ABSTRACT
Prior studies have found that HIV, through the Vpr protein, promotes genome reduplication (polyploidy) in infection-surviving epithelial cells within renal tissue. However, the temporal progression and molecular regulation through which Vpr promotes polyploidy have remained unclear. Here we define a sequential progression to Vpr-mediated polyploidy in human renal tubule epithelial cells (RTECs). We found that as in many cell types, Vpr first initiates G2 cell cycle arrest in RTECs. We then identified a previously unreported cascade of Vpr-dependent events that lead to renal cell survival and polyploidy. Specifically, we found that a fraction of G2-arrested RTECs reenter the cell cycle. Following this cell cycle reentry, two distinct outcomes occur. Cells that enter complete mitosis undergo mitotic cell death due to extra centrosomes and aberrant division. Conversely, cells that abort mitosis undergo endoreplication to become polyploid. We further show that multiple small-molecule inhibitors of the phosphatidylinositol 3-kinase-related kinase (PIKK) family, including those that target ATR, ATM, and mTOR, indirectly prevent Vpr-mediated polyploidy by preventing G2 arrest. In contrast, an inhibitor that targets DNA-dependent protein kinase (DNA-PK) specifically blocks the Vpr-mediated transition from G2 arrest to polyploidy. These findings outline a temporal, molecularly regulated path to polyploidy in HIV-positive renal cells.
IMPORTANCE Current cure-focused efforts in HIV research aim to elucidate the mechanisms of long-term persistence of HIV in compartments. The kidney is recognized as one such compartment, since viral DNA and mRNA persist in the renal tissues of HIV-positive patients. Further, renal disease is a long-term comorbidity in the setting of HIV. Thus, understanding the regulation and impact of HIV infection on renal cell biology will provide important insights into this unique HIV compartment. Our work identifies mechanisms that distinguish between HIV-positive cell survival and death in a known HIV compartment, as well as pharmacological agents that alter these outcomes.
KEYWORDS: G2 arrest, HIV and kidney, HIV reservoir, PIKK family, Vpr, polyploidy
INTRODUCTION
Widespread use of combination antiretroviral therapy (cART) has dramatically altered the landscape of HIV-1-associated morbidity and mortality. The introduction of cART has changed the spectrum of acute kidney injury and chronic kidney disease, including HIV-associated nephropathy (HIVAN). However, the persistence of HIV-1-related renal disease (1, 2) necessitates further investigation into the role of direct HIV-1 infection of the kidney. Such investigation can uncover specific roles of HIV-1 in renal disease pathogenesis and may possibly uncover a role for renal reservoir establishment after HIV-1 infection.
Several observations support the idea that the kidney is a unique HIV-1 infection compartment. HIV-1 mRNA and DNA have been detected in renal epithelial cells in biopsy specimens from seropositive patients with kidney disease (3, 4), and viral sequences within these renal cells diverge from those in peripheral blood cells (5). Further, our demonstration that HIV DNA and RNA persist in renal cells of patients despite therapy suggests that long-term infection of the kidney may play a role in renal disease pathogenesis and the maintenance of a viral reservoir (6).
The pathogenic consequences of direct HIV-1 infection of renal tubule epithelial cells (RTECs) were noted in an early Gag/Pol-deleted HIV-1 transgenic mouse model (Tg26) (7). Subsequent mouse modeling found that both HIV-1 Nef and Vpr contribute to the full pathological findings of HIVAN. Supporting in vitro models demonstrated that Nef expression results in podocyte dedifferentiation and proliferation (reviewed in reference 8), while Vpr expression is associated with HIVAN tubule pathology (9, 10). Vpr expression in RTECs induces DNA damage response activation, G2 arrest, apoptosis, and polyploidy (11–14). While Vpr-induced G2 arrest and apoptosis have been extensively studied in vitro (reviewed in reference 15), the mechanism and physiological role of polyploidy in HIV-positive cells remain unknown. Polyploid cells, which contain multiples of the diploid chromosome number, have been noted in in vitro Vpr model systems (15–20). Additionally, our demonstration of polyploidy in kidneys from Tg26 mice and in biopsy specimens from HIVAN patients indicates that polyploidy is a physiologically relevant aspect of HIV pathology (11).
While many of the molecular mechanisms that generate polyploidy have been identified, relatively little is known about the physiological implications of polyploidy. In addition to being a hallmark of several diseases, polyploidy is associated with the evasion of cell death in numerous contexts (21–24). The ability of the kidney to serve as a unique HIV-1 compartment and the presence of polyploid cells in HIV-positive renal biopsy specimens raise important questions regarding the mechanism of Vpr-induced renal polyploidy and its pathogenic consequences in vivo. These polyploid cells may play a key role in the establishment of a long-term HIV compartment and potential HIV reservoir in the kidney. In this study, we demonstrate that polyploidy is associated with survival in Vpr-expressing RTECs through a bypass of mitotic cell death. We further identify multiple phosphatidylinositol 3-kinase-related kinase (PIKK) family inhibitors, including those that are reported to block ATR, ATM, mammalian target of rapamycin (mTOR), and DNA-dependent protein kinase (DNA-PK), as agents that prevent Vpr-mediated polyploidy. Most of these inhibitors indirectly block polyploidy by blocking G2 arrest, which we find to immediately precede polyploidy. However, the DNA-PK inhibitor NU7441 can specifically block Vpr-mediated polyploidy. Our findings demonstrate a temporal progression to Vpr-induced polyploidy in RTECs, as well as identifying pharmacological agents that block this progression. These findings represent important advances in understanding the role of polyploidy in HIVAN pathogenesis and possible reservoir establishment.
RESULTS
A subset of HIV-1 Vpr+ RTECs escape ATR-dependent G2 arrest to become polyploid.
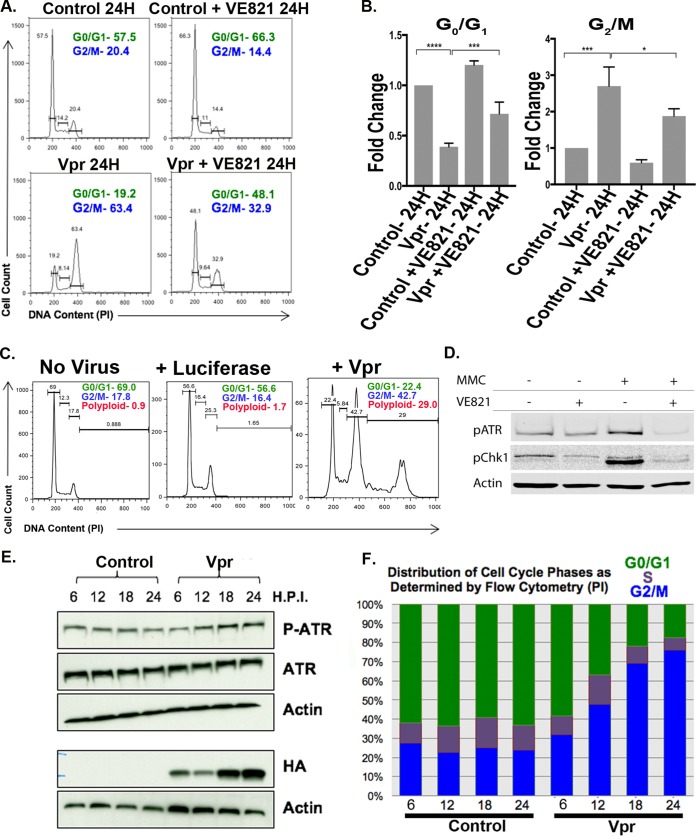
A hallmark of Vpr expression in a number of cell types is G2 cell cycle arrest, mediated by the PIKK family member ATR (15, 16, 18, 25–31). To determine whether ATR-mediated G2 arrest is observed in Vpr+ RTECs, we transduced HK2 human renal tubule epithelial cells with vesicular stomatitis virus glycoprotein (VSV-G)-pseudotyped lentiviral vectors expressing hemagglutinin (HA)-tagged Vpr. The same vector also expressed green fluorescent protein (GFP) from an internal ribosomal entry site (IRES) to allow for the isolation of cells of interest by flow cytometry. Using flow cytometry, we first analyzed DNA content to determine whether infected cells underwent G2 arrest. Twenty-four hours after transduction, we detected a 3-fold increase in the number of cells in G2 (Fig. 1A, B, and F). The increased G2 fraction was not a cellular response to lentiviral infection, since infection with a control (luciferase) vector did not appreciably alter the cell cycle profile (Fig. 1C). As has been shown in other contexts, G2 arrest appeared to depend on ATR, since inhibiting ATR with VE821, an inhibitor of ATR (Materials and Methods), partially relieved this arrest (Fig. 1A, B, and D). VE821 did not fully abrogate G2 arrest in Vpr+ RTECs, possibly reflecting a requirement for other related PIKKs. We concurrently monitored levels of active ATR via Western blotting and the cell cycle phase via flow cytometry over a 24-h period. While control cells had little variation in the phospho-ATR levels and the distribution of cell cycle phases, Vpr+ cells underwent concomitant increases in phospho-ATR levels and G2 accumulation (Fig. 1E and F). These results implicate ATR in the G2 arrest of Vpr-expressing RTECs.
FIG 1.
ATR-dependent G2/M arrest in RTECs. (A) Flow cytometric cell cycle analysis of RTEC line HK2 24 h following transduction with TY2-Vpr-GFP with or without the ATR inhibitor VE821. The x axis shows DNA content, as measured by propidium iodide (PI) staining, and the y axis indicates the relative cell number. The percentages of cells within the G0/G1 and G2/M gates are given in each panel. (B) Quantitation of three replicates of the experiment for which results are shown in panel A. The mean value is plotted for each condition (expressed as fold change), and error bars indicate standard deviations. Asterisks indicate significance by one-way ANOVA: *, P < 0.001; **, P < 0.0002; ***, P < 0.0001. (C) Flow cytometric analysis of uninfected HK2 cells (left) or HK2 cells infected with a lentiviral vector expressing luciferase (center) or Vpr (right). The percentages of cell cycle/ploidy classes are given. (D) VE821 potently inhibits ATR activity in HK2 cells. HK2 cells were either left untreated or treated with mitomycin C (MMC), which induces ATR activity. Active ATR levels were monitored by Western blotting with both a phospho-ATR antibody and an antibody to the phosphorylated (active) form of Chk1 (see Materials and Methods). Actin served as a loading control. (E) Time course of ATR phosphorylation in HK2 cells expressing Vpr from HR-HA-Vpr-GFP (HA-Vpr) as measured by Western blotting. H.P.I., hours postinduction. (F) Corresponding cell cycle phase analysis (by flow cytometry) for the same populations of cells for which results are shown in panel E.
By conducting a longer-term analysis of DNA content, we found that G2 arrest is transient and precedes a doubling of genome content in a substantial subset of Vpr+ RTECs. To determine the duration of G2 arrest, we analyzed DNA contents in Vpr+ and control cells at 24 h, 36 h, and 48 h. While at 24 h posttransduction, a majority of Vpr+ cells were in G2, by 48 h, 25% of the total cell population exhibited polyploidy (Fig. 2A), as evidenced by having >4C DNA content.
FIG 2.
A subset of HIV-1 Vpr+ renal tubule epithelial cells escape ATR-dependent G2 arrest to become polyploid. (A) Flow cytometric analysis of HK2 cells showing the emergence of polyploidy in TY2-Vpr-GFP+ RTECs over time. The same data are averaged for three replicates and are plotted below. (B) (Top left and center) Flow cytometric analysis of HK2 cells 48 h following transduction with TY2-Vpr-GFP (VPR 48H) or TY2-Q65R-GFP (Vpr Q65R 48H) or following treatment with doxorubicin (Doxo) or cisplatin. (Top right) Flow cytometric analysis of HK2 cells 48 h after transduction with Vpr (integrase-positive) or Vpr In- (integrase-deficient) viruses. (Bottom) HK2 cells were harvested 3 days after infection with a lentivirus expressing either HA-tagged wild-type (WT) Vpr or HA-tagged Vpr with a Q65R mutation. Cells were prepared for Western blotting as described in Materials and Methods and were probed with an anti-HA antibody. Actin served as a loading control. −, no-infection control. The HA/actin ratios for WT and Q65R Vpr are plotted, with the average of three replicates for Q65R Vpr shown. n.s., not significant. The mean DNA contents, averaged (± standard deviations) from three separate replicates of the doxorubicin and cisplatin experiments, are also plotted in a bar graph. (C) Flow cytometric analysis of HK2 cells infected with NL4-3.HSA.Vpr+ or Vpr− viruses pseudotyped with the VSV-G envelope. Infections were performed for 3 days. (D) SUP-T1 T cells were either left uninfected or infected with a TY2-Luc-GFP (control) or TY2-Vpr-GFP virus. Cells were harvested at 3 days postinfection and their DNA contents measured by flow cytometry.
Two known causes of polyploidy are DNA damage and a prolonged G2 phase (24, 32). To distinguish between these possibilities, we first treated HK2 cells with the DNA-damaging agent cisplatin, which induces replication stress, or doxorubicin, a potent inducer of double-stranded DNA breaks. Multiple doses and treatment times were tested (data not shown), but in all instances, cells either arrested in G2 phase or underwent cell death, or both, without becoming polyploid (Fig. 2B). Viral integration, which can also cause DNA damage, was not required for Vpr-mediated polyploidy, as evidenced by the fact that an integration-defective Vpr expression vector still induced a comparable level of polyploidy (Fig. 2B, top right, Vpr In-). These results indicate that DNA damage is not sufficient to promote Vpr-induced polyploidy in RTECs. In contrast, we find that an extended G2 phase is required for Vpr-induced polyploidy, since cells expressing the G2 arrest-deficient Vpr Q65R mutant failed to exhibit signs of either G2 arrest or polyploidy by 48 h posttransduction (Fig. 2B). Together, these results indicate that G2 arrest/extension is required, but is not sufficient, for promoting polyploidy in Vpr+ RTECs.
Next, we tested whether the Vpr-mediated cell cycle alterations that we characterized were due to Vpr overexpression and if that can occur in a nonrenal cell type. We found that Vpr-dependent alterations on RTEC cell cycle progression were not due to Vpr overexpression, since expression of Vpr from a molecular clone of HIV (pNL4-3.HSA.Vpr+ or Vpr−) led to similar Vpr-dependent cell cycle alterations (Fig. 2C). Previous work suggested that Vpr can also promote G2 arrest and polyploidy in T cells (16). We confirmed these findings in Sup-T1 cells, although we found that the degree of polyploidy was much less than that in renal cells (Fig. 2D). These findings suggest that the Vpr-mediated cell cycle changes that we observe are not due to overexpression and may occur in other cell types, such as T cells.
Many Vpr+ RTECs undergo aberrant mitosis following a prolonged G2 phase.
In addition to Vpr-mediated G2 arrest, several groups have also described mitotic defects in Vpr+ cells (19, 33, 34). To determine if Vpr+ RTECs enter mitosis following G2, we exposed RTECs to a single thymidine treatment to synchronize a majority of cells (Fig. 3A) and then monitored for both DNA content (by propidium iodide [PI] staining) and the mitotic marker phospho-histone H3 (pH3) every hour for 26 h after transduction with HR-HA-Vpr-ΔGFP. To represent the temporal progression through the cell cycle, the percentage of cells with 4C DNA content, indicating G2/M phase, was charted for each time point. We also charted pH3+ cells within the G2/M population over time to identify mitotic cells (Fig. 3A). After release from the thymidine block, control cells progressed through S phase to accumulate 4C DNA content. A peak in mitotic cells 16 to 18 h postrelease corresponds with a subsequent decrease in the percentage of cells with 4C DNA content, indicating that cells have undergone mitosis. In agreement with a prolonged G2 phase, Vpr+ RTECs with 4C DNA content had a longer delay between arrival at 4C DNA content and the appearance of pH3+ cells (Fig. 3B). Following this prolonged G2 phase, many Vpr+ cells enter mitosis. The higher percentage of pH3+ Vpr+ cells than of control pH3+ cells suggests that Vpr either increases the number of mitotic cells or causes a longer mitotic phase.
FIG 3.
Vpr+ RTECs undergo aberrant mitosis following a prolonged G2 phase. (A) (Left) Schematic indicating how DNA content (measured by propidium iodide [PI] staining) and mitosis (measured by anti-phospho-histone H3 [pH3]) can distinguish between the G0/G1, G2, and M phases. (Center) Flow cytometry plot at 0 min after thymidine release. The percentage of each discernible cell cycle stage is given. (Right) Flow chart showing the steps performed for thymidine synchronization of HK2 cells. (B) Time course of DNA content or percentage of mitotic cells over time in synchronized HK2 control cells and HK2 cells transduced with HR-HA-VprΔGFP. (Left) Graphical representation of flow cytometry analysis of the percentage of cells with 4C DNA content at each time point. (Right) Percentage of mitotic (pH3+) cells within that 4C population at each time point. (C) Immunofluorescence microscopy of mitosis in synchronized HK2 cells 18 h after transduction with HR-HA-VprΔGFP. A total of 22.1% of mitotic Vpr+ cells, had spindle defects, including multipolar spindles (Vpr1, Vpr2), in contrast to only 6.0% of control cells (627 control cells and 461 Vpr cells [P, <0.00001 by the chi-square test]).
We next examined mitotic structures in Vpr+ RTECs via immunofluorescence microscopy. To capture the first round of mitosis following Vpr expression in HK2 cells, we synchronized a majority of cells using a single thymidine treatment and harvested 18 h after transduction with HR-HA-Vpr-ΔGFP. Mitotic Vpr+ cells exhibited a 3.7-fold increase over non-Vpr controls (P < 0.00001) in supernumerary centrosomes and multiple spindle poles, a hallmark of abnormal mitosis (Fig. 3C, Vpr1, Vpr2). Thus, following exit from a prolonged G2 phase, Vpr expression in RTECs leads to multiple spindle poles, which result in aberrant mitosis and can lead to various outcomes, including mitotic cell death and polyploidy.
Bypassing mitosis favors cell survival and polyploidy in Vpr+ RTECs.
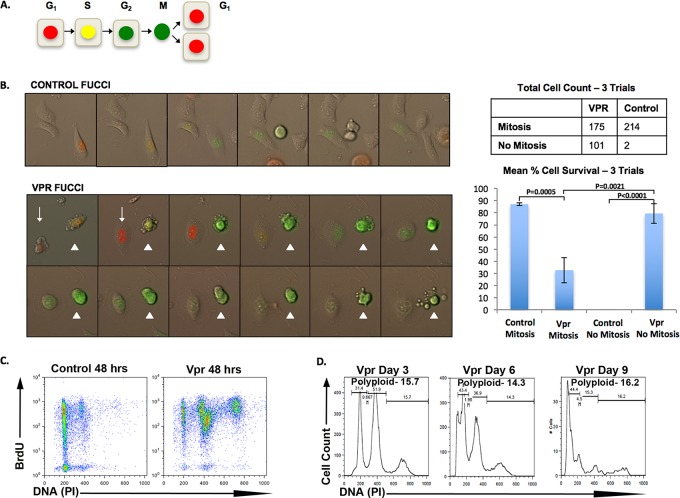
We next sought to determine the outcome of aberrant mitotic progression in Vpr-expressing renal cells. Mitosis in cells with multiple spindle poles is frequently lethal in vivo (reviewed in reference 35). To follow the fate of Vpr+ RTECs during mitosis, we used fluorescent ubiquitination-based cell cycle indicator (FUCCI) probes, which track cell cycle-dependent oscillations of the Cdt1 and Geminin proteins to permit visualization of the G1 (red fluorescence), S/G2 (green fluorescence), and M (nuclear green fluorescence) phases (Fig. 4A) (36). We imaged HK2 cells for 36 h after transduction with HR-HA-Vpr-ΔGFP and counted cells that successfully completed mitosis. Cell counts were done for three separate trials, and mean cell counts were used for analysis. Both control and Vpr+ cells progressed through the G1 and S phases and into G2 phase (Fig. 4B; see also movie S1 in the supplemental materials). Upon exit from G2, however, our imaging revealed two distinct phenotypes in Vpr+ RTECs. As expected, while the majority of HK2 control cells (87%) underwent successful mitosis (Fig. 4B, “Mitosis”), a substantially decreased fraction of Vpr+ cells (63%) progressed through mitosis. The remaining cells (37%) displayed morphological signs of early mitosis, including slight rounding of the cell, but stayed attached to the plate and did not complete mitosis (Fig. 4B, “No Mitosis”). Vpr+ cells that bypassed mitosis had higher survival rates than those that attempted mitosis (79.3% versus 32.7% [P = 0.0021]) (Fig. 4B). Thus, the bypass of mitosis favors cell survival in Vpr+ RTECs that escape G2 arrest.
FIG 4.
Bypassing mitosis favors survival and polyploidy in Vpr+ RTECs. (A) Schematic representation of FUCCI cell cycle indicators. (B) (Top left) Live-cell imaging of HK2 cells transduced with FUCCI vectors. Shown is a representative cell undergoing mitosis and splitting into daughter cells. (Bottom left) Live-cell imaging of HK2 cells cotransduced with HR-HA-VprΔGFP and FUCCI vectors. Cells were counted and were characterized as either mitotic (arrowhead) or not mitotic (arrow). The table at the top right indicates total cell counts for three separate trials. The graph below the table indicates average survival rates for cells that undergo mitosis versus cells that do not complete mitosis in control and Vpr+ cells in 3 separate trials. Error bars represent 95% confidence intervals. (C) Flow cytometry analysis of BrdU incorporation (y axis) and DNA content (PI) (x axis) in control HK2 cells (Control 48 hrs) and HK2 cells transduced with HR-HA-VprΔGFP (Vpr 48 hrs). (D) Flow cytometric analysis showing that polyploidy persists in HK2 cells transduced with TY2-Vpr-GFP over time. The x axis shows DNA content, and the y axis indicates the relative cell number.
Given both the absence of mitosis in many Vpr+ RTECs after G2 and the accumulation of polyploidy in the same cells, we next examined if cells become polyploid by undergoing an additional S phase. We used a single thymidine treatment to synchronize a majority of HK2 cells expressing HR-HA-Vpr-ΔGFP. We then added BrdU to the culture medium 18 h posttransduction, when a majority of Vpr+ cells are in G2/M phase. At 48 h posttransduction, we analyzed cells via flow cytometry for BrdU incorporation and DNA content. Nearly all of the 4C Vpr+ cells incorporated BrdU, further illustrating that RTECs enter a second S phase after escaping Vpr-mediated G2 arrest (Fig. 4C). This second S phase is indicative of endoreplication, a known cell cycle modification that generates polyploidy (24).
We noted that BrdU continued to accumulate in emerging Vpr+ 8C polyploid cells (Fig. 4C). To determine the longer-term fate of polyploid Vpr+ RTECs, we maintained cells in culture for 9 days. Although fewer total cells were recovered at later time points, polyploid cells still persisted in the population and even showed an increased proportion in some experimental replicates (Fig. 4D). The enrichment of polyploid cells in long-term cultures is consistent with the observation that bypassing Vpr-mediated aberrant mitosis favors cell survival and polyploidy in RTECs.
Several PIKK family inhibitors prevent Vpr-mediated polyploidy in RTECs.
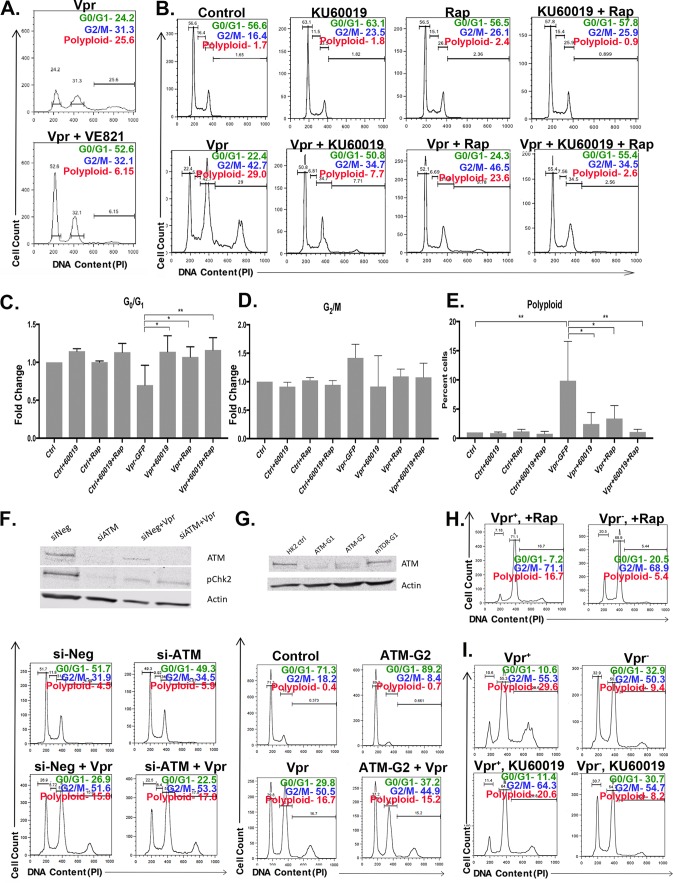
Unlike the established requirement for ATR in Vpr-mediated G2 arrest, the molecular mechanisms behind Vpr-mediated polyploidy are unclear. We first examined the effect of ATR pharmacological inhibition on the accumulation of polyploidy. We thus examined Vpr-expressing RTECs treated with VE821 at the 48-h time point, when polyploidy emerges in Vpr-expressing cells. We continued to observe a lack of G2 arrest in VE821-treated Vpr+ cells at this time point, and we also observed a reduction in Vpr-dependent polyploidy, consistent with our findings that G2 arrest is required for Vpr-mediated polyploidy (Fig. 5A). These findings are consistent with the model that ATR inhibition indirectly blocks Vpr-mediated polyploidy by preventing G2 arrest.
FIG 5.
PIKK inhibitors block multiple steps in the Vpr-mediated progression to polyploidy. (A) Flow cytometric cell cycle analysis of HK2 cells 48 h following transduction with TY2-Vpr-GFP with or without VE821. The x axis shows the DNA content, and the y axis indicates the relative cell number. The relative numbers of cells within the G0/G1, G2/M, and polyploid gates are indicated in each panel. (B) Flow cytometric analysis as in panel A for cells treated with luciferase (control), KU60019, rapamycin (Rap), or KU60019 and rapamycin together. Vpr expression is as in panel A where indicated. (C to E) Quantitation of three replicates of the experiment for which results are shown in panel B. The mean value is plotted for each condition (expressed as the fold change or percentage of cells as indicated), and error bars indicate standard deviations. Asterisks indicate significance by one-way ANOVA (*, P < 0.001; **, P < 0.0002; ***, P < 0.0001). (F) siRNA treatment targeting ATM. (Top) Western blot of HK2 cells treated with a negative control (si-Neg) or ATM (si-ATM) siRNA, with or without Vpr expression. Cells were probed for ATM, phospho-Chk2, and actin as a loading control. (Bottom) Flow cytometric analysis of the DNA contents of cells treated with the indicated siRNA and expressing or not expressing Vpr, as indicated. The relative number of cells is plotted, and the percentage of cells in a given class of DNA content is indicated in each panel. (G) CRISPR/Cas9-mediated ATM deletion in HK2 cells. (Top) Western blot of HK2 cells (ctrl), two different ATM guide RNAs (ATM-G1 and ATM-G2), and an mTOR guide RNA (mTOR-G1). Cells were probed for ATM or for actin as a loading control. (Bottom) Flow cytometric analysis of HK2 or ATM-G2 (ATM-deleted) cell lines upon infection with TY2-Vpr-GFP. (H and I) Flow cytometric analysis of HK2 cells, infected with an NL4-3.HSA.Vpr+ or Vpr− virus pseudotyped with the VSV-G envelope and then treated with rapamycin (H) or KU60019 (I). Infections were performed for 3 days, and the drug was added 1 h after infection. For control data, see Fig. 2C.
VE821 treatment does not completely suppress Vpr-mediated G2 arrest and polyploidy. We reasoned that redundant signaling may act with ATR to alter the cell cycle in response to Vpr expression. ATR belongs to the PIKK family, which includes the related kinase ATM. ATM activation is connected to Vpr pathology (37). To examine the potential role of ATM in Vpr-mediated polyploidy, we transduced HK2 cells with TY2-Vpr-GFP, followed by treatment with KU60019, a known ATM inhibitor (38, 39). We then examined the cell cycle profile of KU60019-treated cells. As with our VE821 treatments, KU60019 treatment reverted the Vpr-mediated G2 arrest (Fig. 5B to D). As with VE821 treatment, this decrease in Vpr-mediated G2 arrest was accompanied by a decrease in the accumulation of polyploid renal cells (Fig. 5B and E). ATM and ATR are highly related kinases, and inhibitors for either kinase could potentially affect both proteins. We next examined whether ATM depletion alone could alter Vpr-mediated cell cycle phenotypes by treating Vpr-expressing HK2 cells either with ATM small interfering RNA (siRNA) (Fig. 5F), or with guide RNAs targeting ATM along with the Cas9 nuclease (ATM clustered regularly interspaced short palindromic repeat [CRISPR] deletion) (Fig. 5G). Despite potent knockdown of ATM, we did not replicate the elimination of polyploidy seen with KU60019 treatment (Fig. 5F and G, bottom). Thus, while KU60019 treatment substantially prevents Vpr-mediated polyploidy, this effect cannot be solely due to ATM inhibition.
Given our ATM inhibitor and knockdown results, we next examined whether other PIKK family members might be involved in Vpr-mediated polyploidy. Unlike ATM and ATR, two other major PIKK family members, mammalian target of rapamycin (mTOR) and DNA-dependent protein kinase (DNA-PK), are often essential for cell survival in culture (40, 41). As expected, when we used established CRISPR reagents to knock out mTOR, we were unable to recover a viable population of cells (data not shown). We thus examined the potential roles of these kinases by treating HK2 cells expressing Vpr with known mTOR and DNA-PK inhibitors. mTOR has been implicated in the pathology of HIVAN in mouse models, human renal cells, and human biopsy specimens (42–46), but the effect of mTOR signaling on Vpr-dependent polyploidy has not been examined. Treatment with rapamycin, a known mTOR inhibitor, substantially reduced polyploidy in Vpr+ cells (Fig. 5B to E). The cell cycle profile of rapamycin-treated cells mirrored that of VE821- or KU60019-treated cells, suggesting that rapamycin treatment blocks Vpr-mediated polyploidy by interfering with Vpr-mediated G2 arrest. Interestingly, cotreatment of Vpr-expressing cells with both KU60019 and rapamycin led to a synergistic reduction of polyploidy, potentially supporting the argument that multiple PIKK family members mediate Vpr-dependent renal G2 arrest/polyploidy (Fig. 5B to E).
Our results with VE821, KU60019, and rapamycin, which are thought to primarily target ATR, ATM, and mTOR, respectively, all suggest an indirect role for PIKK signaling in preventing Vpr-mediated polyploidy by first blocking G2 arrest. We obtained a distinctly different result with an inhibitor of a fourth PIKK family member. NU7441 is considered a selective inhibitor of DNA-PK (47, 48). Unlike treatment with other PIKK inhibitors, only NU7441 treatment substantially decreased the occurrence of Vpr-mediated polyploidy without preventing G2 accumulation (Fig. 6A to D). Our findings with PIKK inhibitors were not due to Vpr overexpression, since Vpr-mediated polyploidy was reduced following treatment with rapamycin, KU60019, or NU7441 when Vpr was expressed from a provirus (Fig. 5H and I and 6E). Interestingly, all PIKK inhibitors appear to specifically block polyploidy and not G2 arrest in our proviral experiments, suggesting that the presence of other HIV proteins can alter the specific cell cycle alterations in response to each inhibitor. Taken together, our findings suggest that PIKK signaling is required at multiple steps during the progression to Vpr-mediated renal polyploidy.
FIG 6.
DNA-PK inhibitors eliminate polyploidy without affecting G2/M arrest. (A) HK2 cells were either left uninfected or infected with TY2-Vpr-GFP. Four hours postinfection, NU7441 (a DNA-PK inhibitor) was added to the medium. Cells were harvested 2 days postinfection and their DNA contents measured by flow cytometry. (B to D) Quantitation of the results for three replicates of the experiment for which results are shown in panel A. (E) Flow cytometric analysis of HK2 cells that were first infected with an NL4-3.HSA.Vpr+ or Vpr− virus pseudotyped with the VSV-G envelope and then treated with NU7441. Infections were performed for 2 days, and the drug was added 1 h after infection.
DISCUSSION
HIV-1 in the kidney may contribute not only to renal pathology but also to the establishment of a long-term viral compartment that could serve as a reservoir seeding subsequent viral rebound. In this study, we used a combination of live imaging, flow cytometry, pharmacology, and genetics in cultured human renal cells to uncover a multistep progression to polyploidy in Vpr-expressing RTECs. We demonstrated that following escape from G2 arrest, polyploidy is a prosurvival outcome associated with the evasion of aberrant multipolar mitosis (Fig. 7). Further, we identified an important role for PIKK signaling in promoting multiple distinct steps of the progression to polyploidy in this context.
FIG 7.
Model for escape from G2 arrest and polyploidy progression in Vpr+ RTECs. In renal cells, expression of HIV-1 Vpr induces G2 arrest. Cells that escape G2 arrest either undergo aberrant mitosis followed by mitotic cell death or endoreplicate to become polyploid. Inhibitors known to target PIKK family members block distinct steps of the progression to polyploidy in renal cells.
Escape from G2 arrest is a critical determinant of cell fate in Vpr+ renal epithelial cells.
A hallmark of Vpr expression is G2/M cell cycle arrest (16, 18, 26, 27). Our work here demonstrates that DNA damage, which can cause G2/M arrest, is not a major trigger of Vpr-mediated polyploidy. In contrast to such genotoxin-induced G2/M arrest, Vpr-mediated G2/M arrest in RTECs can be transient and precedes a critical junction between two distinct outcomes in Vpr+ RTECs: mitotic cell death or polyploidy (Fig. 7). With regard to cell cycle arrest, some previous studies suggest that Vpr-mediated cell cycle arrest and cell death occur independently (49, 50), while many others indicate a temporal and casual relationship between these phenotypes (18, 29, 34, 51, 52). In our study, we find that cell cycle arrest precedes cell death but that cell death depends on progression to mitosis. Vpr+ cells that evade mitosis and become polyploid have a substantially higher survival rate than those that undergo complete mitosis.
While previous studies identified caspase activation and mitochondrial injury as key components of cell death in Vpr+ RTECs (13, 14), mitotic cell death, as we report here, has not been previously attributed to HIV pathogenesis in RTECs. Our fixed and live imaging data suggest that Vpr-mediated renal cell death is, in large part, due to centrosome duplication and subsequent multipolar-spindle formation, a known trigger of mitotic catastrophe (35, 53). Centrosome amplification is a known component of Vpr-associated pathology (19, 33, 34) and occurs only in cells arrested in G2 by Vpr, not by irradiation (34). Given the strong association of mitosis with Vpr-induced cell death, further studies to examine the role of mitotic progression and centrosome regulation in HIV pathogenesis are warranted.
PIKK signaling is required for G2 escape and polyploidy in Vpr+ renal epithelial cells.
Our demonstration that inhibitors of PIKK family signaling can block distinct steps of the Vpr-mediated progression to polyploidy highlights an expanded role for PIKK signaling in Vpr-mediated cell cycle disruption. Future work can determine the exact roles of PIKK family members during the acquisition of Vpr-mediated renal polyploidy. Our results here suggest that DNA-PK may act at a distinct step in the progression to polyploidy, but our data also suggest caution in the interpretation of PIKK family inhibitors and their ability to specifically target a single PIKK family member.
It is intriguing to speculate how DNA-PK may specifically alter the progression to polyploidy in G2-arrested Vpr+ cells. DNA-PK has previously been implicated in HIV infection in T cells, although in this context it appears to promote cell death (54). Perhaps more relevant to the phenotypes we observe here upon NU7441 treatment, DNA-PK has recently been implicated in progression through mitosis. DNA-PK localizes to centrosomes and mitotic spindles, and NU4771 treatment causes defects in microtubule dynamics and chromosome segregation (55, 56). Given our finding that mitotic defects lead to cell death instead of polyploidy in Vpr-expressing renal cells, it is plausible that inhibition of DNA-PK tips the scales toward cell death instead of polyploidy.
Our work also potentially implicates mTOR signaling in Vpr-mediated renal phenotypes. mTOR is already associated with HIVAN pathology, and HIVAN phenotypes in mice can be partially suppressed by rapamycin treatment (46). Although mTOR has not been implicated in Vpr-mediated polyploidy before, mTOR/phosphatidylinositol 3-kinase signaling is linked to the control of endoreplication/polyploidy in Drosophila melanogaster (57, 58). Thus, our work may highlight a potentially conserved role for mTOR and related PIKK members in the acquisition of polyploidy. Future work can pinpoint more-precise functions of PIKK family members. Possible functions of these kinases include controlling the exit from G2 arrest, the amplification of centrosomes, and the aborted mitosis events that lead to polyploidy.
Role of Vpr-induced polyploidy in HIV-positive kidneys.
Our work also illuminates potential host and virus strategies in the kidney. Eradication of HIV-positive renal cells via mitotic catastrophe may benefit the host in terms of viral clearance. As for the virus, there may be benefits of host cell polyploidy in the HIV life cycle. In addition to enabling cell survival by circumventing mitotic cell death, polyploidy may promote long-term HIV survival/propagation through increasing virus production. Polyploidy and associated gene amplification can also increase rates of transcription per cell (59–61), which may increase virus production in surviving polyploid cells. An alternative possibility is that polyploidy enables reconfiguration of the infected cell's genome (62, 63), which, in turn, could facilitate the long-term survival and expansion of an HIV-1-infected cell and the maintenance of an HIV renal compartment. Finally, polyploidy is associated with disease pathogenesis in numerous contexts, including aging, cancer, and drug resistance (reviewed in reference 24). In this context, Vpr-mediated polyploidy may exacerbate renal dysfunction in aging HIV-positive kidneys, possibly by causing cellular hypertrophy. Thus, understanding the physiological implications of Vpr-mediated polyploidy is a critical next step in uncovering the broader consequences of HIV-1 infection in the kidney.
By uncovering the mechanisms and implications of ploidy increase associated with HIV-positive cell survival, we may uncover key strategies to prevent renal disease or to alter a potential HIV renal reservoir. The requirement for PIKK signaling in multiple steps to renal polyploidy suggests that there may be novel therapeutic targets for renal disease prevention and reservoir establishment. Treatment with PIKK inhibitors may effectively eliminate polyploid cells, thereby ameliorating any polyploidy-associated renal pathology and potentially preventing the long-term survival of polyploid HIV-positive RTECs.
In summary, our work defines a temporal progression from G2 arrest to polyploidy in HIV Vpr-expressing renal cells and highlights a role for PIKK signaling at multiple steps in this process. Further, our work highlights how a poorly understood HIV mechanism, ploidy increase, may provide critical insights into long-term reservoir establishment and disease pathogenesis in HIV-positive kidneys.
MATERIALS AND METHODS
Cell culture.
The HK2 and HEK 293T Lenti-X cell lines were obtained from the American Type Culture Collection (ATCC). HK2 cells were cultured in a serum-free keratinocyte medium with growth supplement (Thermo Fisher). HEK 293T Lenti-X cells were cultured in Dulbecco's modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS), penicillin-streptomycin, and l-glutamine. Sup-T1 cells were cultured in RPMI medium supplemented with 10% FBS and were obtained from the NIH AIDS Reagent program.
Generation of pseudotyped virus, transfection, and infection.
Viral particles pseudotyped with the vesicular stomatitis virus glycoprotein (VSV-G) were produced by cotransfection (Polyplus jetPRIME transfection kit) of 293T Lenti-X cells with three plasmids: the VSV envelope plasmid pMD2.G, the packaging plasmid psPAX2, and the desired expression plasmid (pTY2-VPR-IRES-GFP, pHR-VPR-IRES-GFP, pHR-Q65RVPR-IRES-GFP, pHR-HA-Vpr-IRES-GFP, or pHR-HA-Vpr-IRES-ΔGFP). The supernatant was collected and was filtered using a 0.45-μm filter at 48 and 72 h posttransfection. The titer of virus was determined in 293T cells via flow cytometry, and GFP levels were used as a measure of infection. In every case, a multiplicity of infection (MOI) of 1 was used to infect HK2 cells. The pNL4-3.HSA.R-.E- (Vpr-negative) and pNL4-3.HSA.R+E- (Vpr-positive) molecular clones were obtained from the NIH AIDS Reagent Program, from Nathaniel Landau (27, 64). These viruses were pseudotyped with the VSV-G envelope and were used for infecting cells.
siRNA transfections.
ON-TARGETplus SMARTpool siRNAs targeting ATM and a nontargeting control were obtained from Dharmacon (GE Healthcare). HK2 cells were plated at 100,000 per well in 6-well plates and were transfected with 30 pmol of siRNAs against ATM using jetPRIME transfection reagent for 24 h. Cells were infected with TY2-Luc-GFP or TY2-Vpr-GFP for 48 h and harvested for flow cytometry and/or Western blot analyses to analyze the knockdown of gene expression.
Generation of CRISPR knockout cell lines.
Guide RNAs targeting ATM (5′-CTCTATCATGTTCTAGTTGA-3′) were cloned into pLentiCRISPR-v2 (Addgene) as described in reference 65 and confirmed by sequencing. Lentiviral particles were obtained by cotransfecting 293T-Lenti-X cells with plasmids pLentiCRISPR-v2, psPAX2, and pMD2.G as described above. HK2 cells were infected with lentiviruses expressing the guide RNAs targeting ATM, and puromycin selections were performed to remove uninfected cells. Western blot analyses were performed at alternate passages to determine the knockout of gene expression.
Cell cycle analysis.
Cell cycle progression was measured using flow cytometric analysis of DNA content. Infected cells were harvested at the time points indicated on the figures, fixed in 1% paraformaldehyde in phosphate-buffered saline (PBS), and permeabilized with 70% ethanol. DNA staining was performed with 100 mg/ml of propidium iodide (PI) and 0.5 mg/ml of RNAse A prior to flow cytometric analysis with a BD FACSCalibur system (BD Biosciences). Cell cycle parameters were measured using FlowJo (FlowJo LLC). Each experiment was conducted at least three independent times, and representative plots are provided. Statistical analyses were performed using the one-way analysis of variance (ANOVA) test in Prism with uninfected cells as controls. Asterisks stand for P values as follows: *, P < 0.05; **; P < 0.001; ***, P < 0.0002; ****, P < 0.0001.
Drug treatments.
Small-molecule inhibitors against ATM (KU60019 at a working concentration of 500 nM), mTOR (rapamycin at 250 nM), and the catalytic subunit of DNA-PK (DNA-PKcs) (NU7441 at 100 nM) were obtained from Selleck Chemicals. An ATR inhibitor (VE821) was obtained from Axon Medchem and was used at a concentration of 10 μM. Dimethyl sulfoxide (DMSO) was used as a negative control. Cells were treated with the indicated concentrations of drugs (above) in growth media at the time points shown on the figures. Doxorubicin and cisplatin were obtained from Sigma.
Western blot analysis. (i) Whole-cell extract preparation.
Cells were lysed in NP-40 lysis buffer (catalog no. FNN0021; Invitrogen) containing 10 mM phenylmethylsulfonyl fluoride (PMSF) (catalog no. 36978; Thermo Fisher), protease inhibitors (cOmplete Mini protease inhibitor cocktail tablets; catalog no. 11836153001; Roche), and phosphatase inhibitors (Pierce Phosphatase Inhibitor Mini tablets; catalog no. 88667; Thermo Fisher) for 30 min on ice. Cell lysates were run through a QIAshredder column (catalog no. 79654; Qiagen) at 14,000 rpm for 2 min. Each experiment was conducted at least three separate times, and representative blots are provided.
(ii) Standard Western blotting.
Whole-cell extracts were run on a 4- to 20% Tris-glycine gel (catalog no. EC6025; Thermo Fisher) under denaturing conditions and were transferred to polyvinylidene difluoride (PVDF) membranes (catalog no. 1620177; Bio-Rad). Primary antibodies against beta-actin (catalog no. 3700) or the HA tag (catalog no. 3724) and horseradish peroxidase–conjugated anti-rabbit (catalog no. 7074) or anti-mouse (catalog no. 7076) secondary antibodies were obtained from Cell Signaling Technology (CST). Antibodies against various proteins were obtained from CST unless mentioned otherwise. Antibody dilutions were prepared to the manufacturers' specifications, and detection was carried out by chemiluminescence (catalog no. 34080; Thermo Fisher).
(iii) Western blotting for large proteins.
Whole-cell extracts were run on a 3- to 8% Tris-acetate gel (Thermo Fisher) under denaturing conditions and were transferred to PVDF membranes overnight at 4°C. Primary antibodies against ATR (catalog no. 2790), phospho-ATR (S428; catalog no. 2853), ATM (catalog no. 2873), phospho-Chk2 (S19; catalog no. 2666), and phospho-Chk1 (S280; catalog no. 2347) were from CST. Secondary antibodies and detection were as described above. Antibody dilutions were prepared to the manufacturer's specifications.
(iv) Western blot densitometry analysis.
Densitometry of Western blot bands was carried out using ImageJ software. Phosphoprotein densitometry levels were adjusted for the loading control using total nonphosphoprotein densitometry levels. Then the normalized phosphoprotein levels for the control and experimental samples were compared for matched time points to give the values indicated in the figure panels. Loading-control-adjusted densitometric values for Vpr samples were normalized to the densitometric value of the corresponding HK2 time point.
Immunofluorescence flow cytometry.
Staining was performed by flow cytometry (FACSCalibur; BD Biosciences, San Jose, CA, USA). Cells were fixed in 4% paraformaldehyde (PFA) and were then permeabilized with 90% methanol. The fixed and permeabilized cells were stained with primary antibodies against phospho-histone H3 (catalog no. 3377). The secondary antibody was an Alexa Fluor 488-conjugated anti-rabbit antibody (catalog no. A-11034; Thermo Fisher). Cells were treated with 100 mg/ml of propidium iodide and 0.5 mg/ml of RNase A prior to cytometry. FlowJo was used for all analyses.
Thymidine synchronization.
HK2 cells were treated with 2 mM thymidine (Sigma) in HK2 growth medium for 18 h to synchronize growth at the G1 phase of the cell cycle. Cells were treated with a single thymidine block and were released from G1 by thymidine removal followed by infections with TY2-Vpr-GFP (time zero). Cells were harvested every hour for up to 26 h, and their cellular DNA content was measured by propidium iodide staining as described above.
BrdU incorporation assay.
Thymidine-synchronized HK2 cells were transduced with HR-HA-Vpr-ΔGFP for 4 h, at which point the medium was changed, and infection proceeded for 18 h. At 18 h postinfection, the medium was replaced with a medium containing 10 μM bromodeoxyuridine (BrdU) to measure DNA replication. At 48 h postinfection, cells were harvested, and BrdU incorporation was measured using the anti-BrdU antibody (antibody 5292; CST) and Alexa Fluor 488-conjugated anti-mouse secondary antibody (4408) as described by the manufacturer. Cellular DNA was stained with propidium iodide, and flow cytometric analyses of DNA content and BrdU incorporation were performed.
Immunofluorescence microscopy.
Cells were fixed in 4% PFA and were permeabilized in ice-cold 100% methanol. Primary antibodies were to γ-tubulin (catalog no. T3559; Sigma) and α-tubulin (catalog no. 3873). Alexa Fluor 488-conjugated anti-mouse (catalog no. 4408) and Alexa Fluor 647-conjugated anti-rabbit (catalog no. 4414) antibodies were used as secondary antibodies. Cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) (catalog no. 62248; Thermo Fisher), and coverslips were placed on the slide using Antifade mountant (catalog no. P36930; Thermo Fisher). Images were captured using a Leica SP5 inverted confocal microscope. Each experiment was conducted three separate times, and numerous (>50) representative fields of view were analyzed at ×40 magnification; >400 cells/condition were analyzed.
FUCCI probes and live-cell imaging.
Cells were transduced with FUCCI cell cycle sensors according to the manufacturer's instructions at a concentration of 50 particles per cell (catalog no. P36238; Thermo Fisher). Cells were imaged using the Olympus VivaView FL incubator microscope at 10-min intervals. Corresponding survival rates were determined from randomly chosen fields of cells for three separate trials. Cells were scored as surviving if they did not fragment during the course of imaging. Statistical significance was determined using an unpaired t test analysis.
Supplementary Material
ACKNOWLEDGMENTS
E.H.P. and D.R. designed and performed all of the experiments and drafted the manuscript. D.T.F. and M.E.K. oversaw the planning and direction of the project, including the analysis and interpretation of the data and the editing of the manuscript.
We thank the Klotman and Fox laboratory members for discussions or comments on the project; B. Balakumaran, M. Blasi, A. Cara, and D. Negri for technical assistance and reagents; Y. Gao for assistance with microscopy; the Duke Light Microscopy Core Facility (LMCF) for access to microscopes and imaging software; and J. Wong for assistance with flow cytometry. We thank the NIH AIDS Reagent Program for the pNL4-3.R-E- and pNL4-3.R+E- molecular clones.
This research was supported by the Creative and Novel Ideas in HIV Research (CNIHR) Program through a supplement to the University of Alabama at Birmingham (UAB) Center for AIDS Research (P30 AI027767). This funding was made possible by collaborative efforts of the Office of AIDS Research, the National Institute of Allergy and Infectious Diseases, and the International AIDS Society.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01718-17.
REFERENCES
- 1.Rosenberg AZ, Naicker S, Winkler CA, Kopp JB. 2015. HIV-associated nephropathies: epidemiology, pathology, mechanisms and treatment. Nat Rev Nephrol 11:150–160. doi: 10.1038/nrneph.2015.9. [DOI] [PubMed] [Google Scholar]
- 2.Mallipattu SK, Salem F, Wyatt CM. 2014. The changing epidemiology of HIV-related chronic kidney disease in the era of antiretroviral therapy. Kidney Int 86:259–265. doi: 10.1038/ki.2014.44. [DOI] [PubMed] [Google Scholar]
- 3.Bruggeman LA, Ross MD, Tanji N, Cara A, Dikman S, Gordon RE, Burns GC, D'Agati VD, Winston JA, Klotman ME, Klotman PE. 2000. Renal epithelium is a previously unrecognized site of HIV-1 infection. J Am Soc Nephrol 11:2079–2087. [DOI] [PubMed] [Google Scholar]
- 4.Cohen AH, Sun NC, Shapshak P, Imagawa DT. 1989. Demonstration of human immunodeficiency virus in renal epithelium in HIV-associated nephropathy. Mod Pathol 2:125–128. [PubMed] [Google Scholar]
- 5.Marras D, Bruggeman LA, Gao F, Tanji N, Mansukhani MM, Cara A, Ross MD, Gusella GL, Benson G, D'Agati VD, Hahn BH, Klotman ME, Klotman PE. 2002. Replication and compartmentalization of HIV-1 in kidney epithelium of patients with HIV-associated nephropathy. Nat Med 8:522–526. doi: 10.1038/nm0502-522. [DOI] [PubMed] [Google Scholar]
- 6.Winston JA, Bruggeman LA, Ross MD, Jacobson J, Ross L, D'Agati VD, Klotman PE, Klotman ME. 2001. Nephropathy and establishment of a renal reservoir of HIV type 1 during primary infection. N Engl J Med 344:1979–1984. doi: 10.1056/NEJM200106283442604. [DOI] [PubMed] [Google Scholar]
- 7.Hanna Z, Kay DG, Rebai N, Guimond A, Jothy S, Jolicoeur P. 1998. Nef harbors a major determinant of pathogenicity for an AIDS-like disease induced by HIV-1 in transgenic mice. Cell 95:163–175. doi: 10.1016/S0092-8674(00)81748-1. [DOI] [PubMed] [Google Scholar]
- 8.Ross MJ. 2014. Advances in the pathogenesis of HIV-associated kidney diseases. Kidney Int 86:266–274. doi: 10.1038/ki.2014.167. [DOI] [PubMed] [Google Scholar]
- 9.Dickie P, Roberts A, Uwiera R, Witmer J, Sharma K, Kopp JB. 2004. Focal glomerulosclerosis in proviral and c-fms transgenic mice links Vpr expression to HIV-associated nephropathy. Virology 322:69–81. doi: 10.1016/j.virol.2004.01.026. [DOI] [PubMed] [Google Scholar]
- 10.Zuo Y, Matsusaka T, Zhong J, Ma J, Ma L-J, Hanna Z, Jolicoeur P, Fogo AB, Ichikawa I. 2006. HIV-1 genes vpr and nef synergistically damage podocytes, leading to glomerulosclerosis. J Am Soc Nephrol 17:2832–2843. doi: 10.1681/ASN.2005080878. [DOI] [PubMed] [Google Scholar]
- 11.Rosenstiel PE, Gruosso T, Letourneau AM, Chan JJ, LeBlanc A, Husain M, Najfeld V, Planelles V, D'Agati VD, Klotman ME, Klotman PE. 2008. HIV-1 Vpr inhibits cytokinesis in human proximal tubule cells. Kidney Int 74:1049–1058. doi: 10.1038/ki.2008.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenstiel PE, Chan J, Snyder A, Planelles V, D'Agati VD, Klotman PE, Klotman ME. 2009. HIV-1 Vpr activates the DNA damage response in renal tubule epithelial cells. AIDS 23:2054–2056. doi: 10.1097/QAD.0b013e32833088a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snyder A, Alsauskas Z, Gong P, Rosenstiel PE, Klotman ME, Klotman PE, Ross MJ. 2009. FAT10: a novel mediator of Vpr-induced apoptosis in human immunodeficiency virus-associated nephropathy. J Virol 83:11983–11988. doi: 10.1128/JVI.00034-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snyder A, Alsauskas ZC, Leventhal JS, Rosenstiel PE, Gong P, Chan JJK, Barley K, He JC, Klotman ME, Ross MJ, Klotman PE. 2010. HIV-1 viral protein R induces ERK and caspase-8-dependent apoptosis in renal tubular epithelial cells. AIDS 24:1107–1119. doi: 10.1097/QAD.0b013e328337b0ab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersen JL, Le Rouzic E, Planelles V. 2008. HIV-1 Vpr: mechanisms of G2 arrest and apoptosis. Exp Mol Pathol 85:2–10. doi: 10.1016/j.yexmp.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartz SR, Rogel ME, Emerman M. 1996. Human immunodeficiency virus type 1 cell cycle control: Vpr is cytostatic and mediates G2 accumulation by a mechanism which differs from DNA damage checkpoint control. J Virol 70:2324–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimura M, Onozuka Y, Yamaguchi T, Hatake K, Takaku F, Ishizaka Y. 1999. Micronuclei formation with chromosome breaks and gene amplification caused by Vpr, an accessory gene of human immunodeficiency virus. Cancer Res 59:2259–2264. [PubMed] [Google Scholar]
- 18.Poon B, Jowett JB, Stewart SA, Armstrong RW, Rishton GM, Chen IS. 1997. Human immunodeficiency virus type 1 vpr gene induces phenotypic effects similar to those of the DNA alkylating agent, nitrogen mustard. J Virol 71:3961–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang F, Re F, Sebastian S, Sazer S, Luban J. 2004. HIV-1 Vpr induces defects in mitosis, cytokinesis, nuclear structure, and centrosomes. Mol Biol Cell 15:1793–1801. doi: 10.1091/mbc.E03-09-0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimura M, Toyoda Y, Iijima K, Kinomoto M, Tokunaga K, Yoda K, Yanagida M, Sata T, Ishizaka Y. 2011. Epigenetic displacement of HP1 from heterochromatin by HIV-1 Vpr causes premature sister chromatid separation. J Cell Biol 194:721–735. doi: 10.1083/jcb.201010118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ullah Z, Lee CY, Lilly MA, DePamphilis ML. 2009. Developmentally programmed endoreduplication in animals. Cell Cycle 8:1501–1509. doi: 10.4161/cc.8.10.8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehrotra S, Maqbool SB, Kolpakas A, Murnen K, Calvi BR. 2008. Endocycling cells do not apoptose in response to DNA rereplication genotoxic stress. Genes Dev 22:3158–3171. doi: 10.1101/gad.1710208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen H, Maki CG. 2010. Persistent p21 expression after Nutlin-3a removal is associated with senescence-like arrest in 4N cells. J Biol Chem 285:23105–23114. doi: 10.1074/jbc.M110.124990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fox DT, Duronio RJ. 2013. Endoreplication and polyploidy: new insights into development and disease. Development 140:3–12. doi: 10.1242/dev.080531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li G, Park HU, Liang D, Zhao RY. 2010. Cell cycle G2/M arrest through an S phase-dependent mechanism by HIV-1 viral protein R. Retrovirology 7:59. doi: 10.1186/1742-4690-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jowett JB, Planelles V, Poon B, Shah NP, Chen ML, Chen IS. 1995. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J Virol 69:6304–6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He J, Choe S, Walker R, Di Marzio P, Morgan DO, Landau NR. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol 69:6705–6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roshal M, Kim B, Zhu Y, Nghiem P, Planelles V. 2003. Activation of the ATR-mediated DNA damage response by the HIV-1 viral protein R. J Biol Chem 278:25879–25886. doi: 10.1074/jbc.M303948200. [DOI] [PubMed] [Google Scholar]
- 29.Yuan H, Kamata M, Xie Y-M, Chen ISY. 2004. Increased levels of Wee-1 kinase in G2 are necessary for Vpr- and gamma irradiation-induced G2 arrest. J Virol 78:8183–8190. doi: 10.1128/JVI.78.15.8183-8190.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zimmerman ES, Chen J, Andersen JL, Ardon O, DeHart JL, Blackett J, Choudhary SK, Camerini D, Nghiem P, Planelles V. 2004. Human immunodeficiency virus type 1 Vpr-mediated G2 arrest requires Rad17 and Hus1 and induces nuclear BRCA1 and gamma-H2AX focus formation. Mol Cell Biol 24:9286–9294. doi: 10.1128/MCB.24.21.9286-9294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zimmerman ES, Sherman MP, Blackett JL, Neidleman JA, Kreis C, Mundt P, Williams SA, Warmerdam M, Kahn J, Hecht FM, Grant RM, de Noronha CMC, Weyrich AS, Greene WC, Planelles V. 2006. Human immunodeficiency virus type 1 Vpr induces DNA replication stress in vitro and in vivo. J Virol 80:10407–10418. doi: 10.1128/JVI.01212-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davoli T, de Lange T. 2011. The causes and consequences of polyploidy in normal development and cancer. Annu Rev Cell Dev Biol 27:585–610. doi: 10.1146/annurev-cellbio-092910-154234. [DOI] [PubMed] [Google Scholar]
- 33.Minemoto Y, Shimura M, Ishizaka Y, Masamune Y, Yamashita K. 1999. Multiple centrosome formation induced by the expression of vpr gene of human immunodeficiency virus. Biochem Biophys Res Commun 258:379–384. doi: 10.1006/bbrc.1999.0640. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe N, Yamaguchi T, Akimoto Y, Rattner JB, Hirano H, Nakauchi H. 2000. Induction of M-phase arrest and apoptosis after HIV-1 Vpr expression through uncoupling of nuclear and centrosomal cycle in HeLa cells. Exp Cell Res 258:261–269. doi: 10.1006/excr.2000.4908. [DOI] [PubMed] [Google Scholar]
- 35.Vitale I, Galluzzi L, Castedo M, Kroemer G. 2011. Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat Rev Mol Cell Biol 12:385–392. doi: 10.1038/nrm3115. [DOI] [PubMed] [Google Scholar]
- 36.Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, Imamura T, Ogawa M, Masai H, Miyawaki A. 2008. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 37.Nakai-Murakami C, Shimura M, Kinomoto M, Takizawa Y, Tokunaga K, Taguchi T, Hoshino S, Miyagawa K, Sata T, Kurumizaka H, Yuo A, Ishizaka Y. 2007. HIV-1 Vpr induces ATM-dependent cellular signal with enhanced homologous recombination. Oncogene 26:477–486. doi: 10.1038/sj.onc.1209831. [DOI] [PubMed] [Google Scholar]
- 38.Golding SE, Rosenberg E, Valerie N, Hussaini I, Frigerio M, Cockcroft XF, Chong WY, Hummersone M, Rigoreau L, Menear KA, O'Connor MJ, Povirk LF, van Meter T, Valerie K. 2009. Improved ATM kinase inhibitor KU-60019 radiosensitizes glioma cells, compromises insulin, AKT and ERK prosurvival signaling, and inhibits migration and invasion. Mol Cancer Ther 8:2894–2902. doi: 10.1158/1535-7163.MCT-09-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Y, Mao C, Wu J, Li S, Ma R, Cao H, Ji M, Jing C, Tang J. 2014. Improved ataxia telangiectasia mutated kinase inhibitor KU60019 provides a promising treatment strategy for non-invasive breast cancer. Oncol Lett 8:2043–2048. doi: 10.3892/ol.2014.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hart T, Brown KR, Sircoulomb F, Rottapel R, Moffat J. 2014. Measuring error rates in genomic perturbation screens: gold standards for human functional genomics. Mol Syst Biol 10:733–733. doi: 10.15252/msb.20145216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, Mero P, Dirks P, Sidhu S, Roth FP, Rissland OS, Durocher D, Angers S, Moffat J. 2015. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 42.Rai P, Plagov A, Lan X, Chandel N, Singh T, Lederman R, Ayasolla KR, Mathieson PW, Saleem MA, Husain M, Malhotra A, Chander PN, Singhal PC. 2013. mTOR plays a critical role in p53-induced oxidative kidney cell injury in HIVAN. Am J Physiol Renal Physiol 305:F343–F354. doi: 10.1152/ajprenal.00135.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rai P, Lederman R, Haque S, Rehman S, Kumar V, Sataranatrajan K, Malhotra A, Kasinath BS, Singhal PC. 2014. Renin angiotensin system modulates mTOR pathway through AT2R in HIVAN. Exp Mol Pathol 96:431–437. doi: 10.1016/j.yexmp.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng K, Rai P, Plagov A, Lan X, Mathieson PW, Saleem MA, Husain M, Malhotra A, Singhal PC. 2013. Rapamycin-induced modulation of miRNA expression is associated with amelioration of HIV-associated nephropathy (HIVAN). Exp Cell Res 319:2073–2080. doi: 10.1016/j.yexcr.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rehman S, Husain M, Yadav A, Kasinath BS, Malhotra A, Singhal PC. 2012. HIV-1 promotes renal tubular epithelial cell protein synthesis: role of mTOR pathway. PLoS One 7:e30071. doi: 10.1371/journal.pone.0030071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar D, Konkimalla S, Yadav A, Sataranatarajan K, Kasinath BS, Chander PN, Singhal PC. 2010. HIV-associated nephropathy: role of mammalian target of rapamycin pathway. Am J Pathol 177:813–821. doi: 10.2353/ajpath.2010.100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leahy JJ, Golding BT, Griffin RJ, Hardcastle IR, Richardson C, Rigoreau L, Smith GCM. 2004. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg Med Chem Lett 14:6083–6087. doi: 10.1016/j.bmcl.2004.09.060. [DOI] [PubMed] [Google Scholar]
- 48.Ciszewski WM, Tavecchio M, Dastych J, Curtin NJ. 2014. DNA-PK inhibition by NU7441 sensitizes breast cancer cells to ionizing radiation and doxorubicin. Breast Cancer Res Treat 143:47–55. doi: 10.1007/s10549-013-2785-6. [DOI] [PubMed] [Google Scholar]
- 49.Nishizawa M, Kamata M, Mojin T, Nakai Y, Aida Y. 2000. Induction of apoptosis by the Vpr protein of human immunodeficiency virus type 1 occurs independently of G2 arrest of the cell cycle. Virology 276:16–26. doi: 10.1006/viro.2000.0534. [DOI] [PubMed] [Google Scholar]
- 50.Waldhuber MG, Bateson M, Tan J, Greenway AL, McPhee DA. 2003. Studies with GFP-Vpr fusion proteins: induction of apoptosis but ablation of cell-cycle arrest despite nuclear membrane or nuclear localization. Virology 313:91–104. doi: 10.1016/S0042-6822(03)00258-7. [DOI] [PubMed] [Google Scholar]
- 51.Andersen JL, Zimmerman ES, DeHart JL, Murala S, Ardon O, Blackett J, Chen J, Planelles V. 2005. ATR and GADD45α mediate HIV-1 Vpr-induced apoptosis. Cell Death Differ 12:326–334. doi: 10.1038/sj.cdd.4401565. [DOI] [PubMed] [Google Scholar]
- 52.Stewart SA, Poon B, Jowett JB, Chen IS. 1997. Human immunodeficiency virus type 1 Vpr induces apoptosis following cell cycle arrest. J Virol 71:5579–5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Castedo M, Perfettini J-L, Roumier T, Andreau K, Medema R, Kroemer G. 2004. Cell death by mitotic catastrophe: a molecular definition. Oncogene 23:2825–2837. doi: 10.1038/sj.onc.1207528. [DOI] [PubMed] [Google Scholar]
- 54.Cooper A, García M, Petrovas C, Yamamoto T, Koup RA, Nabel GJ. 2013. HIV-1 causes CD4 cell death through DNA-dependent protein kinase during viral integration. Nature 498:376–379. doi: 10.1038/nature12274. [DOI] [PubMed] [Google Scholar]
- 55.Douglas P, Ye R, Trinkle-Mulcahy L, Neal JA, De Wever V, Morrice NA, Meek K, Lees-Miller SP. 2014. Polo-like kinase 1 (PLK1) and protein phosphatase 6 (PP6) regulate DNA-dependent protein kinase catalytic subunit (DNA-PKcs) phosphorylation in mitosis. Biosci Rep 34:257–271. doi: 10.1042/BSR20140051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee K-J, Lin Y-F, Chou H-Y, Yajima H, Fattah KR, Lee S-C, Chen BPC. 2011. Involvement of DNA-dependent protein kinase in normal cell cycle progression through mitosis. J Biol Chem 286:12796–12802. doi: 10.1074/jbc.M110.212969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zielke N, Kim KJ, Tran V, Shibutani ST, Bravo M-J, Nagarajan S, van Straaten M, Woods B, von Dassow G, Rottig C, Lehner CF, Grewal SS, Duronio RJ, Edgar BA. 2011. Control of Drosophila endocycles by E2F and CRL4CDT2. Nature 480:123–127. doi: 10.1038/nature10579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ohhara Y, Kobayashi S, Yamanaka N. 2017. Nutrient-dependent endocycling in steroidogenic tissue dictates timing of metamorphosis in Drosophila melanogaster. PLoS Genet 13:e1006583. doi: 10.1371/journal.pgen.1006583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raslova H, Roy L, Vourc'h C, Le Couedic JP, Brison O, Metivier D, Feunteun J, Kroemer G, Debili N, Vainchenker W. 2003. Megakaryocyte polyploidization is associated with a functional gene amplification. Blood 101:541–544. doi: 10.1182/blood-2002-05-1553. [DOI] [PubMed] [Google Scholar]
- 60.Jones MR, Ravid K. 2004. Vascular smooth muscle polyploidization as a biomarker for aging and its impact on differential gene expression. J Biol Chem 279:5306–5313. doi: 10.1074/jbc.M308406200. [DOI] [PubMed] [Google Scholar]
- 61.Storchova Z, Breneman A, Cande J, Dunn J, Burbank K, O'Toole E, Pellman D. 2006. Genome-wide genetic analysis of polyploidy in yeast. Nature 443:541–547. doi: 10.1038/nature05178. [DOI] [PubMed] [Google Scholar]
- 62.Duncan AW, Taylor MH, Hickey RD, Hanlon Newell AE, Lenzi ML, Olson SB, Finegold MJ, Grompe M. 2010. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature 467:707–710. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duncan AW, Hanlon Newell AE, Bi W, Finegold MJ, Olson SB, Beaudet AL, Grompe M. 2012. Aneuploidy as a mechanism for stress-induced liver adaptation. J Clin Invest 122:3307–3315. doi: 10.1172/JCI64026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Connor RI, Chen BK, Choe S, Landau NR. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 65.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science (New York, NY) 343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.