lipase | Design category | Method(s) | Improvement (°C) | No. in screening library | Reference |
|---|---|---|---|---|---|
| Bacterial | |||||
| Bacillus subtilis lipase A | Irrational | DNA shuffling | 22.0 | 18,000 | 3 |
| Bacillus subtilis lipase LipJ | Irrational | Error-prone PCR | 13.0 | 4,200 | 16 |
| Pseudomonas aeruginosa lipase | Irrational | Error-prone PCR | 2.5 | 2,600 | 19 |
| Bacillus subtilis lipase A | Semirational | B-factor analysis | 45.0 | 8,000 | 4 |
| Geobacillus zalihae T1 lipase | Rational | I-mutant | 2.0 | 2 | 52 |
| Fungal | |||||
| Candida antarctica lipase B | Irrational | DNA shuffling | 6.4 | 2,500 | 17 |
| Rhizopus arrhizus lipase | Irrational | Error-prone PCR, DNA shuffling | 10.0 | 5,500 | 18 |
| Yarrowia lipolytica lipase Lip2 | Semirational | B-factor analysis | 3.0 | 4,200 | 21 |
| Candida antarctica lipase B | Semirational | B-factor analysis | 12.0 | 2,200 | 22 |
| Candida rugosa lipase 1 | Semirational | B-factor analysis | 12.7 | 20,000 | 23 |
| Candida antarctica lipase B | Rational | MODIP, DbD v1.20 | 8.5 | 5 | 38 |
| Candida antarctica lipase B | Rational | B-factor analysis, RosettaDesign | 2.3 | 7 | 31 |
| Rhizomucor miehei lipase | Rational | DbD | 3.0 | 1 | 37 |
| Rhizomucor miehei lipase | Rational | This study | 14.3 | 38 | This study |
It is important to develop rapid and efficient methods for enhancing the thermostability of proteins expressed in eukaryotic systems. In this study, R. miehei lipase, as a model system, was engineered by multiple rational-design methods. Our integrated strategy appears to be a general and efficient way to enhance enzyme thermostability without substantial experimental effort. There were only 36 candidates in the screening library of single point mutations, of which 24 candidates increased stability and 13 mutants increased the apparent melting temperature by at least 1°C. Both of the disulfide bonds predicted by our strategy further enhanced the thermostability of RML. Finally, the apparent melting temperature and the catalytic efficiency of the engineered RML were about 15°C and 40% higher, respectively, than those of the wild-type enzyme. These results demonstrate that rational design of point mutations and disulfide bonds can effectively reduce the number of the screened clones and is thus a good potential strategy for enhancing the thermostability of proteins expressed in eukaryotic systems.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The RML gene was derived from the wild pro-RML gene (GenBank accession no. KP164599.1), and a D3N mutation was introduced to keep the amino acid sequence translated from the gene identical to that from the solved crystal structure (PDB ID 3TGL). E. coli DH5α and E. coli Rosetta-gami2(DE3) (Novagen, Darmstadt, Germany) were cultured in Luria-Bertani medium at 37°C for gene cloning and protein expression, respectively. All strains were stored as glycerol stock at −80°C until use.
Protein X-ray structure treatment.
The X-ray crystal structure of lipase from RML (PDB ID 3TGL) was downloaded from the RCSB PDB database (74). The PDB file was then processed by the clean_pdb script of Rosetta to skip missing residues and remove irrelevant molecules.
Point mutations prediction. (i) Prediction by Rosetta.
The low-resolution mode of the Rosetta ddg_monomer application (E. H. Kellogg and A. Leaver-Fay, https://www.rosettacommons.org [accessed 19 September 2017]) was utilized to predict the energy change upon mutation, since the high-resolution mode had almost the same accuracy and was time-consuming. The options of the protocol were based on the algorithm published by Kellogg et al. (30) (-ddg::weight_file soft_rep_design -fa_max_dis 9.0 -ddg::iterations 50 -ignore_unrecognized_res -ddg::local_opt_only true -ddg:: mean true -ddg::min false -ddg::sc_min_only false).
(ii) Prediction by FoldX.
Before modeling with FoldX, the RepairPDB module (FoldX suite, http://foldxsuite.crg.eu/command/RepairPDB [accessed 19 September 2017]) was used to identify those residues that had bad torsion angles or Van der Waals clashes and to repair them. Subsequently, the output files were submitted to the BuildModel module (FoldX suite, http://foldxsuite.crg.eu/command/BuildModel [accessed 19 September 2017]) to estimate the stability effect. The default settings were used, and the number of runs was set to 5 to get a better average result.
(iii) Prediction by I-Mutant 3.0.
The “Protein Structure” mode (University of Bologna, Bologna, Italy, http://gpcr2.biocomp.unibo.it/cgi/predictors/I-Mutant3.0/I-Mutant3.0.cgi [accessed 19 September 2017]) was selected to predict the protein stability change from the crystal structure. The PDB code, chain label, mutated position, new residue code, temperature in degrees Celsius, and pH value were input to the online server. Binary classification was chosen as the prediction method.
Rational prediction of potential disulfide bonds.
Disulfide by Design 2 (DbD2) (http://cptweb.cpt.wayne.edu/DbD2/index.php [accessed 19 September 2017]) is an upgraded version of the original DbD application (75) for designing disulfide bonds in proteins. Each possible pair of residues in RML was assessed for potential disulfide formation, assuming that the residues were mutated to cysteines. The χ3 torsion angle and Cα-Cβ-Sγ angle of all residue pairs were calculated. The potential residue pairs that meet conformational criteria were collected.
SSBOND (http://hazeslab.med.ualberta.ca/forms/ssbond.html [accessed 19 September 2017]) predicted the presence of disulfide bonds in RML with its PDB structure. The program returned a series of residue pairs that were suitable for the formation of disulfide bonds when mutated into cysteine. The program also gave the dihedral angles of the estimated disulfide bonds and the energy constraints describing their formation. The smaller the energy constraint, the easier it is to form the ideal configuration.
MODIP (http://caps.ncbs.res.in/iws/modip.html [accessed 19 September 2017]) evaluated the sites in RML by a hierarchical system using typical conformational parameters of naturally occurring disulfide bonds. Disulfide bonds with ideal dihedral angles were classified as grade A. Sites that were spatially suitable for the formation of disulfide bonds but slightly distorted were classified as B, and residue pairs that were only spatially close and may form disulfide bonds were classified as C.
BridgeD (http://biodev.cea.fr/bridged/ [accessed 19 September 2017]) used a variety of protein structures to increase the flexibility of the protein backbone to find potential disulfide-forming sites. These protein structures were derived from independent crystal structures of the same protein, multiple structures obtained by nuclear magnetic resonance, or crystal structures of homologous proteins. According to the energy ranking, if the predicted λ equals 1 and ΔE is negative, then it is a good target for protein engineering.
Prediction of stability effect and visual inspection of designed disulfide bonds.
The change of protein stability upon formation of each disulfide bond was predicted by the FoldX plugin (76) in the YASARA program (77). By using the “Mutate multiple residues” command, potential residue pairs were mutated into cysteines, and then the FoldX Stability command optimized the conformations and calculated the difference in stability between mutant and wild-type structures. A negative value indicates that the newly formed SS bond increased the stability of the protein. When the calculation was finished, whether a disulfide bond was formed was easily observed in the optimized model.
Bioinformatic analysis. (i) Identification of residues with critical function.
No residues around 5 Å from the catalytic triad selected by PyMOL (78) were mutated (-select triad, resi 144 + 203 + 257 -select triad5a, byres triad around 5). Residues in functionally important regions (such as the active site, substrate binding pocket, and lid region) have been identified as immutable according to Derewenda et al. (42).
(ii) Evolutionary conservation and correlated mutation analyses.
Evolutionary conservation profiles for proteins of known PDB structure are available from ConSurf-DB (79). Sequences similar to the target sequence were gathered from the Swiss-Prot database (80) using CSI-BLAST, and then a multiple-sequence alignment (MSA) was generated using the MAFFT method. The Rate4Site algorithm (81) calculated the evolutionary conservation at each individual position and then discriminated by different grades using an empirical Bayesian inference. Residues with a conservative scale of ≥8 were regarded as highly conserved and were not mutated. The MSA was used as an input for the 3DM Comulator tool (82) to analyze correlated mutations. Positions with scores of ≥0.8 were treated as evolutionarily correlated and immutable.
Gene cloning, expression, and purification of RML.
Mutants of RML were created with PrimeSTAR HS DNA polymerase (TaKaRa, Otsu, Japan). The mutated gene was inserted between EcoRI and HindIII restriction sites of the vector pET-28a(+), which contains an N-terminal hexahistidine tag. Mutations were validated by DNA sequencing. All mutants were transformed into E. coli Rosetta-gami2(DE3) cells (Novagen, Darmstadt, Germany) for expression. The cells were grown in 800 ml of Luria-Bertani culture medium containing 50 μg/ml kanamycin and 34 μg/ml chloramphenicol at 37°C until the optical density at 600 nm (OD600) reached 0.6 to 0.8, induced with 0.1 mM IPTG (final concentration) at 16°C for 16 h, and then harvested by centrifugation at 4,000 × g for 15 min. Cell lysates were subject to gradient elution with nitrilotriacetic acid (NTA) buffer (0.05 M Tris-HCl [pH 8.0], 0.5 M NaCl) with different concentrations of imidazole (0.03 M, 0.06 M, and 0.5 M) in nickel affinity columns. If needed, additional purification was performed by gel filtration. Proteins were dialyzed against 50 mM sodium phosphate buffer (pH 8.0) for experimental assessment. Samples were centrifuged at 14,000 × g for 2 min to eliminate any aggregated protein.
measurements.
Differential scanning fluorimetry (43) was used to determine the apparent melting temperature values for mutants. The was defined as the maximal value of the relative fluorescence change with respect to the temperature (dRFU/dT). For each analyzed sample, 5 μl of 100× SYPRO Orange solution (Life Technologies, Carlsbad, CA, USA) and 20 μl purified protein were mixed and then centrifuged for 5 min at 4°C. The was determined by heating the samples from 25 to 85°C at 1°C/min in a StepOnePlus real-time PCR system (Life Technologies, USA).
Analysis of enzyme properties.
The optimal enzyme substrate was determined using p-nitrophenyl fatty acyl esters (p-NP esters) with different chain lengths. In a standard assay, the total 1-ml reaction system contained 940 μl of Tris-HCl buffer (50 mM, pH 8.0), 10 μl of p-NP ester (100 mM), 40 μl of ethanol, and 10 μl of diluted enzyme solution (83). In the blank control, the enzyme solution was replaced with the same volume of Tris-HCl buffer. All experiments were conducted in triplicate. One unit (U) of lipase activity was defined as the amount of enzyme that released 1 μmol of p-NP per min under the assay conditions. The optimal pH was determined by examining the enzyme activity at 40°C in Tris-HCl buffer at different pH values. The optimal temperature was determined by varying the temperatures with an interval of 5°C in Tris-HCl buffer (pH 8.0). The thermostability was assessed by testing the residual activity against the optimal substrate after incubating the enzyme solution for different time periods. The kinetic parameters (kcat, Km, and kcat/Km) of the wild-type RML and mutants were determined using different substrate concentrations at their optimum temperature. The kinetic parameters were determined from Lineweaver-Burk plots using Microsoft Excel.
Homology modeling.
Structures of mutated RML were modeled with SWISS-MODEL using the wild-type enzyme (PDB ID 3TGL) as a template. Modeled structures were optimized by the GalaxyRefine server to eliminate steric clashes and disallowed conformations and then assessed by PROCHECK and MolProbity. Structures that had the lowest MolProbity scores and were over 95% in the allowed region of a Ramachandran plot were chosen for analysis.
Analysis of the basis of thermostability change via protein mutations.
Possible interaction changes caused by the mutations were analyzed by the online tool iMutants (59). Ionic interactions and cation-pi interactions within 6 Å of the mutations, hydrophobic interactions within 5 Å of the mutations, aromatic-aromatic interactions with centroid distances 4.5 Å to 7.0 Å, and aromatic-S interactions with a distance cutoff of 5.3 Å were all assessed. Hydrogen bonds were calculated using HBPLUS software (84).
Supplementary Material
ACKNOWLEDGMENTS
We thank Cheng Hong of the Centre of Analysis and Test, Huazhong University of Science and Technology, for enzyme characterization.
We acknowledge the financial support of the National Natural Science Foundation of China (no. 31170078 and J1103514), the National High Technology Research and Development Program of China (no. 2011AA02A204, 2013AA065805, and 2014AA093510), the National Natural Science Foundation of Hubei Province (no. 2015CFA085), and Fundamental Research Funds for HUST (no. 2014NY007).
We declare no competing financial interests.
G.L. designed the experimental scheme and performed the majority of evaluations, comparisons, calculations, and analyses. X.F. and Y.C. performed the enzyme characterization. F.S. and L.X. helped with the experiments. G.L. and Y.Y. wrote and revised the manuscript. All authors reviewed the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02129-17.
REFERENCES
- 1.Su F, Li G, Fan Y, Yan Y. 2016. Enhanced performance of lipase via microcapsulation and its application in biodiesel preparation. Sci Rep 6:29670. doi: 10.1038/srep29670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan Y, Li X, Wang G, Gui X, Li G, Su F, Wang X, Liu T. 2014. Biotechnological preparation of biodiesel and its high-valued derivatives: a review. Appl Energ 113:1614–1631. doi: 10.1016/j.apenergy.2013.09.029. [DOI] [Google Scholar]
- 3.Kamal MZ, Ahmad S, Molugu TR, Vijayalakshmi A, Deshmukh MV, Sankaranarayanan R, Rao NM. 2011. In vitro evolved non-aggregating and thermostable lipase: structural and thermodynamic investigation. J Mol Biol 413:726–741. doi: 10.1016/j.jmb.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Reetz MT, Carballeira JD, Vogel A. 2006. Iterative saturation mutagenesis on the basis of B factors as a strategy for increasing protein thermostability. Angew Chem Int Ed 45:7745–7751. doi: 10.1002/anie.200602795. [DOI] [PubMed] [Google Scholar]
- 5.Demain AL, Vaishnav P. 2009. Production of recombinant proteins by microbes and higher organisms. Biotechnol Adv 27:297–306. doi: 10.1016/j.biotechadv.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt-Dannert C. 1999. Recombinant microbial lipases for biotechnological applications. Bioorgan Med Chem 7:2123–2130. doi: 10.1016/S0968-0896(99)00141-8. [DOI] [PubMed] [Google Scholar]
- 7.Wijma HJ, Floor RJ, Jekel PA, Baker D, Marrink SJ, Janssen DB. 2014. Computationally designed libraries for rapid enzyme stabilization. Protein Eng Des Sel 27:49–58. doi: 10.1093/protein/gzt061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodrigues RC, Fernandez-Lafuente R. 2010. Lipase from Rhizomucor miehei as an industrial biocatalyst in chemical process. J Mol Catal B Enzym 64:1–22. doi: 10.1016/j.molcatb.2010.02.003. [DOI] [Google Scholar]
- 9.Al-Zuhair S. 2007. Production of biodiesel: possibilities and challenges. Biofuels Bioprod Bioref 1:57–66. doi: 10.1002/bbb.2. [DOI] [Google Scholar]
- 10.Noel M, Combes D. 2003. Effects of temperature and pressure on Rhizomucor miehei lipase stability. J Biotechnol 102:23–32. doi: 10.1016/S0168-1656(02)00359-0. [DOI] [PubMed] [Google Scholar]
- 11.Huang J, Xia J, Yang Z, Guan F, Cui D, Guan G, Jiang W, Li Y. 2014. Improved production of a recombinant Rhizomucor miehei lipase expressed in Pichia pastoris and its application for conversion of microalgae oil to biodiesel. Biotechnol Biofuels 7:111. doi: 10.1186/1754-6834-7-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang D, Wang J, Wang B, Yu H. 2012. A new and efficient colorimetric high-throughput screening method for triacylglycerol lipase directed evolution. J Mol Catal B Enzym 82:18–23. doi: 10.1016/j.molcatb.2012.05.021. [DOI] [Google Scholar]
- 13.Su F, Li G, Zhang H, Yan Y. 2014. Enhanced performance of Rhizopus oryzae lipase immobilized on hydrophobic carriers and its application in biorefinery of rapeseed oil deodorizer distillate. Bioenerg Res 7:935–945. doi: 10.1007/s12155-014-9415-y. [DOI] [Google Scholar]
- 14.Stepankova V, Bidmanova S, Koudelakova T, Prokop Z, Chaloupkova R, Damborsky J. 2013. Strategies for stabilization of enzymes in organic solvents. ACS Catal 3:2823–2836. doi: 10.1021/cs400684x. [DOI] [Google Scholar]
- 15.Bommarius AS, Paye MF. 2013. Stabilizing biocatalysts. Chem Soc Rev 42:6534–6565. doi: 10.1039/c3cs60137d. [DOI] [PubMed] [Google Scholar]
- 16.Goomber S, Kumar R, Singh R, Mishra N, Kaur J. 2016. Point mutation Gln121-Arg increased temperature optima of Bacillus lipase (1.4 subfamily) by fifteen degrees. Int J Biol Macromol 88:507–514. doi: 10.1016/j.ijbiomac.2016.04.022. [DOI] [PubMed] [Google Scholar]
- 17.Suen WC, Zhang N, Xiao L, Madison V, Zaks A. 2004. Improved activity and thermostability of Candida antarctica lipase B by DNA family shuffling. Protein Eng Des Sel 17:133–140. doi: 10.1093/protein/gzh017. [DOI] [PubMed] [Google Scholar]
- 18.Niu W-N, Li Z-P, Zhang D-W, Yu M-R, Tan T-W. 2006. Improved thermostability and the optimum temperature of Rhizopus arrhizus lipase by directed evolution. J Mol Catal B Enzym 43:33–39. doi: 10.1016/j.molcatb.2006.04.013. [DOI] [Google Scholar]
- 19.Shinkai A, Hirano A, Aisaka K. 1996. Substitutions of Ser for Asn-163 and Pro for Leu-264 are important for stabilization of lipase from Pseudomonas aeruginosa. J Biochem 120:915–921. doi: 10.1093/oxfordjournals.jbchem.a021506. [DOI] [PubMed] [Google Scholar]
- 20.Nimpiboon P, Kaulpiboon J, Krusong K, Nakamura S, Kidokoro S-i, Pongsawasdi P. 2016. Mutagenesis for improvement of activity and thermostability of amylomaltase from Corynebacterium glutamicum. Int J Biol Macromol 86:820–828. doi: 10.1016/j.ijbiomac.2016.02.022. [DOI] [PubMed] [Google Scholar]
- 21.Wen S, Tan T, Zhao H. 2013. Improving the thermostability of lipase Lip2 from Yarrowia lipolytica. J Biotechnol 164:248–253. doi: 10.1016/j.jbiotec.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 22.Xie Y, An J, Yang G, Wu G, Zhang Y, Cui L, Feng Y. 2014. Enhanced enzyme kinetic stability by increasing rigidity within the active site. J Biol Chem 289:7994–8006. doi: 10.1074/jbc.M113.536045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X-F, Yang G-Y, Zhang Y, Xie Y, Withers SG, Feng Y. 2016. A general and efficient strategy for generating the stable enzymes. Sci Rep 6:33797. doi: 10.1038/srep33797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J-H, Lin Y, Sun Y-F, Ye Y-R, Zheng S-P, Han S-Y. 2012. High-throughput screening of B factor saturation mutated Rhizomucor miehei lipase thermostability based on synthetic reaction. Enzyme Microb Technol 50:325–330. doi: 10.1016/j.enzmictec.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Roth T, Beer B, Pick A, Sieber V. 2017. Thermostabilization of the uronate dehydrogenase from Agrobacterium tumefaciens by semi-rational design. AMB Express 7:103. doi: 10.1186/s13568-017-0405-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Damborsky J, Brezovsky J. 2014. Computational tools for designing and engineering enzymes. Curr Opin Chem Biol 19:8–16. doi: 10.1016/j.cbpa.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Mohammadi M, Sepehrizadeh Z, Ebrahim-Habibi A, Shahverdi AR, Faramarzi MA, Setayesh N. 2016. Enhancing activity and thermostability of lipase A from Serratia marcescens by site-directed mutagenesis. Enzyme Microb Technol 93-94: 18–28. doi: 10.1016/j.enzmictec.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 28.Capriotti E, Fariselli P, Casadio R. 2005. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res 33:W306–W310. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guerois R, Nielsen JE, Serrano L. 2002. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol 320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 30.Kellogg EH, Leaver-Fay A, Baker D. 2011. Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins 79:830–838. doi: 10.1002/prot.22921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim HS, Le QAT, Kim YH. 2010. Development of thermostable lipase B from Candida antarctica (CalB) through in silico design employing B-factor and RosettaDesign. Enzyme Microb Technol 47:1–5. doi: 10.1016/j.enzmictec.2010.04.003. [DOI] [Google Scholar]
- 32.Tidor B, Karplus M. 1993. The contribution of cross-links to protein stability: a normal mode analysis of the configurational entropy of the native state. Proteins 15:71–79. doi: 10.1002/prot.340150109. [DOI] [PubMed] [Google Scholar]
- 33.Hazes B, Dijkstra BW. 1988. Model building of disulfide bonds in proteins with known three-dimensional structure. Protein Eng Des Sel 2:119–125. doi: 10.1093/protein/2.2.119. [DOI] [PubMed] [Google Scholar]
- 34.Dani VS, Ramakrishnan C, Varadarajan R. 2003. MODIP revisited: re-evaluation and refinement of an automated procedure for modeling of disulfide bonds in proteins. Protein Eng Des Sel 16:187–193. doi: 10.1093/proeng/gzg024. [DOI] [PubMed] [Google Scholar]
- 35.Craig DB, Dombkowski AA. 2013. Disulfide by Design 2.0: a web-based tool for disulfide engineering in proteins. BMC Bioinformatics 14:346. doi: 10.1186/1471-2105-14-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pellequer JL, Chen SwW. 2006. Multi-template approach to modeling engineered disulfide bonds. Proteins 65:192–202. doi: 10.1002/prot.21059. [DOI] [PubMed] [Google Scholar]
- 37.Han Z-l, Han S-y, Zheng S-p, Lin Y. 2009. Enhancing thermostability of a Rhizomucor miehei lipase by engineering a disulfide bond and displaying on the yeast cell surface. Appl Microbiol Biotechnol 85:117. doi: 10.1007/s00253-009-2067-8. [DOI] [PubMed] [Google Scholar]
- 38.Le QAT, Joo JC, Yoo YJ, Kim YH. 2012. Development of thermostable Candida antarctica lipase B through novel in silico design of disulfide bridge. Biotechnol Bioeng 109:867–876. doi: 10.1002/bit.24371. [DOI] [PubMed] [Google Scholar]
- 39.Farnoosh G, Khajeh K, Latifi AM, Aghamollaei H. 2016. Engineering and introduction of de novo disulphide bridges in organophosphorus hydrolase enzyme for thermostability improvement. J Biosci 41:577–588. doi: 10.1007/s12038-016-9643-8. [DOI] [PubMed] [Google Scholar]
- 40.Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. 2003. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics 19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 41.Morley KL, Kazlauskas RJ. 2005. Improving enzyme properties: when are closer mutations better? Trends Biotechnol 23:231–237. doi: 10.1016/j.tibtech.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 42.Derewenda ZS, Derewenda U, Dodson GG. 1992. The crystal and molecular structure of the Rhizomucor miehei triacylglyceride lipase at 1.9 Å resolution. J Mol Biol 227:818–839. doi: 10.1016/0022-2836(92)90225-9. [DOI] [PubMed] [Google Scholar]
- 43.Ericsson UB, Hallberg BM, Detitta GT, Dekker N, Nordlund P. 2006. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal Biochem 357:289–298. doi: 10.1016/j.ab.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 44.Rosano GL, Ceccarelli EA. 2014. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol 5:172. doi: 10.3389/fmicb.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baneyx F. 1999. Recombinant protein expression in Escherichia coli. Curr Opin Biotechnol 10:411–421. doi: 10.1016/S0958-1669(99)00003-8. [DOI] [PubMed] [Google Scholar]
- 46.Kane JF. 1995. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr Opin Biotechnol 6:494–500. doi: 10.1016/0958-1669(95)80082-4. [DOI] [PubMed] [Google Scholar]
- 47.Derman AI, Prinz WA, Belin D, Beckwith J. 1993. Mutations that allow disulfide bond formation in the cytoplasm of Escherichia coli. Science 262:1744–1748. doi: 10.1126/science.8259521. [DOI] [PubMed] [Google Scholar]
- 48.Gopal GJ, Kumar A. 2013. Strategies for the production of recombinant protein in Escherichia coli. Protein J 32:419–425. doi: 10.1007/s10930-013-9502-5. [DOI] [PubMed] [Google Scholar]
- 49.Sahdev S, Khattar SK, Saini KS. 2008. Production of active eukaryotic proteins through bacterial expression systems: a review of the existing biotechnology strategies. Mol Cell Biochem 307:249–264. doi: 10.1007/s11010-007-9603-6. [DOI] [PubMed] [Google Scholar]
- 50.Korkegian A, Black ME, Baker D, Stoddard BL. 2005. Computational thermostabilization of an enzyme. Science 308:857–860. doi: 10.1126/science.1107387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bednar D, Beerens K, Sebestova E, Bendl J, Khare S, Chaloupkova R, Prokop Z, Brezovsky J, Baker D, Damborsky J. 2015. FireProt: energy-and evolution-based computational design of thermostable multiple-point mutants. PLoS Comput Biol 11:e1004556. doi: 10.1371/journal.pcbi.1004556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruslan R, Rahman RNZRA, Leow TC, Ali MSM, Basri M, Salleh AB. 2012. Improvement of thermal stability via outer-loop ion pair interaction of mutated T1 lipase from Geobacillus zalihae strain T1. Int J Mol Sci 13:943–960. doi: 10.3390/ijms13010943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khan S, Vihinen M. 2010. Performance of protein stability predictors. Hum Mutat 31:675–684. doi: 10.1002/humu.21242. [DOI] [PubMed] [Google Scholar]
- 54.Malakauskas SM, Mayo SL. 1998. Design, structure and stability of a hyperthermophilic protein variant. Nat Struct Mol Biol 5:470–475. doi: 10.1038/nsb0698-470. [DOI] [PubMed] [Google Scholar]
- 55.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heo L, Park H, Seok C. 2013. GalaxyRefine: protein structure refinement driven by side-chain repacking. Nucleic Acids Res 41:W384–W388. doi: 10.1093/nar/gkt458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 58.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Panigrahi P, Sule M, Ghanate A, Ramasamy S, Suresh CG. 2015. Engineering proteins for thermostability with iRDP web server. PLoS One 10:e0139486. doi: 10.1371/journal.pone.0139486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumar S, Tsai C-J, Nussinov R. 2000. Factors enhancing protein thermostability. Protein Eng Des Sel 13:179–191. doi: 10.1093/protein/13.3.179. [DOI] [PubMed] [Google Scholar]
- 61.Gromiha MM, Thomas S, Santhosh C. 2002. Role of cation-π interactions to the stability of thermophilic proteins. Prep Biochem Biotechnol 32:355–362. doi: 10.1081/PB-120015459. [DOI] [PubMed] [Google Scholar]
- 62.Scheraga HA, Némethy G, Steinberg IZ. 1962. The contribution of hydrophobic bonds to the thermal stability of protein conformations. J Biol Chem 237:2506–2508. [PubMed] [Google Scholar]
- 63.Zhou X-X, Wang Y-B, Pan Y-J, Li W-F. 2008. Differences in amino acids composition and coupling patterns between mesophilic and thermophilic proteins. Amino Acids 34:25–33. doi: 10.1007/s00726-007-0589-x. [DOI] [PubMed] [Google Scholar]
- 64.Chakravarty S, Varadarajan R. 2000. Elucidation of determinants of protein stability through genome sequence analysis. FEBS Lett 470:65–69. doi: 10.1016/S0014-5793(00)01267-9. [DOI] [PubMed] [Google Scholar]
- 65.Ibrahim BS, Pattabhi V. 2004. Role of weak interactions in thermal stability of proteins. Biochem Biophys Res Commun 325:1082–1089. doi: 10.1016/j.bbrc.2004.10.128. [DOI] [PubMed] [Google Scholar]
- 66.Dehnavi E, Fathi-Roudsari M, Mirzaie S, Arab SS, Siadat SOR, Khajeh K. 2017. Engineering disulfide bonds in Selenomonas ruminantium β-xylosidase by experimental and computational methods. Int J Biol Macromol 95:248–255. doi: 10.1016/j.ijbiomac.2016.10.104. [DOI] [PubMed] [Google Scholar]
- 67.Pace CN, Grimsley G, Thomson J, Barnett B. 1988. Conformational stability and activity of ribonuclease T1 with zero, one, and two intact disulfide bonds. J Biol Chem 263:11820–11825. [PubMed] [Google Scholar]
- 68.Hibi T, Kume A, Kawamura A, Itoh T, Fukada H, Nishiya Y. 2016. Hyperstabilization of tetrameric Bacillus sp. TB-90 urate oxidase by introducing disulfide bonds through structural plasticity. Biochemistry 55:724–732. doi: 10.1021/acs.biochem.5b01119. [DOI] [PubMed] [Google Scholar]
- 69.Meinhold D, Beach M, Shao Y, Osuna R, Colón W. 2006. The location of an engineered inter-subunit disulfide bond in factor for inversion stimulation (FIS) affects the denaturation pathway and cooperativity. Biochemistry 45:9767–9777. doi: 10.1021/bi060672n. [DOI] [PubMed] [Google Scholar]
- 70.Dombkowski AA, Sultana KZ, Craig DB. 2014. Protein disulfide engineering. FEBS Lett 588:206–212. doi: 10.1016/j.febslet.2013.11.024. [DOI] [PubMed] [Google Scholar]
- 71.Giver L, Gershenson A, Freskgard P-O, Arnold FH. 1998. Directed evolution of a thermostable esterase. Proc Natl Acad Sci U S A 95:12809–12813. doi: 10.1073/pnas.95.22.12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sheng J, Ji X, Zheng Y, Wang Z, Sun M. 2016. Improvement in the thermostability of chitosanase from Bacillus ehimensis by introducing artificial disulfide bonds. Biotechnol Lett 38:1809–1815. doi: 10.1007/s10529-016-2168-2. [DOI] [PubMed] [Google Scholar]
- 73.Song JK, Rhee JS. 2000. Simultaneous enhancement of thermostability and catalytic activity of phospholipase A1 by evolutionary molecular engineering. Appl Environ Microbiol 66:890–894. doi: 10.1128/AEM.66.3.890-894.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. 2000. The protein data bank. Nucleic Acids Res 28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dombkowski AA. 2003. Disulfide by Design™: a computational method for the rational design of disulfide bonds in proteins. Bioinformatics 19:1852–1853. doi: 10.1093/bioinformatics/btg231. [DOI] [PubMed] [Google Scholar]
- 76.Van Durme J, Delgado J, Stricher F, Serrano L, Schymkowitz J, Rousseau F. 2011. A graphical interface for the FoldX forcefield. Bioinformatics 27:1711–1712. doi: 10.1093/bioinformatics/btr254. [DOI] [PubMed] [Google Scholar]
- 77.Krieger E, Koraimann G, Vriend G. 2002. Increasing the precision of comparative models with YASARA NOVA—a self-parameterizing force field. Proteins 47:393–402. doi: 10.1002/prot.10104. [DOI] [PubMed] [Google Scholar]
- 78.DeLano WL. 2002. The PyMOL molecular graphics system. http://pymol.org.
- 79.Goldenberg O, Erez E, Nimrod G, Ben-Tal N. 2009. The ConSurf-DB: pre-calculated evolutionary conservation profiles of protein structures. Nucleic Acids Res 37:D323–D327. doi: 10.1093/nar/gkn822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boeckmann B, Bairoch A, Apweiler R, Blatter M-C, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, Phan I. 2003. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res 31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pupko T, Bell RE, Mayrose I, Glaser F, Ben-Tal N. 2002. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics 18:S71–S77. doi: 10.1093/bioinformatics/18.suppl_1.S71. [DOI] [PubMed] [Google Scholar]
- 82.Kuipers RK, Joosten HJ, Verwiel E, Paans S, Akerboom J, van der Oost J, Leferink NG, van Berkel WJ, Vriend G, Schaap PJ. 2009. Correlated mutation analyses on super-family alignments reveal functionally important residues. Proteins 76:608–616. doi: 10.1002/prot.22374. [DOI] [PubMed] [Google Scholar]
- 83.Zheng X, Chu X, Zhang W, Wu N, Fan Y. 2011. A novel cold-adapted lipase from Acinetobacter sp. XMZ-26: gene cloning and characterisation. Appl Microbiol Biotechnol 90:971–980. doi: 10.1007/s00253-011-3154-1. [DOI] [PubMed] [Google Scholar]
- 84.McDonald IK, Thornton JM. 1994. Satisfying hydrogen bonding potential in proteins. J Mol Biol 238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.