ABSTRACT
Effectively recognizing invading viruses and subsequently inducing innate antiviral immunity are essential for host antiviral defense. Although these processes are closely regulated by the host to maintain immune balance, viruses have evolved the ability to downregulate or upregulate these processes for their survival. MicroRNAs (miRNAs) are a family of small noncoding RNAs that play vital roles in modulating host immune response. Accumulating evidence demonstrates that host miRNAs as mediators are involved in regulating viral replication and host antiviral immunity in mammals. However, the underlying regulatory mechanisms in fish species are still poorly understood. Here, we found that rhabdovirus infection significantly upregulated host miR-3570 expression in miiuy croaker macrophages. Induced miR-3570 negatively modulated RNA virus-triggered type I interferon (IFN) and antiviral gene production, thus facilitating viral replication. Furthermore, miR-3570 was found to target and posttranscriptionally downregulate mitochondrial antiviral signaling protein (MAVS), which functions as a platform for innate antiviral signal transduction. Moreover, we demonstrated that miR-3570 suppressed the expression of MAVS, thereby inhibiting MAVS-mediated NF-κB and IRF3 signaling. The collective results demonstrated a novel regulation mechanism of MAVS-mediated immunity during RNA viral infection by miRNA.
IMPORTANCE RNA viral infection could upregulate host miR-3570 expression in miiuy croaker macrophages. Induced miR-3570 negatively modulates RNA virus-triggered type I IFN and antiviral gene production, thus facilitating viral replication. Remarkably, miR-3570 could target and inhibit MAVS expression, which thus modulates MAVS-mediated NF-κB and IRF3 signaling. The collective results of this study suggest a novel regulation mechanism of MAVS-mediated immunity during RNA viral infection by miR-3570. Thus, a novel mechanism for virus evasion in fish is proposed.
KEYWORDS: microRNA, miR-3570, MAVS, virus evasion, teleost fish
INTRODUCTION
Innate and adaptive immune responses are essential for host survival during pathogen infection. The host senses viral and bacterial pathogen invasion via pattern recognition receptors, including C-type lectin receptors, nucleotide oligomerization domain-like receptors, Toll-like receptors (TLRs), and RIG-I-like receptors (RLRs). The last two receptors are critical in the detection of viral RNA (1). Different from the mediated antiviral defense of TLRs, RLRs function as cytoplasmic sensors for viral RNA detection (2). Upon effective recognition, RLRs, such as RIG-I and MDA5, interact with the crucial adaptor protein mitochondrial antiviral signaling protein (MAVS; also known as VISA, CARDIF, or IPS-1) and then signal the transcription of type I interferon (IFN) and proinflammatory cytokines, resulting in the production of a family of IFN-stimulated genes (ISGs), which exert a clearance effect on viral invasion (3–6). To achieve the appropriate immune response, the signaling activities of RLRs are tightly controlled by a multistep regulatory mechanism and distinct genes (7, 8). Furthermore, viruses have developed a myriad of regulation strategies for their survival to evade and resist type I IFN production or its signaling therapy (9). As such, antiviral immune response requires an appropriate balance of viral clearance and host preservation.
To date, a series of host regulatory genes has been reported to regulate the RLR pathway and subsequent IFN signal transduction at different layers or at important checkpoints. A recent study that summarized gene regulation at the level of RLRs demonstrated that dihydroxyacetone kinase (DAK) is associated with MDA5 and then negatively regulates MDA5-mediated type I IFN signaling and innate antiviral response (10). Several genes, such as NLRX1 (11), RNF5 (12), MFN1 (13), and MFN2 (14), have been indicated to negatively regulate MAVS, which is a crucial downstream adaptor molecule of RLRs. Although a large number of coding genes modify RLR-mediated antiviral signaling, the role of noncoding gene-mediated regulation is only beginning to emerge.
MicroRNAs (miRNAs), which typically are approximately ∼22 nucleotides (nt) in length, are an extensive class of highly conserved endogenous noncoding RNAs that posttranscriptionally regulate the expression of specific target genes that participate in a wide range of biological processes, including development, apoptosis, proliferation, and differentiation (15, 16). Since the initial discovery, more than 700 miRNAs have been identified in mammals, and as much as 60% of all mRNAs are regulated by miRNAs to some extent (17). Recently, some host miRNAs were identified to function in regulation of viral replication by modulating the RIG-I pathway at multiple levels. miR-146a expression in macrophages that are infected with vesicular stomatitis virus (VSV) is shown in an RIG-I-dependent manner. It sequentially inhibits VSV-triggered type I IFN production by targeting IRAK1, IRAK2, and TRAF6 to promote VSV replication (18). A recent study revealed that miR-466l can directly target and reduce IFN-α expression during VSV infection (19). miRNAs can also modulate antiviral immune response by regulating IFN downstream signals. For example, upon herpes simplex virus 1, Kaposi's sarcoma-associated herpesvirus, or human cytomegalovirus infection, miR-132 is induced and regulates the expression of IFN-stimulated genes by targeting p300, thereby facilitating viral replication (20). Mounting evidence has indicated the vital role of miRNAs in modulating RLR-mediated antiviral immunity, yet the underlying regulation mechanisms of adaptor MAVS by miRNA remain poorly understood. To date, researchers have found that miR-22 (21) and miR-125a (or -b) (22) participate in modulating the expression of MAVS and subsequently the immune response, and only miR-125a (or -b) has been indicated to be involved in regulation of MAVS upon viral infection.
Many molecules and miRNAs reportedly are involved in modulating RLR-mediated antiviral signaling in mammals. However, due to the limitations of research materials and appropriate methods, the regulation mechanism of RLR-mediated antiviral signaling has rarely been reported in fish. Rhabdoviruses are a group of enveloped, single-stranded, and negative-sense RNA viruses. As one of the most significant viral pathogens in teleost fish, they can cause severe hemorrhagic septicemia in freshwater and marine fish (23). In recent decades, several rhabdoviruses have been isolated and identified from aquaculture fish, including hematopoietic necrosis virus (IHNV), Siniperca chuatsi rhabdovirus (SCRV), spring viremia of carp virus (SVCV), viral hemorrhagic septicemia virus (VHSV), and Hirame rhabdovirus (HIRRV) (24). These isolated rhabdoviruses have been reported to cause severe losses to many farmed fish species (24, 25). In the present study, we analyzed the miR-3570 expression profile in miiuy croaker (Miichthys miiuy) macrophages during SCRV infection and found that miR-3570 was upregulated upon SCRV infection. Upregulated miR-3570 inhibited MAVS expression, which eventually repressed the type I IFN-mediated antiviral response, thereby promoting viral replication. Importantly, we demonstrated that miR-3570 enhanced SCRV infection through suppressing the IRF3 and NF-κB signaling pathways. For the first time, we demonstrated miR-3570 as a negative feedback regulator involved in the antiviral immune response in fish.
RESULTS
Viral infection upregulates miR-3570 expression.
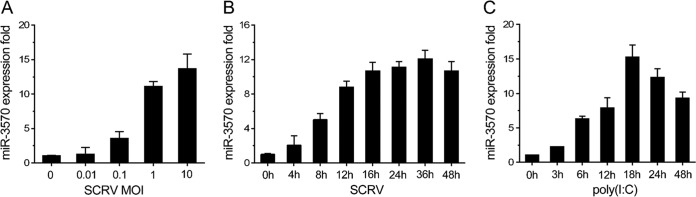
In order to investigate whether any of the host miRNAs are involved in SCRV infection, we analyzed miRNA expression profiles in SCRV-treated miiuy croaker macrophages. Through deep-sequencing analysis, we selected a series of miRNAs that was differentially expressed and focused on miR-3570, which is on the list of the top 10 most abundant miRNAs following viral infection (its fold change was 5.718 and P value was 3.8E−322). Further validation of the miR-3570 expression pattern upon SCRV infection was analyzed in SCRV-challenged macrophages by quantitative reverse transcription-PCR (qRT-PCR). The results revealed that miR-3570 expression levels were significantly increased in a dose- and time-dependent manner (Fig. 1A and B). Additionally, poly(I·C), a synthetic analog of double-stranded RNA (dsRNA), was applied as the stimulus to examine miRNA expression. Similar to the results for SCRV infection in macrophages, miR-3570 was also upregulated in poly(I·C)-stimulated macrophages (Fig. 1C). These results strongly suggest that miR-3570 expression can be upregulated in macrophages in response to RNA viral infection.
FIG 1.
Viral infection upregulates miR-3570 expression in macrophage. Miiuy croaker macrophages were transfected with various MOIs of SCRV for 36 h (A) or different times (MOI, 5) (B), and the miR-3570 expression level was determined with qRT-PCR. (C) Miiuy croaker macrophages were stimulated with poly(I·C) for different times, and the miR-3570 expression level was measured by using qRT-PCR. The results are standardized to 1 in control cells. All data are representative of at least three independent experiments.
miR-3570 suppresses SCRV-triggered production of antiviral genes.
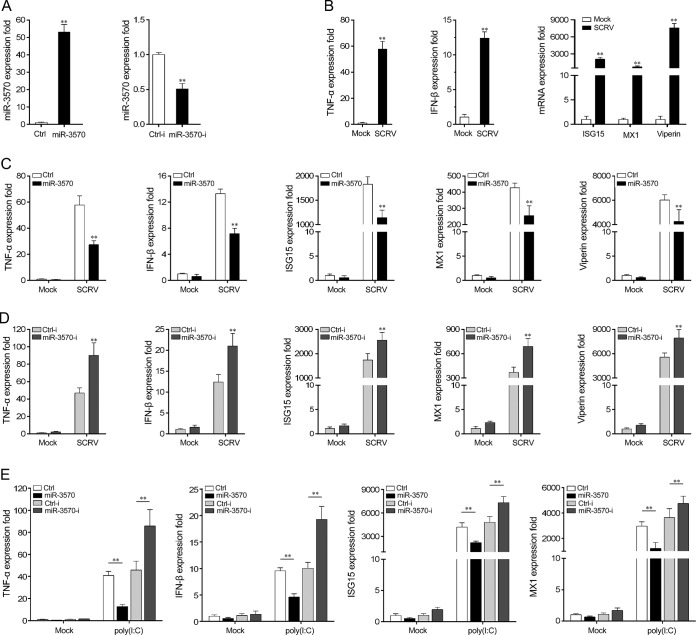
To investigate the underlying mechanisms of miR-3570 in host antiviral immune response, we examined the effects of miR-3570 on inflammatory cytokines and antiviral gene production by using miR-3570 mimics and miR-3570 inhibitors. First, we examined the effect of synthetic miR-3570 mimics and inhibitors on the expression of miR-3570. miRNA mimics are synthetic double-stranded RNAs (dsRNAs) with stimulating naturally occurring mature miRNAs, and miRNA inhibitors are synthetic single-stranded RNAs (ssRNAs) that sequester intracellular miRNAs and block their activity in the RNA-interfering pathway (26). Miiuy croaker macrophages were transfected with miR-3570 mimics or nonspecific control RNA oligonucleotides and miR-3570 inhibitors or control inhibitors. As expected, the miR-3570 mimics enhanced miR-3570 expression sharply, whereas miR-3570 inhibitors decreased miR-3570 expression significantly (Fig. 2A). To examine whether SCRV infection stimulates the expression of inflammatory cytokines and antiviral genes, miiuy croaker macrophages were infected with SCRV at a high multiplicity of infection (MOI; 5) for 24 h, and then inflammatory cytokines and antiviral genes were monitored. As shown in Fig. 2B, the expression of certain cytokines, including tumor necrosis factor alpha (TNF-α), IFN-β, ISG15, MX1, and Viperin, was rapidly induced (Fig. 2B). The results indicated that SCRV can induce certain inflammatory and antiviral gene overexpression in macrophages and trigger antiviral immunity.
FIG 2.
miR-3570 is involved in modulating SCRV-triggered antiviral gene production. (A) Miiuy croaker macrophages were transfected with control mimics (Ctrl), miR-3570 mimics (miR-3570), control inhibitors (Ctrl-i), or miR-3570 inhibitors (miR-3570-i) for 48 h, and then miR-3570 expression was determined by qRT-PCR. (B) Macrophages were infected with SCRV at an MOI of 5 for 36 h, and the mRNA levels of TNF-α, IFN-β, MX1, ISG15, and Viperin were detected by qRT-PCR. (C and D) Macrophages were transfected with Ctrl or miR-3570 (C) and Ctrl-i or miR-3570-i (D) for 48 h and then were infected with SCRV at an MOI of 5 for 36 h. The expression level of miR-3570 was analyzed by qRT-PCR. (E) Ctrl or miR-3570 and Ctrl-i or miR-3570-i were transfected into macrophages for 48 h and then stimulated with poly(I·C) for 12 h. The expression levels of the above-described genes were determined by qRT-PCR. The results are standardized to 1 in control cells. All data are representative of three independent experiments. (**, P < 0.01).
The effects of miR-3570 on the expression of inflammatory cytokines and antiviral genes in macrophages upon viral infection were evaluated next. Macrophages were transfected with either miR-3570 mimics or control mimics and miR-3570 inhibitors or control inhibitors and then infected with SCRV. The results from qRT-PCR analysis showed that certain SCRV-induced inflammatory and antiviral genes, including TNF-α, IFN-β, ISG15, MX1, and Viperin, were significantly decreased by the introduction of miR-3570 mimics in macrophages (Fig. 2C). On the contrary, as shown in Fig. 2D, the inhibition of endogenous miR-3570 significantly elevated SCRV-induced inflammatory cytokines and antiviral gene production compared with transfection with control inhibitors. These results indicated the negative regulation role of miR-3570 in macrophages upon SCRV infection. To determine whether miR-3570 participates in regulating RNA virus-induced immune response, poly(I·C) was used to stimulate macrophages, and then the expression of related cytokines was examined. In agreement with the previous results, the overexpression of miR-3570 significantly decreased poly(I·C)-triggered production of TNF-α, IFN-β, ISG15, and MX1, whereas the inhibition of miR-3570 expression increased this gene expression (Fig. 2E). Taken together, these data strongly demonstrate that miR-3570 negatively regulates a range of responses to RNA viral infection in macrophages.
miR-3570 targets MAVS.
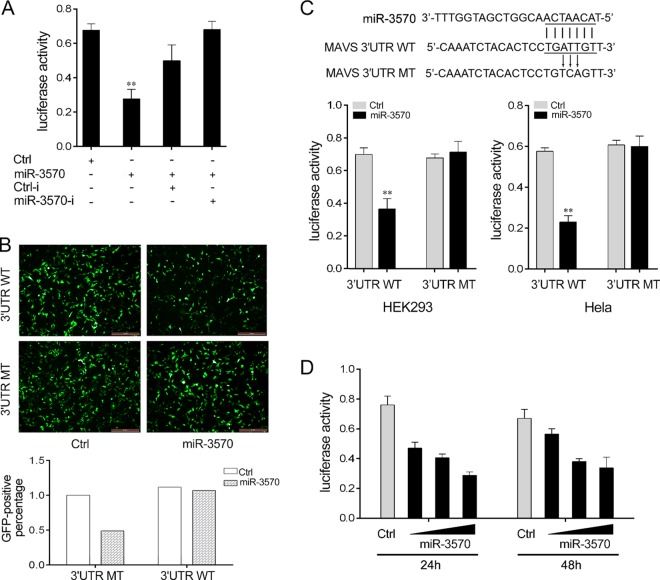
To verify the possible target of miR-3570, we first used bioinformatics software to search for potential miR-3570 targets (data not shown). Among the possible targets of miR-3570, we focused on MAVS, which contributes to antiviral immune response. To evaluate the possibility that MAVS is regulated by miR-3570, we constructed reporter plasmids by cloning the miiuy croaker MAVS 3′-untranslated region (MAVS-3′UTR) into the pmirGLO luciferase reporter vector or pIZ/EGFP vector, and a mutant MAVS-3′UTR luciferase reporter vector containing the miR-3570 target sequence mutations in the seed region was generated as a control. When the luciferase reporter plasmids were transfected into HEK293 cells with miR-3570 mimics or inhibitors, we observed that miR-3570 mimics markedly decreased luciferase activity, whereas the inhibition effect was attenuated after cotransfection with miR-3570 inhibitors (Fig. 3A). miR-3570 mimics also downregulated GFP gene expression when the 3′UTR of MAVS was cloned into the 3′UTR region of the GFP gene (Fig. 3B). On the contrary, luciferase activity could not be regulated by miR-3570 mimics when a mutant-type 3′UTR vector was transfected into HEK293 cells and HeLa cells (Fig. 3C). The dose-dependent effects of miR-3570 mimics on the inhibition of luciferase activity also could be observed at 24 h and 48 h posttransfection into HEK293 cells (Fig. 3D).
FIG 3.
miR-3570 targets miiuy croaker MAVS. (A) HEK293 cells were cotransfected with MAVS-3′UTR (WT), together with Ctrl, miR-3570, Ctrl-i, or miR-3570-I, for 24 h, and the luciferase activity was determined. For each transfection, the total amount of oligonucleotides was controlled and normalized (final concentration, 100 nM). (B) HEK293 cells were cotransfected with pIZ/EGFP-MAVS-3′UTR WT or pIZ/EGFP-MAVS-3′UTR mutant type (MT), together with miR-3570 or Ctrl. At 48 h posttransfection, the fluorescence intensity was evaluated. (C) Schematic diagram of the predicted target sites of miR-3570 in 3′UTR of miiuy croaker MAVS. HEK293 cells or HeLa cells were transfected with miR-3570 or Ctrl, along with wild-type MAVS-3′UTR or the mutant type of MAVS-3′UTR for 24 h, and the luciferase activity was determined. (D) The miR-3570 mimics (0, 30, 60, and 90 nM) together with the control mimics (90, 60, 30, and 0 nM) were cotransfected with WT reporter plasmid into HEK293 cells. After 24 h or 48 h, the luciferase activity was determined. Luciferase activity was normalized to renilla luciferase activity. All data are representative of at least three independent experiments. (**, P < 0.01).
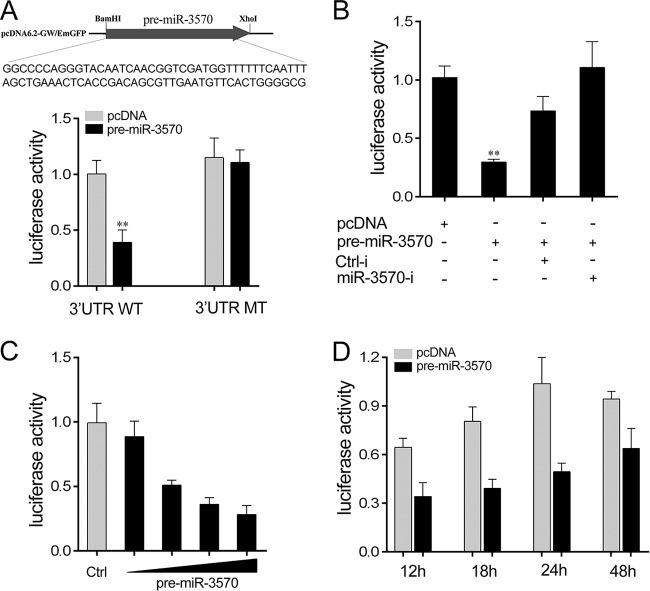
Given that the miRNA processing system is conserved from invertebrates to vertebrates (27), the pre-miR-3570 plasmid was transfected into HEK293 cells for in vitro expression. To construct the pre-miR-3570 plasmid, the pre-miR-3570 sequence was PCR amplified and then cloned into a pcDNA6.2 vector (Fig. 4A). By cotransfection of MAVS's 3′UTR reporter plasmids and the pre-miR-3570 plasmid into HEK293 cells, we observed that pre-miR-3570 plasmids were sufficient to decrease luciferase activity, whereas the mutant type led to a complete abrogation of the negative effect (Fig. 4A). The significantly negative regulation mechanism was further investigated using miR-3570 inhibitors, revealing that the inhibition of luciferase activity was attenuated after cotransfection with miR-3570 inhibitors (Fig. 4B). Concentration and time gradient experiments were likewise conducted. As shown in Fig. 4C and D, the pre-miR-3570 plasmid had an inhibitory effect on luciferase activity in a dose-dependent manner. It showed an increased inhibitory effect on luciferase activity at 24 h posttransfection. Above all, the data adequately demonstrate that the nucleotide sequence in the 3′UTR of MAVS is a potential miR-3570 targeting site.
FIG 4.
pre-miR-3570 regulates miiuy croaker MAVS. (A) Sequence alignment of pre-miR-3570 and the construction of the pre-miR-3570 plasmid. HEK293 cells were transfected with the pre-miR-3570 plasmid, along with wild-type MAVS-3′UTR (WT) or the mutant type of MAVS-3′UTR (MT) for 24 h, and the luciferase activity was determined. (B) HEK293 cells were transfected with MAVS-3′UTR, together with the pre-miR-3570 plasmid and miR-3570 inhibitors or its controls, for 24 h, and the luciferase activity was determined. For each transfection, the total amounts of the transfections were controlled and normalized. (C) HEK293 cells were transfected with the WT, together with the concentration gradient of pre-miR-3570, for 24 h. pcDNA6.2 was used to ensure that the same amounts of plasmid DNA are transfected for each transfection. The luciferase activity value was measured by using the Dual-Luciferase reporter assay system. (D) The time gradient was conducted for transfection with the pre-miR-3570 plasmid. Luciferase activity was normalized to renilla luciferase activity. All data are representative of at least three independent experiments (**, P < 0.01).
miR-3570 suppresses MAVS expression at the posttranscriptional level.
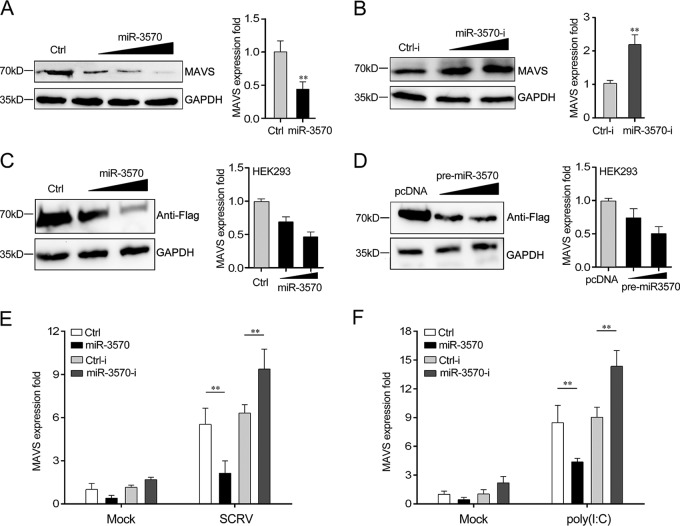
To determine miR-3570 function, the expression of endogenous MAVS was examined in miiuy croaker macrophages treated with miR-3570 mimics or inhibitors. As shown in Fig. 5A and B, the overexpression of miR-3570 significantly inhibited MAVS mRNA and protein level in a dose-dependent manner (Fig. 5A), whereas the inhibition of MAVS expression could be restored in the presence of miR-3570 inhibitors (Fig. 5B). The results suggested that miR-3570 decreased MAVS expression in both the protein and mRNA levels. Additionally, to further examine MAVS as a target of miR-3570, we constructed a MAVS expression plasmid that contains the full-length CDS region and 3′UTR of miiuy croaker MAVS and then cotransfected this with miR-3570 mimics or the pre-miR-3570 plasmid into HEK293 cells. As expected, miR-3570 mimics significantly suppressed the levels of MAVS protein and mRNA in a dose-dependent manner (Fig. 5C), and the pre-miR-3570 plasmid showed a consistent effect (Fig. 5D). In addition, to examine whether inducible miR-3570 suppresses MAVS expression during viral infection, we performed transfection with miR-3570 mimics, control mimics, miR-3570 inhibitors, and control inhibitors into macrophages for 48 h and then infected these with SCRV. As shown in Fig. 5E, the upregulation of MAVS caused by SCRV infection was almost eliminated in miR-3570 mimic-transfected macrophages, whereas its expression could be increased when miR-3570 inhibitors were transfected. A similar regulation of MAVS mRNA levels was observed in poly(I·C)-treated macrophage cells (Fig. 5F). Collectively, these results suggest that MAVS is a direct target of miR-3570, and the induction of miR-3570 represses MAVS expression upon SCRV infection and poly(I·C) stimulation.
FIG 5.
miR-3570 suppresses the expression of MAVS at posttranscriptional level. (A and B) The miiuy croaker macrophages were transfected with miR-3570 or Ctrl (A) and Ctrl-i or miR-3570-i (B). After 48 h of transfection, the protein and mRNA levels of MAVS were determined by Western blotting and qRT-PCR, respectively. (C and D) HEK293 cells were cotransfected with MAVS expression plasmid, along with miR-3570 mimics (C) and the pre-miR-3570 plasmid (D), in a concentration gradient manner, and control mimics (C) and pcDNA6.2 plasmid (D) were used to control the same amount of molecules for transfections. After 48 h, MAVS protein and mRNA levels were determined by Western blotting and qRT-PCR, respectively. (E and F) After transfection of Ctrl, miR-3570, Ctrl-i, and miR-3570-I for 48 h, the macrophages were treated with SCRV for 36 h (E) or poly(I·C) for 12 h (F). The mRNA expression of MAVS was analyzed by qRT-PCR and normalized to that of β-actin. The results are standardized to 1 in control cells. All data are representative of at least three independent experiments (**, P < 0.01; *, P < 0.05).
miR-3570 downregulates MAVS-mediated signaling pathway.
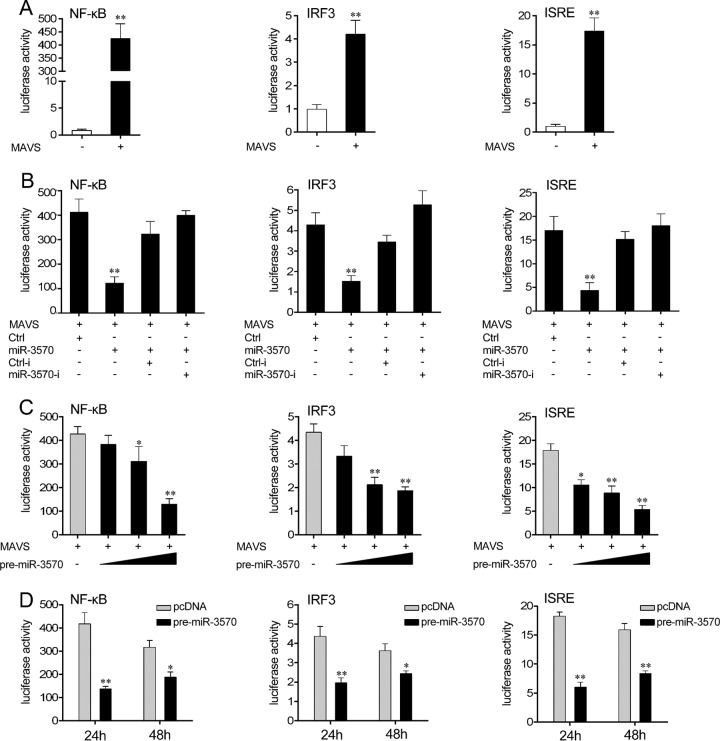
To investigate the underlying regulation mechanisms of miR-3570 upon SCRV infection, we first examined the role of miiuy croaker MAVS in the regulation of type I IFN signaling. A series of assays was performed to monitor the activations of NF-κB, IRF3 promoter, and IFN-stimulated response element (ISRE) after the MAVS expression plasmid was transfected into HEK293 cells. As shown in Fig. 6A, the overexpression of miiuy croaker MAVS efficiently activated the NF-κB, IRF3, and ISRE reporter genes. Given that miR-3570 targets MAVS and negatively regulates its expression, we examined whether miR-3570 could regulate NF-κB, IRF3, and ISRE signaling. When we transfected HEK293 cells with MAVS expression plasmid, together with miR-3570 mimics or control mimics, the miR-3570 mimics sharply suppressed the activations of NF-κB, IRF3, and ISRE reporter genes compared to activation with control mimics, and the inhibitory effect was weakened using the miR-3570 inhibitors (Fig. 6B). Furthermore, the pre-miR-3570 plasmid showed similar inhibition effects on NF-κB, IRF3, and ISRE reporter genes in a dose-dependent manner (Fig. 6C). As shown in Fig. 6D, compared with results at 48 h posttransfection, the pre-miR-3570 plasmid was more effective at 24 h posttransfection. These results demonstrate that miR-3570 suppresses the MAVS-mediated signaling pathway and thus negatively regulates NF-κB and IRF3 signaling.
FIG 6.
Overexpression of miR-3570 inhibits MAVS-mediated NF-κB and IRF3 signaling. (A) HEK293 cells were cotransfected with the MAVS expression plasmid, pRL-TK renilla luciferase plasmid, together with luciferase reporter gene NF-κB, ISRE, or IRF3 for 48 h, and the luciferase activity was measured. (B) HEK293 cells were cotransfected with Ctrl, miR-3570, Ctrl-i, or miR-3570-i, together with pRL-TK renilla luciferase plasmid, MAVS expression plasmid, and luciferase reporter NF-κB, IRF3, or ISRE for 48 h, and the luciferase activity was measured. For each transfection, the total amounts of oligonucleotides were controlled and normalized (final concentration, 100 nM). (C) HEK293 cells were transfected with pre-miR-3570 in a concentration gradient manner, together with NF-κB, ISRE, or IRF3, for 48 h, and pcDNA6.2 plasmid was use to ensure the same amounts of molecules were used for transfections. The luciferase activity was measured by using the Dual-Luciferase reporter assay system. (D) Time gradient experiment of the pre-miR-3570 plasmid. Luciferase activity was normalized to renilla luciferase activity. All data are representative of at least three independent experiments (**, P < 0.01; *, P < 0.05).
Knockdown of MAVS inhibits antiviral inflammatory response.
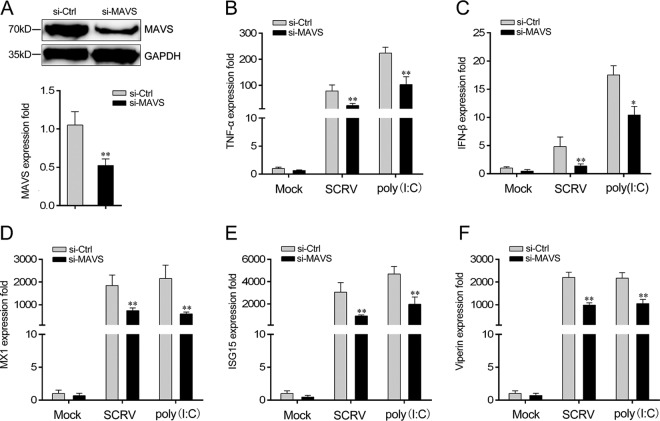
To confirm the contribution of MAVS to the antiviral inflammatory response, we then examined the expression of inflammatory cytokine and antiviral genes after viral infection. After transfection with MAVS-specific small interfering RNA (si-MAVS) into miiuy croaker macrophages, the expression levels of MAVS in protein and mRNA were significantly inhibited (Fig. 7A). We next silenced MAVS and examined the production of inflammatory cytokine and antiviral genes in macrophages treated with SCRV and poly(I·C). As shown in Fig. 7B to F, the knockdown of MAVS significantly downregulated the expression of TNF-α, IFN-β, ISG15, MX1, and Viperin upon SCRV infection, as well as poly(I·C) stimulation. Thus, an effect similar to that of miR-3570 overexpression was obtained.
FIG 7.
Expression levels of MAVS and antiviral genes after MAVS interference. (A) Miiuy croaker macrophages were transfected with control siRNA (si-Ctrl) or siRNA against MAVS (si-MAVS). After 48 h, MAVS mRNA and protein levels were determined by qRT-PCR and Western blotting, respectively. (B to F) After 48 h of transfection with si-Ctrl or si-MAVS, miiuy croaker macrophages were infected with SCRV for 36 h or poly(I·C) for 12 h. The expression levels of TNF-α (B), IFN-β (C), Mx1 (D), ISG15 (E), and Viperin (F) were determined. The results are standardized to 1 in control cells. All data are representative of at least three independent experiments (**, P < 0.01; *, P < 0.05).
miR-3570 regulates the expression of components of MAVS-dependent signaling cascade.
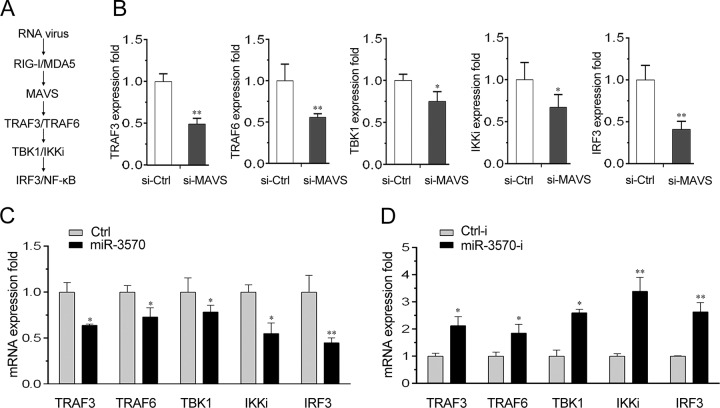
Previous evidence indicated that through interaction with RLRs, adaptor MAVS can further recruit TBK1 and IKK complex to activate the subsequent IFN signal transduction (6) (Fig. 8A). Thus, we then probed whether miR-3570 could modulate the expression of MAVS downstream molecules by regulating MAVS. To the end, we first examined the contribution of MAVS to the expression of its downstream molecules in macrophages. We performed transfection with si-MAVS into macrophages and then tested the expression levels of TRAF6, TRAF3, TBK1, IKKi, and IRF3. As shown in Fig. 8B, the mRNA expression levels of MAVS downstream molecules all are significantly decreased when MAVS is silenced.
FIG 8.
miR-3570 regulates expression of components of MAVS-dependent signaling cascade. (A) Schematic outline of MAVS-mediated pathway in mammals. (B) Miiuy croaker macrophages were transfected with si-Ctrl and si-MAVS for 48 h, and then the mRNA levels of TRAF3, TRAF6, TBK1, IKKi, and IRF3 were detected by qRT-PCR. (C and D) Miiuy croaker macrophages were transfected with Ctrl or miR-3570 (C) or Ctrl-i or miR-3570-i (D) for 48 h, and the mRNA levels of TRAF3, TRAF6, TBK1, IKKi, and IRF3 were detected by qRT-PCR. The results are standardized to 1 in control cells. All data are representative of at least three independent experiments (**, P < 0.01; *, P < 0.05).
As the results have demonstrated that miR-3570 targets MAVS, we subsequently examined the regulation role of miR-3570 on the components of the MAVS-dependent signaling cascade. We performed transfection with miR-3570 mimics into macrophages to overexpress miR-3570 and then detected the expression levels of TRAF6, TRAF3, TBK1, IKKi, and IRF3 mRNA. As shown in Fig. 8C, the mRNA expression levels of MAVS downstream molecules were downregulated by miR-3570 overexpression, which showed an inhibition effect similar to that of si-MAVS. On the contrary, we also evaluated gene expression by transfection with miR-3570 inhibitors. As expected, the miR-3570 inhibitors increased the mRNA expression levels of TRAF6, TRAF3, TBK1, IKKi, and IRF3 (Fig. 8D). These results demonstrate that miR-3570 can modulate the expression of MAVS downstream molecules, indicating miR-3570 is involved in the regulation of the MAVS-dependent signaling pathway.
miR-3570 feedback promotes SCRV replication.
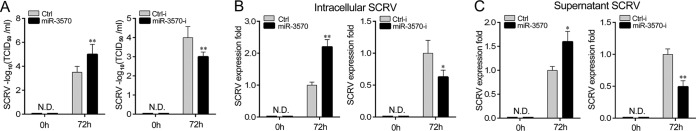
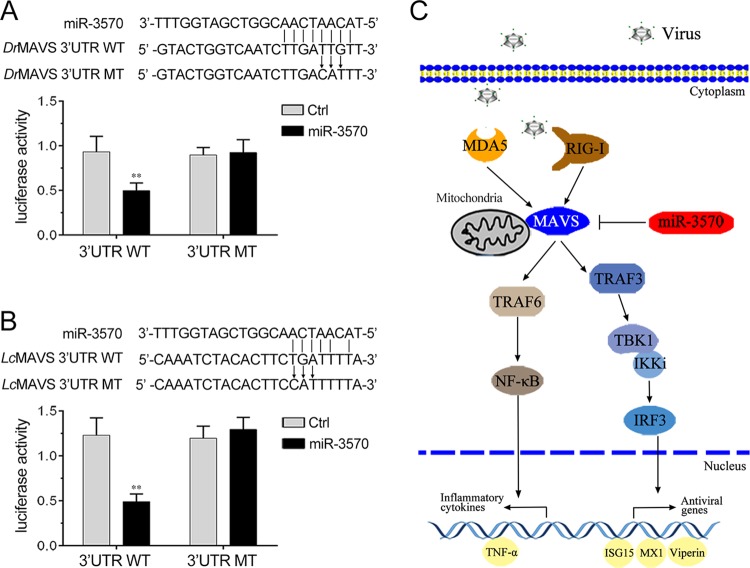
To evaluate the biological significance of upregulated miR-3570 upon viral infection in host cells, we examined the effect of miR-3570 on SCRV replication in Miiuy croaker kidney cells (MKC). The SCRV 50% tissue culture infectious dose (TCID50) levels in the supernatant from the infected MKC cells were measured, and the overexpression of miR-3570 was found to facilitate SCRV replication, whereas the inhibition of miR-3570 suppressed SCRV replication (Fig. 9A). Consistent with these results, the overexpression of miR-3570 increased, whereas the inhibition of miR-3570 decreased the level of intracellular and supernatant SCRV RNA replicates (Fig. 9B and C). In addition, we have also addressed the generality of the findings that miR-3570 targets MAVS in some other model animals, including zebrafish and large yellow croaker, and the results are shown in Fig. 10A and B, indicating that miR-3570 also could target MAVS in either zebrafish or in large yellow croaker, which indicated the generality of our findings. Taken together, these data demonstrate that SCRV-induced miR-3570 inhibits type I IFN and antiviral gene production through the negative regulation of the miR-3570 target MAVS and the suppression of NF-κB and IRF3 signaling, thereby facilitating viral replication (Fig. 10C).
FIG 9.
miR-3570 promotes SCRV replication. (A) Miiuy croaker macrophages were transfected with Ctrl, miR-3570, Ctrl-i, or miR-3570-i, infected by SCRV at an MOI of 5 for 1 h, washed, and then added to fresh medium. After 72 h, SCRV TCID50 in cultural supernatants was measured with MKC cells. (B and C) qRT-PCR analysis was conducted for intracellular SCRV RNA (B) or supernatant SCRV RNA (C). All data are representative of at least three independent experiments (**, P < 0.01; *, P < 0.05; N.D., not detected).
FIG 10.
Proposed model for the miR-3570 regulatory mechanism in fish species. (A) Schematic diagram of the predicted target sites of miR-3570 in the 3′UTR of Danio rerio MAVS. HEK293 cells were transfected with miR-3570 or Ctrl, along with the wild-type Danio rerio MAVS-3′UTR (DrMAVS 3′UTR WT) or the mutant type of Danio rerio MAVS-3′UTR (DrMAVS 3′UTR MT) for 24 h, and the luciferase activity was determined. (B) Schematic diagram of the predicted target sites of miR-3570 in 3′UTR of Larimichthys crocea MAVS. HEK293 cells were transfected with miR-3570 or Ctrl, along with the wild-type Larimichthys crocea MAVS-3′UTR (LcMAVS 3′UTR WT) or the mutant type of Larimichthys crocea MAVS-3′UTR (LcMAVS 3′UTR MT) for 24 h, and the luciferase activity was determined. (C) Proposed model for the mechanism by which induced miR-3570 negatively regulates inflammatory cytokines and antiviral gene production in feedback manner by targeting MAVS and inhibiting NF-κB and IRF3 signaling, thereby promoting virus replication.
DISCUSSION
Fish culture has recently become the fastest and most efficient agricultural production industry. However, aquaculture development has been hampered by various types of viruses, and viral diseases have been frequently reported in aquaculture animals (28). Hence, studying the underlying regulation mechanisms of aquaculture species during viral invasion is vital. miRNAs that emerged as significant and versatile modulators of immune responses regulate the intricate networks of host-pathogen interactions. Recently, several reports documented the molecular mechanisms of endogenous cellular miRNA in regulating virus replication processes in mammals, whereas the underlying mechanisms of miRNAs in fish species are rarely known. Here, we addressed the role of miRNA-regulated pathways in response to viral infection of miiuy croaker, which is an important economic farming fish species that has been studied in depth, from the transcriptome (29) and whole genome (30) to immune genes (31, 32). We found that RNA viral infection upregulates miR-3570 expression, and the upregulation of miR-3570 showed a negative effect on the production of inflammatory cytokines and antiviral genes. We further demonstrated that miR-3570 targets miiuy croaker MAVS, through which miR-3570 inhibited the downstream signaling, including NF-κB and IRF3 signaling. Finally, the overexpression of miR-3570 significantly facilitated virus replication. These findings not only demonstrated that miR-3570 is a negative regulator of the MAVS-mediated antiviral response but also indicated a novel virus-host interaction mechanism for virus evasion of host immune response in fish.
MAVS, as the mitochondrial antiviral signaling adaptor, can bridge the interactions between RIG-I/MDA5 sensing of viral infection and downstream signaling. The activation of MAVS leads to the rapid production of antiviral cytokines, including type I IFNs. Hence, MAVS regulation is essential to prevent excessive immune responses and balance antiviral signaling activity. Previous studies indicated that a series of MAVS-associated molecules could negatively or positively modulate MAVS in various manners. For example, COX5B, a mitochondrial cytochrome c oxidase component, can modulate MAVS aggregation through coordinating with the autophagy pathway, thereby balancing the antiviral signaling activity (33). NLRX1 also interacts with MAVS, resulting in the potent suppression of MAVS-mediated interferon-β promoter activity and the disruption of RLH-MAVS interactions, thereby enhancing antiviral responses (11). Mitofusin 1 (MFN1), as a positive regulator for MAVS, is associated with MAVS on the outer membrane of the mitochondria, positively regulating RLR-mediated innate antiviral responses (13). Meanwhile, several factors also showed negative regulation for MAVS, such as MFN2 (14), PCBP2 (34), and PLK1 (35). Here, we report that miR-3570 negatively regulates MAVS protein availability at the mRNA and protein levels, revealing that the noncoding RNA also could participate in the intricate regulation networks for MAVS.
After sensing viral RNAs by RIG-I/MDA5, MAVS then could recruit the TBK1 and IKK complexes to activate transcription factors IRF3/IRF7 and NF-κB, respectively, leading to the production of inflammatory cytokines and type I IFNs (36, 37). Type I IFNs are crucial for antiviral immune response, and their suitable production induces cellular resistance to viral infection and apoptosis of virus-infected cells. To achieve the appropriate immune response during pathogen infection, type I IFNs are closely regulated by multiple intracellular regulators (7, 8). Our findings and those in previous reports suggest that MAVS-mediated antiviral response and the subsequent IFN signal transduction are modulated by coding RNA and noncoding RNA, both of which contribute to the complex regulatory networks. DNA virus is also a potent activator involved in innate immunity. Double-stranded DNA (dsDNA) could serve as a template for RNA polymerase III and is transcribed into double-stranded RNA (dsRNA), which then triggers host RIG-I activation (37). Several miRNAs modulate host innate immune responses for DNA virus persistence (38). Thus, in addition to the RNA virus, which mediated viral evasion through host MAVS-mediated antiviral response, DNA virus also may utilize a similar regulation mechanism for evasion.
Numerous studies indicated that each gene can be regulated by different miRNAs, and each highly conserved miRNA probably targets several hundred distinct genes. For example, miR-146a targets TRAF6, IRAK1, and IRAK2 (18, 39). Similarly, miR-21 also has been reported to target and downregulate both MyD88 and IRAK1 (40). Our previous studies suggested that miR-3570 could be significantly upregulated in Vibrio anguillarum, which is the causative agent of vibriosis in cultivated fish. It upregulated miR-3570, which suppressed MyD88 expression, subsequently repressing inflammatory cytokine genes through MyD88-mediated NF-κB signaling, thereby avoiding excessive inflammation (41). The present paper revealed that miR-3570 could target and downregulate another gene, MAVS, which enriches the networks of miRNA regulation innate immune response and participates in antiviral response. Highly conserved miRNA can target several distinct mRNAs in mammals as well as in fish species. miR-3570 is also an essential negative regulator involved in one or more physiological processes, which enrich the regulation network of miRNA in immune response. In fish species, the innate immune response is of vital importance for the disease resistance of fish because of the constraints placed on the adaptive immune response by piscine poikilothermic nature and the limited antibody repertoires, affinity maturation, memory, and relatively slow lymphocyte proliferation (42). Due to the significance of innate immunity in fish, the modulators and regulation mechanisms in immune responses are particularly important. Our findings not only enriched our knowledge of the intricate networks of host-pathogen interaction but also demonstrated that miRNA in fish can participate in different regulation signaling upon pathogen exposure.
In conclusion, this study demonstrated that the host miRNA miR-3570 is associated with the SCRV-induced antiviral immune response, and it is a negative regulator in constraining the production of type I IFNs and antiviral genes. Aside from targeting MyD88 and modulating excessive immune response, miR-3570 could target another adapter molecule, MAVS, and subsequently negatively regulate NF-κB and IRF3 pathways. Finally, SCRV upregulates host miR-3570 expression, which then suppresses the expression of antiviral genes and leads to the replication of the invading RNA virus. Our data provide information on new miRNA feedback regulation upon viral infection in fish and insights into the host-virus interaction.
MATERIALS AND METHODS
Cell culture and transfection.
HEK293 and HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) high-glucose medium (HyClone) supplemented with 10% fetal bovine serum (FBS; Gibco), 100 U/ml penicillin, and 100 mg/ml streptomycin at 37°C in 5% CO2. Miiuy croaker kidney cell lines (MKC) were cultured in L-15 medium (HyClone) supplemented with 15% FBS at 26°C in 4% CO2. Macrophages were aseptically isolated from the head kidney of miiuy croaker as previously reported (43) and cultured in L-15 medium supplemented with 20% FBS at 26°C in 4% CO2.
Before transient transfection, cells were seeded into each well of 24-well or 12-well plates and incubated overnight. Subsequently, cells were transfected with plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocols. For each transfection experiment, the pEGFP plasmid was transfected as the positive control to verify transfection efficiency.
Prediction of miR-3570 targeting sites.
The miR-3570 targets were predicted using Targetscan (44), miRanda (45), and miRInspector (46) algorithms. Predictions were ranked based on the predicted efficacy of targeting as calculated using the context and scores of the sites.
Plasmid construction.
In order to construct the MAVS-3′UTR reporter vector, the full-length 3′UTR of the miiuy croaker MAVS gene was amplified using PCR and cloned into pmirGLO luciferase reporter vector (Promega) using NheI and XbaI restriction sites. Similarly, the 3′UTR of the zebrafish (Danio rerio) MAVS gene was amplified and inserted into the SacI and XbaI restriction sites of pmirGLO vector, and the 3′UTR of the large yellow croaker (Larimichthys crocea) MAVS gene was amplified and inserted into the XhoI and SalI restriction sites of pmirGLO vector. The mutant type of the MAVS-3′UTR reporter vector was constructed by using a Mut Express II fast mutagenesis kit, V2 (Vazyme), with specific primers (Table 1). Moreover, the miiuy croaker MAVS-3′UTR or its mutant type was inserted into the pIZ/V5-His vector (Invitrogen), which included the sequence of enhanced green fluorescent protein (GFP). In addition, to construct the pre-miRNA vector for miiuy croaker, the pre-miR-3570 sequence was PCR amplified and then cloned into the pcDNA6.2-GW/EmGFP vector (Invitrogen). The sequence of pre-miR-3570 can be accessed in our previously submitted genome sequence under BioProject accession no. PRJNA272995 (scaffold 73, 626,164 to 626,244). To construct the MAVS expression vector, the full length of the CDS region and 3′UTR of the miiuy croaker MAVS gene (GenBank accession no. MF871620) were amplified by specific primer pairs with the Flag tag and cloned into the pcDNA3.1 vector (Invitrogen) using HindIII and EcoRI restriction sites. Total plasmids were verified by Sanger sequencing and extracted through an endotoxin-free plasmid DNA miniprep kit (Tiangen).
TABLE 1.
Sequences of primers used in the present study
| Primer | Sequences (5′–3′) |
|---|---|
| MAVS-qRT-F | AGGCACCAACAATTCCAG |
| MAVS-qRT-R | ACGGAGCAGGCTTCACTT |
| IFNβ-qRT-F | GCTCTGCCTTCCCTGCTA |
| IFNβ-qRT-R | CAGTTGACTCCGCCCTCT |
| MX1-qRT-F | GCTGCTTGTTTACTCCCA |
| MX1-qRT-R | ACCTGCATCATCTCCCTC |
| TNFa-qRT-F | GTTTGCTTGGTACTGGAATGG |
| TNFa-qRT-R | TGTGGGATGATGATCTGGTTG |
| ISG15-qRT-F | TGAACGGACAGAAGACGC |
| ISG15-qRT-R | TGAGGAATACCTGCATGG |
| Viperin-qRT-F | ACCCGTCCAAGTCCATAC |
| Viperin-qRT-R | TCATGTCAGCTTTGCTCC |
| miR-3570-qRT-F | AGTACAATCAACGGTCGATG |
| miR-3570-qRT-F | GTCCAGTTTTTTTTTTTTTTTAAACCA |
| 5.8S rRNA-qRT-F | AACTCTTAGCGGTGGATCA |
| 5.8SrRNA-qRT-R | GTTTTTTTTTTTTTTTGCCGAGTG |
| β-Actin-qRT-F | GAGCCGCACGCTTCTTT |
| β-Actin-qRT-R | CTGCTGTAGCCGAGGAC |
| Pre-miR-3570-BamHIF | CGCGGATCCGTGGTTTGGAAGCTGGAA |
| Pre-miR-3570-XhoIR | CCGCTCGAGAAGCAGACTGTCATCCCT |
| MAVS-3′UTR-WT-NheIF | CTAGCTAGCCGTATGGTGCCTTATTG |
| MAVS-3′UTR-WT-XbaIR | TGCTCTAGACAGCCTCTGTCCTGTCTACT |
| MAVS-3′UTR-MT-F | CCTGTCGTTAAGTTCTGGGTTGAGATCTCAGGT |
| MAVS-3′UTR-MT-R | CCAGAACTTAACGACAGGAGTGTAGATTTGGTGTCTGC |
| DrMAVS-3′UTR-WT-SacIF | CGCGAGCTCTGAGAATGTCTAACAGGCAC |
| DrMAVS-3′UTR-WT-XbaIR | TGCTCTAGAAAGAACAGGAACAATCAAGA |
| DrMAVS-3′UTR-MT-F | CTTGACATTTCCTGTTCTTAAAGATTTTTACAAATG |
| DrMAVS-3′UTR-MT-R | GAACAGGAAATGTCAAGATTGACCAGTACACAAAAAAG |
| LcMAVS-3′UTR-WT-XhoIF | CCGCTCGAGACCCTCCAGACCTTTGAT |
| LcMAVS-3′UTR-WT-SalIR | GTCGACGTCGACGCTCCTCGTTAATCCTCA |
| LcMAVS-3′UTR-MT-F | CCATTTTTAAGTTCTGGGTTCAGATCTCAGGT |
| LcMAVS-3′UTR-MT-R | CCAGAACTTAAAAATGGAAGTGTAGATTTGGTGTGAGCTCG |
| MAVS-HindIIIF | GACGATGACGACAAGAAGCTTTCGTCTGCCAAAGACAAACTGTA |
| MAVS-EcoRIR | TGATGGATATCTGCAGAATTCCAGCCTCTGTCCTGTCTACTTCATG |
| GFP-MAVS-3′UTR-HindIIIF | CCCAAGCTTGCTAGCGTATGGTGCCTTATTGG |
| GFP-MAVS-3′UTR-BamHIR | CGCGGATCCCAGCCTCTGTCCTGTCTACT |
miRNA mimics and inhibitors.
miR-3570 mimics (dsRNA oligonucleotides), miR-3570 inhibitors (single-stranded chemically modified oligonucleotides), and control oligonucleotides were gained from GenePharma (Shanghai, China). Primer sequences were the following: miR-3570 mimics, 5′-UACAAUCAACGGUCGAUGGUUU-3′ (sense) and 5′-ACCAUCGACCGUUGAUUGUAUU-3′ (antisense); control mimics, 5′-UUCUCCGAACGUGUCACGUTT-3′ (sense) and 5′-ACGUGACACGUUCGGAGAATT-3′ (antisense); miR-3570 inhibitor, 5′-AAACCAUCGACCGUUGAUUGUA-3′ (chemically modified by 2′-Ome); control inhibitor, 5′-CAGUACUUUUGUGUAGUACAA-3′. Macrophages, MKCs, HEK293 cells, or HeLa cells were transfected with 30 to 100 nM each oligonucleotide by using Lipofectamine 2000 (Invitrogen) for 48 h and then infected with SCRV.
RNA interference.
The MAVS-specific short interfering RNAs (siRNAs) were 5′-GAUGAACGUGGUGCAGAUATT-3′ (sense) and 5′-UAUCUGCACCACGUUCAUCTT-3′ (antisense). The scrambled control RNA sequences were 5′-UUCUCCGAACGUGUCACGUTT-3′ (sense) and 5′-ACGUGACACGUUCGGAGAATT-3′ (antisense). Macrophages were transfected with 50 nM each siRNA by using Lipofectamine 2000 (Invitrogen) for up to 48 h and then were stimulated with SCRV.
RNA extraction and real-time quantitative PCR.
Total RNA was isolated with TRIzol reagent (Invitrogen) by following the manufacturer's instructions. Primers of genes then were designed and β-actin was applied as the internal control (Table 1). The expression patterns of each gene were determined by qRT-PCR on a 7500 system (Applied Biosystems, USA) using SYBR premix Ex Taq (TaKaRa) as previously described (47). For the extraction of small RNA, an miRcute miRNA isolation kit (Tiangen) was used. The expression analysis of miR-3570 was executed by using the miRcute miRNA quantitative PCR detection kit (Tiangen) under the following conditions: 95°C for 15 min and 40 cycles of two steps (95°C for 5 s and 60°C for 30 s). 5.8S rRNA was used as the internal control. Sequences of miRNA primers are also listed in Table 1.
Virus yield qualification.
Macrophages were transfected with oligonucleotides and then infected with SCRV as indicated. Volumes of 0.1 ml of the cultural supernatants then were serially diluted on the monolayer of MKCs, and 1 × 104 MKCs were seeded into 96-well plates 1 day before measurement. The 50% tissue culture infectious dose (TCID50) was measured after 3 days. Viral RNA in the supernatant was extracted by using the body fluid viral DNA/RNA miniprep kit (Axygen), and SCRV RNA replicates were qualified as described above.
Prokaryotic expression and polyclonal antiserum.
For prokaryotic expression, the full-length CDS region of miiuy croaker MAVS was cloned into a pGEX-4T-1 vector (GE) with EcoRI/XhoI sites to construct pGEX-4T-1-MAVS plasmid. The plasmid pGEX-4T-1-MAVS then was transformed into the BL21(DE3) Escherichia coli strain and expressed as a protein containing MAVS fused with glutathione S-transferase (GST). The fusion protein was induced by isopropyl β-d-thiogalactoside (IPTG) and purified by GST-bind resin chromatography. The purified fusion protein was applied to immunize New Zealand White rabbits to raise a polyclonal anti-MAVS antiserum (31).
Dual-luciferase reporter assays.
The MAVS-3′UTR luciferase reporter vector was made by amplifying the miiuy croaker MAVS mRNA 3′UTR sequence by PCR and cloned into pmirGLO vector. The mutant type of MAVS-3′UTR luciferase reporter vector was constructed using a Mut Express II fast mutagenesis kit, V2. For miRNA target verification, the wild-type or mutant-type MAVS-3′UTR luciferase reporter vector was cotransfected with miR-3570 mimics, control mimics, or pre-miR-3570 plasmid into HEK293 cells and HeLa cells. Furthermore, HEK293 cells were cotransfected with NF-κB, IRF3, or IFN-stimulated response element (ISRE) luciferase reporter plasmid, MAVS expression plasmid containing the full-length CDS region and 3′UTR of miiuy croaker MAVS, and pRL-TK renilla luciferase plasmid, together with either miR-3570 mimics, inhibitors and control inhibitors, or pre-miR-3570 plasmids for Dual-Luciferase reporter assay. After 24 or 48 h, the cells were lysed for reporter activity testing by using the Dual-Luciferase reporter assay system (Promega). All of the luciferase activity values were achieved against the renilla luciferase control. For each experiment, three independent experiments were conducted, and each experiment was done in triplicate.
Western blotting.
The cells were washed with cold phosphate-buffered saline (PBS) and the cellular lysates were generated by using 1× SDS-PAGE loading buffer. Protein concentrations of the cell lysis extracts were measured with the bicinchoninic acid (BCA) assay (Pierce) and equalized with the extraction reagent. Equal amounts of the extracts were loaded and subjected to SDS-PAGE, which subsequently were transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore) by semidry blotting (Trans-Blot Turbo system; Bio-Rad). Membranes were incubated at 4°C overnight with anti-Flag mouse monoclonal antibody and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) monoclonal antibodies (Beyotime) or polyclonal anti-MAVS antiserum. The following day, the membranes were incubated with the secondary antibody. The immunoreactive proteins were detected by using WesternBright ECL (Advansta). The digital imaging was performed with a cold charge-coupled-device camera.
Statistical analysis.
Generally, all experiments were performed at least three independent times, with three technical replicates for each experiment. The relative gene expression data were acquired using the 2−ΔΔCT method, and comparisons between groups were analyzed by one-way analysis of variance (ANOVA) followed by Duncan's multiple-comparison tests (48). Results are expressed as means ± SE (standard errors), and differences between means with P values of <0.05 were considered to be statistically significant.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (31370049 and 31672682).
T.X., Q.C., J.C., and D.B. carried out the experiments. T.X., Q.C., and J.C. designed the experiments and analyzed the data. T.X., Q.C., and J.C. wrote the manuscript.
We have no conflicts of interest to declare.
REFERENCES
- 1.Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 2.Chiang JJ, Davis ME, Gack MU. 2014. Regulation of RIG-I-like receptor signaling by host and viral proteins. Cytokine Growth Factor Rev 25:491–505. doi: 10.1016/j.cytogfr.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beutler B, Eidenschenk C, Crozat K, Imler JL, Takeuchi O, Hoffmann JA, Akira SZ. 2007. Genetic analysis of resistance to viral infection. Nat Rev Immunol 7:753–766. doi: 10.1038/nri2174. [DOI] [PubMed] [Google Scholar]
- 4.Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 5.Pestka S. 2007. The interferons: 50 years after their discovery, there is much more to learn. J Biol Chem 282:20047–20051. doi: 10.1074/jbc.R700004200. [DOI] [PubMed] [Google Scholar]
- 6.Sadler AJ, Williams BR. 2008. Interferon-inducible antiviral effectors. Nat Rev Immunol 8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui J, Li Y, Zhu L, Liu D, Song Z, Wang H, Wang HY, Wang RF. 2012. NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat Immunol 13:387–395. doi: 10.1038/ni.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiba S, Baghdadi M, Akiba H, Yoshiyama H, Kinoshita I, Dosaka-Akita H, Hirashima M. 2012. Tumor-infiltrating DCs suppress Nucleic acid-mediated innate immune responses through interactions between the receptor TIM-3 and the alarmin HMGB1. Nat Immunol 13:832–842. doi: 10.1038/ni.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iannello A, Debbeche O, Martin E, Attalah LH, Samarani S, Ahmad A. 2006. Viral strategies for evading antiviral cellular immune responses of the host. J Leukoc Biol 79:16–35. doi: 10.1189/jlb.0705397. [DOI] [PubMed] [Google Scholar]
- 10.Diao F, Li S, Tian Y, Zhang M, Xu LG, Zhang Y, Wang RP, Chen DY, Zhai ZH, Zhong B, Tien P, Shu HB. 2007. Negative regulation of MDA5- but not RIG-I-mediated innate antiviral signaling by the dihydroxyacetone kinase. Proc Natl Acad Sci U S A 104:11706–11711. doi: 10.1073/pnas.0700544104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore CB, Bergstralh DT, Duncan JA, Lei Y, Morrison TE, Zimmermann AG, Lich JD. 2008. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature 451:573–577. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- 12.Zhong B, Zhang L, Lei C, Li Y, Mao AP, Yang Y, Shu HB. 2009. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity 30:397–407. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Onoguchi K, Onomoto K, Takamatsu S, Jogi M, Takemura A, Morimoto S, Fujita T. 2010. Virus-infection or 5′ppp-RNA activates antiviral signal through redistribution of IPS-1 mediated by MFN1. PLoS Pathog 6:e1001012. doi: 10.1371/journal.ppat.1001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yasukawa K, Oshiumi H, Takeda M, Ishihara N, Yanagi Y, Seya T, Koshiba T. 2009. Mitofusin 2 inhibits mitochondrial antiviral signaling. Sci Signal 2:ra47. doi: 10.1126/scisignal.2000287. [DOI] [PubMed] [Google Scholar]
- 15.Wienholds E, Plasterk RH. 2005. MicroRNA function in animal development. FEBS Lett 579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 16.Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. 2005. Stem cell division is regulated by the microRNA pathway. Nature 435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 17.Friedman RC, Farh KK, Burge CB, Bartel DP. 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou J, Wang P, Lin L, Liu X, Ma F, An H, Cao X. 2009. MicroRNA-146a feedback inhibits RIG-I-dependent type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J Immunol 183:2150–2158. doi: 10.4049/jimmunol.0900707. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Fan X, He X, Sun H, Zou Z, Yuan H, Shi X. 2012. MicroRNA-466l inhibits antiviral innate immune response by targeting interferon-alpha. Cell Mol Immunol 9:497–502. doi: 10.1038/cmi.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagos D, Pollara G, Henderson S, Gratrix F, Fabani M, Milne RS, Gotch F, Boshoff C. 2010. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat Cell Biol 12:513–519. doi: 10.1038/ncb2054. [DOI] [PubMed] [Google Scholar]
- 21.Wan S, Ashraf U, Ye J, Duan X, Zohaib A, Wang W, Chen Z, Zhu B, Li Y, Chen H, Cao S. 2016. MicroRNA-22 negatively regulates poly(I:C)-triggered type I interferon and inflammatory cytokine production via targeting mitochondrial antiviral signaling protein (MAVS). Oncotarget 7:76667–76683. doi: 10.18632/oncotarget.12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu AC, Dua K, Starkey MR, Haw TJ, Nair PM, Nichol K, Zammit N, Grey ST, Baines JK, Foster PS, Hansbro PM. 2017. MicroRNA-125a and -b inhibit A20 and MAVS to promote inflammation and impair antiviral response in COPD. JCI Insight 2:e90443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang QY, Gui JF. 2015. Virus genomes and virus-host interactions in aquaculture animals. Sci China Life Sci 58:156–169. doi: 10.1007/s11427-015-4802-y. [DOI] [PubMed] [Google Scholar]
- 24.Crane M, Hyatt A. 2011. Viruses of fish: an overview of significant pathogens. Viruses 3:2025–2046. doi: 10.3390/v3112025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tao JJ, Zhou GZ, Gui JF, Zhang QY. 2008. Genomic sequence of mandarin fish rhabdovirus with an unusual small non-transcriptional ORF. Virus Res 132:86–96. doi: 10.1016/j.virusres.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 26.Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. 2008. MicroRNAs: new regulators of immune cell development and function. Nat Immunol 9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 27.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. 2004. Processing of primary microRNAs by the Microprocessor complex. Nature 432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 28.Walker PJ, Winton JR. 2010. Emerging viral diseases of fish and shrimp. Vet Res 41:51. doi: 10.1051/vetres/2010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu Q, Gao Y, Xu G, Wu C, Xu T. 2015. Transcriptome comparative analysis revealed poly(I:C) activated RIG-I/MDA5-mediated signaling pathway in miiuy croaker. Fish Shellfish Immunol 47:168–174. doi: 10.1016/j.fsi.2015.08.032. [DOI] [PubMed] [Google Scholar]
- 30.Xu T, Xu G, Che R, Wang R, Wang Y, Li J, Wang S, Sun Y, Liu T, Liu J, Wang A, Han J, Chu Q, Yang Q. 2016. The genome of the miiuy croaker reveals well-developed innate immune and sensory systems. Sci Rep 6:21902. doi: 10.1038/srep21902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chu Q, Sun Y, Cui J, Xu T. 2017. Inducible microRNA-214 contributes to the suppression of NF-κB-mediated inflammatory response via targeting myd88 gene in fish. J Biol Chem 292:5282–5290. doi: 10.1074/jbc.M117.777078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Bi X, Chu Q, Xu T. 2016. Discovery of Toll-like receptor 13 exists in the teleost fish: miiuy croaker (Perciformes, Sciaenidae). Dev Comp Immunol 61:25–33. doi: 10.1016/j.dci.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 33.Zhao Y, Sun X, Nie X, Sun L, Tang TS, Chen D, Sun Q. 2012. COX5B regulates MAVS-mediated antiviral signaling through interaction with ATG5 and repressing ROS production. PLoS Pathog 8:e1003086. doi: 10.1371/journal.ppat.1003086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.You F, Sun H, Zhou X, Sun W, Liang S, Zhai Z, Jiang Z. 2009. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat Immunol 10:1300–1308. doi: 10.1038/ni.1815. [DOI] [PubMed] [Google Scholar]
- 35.Vitour D, Dabo S, Pour MA, Vilasco M, Vidalain PO, Jacob Y, Tangy F. 2009. Pololike kinase 1 (PLK1) regulates interferon (IFN) induction by MAVS. J Biol Chem 284:21797–21809. doi: 10.1074/jbc.M109.018275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seth RB, Sun L, Ea CK, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF3. Cell 122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 37.Chiu YH, MacMillan JB, Chen ZJ. 2009. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kincaid RP, Burke JM, Cox JC, de Villiers EM, Sullivan CS. 2013. A human torque teno virus encodes a microRNA that inhibits interferon signaling. PLoS Pathog 9:e1003818. doi: 10.1371/journal.ppat.1003818. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Cameron JE, Yin Q, Fewell C, Lacey M, McBride J, Wang X, Flemington EK. 2008. Epstein-Barr virus latent membrane protein 1 induces cellular MicroRNA miR-146a, a modulator of lymphocyte signaling pathways. J Virol 82:1946–1958. doi: 10.1128/JVI.02136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y, Chen J, Wang H, Shi J, Wu K, Liu S, Wu J. 2013. HCV-induced miR-21 contributes to evasion of host immune system by targeting MyD88 and IRAK1. PLoS Pathog 9:e1003248. doi: 10.1371/journal.ppat.1003248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chu Q, Sun Y, Cui J, Xu TJ. 2017. MicroRNA-3570 modulates the NF-κB pathway in teleost fish by targeting MyD88. J Immunol 198:3274–3282. doi: 10.4049/jimmunol.1602064. [DOI] [PubMed] [Google Scholar]
- 42.Magnadóttir B. 2006. Innate immunity of fish (overview). Fish Shellfish Immunol 20:137–151. doi: 10.1016/j.fsi.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 43.Chu Q, Sun Y, Bi D, Cui J, Xu TJ. 2017. Up-regulated of miR-8159-5p and miR-217-5p by LPS stimulation negatively co-regulate TLR1 in miiuy croaker. Dev Comp Immunol 67:117–125. doi: 10.1016/j.dci.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 44.Lewis BP, I-Hung S. 2003. Prediction of mammalian microRNA targets. Cell 115:787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 45.John B, Enright AJ. 2004. Human microRNA targets. PLoS Biol 2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rusinov V, Baev V, Minkov IN, Tabler M. 2005. MicroInspector: a web tool for detection of miRNA binding sites in an RNA sequence. Nucleic Acids Res 33:696–700. doi: 10.1093/nar/gki364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu Z, Wang R, Ren L, Xu T. 2013. Characterization of the CCR3 and CCR9 genes in miiuy croaker and different selection pressures imposed on different domains between mammals and teleosts. Dev Comp Immunol 41:631–643. doi: 10.1016/j.dci.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 48.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]