ABSTRACT
Rabbit hemorrhagic disease virus 2 (RHDV2; Lagovirus GI.2) is a pathogenic calicivirus that affects European rabbits (Oryctolagus cuniculus) and various hare (Lepus) species. GI.2 was first detected in France in 2010 and subsequently caused epidemics in wild and domestic lagomorph populations throughout Europe. In May 2015, GI.2 was detected in Australia. Within 18 months of its initial detection, GI.2 had spread to all Australian states and territories and rapidly became the dominant circulating strain, replacing Rabbit hemorrhagic disease virus (RHDV/GI.1) in mainland Australia. Reconstruction of the evolutionary history of 127 Australian GI.2 isolates revealed that the virus arrived in Australia at least several months before its initial description and likely circulated unnoticed in wild rabbit populations in the east of the continent prior to its detection. GI.2 sequences isolated from five hares clustered with sequences from sympatric rabbit populations sampled contemporaneously, indicating multiple spillover events into hares rather than an adaptation of the Australian GI.2 to a new host. Since the presence of GI.2 in Australia may have wide-ranging consequences for rabbit biocontrol, particularly with the release of the novel biocontrol agent GI.1a/RHDVa-K5 in March 2017, ongoing surveillance is critical to understanding the interactions of the various lagoviruses in Australia and their impact on host populations.
IMPORTANCE This study describes the spread and distribution of Rabbit hemorrhagic disease virus 2 (GI.2) in Australia since its first detection in May 2015. Within the first 18 months following its detection, RHDV2 spread from east to west across the continent and became the dominant strain in all mainland states of Australia. This has important implications for pest animal management and for owners of pet and farmed rabbits, as there currently is no effective vaccine available in Australia for GI.2. The closely related RHDV (GI.1) is used to control overabundant wild rabbits, a serious environmental and agricultural pest in this country, and it is currently unclear how the widespread circulation of GI.2 will impact ongoing targeted wild rabbit management operations.
KEYWORDS: calicivirus, evolution, biocontrol, distribution, establishment, RHDV2, rabbit hemorrhagic disease virus
INTRODUCTION
Rabbit hemorrhagic disease virus 2 (RHDV2) is a calicivirus that causes rabbit hemorrhagic disease (RHD), characterized by necrotizing hepatitis with a high case fatality rate in European rabbits (Oryctolagus cuniculus) (1, 2). Under the newly proposed nomenclature, RHDV2 belongs to the genus Lagovirus, genotype GI.2 (3). GI.2 was first detected in France in 2010 (1) and subsequently spread rapidly throughout Europe (4–11). It was detected in Australia in 2015 (12), and outbreaks have also been reported from Benin in 2015 and Canada in 2016 (13, 14). GI.2 is now considered to be endemic to Europe, where it appears to be replacing classical Rabbit hemorrhagic disease virus (RHDV; Lagovirus GI.1) in some regions (5, 6, 15).
In susceptible adult rabbits, GI.2 causes a fulminant hepatitis similar to that of GI.1 (1). The genomes of these viruses are similarly arranged in two open reading frames (ORFs). ORF1 encodes the nonstructural proteins, including the RNA-dependent RNA polymerase (RdRp), and the major capsid protein, VP60 (16). The second ORF encodes a minor structural protein, VP10 (16). Despite these similarities, GI.2 is genetically and antigenically distinct from GI.1 (5). While infection with GI.1 is widely considered to be restricted to European rabbits (Oryctolagus cuniculus), GI.2 also has been detected in multiple hare species, including Italian hares (Lepus corsicanus), Sardinian cape hares (Lepus capensis mediterraneus), and European brown hares (Lepus europaeus) (2, 17–19). Additionally, GI.2 is able to cause disease in rabbit kittens as young as 11 days old, which are normally highly resistant to clinical GI.1 infection, as well as in GI.1-vaccinated rabbits and in rabbits with natural immunity to GI.1 viruses (1, 8, 20), although cross-reactive antigenic determinants have been identified between GI.1 and GI.2 viruses (5, 18). These features may partially explain why GI.2 has spread so efficiently in Europe.
In May 2015, a GI.2 variant was detected in Australia (12). This variant was closely related to recombinant viruses circulating in Portugal and the Azores (11, 21) and comprised a GI.2 capsid gene with GI.1b (RHDV G1) nonstructural genes (known as GI.1bP-GI.2 under the new nomenclature) (3, 12). Prior to this detection, the only pathogenic rabbit caliciviruses present in Australia were GI.1c (RHDV G2s), descended from the Czech V351 virus released for biocontrol purposes in 1995, and a GI.1a (RHDVa) virus, termed GI.1a-Aus, a recent incursion that was limited to the east coast (Table 1 provides a summary of viral variants) (3, 23, 31, 32). Notably, an additional GI.1a virus, GI.1a-K5, was released nationwide in March 2017 for ongoing control of wild rabbit populations (33), and a benign lagovirus, GI.4, has also been circulating in Australia for many years (27, 34). In light of ongoing rabbit management operations, it is important to understand the spread and distribution of GI.2, as it is currently unclear if and how the increasing genetic diversity of circulating field strains will impact RHD-mediated rabbit biocontrol in Australia. Such studies will also help to better understand the risks to nontarget rabbit populations, such as pet and farmed rabbits, for which an effective vaccine is available to cover GI.1 strains but not GI.2.
TABLE 1.
Known genotypes and variants of lagoviruses infecting Oryctolagus cuniculus
| Capsid genotype | Polymerase genotype | Original nomenclature (polymerase/capsid) | First reference | Location first reported | Prototype/reference strain | Properties | Note |
|---|---|---|---|---|---|---|---|
| GI.1a | GI.1aP | RHDVa | 22 | Italy | DQ205345.1/CHN/JX_97/1997 | Pathogenic, liver tropism | Antigenic variant of GI.1b, c, d; includes GI.1a-K5 |
| GI.1a | GI.4eP | RCV-A1-like/RHDVa | 23 | Australia | KY628309/AUS/NSW/BER_2/2013 | Pathogenic, liver tropism, recombinant | First detected in Australia, although recombination event probably occurred elsewhere; also referred to as GI.1a-Aus |
| GI.1b, GI.1c, GI.1d | GI.1bP, GI.1cP, GI.1dP | RHDV | 24 | China | M67473.1/DEU/FRG/1988 | Pathogenic, liver tropism | Only known virulent genotype circulating globally prior to 1997 |
| GI.2 | GI.2P | RHDV2 | 1, 21 | Spain/Portugal | KM878681.1/ESP/RHDV-N11/2011 | Pathogenic, liver tropism | Genetically and antigenically distinct from GI.1 (RHDV) and GI.1a (RHDVa); GI.2 (capsid) was first detected in 2010 in France (1), but the polymerase genotype was not reported. |
| GI.2 | GI.1bP | RHDV/RHDV2 | 1, 21 | Portugal | KM115714.2/PRT/CBAlgarve14_1/2014 | Pathogenic, liver tropism, recombinant | |
| GI.2 | GI.4Pa | RCV-A1-like/RHDV2 | 1, 21 | Spain/Portugal | KF442963.2/PRT/7-13_Barrancos/2013 | Pathogenic, liver tropism, recombinant | |
| GI.2 | GI.4eP | RCV-A1-like/RHDV2 | 25 | Australia | MF598302/AUS/NSW/CAR-3/2016 | Pathogenic, liver tropism, recombinant | Currently only detected in Australia |
| GI.3 | NDb | RCV-E1 | 26 | France | AM268419.4/FRA/06-11/2006 | Mostly benign, intestinal tropism | Variable pathogenicity and tissue tropism |
| GI.4a, GI.4b, GI.4c | GI.4aP, GI.4bP, GI.4cP | RCV-A1 | 27 | Australia | EU871528.1/AUS/MIC-07(1-4)/2007 | Benign, intestinal tropism | KX357707/NZ/Southland/Gore-425A/2013 and LT708120/PLR56/08-84/2007 are also classified as GI.4 but have not been classified to the variant level |
| GI.4d | ND | RCV-E2 | 3, 28 | France | LT708121.1/BO9/08-117/2008 | Benign, intestinal tropism | Defined in reference 3, but not characterized; characterization in reference 28 |
| Unclassified | |||||||
| UCc | ND | RCV | 29 | Italy | X96868.1/ITA/ItalyRCV/1995 | Benign, intestinal tropism | First reported benign lagovirus |
| UC | UC | MRCV | 30 | USA | GQ166866.1/USA/MRCV/2001 | Pathogenic, liver tropism | Only detected from a single outbreak in the USA |
Unclassified variant of GI.4.
ND, not determined, i.e., no polymerase sequence available.
UC, unclassified.
RESULTS
Between May 2015 and October 2016, pathogenic lagoviruses (GI.1, GI.1a, or GI.2) were detected in 248 cases of lagomorph mortalities in Australia (Table 2 and Fig. 1) in animals ranging from 2 weeks old to mature adults. Multiple samples from the same location within a 2-week period were defined as one case, with one to six samples submitted per case. Sampling was both temporally and spatially biased, dependent on the willingness of rabbit owners and those interested in invasive pest management to participate in collection and submission of samples from wild lagomorphs. A pathogenic lagovirus was detected in 94 cases involving domestic rabbits, 132 cases involving wild rabbits, and five cases involving wild European brown hares (Lepus europaeus). For 17 cases, the source of the sample (i.e., wild or domestic rabbit) was not disclosed.
TABLE 2.
Summary of samples tested for pathogenic lagoviruses in Australia from May 2015 to October 2016
| Sample and genotype | No. of cases per state and territorya |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| ACT | NSW | NT | QLD | SA | TAS | VIC | WA | Total | |
| Wild rabbit | |||||||||
| GI.2 | 10 | 18 | 0 | 2 | 51 | 0 | 8 | 18 | 107 |
| GI.1 | 2 | 1 | 0 | 0 | 19 | 1 | 1 | 1 | 25 |
| GI.1a-Aus | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mixedb | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Negative | 5 | 8 | 0 | 4 | 23 | 0 | 3 | 7 | 46 |
| Domestic rabbit | |||||||||
| GI.2 | 5 | 22 | 1 | 0 | 23 | 1 | 16 | 13 | 81 |
| GI.1 | 0 | 1 | 0 | 0 | 6 | 2 | 0 | 1 | 10 |
| GI.1a-Aus | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| Mixed | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| Negative | 2 | 25 | 0 | 0 | 11 | 5 | 8 | 0 | 14 |
| Unknown | |||||||||
| GI.2 | 0 | 7 | 1 | 0 | 0 | 0 | 3 | 0 | 11 |
| GI.1 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 5 |
| GI.1a-Aus | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mixed | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Negative | 0 | 2 | 0 | 0 | 2 | 0 | 1 | 0 | 4 |
| Hare | |||||||||
| GI.2 | 0 | 0 | 0 | 0 | 4 | 0 | 1 | 0 | 5 |
| Negative | 2 | 1 | 0 | 0 | 4 | 0 | 0 | 0 | 7 |
| Total | 17 | 52 | 2 | 2 | 108 | 4 | 30 | 33 | |
Abbreviations for Australian states and territories: ACT, Australian Capital Territory; NSW, New South Wales; NT, Northern Territory; QLD, Queensland; SA, South Australia; TAS, Tasmania; VIC, Victoria; WA, Western Australia.
Mixed infections in all cases were GI.1 and GI.2 coinfections.
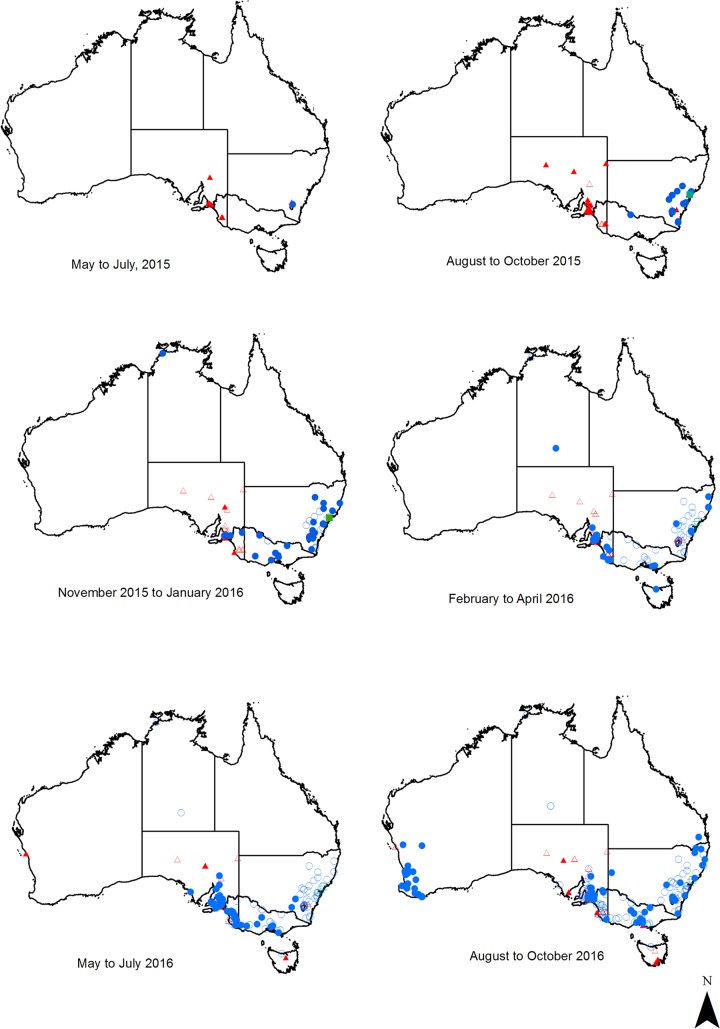
FIG 1.
Pathogenic lagovirus detections in Australia between May 2015 and October 2016. Sites where GI.1 (red triangles), GI.2 (blue circles), and GI.1a-Aus (green squares) were detected are indicated on the map separated into 3-month periods. Filled points indicate detections that were within the respective 3-month period, while hollow points indicate previous detections.
GI.2 was detected in 201 of the 243 total positive cases in rabbits: 15 in the Australian Capital Territory (ACT), 48 in New South Wales (NSW; one as a mixed infection with GI.1), 2 in the Northern Territory (NT), 2 in Queensland (QLD), 74 in South Australia (SA), 1 in Tasmania (TAS), 28 in Victoria (VIC; one as a mixed infection with GI.1), and 31 in Western Australia (WA) (Table 2). An additional five GI.2 cases were detected in European brown hares, as reported previously (17).
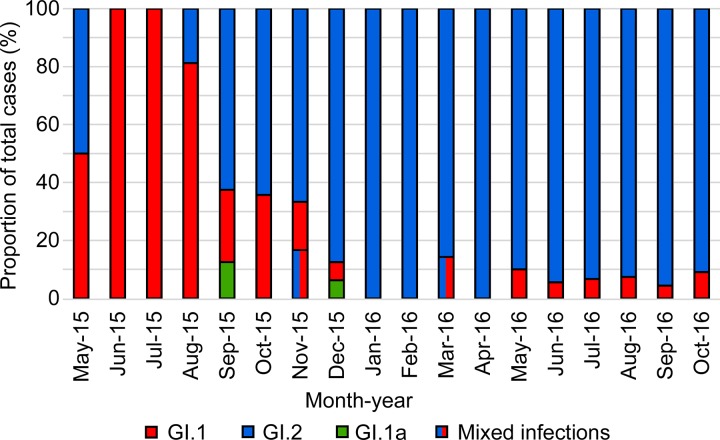
GI.2 spread rapidly across Australia. Surveillance efforts were increased following the initial detection in the ACT in May 2015, and the virus was subsequently detected in NSW in August 2015, VIC in October 2015, SA and the NT in December 2015, TAS in April 2016, WA in August 2016, and QLD in October 2016 (Fig. 1). In contrast to the high detection rate of GI.2, classical GI.1 was detected in only 40 cases during the same sampling period: two in the ACT in June and August 2015, two in NSW in May and October 2015, 30 in SA between June 2015 and May 2016, three in TAS from June to October 2016, one on Phillip Island in VIC in August 2016, and two in WA from May to July 2016, before the first detection of GI.2 in that state (Table 2, Fig. 1 and 2). GI.1a-Aus (23) was detected twice during the sampling period, in September and December 2015 in NSW (Fig. 1 and 2). GI.2 comprised the majority of detections during the study period in all states except Tasmania.
FIG 2.
Proportional detections of GI.1, GI.2, GI.1a-Aus, and mixed infections. Detections of each virus are presented as a proportion (y axis) of total cases per month (x axis) between May 2015 and October 2016.
Of the 81 cases in domestic rabbits that tested positive for GI.2, 15 were reported in rabbits previously vaccinated with the inactivated GI.1 vaccine Cylap RCD (Zoetis Australia), although information regarding the time since last vaccination was not always available. In 15 cases, rabbits were unvaccinated. The vaccination status was unknown for the remaining 51 cases, and sera were not available for antibody screening.
The full genomes of 139 viruses detected during May 2015 to October 2016 were sequenced for further analyses. Prior to sequencing, real-time reverse transcription-PCR was performed to quantify virus load in these samples. The average (geometric mean) viral load in the livers of GI.2-infected rabbits (3 × 108 capsid copies per mg of tissue) was comparable to that observed in GI.1-infected rabbit livers (2 × 108 capsid copies per mg of tissue). Initial genotyping was confirmed by full genome sequencing in all cases. We also explored the deep-sequencing data for evidence of additional mixed infections using an interpretative cutoff of 1%. No cases of mixed infections were detected among the full genomes sequenced.
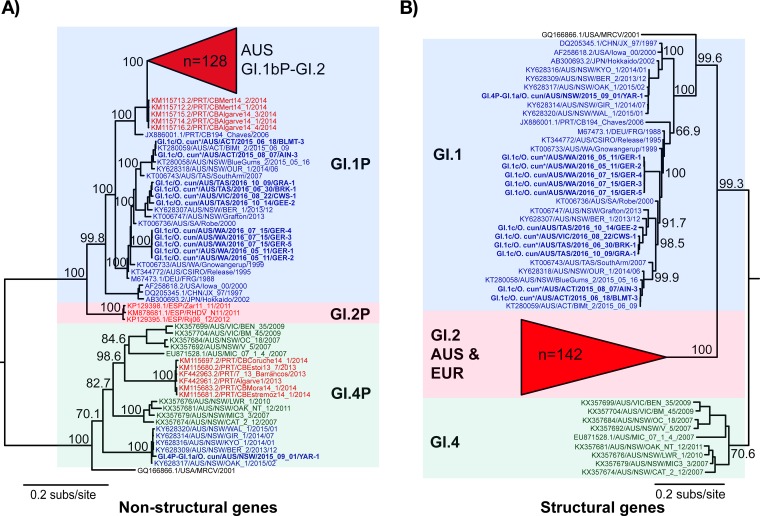
Phylogenetic analyses revealed that classical GI.1 samples clustered with previously published Australian GI.1 sequences, with the WA samples clustering distinctly from those collected in eastern states (VIC, ACT, and NSW) (Fig. 3). One GI.1a-Aus virus from this sampling period was sequenced, and it clustered closely with previously published Australian GI.1a sequences in both the structural and nonstructural gene phylogenies (Fig. 3) (23). The Australian GI.2 sequences formed a monophyletic group in the nonstructural gene phylogeny, clustering closely with European GI.1bP-GI.2 sequences (Fig. 3A). In the structural gene phylogeny, all GI.2 sequences clustered together (Fig. 3B). The European GI.2 samples formed a single clade that clustered within the Australian sequences, although bootstrap support for branching within the GI.2 clade of the structural genes tree was very low (data not shown), likely due in part to the rapid spread and evolution of this virus. As with the previously characterized Australian GI.2 sequence (12), the GI.2 samples sequenced here were all recombinant viruses with structural genes of GI.2 and nonstructural genes related to GI.1b viruses (GI.1bP-GI.2) (Fig. 3A and B).
FIG 3.
Phylogenies of the nonstructural and structural genes of Australian and global lagoviruses. Maximum likelihood phylogenies of the nonstructural genes (n = 184) (A) and structural genes (VP60 and VP10; n = 184) (B) were inferred using the newly sequenced Australian lagovirus strains (shown in boldface) and representative published sequences. The Australian GI.2 clades are collapsed due to their large size. The accession numbers of published sequences are indicated in the taxon names. The species from which the virus was collected is indicated in the taxon names of newly sequenced samples (O. cun., Oryctolagus cuniculus), and collections from wild animals are indicated by an asterisk next to the species name. The genotype of each cluster is indicated by colored boxes: GI.1, blue; GI.2, red; GI.4, green. To illustrate recombinant strains, the taxon labels are colored according to their structural gene genotype. Taxon label coloring that does not match the colored boxes indicates recombination. Phylogenies were rooted using an early European EBHSV isolate (not shown), and the scale bar is proportional to the number of nucleotide substitutions per site. Bootstrap support values are shown for the major nodes.
The Australian GI.2 genome sequences had an average nucleotide identity of 98.4%, with the two most divergent sequences sharing 97.4% nucleotide identity. Group-defining sites, classified here as nonsynonymous mutations shared by ≥80% of all isolates within a distinct phylogenetic group (maintained across both structural and nonstructural phylogenies), were detected in all nonstructural and structural protein-coding regions (Table 3 and Fig. 4). The highest number of group-defining sites in Australian GI.2 strains was detected in the VP60 capsid protein, specifically in the P2 subdomain, the outermost part of the protruding domain.
TABLE 3.
Group-defining sites in Australian GI.2 strains from May 2015 to October 2016
| Virus groupa | Virus protein |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| p16 (143)b | p23/26 (224) | 2C-like protein (351) | p29 (275) | VpGc (114) | Protease (143) | RdRpc (514) | VP60 (579) |
VP10 (117) | ||
| Non-P2 | P2 (163) | |||||||||
| 1a | S130Nd | R170K | I961V | D1062E | K1307R | S2123Ne | ||||
| 1b | ||||||||||
| 1c | I1138V | K1307R M1682V | M87V | |||||||
| 1d | ||||||||||
| 1e | V1391A | |||||||||
| 1f | R2058K | |||||||||
| 2 | A764E | D1287E | A1766V | |||||||
| 3a | E245G D262S | N692Y A709S | A1842S V2255I | S2078N A2143Ve | V62I V108I | |||||
| 3b | V62I | |||||||||
| 4a | L15S F131L | K341R | N928S | D1287E A1360S | V2132Me | |||||
| 4b | L15S N54K F131L | I1117V | A2301T | V2132Me A2143Ve | ||||||
| 4c | L15S F131L | D1287E | S2300L | V2132Me A2143Ve A2126Se | ||||||
Groups are arbitrarily defined in Fig. 4.
The length of the protein (number of amino acid residues) is given in parentheses.
Protein abbreviations: VpG, viral genome-linked protein; RdRp, RNA-dependent RNA polymerase.
Numbers refer to the amino acid residue site number according to numbering in GenBank accession KT280060.1.
Within an extended loop region (35).
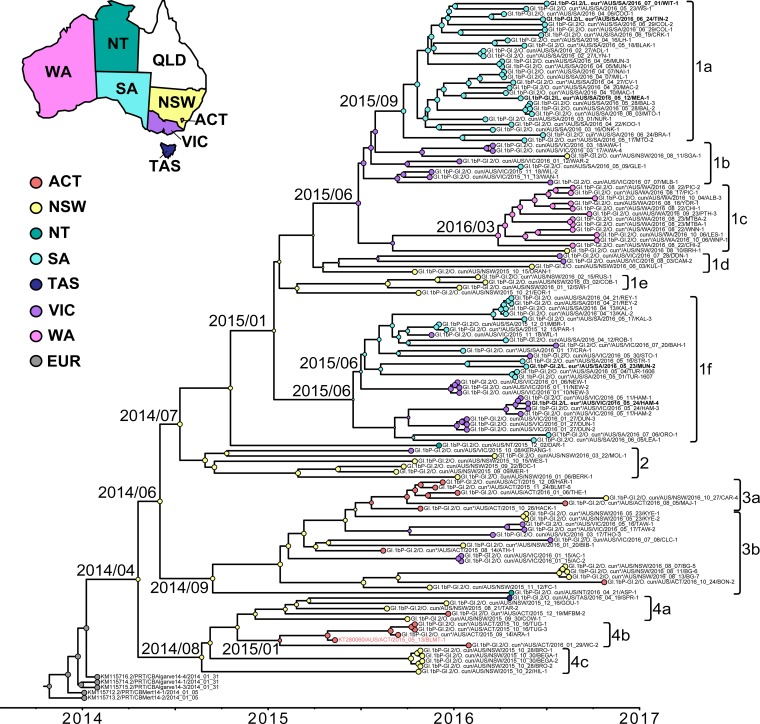
FIG 4.
Inference of the evolutionary history of Australian GI.1bP-GI.2 samples. A Bayesian MCMC time-scaled phylogenetic tree was constructed from 133 GI.1bP-GI.2 nonstructural gene sequences using a relaxed molecular clock (UCLD) and the GMRF Bayesian skyride model of population growth. Circles at tips are color coded to indicate the Australian state from which the virus was collected. ACT, Australian Capital Territory; NSW, New South Wales; NT, Northern Territory; SA, South Australia; TAS, Tasmania; VIC, Victoria; WA, Western Australia. A selection of representative European GI.1bP-GI.2 sequences (EUR) were also included. Circles at the nodes are colored according to their most likely location, as estimated by ancestral state reconstruction. The size of the circles at the node represents the posterior probability that the ancestor occurred in that state, where larger circles represent a higher probability. The taxon name of BlMt-1, the first GI.2 detected in Australia, is highlighted in pale red. The taxon name of viruses isolated from hares are boldfaced. The time to most recent common ancestor is indicated at major nodes (year/month). The accession numbers of sequences obtained from GenBank are included in the taxon names. The species from which the virus was collected is indicated in the taxon name of newly sequenced samples (O. cun, Oryctolagus cuniculus; L. eur, Lepus europaeus). Samples from wild animals are indicated by an asterisk next to the species name. The scale bar is proportional to time in years. Clades and subclades have been defined arbitrarily for reference to Table 3.
The evolutionary history of the Australian GI.2 samples was further explored using a time-scaled phylogenetic analysis (Fig. 4). The results indicate that, based on the sampled GI.2 sequences, the incursion most likely occurred in NSW (posterior probability, >0.85) (Fig. 4) in early 2014 (95% highest posterior density, November 2013 to July 2014), although this inference may change with denser sampling. This result strongly suggests that BlMt-1 (GenBank accession no. KT280060), the initial GI.2 virus detected in Australia in May 2015 (12) was not the index case in Australia and that GI.2 had been circulating in the country for some time prior to its first detection in the ACT. This is further supported by the distance of BlMt-1 from the root of the tree (Fig. 4). It is certainly conceivable that early GI.2 infections were misdiagnosed as GI.1 based on gross pathology. Clinically, GI.1 and GI.2 infections are indistinguishable, such that molecular typing methods are required to differentiate these viruses. Limited surveillance for RHD was being conducted prior to 2015, with an average of 80 samples being tested annually over the preceding 5 years, although this was heavily biased toward domestic rabbits in NSW and wild rabbits in SA. However, since previous diagnostic testing was specific for GI.1, GI.2 infections would have been reported as negative.
The time-structured phylogeny of GI.2 sequences showed some degree of geographical clustering, with the WA sequences forming a distinct group (Fig. 4). Although the majority of the SA sequences clustered within two groups, samples from the eastern states (VIC, TAS, NSW, and ACT) were dispersed throughout multiple clades (Fig. 4), indicating a variable degree of interconnectedness of viruses between these states. The two isolates from the NT did not cluster together, suggesting multiple introductions of GI.2 into this region (Fig. 4). Indeed, the phylogenetic analysis reveals multiple reintroductions of GI.2 into most states (Fig. 4). The GI.2 sequences collected from hares did not form a separate clade; rather, they clustered with virus sequences collected from sympatric rabbit populations (Fig. 4). The viruses sampled here were estimated to evolve at 5.7 × 10−3 substitutions per site per year (95% highest posterior density intervals of 4.8 × 10−3 to 6.5 × 10−3).
DISCUSSION
GI.2 spread rapidly across Australia, being detected in all states and territories and covering a distance of approximately 3,500 km within 18 months of its initial detection in May 2015. We were unable to estimate the precise rate of GI.2 spread through Australia because of a lack of systematic surveillance and the unknown origin of the virus. The dissemination of GI.2 from Europe to geographically isolated locations, such as Canada, Benin, the United Kingdom, the Canary Islands, the Azores, and Australia, highlights the pandemic potential of this virus. Given the stability of caliciviruses in the environment, their efficient transmission, the susceptibility of Australia's rabbit population to GI.2, and the high viral burden present in infected rabbits, GI.2 spread to Australia was probably inevitable. In contrast, the immunological cross-protection between GI.1 strains would limit their dissemination after an incursion, as was observed with GI.1a-Aus, while the competitive advantages of GI.2 allowed this epidemic to develop (23).
During the study period, GI.2 comprised the majority of all pathogenic lagovirus detections in all states except TAS, suggesting it is replacing previously circulating classical GI.1 and GI.1a-Aus viruses on the Australian mainland, as has been reported in Europe (5, 6, 15). Although sampling efforts prior to May 2015 were sporadic, the GI.2 infections occurred instead of, and in addition to, the expected deaths from GI.1. Perhaps the clearest indication of this is seen in the SA data, since sampling efforts were increased in anticipation of the arrival of GI.2, which reached SA in December 2015. From May 2015 to November 2015, 25 cases of GI.1 infection were detected in SA. Thereafter, from December 2015 to October 2016, there were only four cases of GI.1 infection, while 78 cases of GI.2 infection were detected. Based on these data, the GI.2 infection rate was higher than would have been expected with GI.1 alone, and GI.1 detections were well below what would be expected normally. This replacement may have been driven by the lack of immunological cross-protection against GI.2 afforded by the Cylap RCD vaccine (Zoetis Australia) or previous GI.1 infection (20), leading to a large number of susceptible individuals at the start of the epidemic. In addition, the ability of GI.2 to cause lethal infection in young rabbits at a much higher rate than GI.1 (8) implies that new cohorts of rabbits are becoming susceptible to severe outbreaks of GI.2 at an earlier age, which likely provides a strong competitive advantage for virus transmission in the field. A combination of immunological susceptibility of rabbit populations to GI.2 and the ability of the virus to infect young rabbits likely underpins the high epidemiological fitness of GI.2 both in Australia and overseas such that it is able to outcompete GI.1 (36). However, it is possible that other phenotypic differences, such as environmental stability and infectivity, also play a role, although these have not been experimentally assessed for GI.2 specifically.
Our phylogenetic analysis of the nonstructural genes suggested a single GI.2 introduction into Australia from Europe. Based on the available sampling data, the GI.2 incursion most likely occurred in NSW in early 2014, with the virus remaining undetected for several months. GI.2 isolates clustered geographically in a limited fashion, although there was evidence of considerable virus movement between states and multiple reintroductions into most states. Interestingly, the WA sequences were more closely related to sequences from the eastern states than to those from SA, in contrast to what would be expected with natural transmission through wild rabbit populations. This could be explained by (i) human-assisted spread via fomites and the established interstate trade and movement of domestic rabbits, and (ii) transmission on flies as mechanical vectors (37). There is also the possibility of landholders relocating rabbit carcasses in an effort to deliberately spread the virus, similar to what was suggested when GI.1 was originally released in Australia and New Zealand in the mid-1990s (38, 39).
The GI.1bP-GI.2 viruses sampled here evolved at a rate of approximately 5.7 × 10−3 substitutions per site per year, which is comparable to those of GI.1 (2.8 × 10−3) and benign GI.4 (5.3 × 10−3) viruses (34, 40). However, this rate was estimated for a data set with limited temporal spread; therefore, it is not likely to be overly accurate (41). Given the relatively short sampling period, there was considerable genetic diversity between the sampled Australian GI.2 viruses, although this is to be expected from rapidly evolving RNA viruses (42). Group-defining sites in GI.2 were identified in all nonstructural and structural protein-coding regions but were observed at a disproportionally high rate in the VP60 coding region, specifically the P2 domain and its extended loop regions. This is not unexpected, since the P2 domain is the most externally located surface domain and the extended loop regions are known to be inherently variable (43). It has been suggested that the extended loops define the antigenicity of calicivirus virions and are the primary determinants of virus-host interactions, as seen in noroviruses (43–46). Although the biological significance of these mutations with regard to virus transmission, stability, or immune escape has not been determined, it will be of interest to monitor their persistence or loss in Australian GI.2 strains in the future as strains continue to evolve in this environment.
As previously reported, GI.2 was detected in five European brown hares, and these cases have been described previously (17). The GI.2 samples from hares clustered closely with those from sympatric rabbit populations rather than as a hare-specific clade, suggesting that no major evolutionary changes were required for this host jump. In each case, detection in hares was a likely spillover event from rabbits, as reported in Europe (19). It is unknown if GI.2 is capable of transmitting between hares. As the geographical distribution of hares in Australia is limited and overlaps with the much wider distribution of rabbits (47), there are probably far more opportunities for transmission between rabbits or between hares and rabbits than for transmission between hares. Therefore, the opportunity for GI.2 to adapt specifically to the hare host through exclusive hare transmission is likely restricted. Nevertheless, it is important to include hares in future epidemiological studies to better understand the role they are playing in the epidemiology of Australian lagoviruses.
Due to the limited information of the vaccination status of domestic rabbits in this study and, perhaps more importantly, the lack of information about numbers of vaccinated rabbits that have survived GI.2 infection, it is not possible to estimate the effectiveness of the GI.1 vaccine in preventing disease caused by GI.2. However, the detection of GI.2 in 15 vaccinated domestic rabbits supports previous work demonstrating that cross-protection between GI.1 and GI.2 is at best incomplete, and a vaccine covering GI.2 is urgently needed to protect farmed and pet rabbits in Australia (20).
This epizootic once again demonstrates the unique value of Australia's rabbits as “a grand experiment in disease emergence and evolution” (48) and provides a model system for studying RNA virus emergence and evolution in naive populations. Monitoring the interactions of GI.2 with the other genetically and antigenically distinct lagoviruses present in Australia, and assessing the effects of these viruses on wild rabbit populations, is crucial for informing long-term rabbit management strategies. This is especially important considering the recent nationwide release of GI.1a-K5 in Australia in March 2017, as the degree of immunological cross-protection between GI.1a-K5 and GI.2 is not known. It is imperative that surveillance efforts are continued on a national scale to document these interactions. Indeed, it is unknown how widely GI.1 is circulating in Australia since the arrival of GI.2, although serological analyses are under way to address this. Thus far, GI.2 outbreaks have been observed in wild rabbit populations over two sequential years, and this variant has become the dominant strain in the Australian mainland. However, ongoing monitoring is necessary to reveal if this trend will continue or whether other strains will begin to circulate widely again as population immunity or genetic resistance to GI.2 develops.
MATERIALS AND METHODS
Sample collection.
Rabbit or hare liver, kidney, spleen, and bone marrow samples were submitted to either the Elizabeth Macarthur Agricultural Institute diagnostic virology laboratory (EMAI), the Department of Primary Industries and Regions South Australia (PIRSA), or the CSIRO for lagovirus testing. Sampling was conducted from May 2015 to October 2016, inclusive. Samples were obtained from both domestic and wild lagomorphs that had died of unknown causes. Frequently, more than one sample was submitted from the same location within a 2-week period, and these were classified together as a single case. No animal ethics permit is required in Australia for sample collection from rabbits or hares that are found dead.
Virus identification.
RNA was extracted from tissue samples and screened by either reverse transcription-PCR (PIRSA and CSIRO) or real-time reverse transcription-PCR (EMAI) for the detection and specific identification of pathogenic lagoviruses (GI.1, GI.2, or GI.1a; primers are given in Table 4, and additional details are available on request). Selected positive samples were sent to CSIRO for full-genome sequencing. For these samples, RNA was reextracted using the Maxwell simplyRNA tissue kit and extraction robot (Promega) per the manufacturer's instruction. Multiplex reverse transcription-PCR was performed as described previously (25) to confirm the initial diagnosis, and real-time reverse transcription-PCR (25) was performed to quantify virus load in samples and ensure sample suitability for sequencing.
TABLE 4.
Primers and probes used for initial detection of pathogenic lagoviruses
| Laboratory and name | Sense | Sequence (5′-3′) | Strain | Reference |
|---|---|---|---|---|
| EMAI | ||||
| vp60-7_FOR | + | ACYTGACTGAACTYATTGACG | GI.1 | 49 |
| vp60-8_REV | − | TCAGACATAAGAAAAGCCATTGG | 49 | |
| vp60-9_FAM | Probe | CCAARAGCACRCTCGTGTTCAACCT–FAM-BHQ1 | Modified from reference 49 | |
| RHDVXa2010-F1 | + | GCACCCGGCAGTATTCTC | GI.1a | 23 |
| RHDVXa2010-R1 | − | CCCAGCCAGCGTACATCTG | 23 | |
| RHDVXa2010-P1 | Probe | ACTGTCCAACACTCTCCACAGAACA–FAM-BHQ1 | 23 | |
| RHDV2-F | + | CCCGGGCAACATCCTGTA | GI.2 | This study |
| RHDV2-R | − | CCAGCCAGCGTACATTTGAC | This study | |
| RHDV2-P | Probe | CACTGTCCAACACTCGCCACAAA–FAM-BHQ1 | This study | |
| PIRSA | ||||
| RHDVF6793 | + | GGACTTTCGCTCAACAACTACTCGTCAGC | 50 | |
| RHDVR7411 | − | ATAGCTTACTTTAAACTATAAACCCAA | 50 | |
| RHDV2-For | + | ACCACCGAGAACGCGTCCACGTCG | 17 | |
| RHDV2-Rev | − | GGCGGATGTCAACAAGTTCTGA | 17 | |
| CSIRO | ||||
| GI.1a-Aus_fwd | + | GCGTGGCATTGTGCGCAGCATC | GI.1a | 25 |
| GI.1a-Aus_rev | − | TGTTGGTGATAAGCCATAATCGCG | 25 | |
| GI.1c_fwd | + | AGCAAGACTGTTGACTCAATTTCG | GI.1 | 25 |
| GI.1c_rev | − | AGGCCTGCACAGTCGTAACGTT | 25 | |
| GI.2_fwd | + | TTTCCCTGGAAGCAGTTCGTCA | GI.2 | 25 |
| GI.2_rev | − | TGTTGTCTGGTTTATGCCATTTGC | 25 |
Genome sequencing of virus isolates.
First-strand cDNA was prepared using SuperScript III (Life Technologies), and full-length viral genomes were amplified in overlapping fragments using Platinum Taq DNA polymerase high fidelity (Life Technologies), as described previously (31, 44). Amplicons were pooled for each sample, indexed, and sequenced using the Nextera XT DNA sample preparation kit (Illumina) and the Illumina MiSeq platform (300 cycle v2 kit) per the manufacturer's instructions (31, 34). Sequence read quality assessment and trimming were performed as previously described (31). A consensus sequence was generated for each isolate by mapping reads to the GI reference genome sequence (M67473.1/DEU/FRG/1988.50) using Geneious v8.1.5 (51). Primer sequences were trimmed from the genomic termini. Average sequence coverage ranged from 1,117× to 7,787× (mean, 4,696×).
Phylogenetic analysis.
The complete virus genome sequence was obtained for 139 Australian lagovirus samples collected during the sampling period. Genome sequences were aligned with Australian and global representative lagovirus sequences obtained from GenBank (http://www.ncbi.nlm.nih.gov/GenBank/index.html). Maximum likelihood (ML) phylogenies were estimated for the nonstructural genes (n = 184 sequences; 5,283 nucleotides [nt]) and the structural genes (n = 184 sequences; 2,080 nt) using PhyML v3.1 (52). All phylogenies were inferred using the general time reversible + Γ (GTR+Γ) substitution model (as selected using jModelTest v2.1.7 [53]) with five rate categories, an estimated gamma distribution parameter (Γ), and a combination of nearest-neighbor interchange and subtree pruning and regrafting branch-swapping topology searching. Branch support was estimated using 1,000 bootstrap replicates, and trees were rooted using a European brown hare syndrome virus (EBHSV) sequence (accession no. KC832839) as an outgroup.
Origins of GI.2 in Australia.
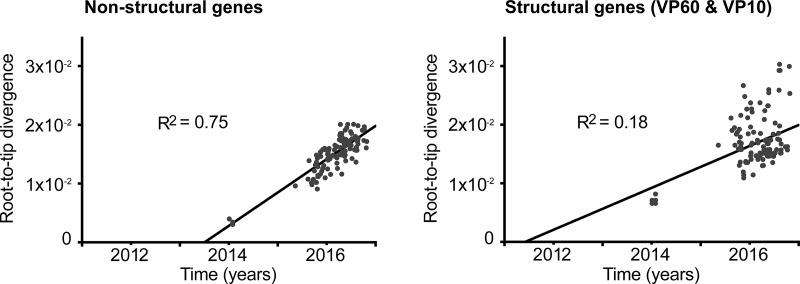
To estimate a time of introduction of GI.2 into Australia and to infer the probable region of introduction, we utilized a Bayesian Markov chain Monte Carlo (MCMC) approach. Of the 139 genomes sequenced for this study, 127 were GI.2. First, these 127 newly sequenced Australian GI.2 genomes and the previously published AUS/ACT/BlMt-1 GI.1bP-GI.2 genome sequence were aligned with five European GI.1bP-GI.2 genome sequences (GI.2 capsid, GI.1b polymerase). These European GI.2 sequences were selected based on similarity to the prototype Australian GI.2, BlMt-1, and because full sequence information was not available for the earliest reported GI.2 isolate, FRA/10-01/2010 (accession no. HE800529.1). Dates of collection were assigned to all sequences in the alignment. Dates were precise to the day for the Australian sequences and for two of the European sequences. The remaining three European sequences (KM115714 to KM115716) were sampled in 2014, with one known to be collected in January 2014 (21). As no precise sampling dates were available for the remaining two sequences, we assumed dates of both January and July 2014, as these represent the maximum possible range of sampling times (i.e., these sequences were originally published in July 2014). Similar rates were obtained using both dating schemes. The genome data set was split into nonstructural genes (n = 133; 5,283 nt) and structural genes (n = 133; 2,071 nt). Maximum likelihood trees were inferred (rooted using the European sequences), and these phylogenies were used as input for linear regression of root-to-tip genetic distances against sampling times, conducted using TempEst (54). This analysis revealed sufficient temporal signal (R2 = 0.75) in the nonstructural gene data set (Fig. 5) to proceed with Bayesian inference using molecular clock models. As no such temporal structure (R2 = 0.18) was observed in the structural genes, we did not use these sequences for molecular clock dating.
FIG 5.
Linear regressions of GI.1bP-GI.2 nonstructural and structural genes. Linear regressions of root-to-tip genetic distances (y axis) against sampling time (x axis) were inferred for GI.1bP-GI.2 Australian (n = 128) and European (n = 5) nonstructural and structural genes.
The time to most recent common ancestor (TMRCA) and most likely geographic location of the common ancestor of Australian GI.2 sampled here was estimated using the Bayesian MCMC method as implemented in the BEAST package, v1.8.2 (55). The GTR+Γ substitution model was implemented for all runs, with five rate categories. The BEAST analyses were initially run using the relaxed uncorrelated lognormal (UCLD) and the strict molecular clocks. However, based on the coefficient of rate variation parameter from the UCLD analyses, the UCLD relaxed clock was deemed most appropriate. The Gaussian Markov random field (GMRF) Bayesian skyride model of population growth was implemented, as the data did not fit a defined parametric demographic function (as determined from the GMRF skyride reconstruction).
The geographic location (state of collection) was also assigned to each sample as a discrete trait, and a separate data partition was created for this trait. The symmetric discrete trait substitution model was assigned to reduce parameter complexity. Ancestral state reconstruction was applied to the trait partition, reconstructing states at all ancestors, and specifically reconstructing states at the most recent common ancestor of the Australian GI.2 samples. Each BEAST analysis was run for 250 million generations until convergence was achieved, and at least two independent runs were performed for each set of priors. A maximum clade credibility (MCC) tree with mean node heights and Bayesian posterior probability values indicating the degree of support for each node was created using the TreeAnnotator program (as available within the BEAST package) from the posterior set of trees.
To further assess the robustness of the BEAST analysis, we performed a Bayesian randomization test in which each sequence was assigned a random date (41) and the BEAST analysis repeated. This date randomization test was completed 10 times and compared to the BEAST results with the correctly dated sequences described above. No overlap of posterior distributions for nucleotide substitution rate or TMRCA was observed between the correctly dated data set and the randomized data sets.
Recombination detection.
Genome sequences generated in this study were screened for recombination using the Recombination Detection Program, v4 (56), in an alignment with 24 reference or potential parent sequences. The RDP, GENECONV, MaxChi, and Bootscan methods were utilized, and significant evidence of recombination was denoted by a P value of <0.05.
Accession number(s).
The sequences generated in this study were deposited into GenBank under accession numbers MF421563 to MF421701.
ACKNOWLEDGMENTS
We thank Alex Thorpe and John Matthews (Agriculture Victoria) and other submitters for sample collection. The assistance of technical staff of the EMAI virology laboratory in undertaking diagnostic screening is greatly appreciated.
Funding for this work was obtained from CSIRO, Biosecurity SA, and the Invasive Animals Cooperative Research Centre. J.E.M. is supported by grant DP140103362 from the Australian Research Council. E.C.H. is supported by an NHMRC Australia Fellowship (GNT1037231).
REFERENCES
- 1.Le Gall-Recule G, Zwingelstein F, Boucher S, Le Normand B, Plassiart G, Portejoie Y, Decors A, Bertagnoli S, Guerin JL, Marchandeau S. 2011. Detection of a new variant of rabbit haemorrhagic disease virus in France. Vet Rec 168:137–138. doi: 10.1136/vr.d697. [DOI] [PubMed] [Google Scholar]
- 2.Camarda A, Pugliese N, Cavadini P, Circella E, Capucci L, Caroli A, Legretto M, Mallia E, Lavazza A. 2014. Detection of the new emerging rabbit haemorrhagic disease type 2 virus (RHDV2) in Sicily from rabbit (Oryctolagus cuniculus) and Italian hare (Lepus corsicanus). Res Vet Sci 97:642–645. doi: 10.1016/j.rvsc.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Le Pendu J, Abrantes J, Bertagnoli S, Guitton JS, Le Gall-Recule G, Lopes AM, Marchandeau S, Alda F, Almeida T, Celio AP, Barcena J, Burmakina G, Blanco E, Calvete C, Cavadini P, Cooke B, Dalton K, Delibes Mateos M, Deptula W, Eden JS, Wang F, Ferreira CC, Ferreira P, Foronda P, Goncalves D, Gavier-Widen D, Hall R, Hukowska-Szematowicz B, Kerr P, Kovaliski J, Lavazza A, Mahar J, Malogolovkin A, Marques RM, Marques S, Martin-Alonso A, Monterroso P, Moreno S, Mutze G, Neimanis A, Niedzwiedzka-Rystwej P, Peacock D, Parra F, Rocchi M, Rouco C, Ruvoen-Clouet N, Silva E, Silverio D, Strive T, Thompson G, Tokarz-Deptula B, Esteves P. 2017. Proposal for a unified classification system and nomenclature of lagoviruses. J Gen Virol 98:1658–1666. doi: 10.1099/jgv.0.000840. [DOI] [PubMed] [Google Scholar]
- 4.Duarte M, Carvalho C, Bernardo S, Barros SV, Benevides S, Flor L, Monteiro M, Marques I, Henriques M, Barros SC, Fagulha T, Ramos F, Luis T, Fevereiro M. 2015. Rabbit haemorrhagic disease virus 2 (RHDV2) outbreak in Azores: disclosure of common genetic markers and phylogenetic segregation within the European strains. Infect Genet Evol 35:163–171. doi: 10.1016/j.meegid.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Le Gall-Recule G, Lavazza A, Marchandeau S, Bertagnoli S, Zwingelstein F, Cavadini P, Martinelli N, Lombardi G, Guerin JL, Lemaitre E, Decors A, Boucher S, Le Normand B, Capucci L. 2013. Emergence of a new lagovirus related to rabbit haemorrhagic disease virus. Vet Res 44:81. doi: 10.1186/1297-9716-44-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin-Alonso A, Martin-Carrillo N, Garcia-Livia K, Valladares B, Foronda P. 2016. Emerging rabbit haemorrhagic disease virus 2 (RHDV2) at the gates of the African continent. Infect Genet Evol 44:46–50. doi: 10.1016/j.meegid.2016.06.034. [DOI] [PubMed] [Google Scholar]
- 7.Westcott DG, Frossard JP, Everest D, Dastjerdi A, Duff JP, Steinbach F, Choudhury B. 2014. Incursion of RHDV2-like variant in Great Britain. Vet Rec 174:333. doi: 10.1136/vr.g2345. [DOI] [PubMed] [Google Scholar]
- 8.Dalton KP, Nicieza I, Balseiro A, Muguerza MA, Rosell JM, Casais R, Alvarez AL, Parra F. 2012. Variant rabbit hemorrhagic disease virus in young rabbits, Spain. Emerging Infect Dis 18:2009–2012. doi: 10.3201/eid1812.120341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baily JL, Dagleish MP, Graham M, Maley M, Rocchi MS. 2014. RHDV variant 2 presence detected in Scotland. Vet Rec 174:411. doi: 10.1136/vr.g2781. [DOI] [PubMed] [Google Scholar]
- 10.Abrantes J, Lopes AM, Dalton KP, Melo P, Correia JJ, Ramada M, Alves PC, Parra F, Esteves PJ. 2013. New variant of rabbit hemorrhagic disease virus, Portugal, 2012-2013. Emerg Infect Dis 19:1900–1902. doi: 10.3201/eid1911.130908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Almeida T, Lopes AM, Magalhaes MJ, Neves F, Pinheiro A, Goncalves D, Leitao M, Esteves PJ, Abrantes J. 2015. Tracking the evolution of the G1/RHDVb recombinant strains introduced from the Iberian Peninsula to the Azores islands, Portugal. Infect Genet Evol 34:307–313. doi: 10.1016/j.meegid.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 12.Hall RN, Mahar JE, Haboury S, Stevens V, Holmes EC, Strive T. 2015. Emerging rabbit hemorrhagic disease virus 2 (RHDVb), Australia. Emerg Infect Dis 21:2276–2278. doi: 10.3201/eid2112.151210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.World Organisation of Animal Health. 2015. Rabbit haemorrhagic disease, Benin–immediate notification report. World Organisation of Animal Health, Paris, France: http://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=19109. [Google Scholar]
- 14.World Organisation of Animal Health. 2016. Rabbit haemorrhagic disease, Canada–immediate notification report. World Organisation of Animal Health, Paris, France: http://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=20799. [Google Scholar]
- 15.Lopes AM, Correia J, Abrantes J, Melo P, Ramada M, Magalhaes MJ, Alves PC, Esteves PJ. 2014. Is the new variant RHDV replacing genogroup 1 in Portuguese wild rabbit populations? Viruses 7:27–36. doi: 10.3390/v7010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalton KP, Abrantes J, Lopes AM, Nicieza I, Alvarez AL, Esteves PJ, Parra F. 2015. Complete genome sequence of two rabbit hemorrhagic disease virus variant b isolates detected on the Iberian Peninsula. Arch Virol 160:877–881. doi: 10.1007/s00705-014-2329-3. [DOI] [PubMed] [Google Scholar]
- 17.Hall RN, Peacock DE, Kovaliski J, Mahar JE, Mourant R, Piper M, Strive T. 2017. Detection of RHDV2 in European brown hares (Lepus europaeus) in Australia. Vet Rec 180:121. doi: 10.1136/vr.104034. [DOI] [PubMed] [Google Scholar]
- 18.Puggioni G, Cavadini P, Maestrale C, Scivoli R, Botti G, Ligios C, Le Gall-Recule G, Lavazza A, Capucci L. 2013. The new French 2010 rabbit hemorrhagic disease virus causes an RHD-like disease in the Sardinian Cape hare (Lepus capensis mediterraneus). Vet Res 44:96. doi: 10.1186/1297-9716-44-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Velarde R, Cavadini P, Neimanis A, Cabezon O, Chiari M, Gaffuri A, Lavin S, Grilli G, Gavier-Widen D, Lavazza A, Capucci L. 11 Sep 2016. Spillover events of infection of brown hares (Lepus europaeus) with rabbit haemorrhagic disease type 2 virus (RHDV2) caused sporadic cases of an European brown hare syndrome-like disease in Italy and Spain. Transbound Emerg Dis doi: 10.1111/tbed.12562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peacock D, Kovaliski J, Sinclair R, Mutze G, Iannella A, Capucci L. 2017. RHDV2 overcoming RHDV immunity in wild rabbits (Oryctolagus cuniculus) in Australia. Vet Rec 180:280. doi: 10.1136/vr.104135. [DOI] [PubMed] [Google Scholar]
- 21.Lopes AM, Dalton KP, Magalhaes MJ, Parra F, Esteves PJ, Holmes EC, Abrantes J. 2015. Full genomic analysis of new variant rabbit hemorrhagic disease virus revealed multiple recombination events. J Gen Virol 96:1309–1319. doi: 10.1099/vir.0.000070. [DOI] [PubMed] [Google Scholar]
- 22.Capucci L, Fallacara F, Grazioli S, Lavazza A, Pacciarini ML, Brocchi E. 1998. A further step in the evolution of rabbit hemorrhagic disease virus: the appearance of the first consistent antigenic variant. Virus Res 58:115–126. doi: 10.1016/S0168-1702(98)00106-3. [DOI] [PubMed] [Google Scholar]
- 23.Mahar JE, Read AJ, Gu X, Urakova N, Mourant R, Piper M, Haboury S, Holmes EC, Strive T, Hall RN. Detection and circulation of a novel RHDVa in Australia. Emerg Infect Dis, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu SJ, Xue HP, Pu BQ, Qian NH. 1984. A new viral disease in rabbits. Anim Husb Vet Med 16:253–255. [Google Scholar]
- 25.Hall RN, Mahar JE, Read AJ, Mourant R, Piper M, Huang N, Strive T. A strain-specific multiplex RT-PCR for Australian rabbit haemorrhagic disease viruses uncovers a new recombinant virus variant in rabbits and hares. Transbound Emerg Dis, in press. [DOI] [PubMed] [Google Scholar]
- 26.Le Gall-Recule G, Zwingelstein F, Fages MP, Bertagnoli S, Gelfi J, Aubineau J, Roobrouck A, Botti G, Lavazza A, Marchandeau S. 2011. Characterisation of a non-pathogenic and non-protective infectious rabbit lagovirus related to RHDV. Virology 410:395–402. doi: 10.1016/j.virol.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 27.Strive T, Wright JD, Robinson AJ. 2009. Identification and partial characterisation of a new lagovirus in Australian wild rabbits. Virology 384:97–105. doi: 10.1016/j.virol.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 28.Le Gall-Recule G, Lemaitre E, Briand F, Marchandeau S. 2015. Characterisation in France of non-pathogenic lagoviruses closely related to the Australian Rabbit calicivirus RCV-A1: confirmation of the European origin of RCV-A1, p 183–185. In Xth International, Congress for, Veterinary Virology, Changing Viruses, in a, Changing World. Centre de Cooperation Internationale en Recherche Agronomique pour le Developpement, Paris, France. [Google Scholar]
- 29.Capucci L, Fusi P, Lavazza A, Pacciarini ML, Rossi C. 1996. Detection and preliminary characterization of a new rabbit calicivirus related to rabbit hemorrhagic disease virus but nonpathogenic. J Virol 70:8614–8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bergin IL, Wise AG, Bolin SR, Mullaney TP, Kiupel M, Maes RK. 2009. Novel calicivirus identified in rabbits, Michigan, U S A. Emerg Infect Dis 15:1955–1962. doi: 10.3201/eid1512.090839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eden JS, Kovaliski J, Duckworth JA, Swain G, Mahar JE, Strive T, Holmes EC. 2015. Comparative phylodynamics of rabbit hemorrhagic disease virus in Australia and New Zealand. J Virol 89:9548–9558. doi: 10.1128/JVI.01100-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooke BD, Fenner F. 2002. Rabbit haemorrhagic disease and the biological control of wild rabbits, Oryctolagus cuniculus, in Australia and New Zealand. Wildl Res 29:689–706. doi: 10.1071/WR02010. [DOI] [Google Scholar]
- 33.World Organisation of Animal Health. 2017. Rabbit haemorrhagic disease, Australia–immediate notification report. World Organisation of Animal Health, Paris, France: http://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?reportid=23377. [Google Scholar]
- 34.Mahar JE, Nicholson L, Eden JS, Duchene S, Kerr PJ, Duckworth J, Ward VK, Holmes EC, Strive T. 2016. Benign rabbit caliciviruses exhibit evolutionary dynamics similar to those of their virulent relatives. J Virol 90:9317–9329. doi: 10.1128/JVI.01212-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leuthold MM, Dalton KP, Hansman GS. 2015. Structural analysis of a rabbit hemorrhagic disease virus binding to histo-blood group antigens. J Virol 89:2378–2387. doi: 10.1128/JVI.02832-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bahl J, Vijaykrishna D, Holmes EC, Smith GJ, Guan Y. 2009. Gene flow and competitive exclusion of avian influenza A virus in natural reservoir hosts. Virology 390:289–297. doi: 10.1016/j.virol.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Asgari S, Hardy JR, Sinclair RG, Cooke BD. 1998. Field evidence for mechanical transmission of rabbit haemorrhagic disease virus (RHDV) by flies (Diptera:Calliphoridae) among wild rabbits in Australia. Virus Res 54:123–132. doi: 10.1016/S0168-1702(98)00017-3. [DOI] [PubMed] [Google Scholar]
- 38.Kovaliski J. 1998. Monitoring the spread of rabbit hemorrhagic disease virus as a new biological agent for control of wild European rabbits in Australia. J Wildl Dis 34:421–428. doi: 10.7589/0090-3558-34.3.421. [DOI] [PubMed] [Google Scholar]
- 39.O'Hara P. 2006. The illegal introduction of rabbit haemorrhagic disease virus in New Zealand. Rev Sci Tech 25:119–123. doi: 10.20506/rst.25.1.1650. [DOI] [PubMed] [Google Scholar]
- 40.Eden JS, Read AJ, Duckworth JA, Strive T, Holmes EC. 2015. Resolving the origin of rabbit hemorrhagic disease virus: insights from an investigation of the viral stocks released in Australia. J Virol 89:12217–12220. doi: 10.1128/JVI.01937-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duchene S, Duchene D, Holmes EC, Ho SY. 2015. The performance of the date-randomization test in phylogenetic analyses of time-structured virus data. Mol Biol Evol 32:1895–1906. doi: 10.1093/molbev/msv056. [DOI] [PubMed] [Google Scholar]
- 42.Duffy S, Shackelton LA, Holmes EC. 2008. Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet 9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Xu F, Liu J, Gao B, Liu Y, Zhai Y, Ma J, Zhang K, Baker TS, Schulten K, Zheng D, Pang H, Sun F. 2013. Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography. PLoS Pathog 9:e1003132. doi: 10.1371/journal.ppat.1003132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elsworth P, Cooke BD, Kovaliski J, Sinclair R, Holmes EC, Strive T. 2014. Increased virulence of rabbit haemorrhagic disease virus associated with genetic resistance in wild Australian rabbits (Oryctolagus cuniculus). Virology 464-465:415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thorne L, Bailey D, Goodfellow I. 2012. High-resolution functional profiling of the norovirus genome. J Virol 86:11441–11456. doi: 10.1128/JVI.00439-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lindesmith LC, Beltramello M, Donaldson EF, Corti D, Swanstrom J, Debbink K, Lanzavecchia A, Baric RS. 2012. Immunogenetic mechanisms driving norovirus GII.4 antigenic variation. PLoS Pathog 8:e1002705. doi: 10.1371/journal.ppat.1002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stott P, Harris S. 2006. Demographics of the European hare (Lepus europaeus) in the Mediterranean climate zone of Australia. Mamm Biol 71:214–226. doi: 10.1016/j.mambio.2006.02.009. [DOI] [Google Scholar]
- 48.Di Giallonardo F, Holmes EC. 2015. Viral biocontrol: grand experiments in disease emergence and evolution. Trends Microbiol 23:83–90. doi: 10.1016/j.tim.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gall A, Hoffmann B, Teifke JP, Lange B, Schirrmeier H. 2007. Persistence of viral RNA in rabbits which overcome an experimental RHDV infection detected by a highly sensitive multiplex real-time RT-PCR. Vet Microbiol 120:17–32. doi: 10.1016/j.vetmic.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 50.Kovaliski J, Sinclair R, Mutze G, Peacock D, Strive T, Abrantes J, Esteves PJ, Holmes EC. 2014. Molecular epidemiology of rabbit haemorrhagic disease virus in Australia: when one became many. Mol Ecol 23:408–420. doi: 10.1111/mec.12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guindon S, Delsuc F, Dufayard JF, Gascuel O. 2009. Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol 537:113–137. doi: 10.1007/978-1-59745-251-9_6. [DOI] [PubMed] [Google Scholar]
- 53.Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 54.Rambaut A, Lam TT, Max Carvalho L, Pybus OG. 2016. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol 2:vew007. doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. 2015. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]