ABSTRACT
Coxsackievirus A6 (CVA6) has recently emerged as one of the predominant causative agents of hand, foot, and mouth disease (HFMD). The structure of the CVA6 mature viral particle has not been solved thus far. Our previous work shows that recombinant virus-like particles (VLPs) of CVA6 represent a promising CVA6 vaccine candidate. Here, we report the first cryo-electron microscopy (cryo-EM) structure of the CVA6 VLP at 3.0-Å resolution. The CVA6 VLP exhibits the characteristic features of enteroviruses but presents an open channel at the 2-fold axis and an empty, collapsed VP1 pocket, which is broadly similar to the structures of the enterovirus 71 (EV71) VLP and coxsackievirus A16 (CVA16) 135S expanded particle, indicating that the CVA6 VLP is in an expanded conformation. Structural comparisons reveal that two common salt bridges within protomers are maintained in the CVA6 VLP and other viruses of the Enterovirus genus, implying that these salt bridges may play a critical role in enteroviral protomer assembly. However, there are apparent structural differences among the CVA6 VLP, EV71 VLP, and CVA16 135S particle in the surface-exposed loops and C termini of subunit proteins, which are often antigenic sites for enteroviruses. By immunological assays, we identified two CVA6-specific linear B-cell epitopes (designated P42 and P59) located at the GH loop and the C-terminal region of VP1, respectively, in agreement with the structure-based prediction of antigenic sites. Our findings elucidate the structural basis and important antigenic sites of the CVA6 VLP as a strong vaccine candidate and also provide insight into enteroviral protomer assembly.
IMPORTANCE Coxsackievirus A6 (CVA6) is becoming one of the major pathogens causing hand, foot, and mouth disease (HFMD), leading to significant morbidity and mortality in children and adults. However, no vaccine is currently available to prevent CVA6 infection. Our previous work shows that recombinant virus-like particles (VLPs) of CVA6 are a promising CVA6 vaccine candidate. Here, we present a 3.0-Å structure of the CVA6 VLP determined by cryo-electron microscopy. The overall architecture of the CVA6 VLP is similar to those of the expanded structures of enterovirus 71 (EV71) and coxsackievirus A16 (CVA16), but careful structural comparisons reveal significant differences in the surface-exposed loops and C termini of each capsid protein of these particles. In addition, we identified two CVA6-specific linear B-cell epitopes and mapped them to the GH loop and the C-terminal region of VP1, respectively. Collectively, our findings provide a structural basis and important antigenic information for CVA6 VLP vaccine development.
KEYWORDS: coxsackievirus A6, cryo-EM, epitope, near-atomic-resolution structure, virus-like particle
INTRODUCTION
Hand, foot, and mouth disease (HFMD) is a common contagious illness throughout the world, particularly in the Asia-Pacific region (1). In China, continuing epidemics of HFMD have resulted in millions of clinical cases and hundreds of deaths annually (2). HFMD can be caused by a number of enteroviruses, including enterovirus 71 (EV71), coxsackievirus A16 (CVA16), coxsackievirus A10 (CVA10), and coxsackievirus A6 (CVA6). Among these viruses, EV71 and CVA16 have been the major pathogens over the last decade (2, 3). However, the incidence of CVA6-associated HFMD has been increasing rapidly around the world in the past 3 years; moreover, clinical surveys reveal that CVA6 has emerged as one of the predominant causative agents of HFMD (4–9). Compared with EV71 and CVA16, CVA6 has a higher rate of infection in adults, and CVA6-infected patients present with some unique clinical manifestations, including widespread vesiculobullous eruption, desquamation, onychomadesis, and epididymitis (10–12).
In general, CVA6 does not grow efficiently in cell cultures (4, 13). Cellular receptors for CVA6 remain elusive. Like other enteroviruses, CVA6 possesses a single-stranded, positive-sense RNA genome of around 7.4 kb (14). Although CVA6 has been visualized as spherical particles of ∼30 nm in diameter by conventional electron microscopy (15), high-resolution structural information for CVA6 mature virus remains unavailable to date.
The increasing prevalence of CVA6 infection suggests that CVA6 should be targeted for vaccine development (4). Indeed, efforts have been made to develop CVA6 vaccines using the traditional inactivated whole-virus vaccine approach (15–18). Recently, our group produced recombinant virus-like particles (VLPs) of CVA6 in insect cells and yeast and evaluated their vaccine potential in preclinical tests (19, 20). The CVA6 VLPs efficiently induced serum antibodies that protected mice against lethal viral challenges (19, 20), indicating that CVA6 VLPs represent an excellent CVA6 vaccine candidate. In the present study, we determined for the first time the atomic-resolution (3.0-Å) structure of CVA6 VLPs by cryo-electron microscopy (cryo-EM) single-particle analysis. In addition, we identified two CVA6-specific conserved linear B-cell epitopes within the VP1 protein by peptide enzyme-linked immunosorbent assay (ELISA) and located these epitopes on the outer surface of the CVA6 VLP structure. Our study elucidates the structural basis of CVA6 VLPs as a strong vaccine candidate and also provides insight into the assembly and stability of CVA6 protomers.
RESULTS
Overall structure of the CVA6 VLP.
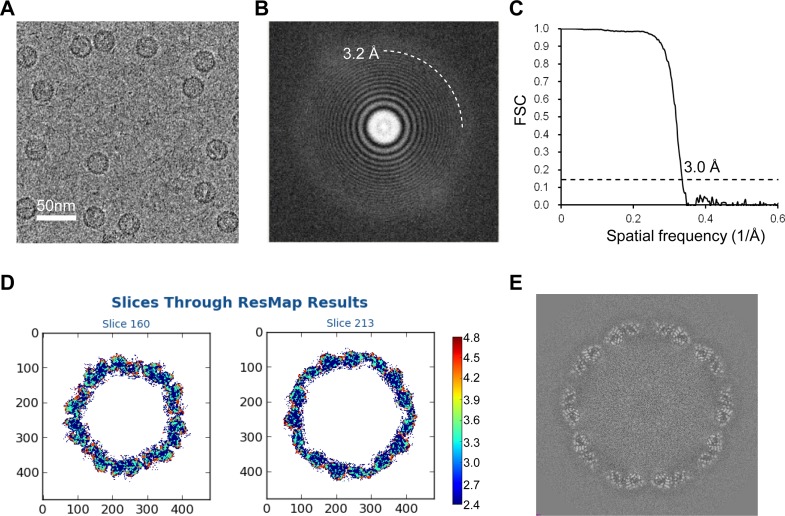
Purified CVA6 VLPs were subjected to cryo-EM analysis using a Titan Krios electron microscope (FEI) equipped with a Gatan K2 Summit camera. Cryo-EM micrographs of CVA6 VLPs revealed spherical particles of uniform size with a diameter of about 30 nm (Fig. 1A). The thon rings could be visible beyond ∼3.2-Å resolution in the two-dimensional (2D) power spectrum of the micrographs (Fig. 1B), and the majority of the data used contain high-resolution information to this level. A total of 11,596 particles were selected from 1,037 movies and used to reconstruct the 3D structure of the CVA6 VLP using the jspr software package (21). The effective resolution of the final reconstruction was estimated at 3.0 Å according to a Fourier shell correlation coefficient (FSC) cutoff criterion of 0.143 (Fig. 1C). The resolution was further confirmed by local resolution evaluation using ResMap (22), which varies between 2.6 Å for the antiparallel β-barrel core and 4.0 Å for several highly dynamic surface loops, such as the VP1 GH loop and VP2 EF loop (Fig. 1D). No density corresponding to RNA was present inside the capsid shell (Fig. 1E).
FIG 1.
Cryo-EM image of CVA6 VLP and resolution evaluation of the cryo-EM map. (A) A representative cryo-EM image of the CVA6 VLP. Bar, 50 nm. (B) Power spectrum of the micrograph shown in panel A. The white dashed curve indicates that the image contains a signal at 3.2-Å resolution. (C) Resolution assessment of the cryo-EM reconstruction by Fourier shell correlation (FSC) at a criterion of 0.143. (D) Local resolution evaluation of the cryo-EM map by ResMap. The resolution color bar (in angstroms) is also labeled. (E) Central section of the cryo-EM map visualized along the 5-fold symmetry axis.
Generally, enterovirus particles have two different states, the expanded state and the native/compact state, with the radii of the former being slightly larger than those of the latter (23–26). The radius of the CVA6 VLP is 161 Å (Fig. 2A), which is similar to those of the expanded particles of EV71 and CVA16 (e.g., 159 Å for the EV71 procapsid, 161 Å for the EV71 VLP, and 162 Å for the CVA16 135S particle) and is slightly larger than those of the compact particles of EV71 and CVA16 (e.g., 155 Å for the EV71 mature virion, 156 Å for the CVA16 mature virion, and 156 Å for the CVA16 VLP) (23, 25–27), suggesting that the CVA6 VLP is likely in an expanded state. However, since no CVA6 mature virus structure is currently available, it is impossible to directly compare the radius of the CVA6 VLP with that of the CVA6 mature virion.
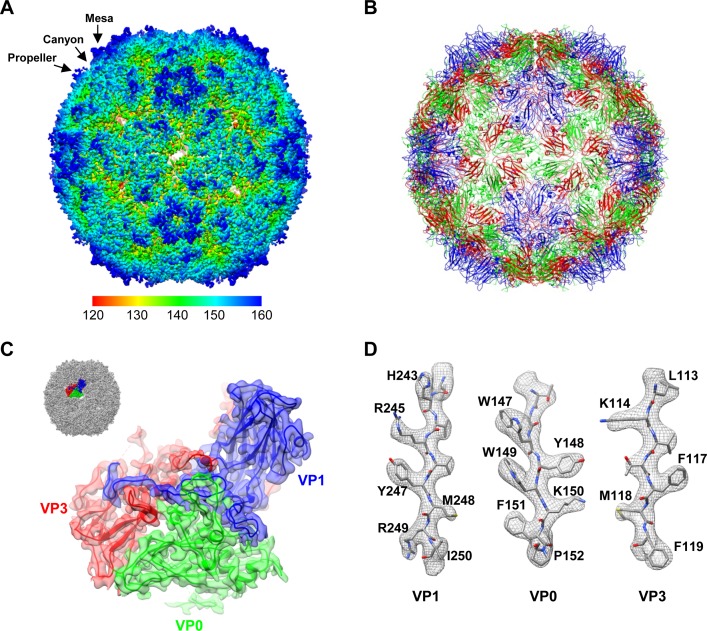
FIG 2.
Overall structure of the CVA6 VLP. (A) Cryo-EM density map of the CVA6 VLP viewed along the icosahedral 2-fold symmetry axis. The map is colored radially. The color bar labels the corresponding radius from the center of the sphere (in angstroms). The mesa, propeller, and canyon are indicated by black arrows. (B) Atomic model of the CVA6 VLP viewed along the 2-fold axis. VP0, VP1, and VP3 are colored in green, blue, and red, respectively. The same color scheme is used throughout, unless otherwise indicated. (C) Good fit between the model and the cryo-EM map at an icosahedral asymmetric unit of the CVA6 VLP. The visualization location with respect to the complete CVA6 VLP map is illustrated in the upper left corner. (D) Good fit between the segmented density (mesh in gray) and the corresponding atomic model (sticks) in the VP1, VP0, and VP3 regions. The well-resolved density for almost all the side chains demonstrates the atomic resolution of the cryo-EM map.
The density map of the CVA6 VLP revealed an icosahedral structure with surface features typical for enteroviruses, including a prominent star-shaped plateau (mesa) at the 5-fold-symmetry axes, a narrow depression (canyon) surrounding the mesa, and a three-bladed propeller-like feature at the 3-fold axes (Fig. 2A). In addition, there is an open channel at the 2-fold axes (Fig. 2A), a typical characteristic of the expanded enteroviral particles. We then built an atomic model of the CVA6 VLP (Fig. 2B) based on the homologous EV71 VLP crystal structure (PDB 4YVS) (27), which fits our cryo-EM map very well after further refinement (Fig. 2C and D). According to the atomic model, the CVA6 VLP capsid contains 60 protomers, each composed of three subunit proteins (VP0, VP1, and VP3) (Fig. 2C). Residues 83 to 92, 99 to 115, and 121 to 321 of VP0, residues 71 to 296 of VP1, and residues 1 to 175 and 188 to 240 of VP3 were well resolved, and almost all side chain densities of these residues were clearly visible in our atomic-resolution cryo-EM map (Fig. 2C and D).
Structural comparison of the CVA6 VLP, EV71 VLP, and CVA16 135S particle.
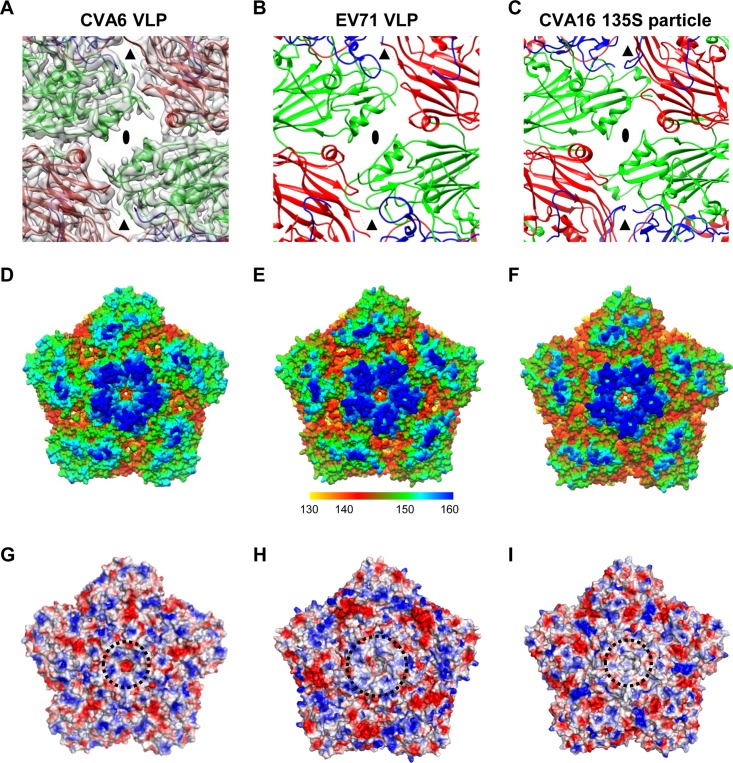
We compared the structure of the CVA6 VLP with those of the EV71 VLP (PDB 4YVS) (27) and CVA16 135S particle (PDB 4JGY) (26). In the CVA6 VLP structure, the 2-fold axis channel is opened, flanked by two α-helices from adjacent VP2 proteins, and measured as about 8 by 26 Å; a smaller channel at the quasi-3-fold axis (called the junction channel) is also evident (Fig. 3A). The two opened channels, typical features of the expanded enteroviral particles, are also clearly seen in the structures of the EV71 VLP and CVA16 135S particle (Fig. 3B and C). In contrast, there are no holes at the 2-fold and quasi-3-fold axes in the structures of the native EV71 and CVA16 mature virions (PDB 3VBF and 5C4W, respectively) (23, 25).
FIG 3.
Surface features of the CVA6 VLP compared to those of the EV71 VLP and CVA16 135S particle. (A) A map and model overlay viewed along the 2-fold symmetry axis (marked as a black ellipse) for the CVA6 VLP. The position of the junction channel is indicated by a black triangle. The density map is shown in transparent gray. (B and C) Atomic models of the outer surfaces of the EV71 VLP (PDB 4YVS) (B) and the CVA16 135S particle (PDB 4JGY) (C) viewed in the same direction as in panel A. (D to F) Surface representation of the pentameric structures of the CVA6 VLP (D), EV71 VLP (E), and CVA16 135S particle (F) viewed along the 5-fold symmetry axis. The surface is radially colored. The color bar labels the corresponding radius from the center of the particle (in angstroms). (G to I) Electrostatic surfaces of pentamers of the CVA6 VLP (G), EV71 VLP (H), and CVA16 135S particle (I). The electrostatic potential was calculated by utilizing PyMOL; the positively and negatively charged areas are shown in blue and red, respectively. The black dashed circle indicates the 5-fold positively charged patch.
A surface canyon surrounding each of the 12 5-fold vertices is a typical structural feature of enteroviruses and is often the site of host cell receptor binding (28). The canyon in the CVA6 VLP appears to be much narrower and shallower than those in the EV71 VLP and in the CVA16 135S particle (Fig. 3D to F). However, the 5-fold plateau of the CVA6 VLP is similar to those of the EV71 VLP and CVA16 135S particle in height (Fig. 3D to F). For all three particles, positively charged surface patches on the 5-fold plateaus are formed; however, the patch in the CVA6 VLP is more compact and closer to the 5-fold axis than that in the EV71 VLP, and it has much stronger positive charges than that in the CVA16 135S particle (Fig. 3G to I).
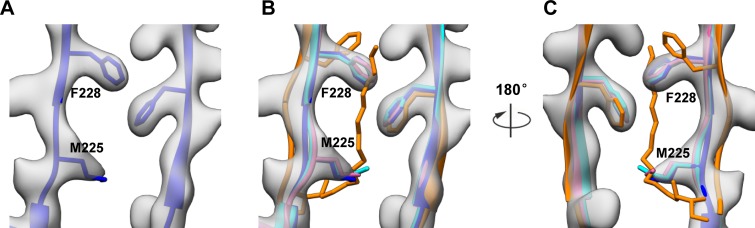
For enteroviruses, a hydrophobic pocket, formed by the VP1 protein and located beneath the canyon floor, often harbors a host-derived lipidic pocket factor, which functions to stabilize virions. Upon binding cellular receptors, enteroviruses may expel their corresponding pocket factors, leading to pocket collapse and subsequent uncoating process (23, 26, 29). Previous studies have demonstrated that the VP1 pocket is empty in the EV71 VLP and CVA16 135S particle (26, 27), whereas it is occupied by a pocket factor in EV71 and CVA16 mature virions (23, 25). In our CVA6 VLP structure, the VP1 pocket is collapsed and empty, and the side chains of residues M225 and F228 of VP1 project inward, partially occupying the potential pocket factor-binding site (Fig. 4).
FIG 4.
Structural comparison of hydrophobic pockets among the CVA6 VLP, EV71 VLP, CVA16 135S particle, and EV71 mature virion. (A) The hydrophobic pocket in VP1 of the CVA6 VLP, with density in transparent gray and model in blue ribbon and stick. (B and C) Superposition of the VP1 pocket regions of the CVA6 VLP (blue), EV71 VLP (pink), CVA16 135S particle (cyan), and EV71 mature virion (PDB 3VBF) (orange) viewed from the front (B) and back (C). The pocket factor in the EV71 mature virion is shown as orange sticks. Shown also are residues M225 and F228 in the CVA6 VLP, whose side chains partially occupy the potential binding site for pocket factor.
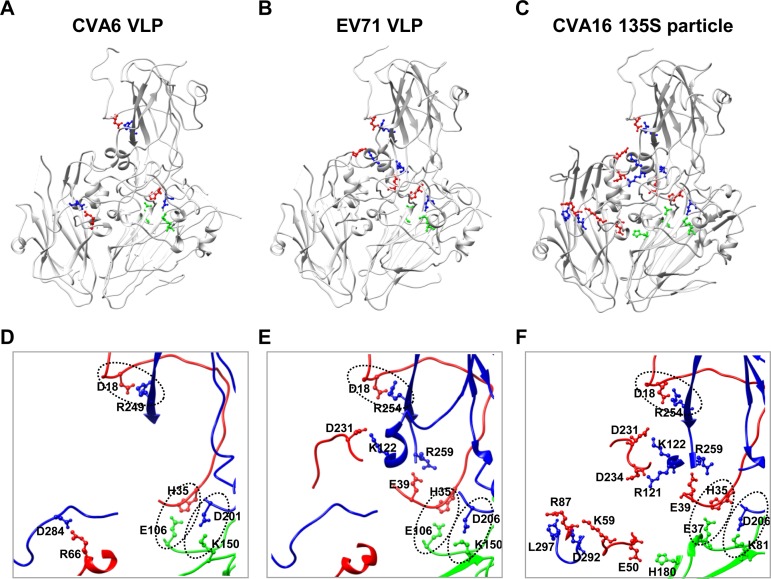
To explore the mechanisms of VLP assembly and stability, we analyzed interactions among the subunit proteins of CVA6 VLP by using the PISA server (30). The results showed that protein-protein interactions exist mainly within, but not between, protomers (data not shown). Apart from numerous hydrogen bonds, four salt bridges are formed between charged amino acids at protein-protein interfaces within a protomer. Specifically, D201 of VP1 interacts with K150 of VP0, R249 of VP1 with D18 of VP3, D284 of VP1 with R66 of VP3, and E106 of VP0 with H35 of VP3 (Fig. 5A and D). It is worth noting that except for the salt bridge between D284 of VP1 and R66 of VP3, the salt bridges are common among the CVA6 VLP, EV71 VLP, and CVA16 135S particle (Fig. 5), as well as mature virions of EV71 and CVA16 (PDB 3VBF and 5C4W, respectively) (23, 25) (data not shown). This finding is consistent with the substantial sequence conservation of these salt bridge-forming residues among the three viruses (data not shown).
FIG 5.
Salt bridges within the protomers of the CVA6 VLP, EV71 VLP, and CVA16 135S particle. (A to C) Salt bridge locations within the protomers of the CVA6 VLP (A), EV71 VLP (B), and CVA16 135S particle (C). Salt bridge-forming residues within VP1, VP0/VP2 (VP0 for the CVA6 VLP and EV71 VLP and VP2 for the CVA16 135S particle), and VP3 are colored in blue, green, and red, respectively, while the other portions of the proteins are in gray. (D to F) Zoom-in view of the salt bridge regions enlarged from panels A, B, and C, respectively. The three common salt bridges observed in the CVA6 VLP, EV71 VLP, and CVA16 135S particle are indicated by oval dashed lines.
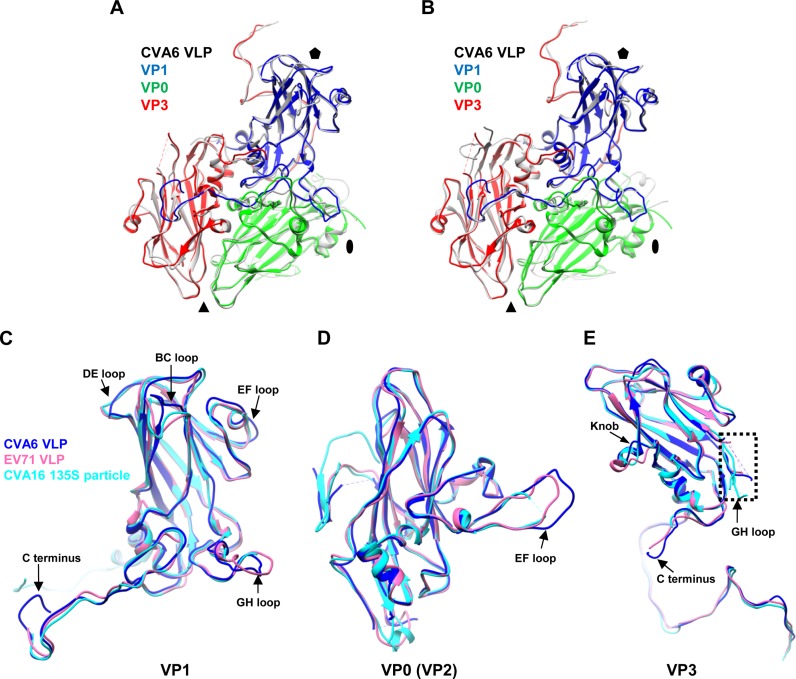
In general, the structures of entire protomers and individual VP1, VP0, and VP3 proteins are similar among the CVA6 VLP, EV71 VLP, and CVA16 135S particle, and each capsid protein consists of an eight-stranded antiparallel β-barrel core (Fig. 6). The overall Cα root mean square deviation (RMSD) values for VP1, VP0 (VP2), and VP3 among the CVA6 VLP, EV71 VLP, and CVA16 135S particle are all less than 1.40 Å, which were calculated by optimizing the overlay of each VP1 of the three structures, optimizing the overlay of each VP0 (VP2) of the three structures, and optimizing the overlay of each VP3 of the three structures separately. It is noteworthy that the main structural differences among the three particles were found in the loop regions and the C termini of subunit proteins, including the BC, DE, and EF loops (near the 5-fold axis), the GH loop (near the pseudo-3-fold junction), and the C terminus of VP1, the EF loop (near the Southern edge of canyon) of VP0 (VP2), and the GH loop (near the pseudo-3-fold junction), the knob (residues 55 to 69 of VP3, VP3/VP0 interface), and the C terminus (VP3/VP1 interface) of VP3 (Fig. 6). Obvious structural differences in these regions were also observed when the CVA6 VLP was compared to the EV71 and CVA16 mature virions (PDB 3VBF and 5C4W, respectively) (23, 25) (data not shown). It is worth noting that these regions are all surface exposed, and most of them were experimentally determined antigenic sites of EV71 or CVA16 (31–37).
FIG 6.
Structural comparison of the protomers and individual capsid proteins of the CVA6 VLP, EV71 VLP, and CVA16 135S particle. (A) Superposition of the protomers of the CVA6 VLP (different capsid proteins in different colors) and EV71 VLP (gray). The black oval, triangle, and pentagon represent the 2-fold, 3-fold, and 5-fold axes, respectively. (B) Superposition of the protomers of the CVA6 VLP and CVA16 135S particle (gray). VP1, VP0, and VP3 of CVA6 VLP are in blue, green, and red, respectively. (C to E) Superposition of individual capsid protein structures of the CVA6 VLP (blue), EV71 VLP (pink), and CVA16 135S particle (cyan). (C) Superposition of VP1. (D) Superposition of VP0/VP2 (VP0 for the CVA6 VLP and EV71 VLP and VP2 for the CVA16 135S particle). (E) Superposition of VP3. The major structural differences of subunit proteins among the CVA6 VLP, EV71 VLP, and CVA16 135S particle are indicated by black arrows. The dashed rectangle indicates the disordered region of the VP3 GH loop.
Mapping of CVA6-specific linear B-cell epitopes within the VP1 protein.
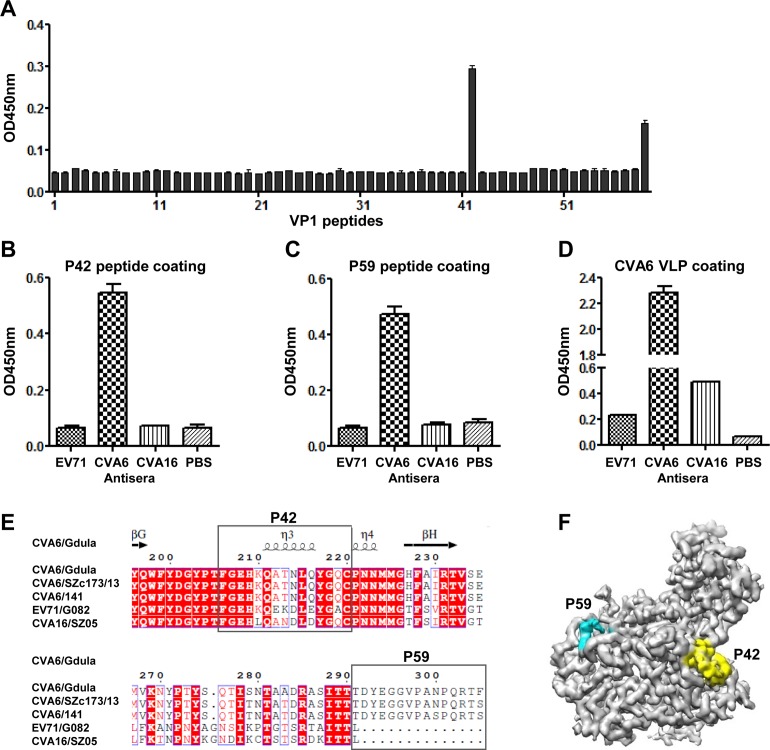
CVA6 VLPs are able to potently elicit protective antibodies in mice (19, 20). To identify linear B-cell epitopes, a panel of 59 synthetic peptides (designated P1 to P59) covering the entire VP1 region of CVA6 was tested for reaction with the anti-CVA6 VLP mouse serum by ELISA. As shown in Fig. 7A, only the peptides P42 and P59, which correspond to residues 206 to 220 and 291 to 305 of VP1, respectively, exhibited significant reactivity, suggesting that P42 and P59 contain linear B-cell epitopes. We next examined whether P42 and P59 could cross-react with the anti-EV71 VLP or anti-CVA16 VLP mouse serum generated in a previous study (38). The ELISA results clearly showed that P42 and P59 reacted with the anti-CVA6 VLP but not the anti-EV71 VLP or anti-CVA16 VLP serum (Fig. 7B and C), indicating that the P42 and P59 epitopes are CVA6 specific. As the control binding antigen, CVA6 VLP exhibited different degrees of cross-reactivity with anti-EV71 VLP or anti-CVA16 VLP serum (Fig. 7D), suggesting that there might be common epitopes for CVA6, EV71, and CVA16.
FIG 7.
Identification and mapping of linear B-cell epitopes within the VP1 protein of CVA6. (A) Reactivity of the anti-CVA6 VLP mouse serum to VP1 peptides determined by peptide ELISA. A set of 59 synthetic peptides covering the entire VP1 region of CVA6 were used as the coating antigens. Mouse anti-CVA6 VLP serum was diluted 1:40 and used in ELISA. (B to D) Cross-reactivities of peptides P42 (B), P59 (C), and CVA6 VLP (D) with mouse polyclonal antibodies against the EV71 VLP or CVA16 VLP determined by ELISA. Anti-CVA6 VLP and anti-PBS mouse sera were used as positive and negative controls, respectively. The antisera were diluted 1:40 (B and C) and 1:100 (D) and used in ELISA. (E) Sequence alignment of VP1 from some representative strains of CVA6, EV71, and CVA16, showing the amino acid sequence variations in the corresponding P42 and P59 regions. The secondary-structure elements for the CVA6 VLP are shown at the top. (F) Locations of the P42 and P59 epitopes on the protomer of the CVA6 VLP. The P42 and P59 epitopes are colored in yellow and cyan, respectively. To better show the densities of P42 and P59, the map was low-pass filtered to 3.5-Å resolution. The electron densities of the nine C-terminal amino acids of P59 remain missing.
Alignment of the VP1 sequences from representative strains of CVA6, EV71, and CVA16 revealed that the P59 region is uniquely present in CVA6 but absent in EV71 and CVA16 (Fig. 7E). In addition, the P42 regions from the three CVA6 strains are identical, while they differ from the counterparts in EV71 and CVA16 by at least four residue variations (Fig. 7E). Based on our structure, P42 is located in the VP1 GH loop at the edge of the canyon and is well exposed on the surface of the CVA6 VLP (Fig. 7F). In addition, P59, which resides in the C terminus of VP1, is also surface exposed (Fig. 7F). Taken together, the above results demonstrate that P42 and P59 represent two CVA6-specific surface-exposed linear B-cell epitopes.
DISCUSSION
CVA6 has recently emerged as one of the predominant pathogens of HFMD (4–9). However, thus far, no vaccine is available for CVA6, and its mature virus structure has not been reported yet. We now alleviate these problems by providing the first atomic-resolution structure of the CVA6 VLP, which has been shown to provide protection against lethal CVA6 infection in preclinical studies (19, 20).
In this study, we found that the overall structure of CVA6 VLP is very similar to those of the expanded EV71 VLP and CVA16 135S particle but differs from those of the native/compact EV71 and CVA16 mature virions in two major aspects. First, the 2-fold axis channel and the junction channel are opened in the CVA6 VLP, EV71 VLP, and CVA16 135S particle (Fig. 3A to C) but closed in EV71 and CVA16 mature virions (23, 25). Second, the VP1 pocket is empty and collapsed in the CVA6 VLP, EV71 VLP, and CVA16 135S particle but is occupied by a pocket factor in EV71 and CVA16 mature virions (Fig. 4) (23, 25). Enlargement of the 2-fold axis channel and loss of the pocket factor are considered two characteristic features of particle expansion which occurs during the mature virion-to-uncoating intermediate transition (26, 39). In addition, the radius of the CVA6 VLP (Fig. 2A) is similar to those of the EV71 VLP and CVA16 135S particle and is slightly larger than those of EV71 and CVA16 mature virions (23, 25–27). Taken together, these findings indicate that the CVA6 VLP most probably adopts an expanded conformation compared to its mature virion.
The present study reveals that the CVA6 VLP consists of 60 protomers, each of which is composed of VP0 (the precursor of VP2 and VP4), VP1, and VP3 subunits. Similar findings have been reported for mature virions, procapsids, and VLPs of other enteroviruses (e.g., EV71 and CVA16) (23, 25, 27, 40). However, the mechanisms underlying the assembly of protomers and particles of enteroviruses remain unclear. A few studies have revealed the critical contributions of salt bridges to particle assembly and/or stability in bacteriophages, influenza virus, HIV-1, and Mason-Pfizer monkey virus (41–46). In this study, four salt bridges located at protein-protein interfaces were identified within each protomer of the CVA6 VLP (Fig. 5A and D). It is worth noting that two of them (salt bridges between R249 of VP1 and D18 of VP3 and between E106 of VP0 and H35 of VP3) also exist inside protomers of other representative viruses from the four species (A, B, C and D) of human enteroviruses, such as EV71 (PDB 3VBF, species A), CVA16 (PDB 5C4W, species A), coxsackievirus B3 (CVB3) (PDB 1COV, species B), poliovirus type 1 (PDB 1VBD, species C), poliovirus type 2 (PDB 1EAH, species C), poliovirus type 3 (PDB 1PVC, species C), and enterovirus D68 (EVD68) (PDB 4WM8, species D) (data not shown), implying that the two highly conserved salt bridges may play a critical role in the assembly and stability of enteroviral protomers.
Although the CVA6 VLP, EV71 VLP, and CVA16 135S particle share a broadly similar overall architecture, there are apparent structural differences in the surface-exposed loops and C termini of their subunit proteins (Fig. 6). Significant structural differences were also found in these regions when the CVA6 VLP structure was compared with those of EV71 and CVA16 mature virions (data not shown). It is noteworthy that some of these regions are experimentally determined antigenic sites of EV71 or CVA16 (31–36). Particularly, the EF and GH loops of VP1, the EF loop of VP2, and the knob and GH loop of VP3 in EV71 were identified to be neutralizing epitopes (31–34), whereas the BC, EF, and GH loops of VP1 are neutralizing epitopes for CVA16 (35). It is reasonable for us to assume that most, if not all, of these regions are also antigenic sites for CVA6. Thus, the differences in these antigenic sites may partially explain the lack of cross-protection by inactivated EV71, CVA16, or CVA6 monovalent vaccines in preclinical trials (17).
We and other research groups have shown that the EV71 procapsid, EV71 VLP, and CVA6 VLP, all of which are in the expanded state (23, 27), could potently elicit the production of neutralizing/protective antibodies in mice and/or monkeys (19, 20, 47–49). These observations are in contrast to previous results that the expanded particles (C or H antigen) of polioviruses could not efficiently induce neutralizing antibody responses in mice (50, 51). It is possible that, unlike in polioviruses, the expanded forms of EV71 and CVA6 may retain their respective immunodominant neutralization epitopes (47).
By screening of a panel of peptides covering the entire VP1 region for reactivity with the anti-CVA6 VLP serum, we identified two CVA6-specific linear B-cell epitopes, represented by the peptides P42 and P59, respectively (Fig. 7A). The P42 and P59 epitopes are located at the GH loop and the C terminus of the CVA6 VP1, respectively (Fig. 7), in agreement with the structure-based epitope prediction. Notably, the P42 epitope (residues 206 to 220) overlaps the EV71 neutralizing epitope SP70 (residues 208 to 222) (31) and the CVA16 neutralizing epitope PEP71 (residues 211 to 225) (35). Thus, it is very likely that P42 also constitutes a neutralizing epitope for CVA6. Another interesting finding is that the P59 region is present in CVA6 but is absent in EV71 and CVA16. Whether P42 and P59 are capable of inducing neutralizing and/or protective antibodies remains to be determined. Nonetheless, the identification of P42 and P59 as CVA6-specific epitopes should have important implications for development of diagnostics and vaccines. For example, the epitopes can be used to design and generate reagents (either antigen or antibody) for assembly of diagnostic kits and for quality control in CVA6 vaccine development.
In summary, our study unravels the structure and unique antigenic sites of the CVA6 VLP, thereby greatly facilitating the development of VLP-based CVA6 vaccines and also enhancing our understanding of the protomer assembly of enteroviruses.
MATERIALS AND METHODS
Expression and purification of CVA6 VLPs.
CVA6 VLPs were produced by infecting Sf9 insect cells with a recombinant baculovirus (Bac-CVA6-P1/3CD) coexpressing P1 (structural protein precursor) and 3CD (protease) derived from the prototype CVA6 strain Gdula (GenBank accession no. AY421764) as described previously (19). The VLP purification was performed as described previously (19).
Cryo-EM imaging.
A 2-μl drop of freshly prepared CVA6 VLP sample was placed onto a glow-discharged Quantifoil holey carbon grid (R1.2x1.3, 200 mesh; Quantifoil Micro Tools). The grid was blotted briefly (1 to 2 s) with filter paper and then quickly plunge-frozen in liquid nitrogen-cooled liquid ethane using an FEI Mark IV Vitrobot. The frozen-hydrated sample was subsequently imaged in an FEI Titan Krios transmission electron microscope operated at 300 kV and equipped with a Cs corrector. Images were collected by using a Gatan K2 Summit direct electron detector in superresolution mode with a pixel size of 0.41 Å. Each movie was dose fractioned into 42 frames. The exposure time was set to be 5.04 s with 0.12 s for each frame, producing a total dose of ∼42 e−/Å2. Defocus values for this data set varied from −0.7 to −1.5 μm. All the images were collected by utilizing the SerialEM automated data collection software package (52).
Cryo-EM single-particle 3D reconstruction.
All 42 frames for each movie were aligned and summed into a single micrograph using Motioncorr (53). A total of 11,596 CVA6 VLPs were semiautomatically selected using the e2boxer.py program in EMAN2.1 (54). Contrast transfer function (CTF) parameters were determined using the fitctf2.py program in the jspr package (21) and then manually adjusted using the EMAN1.9 ctfit program (55). The whole data set was randomly divided into two halves, and each part was independently subjected to reference-free 2D analysis and initial model building using the e2initialmodel.py program in EMAN2.1 (54), and then, the gold-standard 3D reconstruction was performed using jspr (21), including refinements on euler/center, defocus/scale, astigmatism, beamtilt, and anisoscale (56). The effective resolution of our density map was determined using the gold-standard FSC 0.143 cutoff criterion (57). The local resolution was further assessed using ResMap (22).
Model building.
The X-ray crystal structure of the VLP of EV71 (PDB 4YVS), homologous to that of CVA6, was used to build the homology model of the CVA6 VLP through the SWISS-MODEL webserver (58). The well-resolved side chain densities throughout our CVA6 VLP map enabled us to amend and refine the entire atomic model using COOT (59), followed by further refinement using the phenix.real_space_refine program in Phenix (60). The final atomic model was validated using phenix.molprobity (61) and phenix.emringer (62). The validation statistics for the CVA6 VLP atomic model are summarized in Table 1. Figures were generated with either UCSF Chimera (63) or PyMOL (http://www.pymol.org).
TABLE 1.
Cryo-EM data collection and refinement statistics
| Parameter | Value for CVA6 VLP |
|---|---|
| Data collection | |
| EM equipment | Titan Krios |
| Voltage (kV) | 300 |
| Detector | K2 Summit |
| Pixel size (Å) | 0.41 |
| Electron dose (e−/Å2) | 42 |
| Exposure time (s) | 5.04 |
| Frames (no.) | 42 |
| Defocus range (μm) | −0.7 to −1.5 |
| Reconstruction | |
| Software | jspr |
| Raw micrographs (no.) | 1,037 |
| Final particles (no.) | 11,596 |
| Final resolution (Å) | 3.0 |
| Atomic modeling | |
| Software | Phenix and COOT |
| Phenix real-space refinement | |
| Map CC | |
| Whole unit cell | 0.578 |
| Around atoms | 0.849 |
| Avg B factor (Å2) | −126.35 |
| MolProbity | |
| Ramachandran plot (%) | |
| Favored | 94.03 |
| Allowed | 5.67 |
| Outliers | 0.30 |
| Rotamer outliers (%) | 0.0 |
| Cβ deviations | 0 |
| Clashscore | 10.66 |
| RMSD | |
| Bonds | 0.0096 |
| Angles | 1.22 |
| MolProbity score | 1.92 |
| EMRinger score | 5.40 |
ELISA.
A set of 59 peptides spanning the entire amino acid sequence of the VP1 protein of the prototype CVA6 strain Gdula was synthesized by GL Biochem (Shanghai, China). Each peptide was composed of 15 amino acid residues, with 10 residues overlapping with the adjacent peptides. ELISA plates were coated with 400 ng/well of individual peptide in phosphate-buffered saline (PBS) and incubated overnight at 4°C. After three washes with PBS-Tween (PBST), plates were blocked with PBST containing 5% milk at 37°C for 1 h; after washes, 50 μl/well of mouse anti-CVA6 VLP serum (19) (1:40 dilution) was added and incubated at 37°C for 2 h; and after another washing step, horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (1:5,000 dilution; Sigma) was added and incubated at 37°C for 1 h. After color development, the absorbance was measured at 450 nm with a microtiter plate reader. Two selected VP1 peptides were further tested for cross-reactivities with anti-EV71 VLP and anti-CVA16 VLP mouse sera (38) by ELISA as described above except that anti-EV71 VLP and anti-CVA16 VLP mouse sera were used as the primary detection antibodies. In addition, CVA6 VLPs (19) were tested for cross-reactivities with anti-EV71 VLP and anti-CVA16 VLP mouse sera by ELISA as described above except that ELISA plates were coated with CVA6 VLPs (30 ng/well) and anti-EV71 VLP and anti-CVA16 VLP mouse sera were diluted 1:100.
Sequence alignment.
The enterovirus strains used in the analysis were CVA6 Gdula (GenBank accession no. AY421764), CVA6/SZc173/13 (KF682362), CVA6/141 (KR706309), EV71/G082 (64), and CVA16/SZ05 (EU262658). The VP1 protein sequences of these strains were aligned using ESPript (65).
Accession number(s).
The final cryo-EM density map of the CVA6 VLP has been deposited in the Electron Microscopy Data Bank (EMDB) under accession code EMD-6829. The atomic model of CVA6 VLP has been deposited in the Protein Data Bank (PDB) under accession code 5YHQ.
ACKNOWLEDGMENTS
We thank Liangliang Kong and Junrui Li from the Electron Microscopy Facility and staff from the Data Base and Computation Facility of the National Center for Protein Science Shanghai for their assistance with the EM instruments and the parallel computing.
This work was supported by grants from the National Key R&D Program of China (2017YFA0503503), CAS Pilot Strategic Science and Technology Projects B (XDB08030201), the National Natural Science Foundation of China (31370930, 31670754, and 31500153), the Science and Technology Commission of Shanghai Municipality (13431900600 and 15XD1524900), the CAS-Shanghai Science Research Center (CAS-SSRC-YH-2015-01), the TOTAL Foundation, and the State Key Laboratory of Molecular Biology, Shanghai Institute of Biochemistry and Cell Biology, CAS.
ADDENDUM
While our paper was under review, the structures of the CVA6 procapsid and A particle were published (66).
REFERENCES
- 1.Repass GL, Palmer WC, Stancampiano FF. 2014. Hand, foot, and mouth disease: identifying and managing an acute viral syndrome. Cleve Clin J Med 81:537–543. doi: 10.3949/ccjm.81a.13132. [DOI] [PubMed] [Google Scholar]
- 2.Xing W, Liao Q, Viboud C, Zhang J, Sun J, Wu JT, Chang Z, Liu F, Fang VJ, Zheng Y, Cowling BJ, Varma JK, Farrar JJ, Leung GM, Yu H. 2014. Hand, foot, and mouth disease in China, 2008-12: an epidemiological study. Lancet Infect Dis 14:308–318. doi: 10.1016/S1473-3099(13)70342-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong SS, Yip CC, Lau SK, Yuen KY. 2010. Human enterovirus 71 and hand, foot and mouth disease. Epidemiol Infect 138:1071–1089. doi: 10.1017/S0950268809991555. [DOI] [PubMed] [Google Scholar]
- 4.Bian L, Wang Y, Yao X, Mao Q, Xu M, Liang Z. 2015. Coxsackievirus A6: a new emerging pathogen causing hand, foot and mouth disease outbreaks worldwide. Expert Rev Anti Infect Ther 13:1061–1071. doi: 10.1586/14787210.2015.1058156. [DOI] [PubMed] [Google Scholar]
- 5.Osterback R, Vuorinen T, Linna M, Susi P, Hyypia T, Waris M. 2009. Coxsackievirus A6 and hand, foot, and mouth disease, Finland. Emerg Infect Dis 15:1485–1488. doi: 10.3201/eid1509.090438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabrerizo M, Tarrago D, Munoz-Almagro C, Del Amo E, Dominguez-Gil M, Eiros JM, Lopez-Miragaya I, Perez C, Reina J, Otero A, Gonzalez I, Echevarria JE, Trallero G. 2014. Molecular epidemiology of enterovirus 71, coxsackievirus A16 and A6 associated with hand, foot and mouth disease in Spain. Clin Microbiol Infect 20:O150–O156. doi: 10.1111/1469-0691.12361. [DOI] [PubMed] [Google Scholar]
- 7.Wei SH, Huang YP, Liu MC, Tsou TP, Lin HC, Lin TL, Tsai CY, Chao YN, Chang LY, Hsu CM. 2011. An outbreak of coxsackievirus A6 hand, foot, and mouth disease associated with onychomadesis in Taiwan, 2010. BMC Infect Dis 11:346. doi: 10.1186/1471-2334-11-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujimoto T, Iizuka S, Enomoto M, Abe K, Yamashita K, Hanaoka N, Okabe N, Yoshida H, Yasui Y, Kobayashi M, Fujii Y, Tanaka H, Yamamoto M, Shimizu H. 2012. Hand, foot, and mouth disease caused by coxsackievirus A6, Japan, 2011. Emerg Infect Dis 18:337–339. doi: 10.3201/eid1802.111147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han JF, Xu S, Zhang Y, Zhu SY, Wu DL, Yang XD, Liu H, Sun BX, Wu XY, Qin CF. 2014. Hand, foot, and mouth disease outbreak caused by coxsackievirus A6, China, 2013. J Infect 69:303–305. doi: 10.1016/j.jinf.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 10.Laga AC, Shroba SM, Hanna J. 2016. Atypical hand, foot and mouth disease in adults associated with coxsackievirus A6: a clinico-pathologic study. J Cutan Pathol 43:940–945. doi: 10.1111/cup.12775. [DOI] [PubMed] [Google Scholar]
- 11.Ben-Chetrit E, Wiener-Well Y, Shulman LM, Cohen MJ, Elinav H, Sofer D, Feldman I, Marva E, Wolf DG. 2014. Coxsackievirus A6-related hand foot and mouth disease: skin manifestations in a cluster of adult patients. J Clin Virol 59:201–203. doi: 10.1016/j.jcv.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 12.Lott JP, Liu K, Landry ML, Nix WA, Oberste MS, Bolognia J, King B. 2013. Atypical hand-foot-and-mouth disease associated with coxsackievirus A6 infection. J Am Acad Dermatol 69:736–741. doi: 10.1016/j.jaad.2013.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt NJ, Ho HH, Lennette EH. 1975. Propagation and isolation of group A coxsackieviruses in RD cells. J Clin Microbiol 2:183–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang L, Li S, Liu Y, Hou W, Lin Q, Zhao H, Xu L, He D, Ye X, Zhu H, Cheng T, Xia N. 2015. Construction and characterization of an infectious clone of coxsackievirus A6 that showed high virulence in neonatal mice. Virus Res 210:165–168. doi: 10.1016/j.virusres.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Liu CC, Guo MS, Wu SR, Lin HY, Yang YT, Liu WC, Chow YH, Shieh DB, Wang JR, Chong P. 2016. Immunological and biochemical characterizations of coxsackievirus A6 and A10 viral particles. Antiviral Res 129:58–66. doi: 10.1016/j.antiviral.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z, Dong Z, Wei Q, Carr MJ, Li J, Ding S, Tong Y, Li D, Shi W. 13 April 2017. A neonatal murine model of coxsackievirus A6 infection for evaluation of antiviral and vaccine efficacy. J Virol doi: 10.1128/JVI.02450-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caine EA, Fuchs J, Das SC, Partidos CD, Osorio JE. 2015. Efficacy of a trivalent hand, foot, and mouth disease vaccine against enterovirus 71 and coxsackieviruses A16 and A6 in mice. Viruses 7:5919–5932. doi: 10.3390/v7112916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang L, Mao Q, Li S, Gao F, Zhao H, Liu Y, Wan J, Ye X, Xia N, Cheng T, Liang Z. 2016. A neonatal mouse model for the evaluation of antibodies and vaccines against coxsackievirus A6. Antiviral Res 134:50–57. doi: 10.1016/j.antiviral.2016.08.025. [DOI] [PubMed] [Google Scholar]
- 19.Shen C, Ku Z, Zhou Y, Li D, Wang L, Lan K, Liu Q, Huang Z. 2016. Virus-like particle-based vaccine against coxsackievirus A6 protects mice against lethal infections. Vaccine 34:4025–4031. doi: 10.1016/j.vaccine.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Y, Shen C, Zhang C, Zhang W, Wang L, Lan K, Liu Q, Huang Z. 2016. Yeast-produced recombinant virus-like particles of coxsackievirus A6 elicited protective antibodies in mice. Antiviral Res 132:165–169. doi: 10.1016/j.antiviral.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 21.Guo F, Jiang W. 2014. Single particle cryo-electron microscopy and 3-D reconstruction of viruses. Methods Mol Biol 1117:401–443. doi: 10.1007/978-1-62703-776-1_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kucukelbir A, Sigworth FJ, Tagare HD. 2014. Quantifying the local resolution of cryo-EM density maps. Nat Methods 11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Peng W, Ren J, Hu Z, Xu J, Lou Z, Li X, Yin W, Shen X, Porta C, Walter TS, Evans G, Axford D, Owen R, Rowlands DJ, Wang J, Stuart DI, Fry EE, Rao Z. 2012. A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nat Struct Mol Biol 19:424–429. doi: 10.1038/nsmb.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cifuente JO, Lee H, Yoder JD, Shingler KL, Carnegie MS, Yoder JL, Ashley RE, Makhov AM, Conway JF, Hafenstein S. 2013. Structures of the procapsid and mature virion of enterovirus 71 strain 1095. J Virol 87:7637–7645. doi: 10.1128/JVI.03519-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren J, Wang X, Zhu L, Hu Z, Gao Q, Yang P, Li X, Wang J, Shen X, Fry EE, Rao Z, Stuart DI. 2015. Structures of coxsackievirus A16 capsids with native antigenicity: implications for particle expansion, receptor binding, and immunogenicity. J Virol 89:10500–10511. doi: 10.1128/JVI.01102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren J, Wang X, Hu Z, Gao Q, Sun Y, Li X, Porta C, Walter TS, Gilbert RJ, Zhao Y, Axford D, Williams M, McAuley K, Rowlands DJ, Yin W, Wang J, Stuart DI, Rao Z, Fry EE. 2013. Picornavirus uncoating intermediate captured in atomic detail. Nat Commun 4:1929. doi: 10.1038/ncomms2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyu K, He YL, Li HY, Chen R. 2015. Crystal structures of yeast-produced enterovirus 71 and enterovirus 71/coxsackievirus A16 chimeric virus-like particles provide the structural basis for novel vaccine design against hand-foot-and-mouth disease. J Virol 89:6196–6208. doi: 10.1128/JVI.00422-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rossmann MG, He Y, Kuhn RJ. 2002. Picornavirus-receptor interactions. Trends Microbiol 10:324–331. doi: 10.1016/S0966-842X(02)02383-1. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Sheng J, Baggen J, Meng G, Xiao C, Thibaut HJ, van Kuppeveld FJ, Rossmann MG. 2015. Sialic acid-dependent cell entry of human enterovirus D68. Nat Commun 6:8865. doi: 10.1038/ncomms9865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 31.Foo DG, Alonso S, Phoon MC, Ramachandran NP, Chow VT, Poh CL. 2007. Identification of neutralizing linear epitopes from the VP1 capsid protein of enterovirus 71 using synthetic peptides. Virus Res 125:61–68. doi: 10.1016/j.virusres.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 32.Liu CC, Chou AH, Lien SP, Lin HY, Liu SJ, Chang JY, Guo MS, Chow YH, Yang WS, Chang KH, Sia C, Chong P. 2011. Identification and characterization of a cross-neutralization epitope of enterovirus 71. Vaccine 29:4362–4372. doi: 10.1016/j.vaccine.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 33.Kiener TK, Jia Q, Meng T, Chow VT, Kwang J. 2014. A novel universal neutralizing monoclonal antibody against enterovirus 71 that targets the highly conserved “knob” region of VP3 protein. PLoS Negl Trop Dis 8:e2895. doi: 10.1371/journal.pntd.0002895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang L, Fan R, Sun S, Fan P, Su W, Zhou Y, Gao F, Xu F, Kong W, Jiang C. 2015. A new EV71 VP3 epitope in norovirus P particle vector displays neutralizing activity and protection in vivo in mice. Vaccine 33:6596–6603. doi: 10.1016/j.vaccine.2015.10.104. [DOI] [PubMed] [Google Scholar]
- 35.Shi J, Huang X, Liu Q, Huang Z. 2013. Identification of conserved neutralizing linear epitopes within the VP1 protein of coxsackievirus A16. Vaccine 31:2130–2136. doi: 10.1016/j.vaccine.2013.02.051. [DOI] [PubMed] [Google Scholar]
- 36.Foo DG, Ang RX, Alonso S, Chow VT, Quak SH, Poh CL. 2008. Identification of immunodominant VP1 linear epitope of enterovirus 71 (EV71) using synthetic peptides for detecting human anti-EV71 IgG antibodies in Western blots. Clin Microbiol Infect 14:286–288. doi: 10.1111/j.1469-0691.2007.01904.x. [DOI] [PubMed] [Google Scholar]
- 37.Ye X, Fan C, Ku Z, Zuo T, Kong L, Zhang C, Shi J, Liu Q, Chen T, Zhang Y, Jiang W, Zhang L, Huang Z, Cong Y. 2016. Structural basis for recognition of human enterovirus 71 by a bivalent broadly neutralizing monoclonal antibody. PLoS Pathog 12:e1005454. doi: 10.1371/journal.ppat.1005454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ku Z, Liu Q, Ye X, Cai Y, Wang X, Shi J, Li D, Jin X, An W, Huang Z. 2014. A virus-like particle based bivalent vaccine confers dual protection against enterovirus 71 and coxsackievirus A16 infections in mice. Vaccine 32:4296–4303. doi: 10.1016/j.vaccine.2014.06.025. [DOI] [PubMed] [Google Scholar]
- 39.Lyu K, Ding J, Han JF, Zhang Y, Wu XY, He YL, Qin CF, Chen R. 2014. Human enterovirus 71 uncoating captured at atomic resolution. J Virol 88:3114–3126. doi: 10.1128/JVI.03029-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan C, Ye X, Ku Z, Kong L, Liu Q, Xu C, Cong Y, Huang Z. 1 February 2017. Beta-propiolactone inactivation of coxsackievirus A16 induces structural alteration and surface modification of viral capsids. J Virol doi: 10.1128/JVI.00038-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abdurahman S, Youssefi M, Hoglund S, Vahlne A. 2007. Characterization of the invariable residue 51 mutations of human immunodeficiency virus type 1 capsid protein on in vitro CA assembly and infectivity. Retrovirology 4:69. doi: 10.1186/1742-4690-4-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harprecht C, Okifo O, Robbins KJ, Motwani T, Alexandrescu AT, Teschke CM. 2016. Contextual role of a salt bridge in the phage P22 coat protein I-domain. J Biol Chem 291:11359–11372. doi: 10.1074/jbc.M116.716910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gertsman I, Fu CY, Huang R, Komives EA, Johnson JE. 2010. Critical salt bridges guide capsid assembly, stability, and maturation behavior in bacteriophage HK97. Mol Cell Proteomics 9:1752–1763. doi: 10.1074/mcp.M000039-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abdurahman S, Hoglund S, Hoglund A, Vahlne A. 2007. Mutation in the loop C-terminal to the cyclophilin A binding site of HIV-1 capsid protein disrupts proper virus assembly and infectivity. Retrovirology 4:19. doi: 10.1186/1742-4690-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wildova M, Hadravova R, Stokrova J, Krizova I, Ruml T, Hunter E, Pichova I, Rumlova M. 2008. The effect of point mutations within the N-terminal domain of Mason-Pfizer monkey virus capsid protein on virus core assembly and infectivity. Virology 380:157–163. doi: 10.1016/j.virol.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rachakonda PS, Veit M, Korte T, Ludwig K, Bottcher C, Huang Q, Schmidt MF, Herrmann A. 2007. The relevance of salt bridges for the stability of the influenza virus hemagglutinin. FASEB J 21:995–1002. doi: 10.1096/fj.06-7052hyp. [DOI] [PubMed] [Google Scholar]
- 47.Liu CC, Guo MS, Lin FH, Hsiao KN, Chang KH, Chou AH, Wang YC, Chen YC, Yang CS, Chong PC. 2011. Purification and characterization of enterovirus 71 viral particles produced from vero cells grown in a serum-free microcarrier bioreactor system. PLoS One 6:e20005. doi: 10.1371/journal.pone.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chung YC, Ho MS, Wu JC, Chen WJ, Huang JH, Chou ST, Hu YC. 2008. Immunization with virus-like particles of enterovirus 71 elicits potent immune responses and protects mice against lethal challenge. Vaccine 26:1855–1862. doi: 10.1016/j.vaccine.2008.01.058. [DOI] [PubMed] [Google Scholar]
- 49.Lin YL, Yu CI, Hu YC, Tsai TJ, Kuo YC, Chi WK, Lin AN, Chiang BL. 2012. Enterovirus type 71 neutralizing antibodies in the serum of macaque monkeys immunized with EV71 virus-like particles. Vaccine 30:1305–1312. doi: 10.1016/j.vaccine.2011.12.081. [DOI] [PubMed] [Google Scholar]
- 50.Ferguson M, Minor PD. 1990. Differences in conformation of type 3 poliovirus antigenic sites on non-infectious empty particles and infectious virus. J Gen Virol 71:1271–1274. doi: 10.1099/0022-1317-71-6-1271. [DOI] [PubMed] [Google Scholar]
- 51.Von Seefried A, Chun JH, Grant JA, Letvenuk L, Pearson EW. 1984. Inactivated poliovirus vaccine and test development at Connaught Laboratories Ltd. Rev Infect Dis 6(Suppl 2):S345–S349. doi: 10.1093/clinids/6.Supplement_2.S345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 53.Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. 2013. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods 10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 55.Ludtke SJ, Baldwin PR, Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 56.Yu G, Li K, Liu Y, Chen Z, Wang Z, Yan R, Klose T, Tang L, Jiang W. 2016. An algorithm for estimation and correction of anisotropic magnification distortion of cryo-EM images without need of pre-calibration. J Struct Biol 195:207–215. doi: 10.1016/j.jsb.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosenthal PB, Henderson R. 2003. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol 333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 58.Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 59.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barad BA, Echols N, Wang RY, Cheng Y, DiMaio F, Adams PD, Fraser JS. 2015. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat Methods 12:943–946. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goddard TD, Huang CC, Ferrin TE. 2007. Visualizing density maps with UCSF Chimera. J Struct Biol 157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 64.Ku Z, Shi J, Liu Q, Huang Z. 2012. Development of murine monoclonal antibodies with potent neutralization effects on enterovirus 71. J Virol Methods 186:193–197. doi: 10.1016/j.jviromet.2012.06.025. [DOI] [PubMed] [Google Scholar]
- 65.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu L, Zheng Q, Li S, He M, Wu Y, Li Y, Zhu R, Yu H, Hong Q, Jiang J, Li Z, Li S, Zhao H, Yang L, Hou W, Wang W, Ye X, Zhang J, Baker TS, Cheng T, Zhou ZH, Yan X, Xia N. 2017. Atomic structures of coxsackievirus A6 and its complex with a neutralizing antibody. Nat Commun 8:505. doi: 10.1038/s41467-017-00477-9. [DOI] [PMC free article] [PubMed] [Google Scholar]