ABSTRACT
Sucrose and glycogen syntheses in cyanobacteria share the common precursor glucose-1-phosphate. It is generally assumed that lowering glycogen synthesis could drive more carbon toward sucrose synthesis that can be induced by salt stress among cyanobacteria. By using a theophylline-dependent riboswitch system, the expression of glgC, a key gene in glycogen synthesis, was downregulated in a quantitative manner in a sucrose-secreting strain of Synechococcus elongatus PCC 7942. We observed that the stepwise suppression of glycogen synthesis limited rather than stimulated sucrose production in the salt-stressed cells, suggesting that glycogen could serve as a carbon pool for the synthesis of sucrose. Accordingly, we generated glycogen-overproducing strains, but the increased glycogen pool alone did not stimulate sucrose production, indicating that alternative steps limit the carbon flux toward the synthesis of sucrose. Consistent with previous studies that showed that sucrose-phosphate synthase (SPS) catalyzes the rate-limiting step in sucrose synthesis, the combination of glycogen overproduction and sps overexpression resulted in increased sucrose production. Our results indicate that the glycogen and sucrose pools are closely linked in Synechococcus elongatus PCC 7942, and we propose that enhancing the glycogen pool could be a promising strategy for the improvement of sucrose production by cyanobacteria in the presence of a strong sucrose synthesis sink.
IMPORTANCE Many cyanobacteria naturally synthesize and accumulate sucrose when stressed by NaCl, which provides novel possibilities for obtaining sugar feedstock by engineering of cyanobacteria. It has been assumed that glycogen synthesis competes with sucrose synthesis for the carbon flux. However, our results showed that the suppression of glycogen synthesis decreased rather than stimulated sucrose production in a sucrose-secreting strain of Synechococcus elongatus PCC 7942. This result suggests that glycogen could serve as a supportive rather than a competitive carbon pool for the synthesis of sucrose, providing new insights about the relation between glycogen synthesis and sucrose synthesis in cyanobacteria. This finding is also useful to guide metabolic engineering work to optimize the production of sucrose and possibly other products by cyanobacteria.
KEYWORDS: sucrose production, glycogen, riboswitch, Synechococcus elongatus PCC 7942
INTRODUCTION
Microorganisms are increasingly being utilized for the commercial production of a variety of products, such as fine chemicals (1, 2), health products (3, 4), pharmaceuticals (5, 6), and biofuels (7, 8). An important factor affecting the product price is the cost of the sugar feedstock (9, 10). Agricultural crops such as sugarcane, sugar beet, or corn are the main sources for sugar feedstock, and the utilization of cellulosic biomass containing abundant units of glucose is also intensively investigated (11–13). However, due to the competition between food production and sugar feedstock production from agricultural crops and the current high costs of the pretreatment and conversion processes of utilizing cellulosic biomass (14), novel sustainable and economical sources for sugar feedstock and related conversion technologies should be explored.
Cyanobacteria have been attracting increasing research interest for the production of biofuels and biochemicals in recent years. These prokaryotes are capable of converting atmosphere CO2 and light energy to carbohydrates by oxygenic photosynthesis. They show higher photosynthetic efficiency per area than plants and can be easily manipulated for desired product syntheses by genetic and metabolic engineering (15–18). Many freshwater or marine species of cyanobacteria naturally synthesize and accumulate large internal amounts of sucrose as the compatible solute when stressed by NaCl. Alternatively or additionally, other small molecules such as trehalose, glucosylglycerol, glucosylglycerate, glycine betaine, or homoserine betaine can also act as compatible solutes to protect cyanobacterial cells from osmotic stress (19, 20). The main pathway for sucrose synthesis (Fig. 1) starts from fructose-6-phosphate and UDP-glucose and is catalyzed by sucrose-phosphate synthase (SPS) and sucrose-phosphate phosphatase (SPP) (19). Certain cyanobacterial species such as Synechococcus elongatus PCC 7942 (here referred to as Syn7942) express fusion enzymes that catalyze the SPS and SPP reactions by a single protein (21). In recent years, many studies that aim to improve the sucrose-producing capability by metabolic engineering methods have been performed with Synechocystis sp. strain PCC 6803 (here Syn6803) (22), Syn7942 (23, 24), and Synechococcus elongatus UTEX 2973 (here Syn2973) (25). Among these strains, Syn7942 has attracted the most research interest. Due to the limited intracellular space and physiological tolerance, one can hypothesize that the continuous export of sucrose could improve sucrose production. Ducat and colleagues generated the first sucrose-secreting Syn7942 (strain CscB in Table 1) by expressing the Escherichia coli gene cscB, which encodes a proton/sucrose symporter (23). This strain efficiently synthesized and secreted sucrose. Up to ∼80% of the fixed CO2 was channeled into sucrose. Recently, we also generated a cscB-expressing Syn7942 strain (named FL92) and showed that the sucrose productivity was 10-fold higher than that of the wild-type strain (24). Furthermore, we overexpressed the native sps gene in combination with cscB, which almost doubled the sucrose production capability of the resulting strain under salt stress (24). However, the further improvement of sucrose production becomes more difficult. This prompted us to study the metabolic relation between sucrose synthesis and processes such as glycogen synthesis that could possibly interfere or compete with sucrose synthesis.
FIG 1.
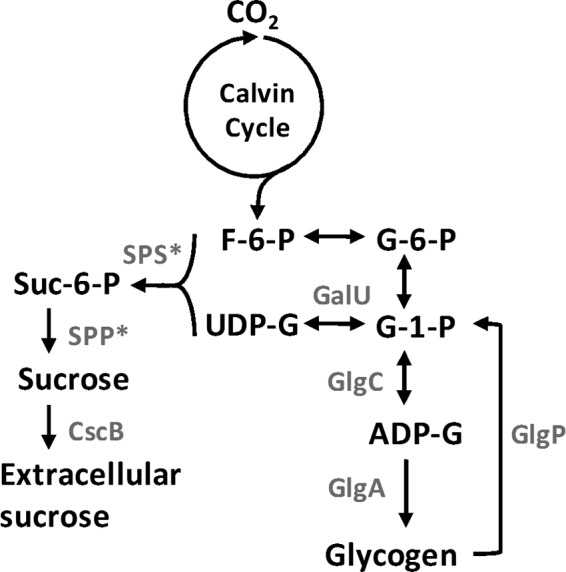
Schematic representation of sucrose and glycogen synthesis in Syn7942. F-6-P, fructose-6-phosphate; G-6-P, glucose-6-phosphate; G-1-P, glucose-1-phosphate; ADP/UDP-G, ADP/UDP-glucose; Suc-6-P, sucrose-6-phosphate; SPS, sucrose-phosphate synthase; SPP, sucrose-phosphate phosphatase; GalU, UDP-glucose pyrophosphorylase; GlgC, glucose-1-phosphate adenylyltransferase; GlgA, glycogen synthase; GlgP, glycogen phosphorylase; CscB, proton/sucrose exporter. *, note that in the genome of Syn7942, sps encodes a combined SPS/SPP fusion protein.
TABLE 1.
Comparison of sucrose production rates in Synechococcus elongatus determined in the present study with published values
| Strain | Genotypea | Cultivation conditions | Titer (mg/liter) | Production rate (mg/liter/h) | Reference |
|---|---|---|---|---|---|
| Syn7942 strains | |||||
| FL92 | NS3::Plac-cscB-Cmr | 30°C, 3% CO2, 100–120 μE/m2/s light, 150 mM NaCl for 3 days | 469.3 | 6.5 | This study |
| FL130 | NS1::Ptrc-sps-Spr NS3::Plac-cscB-Cmr | 30°C, 3% CO2, 100–120 μE/m2/s light, 150 mM NaCl for 3 days | 539.8 | 7.5 | This study |
| QL241 | NS1::Ptrc-sps-Spr NS2::PcpcB1-glgC-Gmr NS3::Plac-cscB-Cmr | 30°C, 3% CO2, 100–120 μE/m2/s light, 150 mM NaCl for 3 days | 577.8 | 8.0 | This study |
| CscB | NS3::Plac-cscB-Cmr | 35°C, 2% CO2, 65 μE/m2/s light, 150 mM NaCl for 3 days | ∼2,016 | ∼28 | 23 |
| CscBΔinvA | NS3::Plac-cscB-Cmr invA::Hygr | 35°C, 2% CO2, 65 μE/m2/s light, 150 mM NaCl for 3 days | ∼2,304 | ∼32 | 23 |
| CscBΔglgC | NS3::Plac-cscB-Cmr glgC::Spr | 35°C, 2% CO2, 65 μE/m2/s light, 150 mM NaCl for 3 days | ∼2,160 | ∼30 | 23 |
| CscBΔglgCΔinvA | NS3::Plac-cscB-Cmr invA::Hygr glgC::Spr | 35°C, 2% CO2, 65 μE/m2/s light, 150 mM NaCl for 3 days | 2,599.2 | 36.1 | 23 |
| Syn2973 strain | |||||
| Syn2973::CscB | M744_RS12430::Plac-cscB-Cmr | 38°C, 3% CO2, 250 μE/m2/s light, 150 mM NaCl for 4 days | 3,340 | 35.5 | 25 |
Plac, lac promoter; Ptrc, trc promoter; PcpcB1, cpcB1 promoter; Hygr, hygromycin resistance; invA, invertase gene of Syn7942; NS1, NS2, and NS3, different neutral sites in the genome of Syn7942.
Glycogen serves as the main carbon sink and energy storage in cyanobacterial cells. It is synthesized in the light and consumed during darkness to provide maintenance energy. However, glycogen can also serve as a reserve of excess carbon under imbalanced nutrient conditions (26). Moreover, the crucial functions of glycogen for cyanobacterial acclimation to unfavorable environments such as nutrient starvation (26–29) and salt and oxidative stress (30) were also reported. Glycogen synthesis is catalyzed by glucose-1-phosphate adenylyltransferase (GlgC) providing ADP-glucose, which is then used by glycogen synthase (GlgA). It is generally assumed that glycogen accumulation represents a major competing pathway for organic carbon with the synthesis of biofuels or feedstock. Sucrose and glycogen biosyntheses both depend on the precursor glucose-1-phosphate (Fig. 1). It has been reported that inhibiting glycogen synthesis improved accumulation of the compatible solutes sucrose and/or glucosylglycerol in Syn6803 and Synechococcus sp. strain PCC 7002 (31–33). However, the effects of similar attempts in Syn7942 differed from case to case (23, 30).
In the present study, we generated Syn7942 strains with reduced and enhanced glycogen production on the background of the cscB-expressing strain FL92 (24). These strains were systematically analyzed to determine the impact of glycogen accumulation on salt-induced sucrose production. Our results indicated that the glycogen and sucrose pools are closely linked in Syn7942, and new insights about the relation between glycogen and sucrose syntheses in cyanobacteria were provided.
RESULTS AND DISCUSSION
Stepwise suppression of glgC expression in sucrose-secreting Syn7942.
Recently, a synthetic theophylline-responsive riboswitch system was developed as a novel genetic tool for the strict regulation of gene expression in Syn7942 (34). It is particularly useful for investigating gene functions in a quantitative manner. To determine the effect of glycogen deficiency on sucrose production, the riboswitch strategy was employed with our cscB-expressing strain FL92 of Syn7942 (24) for a stepwise downregulation of glgC expression. To achieve this goal, we constructed the Syn7942 strain DY126, in which the native glgC gene was placed under the control of the theophylline-dependent riboswitch.
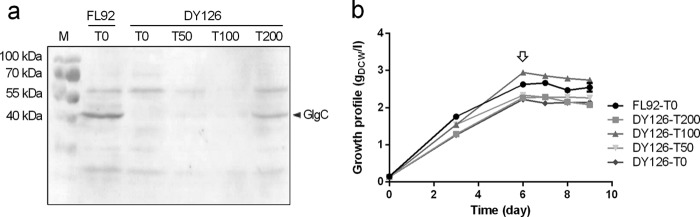
To evaluate the efficiency of the riboswitch, concentrations of theophylline ranging from 0 to 200 μM were applied to DY126 cultures and the expression of glgC was examined by immunoblotting (Fig. 2a). In the medium containing the highest concentration of 200 μM theophylline, DY126 cells accumulated the greatest amount of GlgC. When theophylline concentrations were decreased to 100 and 50 μM, the amounts of GlgC decreased to trace levels. In the absence of theophylline, GlgC was not detectable at all. Thus, glgC expression was successfully stepwise downregulated by theophylline in the medium. During these experiments, we noticed that the control strain FL92 grown in theophylline-free medium expressed noticeably larger amounts of GlgC than DY126 cells grown in medium containing 200 μM theophylline (Fig. 2a, lanes 2 and 6). This finding indicates that the native glgC promoter drives the glgC expression in FL92 more efficiently than the theophylline-dependent riboswitch even if it is switched on in the presence of 200 μM theophylline.
FIG 2.
Analysis of amounts of GlgC in DY126 and the control strain FL92 of Syn7942 (a) and growth profiles of DY126 and FL92 grown in medium containing the indicated concentrations of theophylline (b). Immunoblotting analysis of accumulated GlgC was performed with a specific GlgC antibody in DY126 and FL92. Cyanobacterial cells were grown in BG11 medium containing different concentrations of theophylline for 3 days. T0, T50, T100, and T200 represent 0, 50, 100, and 200 μM theophylline, respectively, supplemented in the medium. Lane M, protein size marker. The growth data are presented as means from three independent replicates. The open arrow indicates the time when 150 mM NaCl was applied.
Compared to the markedly changed level of GlgC, the growth of DY126 was only slightly affected by the suppression of glgC expression compared to that of the control (Fig. 2b). When cultivated in NaCl-free medium containing 0 to 200 μM theophylline, DY126 showed exponential growth like FL92 and reached cell densities in the late exponential phase (day 6) that were similar to those of the control. After adding 150 mM NaCl, the fast growth of DY126 under all theophylline conditions ceased, like that of FL92. The cell densities of DY126 and FL92 remained almost constant during the following 3 days (Fig. 2b).
Glycogen deficiency limits rather than enhances sucrose production.
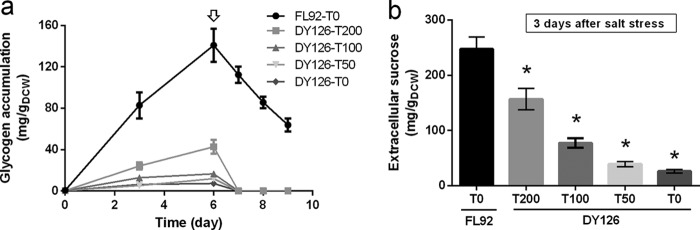
DY126 cells cultivated in medium containing increasing concentrations of theophylline were then examined for their levels of glycogen accumulation and salt-induced sucrose production. As expected, the glycogen contents in Syn7942 cells exhibited a strong correlation with the intracellular GlgC amounts. Under NaCl-free conditions, strain FL92 exhibited the fastest glycogen accumulation and accumulated the highest final cellular glycogen content (140.9 mg/g dry cell weight [gDCW] at day 6) (Fig. 3a). Consistent with the lower GlgC accumulation, the cells of DY126 grown in the presence of 200 μM theophylline accumulated much less glycogen (42.7 mg/gDCW at day 6), which was only 30% of the FL92 level. Further decreased theophylline concentrations led to decreased glycogen accumulation in the cells of DY126 (16.6, 12.0, and 7.2 mg glycogen/gDCW in the presence of theophylline at 100, 50, and 0 μM, respectively, at day 6).
FIG 3.
Effect of glycogen deprivation on sucrose accumulation in strains of Syn7942. Glycogen accumulation (a) and sucrose production (b) by DY126 and FL92 after cultivation in medium containing the indicated concentrations of theophylline are displayed. NaCl (150 mM) and IPTG (1 mM) were added to the medium at day 6, as indicated by the open arrow. T0, T50, T100, and T200 represent 0, 50, 100, and 200 μM theophylline, respectively, supplemented in the medium. Asterisks indicate significant differences from FL92 (Student's t test, P < 0.05).
To determine the effect of decreased glycogen levels on sucrose production, the cells of FL92 and the theophylline-supplemented cells of DY126 were exposed to salt stress with 150 mM NaCl. Contrary to our predictions, the suppression of glycogen accumulation in DY126 decreased rather than enhanced sucrose production (Fig. 3a and b). After 3 days of salt stress, the control strain FL92, which accumulated the largest amount of glycogen, showed the highest level of sucrose in the medium (247.7 mg/gDCW). The extracellular sucrose contents of DY126 were 156.9, 77.1, 39.0, and 26.0 mg/gDCW in the presence of theophylline at 200, 100, 50, and 0 μM, respectively (Fig. 3b). The relationship between the produced sucrose after salt stress and the accumulated glycogen before salt stress was evaluated by Spearman rank order correlation analysis. The result supported that the decrease of sucrose production in DY126 was positively correlated with the reduction of glycogen levels before salt stress (Spearman rank correlation, rs = 0.874 [P < 0.01]). These results indicate a close link between the glycogen pool and compatible solute synthesis in Syn7942 and strongly suggest that the intracellular glycogen serves, at least partially, as a carbon pool supporting sucrose synthesis rather than competing with sucrose synthesis. Similarly, a close link between the turnover of the compatible solute glucosylglycerol and the glycogen pool has been recently shown in salt-stressed cells of Synechococcus sp. strain PCC 7002 (35). Our experiment to examine the level of glycogen after salt stress found that the intracellular glycogen decreased to undetectable levels in all of the DY126 cells after 1 day of salt stress (Fig. 3a), thus indicating that the glycogen consumption was faster than the synthesis. In contrast, the salt-stressed cells of the control strain FL92 maintained significant levels of glycogen even after 3 days of salt stress, which could eventually support sucrose production. Ducat and colleagues also investigated the effect of knocking out glgC on sucrose production in cscB-expressing Syn7942 (23). Their resulting strain exhibited marginally increased sucrose secretion, which is different from the phenotype observed in our strain DY126. The observed phenotypic differences may be due to the different strategies to obtain glgC mutants. In the study by Ducat et al., the glgC deletion completely abolished glycogen synthesis; therefore, sucrose synthesis could be, at least partially, stimulated by organic carbon derived from the de novo assimilation of CO2 via photosynthesis, which was shown to be enhanced (23). In contrast, we aimed to obtain a stepwise-depressed glycogen synthesis. Even in the absence of theophylline, low levels of glycogen still accumulated in DY126 (see above), which might be caused by the leaky expression of glgC. Thus, glycogen could still interfere with sucrose synthesis, for example, directly as a carbon source for sucrose production or indirectly via regulatory impact on carbon metabolism.
Glycogen excess alone does not improve sucrose production.
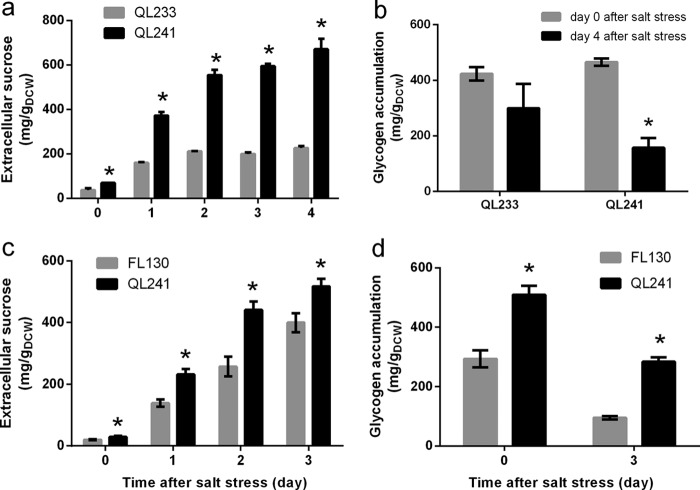
In the experiments described above, it was found that the insufficient glycogen supply limited sucrose production in salt-stressed cells of DY126. Therefore, we assumed that an excess of glycogen might stimulate sucrose production. To prove this hypothesis, we constructed the Syn7942 strain QL233, in which a second copy of glgC was expressed under the control of the strong cpcB1 (encoding c-phycocyanin β subunit) promoter on the background of FL92. Strain QL233 showed growth similar to that of FL92 before and after salt stress (Fig. 4a). Consistent with the enhanced expression of glgC in QL233, this strain accumulated approximately 50% more glycogen than FL92 under control conditions and salt stress (Fig. 4b). However, enhanced sucrose production was not observed in QL233. The amount of extracellular sucrose in the culture of QL233 was comparable to that for FL92 after 2 days of salt stress (Fig. 4c). These results demonstrated that glycogen excess alone could not enhance sucrose production in sucrose-secreting Syn7942. As observed previously, the glycogen contents decreased in salt-stressed cells of QL233 as well as FL92. However, relatively high levels of glycogen were still measured at day 9 in both strains (85% of the day 4 level in QL233 and 57% of the day 4 level in FL92), indicating that the glycogen content was not the limiting factor for sucrose production in these strains. Therefore, we concluded that alternative steps might limit the production of sucrose.
FIG 4.
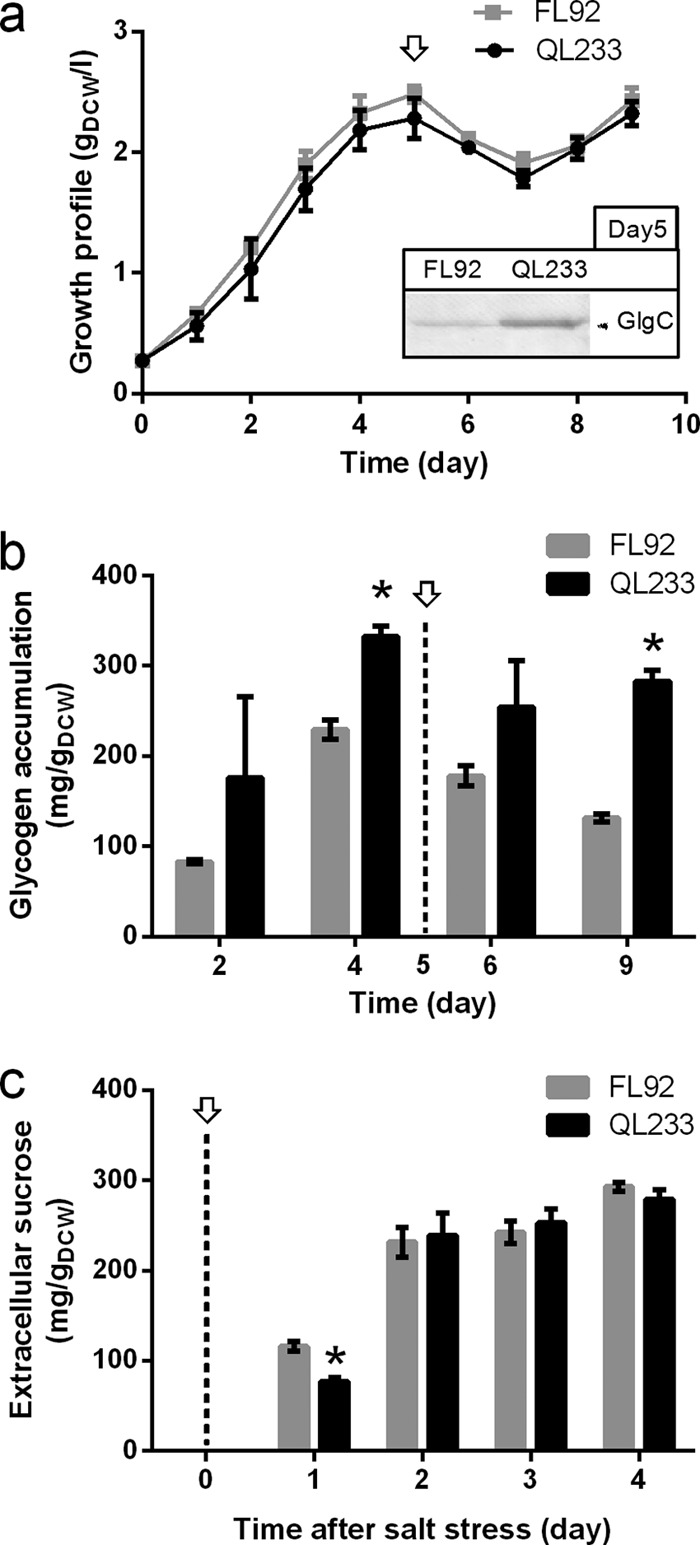
Effect of glycogen overaccumulation on growth and sucrose production in strains of Syn7942. Growth (a), glycogen content (b), and sucrose production (c) of QL233 and FL92 before and after salt stress are displayed. The open arrows and dashed lines indicate the time when 150 mM NaCl was applied. The expression of GlgC in QL233 and FL92 after 5 days of NaCl-free cultivation was examined by immunoblotting, and the result is shown as an inset in panel a. Asterisks indicate significant differences from FL92 (Student's t test, P < 0.05).
It is well known that 4 enzymatic steps are involved in the carbon flow from glycogen into sucrose (Fig. 1). Glycogen phosphorylase (GlgP) catalyzes the initial step of glycogen breakdown, leading to glucose-1-phosphate, which is then converted to UDP-glucose through the action of UDP-glucose pyrophosphorylase (GalU). UDP-glucose and fructose-6-phosphate are the precursors of sucrose synthesis and are catalyzed by the combined action of SPS and SPP. It has been shown that the overexpression of glgP or galU reduced sucrose productivity (10 to 20% less) in cscB-expressing Syn7942 (23). This finding indicated that GlgP and GalU were not the key factors limiting sucrose production. Alternatively, our previous studies demonstrated that overexpressing sps improved sucrose production in Syn7942 and Syn6803 (22, 24). Therefore, we assumed that the overexpression of sps in strain QL233 could relieve the limiting step and further improve sucrose production.
Combined glgC and sps overexpression leads to higher sucrose productivity.
To verify the above assumption, we constructed strain QL241, which cooverexpresses glgC and sps on the background of FL92. As expected, sucrose productivity was significantly improved in salt-stressed cells of QL241 compared to QL233 (2- to 3-fold higher) (Fig. 5a). Whereas the extracellular sucrose amount for QL233 attained a steady level of approximately 225 mg/gDCW after 2 days of salt stress, the sucrose production by QL241 increased further and reached 671.3 mg/gDCW. QL241 and QL233 accumulated similar amounts of glycogen under control conditions before salt was added (day 0). However, the level of glycogen decreased to only 33.8% (compared to day 0) in QL241 but was 70.7% (compared to day 0) in QL233 after 4 days of salt stress (Fig. 5b). The consumption of glycogen in QL241 was faster than that in QL233 and was correlated with the increased sucrose productivity.
FIG 5.
Effect of combined glgC and sps overexpression in strains of Syn7942. Sucrose production (a and c) and glycogen content (b and d) of strains QL241, QL233, and FL130 after salt stress are displayed. Cyanobacterial cells were precultivated in medium containing 1 mM IPTG to induce sps expression and then exposed to 150 mM NaCl to induce sucrose production. Asterisks indicate significant differences from QL233 (a), FL130 (c and d), or day 0 (b) (Student's t test, P < 0.05).
The above result indicated the significant effect of sps overexpression in stimulating sucrose production of Syn7942. This agrees well with our previous observations (24). Previously we constructed the Syn7942 strain FL130 (Table 2), in which the native sps gene was exclusively overexpressed on the background of FL92. The sucrose productivity of strain FL130 was improved by about 2-fold compared to that of FL92. Thus, the effect of glgC overexpression on the improved sucrose production by strain QL241 still remained to be elucidated. To further understand the importance of glgC overexpression in QL241, we compared the sucrose production of QL241 and FL130. We found that QL241 exhibited enhanced glycogen accumulation compared to FL130 (Fig. 5d), and the sucrose production by QL241 was markedly improved (Fig. 5c). Compared to FL130, QL241 cells accumulated approximately 70% more sucrose during the first 2 days of salt stress and 29% more sucrose at day 3 (Fig. 5c). These findings demonstrated that the enhanced glycogen pool could stimulate sucrose production in the presence of a strong sucrose synthesis sink.
TABLE 2.
Strains and oligonucleotides used in this study
| Strain or oligonucleotide | Characteristics or sequence (5′→3′) | Reference |
|---|---|---|
| Syn7942 strains | ||
| FL92 | Starting strain in this study, harboring NS3::cscB, sucrose secreting, Cmr | 24 |
| FL130 | Harboring NS1::Ptrc-sps on the background of FL92, for constructing strain QL241, Cmr Spr | 24 |
| DY126 | Harboring riboswitch-regulated glgC on the background of FL92, Cmr Kmr | This study |
| QL233 | Harboring NS2::PcpcB1-glgC on the background of FL92, Cmr Gmr | This study |
| QL241 | Harboring NS2::PcpcB1-glgC and NS1::Ptrc-sps on the background of FL92, Cmr Gmr Spr | This study |
| Oligonucleotides | ||
| E-F | AAATATTCTGAAATGAGCTGTTGAC | |
| E-glgC-R | ATGATCGCCAGCACGTTTTTCACCTTGTTGCCTCCTTAGCAGGGT | |
| glgC-F | ACCCTGCTAAGGAGGCAACAAGGTGAAAAACGTGCTGGCGATCAT | |
| glgC-R | TTAGATCACCGTGTTGTCGGG | |
| rpe-F | ACTAGTCAGCGTTCACCTCAAGCAACT | |
| rpe-km-R | AGACGTGTAATGCTGCAATCTAATCAATCTCCCCCAAGTCAAG | |
| rpe-km-F | CTTGACTTGGGGGAGATTGATTAGATTGCAGCATTACACGTCT | |
| km-R | ACTAGTGTGACACAGGAACACTTAACGGC | |
| 7942PcpcB1-f | AACACGTGCTACAGCCTGGGTTCCTCATG | |
| 7942PcpcB1-r | CGAGAATGATCGCCAGCACGTTTTTCACTCAACCAGTCTCCTGTTCTCGAC | |
| 7942glgC-f | GTCGAGAACAGGAGACTGGTTGAGTGAAAAACGTGCTGGCGATCATTCTCG | |
| 7942glgC-r | TTAGATCACCGTGTTGTCGG |
We evaluated the sucrose production rates of the strains investigated in the present study and compared them with the recently reported values for Synechococcus elongatus. It was found that our values (6.5 to 8.0 mg/liter/h) were lower than the sucrose production rates of Syn7942 reported by Ducat et al., who obtained 28 to 36.1 mg sucrose/liter/h (Table 1) (23). For example, the production rate of our cscB-expressing strain FL92 is 6.5 mg sucrose/liter/h, whereas strain CscB (the cscB-expressing strain constructed by Ducat et al.) produced sucrose at a much higher rate (∼28 mg/liter/h) (23). Considering the presence of divergent bacterial strains during cell passages in different laboratories, the diverse genetic backgrounds of the parental Syn7942 strains may be responsible for the different sucrose productivities of cscB-expressing Syn7942. Syn2973 is the fastest-growing cyanobacterium reported to date (36). The cscB-expressing strain of Syn2973 (Syn2973::CscB in Table 1) exhibited a much higher sucrose production titer (3,340 mg/liter) and production rate (35.5 mg sucrose/liter/h) than our Syn7942 strains. This might be the result of a higher density of sucrose-producing cells. Whereas the cell density of Syn2973::CscB reached ∼2.55 gDCW/liter under salt stress (25), the cell densities of our strains were markedly lower (1.12 to 1.51 gDCW/liter for FL92, FL130, and QL241). In addition to strain properties, the cultivation conditions used in these studies are also different. Whereas we used 30°C, 3% CO2-containing air, and 100 to 120 μE/m2/s in the present study, cyanobacterial cells were grown at 35°C with 2% CO2-containing air and 65 μE/m2/s in the study by Ducat el al. (23) or at 38°C with 3% CO2-containing air and 250 μE/m2/s in the study by Song el al (25). Thus, the different culture conditions and cultivation vessels limit the direct comparison of production rates from different laboratories. A systematic optimization of cultivation conditions such as temperature, light intensity, CO2 concentration, and bioreactor type may further improve the cell density and sucrose production of the Syn7942 strain QL241.
In conclusion, the present study showed that, in contrast to our expectations, glycogen synthesis and salt-induced sucrose production are positively correlated in sucrose-secreting Syn7942 strains. The stepwise suppression of glycogen synthesis did not stimulate but decreased sucrose production. This indicates that the intracellular glycogen is not a competing carbon pool but rather serves, at least partially, as a carbon pool supporting sucrose synthesis. Based on this finding, sucrose production by Syn7942 could be improved by enhancing glycogen synthesis in combination with sps overexpression.
MATERIALS AND METHODS
Cyanobacterial strains and cultivation conditions.
All Syn7942 strains used in the present study are listed in Table 2. Cyanobacterial cells were grown in 200 ml BG11 medium aerated with 3% (vol/vol) CO2-supplemented air in 350-ml column photobioreactors (580 by 30 mm) at 30°C under constant white-light illumination of 100 to 120 μE/m2/s. To induce sucrose production, cyanobacterial cultures were supplemented by adding NaCl (final concentration, 150 mM) in the late exponential phase. Aliquots of cells were sampled to determine their growth, sucrose production, and glycogen accumulation. Cell growth was monitored by measuring the optical density at a wavelength of 730 nm (OD730) and converted to dry cell weight (DCW) with a preestablished calibration between the OD730 and DCW of Syn7942 cultures (1.0 OD730 unit equals approximately 0.34 gDCW/liter). To induce the trc promoter (Ptrc)-driven expression of sps, 1 mM isopropyl-d-1-thiogalactopyranoside (IPTG) was added in liquid BG11 medium. In addition, 20 μg/ml spectinomycin (Sp), 20 μg/ml kanamycin (Km), 10 μg/ml chloramphenicol (Cm), or 2 μg/ml gentamicin (Gm) was added to the BG11 medium when required. When multiple antibiotics were applied simultaneously, the concentration of each one was reduced.
Chemicals and reagents.
Unless noted otherwise, all of the chemicals used in the present study were from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). Taq and Pfu DNA polymerases for PCR were purchased from Transgene Biotech (Beijing, China). Restriction enzymes, T4 DNA ligase, and pJET1.2/blunt were from ThermoFisher (Waltham, MA, USA). The kits used for molecular cloning were from Omega Bio-tek (Norcross, GA, USA). Oligonucleotide synthesis and DNA sequencing were performed by TsingKe (Qingdao, China).
Construction of plasmids and Syn7942 strains.
The sequences of all oligonucleotides used are listed in Table 2. To construct Syn7942 strain DY126, a 155-bp DNA fragment, ENYC4, containing the trc promoter and the theophylline-dependent riboswitch was synthesized according to the reported sequence (34) and cloned into the cloning vector pMD18-T simple (TaKaRa, Dalian, China). Then, the ENYC4 fragment (amplified by E-F/E-glgC-R) was combined with the glgC open reading frame (ORF) fragment (amplified by glgC-F/glgC-R) by fusion PCR (using primers E-F/glgC-R), yielding fragment ENYC4-glgC. In addition, a 1.3 kb upstream fragment of the glgC ORF (amplified by rpe-F/rpe-km-R) and a 1.56 kb Km resistance cassette from pXT212 (37) (amplified by rpe-km-F/km-R) were amplified and fused, yielding fragment glgCUP-Km. The two fragments glgCUP-Km and ENYC4-glgC were cloned into pJET1.2/blunt, generating pDY121 and pDY122. glgCUP-Km was further excised by SpeI from pDY121 and inserted into the upstream site (XbaI) of the ENYC4-glgC fragment of pDY122. The final construct, pDY123, was used to transform Syn7942 strain FL92. Cm- and Km-resistant transformants were obtained after at least 7 days of selection on BG11 agar plates. The genotypes of the transformants were verified by PCR.
To overexpress glgC in Syn7942, the cpcB1 promoter (PcpcB1, 0.69 kb) and the ORF of glgC (1.3 kb) were amplified by employing primers 7942PcpcB1-f/7942PcpcB1-r, 7942glgC-f/7942glgC-r, and total DNA of Syn7942. Through fusion PCR and restriction digest/ligation, the two fragments were fused and further combined with the Gm-resistant marker aacC1 (1.2 kb) (18). The resulting fragment, aacC1-PcpcB1-glgC, was then cloned into the neutral site 2 (NS2) platform of plasmid pQL224n by Eco91I, yielding pQL225 to transform Syn7942 strain FL92 and FL130 (Table 2) (24). Following a transformation/selection step similar to that described above, the final mutants were obtained and called QL233 and QL241, respectively.
Preparation of GlgC antibodies and immunodetection.
A 121-amino-acid fragment of GlgC (positions 230 to 350) was heterologously expressed in E. coli and purified to prepare rabbit-sourced antibodies against Syn7942 GlgC (Hua'an Bio, Hangzhou, China). Cyanobacterial cells grown in BG11 medium containing gradient concentrations of theophylline were harvested by centrifugation and then resuspended in 50 mM Tris-HCl buffer (pH 8.0). An appropriate amount of quartz sand was added to the cell suspension. The sample was shaken 5 times in a vortex mixer for 3 min at the highest speed with 3-min intervals on ice. After centrifugation, the protein concentration in the supernatant was determined by the Bradford method (38). Two hundred micrograms of total protein was applied to SDS-PAGE, and immunoblotting analysis was performed following a previously described protocol (15).
Determination of glycogen in Syn7942.
The glycogen content in Syn7942 was determined as previously described (25) with minor modifications. Briefly, cyanobacterial cells were harvested by centrifugation and washed three times with sterilized water. Cell pellets were suspended in 30% (wt/vol) KOH and incubated at 95°C for 2 h. Ice-cold absolute ethanol was added to a final concentration of 75% (vol/vol), followed by an incubation at −20°C overnight for glycogen precipitation. After centrifugation at 15,000 × g for 15 min at 4°C, precipitants were washed with 70% and 98% (vol/vol) ethanol sequentially and dried in a SpeedVac (Eppendorf, Hamburg, Germany) at 60°C for 20 min. The dry glycogen precipitants were then suspended in 100 mM sodium acetate (pH 4.5) and hydrolyzed to glucose by treatment with amyloglucosidase at 60°C for 2 h. The amount of glycogen was determined by quantifying the glucose using an SBA-40C biosensor (Shandong Academy of Sciences, Ji'nan, China) according to the manufacturer's instructions.
Determination of sucrose production by Syn7942.
To determine the amount of extracellular sucrose in the cultures of Syn7942, 1 ml of cyanobacterial culture was centrifuged at 10,000 × g. Supernatants were filtered through 0.22-μm polyethersulfone membranes to remove impurities and subsequently analyzed by ion chromatography. Twenty-five microliters of supernatant sample was subjected to ion-exchange chromatography with an ICS-5000+ system equipped with an electrochemical detector and a Dinex CarboPac PA10 analytical column (4 by 250 mm; ThermoFisher, Waltham, MA, USA). The column was equilibrated and eluted with 200 mM NaOH at a flow rate of 1.0 ml/min.
Statistical analyses.
The Student t test and Spearman rank correlation analysis were performed using the statistical software program R 2.13.1 (39).
ACKNOWLEDGMENTS
We acknowledge financial support for this study from the National Science Fund for Distinguished Young Scholars of China (31525002), the joint Sino-German research project (GZ984), the National Science Foundation of China project (31501003), the Shandong Taishan Scholarship to X. Lu, and the Qingdao Innovative Leading Talent project [15-10-3-15-(31)-zch].
REFERENCES
- 1.Willke T, Vorlop KD. 2001. Biotechnological production of itaconic acid. Appl Microbiol Biotechnol 56:289–295. doi: 10.1007/s002530100685. [DOI] [PubMed] [Google Scholar]
- 2.Martinková L, Rucká L, Nešvera J, Pátek M. 2017. Recent advances and challenges in the heterologous production of microbial nitrilases for biocatalytic applications. World J Microbiol Biotechnol 33:8. doi: 10.1007/s11274-016-2173-6. [DOI] [PubMed] [Google Scholar]
- 3.de la Fuente JL, Rodríguez-Sáiz M, Schleissner C, Díez B, Peiro E, Barredo JL. 2010. High-titer production of astaxanthin by the semi-industrial fermentation of Xanthophyllomyces dendrorhous. J Biotechnol 148:144–146. doi: 10.1016/j.jbiotec.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Sallam A, Kast A, Przybilla S, Meiswinkel T, Steinbüchel A. 2009. Biotechnological process for production of β-dipeptides from cyanophycin on a technical scale and its optimization. Appl Environ Microbiol 75:29–38. doi: 10.1128/AEM.01344-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shin HY, Lee JY, Park C, Kim SW. 2011. Utilization of glycerol as cysteine and carbon sources for cephalosporin C production by Acremonium chrysogenum M35 in methionine-unsupplemented culture. J Biotechnol 151:363–368. doi: 10.1016/j.jbiotec.2010.12.021. [DOI] [PubMed] [Google Scholar]
- 6.El-Gendy MM, Al-Zahrani HA, El-Bondkly AM. 2016. Genome shuffling of mangrove endophytic Aspergillus luchuensis MERV10 for improving the cholesterol-lowering agent lovastatin under solid state fermentation. Mycobiology 44:171–179. doi: 10.5941/MYCO.2016.44.3.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sabra W, Dietz D, Tjahjasari D, Zeng AP. 2010. Biosystems analysis and engineering of microbial consortia for industrial biotechnology. Eng Life Sci 10:407–421. doi: 10.1002/elsc.201000111. [DOI] [Google Scholar]
- 8.Kwak S, Jin YS. 2017. Production of fuels and chemicals from xylose by engineered Saccharomyces cerevisiae: a review and perspective. Microb Cell Fact 16:82. doi: 10.1186/s12934-017-0694-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baral NR, Slutzky L, Shah A, Ezeji TC, Cornish K, Christy A. 2016. Acetone-butanol-ethanol fermentation of corn stover: current production methods, economic viability and commercial use. FEMS Microbiol Lett 363: fnw033. doi: 10.1093/femsle/fnw033. [DOI] [PubMed] [Google Scholar]
- 10.Koutinas AA, Wang R, Webb C. 2004. Restructuring upstream bioprocessing: technological and economical aspects for production of a generic microbial feedstock from wheat. Biotechnol Bioeng 85:524–538. doi: 10.1002/bit.10888. [DOI] [PubMed] [Google Scholar]
- 11.Auxenfans T, Crônier D, Chabbert B, Paës G. 2017. Understanding the structural and chemical changes of plant biomass following steam explosion pretreatment. Biotechnol Biofuels 10:36. doi: 10.1186/s13068-017-0718-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brethauer S, Studer MH. 2015. Biochemical conversion processes of lignocellulosic biomass to fuels and chemicals—a review. Chimia (Aarau) 69:572–581. doi: 10.2533/chimia.2015.572. [DOI] [PubMed] [Google Scholar]
- 13.Antonov E, Wirth S, Gerlach T, Schlembach I, Rosenbaum MA, Regestein L, Büchs J. 2016. Efficient evaluation of cellulose digestibility by Trichoderma reesei Rut-C30 cultures in online monitored shake flasks. Microb Cell Fact 15:164. doi: 10.1186/s12934-016-0567-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ziolkowska JR. 2014. Prospective technologies, feedstocks and market innovations for ethanol and biodiesel production in the US. Biotechnol Rep (Amst) 4:94–98. doi: 10.1016/j.btre.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao Z, Zhao H, Li Z, Tan X, Lu X. 2012. Photosynthetic production of ethanol from carbon dioxide in genetically engineered cyanobacteria. Energy Environ Sci 5:9857–9865. doi: 10.1039/C2EE22675H. [DOI] [Google Scholar]
- 16.Tan X, Yao L, Gao Q, Wang W, Qi F, Lu X. 2011. Photosynthesis driven conversion of carbon dioxide to fatty alcohols and hydrocarbons in cyanobacteria. Metab Eng 13:169–176. doi: 10.1016/j.ymben.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Wang W, Liu X, Lu X. 2013. Engineering cyanobacteria to improve photosynthetic production of alka(e)nes. Biotechnol Biofuels 6:69. doi: 10.1186/1754-6834-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu T, Xie X, Li Z, Tan X, Lu X. 2015. Enhancing photosynthetic production of ethylene in genetically engineered Synechocystis sp. PCC 6803. Green Chem 17:421–434. [Google Scholar]
- 19.Hagemann M. 2011. Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol Rev 35:87–123. doi: 10.1111/j.1574-6976.2010.00234.x. [DOI] [PubMed] [Google Scholar]
- 20.Pade N, Michalik D, Ruth W, Belkin N, Hess WR, Berman-Frank I, Hagemann M. 2016. Trimethylated homoserine functions as the major compatible solute in the globally significant oceanic cyanobacterium Trichodesmium. Proc Natl Acad Sci U S A 113:13191–13196. doi: 10.1073/pnas.1611666113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martínez-Noël GM, Cumino AC, Kolman Mde L, Salerno GL. 2013. First evidence of sucrose biosynthesis by single cyanobacterial bimodular proteins. FEBS Lett 587:1669–1674. doi: 10.1016/j.febslet.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 22.Du W, Liang F, Duan Y, Tan X, Lu X. 2013. Exploring the photosynthetic production capacity of sucrose by cyanobacteria. Metab Eng 19:17–25. doi: 10.1016/j.ymben.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Ducat DC, Avelar-Rivas JA, Way JC, Silver PA. 2012. Rerouting carbon flux to enhance photosynthetic productivity. Appl Environ Microbiol 78:2660–2668. doi: 10.1128/AEM.07901-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duan Y, Luo Q, Liang F, Lu X. 2016. Sucrose secreted by the engineered cyanobacterium and its fermentability. J Ocean Univ China 15:890–896. doi: 10.1007/s11802-016-3007-8. [DOI] [Google Scholar]
- 25.Song K, Tan X, Liang Y, Lu X. 2016. The potential of Synechococcus elongatus UTEX 2973 for sugar feedstock production. Appl Microbiol Biotechnol 100:7865–7875. doi: 10.1007/s00253-016-7510-z. [DOI] [PubMed] [Google Scholar]
- 26.Gründel M, Scheunemann R, Lockau W, Zilliges Y. 2012. Impaired glycogen synthesis causes metabolic overflow reactions and affects stress responses in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology 158:3032–3043. doi: 10.1099/mic.0.062950-0. [DOI] [PubMed] [Google Scholar]
- 27.Hickman JW, Kotovic KM, Miller C, Warrener P, Kaiser B, Jurista T, Budde M, Cross F, Roberts JM, Carleton M. 2013. Glycogen synthesis is a required component of the nitrogen stress response in Synechococcus elongatus PCC. 7942. Algal Res 2:98–106. doi: 10.1016/j.algal.2013.01.008. [DOI] [Google Scholar]
- 28.Damrow R, Maldener I, Zilliges Y. 2016. The multiple functions of common microbial carbon polymers, glycogen and PHB, during stress responses in the non-diazotrophic cyanobacterium Synechocystis sp. PCC 6803. Front Microbiol 7:966. doi: 10.3389/fmicb.2016.00966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eisenhut M, Aguirre von Wobeser E, Jonas L, Schubert H, Ibelings BW, Bauwe H, Matthijs HC, Hagemann M. 2007. Long-term response toward inorganic carbon limitation in wild type and glycolate turnover mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Physiol 144:1946–1959. doi: 10.1104/pp.107.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki E, Ohkawa H, Moriya K, Matsubara T, Nagaike Y, Iwasaki I, Fujiwara S, Tsuzuki M, Nakamura Y. 2010. Carbohydrate metabolism in mutants of the cyanobacterium Synechococcus elongatus PCC 7942 defective in glycogen synthesis. Appl Environ Microbiol 76:3153–3159. doi: 10.1128/AEM.00397-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miao X, Wu Q, Wu G, Zhao N. 2003. Sucrose accumulation in salt-stressed cells of agp gene deletion-mutant in cyanobacterium Synechocystis sp PCC 6803. FEMS Microbiol Lett 218:71–77. doi: 10.1111/j.1574-6968.2003.tb11500.x. [DOI] [PubMed] [Google Scholar]
- 32.Guerra LT, Xu Y, Bennette N, McNeely K, Bryant DA, Dismukes GC. 2013. Natural osmolytes are much less effective substrates than glycogen for catabolic energy production in the marine cyanobacterium Synechococcus sp. strain PCC 7002. J Biotechnol 166:65–75. doi: 10.1016/j.jbiotec.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 33.Xu Y, Guerra LT, Li Z, Ludwig M, Dismukes GC, Bryant DA. 2013. Altered carbohydrate metabolism in glycogen synthase mutants of Synechococcus sp. strain PCC 7002: cell factories for soluble sugars. Metab Eng 16:56–67. doi: 10.1016/j.ymben.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 34.Nakahira Y, Ogawa A, Asano H, Oyama T, Tozawa Y. 2013. Theophylline-dependent riboswitch as a novel genetic tool for strict regulation of protein expression in cyanobacterium Synechococcus elongatus PCC 7942. Plant Cell Physiol 54:1724–1735. doi: 10.1093/pcp/pct115. [DOI] [PubMed] [Google Scholar]
- 35.Baran R, Lau R, Bowen BP, Diamond S, Jose N, Garcia-Pichel F, Northen TR. 2017. Extensive turnover of compatible solutes in cyanobacteria revealed by deuterium oxide (D2O) stable isotope probing. ACS Chem Biol 12:674–681. doi: 10.1021/acschembio.6b00890. [DOI] [PubMed] [Google Scholar]
- 36.Yu J, Liberton M, Cliften PF, Head RD, Jacobs JM, Smith RD, Koppenaal DW, Brand JJ, Pakrasi HB. 2015. Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO2. Sci Rep 5:8132. doi: 10.1038/srep08132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan X, Liang F, Cai K, Lu X. 2013. Application of the FLP/FRT recombination system in cyanobacteria for construction of markerless mutants. Appl Microbiol Biotechnol 97:6373–6382. doi: 10.1007/s00253-013-4837-6. [DOI] [PubMed] [Google Scholar]
- 38.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 39.R Development Core Team. 2011. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]