ABSTRACT
Epstein-Barr virus (EBV) is a causative agent of a variety of lymphomas, nasopharyngeal carcinoma (NPC), and ∼9% of gastric carcinomas (GCs). An important question is whether particular EBV variants are more oncogenic than others, but conclusions are currently hampered by the lack of sequenced EBV genomes. Here, we contribute to this question by mining whole-genome sequences of 201 GCs to identify 13 EBV-positive GCs and by assembling 13 new EBV genome sequences, almost doubling the number of available GC-derived EBV genome sequences and providing the first non-Asian EBV genome sequences from GC. Whole-genome sequence comparisons of all EBV isolates sequenced to date (85 from tumors and 57 from healthy individuals) showed that most GC and NPC EBV isolates were closely related although American Caucasian GC samples were more distant, suggesting a geographical component. However, EBV GC isolates were found to contain some consistent changes in protein sequences regardless of geographical origin. In addition, transcriptome data available for eight of the EBV-positive GCs were analyzed to determine which EBV genes are expressed in GC. In addition to the expected latency proteins (EBNA1, LMP1, and LMP2A), specific subsets of lytic genes were consistently expressed that did not reflect a typical lytic or abortive lytic infection, suggesting a novel mechanism of EBV gene regulation in the context of GC. These results are consistent with a model in which a combination of specific latent and lytic EBV proteins promotes tumorigenesis.
IMPORTANCE Epstein-Barr virus (EBV) is a widespread virus that causes cancer, including gastric carcinoma (GC), in a small subset of individuals. An important question is whether particular EBV variants are more cancer associated than others, but more EBV sequences are required to address this question. Here, we have generated 13 new EBV genome sequences from GC, almost doubling the number of EBV sequences from GC isolates and providing the first EBV sequences from non-Asian GC. We further identify sequence changes in some EBV proteins common to GC isolates. In addition, gene expression analysis of eight of the EBV-positive GCs showed consistent expression of both the expected latency proteins and a subset of lytic proteins that was not consistent with typical lytic or abortive lytic expression. These results suggest that novel mechanisms activate expression of some EBV lytic proteins and that their expression may contribute to oncogenesis.
KEYWORDS: whole-genome sequencing, transcriptome, Epstein-Barr virus lytic proteins, gastric cancer
INTRODUCTION
Epstein-Barr virus (EBV) is a common gammaherpesvirus that is the causative agent of a variety of lymphomas as well as nasopharyngeal carcinoma (NPC) and gastric carcinoma (GC) (1). Gastric carcinoma comprises 2% of cancers in Western countries, and 9% of these are EBV infected. EBV-positive GCs have distinct molecular profiles and clinical features that distinguish them from other GCs, reflecting unique mechanisms by which EBV induces cancer (2). However, the mechanisms by which EBV infection promotes GC and other EBV-associated cancers are unclear.
EBV adopts both latent and lytic forms of infection. In latent infection, a small proportion of the EBV genome is expressed, and cells become immortalized. These cells can switch to a lytic infection that involves sequential expression of immediate early, early, and late EBV proteins (∼80 proteins in total), amplification of the viral genomes, and virion production. EBV-induced tumors are monoclonal expansions of latently infected cells, and many studies have focused on the contributions of the few EBV latency proteins expressed in these cells. For example, EBV-positive GCs have been reported to consistently express EBV nuclear antigen 1 (EBNA1) and can also express latent membrane protein 1 (LMP1) and LMP2A proteins (3). However, there have been many reports of some EBV lytic cycle proteins being expressed in EBV-induced tumors (4–10). In addition, mutant viruses that are unable to switch to the lytic cycle have been found to be impaired in their ability to induce EBV-mediated lymphoproliferative disease in mice (11). Together, these reports have led to the suggestion that abortive lytic EBV infection contributes to tumorigenesis (12) although a comprehensive analysis of lytic protein expression in EBV-induced tumors is currently lacking. The importance of expression of specific lytic proteins for cancer induction is consistent with studies on Kaposi's sarcoma-associated herpesvirus (KSHV), the other human gammaherpesvirus, in which specific lytic proteins appear to contribute to oncogenesis (13).
Another outstanding question concerning how EBV infection induces cancer is whether specific EBV variants are more associated with cancer than others. Precedence for this scenario exists for cancer induction by human papillomavirus (HPV), in which there are specific variants (high-risk strains) that promote cancer while most variants (low-risk strains) do not. This difference is due to the differing abilities of the encoded HPV proteins to bind and interfere with the functions of cellular tumor suppressors (14–17). For EBV, it is unclear whether there are high-risk and low-risk variants, but evidence suggests that EBV that is isolated from tumors can differ in sequence and properties from the widely studied blood EBV isolate (18). Efforts to generate EBV genome sequences for comparison are under way and have resulted in EBV genome sequences from NPC, Burkitt's lymphoma (BL), and Hodgkin lymphoma (HL) as well as from the blood or saliva of healthy individuals (19–24). Comparisons of 10 EBV genomes isolated from NPC tumors to those of the few previously sequenced EBV blood isolates suggested that NPC isolates are most similar to each other and identified nonsynonymous mutations of potential biological significance in genes encoding both latent and lytic proteins (22). However, the small number of EBV genome sequences analyzed has limited any conclusions on the association of specific EBV mutations with cancer.
In order to increase the number of EBV genome sequences and examine the relationship of GC EBV isolates to other EBV genomes, we analyzed whole-genome sequencing (WGS) data of GC samples, resulting in the assembly of 13 new EBV genome sequences. These were combined with 15 existing GC isolates and analyzed against 114 EBV genomes from other tumors, blood, and saliva to identify GC-associated EBV nonsynonymous mutations that could potentially impact oncogenesis. In addition, we analyzed whole-transcriptome data for these GC samples to determine EBV expression profiles in GC, identifying distinct sets of EBV lytic proteins that are consistently expressed in GC.
RESULTS
Characterization of newly assembled GC EBV genomes.
We analyzed whole-genome sequencing (WGS) data of 201 gastric adenocarcinoma samples for their EBV content, using 122 WGS samples available from The Cancer Genome Atlas ([TCGA] Stomach Adenocarcinoma, project code STAD-US) (25) and 79 from the new Pan-Cancer Analysis of Whole Genomes ([PCAWG] Stomach Adenocarcinoma, project codes STAD-US and GACA-CN [gastric cancer-China]) from the International Cancer Genome Consortium (ICGC) (26). We identified EBV genomes in 13 of these samples with sufficient read depth (average read depth of 125×) to assemble the EBV genome, which was done using a reference-based approach.
To further assess the quality of our newly assembled GC EBV sequences, we compared them to 15 GC EBV whole-genome sequences downloaded from the NCBI for their average sequence lengths (calculated as the length of the GC-EBV query sequence that aligned to the EBV NC_007605 reference sequence in NCBI), sequence identities, number of single nucleotide variants (SNVs), number of gaps, and number of ambiguous bases. Comparisons of five sequence metrics in Table 1 show that the 13 newly assembled GC EBV genome sequences are of similar quality to those previously reported, with the exception of the number of ambiguous bases, which is significantly lower in our newly assembled sequences.
TABLE 1.
Comparison of the newly assembled EBV genomes to previous GC-derived EBV genomes
| Sequence metric | Value for the GC EBV group (mean ± SD) |
|
|---|---|---|
| Newly assembled genomes | Previously derived genomes in the NCBI nucleotide database | |
| Avg pairwise aligned sequence lengtha | 170,771 ± 3,668 | 164,756 ± 7,820 |
| Avg sequence identity (%) | 98 ± 2 | 95 ± 5 |
| Avg no. of SNVs | 1,245 ± 220 | 1,201 ± 102 |
| Avg no. of gaps | 698 ± 502 | 1,013 ± 848 |
| Avg no. of ambiguous bases | 1,824 ± 2,419 | 6,461 ± 8,152 |
Calculated as the length of the GC-EBV query sequence that aligned to the EBV reference sequence, NCBI accession number NC_007605.
Relationship of GC-derived EBV to other EBV isolates.
We next investigated the relationship between GC-derived EBV and all other currently available EBV genomes. To generate the comparison group, we downloaded 126 EBV genome sequences from NCBI: 72 from tumors and 54 derived from the blood or saliva of people without cancer, as indicated in Table 2. The EBV blood isolates had been used to generate lymphoblastoid cell lines (LCLs), and EBV sequences from three additional LCLs were obtained from the study described in Santpere et al. (27) (cell lines NA19114, NA19315 and NA19384). We then combined our 13 new GC-derived EBV sequences with the 15 GC EBV sequences available in NCBI and compared these to the B95.8-Raji reference EBV sequence commonly used for comparison (NCBI accession number NC_007605) (19). This comparison identified a total of 4,172 SNVs and 112 insertions and 103 deletions (indels).
TABLE 2.
Summary of EBV isolates analyzed
| Sample type | No. of sequences |
|||
|---|---|---|---|---|
| From NCBI | From another sourcea | New assembly | Total | |
| Gastric carcinoma | 15 | 0 | 13 | 28 |
| Hodgkin lymphoma | 8 | 0 | 0 | 8 |
| Nasopharyngeal carcinoma | 22 | 0 | 0 | 22 |
| Burkitt's lymphoma | 27 | 0 | 0 | 27 |
| Lymphoblastoid cell line | 32 | 3 | 0 | 35 |
| Lymphoblastoid cell line from PTLDb | 19 | 0 | 0 | 19 |
| Healthy saliva | 1 | 0 | 0 | 1 |
| Infectious mononucleosis | 2 | 0 | 0 | 2 |
| Total no. of sequences | 126 | 3 | 13 | 142 |
EBV sequences downloaded from Santpere et al. (27).
PTLD, posttransplant lymphoproliferative disease.
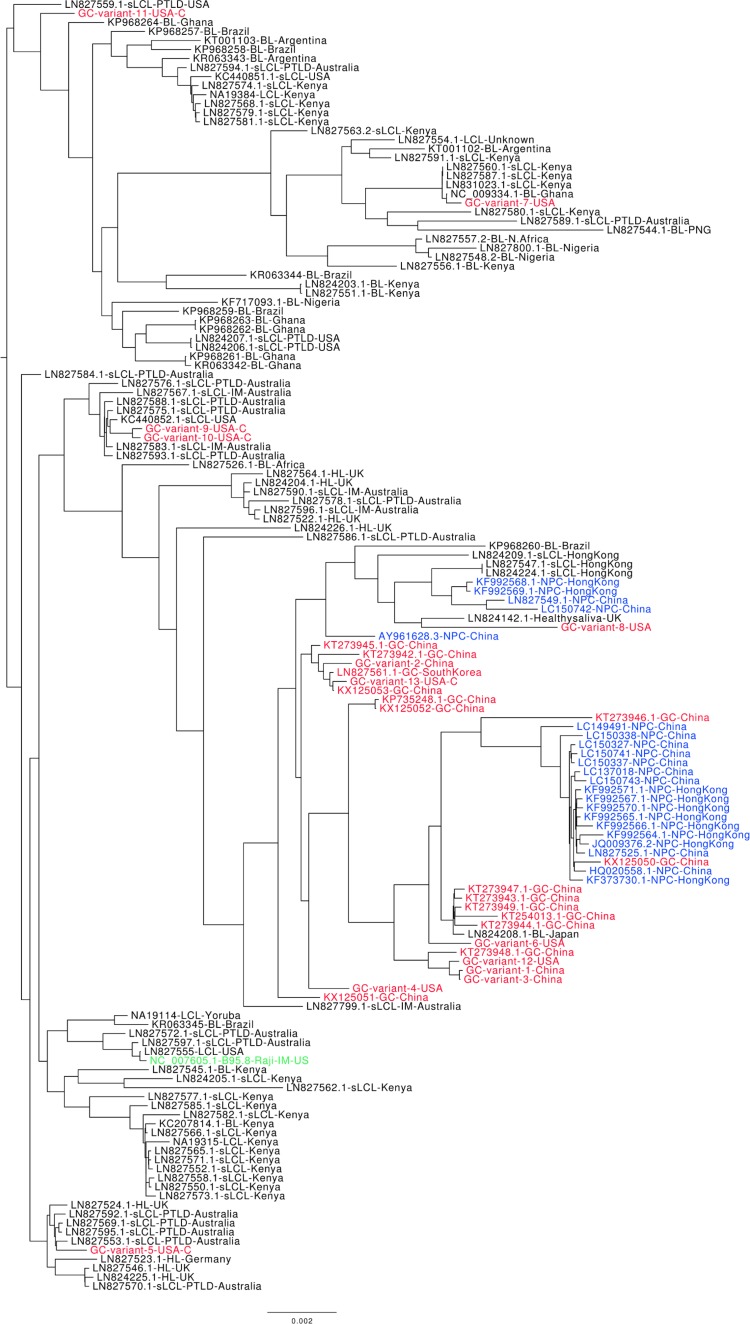
We conducted a phylogenetic analysis on 142 whole EBV genomes, combining our new GC EBV sequences with those from public databases (as indicated above and in Table 2). Figure 1 shows that the majority of GC EBV isolates cocluster with the majority of NPC EBV isolates. The similarity between GC and NPC EBV isolates is also evident in the similar patterns of sequence changes within these isolates, as shown in Fig. 2. One of the issues regarding the clustering and analysis of EBV sequences isolated from specific tumor types is the potential confounding effect due to their geographical localizations. This is particularly an issue for NPC and BL since all NPC-derived EBV genome sequences are from Asia while all BL-derived sequences are from Africa. Prior to this study, all GC-derived EBV whole-genome sequences were also Asian. However, 10 of the EBV sequences we assembled were from American GC samples, and 5 of these were from Caucasians. The phylogenetic tree shown in Fig. 1 shows that all of the Asian GC isolates and a subset of the American GC isolates clustered with the NPC isolates, indicating that they are most closely related. However, four of the five Caucasian American GC isolates were not part of this cluster, suggesting that geography/and or ethnicity is a factor in the close relationship between NPC and GC EBV sequences.
FIG 1.
Phylogenetic analysis on whole EBV genomes. All available EBV genome sequences were compared based on SNVs (disregarding repeat regions). EBV isolates from GC (red), NPC (blue), and the B95.8-Raji reference strain (green) are shown. In black are EBV isolates from LCLs, Burkitt's lymphoma (BL), and Hodgkin's lymphoma (HL). The geographical locations of the EBV isolates are also indicated. U.S. GC isolates from Caucasians are indicated with a “C,” and the remaining samples are from Asians. The phylogenetic tree was rooted using a midpoint rooting.
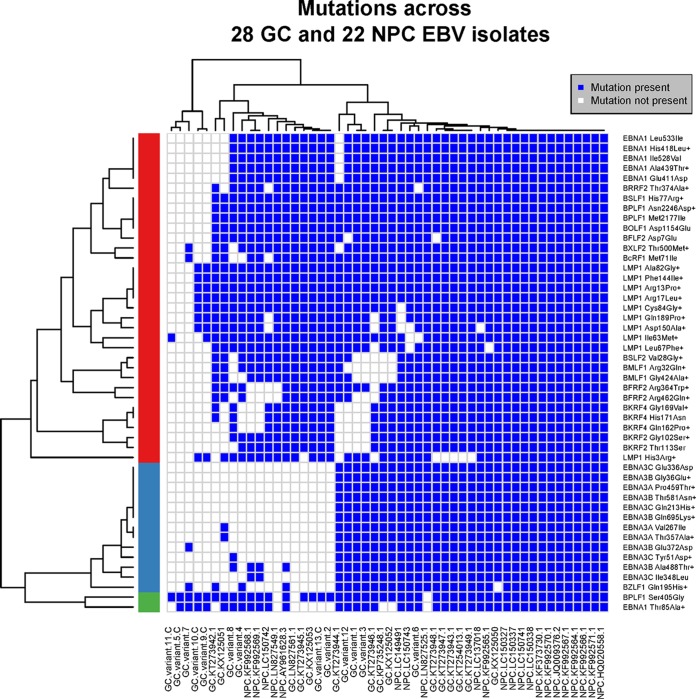
FIG 2.
Comparison of polymorphisms across GC and NPC EBV isolates. Amino acid changes across GC and NPC EBV sequences (compared to all other EBV isolates) as shown in Tables 3 and 4 (columns headed GC + NPC vs all others) were subjected to row-wise (across isolates) and column-wise (across mutation profiles) hierarchical clustering analysis; the Manhattan method was used for obtaining the distance matrix, and the complete-linkage method was used for agglomerating the clusters. Using row-wise clustering, three main clusters were identified; the clusters in red and green indicate amino acid changes that occur in most of the GC and NPC EBV sequences, and the cluster in blue indicates changes that occur in a subset of GC and NPC sequences. The sizes of the cluster in red and green indicate that the majority of amino acid changes observed in GC also occur in NPC isolates. U.S. GC isolates from Caucasians are indicated with a “C,” and the remaining samples are from Asians. Nonconservative amino acid changes are indicated with a plus symbol.
To gain more information about the GC/NPC cluster and how these EBV sequences differ from other EBV sequences, we examined indels and nonsynonymous SNVs that would result in amino acid changes in EBV proteins. One deletion was consistently found: an in-frame deletion of 30 bases in the third exon of the LMP1 gene, resulting in the deletion of 10 amino acids (from Gly343 to His352). This deletion was found in 82% of GC, 61% of NPC, 75% of HL, 48% of BL, and 32% of normal EBV isolates. This deletion has been previously reported in some GC and NPC EBV isolates and was shown not to affect the transforming potential of LMP1 (28–30). This LMP1 deletion appears to be strongly influenced by geography and/or ethnicity as we found it in 100% of the Asian GC isolates but in only one of the five American Caucasian GC isolates.
Nonsynonymous SNVs that are common in both GC (≥50%) and NPC (≥50%) isolates and uncommon in all other EBV isolates (occurring in <25% and significantly different, with an adjusted P value of <0.05) are shown in Tables 3 and 4 (columns headed GC + NPC vs all others) for latent and lytic EBV proteins, respectively. The majority of changes map to the latency proteins, including EBNA1 and LMP1 which are expressed in both NPC and GC. Nonconservative amino acid changes are of particular interest as these are the most likely to affect protein function (shown in boldface in Tables 3 and 4). A graphical representation of the nonconservative amino acids changes common in GC and NPC is shown for individual EBV genomes in Fig. 2, showing that most of these changes occur together. Similar to the phylogenetic tree, these results show coclustering of the Asian GC samples with the NPC samples but less so with American GC samples.
TABLE 3.
EBV latent protein sequence changes occurring in GC-derived EBV isolates
| Protein | Comparison of sequence change(s) by isolate groupsa |
||||
|---|---|---|---|---|---|
| GC + NPC vs all othersb | GC vs LCLc | GC vs B95.8 (≥60%)d | GC vs B95.8 (100%)e | GC vs NPCf | |
| BARF0 | Cys30Arg, Arg375Ser | Cys30Arg | |||
| EBNA1 | Thr85Ala, Glu411Asp, His418Leu, Ala439Thr, Ile528Val, Leu533Ile | Thr85Ala | Glu16Gln, Gly18Glu, Thr85Ala, Thr524Ile, Arg594Lys | Thr42Ile | |
| EBNA3A | Val267Ile, Thr357Ala, Pro459Thr | Leu219Pro, Ile333Leu, Phe492Ser | |||
| EBNA3B | Gly36Glu, Glu372Asp, Ala488Thr, Thr581Asn, Gln695Lys | Val466Ala, Gln815Arg | |||
| EBNA3C | Tyr51Asp, Gln213His, Glu336Asp, Ile348Leu | Glu701Gln | |||
| LMP1 | His3Arg, Arg13Pro, Arg17Leu, Ile63Met, Leu67Phe, Ala82Gly, Cys84Gly, Phe144Ile, Asp150Ala, Gln189Pro | Arg13Pro, Arg17Leu, Leu25Ile, Ala82Gly, Cys84Gly, Phe144Ile, Asp150Ala, Gln189Pro | Arg13Pro, Arg17Leu, Leu25Ile, Asp46Asn, Ala82Gly, Cys84Gly, Ile85Leu, Phe106Tyr, Ile122Leu, Leu126Phe, Met129Ile, Phe144Ile, Asp150Ala, Leu151Ile, Gln189Pro, Gly212Ser, Ser309Asn, Gln322His, Gln334Arg, Leu338Ser, Ser366Thr | Ile85Leu, Phe106Tyr, Met129Ile, Ser309Asn, Ser366Thr | |
| LMP2A | Ser444Thr, Tyr23Asp | Ser444Thr, Tyr23Asp | Ser349Thr | ||
| LMP2B | Ser325Thr | Ser325Thr | Ser229Thr | ||
Nonconservative changes are indicated in bold.
Changes found in 50% of GC and NPC isolates relative to the sequence of B95.8 and in <25% in all other EBV isolates (adjusted P value [FDR], ≤0.05).
Changes found in ≥60% of both Asian and Caucasian GC isolates relative to the sequence of B95.8 but in <25% of LCL isolates (FDR ≤ 0.05).
Changes found in ≥60% of both Asian and Caucasian GC isolates relative to the sequence of B95.8.
Changes found in 100% of GC isolates compared to the sequence of B95.8.
Changes found in ≥75% of Asian GC isolates relative to the sequence of B95.8 and in <25% of Asian NPC isolates (FDR ≤ 0.05).
TABLE 4.
EBV lytic protein sequence changes occurring in GC-derived EBV isolates
| Protein | Comparison of sequence change(s) by isolate groupsa |
||||
|---|---|---|---|---|---|
| GC + NPC vs all othersb | GC vs LCLc | GC vs B95.8 (≥60%)d | GC vs B95.8 (100%)e | GC vs NPCf | |
| BALF4 | Ser416Pro, Pro423Ser, Thr444Ala | ||||
| BBLF2-BBLF3 | Met174Val | ||||
| BBLF4 | Arg37Lys, Ala339Thr | Arg37Lys | |||
| BBRF1 | Lys196Arg | Lys196Arg | |||
| BBRF3 | Leu266Met | ||||
| BcLF1 | Ser651Ala | ||||
| BcRF1 | Met71Ile | Thr33Ala, Gln337His | |||
| BDLF1 | Leu51Ile | ||||
| BDLF2 | Pro79Ala, Thr105Ala, His269Leu, Cys357Ser | ||||
| BDLF3 | Phe83Ser, Ser134Thr | Phe83Ser, Ser134Thr | |||
| BDLF4 | Glu141Asp | ||||
| BdRF1 | Ser222Pro | ||||
| BFLF1 | Ile35Thr, Val246Ile, Leu250Gln | ||||
| BFLF2 | Asp7Glu | ||||
| BFRF2 | Arg364Trp, Arg462Gln | Cys17Gly, Gln34Pro, Gly40Glu, Val130Phe, Asp169Ala | Asp169Ala | ||
| BFRF3 | His165Gln | His165Gln | |||
| BGLF1 | Asp85Glu | Asp85Glu | Ile149Met, Val171Ala | ||
| BGLF3 | Gln150His | Gln150His | |||
| BGLF3.5 | His54Arg | His54Arg | |||
| BGLF5 | Phe46Tyr | Phe46Tyr | |||
| BKRF2 | Gly102Ser, Thr113Ser | Ile10Thr | Ile10Thr | ||
| BKRF4 | Gln162Pro, Gly169Val, His171Asn | ||||
| BLLF1 | Glu201Gln, Trp495Arg, Asn672Ile | ||||
| BLLF2 | His8Arg | ||||
| BLLF3 | Thr247Ala | Thr247Ala | |||
| BMLF1 | Gly424Ala, Arg32Gln | ||||
| BNLF2b | Phe74Cys, Phe74Tyr | ||||
| BNRF1 | Glu36Gln, Asn797Ser, Phe1110Ser | Glu36Gln, Asn797Ser, Phe1110Ser | Gly549Ser | ||
| BOLF1 | Asp1154Glu | His595Arg, Ile968Thr | Ile968Thr | ||
| BORF1 | Val247Gly | Val247Gly | |||
| BPLF1 | Ser405Gly, Met2177Ile, Asn2246Asp | Ser405Gly, Lys515Glu | Ser405Gly, Lys515Glu, Ile1315Val, Gln1428Arg | Ile1315Val, Gln1428Arg | Lys2616Glu |
| BRLF1 | Ser542Asn | ||||
| BRRF2 | Thr374Ala | Gln285Lys, His313Arg, Ser325Leu, Lys392Gln, Phe430Ser, Asp463Ala | Phe430Ser, Asp463Ala | ||
| BSLF1 | His77Arg | ||||
| BSLF2 | Val28Gly | ||||
| BTRF1 | Thr229Ala | Thr229Ala | |||
| BVRF2 | Ser482Pro | ||||
| BXLF2 | Thr500Met | ||||
| BZLF1 | Gln195His | ||||
Nonconservative changes are indicated in bold.
Changes found in ≥50% of GC and NPC isolates relative to the sequence of B95.8 and in <25% in all other EBV isolates (adjusted P value [FDR], ≤0.05).
Changes found in ≥60% of both Asian and Caucasian GC isolates relative to the sequence of B95.8 but in <25% of LCL isolates (FDR ≤ 0.05).
Changes found in ≥60% of both Asian and Caucasian GC isolates relative to the sequence of B95.8.
Changes found in 100% of GC isolates compared to the sequence of B95.8.
Changes found in ≥75% of Asian GC isolates relative to the sequence of B95.8 and in <25% of Asian NPC isolates (FDR ≤ 0.05).
Ten nonsynonymous mutations associated with GC and NPC were identified in LMP1, all resulting in nonconservative amino acid changes within the first 189 amino acids (Table 3). Six nonsynonymous mutations were identified in EBNA1, including three nonconservative changes (Thr85Ala, His418Leu, and Ala439Thr) (Table 3). These changes occur, on average, in 71% of GC and 99% of NPC isolates but are much less common in lymphoma and normal EBV isolates (10% of BL, 0% of HL, and 7% of normal EBV isolates; P value of <2e−10). Five of the nonsynonymous changes have been previously reported to be associated with EBV isolated from NPC (30). Our analysis shows that these changes are also significantly associated (P value of 0.05) with Asian GC samples (19/23) but not with U.S. Caucasian GC samples (1/5), suggesting that these changes may reflect strain prevalence in Asia. In addition, our analysis identified a previously unreported nonconservative change in Thr85Ala in EBNA1 that is found in 91% of NPC and 68% of GC isolates but in only 8% of other EBV isolates. Interestingly, this change is found with similar frequencies in Asian (16/23) and American Caucasian (3/5) GC isolates.
GC-associated EBV genome alterations.
In order to identify EBV protein sequence changes that might be involved in induction of GC, we investigated EBV protein sequence changes that are common in GC and uncommon in LCLs. Since few of the LCLs are from Asia whereas most of GC samples are Asian, there is the potential for a strong geographical bias. To address this issue, we looked for EBV sequence changes that were common in both Asian and American Caucasian GC isolates (≥60% for each group) and uncommon (<25%) in LCL isolates. This identified 11 significant nonsynonymous SNV changes (adjusted P value of <0.05) as shown in Fig. 3. As shown in Tables 3 and 4 (columns headed GC vs LCLs), these SNVs corresponded to amino acid changes in one EBV lytic protein (BPLF1) as well as eight nonconservative changes in LMP1 and one nonconservative change in EBNA1. The EBNA1 sequence change (Thr85Ala) and seven of the LMP changes are the same as those reported above to be altered in the GC/NPC cluster compared to sequences of other EBV isolates. This suggests that there are a small number of EBV sequence alterations in GC EBV isolates that are independent of the geographical origin or ethnicity of the GC.
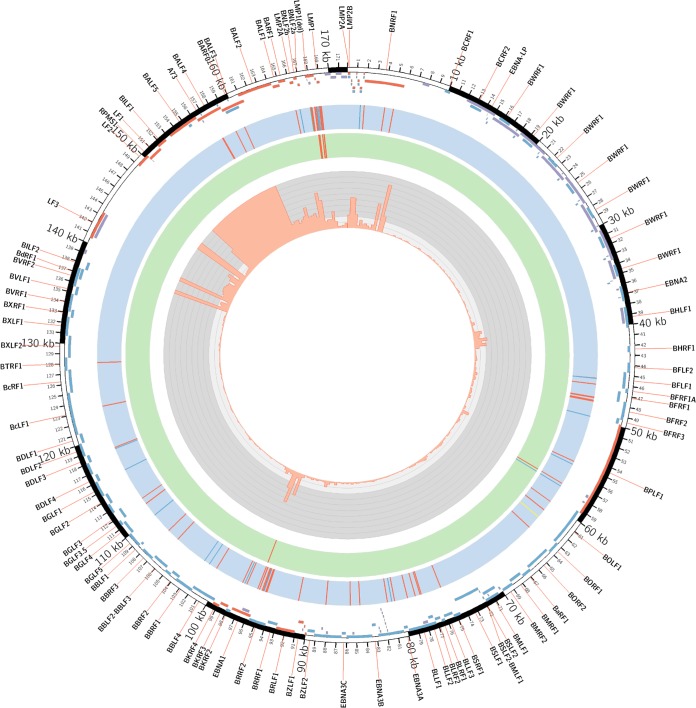
FIG 3.
Genome-wide analysis of EBV sequences and gene expression associated with gastric adenocarcinoma. An annotated Circos plot depicting EBV amino acid changes common (≥60%) in both Asian and Caucasian GC isolates relative to the sequence of B95.8 (blue track) or common (≥60%) in both Asian and Caucasian GC samples but uncommon in LCLs (green track) and RNA-Seq read coverage across the EBV (NCBI accession number NC_007605) reference genome (gray track). For the EBV genome changes, nonconservative and conservative amino acid changes are indicated in red and blue, respectively; the BOLF1 insertion is shown in yellow, and the LMP1 deletion is marked in green and labeled. In the outer track, the positions of EBV genes that are expressed (red) or not expressed (blue), as well as the repeat regions excluded from the analysis (violet), are indicated.
We also asked whether there are changes in GC isolates that do not occur in NPC isolates. To avoid a bias due to geography and/or ethnicity, we included only Asian GC samples in this comparison since all NPC samples were Asian. We looked for amino acid changes found in >75% of Asian GC isolates but in <25% of NPC (Tables 3 and 4, GC vs NPC). The results showed only two conservative changes in the latency proteins (in LMP2A and LMP2B) and several changes in 10 lytic proteins. This is consistent with the above conclusions that Asian NPC and GC EBV isolates are very similar.
Finally, since the B95.8 EBV isolate is the most commonly studied and since protein expression clones are typically based on this variant, it was of interest to determine what changes in EBV proteins are common in GC isolates relative to the B95.8 sequence. Again, we looked for changes that occurred in ≥60% of Asian and in ≥60% of Caucasian GC isolates to counter any geographical/ethnicity effects. Figure 3 shows a graphic summary of the genetic variants from this comparison, which identified 87 nonsynonymous SNVs and two indels. Amino acid changes in 9 latency and 25 lytic proteins are shown in Tables 3 and 4 (columns headed GC vs B95.8, ł60%), respectively. The indels were the LMP1 deletion indicated above and an insertion in BOLF1, consisting of a glycine at amino acid 261 relative to B95.8 sequence. We also asked whether there were any changes that occurred in 100% of GC samples relative to the sequence of B95.8. This showed that a subset of the above GC-associated changes in both latent and lytic proteins occurred in all GC-derived EBVs regardless of geography (Tables 3 and 4, GC vs B95.8, 100%), including one change in EBNA1 (Thr524Ile). This change is common in many EBV isolates, indicating that the B95.8 reference sequence is unusual at this position (30, 31). Thr524 is in the DNA binding/dimerization domain in an α-helix important for contacting the DNA (32, 33) although the contribution of Thr524 in DNA recognition has not been determined. In addition, all GC isolates have the BOLF1 insertion at amino acid 261 indicated above. These sequence changes will be important to incorporate when expression clones are generated to study the functions and protein interactions of these EBV proteins in the context of gastric infections.
EBV gene expression in gastric carcinoma.
Another important question for understanding the mechanism of cancer induction by EBV is which EBV proteins are expressed. Although many studies have focused on the EBV latent proteins that are consistently expressed in tumors, there have been many reports of detection of lytic proteins in a variety of EBV tumors although the profile of which EBV lytic proteins are expressed and of their frequencies of expression is not clear. Of the 13 EBV-positive gastric tumor samples that we used to assemble EBV genome sequences, eight had whole-transcriptome data [on purified poly(A)-containing RNA] which we used to determine the level of each EBV transcript (in reads per kilobase of transcript per million mapped reads [RPKM]) using the EBV NC_007605 in NCBI as the reference genome. A previous paper had also analyzed EBV transcripts in four GC samples but had not reported the complete profile of EBV lytic protein transcripts (12). Therefore, we attempted to reanalyze these data as well. However, we learned that TCGA had determined that all four samples were from the same patient, and hence three out of four of these duplicate samples (BR-4298, BR-4376, and BR-4271) were removed from the TCGA database (https://portal.gdc.cancer.gov/). Analysis of the remaining sample (BR-4253) showed a lower overall number of reads mapping to the EBV transcriptome (4- to 18-fold less) than for the eight samples we analyzed, which would hinder identification of low-abundance transcripts; hence, this sample was not included in our further analyses.
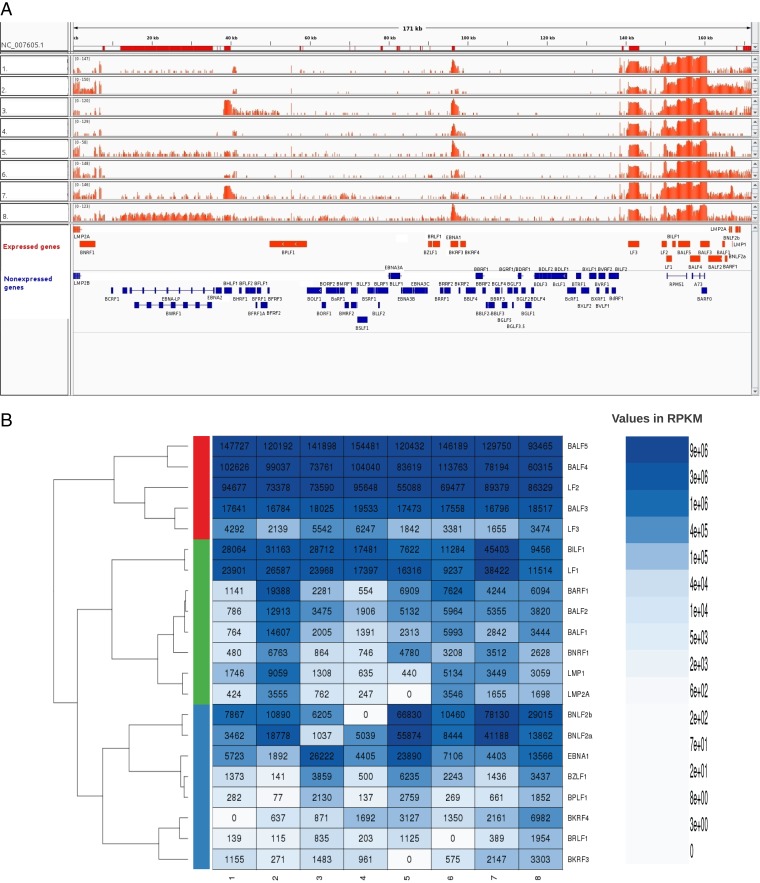
The EBV transcriptome profiles for each of the eight GC samples are shown in Fig. 4A. Since it is well established that EBNA2, -3A, -3B, and -3C are not expressed in GC, the average RPKM value for these transcripts was used as background in each sample, and transcripts that were above this level in 7 or 8 of the 8 samples are shown in Table 5. As expected based on previous reports, EBNA1 transcripts were readily detected in all samples. We also detected variable levels of LMP1 transcripts and low levels of LMP2A transcripts in 7 out of 8 samples.
FIG 4.
EBV gene expression profiles for GC samples. (A) EBV transcriptome read coverage is shown for eight individual GC samples relative to the reference EBV genome (NCBI accession number NC_007605; represented using a log2 scale). The red bars in the top track indicate the location of the EBV repeat regions. The annotation track at the bottom shows EBV expressed genes in red and nonexpressed genes in blue. (B) Expressed genes shown in Table 5 were subjected to the row-wise hierarchical clustering analysis across eight EBV-positive GC transcriptome samples. Pearson correlation was used for obtaining the distance matrix, and the complete-linkage method was used for agglomerating the clusters. The three main clusters shown in red, green, and blue indicate three different groups of genes with highest correlations between their gene expression levels across samples.
TABLE 5.
EBV transcripts detected in GC samplesa
| Protein name | Protein classificationb | Fold change in expressionc | Mean expression level (RPKM [95% CI])c,d | No. of GC samplesc | P value | FDRe |
|---|---|---|---|---|---|---|
| BALF5 | Lytic (E) | 444.8 | 131,767 (117,871, 145,663) | 8 | 1.63E-007 | 1.71E-006 |
| BALF4 | Lytic (L) | 301.8 | 89,419 (76,790, 102,049) | 8 | 1.21E-006 | 6.35E-006 |
| LF2 | Lytic (E) | 269 | 79,696 (69,882, 89,509) | 8 | 4.74E-007 | 3.32E-006 |
| BNLF2b | Lytic (E) | 88.4 | 26,175 (5,452, 46,897) | 7 | 2.21E-002 | 2.92E-002 |
| BILF1 | Lytic (E) | 75.6 | 22,398 (13,281, 31,515) | 8 | 1.04E-003 | 3.11E-003 |
| LF1 | Lytic (E) | 70.6 | 20,918 (14,421, 27,415) | 8 | 2.16E-004 | 9.06E-004 |
| BNLF2a | Lytic (E) | 62.3 | 18,461 (4,734, 32,187) | 8 | 1.79E-002 | 2.68E-002 |
| BALF3 | Lytic (E) | 60.1 | 17,791 (17,160, 18,421) | 8 | 1.00E−008 | 1.71E−006 |
| EBNA1 | Latent | 36.8 | 10,901 (4,393, 17,409) | 8 | 7.59E−003 | 1.59E−002 |
| BARF1 | Lytic (E) | 20.4 | 6,029 (1,864, 10,195) | 8 | 1.54E−002 | 2.48E−002 |
| BALF2 | Lytic (E) | 16.6 | 4,919 (2,370, 7,468) | 8 | 4.59E−003 | 1.20E−002 |
| BALF1 | Lytic (E) | 14.1 | 4,170 (1,049, 7,291) | 8 | 2.27E−002 | 2.92E−002 |
| LF3 | Lytic (L) | 12.1 | 3,572 (2,391, 4,752) | 8 | 3.93E−004 | 1.38E−003 |
| LMP1 | Latent | 10.5 | 3,104 (1,109, 5,098) | 7 | 1.41E−002 | 2.46E−002 |
| BNRF1 | Lytic (L) | 9.7 | 2,873 (1,354, 4,392) | 8 | 6.23E−003 | 1.45E−002 |
| BZLF1 | Lytic (IE) | 8.1 | 2,403 (1,002, 3,804) | 8 | 1.07E−002 | 2.05E−002 |
| BKRF4 | Lytic (E) | 7.1 | 2,103 (582, 3,623) | 7 | 2.68E−002 | 2.96E−002 |
| LMP2A | Latent | 5 | 1,486 (506, 2,466) | 7 | 2.56E−002 | 2.96E−002 |
| BKRF3 | Lytic (E) | 4.2 | 1,237 (490, 1,984) | 7 | 2.36E−002 | 2.92E−002 |
| BPLF1 | Lytic (L) | 3.4 | 1,021 (287, 1,755) | 8 | 5.30E−002 | 5.57E−002 |
| BRLF1 | Lytic (IE) | 2 | 595 (129, 1,061) | 7 | 1.56E−001 | 1.56E−001 |
Transcripts that are present at levels higher than the average values for EBNAs 2, 3A, 3B, and 3C in 7 or 8 samples are shown. P values were calculated by comparing for each gene its gene expression across 8 samples to the baseline (mean, 296 [95% confidence interval = 91, 502] RPKM).
Phase of infection in which protein expression is expected. IE, immediate early; E, early; L, late lytic phase.
Above baseline value.
CI, confidence interval.
The false discovery rate (FDR) is the P value adjusted for multiple testing.
In addition to latency genes, we identified 18 EBV lytic genes that were consistently expressed (in 7 or 8 out of 8 samples) (Table 5). The expression pattern was not consistent with a lytic or abortive lytic infection because subsets of both early and late EBV proteins were expressed. For example, a subset of only early genes necessary for viral DNA replication were expressed; transcripts for BALF5 (DNA polymerase) and BALF2 (single-stranded DNA binding protein) were readily detectable in all samples, whereas transcripts for BMRF1 (polymerase processivity factor), BSLF1 (primase), BBLF4 (helicase), and BBLF2/BBLF3 (primase accessory protein) were not consistently detected. These results suggest that lytic DNA replication would not occur. However, this does not prevent the expression of specific late lytic genes that would normally be transcribed after DNA replication as transcripts for four late genes (BALF4, LF3, BNRF1, and BPLF1) were consistently detected. Together, the expression profiles suggest that EBV lytic gene expression in GC is regulated differently than in a lytic infection.
For the expressed genes shown in Table 5, we show in Fig. 4B the agglomerative hierarchical clustering of their expression patterns across 8 samples using the Pearson correlation distance measure. Three clusters (shown in red, green, and blue) were identified. All 13 genes in the red and green clusters map to a ∼40-kb region of the genome between LF3 and BNRF1, which includes LMP1 and LMP2A (Fig. 4A). This region contains the most highly expressed genes (i.e., BALF5, BALF4, BALF3, BALF2, BILF1, LF1, LF2, BNLF2a, and BNLF2b) (Table 5), defining a region of the genome that is activated. We note that the read coverage across the BALF4 and BALF5 transcripts could also be associated with the expression of the RPMS1 and A73 genes (situated on the opposite strand from BAFL4/5); however, our analysis shows that portions of BALF4 and BALF5 transcripts that do not overlap the RPMS1 and A73 (BART) transcripts are expressed. Among genes in the green cluster, a subset of six genes (BARF1, BALF2, BALF1, BNRF1, LMP1, and LMP2A) have the strongest positive correlation (mean Pearson's r = 0.85 versus r = 0.53 for overall correlations of genes in the green cluster), suggesting that they may be coordinately regulated. The blue cluster includes EBNA1 and seven lytic genes, five of which (BPLF1, BZLF1, BRLF1, BKRF3, and BKRF4) are spread over the EBV genome outside the activated 40-kb region and two genes (BNLF2a and BNLF2b) that belong to it. BNLF2a and BNLF2b have high positive correlation (mean Pearson's r = 0.92 versus r = 0.45 for overall correlations of genes in the blue cluster) and cluster apart from the rest of the genes in this cluster. Among the genes outside the 40-kb activated region, three genes (BZLF1, BPLF1, and EBNA1) show the highest expression correlation (Pearson's r = 0.93). In contrast, BKRF3 and BKRF4, which are localized close to the EBNA1 gene, show poor correlation with EBNA1 expression (mean Pearson's r = 0.1), which suggests that they are not coregulated with EBNA1.
Association of GC sequence changes with T cell epitopes.
Previous studies have identified epitopes in EBV proteins recognized by CD4+ and CD8+ T cells (34). To determine how immune recognition might be altered in GC EBV isolates, we looked at the sequence changes in all of the EBV proteins that we found to be expressed in GC to determine if the amino acid changes correspond to known T cell epitopes. We included all of the changes identified in the second to fourth columns of Tables 3 and 4. By using the NCBI entry NC_007605 (B95.8-Raji) as the reference sequence and the epitope information from Taylor et al. (34), this analysis showed that 21 of these amino acid changes within five different expressed EBV proteins mapped to known epitopes (Table 6). In particular, most of the amino acid changes found to be common in both Asian and American Caucasian GC but uncommon in LCLs (Tables 3 and 4, GC vs LCL) mapped to T cell epitopes.
TABLE 6.
Association of GC sequence changes with T cell epitopes
| Protein group and name | Amino acid change | CD4 HLA restriction affected (position or locus)a | CD8 HLA restriction affected (position or locus)a |
|---|---|---|---|
| Latent proteins | |||
| EBNA1 | Thr85Ala | 71–85 | |
| Glu411Asp | 403–417 | B53, B35.01 | |
| His418Leu | DR4 | ||
| Ala439Thr | 429–448, 434–458 | ||
| Thr524Ile | DP3, DR1, DR7/DR11, 519–533, 519–543, 518–530, 515–528, 509–528 | B8 | |
| Ile528Val | DP3, DR1, DR13, DR7/DR11, 519–533, 519–543 | B7 | |
| Leu533Ile | DR13, DR14, 519–533, 519–543 | B7 | |
| Arg594Lys | 589–613, 594–613 | ||
| LMP1 | Arg13Pro | DR7, DR9 | |
| Arg17Leu | DR7, DR9 | ||
| Leu25Ile | DR7, DR9 | ||
| Asp46Asn | 38–46 | ||
| Ala82Gly | 68–83 | B40 | |
| Leu126Phe | A2 | ||
| Met129Ile | A2 | ||
| Phe144Ile | 130–144 | ||
| Gln189Pro | DR16 | ||
| Gly212Ser | DQ2, DQB1*0601, 212–226 | ||
| LMP2A | Ser444Thr | A25 | |
| Lytic protein | |||
| BZLF1 | Gln195His | C6, B8 | |
| Ser542Asn | B61 |
Epitope coordinates (amino acids) are used to indicate affected epitopes that are lacking HLA restriction names.
DISCUSSION
Whether particular EBV variants are more oncogenic than others is an important question and one that requires analysis of many EBV whole-genome sequences from tumors and healthy people in different geographical locations. We have contributed to this question by generating 13 new EBV sequences from gastric carcinoma, including 10 samples from the United States, 5 of which are from Caucasians. These are the first reported non-Asian GC-derived EBV genome sequences, enabling initial studies on the effect of geography/ethnicity on GC-derived EBV sequences. We have combined these new EBV sequences with all preexisting EBV whole-genome sequences to conduct the most extensive sequence analysis to date of EBV isolates.
Whole-genome phylogenetic tree analysis showed that most GC isolates are most closely related to NPC isolates; however, there was a strong geographical or/and ethnic component in that few American Caucasian EBV isolates were part of this cluster. We also identified a variety of amino acid sequence changes common to GC and NPC isolates but uncommon in other EBV isolates. Many of these result in nonconservative amino acid changes in subsets of the EBV proteins, which might affect their functions and/or host protein interactions, thereby promoting oncogenesis. Of particular interest are the multiple nonconservative amino acid changes in LMP1 and EBNA1, both of which are expressed in NPC and GC. All of the 10 nonconservative changes in LMP1 are within the first 189 amino acids of LMP1, corresponding to the transmembrane region, which can impact the ability of LMP1 to activate the NF-κB pathway (35). EBNA1 was found to have three nonconservative sequence changes (Thr85Ala, His418Leu, and Ala439Thr, all of which are outside the DNA binding domain) that are common in both NPC and GC but uncommon in other EBV isolates. His418Leu and Ala439Thr were previously reported as common changes in NPC isolates (30), but the Thr85Ala change has not been previously identified.
We also identified EBV sequences that are usually different in GC isolates, regardless of geography or ethnicity, compared to EBV genomes that are not from tumors (LCLs). These changes were seen in only three EBV proteins (EBNA1, LMP1, and BPLF1) and, according to our transcriptome analysis, all are consistently expressed in GC. This raises the intriguing possibility that these changes may impact GC by altering the functions or host interactions of these proteins. Interestingly, the changes in EBNA1 and LMP1 largely fell within known T cell epitopes, suggesting that immune pressure could be partially responsible for the sequence changes.
The EBNA1 change is Thr85Ala, which falls in a region of EBNA1 required for transcriptional activation of other EBV latency genes (36–38) and therefore could affect this important EBNA1 function. Eight nonconservative amino acid changes were identified in the transmembrane region in the first 189 amino acids of LMP1, all of which were also found to be common in NPC (identified in the isolate analysis of GC plus NPC versus other EBV isolates). BPLF1 is a deubiquitinase that has been found to contribute to innate immune evasions by interfering with Toll-like receptor signaling (39), as well as by contributing to B cell transformation (40). The roles of the two BPLF1 amino acids that are commonly altered in GC isolates (Lys515Gly and Ser405GLy) have yet to be determined.
Another important question in understanding how EBV induces GC is determining which EBV proteins are expressed. While the EBV latency proteins that are expressed in GC have been well characterized, there have also been reports of the presence of EBV lytic transcripts and proteins in the absence of a full lytic infection (12). However, a comprehensive analysis of EBV transcripts in GC has not been reported. For eight of the GC WGS samples from which we generated EBV genome sequences, transcriptome sequencing data (RNA-Seq) were also available, enabling the determination of which EBV genes are transcribed in the context of GC. We detected consistent expression of three latency proteins, EBNA1, LMP1, and LMP2A. This was expected although the frequency of LMP2A expression was higher than the previously reported 50% detection rate (41).
In addition, we identified specific subsets of lytic genes that are consistently expressed in GC. The expression profiles do not fit with conventional EBV lytic infection or abortive lytic infection since specific subsets of early and late genes are expressed. Consistent with previous studies on EBV expression in GC (2, 12), we observed a cluster of genes (BAFL3, BALF4, BALF5, BILF1, LF1, LF2, and BNLF2a) that were highly activated. This cluster of highly activated genes has also been reported in NPC and Burkitt's lymphoma although BALF5 transcripts are not detected in Burkitt's lymphoma (6–8). However, in addition to this cluster, we now identify several other transcripts from lytic genes, from immediate early, early, and late gene classes, that are consistently expressed at levels similar to those of LMP1 and LMP2A. In a lytic infection, expression of the late genes requires viral DNA replication. However, in the GC samples analyzed here, several of the early viral proteins needed for viral DNA replication are not expressed, and yet a subset of late genes (5) are expressed. Our data suggest that there are novel mechanisms of regulating expression of specific lytic genes in the context of GC. Interestingly, the transcription of BPLF1 was previously shown to be regulated in a manner distinct from that of most late genes (42), which may enable its expression in the absence of lytic infection, as we have observed in the GC samples.
Several lytic EBV proteins that are expressed in GC have functions that could contribute to tumorigenicity. As mentioned above, BPLF1 interferes with Toll-like receptor signaling in innate immunity and can promote cell transformation (39, 40). In addition, BARF1 is known to stimulate the proliferation of GC cells (43, 44). The highly expressed BILF1 is a seven-transmembrane, constitutively active, G protein-coupled receptor with transforming activity (45, 46). BILF1 and BNLF2a have also been shown to cooperate in immune evasion by inhibiting the presentation of viral antigens (47). Similarly, LF2 has been found to antagonize type I interferon signaling, suggesting that it would be important for avoiding host immune responses (48). BALF1 is a Bcl-2 homologue that increases tumorigenicity and cell survival (49) and has also been reported to be expressed in Burkitt's lymphoma cell lines and NPC samples (50). Finally, BNRF1 induces centrosome amplification leading to chromosome instability and therefore would be expected to increase the risk of oncogenesis (51). Overall our data support a model in which expression of specific EBV lytic proteins contributes to tumorigenesis in gastric and perhaps other cancers.
MATERIALS AND METHODS
EBV identification using whole-genome sequencing data.
The CaPSID (Computational Pathogen Sequence Identification) bioinformatics platform (52) (developed by our group) was used to identify EBV in whole-genome sequencing data of gastric adenocarcinoma samples, with additional filtering and alignment steps described below. For each whole-genome-sequenced sample, BAM (53) files containing reads aligned to the human reference sequence (GRCh37/hg19) were downloaded from The Cancer Genome Atlas ([TCGA] Stomach Adenocarcinoma, project code STAD) (25) and from the new PanCancer Analysis of Whole Genomes ([PCAWG] Stomach Adenocarcinoma, project codes STAD-US and GACA-CN) from the International Cancer Genome Consortium (ICGC) (26). Reads that did not map to the human reference were extracted and filtered for low complexity and quality and then aligned in single-end mode using the Bowtie2 aligner (54) to a database containing a complete set of 5,652 NCBI RefSeq viral reference sequences (including the EBV reference sequences NC_007605 and NC_009334) and a filter reference database composed of 5,242 bacterial and 1,138 fungal reference sequences that was downloaded from the NCBI (55). In order to improve the sensitivity and specificity with which viral sequences were detected, reads that did not map to any reference with Bowtie2 were realigned against the same RefSeq viral reference database, using a more sensitive SHRiMP2 aligner with the ability to perform local alignments (56). To reduce the number of potential false positives, we then applied filtering criteria using CaPSID's average gene coverage metric (average gene coverage of >90%) to identify samples in which EBV was present with the highest confidence.
EBV genome assembly.
For each individual sample that was identified as harboring EBV (as described in the previous section), reads that did not map to the human reference were realigned (using Bowtie2 and SHRiMP2 as described above) to an initial reference sequence database composed of 57 complete EBV sequences downloaded from the NCBI. Following this realignment step, reads with ambiguous alignments were reassigned to the most probable EBV genome of origin using a statistical model based on the read alignment scores as described in Hong et al. (57). Based on the information about the most probable EBV genome of origin, the read depth coverage, and the overall EBV genome coverage, 13 samples in total were selected for the EBV genome reference-based assembly. Twelve of these adenocarcinoma samples harboring EBV can be downloaded from the ICGC data portal (https://dcc.icgc.org/repositories/) using their unique file identification numbers: FI13619, FI49302, FI24570, FI28165, FI35962, FI48909, FI31442, FI33260, FI19435, FI49266, FI17300, and FI51320; the additional TCGA sample, TCGA-D7-5577-01A-01D-1598-02, can be downloaded from the Genomic Data Commons (GDC) data portal (https://portal.gdc.cancer.gov/). Of these 13 samples, 11 had read depth of coverage ranging between 54× and 176×, 1 sample had a read depth of 14×, and 1 sample had a read depth of 3.4× but even genome coverage (3.4× ± 1.4×). For each of these 13 samples, the top-ranked EBV reference sequence to which the majority of the reads aligned was then used as the input reference sequence for the reference-based assembly. The reference-based assembly was performed using SAMtools (53) for variant calls (with the read base quality parameter threshold set to -Q15), followed by the FastaAlternateReferenceMaker (a tool available from the Genome Analysis Toolkit [GATK]) (58) to generate the newly assembled EBV reference sequence.
EBV gene expression analysis.
Additional transcriptome sequencing (RNA-Seq) data [on purified poly(A)-containing RNA] available for 8 out of 13 whole-genome gastric adenocarcinoma (primary solid) sequenced samples that tested positive for EBV were downloaded from PCAWG (26) (Stomach Adenocarcinoma, project code STAD-US [25]). RNA-Seq samples used in this study can be downloaded from the ICGC data portal (https://dcc.icgc.org/repositories/) using their unique file identification numbers: FI35960, FI33258, FI17298, FI31440, FI19433, FI48907, FI28163, and FI49264. Reads that did not map to the human reference were extracted and filtered for low complexity and quality and then aligned to the NCBI EBV reference genome NC_007605 using the Bowtie2 alignment algorithms in single-end mode as previously described in Borozan et al. (59). RNA-Seq analysis and transcript read quantification were performed using the R Biocondoctor packages (60). Levels of gene expression (in units of reads per kilobase of transcript per million mapped reads [RPKM]) were calculated using the formula RPKM = (109 × C)/(N × L), where C is the number of reads mapped to a gene, N is the total number of mapped reads in the experiment, and L is the transcript length in base pairs. For each gene, transcripts in this study were defined over gene coding sequence (CDS) regions. P values were calculated using a two-sample t test with the alternative hypothesis set to “greater” as implemented in the R function t.test (61). P values were then adjusted for multiple testing in order to control for the false discovery rate (FDR) using the Benjamini-Hochberg method as implemented in the R stats package (61).
Mutation analysis.
For each EBV sequence, the lists of single nucleotide variants (SNVs) and insertions and deletions (indels) was generated by performing pairwise sequence alignments to the NCBI reference EBV genome (NC_007605) using the EMBOSS Stretcher algorithm (62). Genetic variations among EBV genomes were determined by considering the complete set of variants (i.e., substitutions, insertions, and deletions) using a combination of bioinformatics tools including the VCFtools (63) and custom Python scripts. The statistical significance of the number of occurrences of each variant found in EBV sequences isolated from GC samples was evaluated by comparing it to the number of occurrences of the same variant across EBV sequences found in other cancers or healthy blood and saliva using Fisher's exact test as implemented in the R stats package (61). P values calculated using Fisher's exact test were then adjusted for multiple testing in order to control for the false discovery rate (FDR) using the Benjamini-Hochberg method as implemented in the R stats package (61). Variants considered significant were annotated using the genetic variant annotation and effect prediction toolbox (snpEff) (64) using the NCBI NC_007605 genome as the reference database. Variants that occurred in the repeat regions of the NCBI reference sequence NC_007605 were discarded from further analysis. Phylogenetic analysis and visualization were performed using FastTree-2 (65) and FigTree software (http://tree.bio.ed.ac.uk/software/figtree). The phylogenetic tree (Fig. 1) was rooted using a midpoint rooting. The annotated circular plot of the EBV genome (Fig. 3) was made by using the Circos visualization tool (66).
Accession number(s).
Sequence data for the 13 GC EBV genomes were submitted to GenBank under accession numbers MG021314 (GC-variant-1), MG021305.1 (GC-variant-2), MG021315 (GC-variant-3), MG021317 (GC-variant-4), MG021308 (GC-variant-5), MG021307 (GC-variant-6), MG021312 (GC-variant-7), MG021316 (GC-variant-8), MG021310 (GC-variant-9) MG021311 (GC-variant-10), MG021309 (GC-variant-11), MG021313 (GC-variant-12), and MG021306 (GC-variant-13).
ACKNOWLEDGMENTS
V.F. and I.B. received support for their work from the Ontario Institute for Cancer Research (OICR) through funding provided by the government of Ontario. I.B., M.Z., and V.F. performed the work on behalf of the WGS pan-cancer consortium. L.F. is a tier 1 Canada Research Chair in Molecular Virology, and her work is supported by Canadian Institutes of Health Research project grant 153014. We also thank the Stomach Adenocarcinoma project groups (TCGA/STAD-US and ICGC/GACA-CN) that have contributed their data to the PCAWG.
REFERENCES
- 1.Jha HC, Pei Y, Robertson ES. 2016. Epstein-Barr virus: diseases linked to infection and transformation. Front Microbiol 7:1602. doi: 10.3389/fmicb.2016.01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Camcer Genome Atlas Research Network. 2014. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niller HH, Minarovits J. 2016. Patho-epigenetics of infectious diseases caused by intracellular bacteria. Adv Exp Med Biol 879:107–130. doi: 10.1007/978-3-319-24738-0_6. [DOI] [PubMed] [Google Scholar]
- 4.Murata T. 2014. Regulation of Epstein-Barr virus reactivation from latency. Microbiol Immunol 58:307–317. doi: 10.1111/1348-0421.12155. [DOI] [PubMed] [Google Scholar]
- 5.Martel-Renoir D, Grunewald V, Touitou R, Schwaab G, Joab I. 1995. Qualitative analysis of the expression of Epstein-Barr virus lytic genes in nasopharyngeal carcinoma biopsies. J Gen Virol 76:1401–1408. doi: 10.1099/0022-1317-76-6-1401. [DOI] [PubMed] [Google Scholar]
- 6.Abate F, Ambrosio MR, Mundo L, Laginestra MA, Fuligni F, Rossi M, Zairis S, Gazaneo S, De Falco G, Lazzi S, Bellan C, Rocca BJ, Amato T, Marasco E, Etebari M, Ogwang M, Calbi V, Ndede I, Patel K, Chumba D, Piccaluga PP, Pileri S, Leoncini L, Rabadan R. 2015. Distinct viral and mutational spectrum of endemic Burkitt lymphoma. PLoS Pathog 11:e1005158. doi: 10.1371/journal.ppat.1005158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu L, Lin Z, Wu Y, Dong J, Zhao B, Cheng Y, Huang P, Xu L, Xia T, Xiong D, Wang H, Li M, Guo L, Kieff E, Zeng Y, Zhong Q, Zeng M. 2016. Comprehensive profiling of EBV gene expression in nasopharyngeal carcinoma through paired-end transcriptome sequencing. Front Med 10:61–75. doi: 10.1007/s11684-016-0436-0. [DOI] [PubMed] [Google Scholar]
- 8.Tierney RJ, Shannon-Lowe CD, Fitzsimmons L, Bell AI, Rowe M. 2015. Unexpected patterns of Epstein-Barr virus transcription revealed by a high throughput PCR array for absolute quantification of viral mRNA. Virology 474:117–130. doi: 10.1016/j.virol.2014.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin Z, Xu G, Deng N, Taylor C, Zhu D, Flemington EK. 2010. Quantitative and qualitative RNA-Seq-based evaluation of Epstein-Barr virus transcription in type I latency Burkitt's lymphoma cells. J Virol 84:13053–13058. doi: 10.1128/JVI.01521-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strong MJ, Laskow T, Nakhoul H, Blanchard E, Liu Y, Wang X, Baddoo M, Lin Z, Yin Q, Flemington EK. 2015. Latent expression of the Epstein-Barr virus (EBV)-encoded major histocompatibility complex class I TAP inhibitor, BNLF2a, in EBV-positive gastric carcinomas. J Virol 89:10110–10114. doi: 10.1128/JVI.01110-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong GK, Gulley ML, Feng W-H, Delecluse H-J, Holley-Guthrie E, Kenney SC. 2005. Epstein-Barr virus lytic infection contributes to lymphoproliferative disease in a SCID mouse model. J Virol 79:13993–14003. doi: 10.1128/JVI.79.22.13993-14003.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strong MJ, Xu G, Coco J, Baribault C, Vinay DS, Lacey MR, Strong AL, Lehman TA, Seddon MB, Lin Z, Concha M, Baddoo M, Ferris M, Swan KF, Sullivan DE, Burow ME, Taylor CM, Flemington EK. 2013. Differences in gastric carcinoma microenvironment stratify according to EBV infection intensity: implications for possible immune adjuvant therapy. PLoS Pathog 9:e1003341. doi: 10.1371/journal.ppat.1003341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giffin L, Damania B. 2014. KSHV: pathways to tumorigenesis and persistent infection. Adv Virus Res 88:111–159. doi: 10.1016/B978-0-12-800098-4.00002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gage JR, Meyers C, Wettstein FO. 1990. The E7 proteins of the nononcogenic human papillomavirus type 6b (HPV-6b) and of the oncogenic HPV-16 differ in retinoblastoma protein binding and other properties. J Virol 64:723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huibregtse JM, Scheffner M, Howley PM. 1991. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J 10:4129–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 17.Scheffner M, Münger K, Byrne JC, Howley PM. 1991. The state of the p53 and retinoblastoma genes in human cervical carcinoma cell lines. Proc Natl Acad Sci U S A 88:5523–5527. doi: 10.1073/pnas.88.13.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai M-H, Raykova A, Klinke O, Bernhardt K, Gärtner K, Leung CS, Geletneky K, Sertel S, Münz C, Feederle R, Delecluse H-J. 2013. Spontaneous lytic replication and epitheliotropism define an Epstein-Barr virus strain found in carcinomas. Cell reports 5:458–470. doi: 10.1016/j.celrep.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 19.Palser AL, Grayson NE, White RE, Corton C, Correia S, Ba Abdullah MM, Watson SJ, Cotten M, Arrand JR, Murray PG, Allday MJ, Rickinson AB, Young LS, Farrell PJ, Kellam P. 2015. Genome diversity of Epstein-Barr virus from multiple tumor types and normal infection. J Virol 89:5222–5237. doi: 10.1128/JVI.03614-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu P, Fang X, Feng Z, Guo Y-M, Peng R-J, Liu T, Huang Z, Feng Y, Sun X, Xiong Z, Guo X, Pang S-S, Wang B, Lv X, Feng F-T, Li D-J, Chen L-Z, Feng Q-S, Huang W-L, Zeng M-S, Bei J-X, Zhang Y, Zeng Y- X. 2011. Direct sequencing and characterization of a clinical isolate of Epstein-Barr virus from nasopharyngeal carcinoma tissue by using next-generation sequencing technology. J Virol 85:11291–11299. doi: 10.1128/JVI.00823-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwok H, Tong AH, Lin CH, Lok S, Farrell PJ, Kwong DL, Chiang AK. 2012. Genomic sequencing and comparative analysis of Epstein-Barr virus genome isolated from primary nasopharyngeal carcinoma biopsy. PLoS One 7:e36939. doi: 10.1371/journal.pone.0036939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwok H, Wu CW, Palser AL, Kellam P, Sham PC, Kwong DLW, Chiang AKS. 2014. Genomic diversity of Epstein-Barr virus genomes isolated from primary nasopharyngeal carcinoma biopsy samples. J Virol 88:10662–10672. doi: 10.1128/JVI.01665-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Yang W, Pan Y, Ji J, Lu Z, Ke Y. 2016. Genome-wide analysis of Epstein-Barr virus (EBV) isolated from EBV-associated gastric carcinoma (EBVaGC). Oncotarget 7:4903–4914. doi: 10.18632/oncotarget.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lei H, Li T, Li B, Tsai S, Biggar RJ, Nkrumah F, Neequaye J, Gutierrez M, Epelman S, Mbulaiteye SM, Bhatia K, Lo SC. 2015. Epstein-Barr virus from Burkitt lymphoma biopsies from Africa and South America share novel LMP-1 promoter and gene variations. Sci Rep 5:16706. doi: 10.1038/srep16706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. 2013. The Cancer Genome Atlas pan-cancer analysis project. Nat Genet 45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.International Cancer Genome Consortium, Hudson TJ, Anderson W, Artez A, Barker AD, Bell C, Bernabé RR, Bhan MK, Calvo F, Eerola I, Gerhard DS, Guttmacher A, Guyer M, Hemsley FM, Jennings JL, Kerr D, Klatt P, Kolar P, Kusada J, Lane DP, Laplace F, Youyong L, Nettekoven G, Ozenberger B, Peterson J, Rao TS, Remacle J, Schafer AJ, Shibata T, Stratton MR, Vockley JG, Watanabe K, Yang H, Yuen MM, Knoppers BM, Bobrow M, Cambon-Thomsen A, Dressler LG, Dyke SO, Joly Y, Kato K, Kennedy KL, Nicolás P, Parker MJ, Rial-Sebbag E, Romeo-Casabona CM, Shaw KM, Wallace S, Wiesner GL, Zeps N, et al. 2010. International network of cancer genome projects. Nature 464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santpere G, Darre F, Blanco S, Alcami A, Villoslada P, Mar Albà M, Navarro A. 2014. Genome-wide analysis of wild-type Epstein-Barr virus genomes derived from healthy individuals of the 1,000 Genomes Project. Genome Biol Evol 6:846–860. doi: 10.1093/gbe/evu054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayashi K, Chen WG, Chen YY, Murakami I, Chen HL, Ohara N, Nose S, Hamaya K, Matsui S, Bacchi MM, Bacchi CE, Chang KL, Weiss LM. 1998. Deletion of Epstein-Barr virus latent membrane protein 1 gene in Japanese and Brazilian gastric carcinomas, metastatic lesions, and reactive lymphocytes. Am J Pathol 152:191–198. [PMC free article] [PubMed] [Google Scholar]
- 29.BenAyed-Guerfali D, Ayadi W, Miladi-Abdennadher I, Khabir A, Sellami-Boudawara T, Gargouri A, Mokdad-Gargouri R. 2011. Characteristics of Epstein-Barr virus variants associated with gastric carcinoma in southern Tunisia. Virol J 8:500. doi: 10.1186/1743-422X-8-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang JT, Sheeng TS, Su IJ, Chen JY, Chen MR. 2003. EBNA-1 sequence variations reflect active EBV replication and disease status or quiescent latency in lymphocytes. J Med Virol 69:417–425. doi: 10.1002/jmv.10305. [DOI] [PubMed] [Google Scholar]
- 31.Correia S, Palser A, Elgueta Karstegl C, Middeldorp JM, Ramayanti O, Cohen JI, Hildesheim A, Fellner MD, Wiels J, White RE, Kellam P, Farrell PJ. 2017. Natural variation of Epstein-Barr virus genes, proteins, and primary microRNA. J Virol 91:e00375-. doi: 10.1128/JVI.00375-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bochkarev A, Barwell JA, Pfuetzner RA, Furey W Jr, Edwards AM, Frappier L. 1995. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein EBNA 1. Cell 83:39–46. doi: 10.1016/0092-8674(95)90232-5. [DOI] [PubMed] [Google Scholar]
- 33.Cruickshank J, Shire K, Davidson AR, Edwards AM, Frappier L. 2000. Two domains of the Epstein-Barr virus origin DNA-binding protein, EBNA1, orchestrate sequence-specific DNA binding. J Biol Chem 275:22273–22277. doi: 10.1074/jbc.M001414200. [DOI] [PubMed] [Google Scholar]
- 34.Taylor GS, Long HM, Brooks JM, Rickinson AB, Hislop AD. 2015. The immunology of Epstein-Barr virus-induced disease. Annu Rev Immunol 33:787–821. doi: 10.1146/annurev-immunol-032414-112326. [DOI] [PubMed] [Google Scholar]
- 35.Miller WE, Cheshire JL, Baldwin AS, Raab-Traub N. 1998. The NPC derived C15 LMP1 protein confers enhanced activation of NF-κB and induction of the EGFR in epithelial cells. Oncogene 16:1869–1877. doi: 10.1038/sj.onc.1201696. [DOI] [PubMed] [Google Scholar]
- 36.Wu H, Kapoor P, Frappier L. 2002. Separation of the DNA replication, segregation, and transcriptional activation functions of Epstein-Barr nuclear antigen 1. J Virol 76:2480–2490. doi: 10.1128/jvi.76.5.2480-2490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kennedy G, Sugden B. 2003. EBNA-1, a bifunctional transcriptional activator. Mol Cell Biol 23:6901–6908. doi: 10.1128/MCB.23.19.6901-6908.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altmann M, Pich D, Ruiss R, Wang J, Sugden B, Hammerschmidt W. 2006. Transcriptional activation by EBV nuclear antigen 1 is essential for the expression of EBV's transforming genes. Proc Natl Acad Sci U S A 103:14188–14193. doi: 10.1073/pnas.0605985103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Gent M, Braem SGE, de Jong A, Delagic N, Peeters JGC, Boer IGJ, Moynagh PN, Kremmer E, Wiertz EJ, Ovaa H, Griffin BD, Ressing ME. 2014. Epstein-Barr virus large tegument protein BPLF1 contributes to innate immune evasion through interference with toll-like receptor signaling. PLoS Pathog 10:e1003960. doi: 10.1371/journal.ppat.1003960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whitehurst CB, Li G, Montgomery SA, Montgomery ND, Su L, Pagano JS. 2015. Knockout of Epstein-Barr virus BPLF1 retards B-cell transformation and lymphoma formation in humanized mice. mBio 6:e01574-. doi: 10.1128/mBio.01574-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cen O, Longnecker R. 2015. Latent membrane protein 2 (LMP2). Curr Top Microbiol Immunol 391:151–180. doi: 10.1007/978-3-319-22834-1_5. [DOI] [PubMed] [Google Scholar]
- 42.McKenzie J, Lopez-Giraldez F, Delecluse H-J, Walsh A, El-Guindy A. 2016. The Epstein-Barr virus immunoevasins BCRF1 and BPLF1 are expressed by a mechanism independent of the canonical late pre-initiation complex. PLoS Pathog 12:e1006008. doi: 10.1371/journal.ppat.1006008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang MS, Kim DH, Roh JK, Middeldorp JM, Kim YS, Kim S, Han S, Kim CW, Lee BL, Kim WH, Woo JH. 2013. Epstein-Barr virus-encoded BARF1 promotes proliferation of gastric carcinoma cells through regulation of NF-κB. J Virol 87:10515–10523. doi: 10.1128/JVI.00955-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakka E, Zur Hausen A, Houali K, Liu H, Fiorini S, Ooka T. 2013. Cellular localization of BARF1 oncoprotein and its cell stimulating activity in human epithelial cell. Virus Res 174:8–17. doi: 10.1016/j.virusres.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 45.Lyngaa R, Nørregaard K, Kristensen M, Kubale V, Rosenkilde MM, Kledal TN. 2010. Cell transformation mediated by the Epstein-Barr virus G protein-coupled receptor BILF1 is dependent on constitutive signaling. Oncogene 29:4388–4398. doi: 10.1038/onc.2010.173. [DOI] [PubMed] [Google Scholar]
- 46.Beisser PS, Verzijl D, Gruijthuijsen YK, Beuken E, Smit MJ, Leurs R, Bruggeman CA, Vink C. 2005. The Epstein-Barr virus BILF1 gene encodes a G protein-coupled receptor that inhibits phosphorylation of RNA-dependent protein kinase. J Virol 79:441–449. doi: 10.1128/JVI.79.1.441-449.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quinn LL, Zuo J, Abbott RJM, Shannon-Lowe C, Tierney RJ, Hislop AD, Rowe M.. 2014. Cooperation between Epstein-Barr virus immune evasion proteins spreads protection from CD8+ T cell recognition across all three phases of the lytic cycle. PLoS Pathog 10:e1004322. doi: 10.1371/journal.ppat.1004322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu L, Fossum E, Joo CH, Inn K-S, Shin YC, Johannsen E, Hutt-Fletcher LM, Hass J, Jung JU. 2009. Epstein-Barr virus LF2: an antagonist to type I interferon. J Virol 83:1140–1146. doi: 10.1128/JVI.00602-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsu W-L, Chung P-J, Tsai M-H, Chang CL-T, Liang C-L. 2012. A role for Epstein-Barr viral BALF1 in facilitating tumor formation and metastasis potential. Virus research 163:617–627. doi: 10.1016/j.virusres.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 50.Cabras G, Decaussin G, Zeng Y, Djennaoui D, Melouli H, Broully P, Bouguermouh AM, Ooka T. 2005. Epstein-Barr virus encoded BALF1 gene is transcribed in Burkitt's lymphoma cell lines and in nasopharyngeal carcinoma's biopsies. J Clin Virol 34:26–34. doi: 10.1016/j.jcv.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 51.Shumilov A, Tsai MH, Schlosser YT, Kratz AS, Bernhardt K, Fink S, Mizani T, Lin X, Jauch A, Mautner J, Kopp-Schneider A, Feederle R, Hoffmann I, Delecluse HJ. 2017. Epstein-Barr virus particles induce centrosome amplification and chromosomal instability. Nat Commun 8:14257. doi: 10.1038/ncomms14257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borozan I, Wilson S, Blanchette P, Laflamme P, Watt SN, Krzyzanowski PM, Sircoulomb F, Rottapel R, Branton PE, Ferretti V. 2012. CaPSID: a bioinformatics platform for computational pathogen sequence identification in human genomes and transcriptomes. BMC Bioinformatics 13:206. doi: 10.1186/1471-2105-13-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pruitt KD, Tatusova T, Klimke W, Maglott DR. 2009. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res 37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.David M, Dzamba M, Lister D, Ilie L, Brudno M. 2011. SHRiMP2: sensitive yet practical SHort Read Mapping. Bioinformatics 27:1011–1012. doi: 10.1093/bioinformatics/btr046. [DOI] [PubMed] [Google Scholar]
- 57.Hong C, Manimaran S, Shen Y, Perez-Rogers JF, Byrd AL, Castro-Nallar E, Crandall KA, Johnson WE. 2014. PathoScope 2.0: a complete computational framework for strain identification in environmental or clinical sequencing samples. Microbiome 2:33. doi: 10.1186/2049-2618-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Borozan I, Watt SN, Ferretti V. 2013. Evaluation of alignment algorithms for discovery and identification of pathogens using RNA-Seq. PLoS One 8:e76935. doi: 10.1371/journal.pone.0076935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MacDonald J, Obenchain V, Oleœ AK, Pagès H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron L, Morgan M. 2015. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.R Development Core Team. 2004. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 62.Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 63.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R, 1000 Genome Project Analysis Group. 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]