ABSTRACT
Chickens with good or poor feed efficiency (FE) have been shown to differ in their intestinal microbiota composition. This study investigated differences in the fecal bacterial community of highly and poorly feed-efficient chickens at 16 and 29 days posthatch (dph) and evaluated whether a fecal microbiota transplant (FMT) from feed-efficient donors early in life can affect the fecal microbiota in chickens at 16 and 29 dph and chicken FE and nutrient retention at 4 weeks of age. A total of 110 chickens were inoculated with a FMT or a control transplant (CT) on dph 1, 6, and 9 and ranked according to residual feed intake (RFI; the metric for FE) on 30 dph. Fifty-six chickens across both inoculation groups were selected as the extremes in RFI (29 low, 27 high). RFI-related fecal bacterial profiles were discernible at 16 and 29 dph. In particular, Lactobacillus salivarius, Lactobacillus crispatus, and Anaerobacterium operational taxonomic units were associated with low RFI (good FE). Multiple administrations of the FMT only slightly changed the fecal bacterial composition, which was supported by weighted UniFrac analysis, showing similar bacterial communities in the feces of both inoculation groups at 16 and 29 dph. Moreover, the FMT did not change the RFI and nutrient retention of highly and poorly feed-efficient recipients, whereas it tended to increase feed intake and body weight gain in female chickens. This finding suggests that host- and environment-related factors may more strongly affect chicken fecal microbiota and FE than the FMT.
IMPORTANCE Modulating the chicken's early microbial colonization using a FMT from highly feed-efficient donor chickens may be a promising tool to establish a more desirable bacterial profile in recipient chickens, thereby improving host FE. Although FE-associated fecal bacterial profiles at 16 and 29 dph could be established, the microbiota composition of a FMT, when administered early in life, may not be a strong factor modulating the fecal microbiota at 2 to 4 weeks of life and reducing the variation in chicken's FE. Nevertheless, the present FMT may have potential benefits for growth performance in female chickens.
KEYWORDS: fecal microbiota transplant, chicken, fecal microbiota, nutrient retention, feed efficiency
INTRODUCTION
The chicken gastrointestinal tract harbors complex microbial communities, with the highest bacterial diversity found in the ceca (1). The ceca are dominated by strictly anaerobic bacteria, mainly Ruminococcaceae and Lachnospiraceae (1, 2), whereas in the crop, small intestine, and feces Lactobacillus is highly abundant (1, 3). The establishment of the chicken's intestinal microbiota begins immediately posthatch and is influenced by internal (e.g., host genetics) and external factors (e.g., diet and encountered environmental microbes) (4). The development of a mature microbial community mostly occurs until 3 weeks of age (1). Maturational changes within the gastrointestinal microbiota significantly impact host's phenotype by modulating the development of the digestive tract and the secretion of bile acids and digestive enzymes, which influences nutrient digestion and absorption and consequently may affect growth performance and feed efficiency (FE) (5). Moreover, the commensal intestinal microbiota play important roles in stimulating intestinal immune functions and in colonization resistance against pathogens (4, 5).
Although different FE-associated intestinal microbiota have been reported for chickens, the identification of target bacteria proves to be challenging, since bacteria associated with good FE vary greatly both within and between studies (6, 7, 8). Moreover, many of the FE-associated phylotypes failed to be classified at the genus or species level, rendering it difficult to cultivate or nutritionally target these bacteria (7, 8). Also, the complex interactions with the microbial communities cannot be considered by administrating single-strain inocula. In human medicine, complex fecal microbiota transplants (FMT) have been effectively used to treat severe intestinal dysfunctions, such as recurrent Clostridium difficile infections (9, 10). Similarly, in chickens, administration of the fecal microbiota from healthy adults has been used to transfer colonization resistance against Salmonella to newly hatched chickens (11). Inoculating the surfaces of incubating eggs with cecal contents from highly or poorly feed-efficient donor chickens has been shown to reduce bird-to-bird variation in microbiota composition but did not impact FE in the growing chicks (12). In contrast, previous work in mice colonized with human microbiota indicated that a FMT can induce obesity (13) and hence may modify the FE of the host. Whether an FMT may be more efficacious to improve chicken FE has not been sufficiently investigated so far. Since differences in the fecal microbiota between poorly and highly feed-efficient chickens exist (6, 14), we hypothesized that excreta collected from highly feed-efficient chickens may influence the chicken's early microbial colonization and subsequently improve FE.
Because improvements in chicken FE can reduce nutrient excretion (15), we further hypothesized that, if the FMT can improve chicken FE, this should be associated with enhanced nutrient retention and reduced excretion of environmental pollutants. The objectives of the present study were to investigate (i) the FE-associated bacterial profiles in feces of chickens at 16 and 29 days posthatch (dph), (ii) the effect of administrating a FMT from highly feed-efficient donors early in life on the fecal microbiota of highly and poorly feed-efficient chickens at 16 and 29 dph, and (iii) whether the FMT could modify chicken FE and nutrient retention at slaughter age. To assess whether the fecal microbiota generally impacted host FE and physiological traits, correlations between fecal bacterial abundances and performance traits and excreta characteristics were calculated.
RESULTS
Performance, excreta characteristics, and nutrient retention.
The effects of residual feed intake (RFI) rank and inoculation treatment on FE and performance traits between 9 and 30 dph, combined for both sexes and separately for females and males, are summarized in Table 1. Across sexes, the RFI was 289 g lower in low-RFI (highly feed-efficient) compared to high-RFI (poorly feed-efficient) chickens (P < 0.001). Furthermore, chickens with a low RFI showed a 278.5-g-lower total feed intake (TFI) than their high-RFI counterparts (P < 0.001). Male chickens ate more feed, were heavier (P < 0.001), and showed a better FE (P < 0.05) than female chickens. The FMT had no effect on RFI in both sexes and, in males, on TFI and total body weight gain (TBWG) (P > 0.10). However, in females, the FMT tended to increase TFI and TBWG by 147 and 82 g, respectively, compared to the control transplant (CT) (P < 0.10).
TABLE 1.
Total feed intake, total body weight gain, and residual feed intake values of low- and high-RFI broiler chickens receiving either a fecal microbiota transplant or a control transplant
| Parameter | Mean wt (g) and pooled SEMa |
P |
||||||
|---|---|---|---|---|---|---|---|---|
| FMT |
CT |
SEM | ||||||
| Low RFI | High RFI | Low RFI | High RFI | FMT | RFI | FMT×RFI | ||
| Both sexes | ||||||||
| TFI | 2315 | 2631 | 2327 | 2568 | 57.3 | 0.665 | <0.001 | 0.517 |
| TBWG | 1641 | 1654 | 1656 | 1640 | 39.8 | 0.980 | 0.964 | 0.724 |
| RFI | −108 | 198 | −117 | 155 | 25.2 | 0.310 | <0.001 | 0.506 |
| Females | ||||||||
| TFI | 2233 | 2544 | 2117 | 2366 | 82.2 | 0.087 | 0.002 | 0.707 |
| TBWG | 1564 | 1516 | 1472 | 1444 | 45.0 | 0.081 | 0.407 | 0.837 |
| RFI | −104 | 259 | −103 | 192 | 38.8 | 0.405 | <0.001 | 0.390 |
| Males | ||||||||
| TFI | 2398 | 2694 | 2537 | 2770 | 85.7 | 0.221 | 0.005 | 0.713 |
| TBWG | 1710 | 1789 | 1841 | 1837 | 63.2 | 0.163 | 0.551 | 0.517 |
| RFI | −105 | 128 | −130 | 118 | 31.7 | 0.591 | <0.001 | 0.814 |
Total feed intake (TFI), total body weight gain (TBWG) and residual feed intake (RFI) were calculated for the experimental period from 9 to 30 dph. Data are presented as least-square means and pooled SEM. FMT, fecal microbiota transplant; CT, control transplant. Low-RFI FMT females, n = 8; low-RFI FMT males, n = 7; high-RFI FMT females, n = 7; high-RFI FMT males, n = 6; low-RFI CT females, n = 7; low-RFI CT males, n = 7; high-RFI CT females, n = 7; high-RFI CT males, n = 7. Sex affected both TFI and TBWG (P < 0.001) and RFI (P < 0.05).
Excreta characteristics and nutrient retention data across sexes are presented according to RFI rank and inoculation treatment in Table 2. Chickens with a low RFI had a 4.15 and 7.05% higher protein (P < 0.05) and phosphorus (P < 0.10) retention, respectively, compared to their high-RFI counterparts. The FMT did not affect nutrient retention and excreta characteristics across RFI ranks (P > 0.10). However, a FMT×RFI interaction (P < 0.05) indicated that the FMT compared to the CT increased the dry matter (DM) content in excreta of low-RFI chickens, but not in high-RFI chickens.
TABLE 2.
Excreta characteristics and retention of nutrients in low- and high-RFI broiler chickens receiving either a FMT or a CT
| Parameter | Mean or SEMa |
P |
||||||
|---|---|---|---|---|---|---|---|---|
| FMT |
CT |
SEM | ||||||
| Low RFI | High RFI | Low RFI | High RFI | FMT | RFI | FMT×RFI | ||
| Dry matter content (%) | 18.4a | 17.0ab | 16.6bB | 18.2abA | 0.63 | 0.592 | 0.926 | 0.024 |
| pH | 6.8 | 6.7 | 7.0 | 6.7 | 0.15 | 0.486 | 0.243 | 0.629 |
| Ammonia (μmol/g of fresh sample) | 51.0 | 41.9 | 42.1 | 47.6 | 3.68 | 0.666 | 0.631 | 0.052 |
| Retention (%) | ||||||||
| Dry matter | 82.3 | 78.7 | 81.8 | 80.9 | 1.68 | 0.625 | 0.181 | 0.434 |
| Crude ash | 48.9 | 42.6 | 50.1 | 45.1 | 4.56 | 0.682 | 0.220 | 0.885 |
| Crude protein | 79.5 | 74.7 | 79.5 | 76.0 | 1.94 | 0.727 | 0.037 | 0.739 |
| Phosphorus | 58.0 | 50.4 | 56.7 | 50.2 | 3.93 | 0.858 | 0.081 | 0.880 |
Data are presented as least-square means and pooled SEM. Low-RFI FMT females, n = 8; low-RFI FMT males, n = 7; high-RFI FMT females, n = 7; high-RFI FMT males, n = 6; low-RFI CT females, n = 7; low-RFI CT males, n = 7; high-RFI CT females, n = 7; high-RFI CT males, n = 7. Different lowercase superscript letters within a row indicate a significant difference (P≤ 0.05); different uppercase superscript letters within a row indicate a tendency (P ≤ 0.10).
16S rRNA sequencing metrics.
After quality control and chimera check, a total of 3,266,165 sequencing reads with a mean of 25,516 (standard deviation [SD], ±7,616) sequences per sample were obtained for the 112 fecal (FMT chickens, n = 28/time point; and CT chickens, n = 28/time point), 8 FMT inoculum, 6 diet, and 2 water samples. Rarefaction curves, using a maximum rarefaction depth of 10,000 sequences and the observed operational taxonomic unit (OTU) index, are presented in Fig. S1 in the supplemental material.
Bacterial composition of the FMT, diet, and water samples.
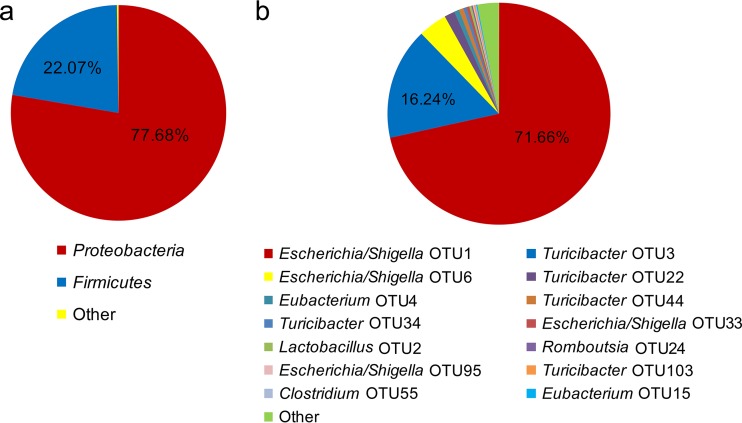
The FMT comprised mainly the phyla Proteobacteria (77.7%) and Firmicutes (22.1%, Fig. 1a), whereby the Escherichia/Shigella OTU (OTU1; 71.7%) and Turicibacter OTU (OTU3; 16.2%) dominated (Fig. 1b). The bacteria found in diet and water samples are presented at the phylum and OTU levels in Tables S1 and S2 in the supplemental material. The bacterial communities of diets and feces formed separate clusters, as indicated by weighted UniFrac β-diversity analysis (see Fig. S2 in the supplemental material).
FIG 1.
Bacterial composition of the fecal microbiota transplant: phyla (a) and operational taxonomic units (OTU) (b). FMT inoculum, n = 8 (FMT inoculum of the three individual inoculation days of the two batches and pooled samples of the FMT inocula across the three inoculation days per batch).
Structure- and time-related shifts in the fecal bacterial community.
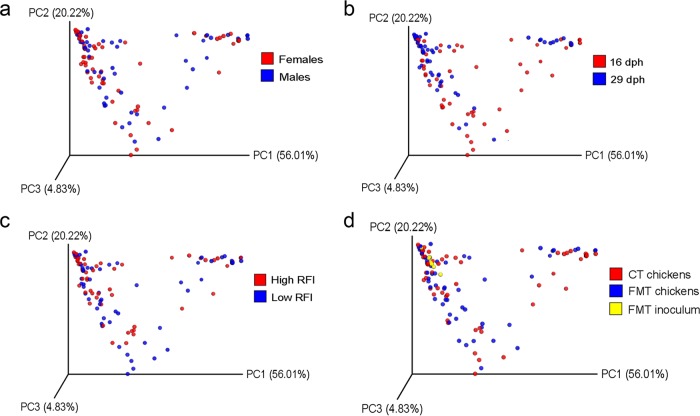
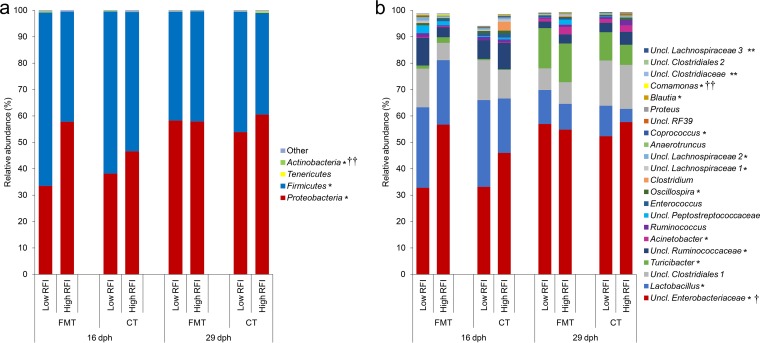
The overall bacterial community structure was similar between sexes, as indicated by β-diversity analysis (Fig. 2a). At the phylum level, Proteobacteria (50.8%) and Firmicutes (48.6%) were clearly predominant in chicken feces across inoculation treatment groups, RFI ranks, and time points, with all other phyla showing much lower abundance (<0.5%; Fig. 3a). At the genus level, an unclassified Enterobacteriaceae genus was predominant (48.8%), followed by Lactobacillus (18.4%), an unclassified Clostridiales genus (12.2%), Turicibacter (6.5%), an unclassified Ruminococcaceae genus (5.7%), Acinetobacter (1.1%), and Ruminococcus (1.0%), while all other genera showed abundances of <1.0% (Fig. 3b). Comparisons of α-diversity and the microbiota composition at all taxonomic levels (i.e., phylum, family, genus, and OTU levels) indicated maturational changes in the fecal community from 16 to 29 dph (Fig. 3 and Tables 3 and 4; see also Table S3 in the supplemental material). A β-diversity analysis using weighted UniFrac distances, in turn, did not show differences in the overall bacterial community structure between the two sampling time points (Fig. 2b). The α-diversity indices based on species richness and evenness (Shannon and Simpson) tended to decrease from 16 to 29 dph (P < 0.10; Table 3). This was accompanied by a decrease in the fecal abundance of highly abundant Firmicutes and low-abundance Actinobacteria, whereas highly abundant Proteobacteria increased from 16 to 29 dph (P < 0.05; Fig. 3a). Moreover, time point-related effects existed within the highly abundant Lactobacillus genus and for two Eubacterium OTU types (OTU4 and OTU15), which were 2.7-, 3.2-, and 3.1-fold more abundant at 16 dph than at 29 dph (P < 0.05; Fig. 3b; se also Table S3 in the supplemental material). In contrast, highly abundant Escherichia/Shigella OTU (OTU1 and OTU6) and Turicibacter OTU (OTU3 and OTU22) increased by 1.3-, 1.4-, 11.5-, and 13.1-fold from 16 to 29 dph, respectively (P < 0.05).
FIG 2.
Principal-coordinate analysis plots of weighted UniFrac analysis. (a) Fecal samples of females and males; (b) fecal samples at 16 and 29 dph; (c) fecal samples of low- and high-RFI broiler chickens; (d) broiler chickens receiving either a fecal microbiota transplant (FMT chickens) or a control transplant (CT chickens) and the FMT inoculum. The yellow circles in panel d represent the FMT inoculum of the different inoculation days of the two batches and pooled samples of the FMT inocula across the three inoculation days per batch. Low-RFI FMT females, n = 8/time point; low-RFI FMT males, n = 7/time point; high-RFI FMT females, n = 7/time point; high-RFI FMT males, n = 6/time point; low-RFI CT females, n = 7/time point; low-RFI CT males, n = 6 at 16 dph and n = 7 at 29 dph; high-RFI CT females, n = 7/time point; high-RFI CT males, n = 7/time point; FMT inoculum, n = 8. A rarefaction depth of 10,000 sequences per sample removed one sample from the data set (male, low RFI, CT, 16 dph).
FIG 3.
Relative abundances (%): bacterial phyla (a) and most abundant bacterial genera (relative abundance > 0.5%) (b) in feces at 16 and 29 dph in low- and high-RFI broiler chickens receiving either a FMT or a CT. *, P ≤ 0.05 (effect of time point); **, P ≤ 0.10 (trend for time point effect); †, P ≤ 0.10 (trend for RFI rank effect); ††, P ≤ 0.10 (trend for FMT effect). Low-RFI FMT females, n = 8/time point; low-RFI FMT males, n = 7/time point; high-RFI FMT females, n = 7/time point; high-RFI FMT males, n = 6/time point; low-RFI CT females, n = 7/time point; low-RFI CT males, n = 7/time point; high-RFI CT females, n = 7/time point; high-RFI CT males, n = 7/time point. Uncl., unclassified.
TABLE 3.
Differences in α-diversity indices in feces at 16 and 29 dph in low- and high-RFI broiler chickens receiving either a FMT or a CT
| Index | Mean or SEMa |
P |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16 dph |
29 dph |
SEM | ||||||||||||
| FMT |
CT |
FMT |
CT |
|||||||||||
| Low RFI | High RFI | Low RFI | High RFI | Low RFI | High RFI | Low RFI | High RFI | Tb | FMT | RFI | FMT×RFI | T×FMT×RFI | ||
| Shannon | 4.0 | 2.8 | 3.4 | 3.6 | 2.8 | 2.8 | 3.0 | 3.2 | 0.42 | 0.076 | 0.568 | 0.489 | 0.167 | 0.475 |
| Simpson | 0.77 | 0.61 | 0.69 | 0.68 | 0.61 | 0.58 | 0.62 | 0.62 | 0.060 | 0.053 | 0.811 | 0.248 | 0.306 | 0.692 |
| Chao1 | 972 | 521 | 707 | 842 | 577 | 578 | 652 | 792 | 143.1 | 0.248 | 0.425 | 0.686 | 0.096 | 0.355 |
Data are presented as least-square means and pooled SEM. Low-RFI FMT females, n = 8/time point; low-RFI FMT males, n = 7/time point; high-RFI FMT females, n = 7/time point; high-RFI FMT males, n = 6/time point; low-RFI CT females, n = 7/time point; low-RFI CT males, n = 6 at 16 dph and n = 7 at 29 dph; high-RFI CT females, n = 7/time point; high-RFI CT males, n = 7/time point. A rarefaction depth of 10,000 sequences per sample removed one sample from the data set (male, low RFI, CT, 16 dph).
T, time point.
TABLE 4.
Differences in relative abundance (%) of most abundant bacterial families in feces at 16 and 29 dph in low- and high-RFI chickens receiving either a FMT or a CT
| Parameter | Mean or SEMa |
P |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16 dph |
29 dph |
SEM | ||||||||||||
| FMT |
CT |
FMT |
CT |
|||||||||||
| Low RFI | High RFI | Low RFI | High RFI | Low RFI | High RFI | Low RFI | High RFI | Tb | FMT | RFI | FMT×RFI | T×FMT×RFI | ||
| Enterobacteriaceae | 32.80 | 56.79 | 33.19 | 46.05 | 57.02 | 54.92 | 52.38 | 58.07 | 8.186 | 0.021 | 0.621 | 0.095 | 0.889 | 0.403 |
| Lactobacillaceae | 30.53 | 24.41 | 32.87 | 20.54 | 12.83 | 9.73 | 11.60 | 4.99 | 6.419 | 0.001 | 0.663 | 0.107 | 0.572 | 0.964 |
| Ruminococcaceae | 13.24 | 4.84 | 9.14 | 12.39 | 3.41 | 4.91 | 4.77 | 7.91 | 3.476 | 0.030 | 0.486 | 0.964 | 0.238 | 0.424 |
| Turicibacteraceae | 1.24 | 2.14 | 0.45 | 0.26 | 15.17 | 14.56 | 10.71 | 7.59 | 3.619 | <0.001 | 0.176 | 0.769 | 0.727 | 0.809 |
| Lachnospiraceae | 2.36 | 0.64 | 1.42 | 2.31 | 0.44 | 0.77 | 0.55 | 0.79 | 0.519 | 0.004 | 0.583 | 0.867 | 0.109 | 0.196 |
| Moraxellaceae | 0.27 | 0.29 | 0.05 | 0.24 | 1.10 | 2.86 | 1.43 | 2.43 | 1.027 | 0.018 | 0.902 | 0.319 | 0.843 | 0.821 |
| Unclassified RF39 | 0.28 | 0.01 | 0.11 | 0.07 | 0.04 | 0.05 | 0.05 | 0.47 | 0.127 | 0.692 | 0.386 | 0.743 | 0.084 | 0.095 |
| Comamonadaceae | 0.19 | 0.40 | 0.21 | 0.14 | 0.005 | 0.007 | 0.006 | 0.01 | 0.100 | 0.002 | 0.421 | 0.611 | 0.332 | 0.589 |
Data are presented as least-square means and pooled SEM. Low-RFI FMT females, n = 8/time point; low-RFI FMT males, n = 7/time point; high-RFI FMT females, n = 7/time point; high-RFI FMT males, n = 6/time point; low-RFI CT females, n = 7/time point; low-RFI CT males, n = 7/time point; high-RFI CT females, n = 7/time point; high-RFI CT males, n = 7/time point. Sex affected Ruminococcaceae (P < 0.10).
T, time point.
RFI-associated differences in the fecal microbiota.
Weighted UniFrac-based distances showed no difference in the fecal microbiota composition between RFI ranks (Fig. 2c). Compositional differences in the fecal bacterial abundances between low- and high-RFI chickens at the phylum, family, and genus levels were hardly detectable (Fig. 3 and Table 4). However, within the Enterobacteriaceae an unclassified genus tended to be 1.2-fold less abundant in low-RFI than in high-RFI chickens (P < 0.10). At the OTU level, a low RFI was significantly associated with increased abundance of four Lactobacillus OTU, with the closest reference strains being Lactobacillus salivarius (OTU47 and OTU43), Lactobacillus crispatus (OTU51 and OTU67), and Anaerobacterium OTU81 (P ≤ 0.05; see Tables S3 and S4 in the supplemental material). Furthermore, six other Lactobacillus crispatus OTU (OTU5, OTU8, OTU21, OTU30, OTU42, and OTU98) tended to be increased, whereas Klebsiella OTU18 tended to be decreased in low-RFI compared to high-RFI chickens (P < 0.10).
FMT-related microbiota shifts.
Weighted UniFrac distances indicated high similarities in the structures of fecal bacterial communities between treatment groups and the FMT (Fig. 2d). The high-abundance Escherichia/Shigella OTU1 in the FMT also predominated in chicken's feces at 16 (38.6%) and 29 dph (50.1%) but did not differ between the two inoculation groups (P > 0.10; see Table S3 in the supplemental material). Likewise, the second most abundant OTU in the FMT (Turicibacter OTU3) was predominant in feces across time points (0.83% at 16 dph and 9.6% at 29 dph) but again was equally abundant between both inoculation groups. Administration of the FMT decreased the abundance of one Anaerobacterium OTU (OTU56) and Klebsiella OTU18 by 2.8- and 2.6-fold (P < 0.05). Moreover, the FMT tended (P < 0.10) to decrease the abundances of five Anaerobacterium OTU (OTU16, OTU28, OTU58, OTU119, and OTU130) and one Acetivibrio OTU (OTU80) compared to the CT, whereas one Comamonas OTU (OTU113) tended (P < 0.10) to be more abundant in the FMT compared to the CT chicken group. Likewise, the FMT tended (P < 0.10) to increase Comamonas at the genus level (Fig. 3b). At the phylum level, low-abundance Actinobacteria tended to be less abundant in the FMT chicken group than in the CT chicken group (P < 0.10; Fig. 3a).
Correlations between fecal bacterial abundances at 29 dph and RFI, TBWG, TFI, and excreta characteristics.
Multiple fecal bacterial associations with FE, growth performance, and excreta characteristics were observed across treatment groups (Fig. 4). Only positive correlations between OTU abundances at 29 dph and RFI existed (P < 0.05). Accordingly, three OTU positively correlated with high RFI; two Gracilibacter OTU (OTU50 [r = 0.33] and OTU88 [r = 0.34]) and one Clostridium OTU (OTU39 [r = 0.38]). One Anaerobacterium OTU (OTU29) negatively correlated with TFI (r = −0.37; P < 0.05). Anaerobacterium OTU29 further negatively correlated with TBWG (r = −0.36), as well as Hespellia OTU64 (r = −0.33; P < 0.05). Furthermore, two Lactobacillus OTU were negatively correlated with fecal pH (OTU7 [r = −0.36] and OTU46 [r = −0.35]; P < 0.05). Negativibacillus OTU70 positively correlated with fecal DM (r = 0.33; P < 0.05). Furthermore, three Turicibacter OTU (OTU3 [r = 0.36], OTU22 [r = 0.36], and OTU44 [r = 0.38]) positively correlated with fecal NH3 (P < 0.05).
FIG 4.
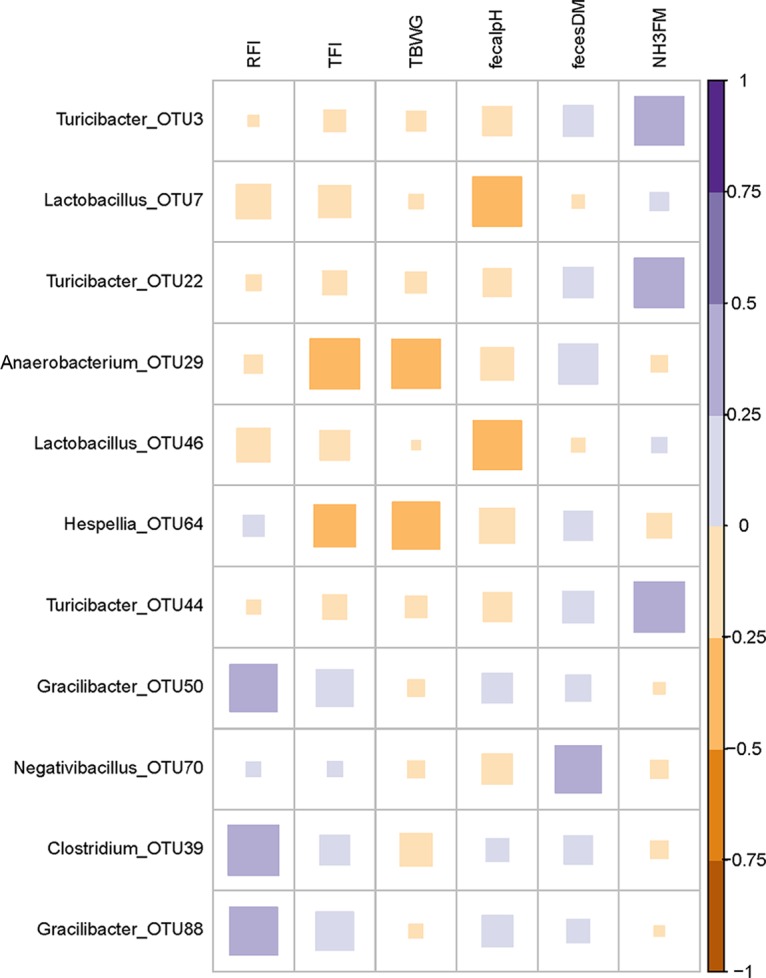
Correlations between OTU at 29 dph and feed efficiency, performance traits, and excreta characteristics. Correlations were significant (P ≤ 0.05) if correlation coefficients were ≤−0.33 or ≥0.33. The OTU were included in the matrix if they occurred in at least half of the chickens and if they were significant for at least one of the parameters. Low-RFI FMT females, n = 8; low-RFI FMT males, n = 7; high-RFI FMT females, n = 7; high-RFI FMT males, n = 6; low-RFI CT females, n = 7; low-RFI CT males, n = 7; high-RFI CT females, n = 7; high-RFI CT males, n = 7. RFI, residual feed intake; TFI, total feed intake; TBWG, total body weight gain; DM, dry matter; NH3, ammonia; FM, fresh matter.
DISCUSSION
The present study investigated differences in the fecal bacterial community in chickens divergent for FE and evaluated whether the application of a FMT in the first week posthatch would modify the fecal microbiota and promote FE and nutrient retention in recipient chickens. Differences in the fecal bacterial abundances at 16 and 29 dph and Pearson's correlations supported that especially Lactobacillus and Anaerobacterium species could be used as indicators for low RFI in the present chicken population. Moreover, the results support that low-RFI chickens more efficiently used the dietary protein than high-RFI chickens (15). However, the FMT from highly feed-efficient donor chickens led only to a few compositional changes in the fecal microbiota of recipient chickens and had little effect on RFI-associated bacterial abundances at 16 and 29 dph. Also, the current microbiota profile used as the FMT did not improve the chicken RFI or nutrient utilization at 4 weeks of age, indicating, together with the bacterial data, that other probably host-related and environment (e.g., diet)-related factors more strongly affected chicken's microbiota maturation and FE than did the application of an external microbial inoculum within the first days of life (16). In contrast, the FMT effectively reduced the time taken to reach slaughter weight in female chickens, as it tended to stimulate feed intake and, as a consequence, TBWG in females, which may be associated with differences in microbe-host signaling between females receiving the FMT and those receiving the CT.
In contrast to our previous study using the same chicken line, housing, and dietary formulation (17), different FE-associated bacterial taxa were found in the present study. This may be partly ascribed to maturational changes in the intestinal microbiota, since the age of the chickens at fecal sampling between the two studies differed, with the current chickens being 6 days younger than in our previous experiment (17). Maturational changes also tended to decrease the diversity of the fecal bacterial community from 16 to 29 dph in the present study, which was in contrast to previous findings for the fecal bacterial community in chickens 7, 21, and 42 dph (18). Similar to the maturational changes reported by Ranjitkar et al. (1) for the chicken cecum at 15 and 29 dph, Lactobacillus was more abundant in feces at 16 dph than at 29 dph in the present study. As the chickens aged, we observed an increase in Turicibacter and Escherichia/Shigella phylotypes in feces, which contrasted with previous findings for the cecum (1, 19) and may be related to the intestinal site and differences in the succession of diets fed to the chickens during the respective experiments.
Despite the age-related changes in the fecal bacterial composition, similar RFI-associated bacteria could be identified at 16 and 29 dph. In particular, mainly L. salivarius and L. crispatus OTU types were associated with low RFI and thus improved FE. Both L. salivarius and L. crispatus are resistant to bile salts and acidic pH, inhibit the growth of potential pathogens such as Escherichia coli and Salmonella enteritidis, and have immunomodulatory properties (20, 21). Moreover, L. salivarius and Lactobacillus agilis may enhance intestinal butyrate production, which may be beneficial for intestinal health via cross-feeding, as shown for an in vitro-simulated chicken cecum (22). Nevertheless, inconsistent findings were reported for the association between lactobacilli and FE in chickens (7, 8, 23, 24), which may be due to particularities in experimental design, microbiota composition, diet, and the intestinal site investigated. When correlating individual RFI values with bacterial abundances in feces at 29 dph, the results indicated that RFI associations existed with other low-abundance phylotypes (relative abundances of 0.09 to 0.18% at 29 dph). These phylotypes were classified as one Clostridium saccharolyticum and two Gracilibacter OTU using the NCBI database. C. saccharolyticum has been shown to possess saccharolytic activities (25). An association of this taxa with a poor RFI might be therefore indicative for enhanced intestinal fermentation (4) in high-RFI chickens, which may reduce the nutrient availability for the host. This may also explain the positive relationships between RFI and the fecal abundance of proteolytic Gracilibacter (26). However, the fecal bacterial community is determined by the microbiota originating from different intestinal sections (27); therefore, caution should be taken when using the microbiota composition in feces to infer the microbiota in other intestinal segments (28). It was probably the complex interplay between all these taxa together that influenced chicken intestinal homeostasis and FE. However, it cannot be ignored that the driving force behind these differences in taxa abundances may have been the higher feed intake in high-RFI chickens, thereby altering the intestinal substrate availability with consequences for host-microbiota interactions and chicken FE.
The current FMT represented the fecal microbial community of highly feed-efficient chickens, with Escherichia/Shigella and Turicibacter being the two most abundant OTU types in the FMT. These phylotypes were previously associated with improved FE in chickens (14, 17). Therefore, the present FMT may have been an appropriate inoculum to influence chicken's early microbial colonization and concurrently its FE. Although some bacterial losses due to the FMT preparation steps likely occurred and low pH in the gizzard may have further decreased bacterial numbers of particular taxa (1), weighted UniFrac distance analyses supported that the bacterial community in the feces of FMT chickens at 16 and 29 dph closely resembled the bacterial community in the FMT inoculum. Nevertheless, the current microbiota profile in the FMT was not able to strongly modify fecal bacterial abundances at 16 and 29 dph, chickens' RFI, or nutrient retention in low- and high-RFI recipient chickens. Accordingly, inoculating incubating eggs with cecal contents from highly feed-efficient donors did not transfer the dominant bacterial population from donors to the ceca of recipient birds, nor did it improve chickens' FE (12).
Considering that the FMT was prepared from feces of 30-dph chickens, it is conceivable that not all bacteria from the more mature FMT inoculum may have been able to successfully colonize the intestinal environment during the first week of life, thereby indicating a potential mismatch between donor and recipient birds. Due to the high dynamics in the reorganization of the intestinal bacterial community with time, normal maturational processes within the chicken microbiota may have therefore been more influential in shaping the host's intestinal microbial community than the FMT (12). In particular, diet is a major factor shaping the intestinal microbiota (29), and all chickens received the same starter, grower, and finisher diets. Therefore, the equal fecal abundance of the predominant bacterial taxa in the FMT, both Escherichia/Shigella and Turicibacter, between chickens of the FMT and CT groups may be explained by the fact that the diet and possibly the chickens' feed intake more strongly affected their fecal abundances in the grower-finisher period than did the administration of the FMT early in life. Moreover, since the fecal samples were first collected at 16 dph, whereas the FMT was administered only during the first few days posthatch, it is possible that the effects of the FMT on early bacterial colonization were missed, since they may have disappeared until 16 dph. This possibility may be supported by the trend of the higher abundance of the early colonizer Comamonas in FMT chickens compared to CT chickens, which was evident at 16 dph but no longer evident at 29 dph. However, we cannot draw any conclusion regarding whether the influence of the FMT on bacterial colonization may have been more apparent and more permanent in other intestinal segments. Nonetheless, the FMT application consistently decreased the fecal abundance of several cellulolytic Anaerobacterium species within the Ruminococcaceae family (30) over the two sampling time points, thus supporting that a certain effect of the FMT application on the intestinal community was measurable in feces at 2 and 4 weeks post-application.
Although sex-related differences in fecal microbiota profiles (17, 31) were small in the present study, differences in intestinal microbe-host signaling between males and females associated with the FMT may have mediated the higher TFI and TBWG values of females in the present study. The intestinal microbiota has been reported to alter the feeding behavior of the host via fermentation metabolites (e.g., short-chain fatty acids), the production of toxins, receptor recognition, and stimulation of the vagus nerve, thereby affecting the secretion of satiety-regulating hormones and controlling satiety and feed intake (32, 33).
In conclusion, we demonstrated here RFI-related differences in the fecal bacterial community in chickens at 16 and 29 dph, with mainly L. salivarius and L. crispatus OTU being indicative of good FE. However, multiple applications of a FMT within the first 9 dph only slightly modified the fecal bacterial community in recipient chickens and did not improve the RFI and nutrient retention in chickens. This indicates that other probably host- and environment-related factors were more important for chicken fecal microbiota composition at 16 and 29 dph and variation in RFI than the administration of an FMT early in chickens' lives.
MATERIALS AND METHODS
Ethical approval.
This study was conducted at the Institute of Animal Nutrition and Functional Plant Compounds (University of Veterinary Medicine, Vienna, Austria). The animal procedures were approved by the institutional ethics committee of the University of Veterinary Medicine Vienna and the Austrian national authority according to paragraph 26 of Law for Animal Experiments, Tierversuchsgesetz 2012–TVG 2012 (GZ 68.205/0131-II/3b/2013).
Animals and diets.
A total of 110-day-old Cobb 500 broiler chicks of both sexes were used in two replicate batches (batch 1, n = 54; batch 2, n = 56). One more female and one more male were used in batch 2 compared to batch 1. From 1 to 8 dph, chicks of the same sex were housed together (n = 5 to 6 chicks/cage). On 9 dph until the end of the experiment (30 dph), chickens were randomly allocated to individual cages to determine their individual feed intakes. Housing and environmental conditions were as previously described (15). Chickens were housed in stainless steel metabolic cages throughout the study, with flooring made of wire mash (10 × 10 mm) and padded with rubber tubing. A tray was put under each cage and was laid out with parchment paper to facilitate excreta collection. Each cage was equipped with one manual feeder and drinker. All chickens had ad libitum access to starter (1 to 8 dph), grower (9 to 20 dph), and finisher (21 to 30 dph) corn-soybean meal-based diets and demineralized water. The detailed dietary ingredient and chemical composition can be found in Table S5 in the supplemental material. Diets were free of antimicrobials and coccidiostats. Fresh feed was provided at 9:00 a.m. every morning, and feeders were refilled with feed at 3:00 p.m. to ensure ad libitum access to feed.
Inoculation and preparation of the FMT.
Immediately upon arrival before having access to feed and water and on 6 and 9 dph, chickens were either inoculated with 100 μl of the FMT (104 CFU) or a CT (sterile phosphate-buffered saline [PBS]). Chickens housed together received the same transplant. The transplant was orally administered at the back of the tongue and chicks were supervised to ensure that they swallowed. On 6 and 9 dph, feed was withheld for 15 min before and after the administration.
For preparation of the FMT, freshly dropped excreta from low-RFI (good-FE) chickens (females, n = 4; males, n = 2) were aseptically collected on 30 dph in a previous chicken experiment. Fecal droppings were immediately processed per bird under anaerobic conditions and were kept on ice throughout the procedure. The white portion of the excreta, mainly comprising uric acid, was removed. Twice the amount of PBS was added, and the mixture was thoroughly homogenized. To separate undigested feed and particulate material from the microbial fraction, the slurry was centrifuged at low speed (800 × g for 3 min at 4°C; centrifuge 5810 R; Eppendorf, Hamburg, Germany). To ensure microbial survival during storage (−80°C), the supernatant from each chicken was mixed with sterile glycerol (10% [vol]) and kept on ice for 60 min to allow the glycerol to penetrate the bacterial cells. The fecal suspension from each chicken was then divided into aliquots to avoid multiple thawing and freezing cycles for the single inoculations. On inoculation days, one aliquot of the fecal suspension from each low-RFI female and male chicken was thawed on ice, and equal volumes of the single suspensions were combined and homogenized to form the FMT stock.
Anaerobic and aerobic culturing and quantitative PCR (qPCR) was used to estimate the bacterial numbers in the prepared FMT stock before the start of the chicken experiment. Analysis by qPCR was also used to verify the administered bacterial gene copies on each inoculation day. For aerobic and anaerobic cultivation, a 1:10 dilution series of the FMT stock using Ringer solution (Fresenius Kabi, Graz, Austria) was prepared and plated onto tryptic soy agar plates. Plates were either aerobically or anaerobically incubated in an anaerobic jar (Oxoid, Wesel, Germany) containing one sachet of anaerobic atmosphere generator (bioMérieux, Marcy l'Etoile, France) for 48 h at 37°C. The DNA extraction and qPCR amplification protocol are described below. The FMT stock contained 7.28 × 107 CFU of culturable aerobic and anaerobic bacteria and 8.4 log10 total bacterial 16S rRNA gene copies per milliliter. For administration, a 1:100 dilution of the FMT stock was prepared using PBS. All female and male chickens in the FMT group were inoculated with the same FMT dilution.
Determination of FE.
For the calculation of chickens' TFI, the individual feed intake of each chicken was determined weekly. For this, the amount of feed provided and feed refusals and spills were recorded. Feed refusals were collected before feeding at 9:00 a.m. daily, and spills were collected before recording feed intake on 9, 14, 21, 28, and 30 dph. Body weight was measured upon arrival and at 6, 9, 14, 21, 28, and 30 dph. The RFI was determined for the experimental period from 9 to 30 dph. A nonlinear mixed model (SAS/STAT, version 9.4; SAS, Inc., Cary, NC) based on data for TFI, metabolic mid-test body weight, and TBWG from 9 to 30 dph was used to estimate each chicken's RFI as the residuals over the test period (15). Regression analysis was performed for each batch individually. In order to investigate whether the FMT could improve the RFI of poorly feed-efficient (high-RFI) chickens without impairing the RFI of highly feed-efficient (low-RFI) chickens, chickens with the most extreme RFI values were selected. A total of 15 low-RFI (females, n = 8; males, n = 7) and 13 high-RFI (females, n = 7; males, n = 6) chickens receiving the FMT and 14 low-RFI (n = 7/sex) and 14 high-RFI (n = 7/sex) chickens receiving the CT were selected. Only fecal samples from these selected chickens were analyzed for nutrient content and microbiota composition.
Sample collection.
The gastrointestinal origin of the chicken feces determines the fecal bacterial composition (27). Therefore, for the microbiota analysis, freshly dropped excreta of paste-like texture without the uric acid-containing white part were predominantly collected on 16 and 29 dph. Within 5 to 10 min after defecation, feces were aseptically collected, placed into sterile 2-ml cryotubes (Sarstedt, Nümbrecht, Germany), snap-frozen in liquid N2, and stored at −80°C until DNA extraction. Moreover, water and diet samples (n = 2 per starter, grower, and finisher diet) were collected for microbial analysis. At 28 dph, freshly dropped excreta samples were collected and stored at −20°C until analysis for NH3 and pH. To determine fecal DM concentration and retention of nutrients, excreta were collected at 29 and 30 dph and stored at −20°C.
DNA extraction.
Total DNA was extracted from 300 μl of the prepared FMT stocks, 250 mg of fecal and water samples, and 150 mg of diet samples using a PowerSoil DNA isolation kit (MoBio Laboratories, Inc., Carlsbad, CA) as described previously (34). A Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA) with a Qubit dsDNA HS assay kit (Life Technologies) was used to quantify the DNA concentration.
16S rRNA sequencing and bioinformatic analysis.
An aliquot of each of the extracted DNA samples (fecal samples, n = 112; FMT inoculum, n = 8; water samples, n = 2; diet samples, n = 6) was sent for PCR amplification of the 16S rRNA gene, library preparation, and DNA sequencing to a commercial provider (Microsynth AG, Balgach, Switzerland). Primers 357F-HMP (CCT ACG GGA GGC AGC AG) and 926R-HMP (CCG TCA ATT CMT TTR AGT) targeting the V3-5 region of the 16S rRNA gene were used for amplification to generate an approximate amplicon size of 570 bp (35). A Nextera XT sample preparation kit (Illumina, Inc., San Diego, CA) was used according to the manufacturer's recommendations. For each library, equimolar amounts were pooled and sequenced on an Illumina MiSeq personal sequencer using a 300-bp read length paired-end protocol. The resultant overlapping paired-end reads were stitched and quality filtered by Microsynth.
The prefiltered and stitched reads were processed using the Quantitative Insights Into Microbial Ecology (QIIME) package (v1.9.1) (36). Fastq files were quality trimmed using the “multiple_split_libraries_fastq” script for demultiplexed Illumina fastq data using a quality threshold of q < 15. Chimeric sequences were removed with the UCHIME method using the 64-bit version of USEARCH (37, 38) and the GOLD database (drive5). Sequences were clustered into OTU types (97% similarity) using open-reference OTU picking and UCLUST (37). Taxonomy was assigned against the 13_8 Greengenes default database in QIIME (v1.9.1; http://qiime.org/home_static/dataFiles.html) (39). OTU with fewer than 10 sequences were removed. The most abundant OTU in the FMT, as well as OTU differently affected by time point, FMT, and RFI, were additionally classified against the National Center for Biotechnology Information (NCBI) nucleotide database using BLASTN for taxonomic classification and the database limited to the 16S rRNA target (https://blast.ncbi.nlm.nih.gov/). A rarefaction depth of 10,000 sequences was used for α- and β-diversity analyses of diet, fecal, and FMT samples, thereby excluding one fecal sample with fewer reads (low RFI, male, CT, 16 dph). The β-diversity was determined using the unweighted and weighted UniFrac distance (40, 41). In addition, rarefaction curves for all diet, fecal, and FMT samples were calculated using a maximum rarefaction depth of 10,000 sequences and the observed OTU index.
qPCR.
The DNA concentrations of the FMT stocks were adjusted. qPCR was performed on a Stratagene Mx3000P qPCR system (Agilent Technologies, Santa Clara, CA) in 20-μl reaction volume using 10 μl of the Fast-Plus EvaGreen master mix with Low ROX (Biotium, Hayward, CA), the forward and reverse primers 341-357F and 518-534R (62.5 nmol of each), and 0.3 ng DNA extract as previously described (34). The amplification specificity was determined by melting curve analysis. Standard curves were generated using 10-fold serial dilutions (107 to 103 molecules/μl) of the purified and quantified 16S rRNA gene PCR product generated by standard PCR (PCR efficiencies of 95 to 102% [R2 = 0.999]) (39).
Chemical analyses.
To determine NH3 in excreta, the indophenol method was used (42). The pH in fresh excreta was measured in a 1:9 (vol/vol) dilution, and the DM content was determined by oven drying at 105°C overnight (43). Prior to proximate analysis (DM, crude protein [protein], crude ash [ash], and phosphorus) as described previously (15), total excreta samples were pooled per chicken, freeze-dried, and ground through a 0.5-mm-pore-size screen. Acid-insoluble ash, analyzed in feed and feces (43), was used as an inert marker to calculate nutrient retention.
Statistical analyses.
Descriptive statistics on the bacterial composition of the FMT inoculum, diet, and water samples at the phylum and OTU level were assessed using the MEANS procedure in SAS. To test for normality, the FE, performance traits, excreta parameters, nutrient retention, and microbiota data were first analyzed using a Shapiro-Wilk test with the UNIVARIATE procedure in SAS. After we established the normal distribution of our parameters, we evaluated the data by analysis of variance using the MIXED procedure in SAS. To analyze FE, performance traits, excreta parameters, and nutrient retention data, the fixed effects of batch, sex, FMT, RFI, and the two-way-interaction FMT×RFI were considered in the main model. Because batch affected some of the performance traits, batch was considered a random effect in the final model. Chicken was the experimental unit. Since sex was significant for the FE and performance data, a second model for these parameters was adjusted, and data were additionally separately analyzed for females and males. The second model included the fixed effects of FMT, RFI, and the two-way-interaction FMT×RFI. For the microbiota data, fixed effects also included the time point of excreta collection and the three-way-interaction time point×FMT×RFI. Measurements made for the same chicken at different time points were considered repeated measures in the model. The experimental unit was chicken nested within batch. The degrees of freedom were approximated by the Kenward-Roger method. Least-squares means were computed using the pdiff statement. A P value of ≤0.05 was considered significant, whereas a P value between 0.05 and 0.10 was considered a trend. Bacterial families, genera, and OTU types comprising a relative abundance of >0.05% across both sampling time points and sexes were statistically analyzed.
Pearson's correlation analysis (CORR procedure of SAS) was used to establish and quantify the relationships between fecal abundances of OTU types at 29 dph and individual RFI, TFI, TBWG, fecal pH, fecal DM, and fecal NH3 values. Correlations were visualized using the R packages corrplot and RColorBrewer (v3.4).
Accession number(s).
Raw sequencing data are available in the NCBI BioProject SRA database under accession no. PRJNA392215.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Sener, M. Hollmann, A. Dockner, M. Wild, S. Eisen, S. Koger, and S. Leiner for assistance with laboratory analysis and the animal experiment.
This project (ECO-FCE) received funding from the European Union's Seventh Framework Programme for Research, Technological Development, and Demonstration under grant 311794.
B.M.Z., E.M., P.L., and N.O. conceived and designed the study. S.C.S. and B.M.Z. conducted the animal study and, together with R.P., collected samples. S.C.S. and B.M.Z. performed laboratory analyses. S.C.S., B.M.Z., and R.P. performed bioinformatic analyses. Q.Z. provided resources. B.M.Z. wrote the codes for statistical analysis, and S.C.S. statistically analyzed all data. S.C.S. and B.M.Z. collated and interpreted the data and wrote and edited the manuscript. R.P., P.L., and Q.Z. revised the manuscript. All authors read and approved the final version of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02330-17.
REFERENCES
- 1.Ranjitkar S, Lawley B, Tannock G, Engberg RM. 2016. Bacterial succession in the broiler gastrointestinal tract. Appl Environ Microbiol 82:2399–2410. doi: 10.1128/AEM.02549-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mancabelli L, Ferrario C, Milani C, Mangifesta M, Turroni F, Duranti S, Lugli GA, Viappiani A, Ossiprandi MC, van Sinderen D, Ventura M. 2016. Insights into the biodiversity of the gut microbiota of broiler chickens. Environ Microbiol 18:4727–4738. doi: 10.1111/1462-2920.13363. [DOI] [PubMed] [Google Scholar]
- 3.Stanley D, Geier MS, Chen H, Hughes RJ, Moore RJ. 2015. Comparison of fecal and cecal microbiotas reveals qualitative similarities but quantitative differences. BMC Microbiol 15:51. doi: 10.1186/s12866-015-0388-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rinttilä T, Apajalahti J. 2013. Intestinal microbiota and metabolites: implications for broiler chicken health and performance. J Appl Poult Res 22:647–658. doi: 10.3382/japr.2013-00742. [DOI] [Google Scholar]
- 5.Schokker D, Veninga G, Vastenhouw SA, Bossers A, de Bree FM, Kaal-Lansbergen LM, Rebel JM, Smits MA. 2015. Early life microbial colonization of the gut and intestinal development differ between genetically divergent broiler lines. BMC Genomics 16:418. doi: 10.1186/s12864-015-1646-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh KM, Shah TM, Reddy B, Deshpande S, Rank DN, Joshi CG. 2014. Taxonomic and gene centric metagenomics of the fecal microbiome of low and high feed conversion ratio (FCR) broilers. J Appl Genet 55:145–154. doi: 10.1007/s13353-013-0179-4. [DOI] [PubMed] [Google Scholar]
- 7.Stanley D, Geier MS, Denman SE, Haring VR, Crowley TM, Hughes RJ, Moore RJ. 2013. Identification of chicken intestinal microbiota correlated with the efficiency of energy extraction from feed. Vet Microbiol 164:85–92. doi: 10.1016/j.vetmic.2013.01.030. [DOI] [PubMed] [Google Scholar]
- 8.Stanley D, Hughes RJ, Geier MS, Moore RJ. 2016. Bacteria within the gastrointestinal tract microbiota correlated with improved growth and feed conversion: challenges presented for the identification of performance enhancing probiotic bacteria. Front Microbiol 7:187. doi: 10.3389/fmicb.2016.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta S, Allen-Vercoe E, Petrof EO. 2016. Fecal microbiota transplantation: in perspective. Ther Adv Gastroenterol 9:229–239. doi: 10.1177/1756283X15607414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelly CR. 2015. Update on fecal microbiota transplantation 2015: indications, methodologies, mechanisms, and outlook. Gastroenterology 149:223–237. doi: 10.1053/j.gastro.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nurmi E, Rantala M. 1973. New aspects of Salmonella infection in broiler production. Nature 241:210–211. doi: 10.1038/241210a0. [DOI] [PubMed] [Google Scholar]
- 12.Donaldson EE, Stanley D, Hughes RJ, Moore RJ. 2017. The time-course of broiler intestinal microbiota development after administration of cecal contents to incubating eggs. PeerJ 5:e3587. doi: 10.7717/peerj.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, Muehlbauer MJ, Ilkayeva O, Semenkovich CF, Funai K, Hayashi DK, Lyle BJ, Martini MC, Ursell LK, Clemente JC, Van Treuren W, Walters WA, Knight R, Newgard CB, Heath AC, Gordon JI. 2013. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh KM, Shah T, Deshpande S, Jakhesara SJ, Koringa PG, Rank DN, Joshi CG. 2012. High-throughput 16S rRNA gene-based pyrosequencing analysis of the fecal microbiota of high FCR and low FCR broiler growers. Mol Biol Rep 39:10595–10602. doi: 10.1007/s11033-012-1947-7. [DOI] [PubMed] [Google Scholar]
- 15.Metzler-Zebeli BU, Molnár A, Hollmann M, Magowan E, Hawken RJ, Lawlor PG, Zebeli Q. 2016. Comparison of growth performance and excreta composition in broiler chickens when ranked according to various feed efficiency metrics. J Anim Sci 94:2890–2899. doi: 10.2527/jas.2016-0375. [DOI] [PubMed] [Google Scholar]
- 16.Bottje WG, Carstens GE. 2009. Association of mitochondrial function and feed efficiency in poultry and livestock species. J Anim Sci 87:E48–E63. doi: 10.2527/jas.2008-1379. [DOI] [PubMed] [Google Scholar]
- 17.Siegerstetter S-C, Petri RM, Magowan E, Zebeli Q, Lawlor PG, Metzler-Zebeli BU. 2016. Feed efficiency related gut microbiota profiles vary in chickens raised at two locations, p 200. In Abstr 67th Annu Meet Eur Fed Anim Sci. Wageningen Academic Publishers, Wageningen, The Netherlands. [Google Scholar]
- 18.Oakley BB, Kogut MH. 2016. Spatial and temporal changes in the broiler chicken cecal and fecal microbiomes and correlations of bacterial taxa with cytokine gene expression. Front Vet Sci 3:11. doi: 10.3389/fvets.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oakley BB, Buhr RJ, Ritz CW, Kiepper BH, Berrang ME, Seal BS, Cox NA. 2014. Successional changes in the chicken cecal microbiome during 42 days of growth are independent of organic acid feed additives. BMC Vet Res 10:282. doi: 10.1186/s12917-014-0282-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nouri M, Rahbarizadeh F, Ahmadvand D, Moosakhani F, Sadeqzadeh E, Lavasani S, Vishteh VK. 2010. Inhibitory effects of Lactobacillus salivarius and Lactobacillus crispatus isolated from chicken gastrointestinal tract on Salmonella enteritidis and Escherichia coli growth. Iran J Biotech 8:32–37. [Google Scholar]
- 21.Brisbin JT, Davidge L, Roshdieh A, Sharif S. 2015. Characterization of the effects of three Lactobacillus species on the function of chicken macrophages. Res Vet Sci 100:39–44. doi: 10.1016/j.rvsc.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Meimandipour A, Shuhaimi M, Hair-Bejo M, Azhar K, Kabeir BM, Rasti B, Yazid AM. 2009. In vitro fermentation of broiler cecal content: the role of lactobacilli and pH value on the composition of microbiota and end products fermentation. Lett Appl Microbiol 49:415–420. doi: 10.1111/j.1472-765X.2009.02674.x. [DOI] [PubMed] [Google Scholar]
- 23.Crisol-Martínez E, Stanley D, Geier MS, Hughes RJ, Moore RJ. 2017. Sorghum and wheat differentially affect caecal microbiota and associated performance characteristics of meat chickens. PeerJ 5:e3071. doi: 10.7717/peerj.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konsak BM, Stanley D, Haring VR, Geier MS, Hughes RJ, Howarth GS, Crowley TM, Moore RJ. 2013. Identification of differential duodenal gene expression levels and microbiota abundance correlated with differences in energy utilisation in chickens. Anim Prod Sci 53:1269–1275. doi: 10.1071/AN12426. [DOI] [Google Scholar]
- 25.Murray WD, Khan AW, van den Berg L. 1982. Clostridium saccharolyticum sp. nov., a saccharolytic species from sewage sludge. Int J Syst Evol Microbiol 32:132–135. doi: 10.1099/00207713-32-1-132. [DOI] [Google Scholar]
- 26.Ma H, Liu H, Zhang L, Yang M, Fu B, Liu H. 2017. Novel insight into the relationship between organic substrate composition and volatile fatty acids distribution in acidogenic cofermentation. Biotechnol Biofuels 10:137. doi: 10.1186/s13068-017-0821-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sekelja M, Rud I, Knutsen SH, Denstadli V, Westereng B, N[ligae]s T, Rudi K. 2012. Abrupt temporal fluctuations in the chicken fecal microbiota are explained by its gastrointestinal origin. Appl Environ Microbiol 78:2941–2948. doi: 10.1128/AEM.05391-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan W, Sun C, Yuan J, Yang N. 2017. Gut metagenomic analysis reveals prominent roles of Lactobacillus and cecal microbiota in chicken feed efficiency. Sci Rep 7:45308. doi: 10.1038/srep45308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan D, Yu Z. 2014. Intestinal microbiome of poultry and its interaction with host and diet. Gut Microbes 5:108–119. doi: 10.4161/gmic.26945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horino H, Fujita T, Tonouchi A. 2014. Description of Anaerobacterium chartisolvens gen. nov., sp. nov., an obligately anaerobic bacterium from Clostridium rRNA cluster III isolated from soil of a Japanese rice field, and reclassification of Bacteroides cellulosolvens Murray et al. 1984 as Pseudobacteroides cellulosolvens gen. nov., comb. nov. Int J Syst Evol Microbiol 64:1296–1303. doi: 10.1099/ijs.0.059378-0. [DOI] [PubMed] [Google Scholar]
- 31.Zhao L, Wang G, Siegel P, He C, Wang H, Zhao W, Zhai Z, Tian F, Zhao J, Zhang H, Sun Z, Chen W, Zhang Y, Meng H. 2013. Quantitative genetic background of the host influences gut microbiomes in chickens. Sci Rep 3:1163. doi: 10.1038/srep01163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alcock J, Maley CC, Aktipis CA. 2014. Is eating behavior manipulated by the gastrointestinal microbiota? Evolutionary pressures and potential mechanisms. Bioessays 36:940–949. doi: 10.1002/bies.201400071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaiko GE, Stappenbeck TS. 2014. Host-microbe interactions shaping the gastrointestinal environment. Trends Immunol 35:538–548. doi: 10.1016/j.it.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Metzler-Zebeli BU, Lawlor PG, Magowan E, Zebeli Q. 2016. Effect of freezing conditions on fecal bacterial composition in pigs. Animals (Basel) 6:18. doi: 10.3390/ani6030018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, Baker CC, Di Francesco V, Howcroft TK, Karp RW, Lunsford RD, Wellington CR, Belachew T, Wright M, Giblin C, David H, Mills M, Salomon R, Mullins C, Akolkar B, Begg L, Davis C, Grandison L, Humble M, Khalsa J, Little AR, Peavy H, Pontzer C, Portnoy M, Sayre MH, Starke-Reed P, Zakhari S, Read J, Watson B, Guyer M. 2009. The NIH human microbiome project. Genome Res 19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 38.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. 2001. UniFrac: an effective distance metric for microbial community comparison. ISME J 5:169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lozupone CA, Hamady M, Kelley ST, Knight R. 2007. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weatherburn MW. 1967. Phenol-hypochlorite reaction for determination of ammonia. Anal Chem 39:971–974. doi: 10.1021/ac60252a045. [DOI] [Google Scholar]
- 43.Naumann C, Basler R. 2012. Die Chemische Untersuchung von Futtermitteln, 3rd ed VDLUFA Verlag, Darmstadt, Germany. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.