Abstract
Tumor metastasis portrayed the most serious challenges of cancer as it is the major cause of mortality in patients with solid tumors, including hepatocellular carcinoma (HCC). In this regard, anti-metastatic genes have a great potential for metastasis inhibition. Recent evidence pointed to a role of Breast cancer metastasis suppressor 1 (BRMS1) in suppression of metastasis of several types of cancers, whereas the regulation of BRMS1 in HCC remains unknown. Here, we used bioinformatics analyses to predict BRMS1-targeting microRNAs (miRNAs), and evaluated the functional binding of miRNAs to BRMS1 mRNA using a dual luciferase reporter assay. Among all BRMS1-targeting miRNAs, we only detected significant expression of miR-626, miR-1289 and miR-423 in HCC specimens. Specifically, we found that only miR-423 significantly inhibited protein translation of BRMS1 via specific binding to 3’-UTR of the BRMS1 mRNA. Moreover, we detected significantly lower levels of BRMS1 and significantly higher levels of miR-423 in the HCC specimens, relative to paired adjacent non-tumor liver tissue. Furthermore, BRMS1 and miR-423 levels were correlated inversely. Overexpression of miR-423 significantly decreased BRMS1 levels and promoted HCC cell invasion, while depletion of miR-423 significantly increased BRMS1 levels and inhibited HCC cell invasion. This study sheds light on miR-423 as a crucial factor that enhances HCC cell invasiveness, and suggests miR-423 as a promising therapeutic target for HCC treatment.
Keywords: Hepatocellular carcinoma (HCC), miR-423, breast cancer metastasis suppressor 1 (BRMS1)
Introduction
Hepatocellular carcinoma (HCC) is a primary malignant liver tumor in humans, and HCC is notorious partially from its invasiveness and distal metastases [1]. Despite years of continuous progresses on cancer therapy, metastasis remains the major driver of cancer mortality and is responsible for approximately 90% of cancer-associated deaths. Metastasis is a complex, multi-step process by which primary tumor cells invade adjacent tissues [2]. Each of the steps for cancer metastasis requires a complex molecular regulation.
Breast cancer metastasis suppressor 1 (BRMS1) was originally identified in breast carcinoma cell line [3]. Afterward, BRMS1 was shown to suppress metastasis of multiple tumor types [4-7], through regulation of the expression of some metastasis-associated genes including osteopontin, urokinase plasminogen activator, fascin, epidermal growth factor receptor and CXCR4 [8-11]. Hence, BRMS1 affects multiple phenotypes implicated in cancer metastasis, e.g. gap junctional intercellular communication, migration, invasion, anoikis, and growth factor signaling [8-11]. Nevertheless, the regulation of BRMS1 during tumorigenesis remains largely unknown.
MicroRNAs (miRNAs) are known as master regulators of gene expression, are small non coding RNAs capable of post-transcriptional suppression of gene expression by sequence-specific interactions with the miRNA response elements (MREs) on the 3’ un-translated regions (3’-UTRs) of related mRNA targets [12,13]. Abnormal expression of miRNAs in different steps of malignancies has been discovered in a variety of cancers by genome-wide survey techniques, various microarray platforms and bead-based flow cytometry.
Here, we studied the regulation of BRMS1 by miRNAs in HCC specimens and HCC cells. We used bioinformatics analyses to predict BRMS1-targeting miRNAs, and evaluated functional binding of miRNAs to BRMS1 mRNA using a dual luciferase reporter assay. Moreover, we examined and compared BRMS1 and miR-423 levels in the HCC specimens. Furthermore, we used a set of gain-of-function and loss-of-function experiments to study the effects of miR-423-modification on BRMS1 and on HCC cell invasion.
Materials and methods
Experimental protocol approval and patient specimens
All experimental protocols were approved by the Research Bureau of Linyi People’s Hospital. Surgical resected specimens were obtained from 36 HCC patients and paired adjacent non-tumor liver tissues (NT) (Table 1). All patients obtained Informed consent and provided signed agreement about this study. The histology of the resected tissue were examined and determined independently by 2 senior pathologists.
Table 1.
Clinical-pathological characteristics (total)
| Patients (n; %) | p | |
|---|---|---|
| HCC tissue/Normal tumor-adjacent tissue (NT) | 36 (100%)/36 (100%) | |
| Age (<60/≥60 years old) | 12 (33%)/24 (67%) | 0.52 |
| Gender (male/female) | 24 (67%)/12 (33%) | |
| Tumor site (liver) | 36 (100%) | |
| Tumor grade (well or moderate/poor) | 0 (0%)/15 (42%)/21 (58%) | 0.009 |
| Tumor stage (I/II/III/IV) | 0 (0%)/0 (0%)/18 (50%)/18 (50%) | 0.007 |
| Lymph node metastasis (no/yes) | 0 (0%)/36 (100%) | 0.006 |
| Distal metastasis at diagnosis (no/yes) | 32 (89%)/4 (11%) | 0.008 |
Culturing and transfection of cells
SNU-398 and HepG2 are two widely used human HCC cell lines purchased from American Type Culture Collection (ATCC, Rockville, MD, USA), and were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich, St Louis, MO, USA) in a humidified chamber with 5% CO2 at 37°C. We chose these two lines since HepG2 cells express relatively low miR-423 and relatively high BRMS1, while SNU-398 cells express relatively high miR-423 and relatively low BRMS1. All constructs used for cell transfection were purchased from Origene (Beijing, China). Transfection was performed with 50 nmol/l plasmids, using Lipofectamine 2000 (Invitrogen). The transfection efficiency (>95%) was determined based on expression of GFP in the transfected cells.
Transwell cell invasion assay
Transwell cell invasion assay was analyzed using Matrigel-coated Transwell cell culture chambers (8 μm pore size). Briefly, transfected cells (20000 cells/well) were serum starved for 24 h and plated in the upper insert of a 24-well chamber in a serum-free medium. A medium containing 10% serum was added to the lower wells. The cells were incubated for 24 h. Cells on the upper side of the filters were mechanically removed by scrubbing with a cotton swab, after which the membrane was fixed with 4% formaldehyde for 30 min and stained with 0.5% crystal violet for 10 min. Ultimately, invasive cells were counted at ×200 magnification from 10 different fields of each filter.
Cell growth assay
An MTT Kit (MTT, Roche, USA) was used for analyzing cell growth.
MiRNA target prediction and 3’-UTR luciferase-reporter assay
MiRNAs targets were predicted using the algorithms from TargetScan, using context + scoring system [14]. The data were analyzed as previously described [15]. The BRMS1 3’-UTR reporter plasmid (pRL-BRMS1) and the BRMS1 3’-UTR reporter plasmid with a mutant at the corresponding miRNA binding site (pRL-BRMS1-mut) were purchased from Origene (Beijing, China). Dual-luciferase reporter assay (Promega, Fitchburg, WI, USA) was performed according to the instructions from manufacturer.
Quantitative RT-PCR (RT-qPCR)
Quantitative RT-PCR (RT-qPCR) was performed as routine. Briefly, total RNA, with efficient recovery of small RNAs, was using miRCURY RNA isolation kit (Exiqon, Denmark) and cDNA was synthesized by Universal cDNA Synthesis Kit (Exiqon). Detection of the mature form of miRNAs was performed using the miRCURY LNA Universal RT microRNA PCR Kit and LNA microRNA Primer Sets, according to the manufacturer’s instructions (Exiqon). Relative expression levels were determined using the comparative quantification characteristic of the Rotorgene software. The U6 small nuclear RNA was used as an internal control and the comparative Ct (ΔΔCt) method was used to determine the expression fold change. A melting curve analysis was done for each of the primer sets utilized, and each showed a single peak indicating the specificity of each of the primers tested.
ELISA
Protein levels for human BRMS1 were determined using a human BRMS1 ELISA kit (LSBio, Seattle, USA).
Statistical analysis
All statistical analyses were performed using the GraphPad Prism 6 (GraphPad Software, San Diego, CA, USA). Statistical analysis of group differences was carried out using a one-way analysis of variance (ANOVA) test followed by followed by Turkey multiple comparison post-hoc analysis. The relationship between miR-423 levels and clinicopathological characteristics was evaluated using multivariate Cox regression analysis. Bivariate correlations were calculated by Spearman’s Rank Correlation Coefficients. All values represent the mean ± standard deviation (SD). A value of P<0.05 was considered statistically significant.
Results
MiR-423 targets 3’-UTR of BRMS1 mRNA to inhibit its translation in HCC cells
First, we examined whether BRMS1 may be regulated by miRNAs in HCC cells. Thus, we performed bioinformatics analyses to identify the candidate miRNAs that bind to the 3’-UTR of BRMS1 mRNA. Among all BRMS1-targeting miRNAs (Supplementary Table 1), we found 3 miRNAs (miR-626, miR-1289 and miR-423) that were expressed in HCC cell line SNU-398. To determine whether the binding of these 3 miRNAs to BRMS1 mRNA may functionally inhibit protein translation of BRMS1, we transfected SNU-398 cells with either miR-626, miR-1289 and miR-423 or antisense for them (as-miR-626, as-miR-1289 and as-miR-423). The SNU-398 cells were also transfected with a null sequence as a control (null). Changes in the miRNA levels in cells was confirmed by RT-qPCR (Figure 1A-C). Then, the intact 3’-UTR of BRMS1 mRNA (BRMS1 3’-UTR), together with a 3’-UTR with mutant at either miRNA binding site of BRMS1 mRNA (BRMS1 3’-UTR mut), was then cloned into luciferase reporter plasmids. SNU-398 cells were co-transfected with as-miRNA/null plasmids and BRMS1 3’-UTR or BRMS1 3’-UTR mut plasmids or co-transfected with miRNA/null plasmids and BRMS1 3’-UTR or BRMS1 3’-UTR mut plasmids. The results suggest that among all 3 miRNAs, only miR-423 may specifically target 3’-UTR of BRMS1 mRNA to inhibit its translation in HCC cells (Figure 1D-G), as shown by bioinformatics analyses for the binding site of miR-423 on the 3’-UTR of BRMS1 mRNA (Figure 1G).
Figure 1.
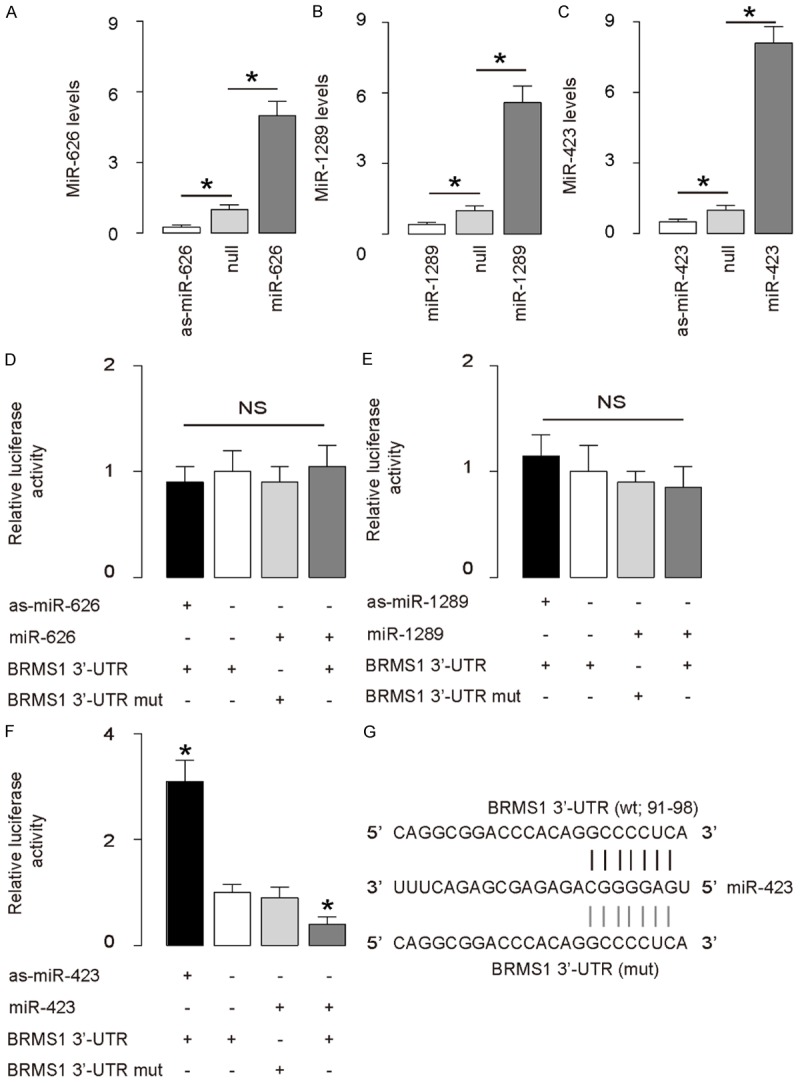
MiR-423 targets 3’-UTR of BRMS1 mRNA to inhibit its translation in HCC cells. (A-C) SNU-398 cells was transfected with either miR-626, miR-1289 and miR-423 or antisense for them (as-miR-626, as-miR-1289 and as-miR-423). The SNU-398 cells were also transfected with a null sequence as a control (null). RT-qPCR showed changes in the miR-626 levels in miR-626-modified cells (A), changes in the miR-1289 levels in miR-1289-modified cells (B), and changes in the miR-423 levels in miR-423-modified cells (C). (D-F) The intact 3’-UTR of BRMS1 mRNA (BRMS1 3’-UTR), together with a 3’-UTR with mutant at either miRNA binding site of BRMS1 mRNA (BRMS1 3’-UTR mut), was then cloned into luciferase reporter plasmids. SNU-398 cells were co-transfected with 1 μg as-miRNA/null plasmids and 1 μg BRMS1 3’-UTR or BRMS1 3’-UTR mut plasmids or co-transfected with 1 μg miRNA/null plasmids and 1 μg BRMS1 3’-UTR or BRMS1 3’-UTR mut plasmids. (D) Assay for miR-626. (E) Assay for miR-1289 (F) Assay for miR-423. (G) Bioinformatics analyses for binding site of miR-423 on 3’-UTR of BRMS1 mRNA. *P<0.05. NS: non-significant. N=5.
Association of miR-423 and BRMS1 levels in HCC specimens
The levels of BRMS1 and miR-423 in 36 pairs of resected HCC tissues and adjacent non-tumor liver tissues (NT) were measured by ELISA and RT-qPCR, respectively (Table 1). We found that HCC specimens contained significantly lower levels of BRMS1 (Figure 2A), and significantly higher levels of miR-423 (Figure 2B). We then performed a correlation test using these 36 HCC specimens, and detected a strong inverse correlation between BRMS1 and miR-423 (Figure 1C, γ=-0.69, P<0.0001), indicating a possible regulatory relationship between miR-423 and BRMS1 in HCC.
Figure 2.
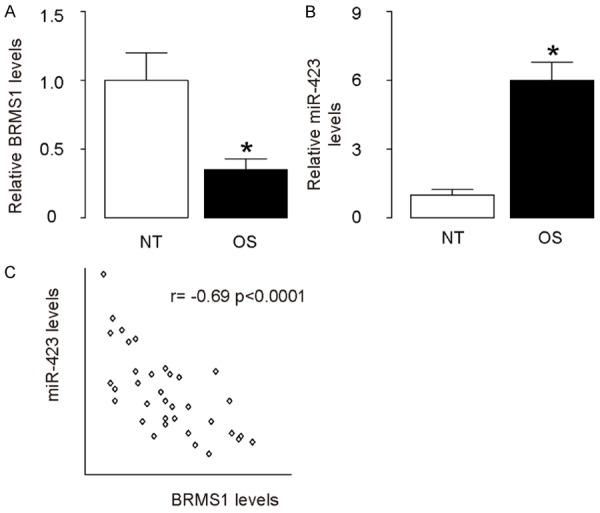
Association of miR-423 and BRMS1 levels in HCC specimens. A. The levels of BRMS1 in 36 pairs of HCC tissues and adjacent non-tumor liver tissues (NT) were measured by ELISA. B. The levels of miR-423 in 36 pairs of HCC tissues and NT were measured by RT-qPCR. C. A correlation test was performed between BRMS1 and miR-423, using the 36 HCC specimens. *P<0.05. N=36.
Modification of miR-423 levels in HCC cells
Next, we examined the effects of miR-423 on BRMS1 and cell invasion in HCC cells. We chose SNU-398 and HepG2 cell lines for this study, since HepG2 cells express relatively low miR-423 and relatively high BRMS1, while SNU-398 cells express relatively high miR-423 and relatively low BRMS1 (Figure 3A, 3B). We then transfected HepG2 cells with miR-423 and SNU-398 cells with antisense for miR-423 (as-miR-423), and confirmed the alteration of miR-423 levels in these cells by RT-qPCR (Figure 3C, 3D).
Figure 3.
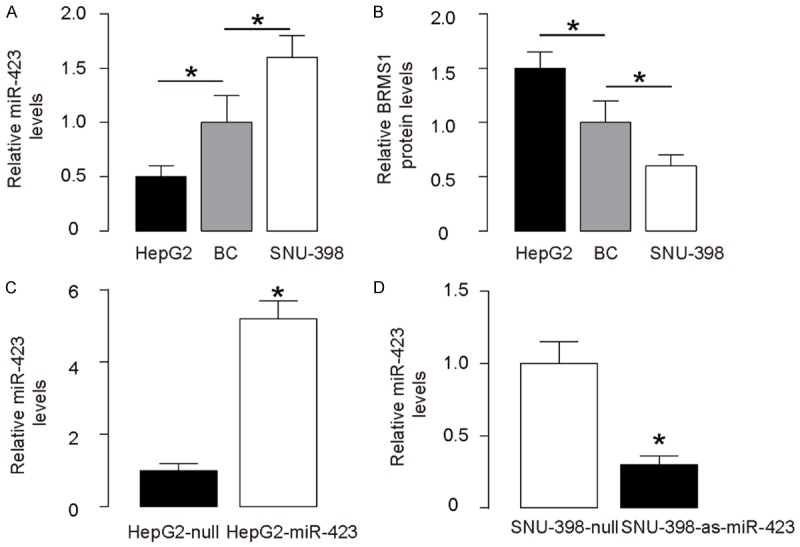
Modification of miR-423 levels in HCC cells. A. MiR-423 levels by RT-qPCR in SNU-398 and HepG2 cells, compared to HCC specimens. B. BRMS1 protein levels by ELISA in SNU-398 and HepG2 cells, compared to HCC specimens. C. Transfection of HepG2 cells with miR-423 or null control plasmids. MiR-423 levels were determined by RT-qPCR. D. Transfection of SNU-398 cells with as-miR-423 or null control plasmids. MiR-423 levels were determined by RT-qPCR. *P<0.05. N=5.
Overexpression of miR-423 in HCC cells promotes cell invasion via BRMS1 suppression
We found that overexpression of miR-423 did not alter BRMS1 mRNA (Figure 4A), but decreased BRMS1 protein in HepG2 cells (Figure 4B). The effects of miR-423 modification on cell growth and invasiveness were then investigated. We found that miR-423 overexpression in HepG2 cells did not alter cell growth in an MTT assay (Figure 4C), but significantly increased cell invasion in a transwell cell invasion assay (Figure 4D, 4E). In order to ascertain whether miR-423 promotes HCC cell invasion through suppressing BRMS1, we prepared plasmids for BRMS1 overexpression (BRMS1) and depletion (shBRMS1). HepG2-miR-423 cells were further transfected with BRMS1, which rescued expression of BRMS1 mRNA (Figure 4A) and protein (Figure 4B) in these cells. We found that overexpression of BRMS1 abrogated the effects of miR-423 on cell invasion in HepG2 cells (Figure 4D, 4E), without affected cell growth (Figure 4C). Hence, overexpression of miR-423 in HCC cells promotes cell invasion via BRMS1 suppression.
Figure 4.
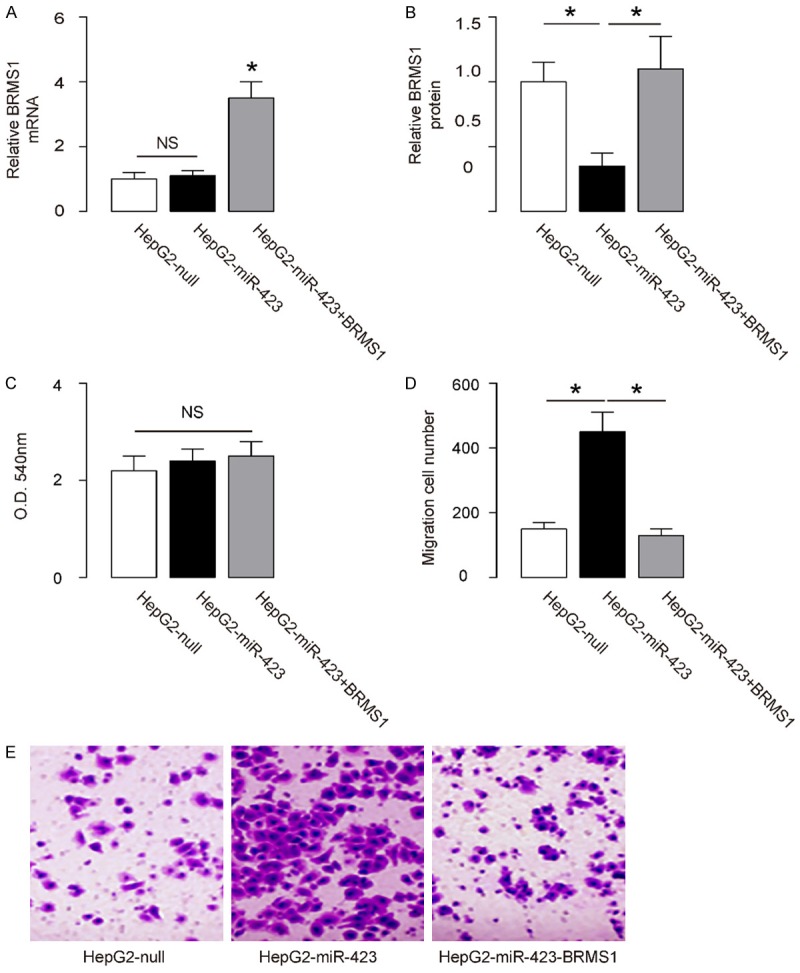
Overexpression of miR-423 in HCC cells promotes cell invasion via BRMS1 suppression. (A, B) The BRMS1 levels in miR-423-overexpressing (and BRMS1-overexpressing) HepG2 cells by RT-qPCR (A) and by ELISA (B). (C) MTT assay. (D, E) HepG2 cell invasion by miR-423 overexpression (and BRMS1 overexpression) in a transwell cell invasion assay, shown by quantification (D), and by representative images (E). *P<0.05. NS: non-significant. N=5.
Depletion of miR-423 in HCC cells inhibits cell invasion via BRMS1 augmentation
Next, we found that depletion of miR-423 did not alter BRMS1 mRNA (Figure 5A), but increased BRMS1 protein in SNU-398 cells (Figure 5B). The effects of miR-423 modification on cell growth and invasiveness were then investigated. We found that miR-423 depletion in SNU-398 cells did not alter cell growth in an MTT assay (Figure 5C), but significantly decreased cell invasion in a transwell cell invasion assay (Figure 5D, 5E). In order to ascertain whether miR-423 suppression inhibits HCC cell invasion through increasing BRMS1, SNU-398-as-miR-423 cells were further transfected with shBRMS1, which abolished the induction of expression of BRMS1 mRNA (Figure 5A) and protein (Figure 5B) in these cells. We found that depletion of BRMS1 abrogated the effects of as-miR-423 on cell invasion in SNU-398 cells (Figure 5D, 5E), without affected cell growth (Figure 5C). Hence, depletion of miR-423 in HCC cells inhibits cell invasion via BRMS1 augmentation. A schematic was thus made to summarize the current study, showing that miR-423 may inhibit protein translation of BRMS1 via pairing to the 3’-UTR of the BRMS1 mRNA in HCC cells to promote cell invasion.
Figure 5.
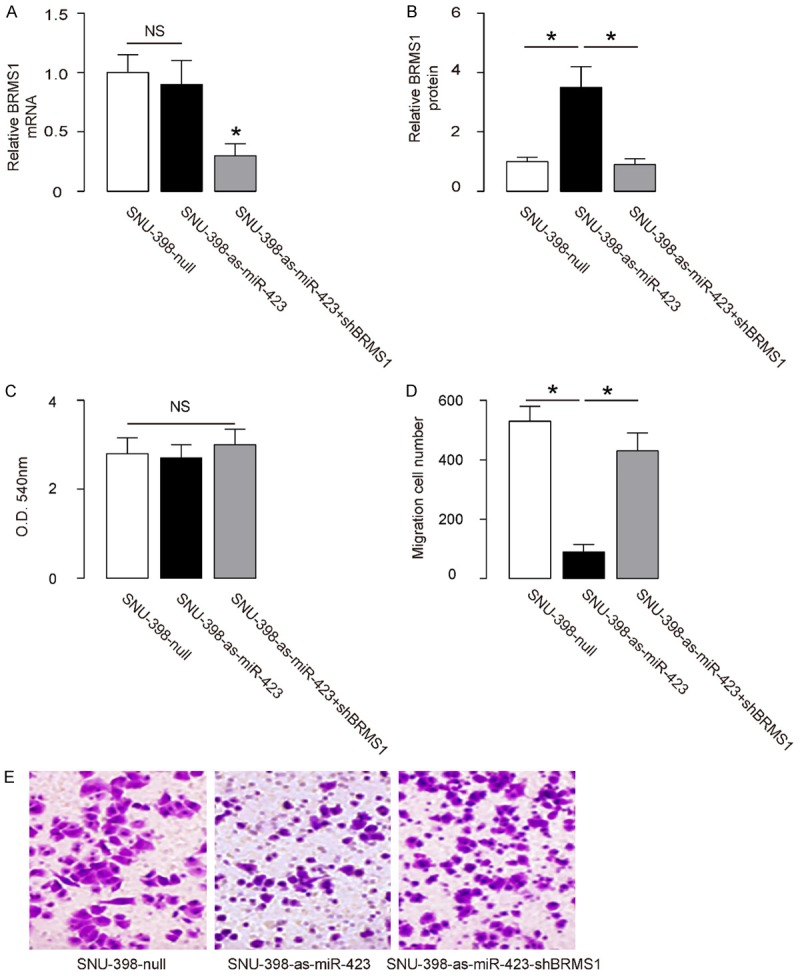
Depletion of miR-423 in HCC cells inhibits cell invasion via BRMS1 augmentation. (A, B) The BRMS1 levels in miR-423-depleted (and BRMS1-depleted) SNU-398 cells by RT-qPCR (A) and by ELISA (B). (C) MTT assay. (D, E) SNU-398 cell invasion by miR-423 depletion (and BRMS1 depletion) in a transwell cell invasion assay, shown by quantification (D), and by representative images (E). *P<0.05. NS: non-significant. N=5.
Discussion
The inhibitory role of BRMS1 in cancer invasion has been well documented in the past studies. For example, Roesley et al. have shown that BRMS1 is a substrate of Cyclin-Dependent Kinase 2 (CDK2), by which it is phosphorylated on serine 237 in breast cancer cells [11]. Although the mutation of BRMS1 on serine 237 did not affect cell cycle progression and proliferation of cancer cells, it indeed changed the cell migration [11]. However, the regulation of BRMS1 by miRNAs was not reported in breast cancer, but in nasopharyngeal carcinoma cells [16]. In this study, Yan et al. showed that miR-346, a BRMS1-targeting miRNA, was upregulated in nasopharyngeal carcinoma tissues compared with adjacent non-tumorous nasopharyngeal tissues. Inhibition of miR-346 significantly attenuated the migration and invasion of nasopharyngeal carcinoma cells [16]. Nevertheless, a role of BRMS1 in HCC invasion and metastasis, as well as its regulation by miRNAs, has not been documented so far.
Here, we used bioinformatics analyses to screen all miRNAs that target BRMS1 in HCC cells, and we focused on the expression levels of those that were detectable in HCC cells. From the 3 candidates, we only found miR-423 to be the miRNA that has a functional binding to 3’-UTR of BRMS1 mRNA. To the best of our knowledge, this is the first study showing that BRMS1 protein levels could be regulated by a specific miRNA in HCC. High level of miR-423 in HCC specimens was associated with low BRMS1 levels. We thus designed in vitro experiments to show a regulatory relationship between miR-423 and BRMS1 in HCC cells, which was consistence with the clinic findings showing an inverse correlation of these two factors in HCC specimens.
In addition to regulation of BRMS1 by miRNAs, BRMS1 protein levels may be modulated at the level of degradation, such as through protein ubiquitination. Moreover, miR-423 may have targets other than BRMS1, and these targets should be analyzed in future to have an overview of the effects of miR-423 in the HCC cell invasion. Furthermore, future studies may also address the regulation of miR-423 in HCC and confirm this model in vivo.
Compared with overexpression of BRMS1, using as-miR-423 to increase BRMS1 levels has an advantage, since overexpression of BRMS1 in HCC cells may result in a further increase in the levels of miR-423 as a feedback, to level down the efficacy of the treatment. To summarize, the current study sheds light on miR-423 as a crucial factor that enhances HCC cell invasiveness, and provides evidence for using miR-423 as a promising therapeutic target for HCC treatment.
Acknowledgements
This study was supported by internal funding from Linyi People’s Hospital.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Ren Y, Dai C, Zheng H, Zhou F, She Y, Jiang G, Fei K, Yang P, Xie D, Chen C. Prognostic effect of liver metastasis in lung cancer patients with distant metastasis. Oncotarget. 2016;7:53245–53253. doi: 10.18632/oncotarget.10644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Catalano V, Turdo A, Di Franco S, Dieli F, Todaro M, Stassi G. Tumor and its microenvironment: a synergistic interplay. Semi Cancer Biol. 2013;23:522–532. doi: 10.1016/j.semcancer.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 3.Seraj MJ, Samant RS, Verderame MF, Welch DR. Functional evidence for a novel human breast carcinoma metastasis suppressor, BRMS1, encoded at chromosome 11q13. Cancer Res. 2000;60:2764–2769. [PubMed] [Google Scholar]
- 4.Smith PW, Liu Y, Siefert SA, Moskaluk CA, Petroni GR, Jones DR. Breast cancer metastasis suppressor 1 (BRMS1) suppresses metastasis and correlates with improved patient survival in non-small cell lung cancer. Cancer Lett. 2009;276:196–203. doi: 10.1016/j.canlet.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seraj MJ, Harding MA, Gildea JJ, Welch DR, Theodorescu D. The relationship of BRMS1 and RhoGDI2 gene expression to metastatic potential in lineage related human bladder cancer cell lines. Clin Exp Metastasis. 2000;18:519–525. doi: 10.1023/a:1011819621859. [DOI] [PubMed] [Google Scholar]
- 6.Nagji AS, Liu Y, Stelow EB, Stukenborg GJ, Jones DR. BRMS1 transcriptional repression correlates with CpG island methylation and advanced pathological stage in non-small cell lung cancer. J Pathol. 2010;221:229–237. doi: 10.1002/path.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samant RS, Clark DW, Fillmore RA, Cicek M, Metge BJ, Chandramouli KH, Chambers AF, Casey G, Welch DR, Shevde LA. Breast cancer metastasis suppressor 1 (BRMS1) inhibits osteopontin transcription by abrogating NFkappaB activation. Mol Cancer. 2007;6:6. doi: 10.1186/1476-4598-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huo X, Li S, Shi T, Suo A, Ruan Z, Guo H, Yao Y. Cullin3 promotes breast cancer cells metastasis and epithelial-mesenchymal transition by targeting BRMS1 for degradation. Oncotarget. 2015;6:41959–41975. doi: 10.18632/oncotarget.5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meehan WJ, Samant RS, Hopper JE, Carrozza MJ, Shevde LA, Workman JL, Eckert KA, Verderame MF, Welch DR. Breast cancer metastasis suppressor 1 (BRMS1) forms complexes with retinoblastoma-binding protein 1 (RBP1) and the mSin3 histone deacetylase complex and represses transcription. J Biol Chem. 2004;279:1562–1569. doi: 10.1074/jbc.M307969200. [DOI] [PubMed] [Google Scholar]
- 10.Hurst DR, Xie Y, Vaidya KS, Mehta A, Moore BP, Accavitti-Loper MA, Samant RS, Saxena R, Silveira AC, Welch DR. Alterations of BRMS1-ARID4A interaction modify gene expression but still suppress metastasis in human breast cancer cells. J Biol Chem. 2008;283:7438–7444. doi: 10.1074/jbc.M709446200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roesley SN, Suryadinata R, Morrish E, Tan AR, Issa SM, Oakhill JS, Bernard O, Welch DR, Sarcevic B. Cyclin-dependent kinase-mediated phosphorylation of breast cancer metastasis suppressor 1 (BRMS1) affects cell migration. Cell Cycle. 2016;15:137–151. doi: 10.1080/15384101.2015.1121328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang W, Zhou X, Wei M. MicroRNA-144 suppresses osteosarcoma growth and metastasis by targeting ROCK1 and ROCK2. Oncotarget. 2015;6:10297–10308. doi: 10.18632/oncotarget.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han K, Chen X, Bian N, Ma B, Yang T, Cai C, Fan Q, Zhou Y, Zhao TB. MicroRNA profiling identifies MiR-195 suppresses osteosarcoma cell metastasis by targeting CCND1. Oncotarget. 2015;6:8875–8889. doi: 10.18632/oncotarget.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coronnello C, Benos PV. ComiR: combinatorial microRNA target prediction tool. Nucleic Acids Res. 2013;41:W159–164. doi: 10.1093/nar/gkt379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan HL, Li L, Li SJ, Zhang HS, Xu W. miR-346 promotes migration and invasion of nasopharyngeal carcinoma cells via targeting BRMS1. J Biochem Mol Toxicol. 2016;30:602–607. doi: 10.1002/jbt.21827. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


