ABSTRACT
Host cells harbor various intrinsic mechanisms to restrict viral infections as a first line of antiviral defense. Viruses have evolved various countermeasures against these antiviral mechanisms. Here we show that N-Myc downstream-regulated gene 1 (NDRG1) limits productive hepatitis C virus (HCV) infection by inhibiting viral assembly. Interestingly, HCV infection downregulates NDRG1 protein and mRNA expression. The loss of NDRG1 increases the size and number of lipid droplets, which are the sites of HCV assembly. HCV suppresses NDRG1 expression by upregulating MYC, which directly inhibits the transcription of NDRG1. The upregulation of MYC also leads to the reduced expression of the NDRG1-specific kinase serum/glucocorticoid-regulated kinase 1 (SGK1), resulting in a markedly diminished phosphorylation of NDRG1. The knockdown of MYC during HCV infection rescues NDRG1 expression and phosphorylation, suggesting that MYC regulates NDRG1 at both the transcriptional and posttranslational levels. Overall, our results suggest that NDRG1 restricts HCV assembly by limiting lipid droplet formation. HCV counteracts this intrinsic antiviral mechanism by downregulating NDRG1 via a MYC-dependent mechanism.
IMPORTANCE Hepatitis C virus (HCV) is an enveloped single-stranded RNA virus that targets hepatocytes in the liver. HCV is a leading cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma, and estimates suggest a global prevalence of 2.35%. Up to 80% of acutely infected individuals will develop chronic infection, and as many as 5% eventually progress to liver cancer. An understanding of the mechanisms behind virus-host interactions and viral carcinogenesis is still lacking. The significance of our research is that it identifies a previously unknown relationship between HCV and a known tumor-associated gene. Furthermore, our data point to a new role for this gene in the liver and in lipid metabolism. Thus, HCV infection serves as a great biological model to advance our knowledge of liver functions and the development of liver cancer.
KEYWORDS: host restriction factors, viral assembly, lipid droplets, phosphorylation, oncogene, tumor suppressor gene, liver cancer, hepatitis C virus
INTRODUCTION
Hepatitis C virus (HCV) is a member of the family Flaviviridae and the prototypical member of the genus Hepacivirus. It infects approximately 160 million individuals worldwide (1). Up to 80% of acutely infected individuals will develop chronic infection, and many eventually progress to cirrhosis and liver cancer (2). Like all viruses, HCV depends upon the host cell for propagation, and many host dependencies have been uncovered by using yeast two-hybrid assays, small interfering RNA (siRNA) screens, and other approaches. HCV entry alone includes interactions with multiple host factors (3). After entry, the viral RNA (vRNA) binds to the host ribosome through the internal ribosome entry site (IRES) and translates into a single large polypeptide. The polypeptide is cleaved by both cellular and viral proteases to yield mature structural (E1, E2, and core) and nonstructural (NS) (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) viral proteins. The NS proteins form replication complexes on modified host endoplasmic reticulum (ER) membranes, and the RNA-dependent RNA polymerase NS5B replicates the viral RNA. HCV core and NS5A associate with lipid droplets (LDs), which are thought to be the major sites for assembly (4). Finally, the virus particle is enveloped in an ER-derived compartment and traffics to the cell surface via vesicular transport for release. Despite the significant advances in our knowledge of replication, numerous gaps still exist, especially for assembly and release. Further investigation of these host-virus interactions is vital to gain a better understanding of viral replication and cellular biology.
Whole-genome siRNA screens have produced an extensive list of potentially important host-virus interactions for several different viruses (5–9). Our laboratory previously performed a siRNA screen to identify proteins and pathways important for the complete HCV life cycle (10). While this is an important first step to represent potential candidates, a more extensive analysis is required to understand the significance of these interactions in vivo. This approach allows us to fully understand the complex interactions during infection of hepatocytes, which can lead to cirrhosis and cancer. Additionally, these pathways may be shared by other viral pathogens and identify key components that viruses have evolved to utilize or counteract for efficient propagation.
The above-mentioned RNA interference (RNAi) screen identified N-Myc downstream-regulated gene 1 (NDRG1) as a potential antiviral gene since silencing of its expression increased HCV replication. NDRG1 is a widely expressed protein and is highly conserved (>80% identical) in a variety of species (11). The NDRG family of proteins (NDRG1 to -4) belongs to the α/β-hydrolase family, but NDRG1 appears to lack catalytic activity (12). As indicated by its name, NDRG1 expression is suppressed by both v-myc avian myelocytomatosis viral oncogene neuroblastoma-derived homolog (MYCN) and v-myc avian myelocytomatosis viral oncogene homolog (MYC) overexpression (13, 14). In addition, NDRG1 is upregulated by numerous cellular stimuli, including heavy metals, hypoxia, oxidative stress, and several other stress-inducing compounds (11, 15). Evidence in various tissues suggests that NDRG1 plays an important role in several pathways, including the stress response (16), cell differentiation (17, 18), the cell cycle (19), vesicle transport (20), autophagy (21), and cancer metastasis (22, 23). The precise functions of NDRG1 in these pathways are still under investigation.
NDRG1 has been suggested to be a potential tumor suppressor (24–27), while others indicate that it may promote tumor growth and metastasis (28–30). It is possible that these conflicting results are due to the different tissues and/or cell types examined in each study. Thus, it is important to gain a better understanding of the functions of NDRG1 in the liver and the role that it plays in the replication of HCV. Furthermore, these studies may shed light on the pathogenesis of HCV-associated hepatocellular carcinoma (HCC), which has emerged as an urgent global public health problem in light of the high disease burden and possibly unexpected acceleration of HCC development in direct-acting antiviral (DAA)-treated patients (2, 31).
In the present study, we show that NDRG1 restricts productive HCV infection by inhibiting viral assembly at lipid droplets and demonstrate that HCV downregulates NDRG1 to increase viral production via a MYC-dependent mechanism.
RESULTS
NDRG1 restricts HCV replication at the stage of viral assembly.
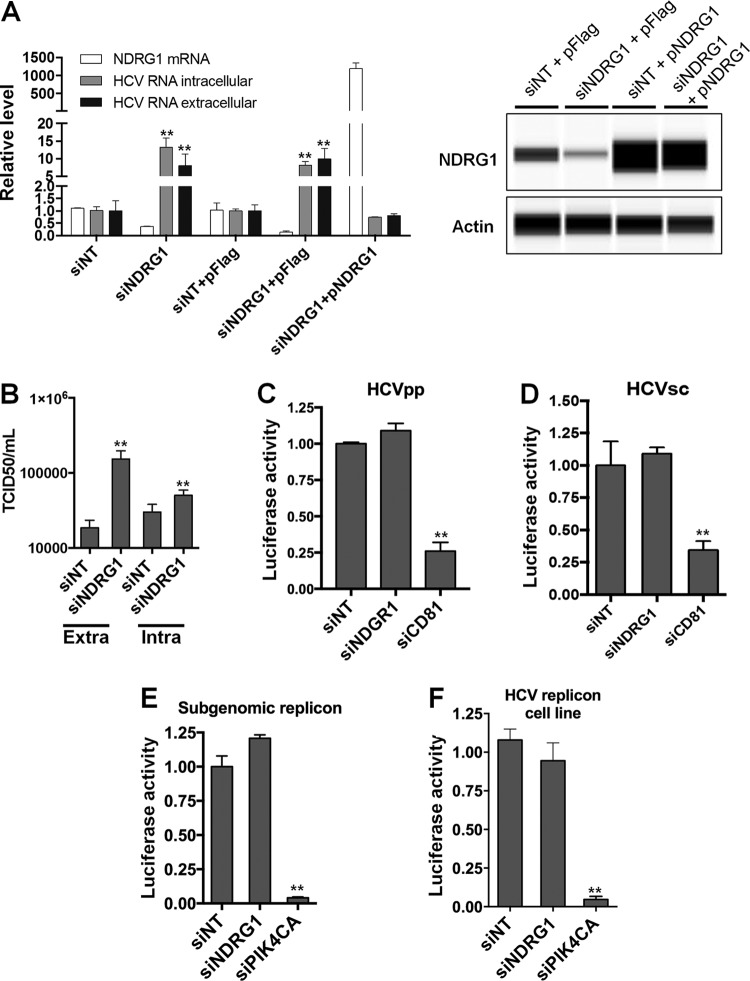
We confirmed the phenotype observed in a previously reported siRNA genome-wide screen (10). As mentioned above, the screen identified NDRG1 as an antiviral gene since the knockdown of its expression increased HCV replication (approximately 2- or 3-fold). Huh7.5.1 cells were transfected with a pool of 4 siRNAs directed at NDRG1 (siNDRG1) and a pool of nontargeting siRNAs (siNT) as a negative control for this experiment. Knockdown was confirmed by Western blotting and reverse transcription-quantitative PCR (RT-qPCR) (Fig. 1A), which indicated 70 to 80% knockdown at 72 h posttransfection. After siRNA transfection, the cells were infected with HCV for another 48 h. To examine HCV replication, the intracellular and extracellular vRNAs were isolated and quantified by using RT-qPCR. We confirmed that siRNA knockdown significantly increases HCV RNA levels in both the intracellular and extracellular samples (Fig. 1A). The increase in HCV production was also validated by measuring the infectious titers of the virus both intracellularly and extracellularly in cells treated with siNDRG1 (Fig. 1B). Notably, the increases in the infectious HCV titers were much more pronounced for the extracellular than for the intracellular levels, which is suggestive of an effect on assembly.
FIG 1.
Loss of NDRG1 enhances HCV infection. (A) Knockdown of NDRG1. Huh7.5.1 cells were transfected with a pool of NDRG1-specific, no-target control siRNAs (siNT), and/or the NDRG1 plasmid (pNDRG1) for 72 h. Cells were infected with HCV for 48 h, and total intracellular and extracellular RNA was isolated and quantified by RT-qPCR using gene-specific primers. NDRG1 knockdown and overexpression were also detected by Western blotting using the Wes system. (B) HCV assembly is increased in NDRG1-depleted cells. Huh7.5.1 cells were treated as described above for panel A and infected with HCV. At 48 h postinfection, extracellular and intracellular virus was isolated and used to infect naive Huh7.5.1 cells. Infectivity (50% tissue culture infective dose [TCID50] per milliliter) was calculated by limiting dilution and detection of the HCV core protein by immunofluorescence. (C and D) NDRG1 depletion does not affect HCV entry. Knockdown in Huh7.5.1 cells was carried out as described above, and two different viral luciferase reporter constructs were utilized to examine entry (HCVpp and HCVsc). Cells were lysed, and luciferase activity was measured 48 h later by using a luminometer. CD81 knockdown was used as a positive control for entry. (E and F) NDRG1 knockdown does not alter HCV RNA replication. Huh7.5.1 cells were treated with siRNA for 72 h and transfected with the HCV SGR (E), while Huh7-SGR cells (F) were treated with siRNAs only. Knockdown of PI4KCA was used as a positive control in each system. Luciferase activity was measured 72 h after treatment. All error bars denote standard deviations. All experiments were performed three times, and the data from a representative experiment are shown. **, P ≤ 0.01; *, P ≤ 0.05 (comparison to the negative controls).
Transfection of the NDRG1 expression construct did not lead to the expected decrease in HCV levels in infected cells. Because of its important role in cell growth and differentiation, NDRG1's functions may be tightly regulated (11), resulting in a lack of a suppressive effect on HCV by overexpressing NDRG1. To further study the role of NDRG1 in HCV replication, we overexpressed a siRNA-resistant NDRG1 construct lacking the 3′ untranslated region (UTR) (pNDRG1) in cells transfected with siNDRG targeting the 3′ UTR to knock down endogenous NDRG1. The overexpression of this construct abrogated the increase in the HCV RNA level by siNDRG1 treatment (siNDRG1 plus pFLAG versus siNDRG1 plus pNDRG1, and siNT plus pFLAG versus siNDRG1 plus pNDRG1) (Fig. 1A, left). Overall, these data confirm that NDRG1 restricts productive HCV infection.
Next, we sought to understand the step of viral replication affected by NDRG1 expression. First, we checked the effect of NDRG1 knockdown on HCV entry in NDRG1 knockdown cells. We utilized HCV pseudoparticles (HCVpp) and HCV single-cycle (HCVsc) assays (32). Knockdown of CD81 was used as a positive control. As expected, we did not see any significant change in either assay but saw a strong reduction of infection in cells depleted of CD81 (Fig. 1C and D). Next, we used a HCV subgenomic replicon (SGR) luciferase reporter system that directly mimics vRNA replication. We tested two separate systems: transfection of SGR RNA into Huh7.5.1 cells and a Huh7 cell line stably expressing the HCV SGR (Huh7-SGR) (33). NDRG1 knockdown was performed as described above, and 48 h after transfection, the cells were assayed for luciferase activity. Knockdown of PI4KCA was performed as a positive control. We observed no significant differences in HCV replicon activity in NDRG1 knockdown cells compared to the control in either system (Fig. 1E and F). Meanwhile, PI4KCA-depleted cells had severely decreased HCV RNA replication. Overall, our data suggest that NDRG1 depletion affects predominantly the late stages, probably the assembly step, of the HCV life cycle.
Interferon induces many interferon-stimulated genes (ISGs) that are antiviral, and NDRG1 is not known to be an ISG (34). We also indeed showed that interferon alpha treatment of Huh7.5.1 cells did not affect the expression level of NDRG1 (data not shown), indicating that NDRG1 is an intrinsic cellular restriction factor of HCV.
HCV infection downregulates NDRG1 expression.
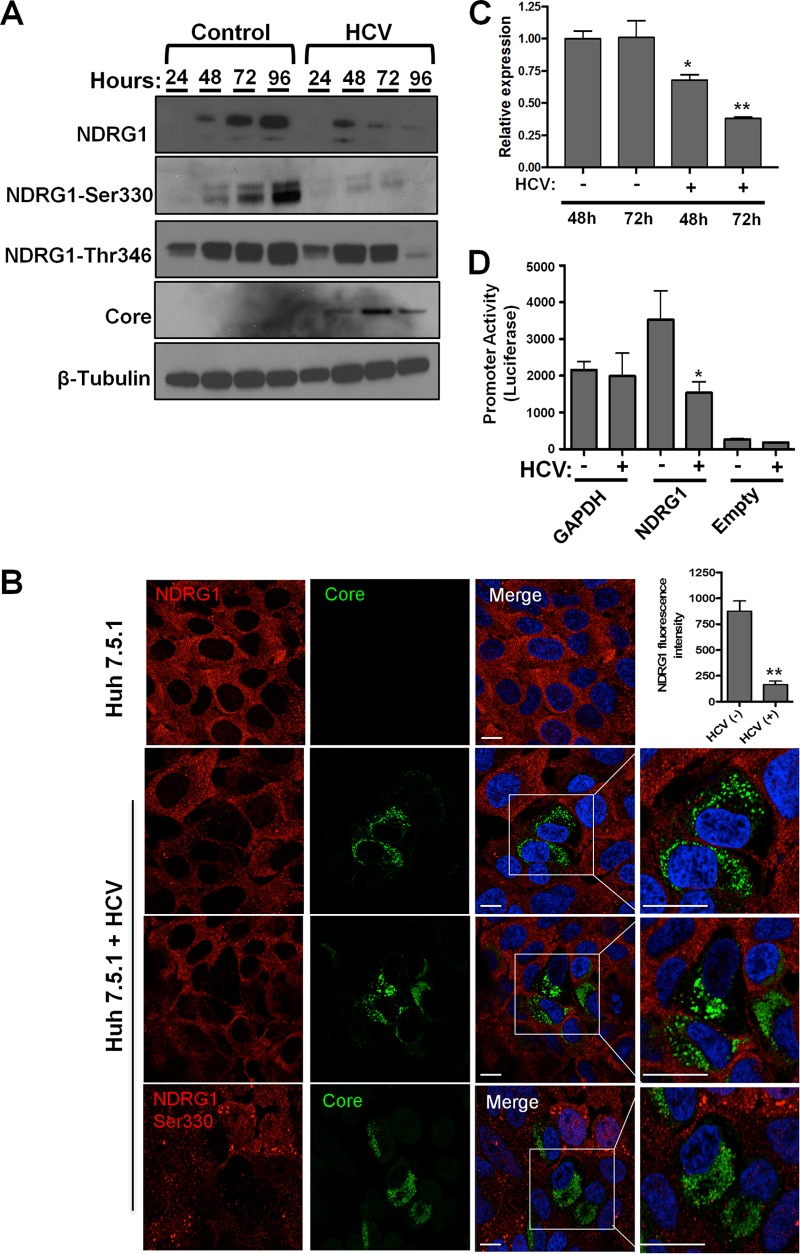
Previous studies reported that NDRG1 expression was reduced in liver tissues of patients with chronic HCV infection (35). Whether this decrease is a result of HCV infection or another indirect effect is not known. Thus, we examined the NDRG1 protein and mRNA levels in HCV-infected Huh7.5.1 cells (Fig. 2). We observed a significant decrease in NDRG1 levels compared to those for uninfected controls, and this finding was confirmed with three different NDRG1-specific antibodies, including two that are specific for the phosphorylated forms of NDRG1 (Ser330 and Thr346) (Fig. 2A). While control cells accumulated NDRG1 for the duration of culture, HCV-infected cells displayed strongly decreased NDRG1 levels beginning at 48 h postinfection, which were nearly undetectable by 96 h postinfection. The HCV core protein was detectable at 48 h postinfection, and its levels peaked at 72 h, which correlated with the observed decrease of the NDRG1 levels.
FIG 2.
HCV infection reduces NDRG1 mRNA and protein levels. (A) HCV infection reduces NDRG1 protein levels. Huh7.5.1 cells were infected in 12-well plates with HCV, and total protein was isolated at the indicated time points. The indicated proteins were detected by Western blotting (film) using protein-specific antibodies and HRP-conjugated species-specific secondary antibodies. β-Tubulin was used as a loading control. (B) NDRG1 staining is reduced in HCV-positive cells. Knockdown or control uninfected cells were infected with HCV for 48 h and then fixed and stained by using NDRG1-specific antibodies (NDRG1 [top three panels] and phosphospecific NDRG1 Ser330 [bottom panel]) and Alexa Fluor 568 secondary antibody (red). HCV-infected cells were detected with anticore antibody and Alexa Fluor 488 secondary antibody (green). White boxes denote areas magnified three times from the original images. The fluorescent signal in both uninfected and HCV-positive cells (n = 15) was quantified by using ImageJ. (C) NDRG1 mRNA expression is diminished by HCV. Huh7.5.1 cells were infected with HCV, and total RNA was isolated at 48 and 72 h postinfection. NDRG1 mRNA was detected by using RT-qPCR, and the values were normalized to the GAPDH mRNA level. (D) HCV infection reduces NDRG1 promoter activity. Huh7.5.1 cells were transfected with the indicated promoter luciferase constructs for 24 h and infected with HCV for 48 h. Cells were lysed and measured for luciferase activity on a luminometer. Uninfected cells were treated and measured in parallel. The GAPDH promoter was used as a normalization control. In panel A, the image is of one of three independent blots. For panel B, n = 15, and for panels A to D, error bars denote standard deviations. All experiments were performed three times, and the data from a representative experiment are shown. **, P ≤ 0.01; *, P ≤ 0.05 (comparison to the negative controls). Bars, 10 μm.
We confirmed this phenotype by immunofluorescence confocal microscopy and observed substantial decreases in NDRG1 and phosphorylated NDRG1 staining in cells positive for the HCV core protein (Fig. 2B, highlighted boxes). Quantification of the mean fluorescence intensity revealed a significant reduction in NDRG1 staining in core-positive cells (n = 15) (Fig. 2B).
We next examined the NDRG1 mRNA levels in HCV-infected cells and observed a significant decrease at 72 h postinfection compared to the levels in uninfected cells (Fig. 2C). To validate that the effect is indeed at the transcriptional level, we examined NDRG1 promoter activity in HCV-infected cells. To test this, we used a luciferase reporter under the control of the NDRG1 promoter and a plasmid containing the promoter of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a control. Huh7.5.1 cells were transfected with the indicated plasmids and infected with HCV for 48 h. We observed a significant decrease in the promoter activity of NDRG1 in HCV-infected cells, while there was no significant change detected for the GAPDH promoter construct (Fig. 2D). These data together indicate that HCV infection suppresses NDRG1 expression at the transcriptional level.
Reduced NDRG1 expression in HCV-infected primary human hepatocytes.
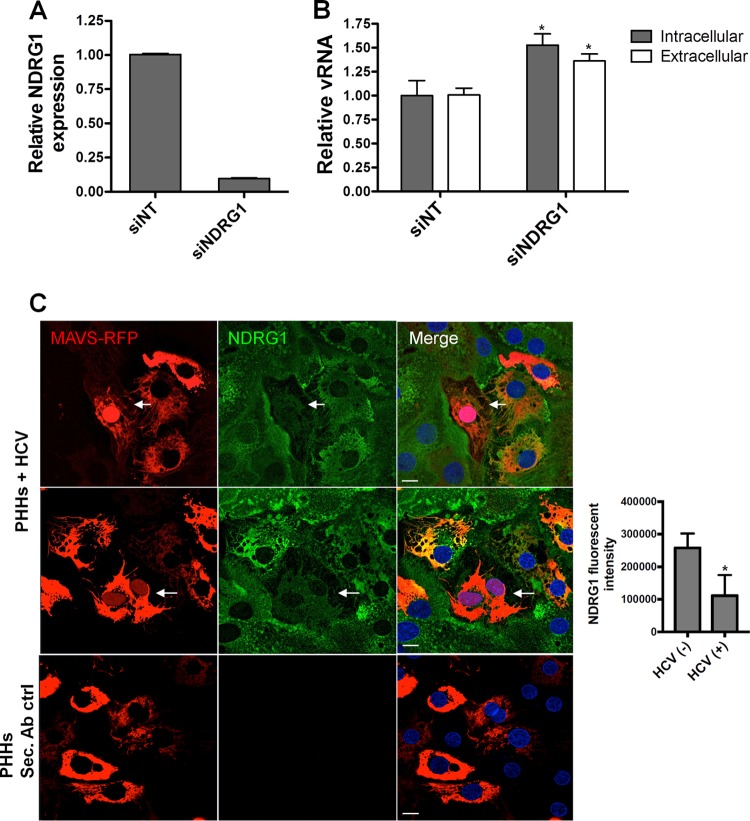
While the Huh7.5.1 cell line represents a useful and robust system to study the HCV life cycle, it does not directly mimic natural HCV infection in the liver. The cells support high levels of infectivity because of a defect in the antiviral sensor RIG-I, which normally recognizes viral RNA and stimulates the production of interferon and numerous antiviral genes (36). To confirm our results in a more physiologically relevant model, we performed NDRG1 knockdown in primary human hepatocytes (PHHs) (Fig. 3A). After the detection of vRNA as outlined above, we observed an ∼1.5-fold enhancement of HCV replication in NDRG1 knockdown cells compared to siNT-treated cells (Fig. 3B). Although the effect was less pronounced than that in Huh7 cells, this increase was still significant (P < 0.02). This reduced effect was likely attributed to a much lower level of infection observed in PHHs (at most 10% were infected) (37).
FIG 3.
NDRG1 knockdown alters HCV infection in primary human hepatocytes (PHHs). (A) Efficient knockdown of NDRG1 in PHHs. Knockdown was performed by transfecting PHHs with a pool of 4 NDRG1-specific siRNAs. Control cells were transfected with scrambled nontargeting siRNAs (siNT). At 72 h posttransfection, total RNA was harvested, and the NDRG1 mRNA level was measured by RT-qPCR. Expression was normalized to GAPDH mRNA detection. (B) HCV infection is enhanced in NDRG1-depleted PHHs. Knockdown was performed as described above for panel A, and both control and knockdown cells were infected with HCV. Total intracellular and extracellular RNAs were harvested 48 h later, and HCV RNA was quantified by RT-qPCR. (C) NDRG1 levels are reduced in HCV-infected PHHs. HCV-dependent fluorescence relocalization (HDFR) was used to detect HCV infection of PHHs. PHHs were transduced and incubated with MAVS-RFP reporter lentivirus for 24 h and then infected with HCV for 48 h. NDRG1 staining was detected by using an anti-NDRG1 antibody and anti-mouse Alexa Fluor 488. Cells with red nuclei indicate HCV-infected cells. Sec. Ab ctrl (secondary antibody control) indicates the absence of primary antibody incubation. Error bars denote standard deviations. All experiments were performed three times, and the data from a representative experiment are shown. **, P ≤ 0.01; *, P ≤ 0.05 (compared to the negative controls). Bars, 10 μm.
Next, we sought to confirm the negative effect of HCV infection on NDRG1 expression in the PHH culture system. However, due to the fewer number of infected cells and lower levels of HCV replication in PHHs than in Huh7.5.1 cells, we could not detect any change in NDRG1 protein or mRNA levels. To overcome this limitation, we used a HCV-dependent fluorescence relocalization system (HDFR) (38). We transduced PHH cultures with the MAVS-red fluorescent protein (RFP) construct and infected the cells with HCV 24 h later. The cells were fixed and stained for NDRG1 at 72 h postinfection. We confirmed that cells containing RFP-positive nuclei demonstrated reduced NDRG1 staining (Fig. 3C, top two panels). Figure 3C (third panel) shows that staining was specific for NDRG1, as there was no fluorescence detected in control cells without the primary NDRG1 antibody. We also quantified the fluorescence intensity of NDRG1 in many HCV-infected cells compared to uninfected cells (Fig. 3C, graph on the right). Overall, these data confirm that HCV infection negatively regulates NDRG1 expression.
NDRG1 restricts HCV assembly via regulation of lipid droplet formation and metabolism.
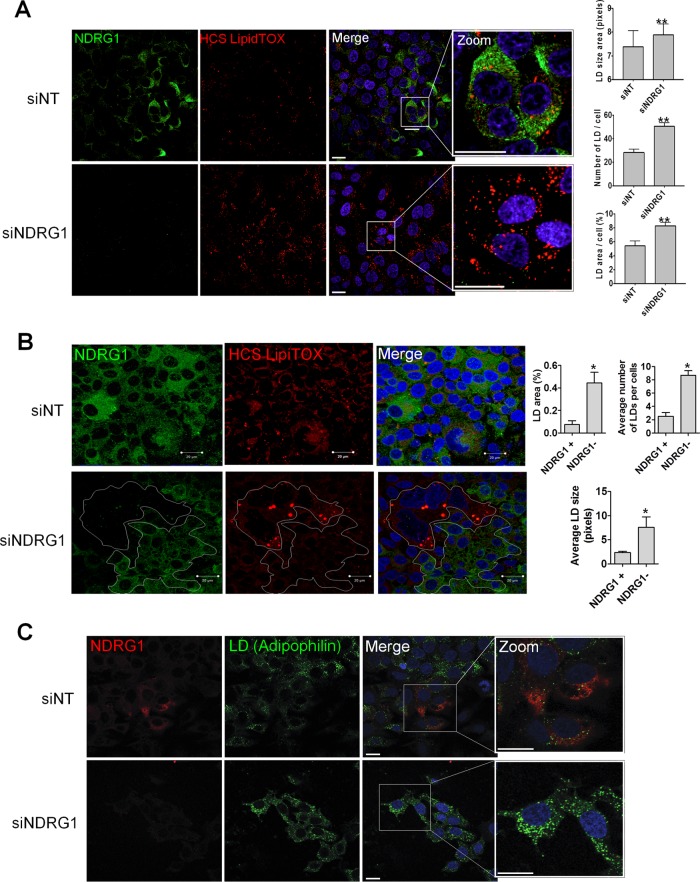
HCV assembly is known to occur in close association with lipid droplets (39). We next examined the effect of NDRG1 knockdown on the formation of lipid droplets by confocal microscopy. Our data showed that the formation of lipid droplets was substantially increased in NDRG1 knockdown cells (Fig. 4). This increase was demonstrated by using two different markers for lipid droplets, HCS LipidTOX (Fig. 4A and B) and adipophilin (Fig. 4C). HCS LipidTOX is a commercially available cell-permeable stain that has a high affinity for neutral lipid droplets. Interestingly, in NDRG knockdown cells, we noted some variable suppression of NDRG1 levels. We thus examined cells with no NDRG1 expression (NDRG1 negative [NDRG−]) or detectable NDRG1 expression (NDRG1 positive [NDRG+]) for LD formation in the NDRG1 knockdown sample. NDRG− cells again showed substantially higher LD levels than did NDRG+ cells (Fig. 4B, white).
FIG 4.
The effects of NDRG1 knockdown on lipid droplet (LD) formation. (A) NDRG1 depletion increases lipid droplet formation. Huh7.5.1 cells were treated with a pool of 4 NDRG1-specific siRNAs for 72 h and prepared for staining as described in the legend of Fig. 2B. Cells were fixed and incubated with NDRG1 mouse antibody and HCS LipidTOX followed by Alexa Fluor 488 anti-mouse secondary antibody. Nuclei were stained with Hoechst dye. Panels on the far right are magnified three times to show greater detail. Lipid droplets were quantified for 849 cells in the siNT control-treated sample and 1,514 cells in the siNDRG1-treated sample. The LD area, number of LDs per cell, and LD area per cell are shown on the right. (B) The lipid droplet size/intensity negatively correlates with NDRG1 levels. Cells were fixed, incubated with NDRG1 antibodies followed by Alexa Fluor 488 secondary antibodies (green), and stained for lipid droplets with HCS LipidTOX stain. In siNDRG1-treated samples, clusters of cells with or without NDRG1 expression are demarcated (in white). Cells with undetectable NDRG1 showed much higher levels of lipid droplet formation, and quantifications of the average LD size, average number of LDs per cell, and LD area per cell are shown on the right. (C) NDRG1 knockdown increases the lipid droplet size and intensity. Knockdown was performed by using a pool of 4 NDRG1-specific siRNAs as described above for panel A. Cells were fixed and incubated with an NDRG1 primary mouse antibody followed by Alexa Fluor 488 anti-mouse antibody (green). Lipid droplets were stained by using an adipophilin primary rabbit antibody and Alexa Fluor 568 anti-rabbit second antibody (red). Nuclei were stained with Hoechst dye (blue). The far-right panels are zoomed three times to show more details. The fluorescence intensity of lipid droplets was determined by using ImageJ software. All error bars denote standard deviations. All experiments were performed three times, and the data from a representative experiment are shown. **, P ≤ 0.01; *, P ≤ 0.05 (compared to the negative controls). Bars, 10 mm.
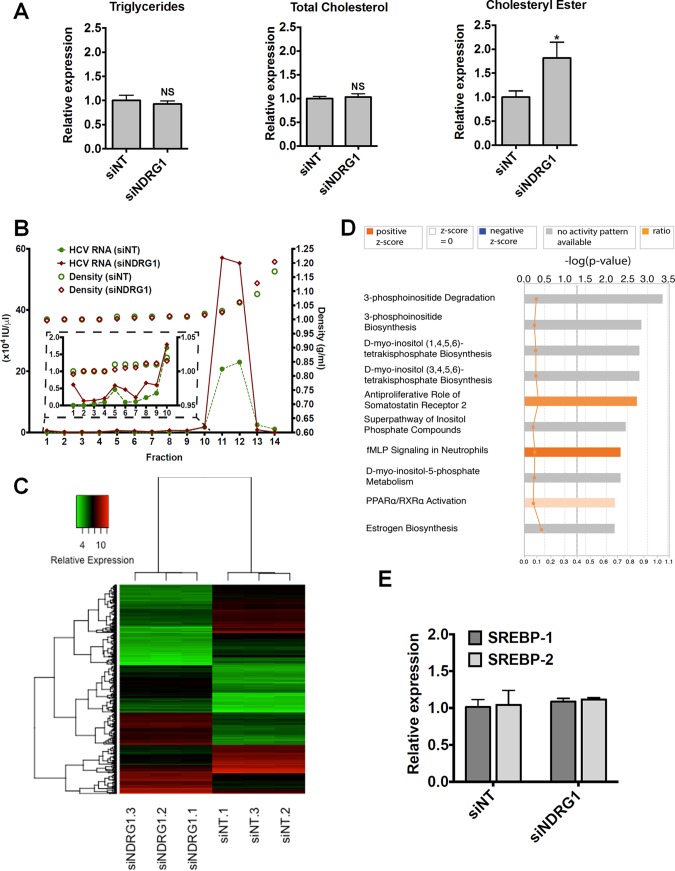
Next, we showed that the total cellular triglyceride and cholesterol levels were not affected by NDRG1 knockdown, but the cholesteryl ester level was elevated in NDRG1 knockdown cells (Fig. 5A). This finding suggests that NDRG1 may affect the redistribution of cellular lipids by regulating lipid droplet formation, which contains predominantly cholesterol esters.
FIG 5.
Evaluation of the effect of NDRG1 depletion on lipid metabolism. (A) Effect of NDRG1 knockdown on the intracellular lipid composition. Huh7.5.1 cells were transfected with siNT or siNDRG1 pools as described in Materials & Methods. At 72 h posttransfection, the contents of triglyceride, total cholesterol, and cholesteryl ester were determined. NS, not significant. (B) Density gradient analysis of secreted HCV particles. The viral supernatants from NDRG1 knockdown and control infected Huh7.5.1 cells producing HCV were subjected to isopycnic centrifugation through iodixanol gradients, as described in Materials and Methods. Representative profiles are shown. Fractions were harvested from the gradients, and the relative HCV RNA level was calculated by RT-qPCR. RNA levels are expressed as HCV RNA international units (IU) per microliter. The inset graph shows an enlarged view of fractions 1 to 10, where the low-density or highly lipidated viral particles are located. (C) Heat map displaying the normalized gene expression levels of the top genes identified in the analysis. Differentially expressed genes were filtered by a P value of less than 0.0001 and a log 2-fold change of more than 2 or less than −2. The x axis dendrogram shows hierarchical clustering between samples, and the y axis dendrogram shows hierarchical clustering between genes. The range of relative expression levels is represented by green for low expression, through black, to red for high expression levels. (D) Bar chart representing the top canonical pathways identified by IPA. The top x axis shows the significance [−log 10 (P value)] of the pathways as determined by the right-tailed Fisher exact test. The length of each bar denotes the significance of the pathway. The bottom x axis refers to the orange squares connected by a thin line and represents the ratio (the number of genes in a given pathway from the data set, divided by the total number of genes that make up that pathway). The gray dotted line shows the significance cutoff for pathways (P value of less than 0.05). Data were analyzed by using Qiagen IPA. fMLP, formyl Met-Leu-Phe receptor. (E) NDRG1 depletion does not affect SREBP1 or SREBP2 expression. Huh7.5.1 cells were transfected with siNDRG1 or siNT pools for 72 h. Total intracellular RNA was isolated, and SREBP1 and SREBP2 expression was detected by using RT-qPCR. Error bars denote standard deviations. All experiments were performed three times, and the data from a representative experiment are shown. *, P ≤ 0.05 compared to the negative controls.
To examine whether NDRG1 knockdown affects the lipid composition of assembled HCV particles, we performed density gradient centrifugation of secreted HCV in the supernatant. We noted no differences in the density profiles of HCV particles from control and NDRG1-depleted cells (Fig. 5B). As expected, we noted a significant increase in the total amount of viral particles as measured by viral RNA in the fractions that contain HCV. These data suggest that NDRG1 probably targets HCV assembly at the site of lipid droplet biogenesis and does not affect the lipid composition of the secreted virions. To further ascertain the cellular function of NDRG1, we performed microarray transcriptome analysis of NDRG1 knockdown cells. Figure 5C displays a heat map of the normalized gene expression values for the top differentially expressed genes identified in this analysis. By using Ingenuity Pathway Analysis (IPA), the top 10 canonical pathways shown in Fig. 5D include various pathways involved in lipid metabolism and biogenesis. This finding is consistent with the function of NDRG1 in HCV assembly. In particular, the peroxisome proliferator-activated receptor alpha/retinoid X receptor alpha (PPARα/RXRα) pathway, shown previously to be involved in HCV infection (40), is highlighted here (Fig. 5D). By using Gene Set Enrichment Analysis (GSEA) software, leading-edge analysis also identified the peroxisome pathway as a key pathway affected by NDRG1 knockdown. Upregulation of sterol regulatory element binding protein (SREBP) has been implicated in increased lipid droplet formation in HCV-infected cells (34). We showed here that NDRG1 knockdown does not affect the expression levels of SREBP1 and -2 (Fig. 5E).
Reduction of NDRG1 expression by HCV is MYC dependent.
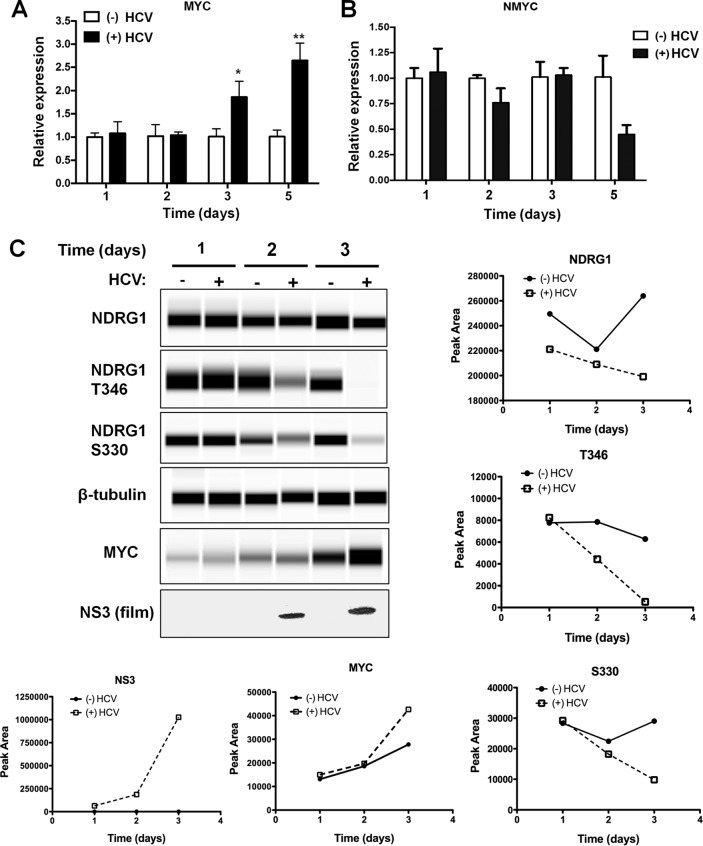
One plausible explanation for the suppression of NDRG1 by HCV is through the induction of the MYC or MYCN oncogenes, whose proteins have been shown to directly bind to the NDRG1 core promoter and downregulate its expression (13, 14). This is consistent with recent data demonstrating that HCV replication upregulates MYC (41). To examine if HCV replication could induce MYC or MYCN expression in our system, we performed a HCV infection time course. Samples were collected for both protein and RNA. Using RT-qPCR, we observed a gradual increase in MYC RNA levels at 72 h postinfection (Fig. 6A). The MYC RNA level did not show appreciable changes at 24 or 48 h postinfection, which appears to be somewhat different from the time course of NDRG1 expression (decreased level at the 48-h time point) after HCV infection. We did not detect any increase in the RNA level of the other MYC-related gene, MYCN, by HCV infection (Fig. 6B). In contrast, there was a modest reduction of the MYCN RNA level on day 5.
FIG 6.
HCV infection upregulates MYC and reduces NDRG1 phosphorylation. (A) HCV replication upregulates MYC. Huh7.5.1 cells were infected with HCV, and total RNA was harvested at the indicated intervals. MYC expression was measured by RT-qPCR. Uninfected cells were used as a control. (B) HCV replication does not upregulate NMYC. Huh7.5.1 cells were infected with HCV and harvested as described above for panel A. NMYC expression was detected by RT-qPCR. Uninfected cells were used as a control. (C) Quantitative Western blot analysis of HCV-infected cells. Huh7.5.1 cells were infected with HCV, and total protein was harvested at the indicated time points. Protein levels were quantified by using the Wes system, except for NS3, which was quantified by a traditional Western method. Uninfected cells were harvested in parallel as a control. Error bars denote standard deviations. All experiments were performed three times, and the data from a representative experiment are shown. **, P ≤ 0.01; *, P ≤ 0.05 (compared to the negative controls). In panel C, graphs represent total peak intensity as measured by the instrument.
We next examined the protein levels of MYC during HCV infection. We utilized a new Western blot technology (Proteinsimple Wes) to directly quantify protein blots. This instrumentation uses microcapillary matrix separation of proteins followed by primary and secondary detection of proteins by chemiluminescence. The signal is directly read by a camera in real time, and the software generates the “bands” or lane view seen in Fig. 6C. Lysates from infected and uninfected cells were prepared and run according to the manufacturer's protocol. We monitored HCV infection by the detection of NS3 and once again observed a decrease in the NDRG1 level during HCV infection (Fig. 6C, bottom panel). Using this sensitive system, we detected an even greater reduction in the levels of the phosphorylated forms of NDRG1 (Fig. 6C, second and third panels). This finding suggests that HCV infection may have an additional effect on the phosphorylation of NDRG1, possibly by altering NDRG1-specific kinases.
We observed no change in MYC protein at 48 h postinfection when the reduction in the NDRG1 protein level was evident; however, there was an increase in the MYC protein level observed at 72 h postinfection (Fig. 6C, fifth panel). This is in agreement with the mRNA data described above and confirms previously reported data for a mouse model (41). MYC is an unstable protein and is stabilized by binding to Max, which then leads to translocation into the nucleus to activate (42) or repress (43) transcription. It is possible that HCV infection, in addition to increasing MYC transcription at a later time, causes an earlier functional alteration of MYC that is mediated through alternate mechanisms such as posttranscriptional modification or degradation (44) and suppresses NDRG1 expression.
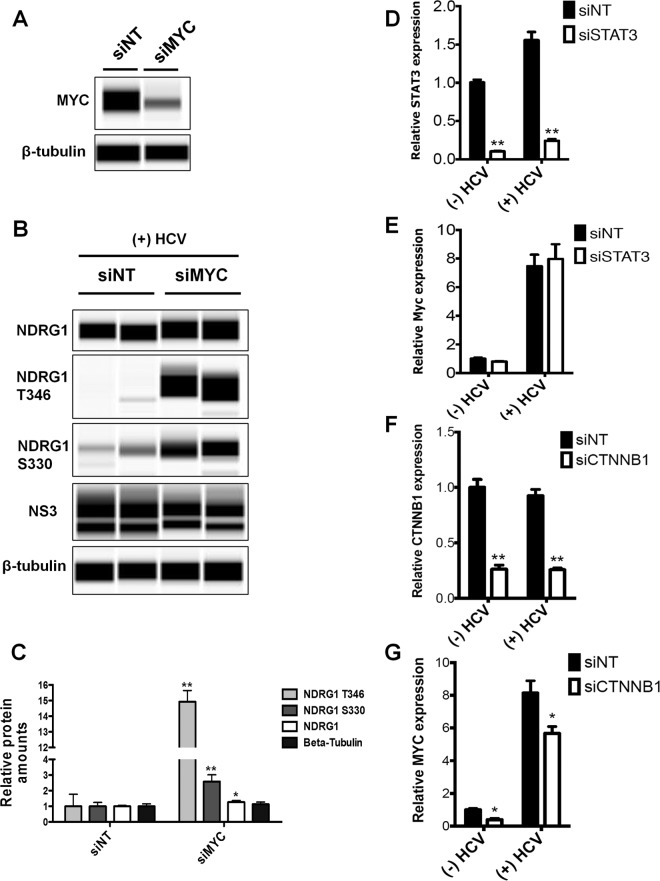
To further discern whether the reduction of the NDRG1 level by HCV is dependent on MYC, we performed siRNA knockdown of MYC before infection with HCV. Efficient knockdown was shown by Western blotting at 72 h posttransfection and indicated an approximately 2.5-fold reduction in the level of the endogenous MYC protein (Fig. 7A). We then examined NDRG1, phospho-NDRG1 T346, phospho-NDRG1 S330, NS3, and β-tubulin levels in control and knockdown cells at 72 h post-HCV infection. MYC knockdown was able to substantially increase the phosphorylated forms of NDRG1 and moderately increase total NDRG1 levels (Fig. 7B and C). We observed a slight decrease in the HCV NS3 levels after knockdown of MYC (Fig. 7B), consistent with the primary effect of NDRG1 on viral assembly but not replication. Overall, these data support that the reduction of the NDRG1 level observed during HCV infection is MYC dependent.
FIG 7.
Roles of MYC and CTNNB1 in regulating NDRG1 levels. (A) Knockdown of MYC. Huh7.5.1 cells were transfected with siRNAs targeting MYC or the control siNT, total protein was isolated 72 h later, and the MYC expression level was measured by Western blotting. (B) HCV-mediated reduction of NDRG1 levels is dependent on MYC. Huh7.5.1 cells were transfected with MYC-specific siRNAs and infected with HCV 48 h later. Total protein was isolated at 72 h postinfection. Proteins were detected by using the indicated antibodies. (C) Quantitation of Western blots. Data from panel B were quantified and normalized to the value for the β-tubulin control. (D and E) Knockdown of STAT3. Huh7.5.1 cells were transfected with control and STAT3-specific siRNAs and infected with HCV. STAT3 and MYC mRNA levels were quantified by RT-qPCR. (F and G) CTNNB1 knockdown. Huh7.5.1 cells were transfected with control and CTNNB1-specific siRNAs and infected with HCV. CTNNB1 and MYC mRNA levels were quantified by RT-qPCR. Error bars denote standard deviations. All experiments were performed three times, and the data from a representative experiment are shown. **, P ≤ 0.01; *, P ≤ 0.05 (compared to the negative controls).
HCV has been shown to activate STAT3 and the WNT/β-catenin pathway, both of which can induce MYC gene expression (45). To examine if either of these pathways is involved in the upregulation of MYC here, we performed knockdown of both genes in HCV-infected cells and demonstrated that the knockdown of β-catenin (CTNNB1) but not STAT3 gene expression can partially block the increased expression of MYC by HCV infection (Fig. 7D to G).
MYC and HCV infection suppress expression of the NDRG1-specific kinase SGK1.
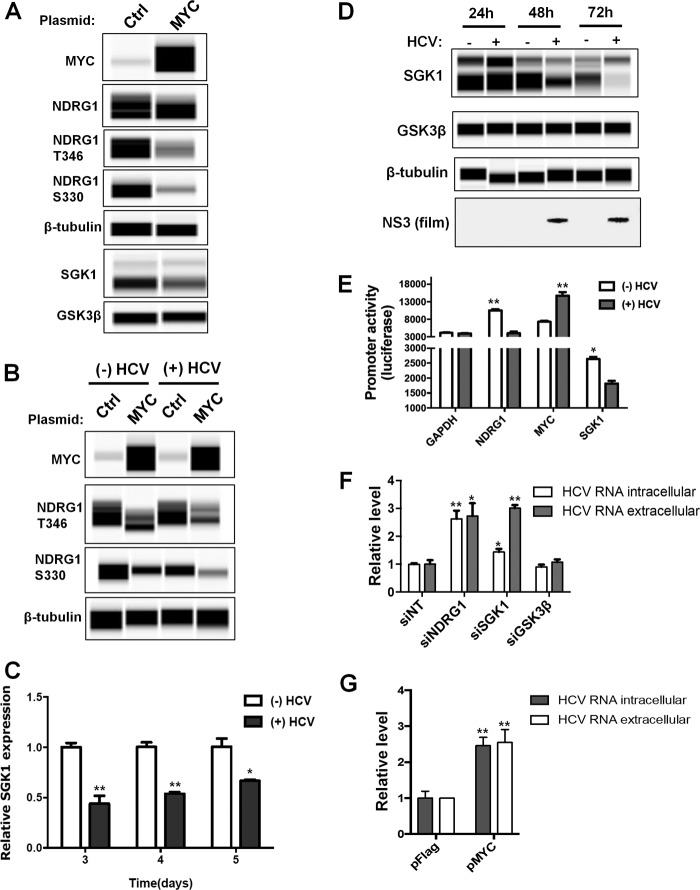
Next, we overexpressed exogenous MYC and determined the effect on NDRG1 and its kinases. We confirmed the overexpression of MYC in uninfected cells by Wes detection (Fig. 8A). Additionally, we detected a modest decrease in the total amount of NDRG1, and similar to the HCV time course, there was a substantial decrease in the level of phosphorylated forms of NDRG1 (Fig. 8A). This effect was enhanced in HCV-infected cells (Fig. 8B). A previous report showed that NDRG1 is phosphorylated by both serum/glucocorticoid-regulated kinase 1 (SGK1) and glycogen synthase kinase 3β (GSK3β) (46). NDRG1 is thought to be phosphorylated initially by SGK1 and then phosphorylated further by GSK3β, although it is still unclear how phosphorylation affects NDRG1 function. We next tested the effect of MYC overexpression on SGK1 or GSK3β levels. We observed a significant reduction in SGK1 protein levels but only a slight effect on GSK3β (Fig. 8A). The reduction of SGK1 was also observed at the mRNA level (Fig. 8C). To our knowledge, this is the first report demonstrating a suppressive effect of MYC on SGK1 expression.
FIG 8.
MYC reduces NDRG1 phosphorylation by downregulating SGK1 expression. (A) MYC overexpression. Huh7.5.1 cells were transfected with either control or MYC overexpression vectors, and protein was isolated at 48 h posttransfection. Protein was detected by using quantitative Western blotting (Wes) with the indicated antibodies. (B) MYC overexpression further reduces NDRG1 levels in HCV-infected cells. Huh7.5.1 cells were transfected with control or MYC expression vectors as described above for panel A. At 24 h posttransfection, the cells were HCV infected for an additional 72 h. The cells and proteins were then prepared as described above for panel A. Uninfected control cells were harvested in parallel. (C) HCV infection decreases SGK1 mRNA expression. A HCV time course was performed as described in the legend of Fig. 2, and SGK1 mRNA levels were monitored by RT-qPCR. An uninfected control was performed in parallel. (D) HCV infection reduces SGK1 protein levels. A HCV time course was performed as described in the legend of Fig. 2. SGK1 and GSK3β levels were monitored with the indicated primary antibodies and secondary HRP conjugates. Uninfected controls were run in parallel. (E) HCV alters the promoter activity of NDRG1, MYC, and SGK1. Promoter constructs containing a luciferase reporter under the control of NDRG1, MYC, and SGK1 promoter regions were transfected into Huh7.5.1 cells. At 6 h posttransfection, the cells were infected with HCV. Luciferase activity was measured at 48 h postinfection. The GAPDH promoter construct was used as a control. (F) Knockdown of SGK1 increases HCV RNA levels. Knockdown was performed as described in the text, using a pool of siRNAs. Seventy-two hours later, cells were infected with HCV, total intracellular and extracellular RNAs were harvested at 48 h postinfection, and HCV RNA was detected by RT-qPCR. (G) Overexpression of MYC increases HCV RNA levels. Cells were transfected with control or MYC expression plasmids for 24 h and infected with HCV. Total intracellular and extracellular RNAs were harvested at 48 h postinfection, and HCV RNA was detected by RT-qPCR. Error bars indicate standard deviations. All experiments were performed three times, and the data from a representative experiment are shown. **, P ≤ 0.01; *, P ≤ 0.05 (compared to the negative controls).
Since we observed a significant reduction in the level of the phosphorylated forms of NDRG1 after infection (Fig. 2), we next examined SGK1 and GSK3β levels in HCV-infected cells. We performed a HCV infection time course and examined the levels of these proteins. We observed a reduction in SGK1 mRNA levels (Fig. 8C) as well as a significant decrease in SGK1 protein levels beginning at 48 h postinfection; the protein was nearly undetectable by 72 h postinfection compared to the uninfected control (Fig. 8D). Conversely, there was no change in the GSK3β level at any time point. These data suggest that HCV infection reduces NDRG1 phosphorylation by downregulating SGK1 levels through enhanced MYC expression.
To validate the effects of HCV infection on the transcription of these genes, we transfected cells with various promoter-driven reporter constructs, including the promoters of MYC, SGK1, and NDRG1 (Fig. 8E). As expected, HCV infection increased the MYC reporter activity and decreased the activities of the SGK1 and NDRG1 reporters.
To further define the role of the MYC-SGK1-NDRG1 axis in HCV infection, we tested the effects of siRNA against SGK1 or GSK3β as well as MYC overexpression on HCV infection (Fig. 8F and G). We showed that these treatments indeed produced an effect similar to that of NDRG1 knockdown on enhancing HCV production.
DISCUSSION
By a genome-wide RNAi screen, we previously identified an antiviral role for NDRG1 in HCV infection. In this study, we confirmed that NDRG1 restricts the HCV life cycle at the stage of viral assembly. Conversely, HCV infection was able to reduce NDRG1 expression at the transcriptional level as well as diminishing NDRG1 phosphorylation by downregulating SGK1 (a NDRG1 kinase). Our data indicate that NDRG1 affects the late stages of HCV infection, and it is likely at the stage of viral assembly instead of secretion, although we cannot rule out a minor effect on viral secretion. We showed a 10-fold increase in the extracellular viral titer and also a modest increase in the intracellular viral titer. The significant increase in extracellular infectious viral titers supports an increase in viral assembly in NDRG1 knockdown cells. This effect was dependent on MYC, which was upregulated in HCV-infected cells. The siRNA-mediated knockdown of MYC rescued NDRG1 expression and phosphorylation. The marked loss of the phosphorylated NDRG1 forms in HCV-infected cells is a result of reduced SGK1 expression, which is also reduced by MYC upregulation.
NDRG1 has been implicated in several forms of cancer and is generally considered a tumor suppressor (11). Thus, in many malignancies, a loss of NDRG1 expression is associated with an aggressive phenotype (26–28, 47, 48). However, the precise mechanisms involved in this suppressive action are currently unknown. Furthermore, HCV infection is strongly associated with HCC, but the exact mechanisms involved in tumorigenesis are unclear. While chronic injury and inflammation are probably the predominant reasons for HCC development, whether HCV can directly induce a pro-oncogenic state in infected hepatocytes is unknown. A previous study indicated that HCV infection is associated with an increase in the expression level of the proto-oncogene MYC (41). In accordance with those results, we too observed increases in both RNA and protein levels of MYC during HCV infection. This observation coincided with a suppression of the NDRG1 transcript and protein, especially the phosphorylated forms. Thus, MYC upregulation by HCV may not only activate growth and oncogenic genes but also downregulate genes that suppress growth.
MYC was identified over 30 years ago, but the precise mechanisms of regulation are still largely unknown. MYC is frequently activated in human solid tumors and is a gene of significance in human HCCs (49, 50). MYC expression appears to respond to nearly all pathways in different cell types, but the specific locations of these regulatory elements are unclear (45). In addition, MYC is subject to multiple layers of regulation at both the mRNA and protein levels (44). Previous data indicated that several pathways involved in the transcriptional regulation of MYC could be affected by HCV infection, including STAT3 and WNT/β-catenin (41, 45, 51). We showed here that the activation of β-catenin by HCV likely mediates the upregulation of MYC expression, which in turn blocks NDRG1 function and facilitates viral assembly. Additionally, the activation of MYC by HCV renders infected cells susceptible to further oncogenic events.
MYC has been shown to bind the NDRG1 promoter and hinder its transcription (13, 14). MYC is generally studied as a transcriptional activator, but it can also function as a transcriptional repressor (43). NDRG1 expression is highly regulated, and numerous stimuli that alter NDRG1 expression have been discovered. Consistent with data from a previous study (34), NDRG1 is not an ISG. In our system, we found that the reduction of the NDRG1 level by HCV was indeed mediated by MYC induction. Previously reported evidence indicates that MYC directly binds the NDRG1 promoter and decreases NDRG1 expression but also requires histone deacetylases (13, 14). We noted a decrease in the NDRG1 level prior to the increase in the expression level of MYC (Fig. 2); however, it is possible that HCV also stabilizes the already present MYC protein prior to upregulation. As evidence supporting an antitumor role for NDRG1 continues to accumulate, this finding is in agreement and suggests that MYC not only upregulates cancer-related genes but also downregulates tumor-suppressing genes at the same time.
The reductions in the levels of the phosphorylated forms of NDRG1 suggest that HCV may also alter the kinases linked to NDRG1. Indeed, we found that HCV infection drastically decreased SGK1 protein levels, but GSK3β was unaffected. The C-terminal motif of NDRG1 contains a unique decapeptide sequence that is repeated three times. The current model suggests that SGK1 phosphorylates NDRG1 at Thr328 and Ser330 but also at each threonine residue located on the decapeptide repeat (Thr346, Thr356, and Thr366) (46). This phosphorylation was detected in the liver, lung, and spleen of wild-type mice but not in SGK1−/− mice (52). This phosphorylation by SGK1 then makes NDRG1 an excellent substrate for GSK3β, which further phosphorylates at Ser342, Ser352, and Ser362. Our results indicate that the initial phosphorylation by SGK1 was disrupted in HCV-infected Huh7.5.1 cells. This was directly related to a decrease in the SGK1 protein level in HCV-infected cells. We also showed that MYC overexpression reduced SGK1 levels, and to our knowledge, this is the first report showing that MYC suppresses SGK1 expression. These observations led us to hypothesize that HCV not only induces MYC to directly suppress NDRG1 transcription but also suppresses SGK1 expression to further perturb NDRG1 function by blocking its phosphorylation. The precise function of phosphorylated NDRG1 is unknown, although a previous report suggests that its phosphorylation is required to suppress the NF-κB pathway in pancreatic cancer cells (53). Here phosphorylated NDRG1 appears to be important for the downstream function of NDRG1: the regulation of lipid droplet formation and metabolism.
Our data showed that HCV infection perturbs normal NDRG1 expression/phosphorylation and that its knockdown increases HCV assembly. In this study, we observed significant increases in the size and fluorescence intensity of the LDs in NDRG1 knockdown cells. NDRG1 has been reported to be functionally involved in lipid metabolism, specifically cholesterol (54). Examination of its amino acid sequence reveals a phosphopantetheine attachment site, which allows a protein to interact with fatty acid substrates. Although NDRG1 does not appear to directly colocalize with LDs, this site may be important for upstream steps in the lipid-trafficking cycle. Low-density lipoproteins are transported to late endosomes for hydrolysis to free cholesterol and then released by Niemann-Pick type C1 (NPC1) and NPC2. A genome-wide expression analysis of NPC-defective fibroblasts found that the NDRG1 expression level was elevated (55). Furthermore, an RNAi screen focused on lipid biogenesis identified NDRG1 as a potential gene affecting cholesterol homeostasis (56). Additional evidence demonstrated that NDRG1 negatively regulates ceramide-rich microvesicles (54). Finally, there is increasing evidence linking lipid droplets to the endomembrane system (57). We showed here that the function of NDRG1 in lipid metabolism does not seem to be mediated by an effect on SREBP expression. These observations fit our hypothesis that NDRG1 seems to affect the distribution and sizes of LDs rather than overall increased lipogenesis. Further supporting this conclusion are our data indicating that there were no increases in the levels of triglycerides but that there were increase in the levels of cholesterol esters (the predominant form of cholesterol in lipid droplets) in our depleted cells.
Consistent with a link between lipid droplets and NDRG1, our microarray expression profiling results suggest the involvement of the PPARα pathway. PPARα is an important regulator of lipid oxidation and lipogenesis in the liver (58) Thus, the loss of NDRG1 and the increased number and size of lipid droplets may be functionally linked to PPARα-related pathways. The link of HCV infection to lipids and cholesterol has been well documented (59), so NDRG1 is likely a crucial host factor in the nexus of HCV infection and lipid metabolism.
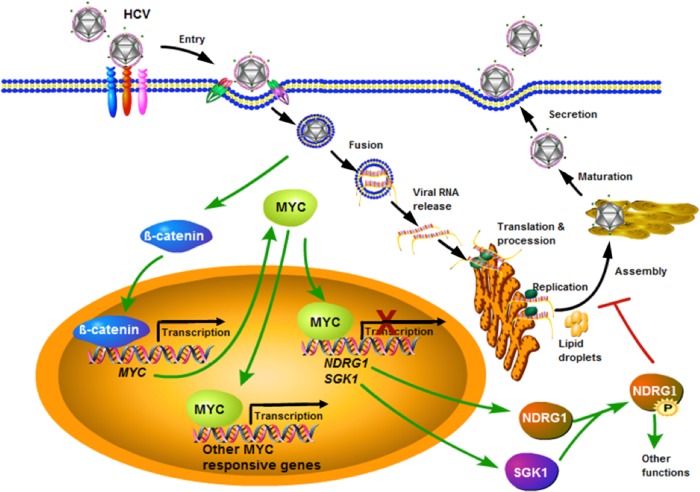
Overall, we describe a previously unrecognized relationship between HCV, NDRG1, and MYC in the context of viral infection (Fig. 9). Our working model suggests that HCV coopts the MYC pathway responsible for NDRG1 expression and phosphorylation that regulates lipid droplet formation and metabolism. NDRG1 appears to restrict HCV by suppressing the lipid droplet formation that is necessary for HCV assembly. HCV has evolved a mechanism to counteract this restriction by upregulating MYC expression, resulting in reduced NDRG1 functions. Interestingly, an unintended consequence of this effect is the inception of a pro-oncogenic condition that predisposes HCV-infected cells to malignant transformation. Until now, NDRG1 has not been implicated in viral disease. Further work is needed to determine the relationship between NDRG1 phosphorylation and the lipid homeostasis of hepatocytes. We have previously shown that IκB kinase α (IKKα), also identified by our genome-wide siRNA screen, stimulates lipid biogenesis via an alternative mechanism of upregulating SREBP1 expression and, thus, increases viral assembly (60). HCV has cleverly evolved multiple mechanisms to affect host lipid metabolism for its propagation and serves as an invaluable model to study human biology.
FIG 9.
Model for HCV, MYC, and NDRG1 interactions. HCV infection induces MYC expression probably via the activation of β-catenin and concomitantly decreases SGK1 and NDRG1 expression levels. The loss of SGK1 further diminishes the phosphorylation of NDRG1, and this results in increased lipid droplet formation. See Discussion for more details.
MATERIALS AND METHODS
Plasmids and siRNAs.
The NDRG1 (entire open reading frame [ORF]) Flag-Myc-tagged vector, the MYC vector (untagged), and the empty parental vector were obtained commercially (Origene). The ON-TARGETplus siRNA pools (4 different siRNAs) targeting NDRG1, SGK1, GSK3β, and MYC were obtained from Dharmacon.
Cell culture and virus.
The Huh7.5.1 cell line was provided by Francis Chisari (The Scripps Research Institute, La Jolla, CA). Cells were maintained in complete Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) (Omega Scientific), 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C with 5% CO2. Primary human hepatocytes were obtained from Invitrogen (lot number 1457) and maintained in Williams medium E containing cell maintenance supplement (Invitrogen). The HCV JFH-1 strain was propagated and titrated as previously described (61, 62).
Antibodies.
The following antibodies were obtained commercially: anti-NDRG1 (catalog number D8G9; Cell Signaling) (for immunofluorescence assays [IFAs]), anti-phospho-NDRG1 Ser330 (catalog number D3A12; Cell Signaling), anti-phospho-NDRG1 Thr346 (catalog number 3217; Cell Signaling), anti-NDRG1 (catalog number EPR5593; Novus Biologicals) (Western blots), anti-SGK1 (catalog number D27C11; Cell Signaling), anti-GSK3β (catalog number H-76; Santa Cruz), anti-β-tubulin (catalog number T5201; Sigma), and anti-Flag (catalog number F1804; Sigma). Anti-HCV core monoclonal antibody was produced from anticore 6G7 hybridoma cells, provided by Harry Greenberg and Xiaosong He (Stanford University, Palo Alto, CA). HCV anti-NS3 (8G-2) antibody was obtained from Abcam. Lipid droplets were stained with antiadipophilin (Progen) and HCS LipidTOX (Thermo Fisher). All secondary antibodies were obtained from Thermo Fisher.
siRNA knockdown, overexpression, and HCV replication assays.
Huh7.5.1 cells were transfected with siRNAs, as described previously, by reverse transfection in a 12-well plate (60). To monitor efficient siRNA-mediated knockdown, total RNA (RNeasy; Qiagen) and total protein (lithium dodecyl sulfate buffer; Life Technologies) were collected at 72 h. Knockdown was confirmed by RT-qPCR using NDRG1 primers/probes and normalized to the GAPDH mRNA level. Protein knockdown was confirmed by SDS-PAGE and Western blotting. At 72 h posttransfection, the cells were infected with HCV (multiplicity of infection [MOI] of 1 as calculated by a focus-forming assay prior to the experiment) for 48 h. Total intracellular RNA was isolated by using the RNeasy kit (Qiagen), and extracellular RNA was harvested by using the QIAamp viral RNA kit (Qiagen) according to the manufacturer's protocol. Intracellular and extracellular copy numbers of HCV RNA were determined by RT-qPCR with the probe, primers, and parameters described previously (10).
For HCV replicon assays, siRNA knockdown was performed as described above. At 72 h, the HCV renilla luciferase (R-Luc) subgenomic replicon was transfected into Huh7.5.1 cells by using DMRIE-C (2,3-di(tetradecoxy)propyl-(2-hydroxyethyl)-dimethylazanium bromide; Thermo Fisher) according to the manufacturer's protocol. Cells were lysed, and luciferase activity was measured 48 h later by using the luciferase assay system (Promega). The Huh7 cell line expressing the HCV subgenomic replicon was described previously (43). HCVsc assays were performed by infecting Huh7.5.1 cells with single-cycle infectious virus at an MOI of 1. Cells were lysed, and luciferase activity was measured 48 h later by using the luciferase assay system (Promega).
A commercial eukaryotic expression vector containing the ORF of NDRG1 plus a C-terminal Flag tag (pCMV6-NDRG1), a full-length Myc tag (pCMV6-Myc), and the empty vector (pCMV6-Empty) was obtained commercially (Origene). The plasmids were transfected into Huh7.5.1 cells by using TurboFect (Thermo Fisher) according to the manufacturer's protocol. At 48 h posttransfection, the cells were infected with HCV at an MOI of 1, and total intracellular and extracellular RNA was isolated and quantified as described above.
Viral titer assays were performed as described previously (63).
Western blot and immunofluorescence assays.
We used both traditional and fully automated Western blot assays to determine protein amounts. Traditional Western blotting was carried out as described previously (64). Automated quantitative Western blotting was performed by using the Wes simple Western instrument (Proteinsimple) (65, 66). The assays were carried out according to the manufacturer's protocol, and the data were analyzed by using Compass software (Proteinsimple). Briefly, protein samples were quantified via a bicinchoninic acid (BCA) assay (Thermo Fisher), mixed with 1× sample buffer and 40 mM dithiothreitol (DTT), and then heated at 95°C for 5 min. The 1× fluorescent molecular weight markers were prepared as described above. Samples, molecular weight markers, blocking reagent, primary antibodies, secondary horseradish peroxidase (HRP)-conjugated antibodies, and luminol-peroxide were loaded into the supplied assay plate. The default parameters for the instrument were used to carry out all experiments. This technique uses microcapillary SDS-PAGE and fixes the proteins in the capillary tube by using UV cross-linking. After fixation, the tubes are flushed, and all antibody incubations and washes are carried out in the capillary tube. Simple Western systems capture the data as a chemiluminescent image of the capillary. Compass software then analyzes the images, processes all the data, and provides the option to view the data as an electropherogram or as a lane view. This method is similar to traditional Western blotting but uses the microfluidic technology.
Immunofluorescence assays were performed as previously described (60). HCV-dependent fluorescence relocalization (HDFR) was performed as previously described (67). Briefly, PHHs were transduced and incubated with MAVS-RFP reporter lentivirus for 24 h and then infected with HCV for 48 h. Cells grown on Lab-Tek II borosilicate 4184 well chamber coverslips (Nunc) were fixed with 4% paraformaldehyde, permeabilized in 0.1% Triton X-100, and incubated with a blocking solution in phosphate-buffered saline (PBS) containing 3% bovine serum albumin (BSA) and 10% normal goat serum (Vector Laboratories). Cells were then labeled with appropriate primary antibodies diluted in PBS with 1% BSA and subsequently incubated with Alexa Fluor 488 or 568 secondary antibodies (Thermo Fisher) in PBS with 1% BSA. Nuclei were counterstained with Hoechst 33342 (Thermo Fisher) at a 1:5,000 dilution in PBS. Each step was followed by three washes with PBS. Confocal laser scanning microscopy analysis was performed with an Axio Observer Z1 microscope equipped with a Zeiss LSM 5 Live DuoScan system under an oil immersion 1.4-numerical-aperture (NA) 63× lens objective (Carl Zeiss). Images were acquired by using ZEN 2009 software (Carl Zeiss). Dual- or triple-color images were acquired by consecutive scanning with only one laser line active per scan to avoid cross-excitation. Image quantification was performed by using ImageJ software.
Promoter assays.
The promoter constructs were obtained commercially (Active Motif). Cells were plated into white 96-well plates and transfected according to the manufacturer's protocol. At 6 h posttransfection, the medium was replaced with fresh medium containing HCV at an MOI of 10. Infection was carried out for 48 h, and promoter activity was detected by using a luminometer according to the manufacturer's instructions.
Analysis of HCV by iodixanol gradient ultracentrifugation.
Discontinuous 5 to 50% iodixanol density gradients (OptiPrep; Sigma) were prepared from five buffered solutions of iodixanol in HEPES-buffered saline (Sigma). Samples were overlaid onto the gradients and centrifuged for 24 h at 4°C and at 50,000 rpm in an SW60Ti rotor in a Beckman Coulter instrument as previously described (68). Fractions (250 μl each) were collected, and their density was determined by the absorbance at 340 nm and calculated by using a table provided by the manufacturer (OptiPrep; Sigma). Total RNA was extracted, and HCV RNA was detected by using RT-qPCR. The HCV RNA level was quantified with a HCV RNA quantification panel from AcroMetrix (AcroMetrix HCV-S panel, catalog number 950350). RNA levels are expressed as HCV RNA international units per microliter. We arbitrarily defined the lowest threshold cycle (CT) as the point of reference and calculated the relative amount of each fraction compared to this point.
Microarray data analysis.
A microarray scan was performed by using the Affymetrix GeneChip Human Genome U133 Plus 2.0 array. Raw CEL files from the scan were analyzed by using a microarray pipeline written in “R” software. Several packages were used, including the limma and oligo packages by Bioconductor (69, 70). Data were normalized by using the robust multichip average (RMA) method as previously described (71). Linear modeling was performed by using the limma lmFit function, and differential gene expression was determined by using the contrasts.fit and eBayes functions (69). By using the toptable function, false discovery rates (FDRs) were calculated to adjust P values for multiple testing, and the top-ranked differentially expressed genes were extracted for pathway analysis (69).
Pathway analysis.
Differentially expressed genes were filtered by a FDR of less than 0.05 and a log fold change of more than 2 or less than −2. The filtered list was analyzed by using the Core Analysis feature of IPA. The IPA Ingenuity Knowledge Base was used as a reference set for the analysis. The following parameters were set: direct and indirect relationships were considered, all nonchemical molecule types were selected, all available data sources were used, confidence was set to experimentally observed relationships, species were filtered to humans, and all cell lines and mutations were considered. Data were analyzed through the use of IPA (Qiagen, Redwood City, CA). Additionally, the data set was analyzed by using GSEA in order to provide validation for the pathways discovered by IPA (72). KEGG gene sets were used for the analysis, data were normalized by using RMA background correction, and parameters were set to the default recommended settings.
Lipid assays.
Huh7.5.1 cells were transfected with siNT or siNDRG1 and challenged with HCV generated in cell culture (HCVcc) at 72 h posttransfection. Cells were then harvested at 48 h postinfection, washed with PBS, scraped, and lysed with Tris-buffered saline (TBS) supplemented with 5% Triton X-100. Triglyceride contents were determined with a triglyceride quantification kit (BioVision) according to the manufacturer's instructions. Total intracellular cholesterol and cholesteryl ester levels were quantified by using the Cholesterol/Cholesteryl Ester assay kit (Abcam) according to the manufacturer's protocol.
Accession number(s).
The microarray data were deposited in the GEO repository under accession number GSE106988.
ACKNOWLEDGMENTS
We thank Qisheng Li for the thoughtful discussion during the project and aid with technical details on methods. We also thank Fathi Elloumi of the National Cancer Institute for help with the microarray analysis.
This work was supported by the intramural research program of the NIDDK, NIH.
REFERENCES
- 1.Lavanchy D. 2011. Evolving epidemiology of hepatitis C virus. Clin Microbiol Infect 17:107–115. doi: 10.1111/j.1469-0691.2010.03432.x. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB. 2012. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 142:1264.e1–1273.e1. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeisel MB, Felmlee DJ, Baumert TF. 2013. Hepatitis C virus entry. Curr Top Microbiol Immunol 369:87–112. doi: 10.1007/978-3-642-27340-7_4. [DOI] [PubMed] [Google Scholar]
- 4.Shulla A, Randall G. 2012. Hepatitis C virus-host interactions, replication, and viral assembly. Curr Opin Virol 2:725–732. doi: 10.1016/j.coviro.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 6.Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, Seidel S, Opaluch AM, Caldwell JS, Weitzman MD, Kuhen KL, Bandyopadhyay S, Ideker T, Orth AP, Miraglia LJ, Bushman FD, Young JA, Chanda SK. 2008. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y. 2008. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454:890–893. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, Brass AL, Adametz R, Tsui M, Qian F, Montgomery RR, Lev S, Mason PW, Koski RA, Elledge SJ, Xavier RJ, Agaisse H, Fikrig E. 2008. RNA interference screen for human genes associated with West Nile virus infection. Nature 455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, Rodgers MA, Ramirez JL, Dimopoulos G, Yang PL, Pearson JL, Garcia-Blanco MA. 2009. Discovery of insect and human dengue virus host factors. Nature 458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. 2009. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci U S A 106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun J, Zhang D, Bae DH, Sahni S, Jansson P, Zheng Y, Zhao Q, Yue F, Zheng M, Kovacevic Z, Richardson DR. 2013. Metastasis suppressor, NDRG1, mediates its activity through signaling pathways and molecular motors. Carcinogenesis 34:1943–1954. doi: 10.1093/carcin/bgt163. [DOI] [PubMed] [Google Scholar]
- 12.Shaw E, McCue LA, Lawrence CE, Dordick JS. 2002. Identification of a novel class in the alpha/beta hydrolase fold superfamily: the N-myc differentiation-related proteins. Proteins 47:163–168. doi: 10.1002/prot.10083. [DOI] [PubMed] [Google Scholar]
- 13.Li J, Kretzner L. 2003. The growth-inhibitory Ndrg1 gene is a Myc negative target in human neuroblastomas and other cell types with overexpressed N- or c-myc. Mol Cell Biochem 250:91–105. doi: 10.1023/A:1024918328162. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J, Chen S, Zhang W, Liu X, Shi H, Che H, Wang W, Li F, Yao L. 2008. Human differentiation-related gene NDRG1 is a Myc downstream-regulated gene that is repressed by Myc on the core promoter region. Gene 417:5–12. doi: 10.1016/j.gene.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Melotte V, Qu X, Ongenaert M, van Criekinge W, de Bruine AP, Baldwin HS, van Engeland M. 2010. The N-myc downstream regulated gene (NDRG) family: diverse functions, multiple applications. FASEB J 24:4153–4166. doi: 10.1096/fj.09-151464. [DOI] [PubMed] [Google Scholar]
- 16.Chen B, Nelson DM, Sadovsky Y. 2006. N-myc down-regulated gene 1 modulates the response of term human trophoblasts to hypoxic injury. J Biol Chem 281:2764–2772. doi: 10.1074/jbc.M507330200. [DOI] [PubMed] [Google Scholar]
- 17.Xu B, Lin L, Rote NS. 1999. Identification of a stress-induced protein during human trophoblast differentiation by differential display analysis. Biol Reprod 61:681–686. doi: 10.1095/biolreprod61.3.681. [DOI] [PubMed] [Google Scholar]
- 18.Gratton RJ, Gluszynski M, Nygard K, Mazzuca DM, Graham CH, Han VK. 2004. Reducing agent and tunicamycin-responsive protein (RTP) mRNA expression in the placentae of normal and pre-eclamptic women. Placenta 25:62–69. doi: 10.1016/S0143-4004(03)00216-9. [DOI] [PubMed] [Google Scholar]
- 19.McCaig C, Potter L, Abramczyk O, Murray JT. 2011. Phosphorylation of NDRG1 is temporally and spatially controlled during the cell cycle. Biochem Biophys Res Commun 411:227–234. doi: 10.1016/j.bbrc.2011.06.092. [DOI] [PubMed] [Google Scholar]
- 20.Askautrud HA, Gjernes E, Gunnes G, Sletten M, Ross DT, Borresen-Dale AL, Iversen N, Tranulis MA, Frengen E. 2014. Global gene expression analysis reveals a link between NDRG1 and vesicle transport. PLoS One 9:e87268. doi: 10.1371/journal.pone.0087268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sahni S, Bae DH, Lane DJ, Kovacevic Z, Kalinowski DS, Jansson PJ, Richardson DR. 2014. The metastasis suppressor, N-myc downstream-regulated gene 1 (NDRG1), inhibits stress-induced autophagy in cancer cells. J Biol Chem 289:9692–9709. doi: 10.1074/jbc.M113.529511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang BA, Kovacevic Z, Park KC, Kalinowski DS, Jansson PJ, Lane DJ, Sahni S, Richardson DR. 2014. Molecular functions of the iron-regulated metastasis suppressor, NDRG1, and its potential as a molecular target for cancer therapy. Biochim Biophys Acta 1845:1–19. doi: 10.1016/j.bbagrm.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Song Y, Cao L. 2013. N-myc downstream-regulated gene 1: diverse and complicated functions in human hepatocellular carcinoma (review). Oncol Lett 6:1539–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Z, Zhang D, Yue F, Zheng M, Kovacevic Z, Richardson DR. 2012. The iron chelators Dp44mT and DFO inhibit TGF-beta-induced epithelial-mesenchymal transition via up-regulation of N-Myc downstream-regulated gene 1 (NDRG1). J Biol Chem 287:17016–17028. doi: 10.1074/jbc.M112.350470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu W, Xing F, Iiizumi-Gairani M, Okuda H, Watabe M, Pai SK, Pandey PR, Hirota S, Kobayashi A, Mo YY, Fukuda K, Li Y, Watabe K. 2012. N-myc downstream regulated gene 1 modulates Wnt-beta-catenin signalling and pleiotropically suppresses metastasis. EMBO Mol Med 4:93–108. doi: 10.1002/emmm.201100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang B, Li J, Ye Z, Li Z, Wu X. 2014. N-myc downstream regulated gene 1 acts as a tumor suppressor in ovarian cancer. Oncol Rep 31:2279–2285. doi: 10.3892/or.2014.3072. [DOI] [PubMed] [Google Scholar]
- 27.Mao Z, Sun J, Feng B, Ma J, Zang L, Dong F, Zhang D, Zheng M. 2013. The metastasis suppressor, N-myc downregulated gene 1 (NDRG1), is a prognostic biomarker for human colorectal cancer. PLoS One 8:e68206. doi: 10.1371/journal.pone.0068206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishio S, Ushijima K, Tsuda N, Takemoto S, Kawano K, Yamaguchi T, Nishida N, Kakuma T, Tsuda H, Kasamatsu T, Sasajima Y, Kage M, Kuwano M, Kamura T. 2008. Cap43/NDRG1/Drg-1 is a molecular target for angiogenesis and a prognostic indicator in cervical adenocarcinoma. Cancer Lett 264:36–43. doi: 10.1016/j.canlet.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 29.Yan X, Chua MS, Sun H, So S. 2008. N-Myc down-regulated gene 1 mediates proliferation, invasion, and apoptosis of hepatocellular carcinoma cells. Cancer Lett 262:133–142. doi: 10.1016/j.canlet.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 30.Lu WJ, Chua MS, So SK. 2014. Suppressing N-Myc downstream regulated gene 1 reactivates senescence signaling and inhibits tumor growth in hepatocellular carcinoma. Carcinogenesis 35:915–922. doi: 10.1093/carcin/bgt401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reig M, Marino Z, Perello C, Inarrairaegui M, Ribeiro A, Lens S, Diaz A, Vilana R, Darnell A, Varela M, Sangro B, Calleja JL, Forns X, Bruix J. 2016. Unexpected high rate of early tumor recurrence in patients with HCV-related HCC undergoing interferon-free therapy. J Hepatol 65:719–726. doi: 10.1016/j.jhep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 32.Masaki T, Suzuki R, Saeed M, Mori K, Matsuda M, Aizaki H, Ishii K, Maki N, Miyamura T, Matsuura Y, Wakita T, Suzuki T. 2010. Production of infectious hepatitis C virus by using RNA polymerase I-mediated transcription. J Virol 84:5824–5835. doi: 10.1128/JVI.02397-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ikeda M, Yi M, Li K, Lemon SM. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J Virol 76:2997–3006. doi: 10.1128/JVI.76.6.2997-3006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas E, Gonzalez VD, Li Q, Modi AA, Chen W, Noureddin M, Rotman Y, Liang TJ. 2012. HCV infection induces a unique hepatic innate immune response associated with robust production of type III interferons. Gastroenterology 142:978–988. doi: 10.1053/j.gastro.2011.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hagist S, Sultmann H, Millonig G, Hebling U, Kieslich D, Kuner R, Balaguer S, Seitz HK, Poustka A, Mueller S. 2009. In vitro-targeted gene identification in patients with hepatitis C using a genome-wide microarray technology. Hepatology 49:378–386. doi: 10.1002/hep.22677. [DOI] [PubMed] [Google Scholar]
- 36.Sumpter R Jr, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M Jr. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol 79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu H, Elyar J, Foss R, Hemming A, Hall E, Lecluyse EL, Liu C. 2009. Primary human hepatocyte culture for HCV study. Methods Mol Biol 510:373–382. doi: 10.1007/978-1-59745-394-3_28. [DOI] [PubMed] [Google Scholar]
- 38.Jones CT, Catanese MT, Law LM, Khetani SR, Syder AJ, Ploss A, Oh TS, Schoggins JW, MacDonald MR, Bhatia SN, Rice CM. 2010. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat Biotechnol 28:167–171. doi: 10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alvisi G, Madan V, Bartenschlager R. 2011. Hepatitis C virus and host cell lipids: an intimate connection. RNA Biol 8:258–269. doi: 10.4161/rna.8.2.15011. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka N, Moriya K, Kiyosawa K, Koike K, Gonzalez FJ, Aoyama T. 2008. PPARalpha activation is essential for HCV core protein-induced hepatic steatosis and hepatocellular carcinoma in mice. J Clin Invest 118:683–694. doi: 10.1172/JCI33594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Higgs MR, Lerat H, Pawlotsky JM. 2013. Hepatitis C virus-induced activation of beta-catenin promotes c-Myc expression and a cascade of pro-carcinogenetic events. Oncogene 32:4683–4693. doi: 10.1038/onc.2012.484. [DOI] [PubMed] [Google Scholar]
- 42.Dang CV. 1999. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol 19:1–11. doi: 10.1128/MCB.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kleine-Kohlbrecher D, Adhikary S, Eilers M. 2006. Mechanisms of transcriptional repression by Myc. Curr Top Microbiol Immunol 302:51–62. [DOI] [PubMed] [Google Scholar]
- 44.Farrell AS, Sears RC. 2014. MYC degradation. Cold Spring Harb Perspect Med 4:a014365. doi: 10.1101/cshperspect.a014365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kress TR, Sabo A, Amati B. 2015. MYC: connecting selective transcriptional control to global RNA production. Nat Rev Cancer 15:593–607. doi: 10.1038/nrc3984. [DOI] [PubMed] [Google Scholar]
- 46.Murray JT, Campbell DG, Morrice N, Auld GC, Shpiro N, Marquez R, Peggie M, Bain J, Bloomberg GB, Grahammer F, Lang F, Wulff P, Kuhl D, Cohen P. 2004. Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. Biochem J 384:477–488. doi: 10.1042/BJ20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee JC, Chung LC, Chen YJ, Feng TH, Juang HH. 2014. N-myc downstream-regulated gene 1 downregulates cell proliferation, invasiveness, and tumorigenesis in human oral squamous cell carcinoma. Cancer Lett 355:242–252. doi: 10.1016/j.canlet.2014.08.035. [DOI] [PubMed] [Google Scholar]
- 48.Chang X, Xu X, Ma J, Xue X, Li Z, Deng P, Zhang S, Zhi Y, Chen J, Dai D. 2014. NDRG1 expression is related to the progression and prognosis of gastric cancer patients through modulating proliferation, invasion and cell cycle of gastric cancer cells. Mol Biol Rep 41:6215–6223. doi: 10.1007/s11033-014-3501-2. [DOI] [PubMed] [Google Scholar]
- 49.Meyer N, Penn LZ. 2008. Reflecting on 25 years with MYC. Nat Rev Cancer 8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 50.Chan KL, Guan XY, Ng IO. 2004. High-throughput tissue microarray analysis of c-myc activation in chronic liver diseases and hepatocellular carcinoma. Hum Pathol 35:1324–1331. doi: 10.1016/j.humpath.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 51.Li Q, Sodroski C, Lowey B, Schweitzer CJ, Cha H, Zhang F, Liang TJ. 2016. Hepatitis C virus depends on E-cadherin as an entry factor and regulates its expression in epithelial-to-mesenchymal transition. Proc Natl Acad Sci U S A 113:7620–7625. doi: 10.1073/pnas.1602701113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zarrinpashneh E, Poggioli T, Sarathchandra P, Lexow J, Monassier L, Terracciano C, Lang F, Damilano F, Zhou JQ, Rosenzweig A, Rosenthal N, Santini MP. 2013. Ablation of SGK1 impairs endothelial cell migration and tube formation leading to decreased neo-angiogenesis following myocardial infarction. PLoS One 8:e80268. doi: 10.1371/journal.pone.0080268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murakami Y, Hosoi F, Izumi H, Maruyama Y, Ureshino H, Watari K, Kohno K, Kuwano M, Ono M. 2010. Identification of sites subjected to serine/threonine phosphorylation by SGK1 affecting N-myc downstream-regulated gene 1 (NDRG1)/Cap43-dependent suppression of angiogenic CXC chemokine expression in human pancreatic cancer cells. Biochem Biophys Res Commun 396:376–381. doi: 10.1016/j.bbrc.2010.04.100. [DOI] [PubMed] [Google Scholar]
- 54.Pietiainen V, Vassilev B, Blom T, Wang W, Nelson J, Bittman R, Back N, Zelcer N, Ikonen E. 2013. NDRG1 functions in LDL receptor trafficking by regulating endosomal recycling and degradation. J Cell Sci 126:3961–3971. doi: 10.1242/jcs.128132. [DOI] [PubMed] [Google Scholar]
- 55.Reddy JV, Ganley IG, Pfeffer SR. 2006. Clues to neuro-degeneration in Niemann-Pick type C disease from global gene expression profiling. PLoS One 1:e19. doi: 10.1371/journal.pone.0000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bartz F, Kern L, Erz D, Zhu M, Gilbert D, Meinhof T, Wirkner U, Erfle H, Muckenthaler M, Pepperkok R, Runz H. 2009. Identification of cholesterol-regulating genes by targeted RNAi screening. Cell Metab 10:63–75. doi: 10.1016/j.cmet.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 57.Goodman JM. 2008. The gregarious lipid droplet. J Biol Chem 283:28005–28009. doi: 10.1074/jbc.R800042200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pawlak M, Lefebvre P, Staels B. 2015. Molecular mechanism of PPARalpha action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J Hepatol 62:720–733. doi: 10.1016/j.jhep.2014.10.039. [DOI] [PubMed] [Google Scholar]
- 59.Gao Q, Goodman JM. 2015. The lipid droplet—a well-connected organelle. Front Cell Dev Biol 3:49. doi: 10.3389/fcell.2015.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Q, Pene V, Krishnamurthy S, Cha H, Liang TJ. 2013. Hepatitis C virus infection activates an innate pathway involving IKK-alpha in lipogenesis and viral assembly. Nat Med 19:722–729. doi: 10.1038/nm.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kato T, Matsumura T, Heller T, Saito S, Sapp RK, Murthy K, Wakita T, Liang TJ. 2007. Production of infectious hepatitis C virus of various genotypes in cell cultures. J Virol 81:4405–4411. doi: 10.1128/JVI.02334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Q, Zhang YY, Chiu S, Hu Z, Lan KH, Cha H, Sodroski C, Zhang F, Hsu CS, Thomas E, Liang TJ. 2014. Integrative functional genomics of hepatitis C virus infection identifies host dependencies in complete viral replication cycle. PLoS Pathog 10:e1004163. doi: 10.1371/journal.ppat.1004163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schweitzer CJ, Matthews JM, Madson CJ, Donnellan MR, Cerny RL, Belshan M. 2012. Knockdown of the cellular protein LRPPRC attenuates HIV-1 infection. PLoS One 7:e40537. doi: 10.1371/journal.pone.0040537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harris VM. 2015. Protein detection by simple Western analysis. Methods Mol Biol 1312:465–468. doi: 10.1007/978-1-4939-2694-7_47. [DOI] [PubMed] [Google Scholar]
- 66.Chen JQ, Heldman MR, Herrmann MA, Kedei N, Woo W, Blumberg PM, Goldsmith PK. 2013. Absolute quantitation of endogenous proteins with precision and accuracy using a capillary Western system. Anal Biochem 442:97–103. doi: 10.1016/j.ab.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carpentier A, Tesfaye A, Chu V, Nimgaonkar I, Zhang F, Lee SB, Thorgeirsson SS, Feinstone SM, Liang TJ. 2014. Engrafted human stem cell-derived hepatocytes establish an infectious HCV murine model. J Clin Invest 124:4953–4964. doi: 10.1172/JCI75456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maillard P, Walic M, Meuleman P, Roohvand F, Huby T, Le Goff W, Leroux-Roels G, Pecheur EI, Budkowska A. 2011. Lipoprotein lipase inhibits hepatitis C virus (HCV) infection by blocking virus cell entry. PLoS One 6:e26637. doi: 10.1371/journal.pone.0026637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carvalho BS, Irizarry RA. 2010. A framework for oligonucleotide microarray preprocessing. Bioinformatics 26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 72.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]