Abstract
The expression of intron‐containing genes in eukaryotes requires generation of protein‐coding messenger RNAs (mRNAs) via RNA splicing, whereby the spliceosome removes non‐coding introns from pre‐mRNAs and joins exons. Spliceosomes must ensure accurate removal of highly diverse introns. We show that Sde2 is a ubiquitin‐fold‐containing splicing regulator that supports splicing of selected pre‐mRNAs in an intron‐specific manner in Schizosaccharomyces pombe. Both fission yeast and human Sde2 are translated as inactive precursor proteins harbouring the ubiquitin‐fold domain linked through an invariant GGKGG motif to a C‐terminal domain (referred to as Sde2‐C). Precursor processing after the first di‐glycine motif by the ubiquitin‐specific proteases Ubp5 and Ubp15 generates a short‐lived activated Sde2‐C fragment with an N‐terminal lysine residue, which subsequently gets incorporated into spliceosomes. Absence of Sde2 or defects in Sde2 activation both result in inefficient excision of selected introns from a subset of pre‐mRNAs. Sde2 facilitates spliceosomal association of Cactin/Cay1, with a functional link between Sde2 and Cactin further supported by genetic interactions and pre‐mRNA splicing assays. These findings suggest that ubiquitin‐like processing of Sde2 into a short‐lived activated form may function as a checkpoint to ensure proper splicing of certain pre‐mRNAs in fission yeast.
Keywords: deubiquitinating enzymes, intron‐specific pre‐mRNA splicing, N‐end rule pathway, telomeric silencing, ubiquitin‐like processing
Subject Categories: DNA Replication, Repair & Recombination; Post-translational Modifications, Proteolysis & Proteomics; RNA Biology
Introduction
Protein‐coding genes in eukaryotes are often interrupted by non‐coding introns. Following their transcription into precursor messenger RNAs (pre‐mRNAs), the splicing process removes the introns and joins the exons to generate translatable mRNAs. The pre‐mRNAs can also undergo alternative RNA splicing to generate more than one protein‐coding mRNAs from a gene. The majority of human genes have introns and undergo constitutive as well as alternative RNA splicing (Ast, 2004; Kim et al, 2006). The spliceosome is a large ribonucleoprotein (RNP) complex that assembles with five small‐nuclear RNAs (snRNAs) and nearly two hundred proteins to perform pre‐mRNA splicing. The snRNP complexes form the spliceosomal core, while RNA‐binding proteins, RNP modifying enzymes (ATPases, helicases) and modifications of splicing factors (phosphorylation, methylation) regulate activity of this complex (Wahl et al, 2009).
Ubiquitin and ubiquitin‐like proteins (collectively referred to as UBLs) share the ubiquitin‐fold and act as key regulators of a variety of cellular processes including protein homoeostasis, signalling, trafficking and DNA transactions (Welchman et al, 2005; Hochstrasser, 2009; van der Veen & Ploegh, 2012). The canonical ubiquitin is covalently conjugated to proteins by a set of enzymes in a process termed ubiquitination. Ubiquitin itself is not encoded as a single polypeptide, but rather is translated as a precursor, fused either to ribosomal protein genes, or with multiple ubiquitin moieties in tandem (Finley et al, 1989; Hochstrasser, 2009; Kimura & Tanaka, 2010). These precursors are processed by ubiquitin‐specific hydrolases (also referred to as deubiquitinating enzymes or DUBs) immediately after di‐glycine (GG) motifs to generate functional ubiquitin (Turcu et al, 2009).
Whereas several UBLs follow ubiquitin's mode of processing and conjugation by employing respective UBL‐specific enzymes, Hub1/UBL5 lacks the di‐glycine motif, and acts through a non‐covalent process. Hub1 binds to the HIND domain‐containing splicing factors Snu66 and/or Prp38 and functions in pre‐mRNA splicing by promoting the usage of non‐canonical 5′ splice sites in introns. Hub1 thereby promotes alternative splicing of the SRC1 pre‐mRNA in Saccharomyces cerevisiae (Mishra et al, 2011). Specific roles of Hub1 in pre‐mRNA splicing have also been reported in both Schizosaccharomyces pombe and mammalian cells (Wilkinson et al, 2004; Mishra et al, 2011; Ammon et al, 2014). Whereas HUB1 is a non‐essential gene in S. cerevisiae, its orthologs in S. pombe and mammalian cells are essential for viability, perhaps because of the increased prevalence of introns and alternative splicing in S. pombe and humans.
The process of pre‐mRNA splicing has been linked to heterochromatin formation at the centromeres and telomeres in S. pombe (Bayne et al, 2008; Huang & Zhu, 2014; Kallgren et al, 2014; Wang et al, 2014). Earlier splicing factors were thought to provide a platform for the generation of small interfering siRNAs in the RNAi pathway of heterochromatin formation, since their mutants were defective in heterochromatin formation, but appeared normal in pre‐mRNA splicing (Bayne et al, 2008). Recent reports however connect the heterochromatin defects observed in splicing factor mutants with reduced splicing of mRNAs encoding bona fide components of the heterochromatin pathway (Bayne et al, 2014; Kallgren et al, 2014; Wang et al, 2014). Schizosaccharomyces pombe lacking the sde2 gene, named for silencing defective, showed defective telomeric silencing (Sugioka‐Sugiyama & Sugiyama, 2011). Strains lacking sde2 also showed defects in centromeric silencing and defects in splicing of cytoskeleton constituents and centromeric outer repeat transcripts (Bayne et al, 2014). The human ortholog of Sde2, C1orf55, was present in spliceosomal preparations (Bessonov et al, 2008), and recently, the S. pombe protein was also shown to associate with splicing factors (Bayne et al, 2014; Chen et al, 2014).
In this study, we report that sde2 genetically interacts with hub1 in S. pombe. The Sde2 protein has a ubiquitin‐fold at its N‐terminus, which must be cleaved by the ubiquitin‐specific proteases (USPs) Ubp5 and Ubp15. After cleavage, the C‐terminal domain of Sde2, which starts with a lysine, gets incorporated into the spliceosome. Loss of Sde2 results in inefficient removal of selected introns from a subset of genes having functions in telomeric silencing, DNA replication and transcription. Furthermore, the N‐end rule pathway of proteasomal degradation controls the level of Sde2 protein in the cell. The intron‐specific pre‐mRNA splicing activity of the ubiquitin‐fold‐containing Sde2 protein becomes critical for chromatin silencing and genomic stability in S. pombe.
Results
Sde2 is processed like ubiquitin precursors
We have previously reported the role of the UBL Hub1 in alternative RNA splicing through usage of non‐canonical 5′ splice sites in the budding yeast S. cerevisiae (Mishra et al, 2011). Orthologs of Hub1 in S. pombe and humans are essential for viability and play specific roles in pre‐mRNA splicing (Mishra et al, 2011; Ammon et al, 2014). To identify spliceosomal regulators in an intron prevalent organism, we screened for genetic interactors of hub1 by combining S. pombe hub1‐1 mutant (Yashiroda & Tanaka, 2004) with the haploid deletion library of non‐essential genes (Kim et al, 2010). As expected from Hub1's role in pre‐mRNA splicing, deletion mutants of multiple splicing factors were synthetically sick with hub1‐1 (Fig EV1A). Among them, the deletion of sde2 gene was also synthetically sick with hub1‐1. We confirmed this interaction by generating double mutants of hub1‐1 with Δsde2 independently (Fig EV1B). As reported previously in high‐throughput studies (Kennedy et al, 2008; Zhang et al, 2013), the Δsde2 strain grew slowly under standard conditions, and the growth defect was more pronounced at elevated temperatures and under genotoxic stress conditions (hydroxyurea, valproic acid, sodium butyrate, cadmium) (Appendix Fig S1A).
Figure EV1. Sde2 is processed like ubiquitin and genetically interacts with ubiquitin‐like protein Hub1.
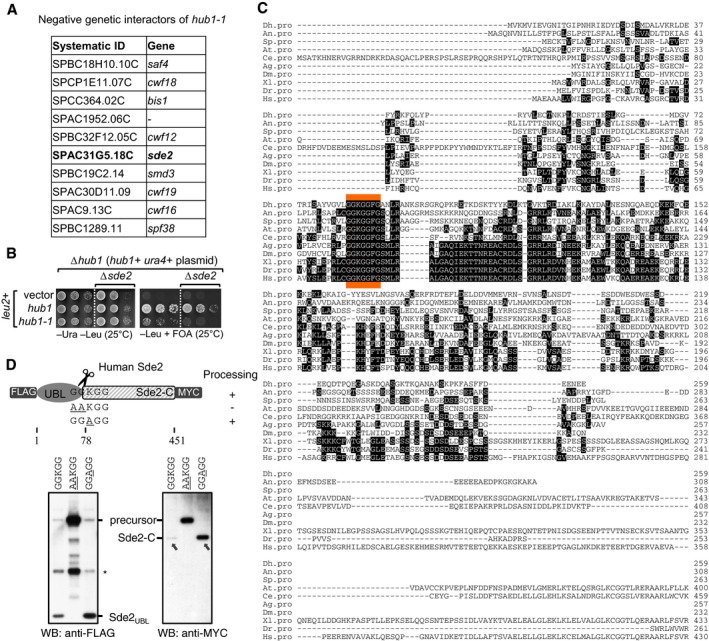
- Genetic interactors of hub1. hub1‐1 is the temperature‐sensitive hub1(I42S) mutant reported by Yashiroda and Tanaka (2004). A genetic screen was performed with hub1‐1 and the haploid deletion library of non‐essential genes in Schizosaccharomyces pombe. hub1‐1 was synthetically sick with the deletion mutants of given genes including sde2. Among the top hits, genes with relevance to pre‐mRNA splicing are shown.
- Confirmation of the negative genetic interaction between hub1‐1 and Δsde2 mutants. sde2 gene, from ATG to the stop codon, was deleted in a S. pombe Δhub1 strain, which was kept viable with a ura4 + marked hub1 expression plasmid with its own promoter and terminator. The resultant strain was transformed with leu2 + marked expression plasmids expressing wild‐type (WT) hub1 or the hub1‐1 mutant. Fivefold serial dilutions of cells from these transformations were spotted on indicated agar plates. Plates were incubated at 25°C. 5‐fluoroorotic acid (FOA) (1 g/l of media) was used to shuffle‐out ura4 + plasmid. Thus, cells growing on −Leu + FOA plates will have hub1 mutants in Δsde2 background.
- Alignment of Sde2 protein orthologs from different eukaryotes. Abbreviations used are (respective NCBI protein accession numbers are given in parentheses) as follows: Dh, Debaryomyces hansenii (XP_458854); An, Aspergillus niger (XP_001391007); Sp, Schizosaccharomyces pombe (NP_594019); At, Arabidopsis thaliana (NP_192009); Ce, Caenorhabditis elegans (NP_506378); Ag, Anopheles gambiae (XP_321833); Dm, Drosophila melanogaster (NP_651207); Xl, Xenopus laevis (NP_001084858); Dr, Danio rario (AAH93198); Hs, Homo sapiens (NP_689821). The sequence marked in orange indicates most conserved region in Sde2 orthologs.
- Expression of human Sde2 (C1orf55) protein in U2OS cells. Constructs with sequences encoding 3FLAG epitope tag at the N‐terminus of HsSDE2 gene and single MYC epitope tag at its C‐terminus under CMV (cytomegalovirus) promoter were used. With WT Sde2, Sde2‐C would start with lysine (KGG…), whereas with Sde2 GGAGG mutant, Sde2‐C would start with alanine (AGG…). Asterisk indicates antibody cross reactivity signal. Arrows indicate the HsSde2‐C protein formed after processing of the GGAGG mutant precursor accumulated to higher levels than the protein formed from the wild‐type precursor.
Putative orthologs of Sde2 exist in organisms from yeast to humans (Fig EV1C). However, in the Fungi subphylum Saccharomycotina, an Sde2‐like protein is observed in Debaryomyces, but is absent in S. cerevisiae, Candida albicans and Pichia pastoris. The protein structure prediction program i‐TASSER (Yang et al, 2015) predicted the presence of a ubiquitin‐fold at the N‐terminus and a C‐terminal helical domain in Sde2 (henceforth referred to as Sde2UBL and Sde2‐C, respectively). An invariant signature motif, GGKGG, separates the moderately conserved Sde2UBL and Sde2‐C (Figs 1A and EV1C). This motif flanks Sde2UBL and resembles the di‐glycine (GG) motif typical of UBL precursors processed by the UBL‐specific proteases.
Figure 1. Sde2 is a conserved protein with ubiquitin‐fold.
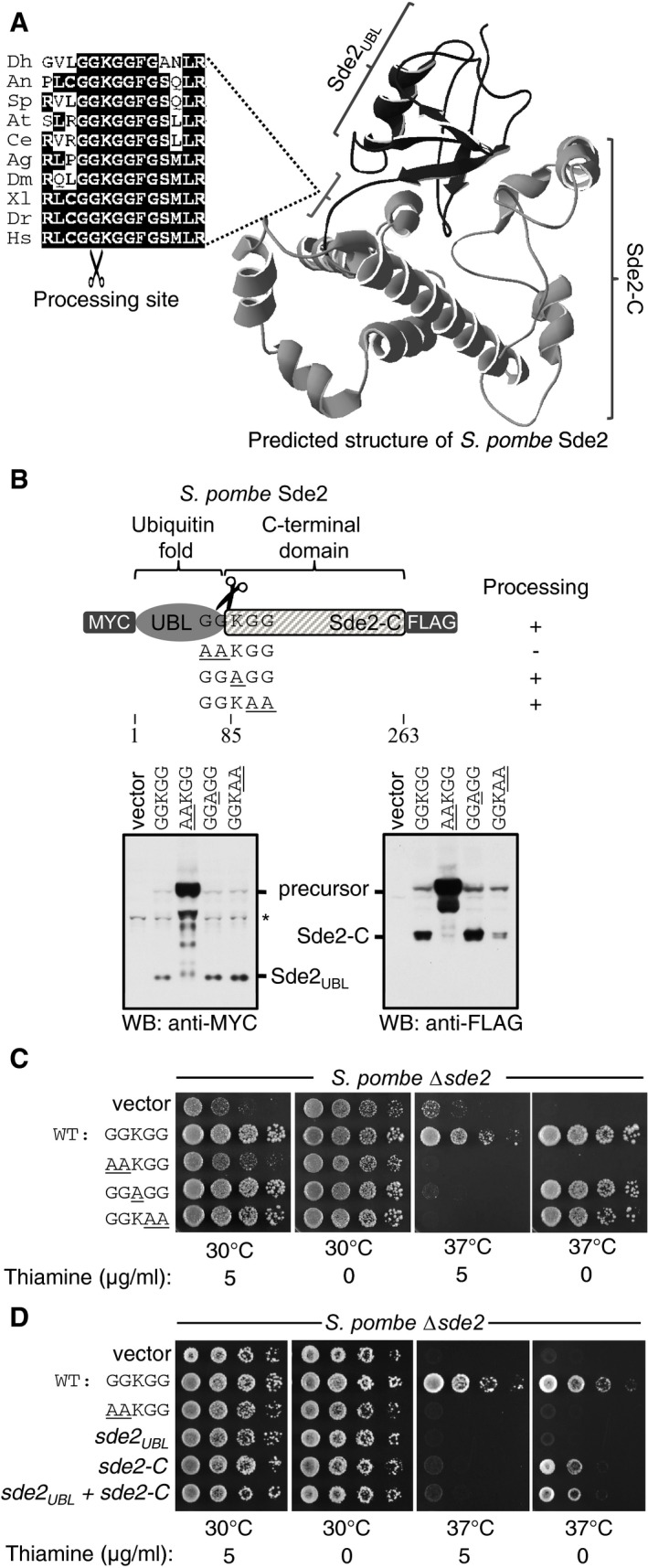
- Predicted structure of Schizosaccharomyces pombe Sde2 protein shows the presence of a ubiquitin‐fold (Sde2UBL) and a helical C‐terminal domain (Sde2‐C). The alignment on the left shows the linker GGKGG motif between Sde2UBL and Sde2‐C in Sde2 orthologs. Dh, Debaryomyces hansenii; An, Aspergillus niger; Sp, Schizosaccharomyces pombe; At, Arabidopsis thaliana; Ce, Caenorhabditis elegans; Ag, Anopheles gambiae; Dm, Drosophila melanogaster; Xl, Xenopus laevis; Dr, Danio rario; Hs, Homo sapiens.
- Expression and processing of Sde2 protein in S. pombe detected by immunoblot analysis (Western blot, WB) using epitope tag‐specific antibodies. Underlined residues mark changes from wild‐type (WT) Sde2. Asterisk indicates antibody cross reactivity signal.
- Complementation of S. pombe Δsde2 by GGKGG mutants of Sde2. Constructs are as in (B). Fivefold serial dilution spotting was done on indicated agar plates. Plates were incubated at 30°C and 37°C until growth appeared.
- Complementation of S. pombe Δsde2 by Sde2 domains. The experiment is as in (C). Expression constructs encoding Sde2 WT, the processing‐defective mutant sde2( AA KGG), Sde2UBL, Sde2‐C, and Sde2UBL and Sde2‐C together were used.
Source data are available online for this figure.
We generated a tagged version of Sde2, which contained epitope tags at both its N‐ and C‐termini, to detect the protein in S. pombe by immunoblot assays. These experiments revealed that the full‐length protein was cleaved, separating it into Sde2UBL and Sde2‐C (Fig 1B). After processing, Sde2UBL was diffusely localized in S. pombe whereas Sde2‐C was predominantly nuclear (Appendix Fig S1B). Processing of Sde2 is presumed to occur at the GG^KGG sequence as alanine substitutions of the first GG residues abolished cleavage (Fig 1B) and a processing‐defective Sde2 was nuclear (Appendix Fig S1B). Importantly, these alanine substitutions also failed to rescue growth defects of a Δsde2 strain (Fig 1C), suggesting that cleavage of Sde2 is essential for its function. By contrast, alanine mutations of the lysine or the second GG residues had negligible effects on processing and these mutants also rescued growth defects of Δsde2, albeit to a lesser extent than the wild type. Sde2 must be synthesized in the precursor form that should get processed for function, as de novo expressions of Sde2UBL did not complement growth defects in Δsde2 strain whereas Sde2‐C complemented the defects partially after overexpression (level of Sde2‐C when expressed from a plasmid using methionine codon was higher than Sde2‐C generated from its chromosomal location; Appendix Fig S1C). Co‐expression of the two domains did not improve growth of the deletion strain (Fig 1D). An extension with a non‐cleavable peptide upstream of Sde2‐C did not result in any complementation (Appendix Fig S1D). Thus, Sde2‐C is the functional unit whereas Sde2UBL plays a regulatory role.
To examine evolutionary conservation of Sde2 processing, an epitope‐tagged version of the human ortholog, C1orf55 (herein referred to as HsSde2) was generated. Similar to S. pombe, HsSde2 precursor was also cleaved into HsSde2UBL and HsSde2‐C in mammalian cells, alanine mutations of the first GG residues in the GGKGG motif of HsSde2 abolished its processing, and mutation of the lysine did not have any visible effect on processing (Fig EV1D).
Interestingly, the HsSde2‐C protein formed after processing of the GGAGG mutant precursor accumulated to higher levels than the protein formed from the wild‐type precursor (Fig EV1D; see below). Nevertheless, despite the similar post‐translational processing of Sde2 in S. pombe and humans, these orthologs retain organism‐specific features, as expression of HsSde2 in S. pombe Δsde2 strain failed to complement its growth defects. Furthermore, the two domains of HsSde2 were not interchangeable with the S. pombe counterparts, as a domain‐swapped S. pombe–human Sde2 chimera could not complement Δsde2 (Appendix Fig S1E).
Ubiquitin‐specific proteases Ubp5 and Ubp15 process Sde2
To identify Sde2 processing enzymes, we monitored accumulation of its precursor in selected mutants of S. pombe proteases by immunoblot assays. The precursor accumulated in a strain lacking ubp15, a USP domain‐containing DUB (Appendix Fig S2A). Residual Sde2‐C in the Δubp15 strain suggested involvement of additional enzymes for the processing. It was previously reported that ubp15 genetically interacts with ubp5, with the double mutant showing synthetic sickness (Richert et al, 2002). Processing of Sde2 was completely abolished in a Δubp5 Δubp15 double mutant (Fig 2A and B). To monitor sub‐cellular location of Sde2 processing, we generated Sde2UBL–GFP chimera, with a nuclear localization signal (NLS) or a nuclear export signal (NES) at the C‐terminus of GFP. An inefficient processing of the Sde2UBL–GFP–NES chimera in S. pombe suggests that Sde2 processing takes place in the nucleus (Fig 2C). Ubp5 and Ubp15 are generally involved in processing of ubiquitin and ubiquitin conjugates (Kouranti et al, 2010), and clearly, Sde2 is not the only substrate of Ubp5 and Ubp15; accordingly, the Δubp5 Δubp15 strain showed more severe growth defects than the Δsde2 strain (Appendix Fig S2B).
Figure 2. Ubiquitin‐specific proteases Ubp5 and Ubp15 process Sde2.
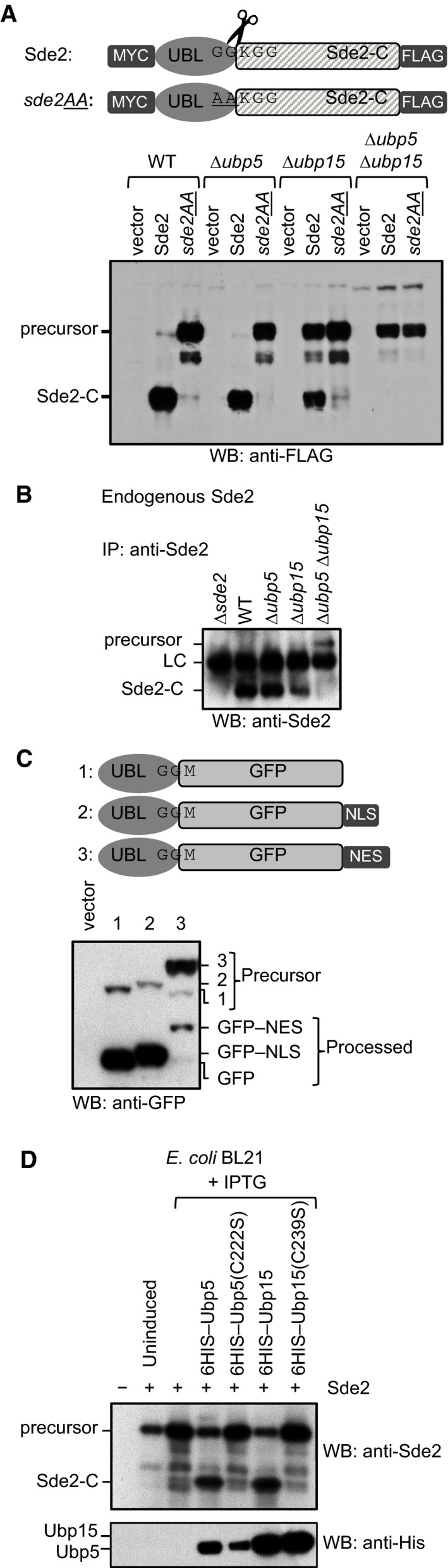
- Sde2 processing in Schizosaccharomyces pombe. Constructs from Fig 1B were expressed in S. pombe WT, ∆ubp5, ∆ubp15 and ∆ubp5 ∆ubp15 strains. The protein expression was analysed by anti‐FLAG immunoblotting.
- Endogenous S. pombe Sde2‐C and the full‐length Sde2 precursor in ∆ubp5 ∆ubp15 strain. Sde2 protein from indicated strains was immunoprecipitated using a polyclonal antibody against recombinant Sde2‐C, followed by Western blot analysis using same antibody.
- Sde2 is processed in the nucleus. Anti‐GFP immunoblot analysis of S. pombe expressing Sde2UBL–GFP, and its variants with a nuclear localization signal (NLS) or a nuclear export signal (NES). The non‐nuclear NES version was poorly processed.
- Processing of S. pombe Sde2 in bacteria. Expression constructs harbouring indicated cDNAs were co‐transformed in Escherichia coli BL21(DE3) strain. Following protein expression, total cell lysates were processed by immunoblotting using anti‐Sde2‐C antibody. Anti‐HIS immunoblotting was done to monitor expression of the proteases and its mutants.
Source data are available online for this figure.
To further test whether Ubp5 and Ubp15 acted directly as the proteases, we examined processing of SpSde2 by these DUBs in the recombinant system of Escherichia coli. Efficient processing of Sde2 was readily observed in E. coli upon co‐expression of Ubp5 and Ubp15, but not their catalytically inactive cysteine mutants (Fig 2D). Activity of these proteases on Sde2 is highly specific, as another USP domain‐containing DUB, Ubp16, did not process Sde2 (Fig EV2A). Although S. cerevisiae lacks Sde2, it contains an Ubp15 ortholog. Nevertheless, SpSde2 was poorly processed in wild‐type S. cerevisiae, presumably because SpUbp15 and ScUbp15 share only 44% identity. Overexpression of ScUbp15 enabled detection of processed SpSde2‐C in S. cerevisiae. Importantly, increased efficiency of SpSde2 processing was apparent upon co‐expression of either SpUbp5 or SpUbp15 in S. cerevisiae, highlighting their roles in processing of SpSde2 (Appendix Fig S2C).
Figure EV2. Replacement of Sde2 UBL fold with other ubiquitin‐like folds.
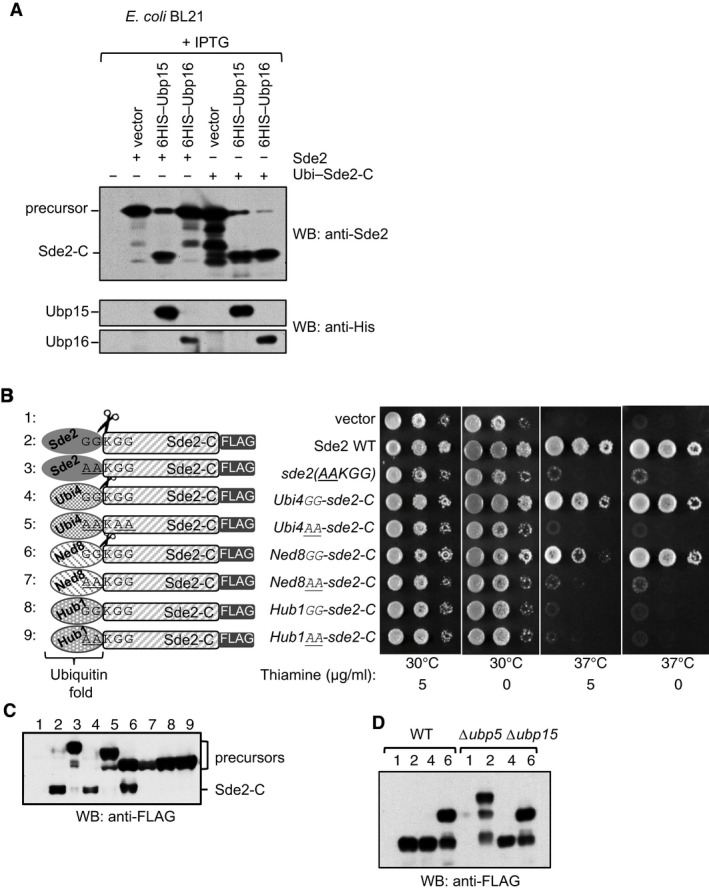
- A USP domain‐containing DUB Ubp16 does not process Sde2. Experiment similar to Fig 2D. Processing of ubiquitin–Sde2‐C fusion by the proteases was used as control. Anti‐HIS immunoblotting was used to monitor expression of the proteases.
- Sde2UBL can be replaced with the UBLs ubiquitin and Ned8. Complementation of growth defects of Schizosaccharomyces pombe Δsde2 strain with indicated fusion constructs. All constructs with a C‐terminal 3FLAG tag were expressed under the thiamine‐repressible nmt81 promoter. Expressed proteins are N‐terminal fusions of the ubiquitin‐folds of various UBLs (Ubiquitin/Ubi4, Ned8 or Hub1) with Sde2‐C. The GGKGG motif was kept at the junctions in the fusion proteins. After processing of the UBLs, Sde2‐C would start with a lysine.
- Anti‐FLAG immunoblot assay of fusion proteins shown in (B).
- Processing of ubiquitin‐ and Ned8–Sde‐C chimeras was not affected in Δubp5 Δubp15 double mutant. Expression of the clones used in (B) and indicated with numerical was monitored by immunoblotting using anti‐FLAG antibody.
Source data are available online for this figure.
We reasoned that if Sde2UBL was processed at the GG motif mainly for generating LysSde2‐C, then it should be replaceable with UBLs that are similarly processed. Therefore, we generated fusions of Sde2‐C with S. pombe ubiquitin, Ned8 (NEDD8 ortholog) or Hub1 (used here as a control UBL that is not processed). The GGKGG sequence was kept at the junction of the chimera. Ubiquitin and Ned8, but not Hub1, chimeras of LysSde2‐C complemented growth defects of the Δsde2 strain (Fig EV2B). Ubiquitin‐ and Ned8‐specific proteases processed respective fusions to form the functional LysSde2‐C (Fig EV2C). Processing of ubiquitin–Sde2‐C and Ned8–Sde2‐C fusions was not affected in Δubp5 Δubp15 double mutant (Fig EV2D). Since Hub1 does not get processed, its chimera was unable to generate functional Sde2‐C. Similar to the processing‐defective mutants of Sde2, alanine mutations of the GG in the chimeras were not processed and remained non‐functional.
Sde2‐C is a substrate of the N‐end rule pathway
To monitor steady‐state levels of wild‐type and mutant Sde2 proteins, sequence encoding 6HA epitope tag was inserted chromosomally at the C‐terminus of sde2 variants. MetSde2‐C formed after processing of the GGMGG mutant precursor accumulated to higher levels than the LysSde2‐C formed from the wild‐type GGKGG protein (Fig 3A and B). Interestingly, the level of MetSde2‐C after de novo translation using methionine codon was lower than the MetSde2‐C formed after processing of the GGMGG mutant precursor. However, the differences in protein levels were not due to variations in transcription, as all strains showed comparable levels of sde2 mRNA in reverse transcription quantitative PCR (RT–qPCR) assays (Appendix Fig S3A). Thus, the presence of Sde2UBL facilitates the expression of Sde2‐C protein. Both strains with MetSde2‐C showed strong growth defects; however, growth of the strain with MetSde2‐C formed after processing of the GGMGG mutant was better than with MetSde2‐C from de novo translation (Fig 3C).
Figure 3. Sde2‐C is made short‐lived by the N‐end rule pathway.
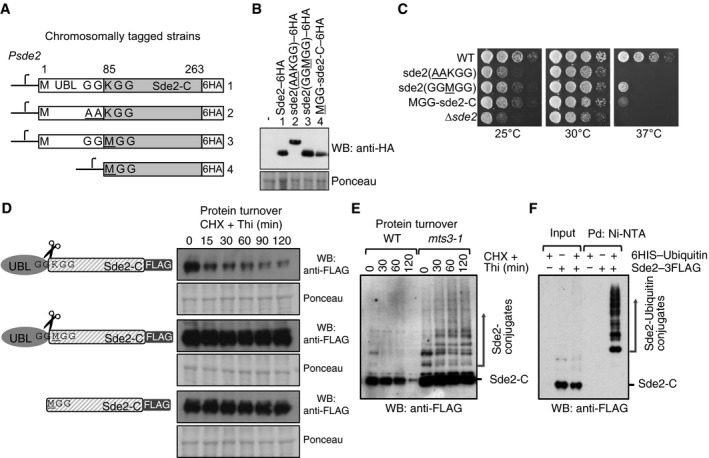
- Chromosomal 6HA epitope‐tagged sde2 variants. The sde2 promoter is common to all variants.
- Western blot with anti‐HA antibody to detect steady‐state levels of wild‐type and mutant Sde2 proteins.
- Growth phenotype of strains in (A). Strains with MetSde2‐C proteins, formed either after processing of GGMGG mutant or translated de novo, show ∆sde2‐like growth defects.
- Constructs used to monitor steady‐state level and turnover of the proteins. Plasmids contain 3FLAG epitope tags at the C‐termini of Sde2 under nmt81 promoter. Protein turnover assays. The Sde2 variants with C‐terminal 3FLAG tag were expressed in Δsde2 strain from a plasmid under nmt81 promoter. In Sde2 WT, after processing Sde2‐C starts with lysine, in GGMGG mutant with methionine and Met–Sde2‐C is de novo translated. Total proteins from 1.0 OD600 cells for the given time points were run on SDS–PAGE followed by anti‐FLAG Western blotting.
- WT Sde2 is stable in the proteasome mutant mts3‐1. Assay is similar to (B). Higher molecular weight adducts detected with anti‐FLAG antibody likely represent ubiquitinated Sde2.
- Denaturing Ni‐NTA pull‐down (Pd) of ubiquitin conjugates from Schizosaccharomyces pombe expressing Sde2–3FLAG. Ubiquitination of Sde2 is detected by anti‐FLAG immunoblotting.
Source data are available online for this figure.
The lower levels of S. pombe and human LysSde2‐C (Figs 3D and EV1D) could be because lysine residue at the amino‐terminus of proteins is descriptive of a type 1 degron in the N‐end rule pathway (Bachmair et al, 1986; Sriram et al, 2011). Thus, we tested whether Sde2‐C was a substrate of this pathway by measuring its turnover rates. Indeed, the LysSde2‐C protein was short‐lived, whereas the MetSde2‐C proteins were significantly stabilized (Fig 3D). Schizosaccharomyces pombe deletion of the N‐end rule ligase ubr11 (Fujiwara et al, 2013) partially stabilized LysSde2‐C (Appendix Fig S3B). The mutation of the 19S proteasome regulatory particle subunit mts3 (Gordon et al, 1996) stabilized LysSde2‐C, and higher molecular weight adducts of Sde2 observed in this mutant (Fig 3E) were confirmed to be ubiquitin conjugates of Sde2 by pull‐down assays of the conjugates under denaturing conditions (Fig 3F). Ubiquitination of Sde2 was diminished but not abolished in a Δubr11 strain (Appendix Fig S3C), indicating involvement of additional ligase(s) for degradation of Sde2‐C.
To elucidate role of the conserved lysine in GGKGG motif in Sde2 orthologs, we generated substitution mutations with each of the other 19 amino acids and measured the processing efficiency, half‐life and complementation capacity of each of the resulting proteins. All amino acid variants, excluding the version with a proline GGPGG, underwent efficient processing (Fig EV3; Appendix Fig S3D and E), indicating that the lysine is not required for processing. In assays to measure proteins half‐life, the MetSde2‐C variant was found to be most stable, the ArgSde2‐C variant was the least stable, and the wild‐type LysSde2‐C had an intermediate stability. Function‐wise, growth defects of the Δsde2 strain could be best complemented by the wild‐type LysSde2‐C followed by the TrpSde2‐C and GlySde2‐C variants. By contrast, the CysSde2‐C, GluSde2‐C and AspSde2‐C variants showed the weakest complementation. The GGPGG mutant of Sde2 was defective for processing and failed to complement Δsde2 (Fig EV3; Appendix Fig S3D and E). Similar defects in the processing of a ubiquitin–proline–β‐galactosidase chimera were reported in S. cerevisiae (Bachmair et al, 1986). Thus, the N‐end rule pathway controls the levels of LysSde2‐C by its turnover in the proteasome.
Figure EV3. Sde2‐C is a substrate of the N‐end rule pathway.
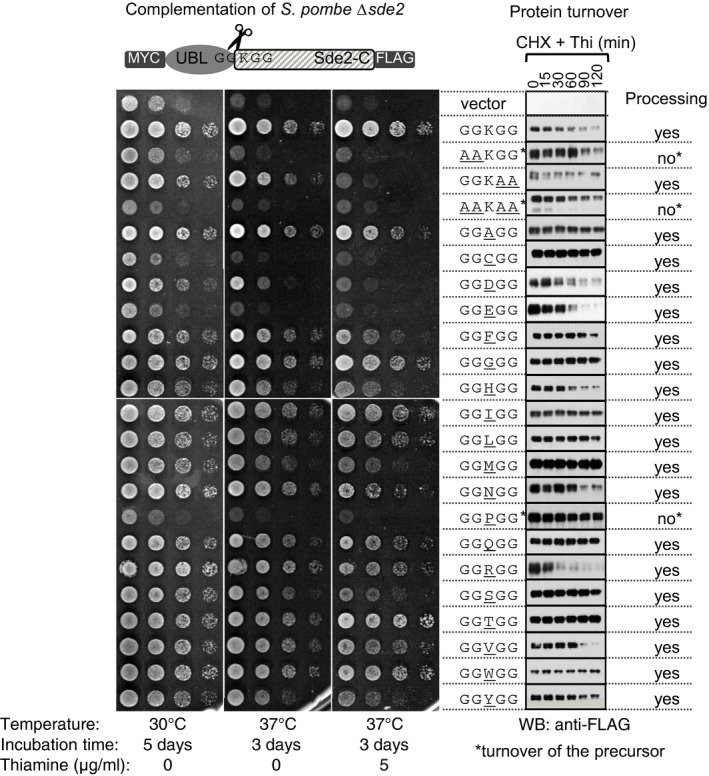
Complementation of growth defects of Schizosaccharomyces pombe Δsde2 strain (left) with indicated constructs (middle), and protein turnover assays (right). The constructs are as in Fig 1B. Spot assay was performed as in Fig 1C. For protein turnover assays, transcription from the nmt81 promoter was repressed by adding 1.7 μg/ml thiamine, and translation was halted by 100 μg/ml of cycloheximide. Cells harvested from equal volume of cultures at indicated time intervals were processed for immunoblot analysis with anti‐FLAG antibody. Asterisks mark Sde2 variants defective in processing; therefore, in these cases, turnover of the precursor has been analysed. Source data are available online for this figure.
Processing of Sde2UBL promotes association of Sde2‐C with the spliceosome
To better understand the cellular role of Sde2, mass spectrometry was used to identify proteins that co‐immunoprecipitated (Co‐IP) with an epitope‐tagged version of Sde2 (Appendix Fig S4A). Here, we confirmed association between Sde2 and components of the spliceosome, as recently reported by others (Bayne et al, 2014; Chen et al, 2014). Co‐IP assays using chromosomally epitope‐tagged versions of multiple Cdc5/Prp19 complex subunits, including Cdc5, Prp19 and Isy1, further validated these interactions (Fig 4A). Notably, the level of Sde2 protein detected in these experiments appears lower than the subunits of the Cdc5 complex (Appendix Fig S4B), suggesting that Sde2 functions substoichiometrically within the spliceosome.
Figure 4. Processing of Sde2UBL promotes interaction of Sde2‐C with the spliceosomal factors.
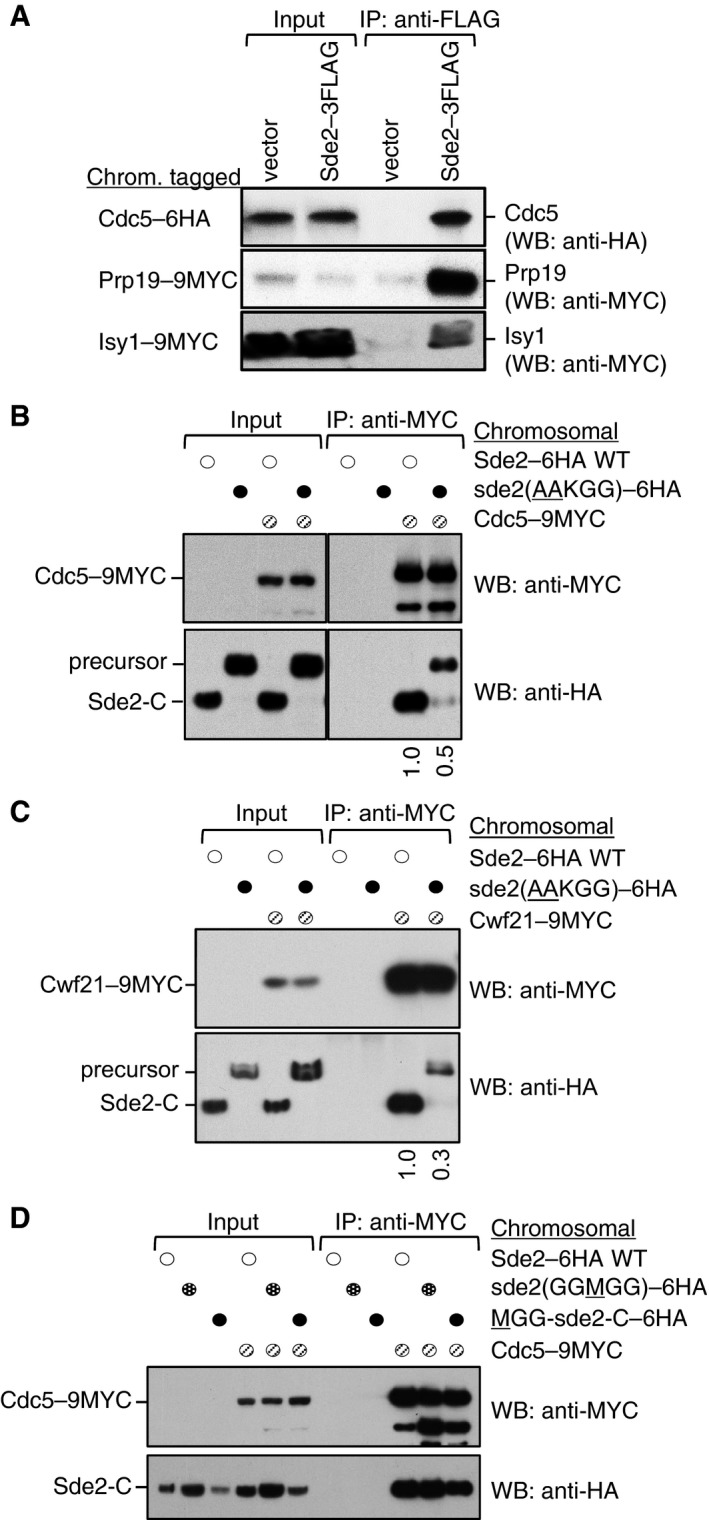
- Sde2 interacts with the spliceosomal Prp19 complex factors Cdc5, Prp19 and Isy1 in vivo. sde2 gene was 3FLAG‐tagged, and cdc5–6HA, prp19–9MYC and isy1–9MYC genes were tagged chromosomally.
- Co‐IP assay to monitor association of the processing‐defective Sde2 with Cdc5. Experiments are performed with proteins expressed at their endogenous levels by using chromosomally tagged strains with given epitopes at the C‐termini of indicated genes. IP was performed using anti‐MYC agarose resins followed by immunoblot assays with anti‐MYC antibody to monitor IP efficiency of Cdc5, and with anti‐HA antibody to monitor Co‐IP of Sde2 versions. Numbers below anti‐HA blot indicate ratio of Sde2 to Cdc5 (HA/MYC) signals obtained from ImageJ quantification of immunoblot signals. Cdc5 association of the processing‐defective Sde2 precursor is diminished to half.
- Co‐IP assay to monitor association of the processing‐defective Sde2 with Cwf21. Assay is similar to (B). Cwf21 association of the processing‐defective Sde2 precursor is diminished to one‐third.
- Co‐IP assay to monitor association of MetSde2‐C with Cdc5. Assay and quantitation is similar to (B). MetSde2‐C generated from processing of GGMGG mutant precursor or after translation de novo does not show obvious defects in spliceosomal association.
Source data are available online for this figure.
We tested the ability of processing‐defective and lysine mutants of Sde2 for interaction/incorporation in spliceosomes. For this purpose, we performed Co‐IP assays of these mutants with core spliceosomal factors Cdc5 and Cwf21. To test the interaction at the endogenous levels of the proteins, we chromosomally inserted a sequence encoding 9MYC epitope tag at the C‐terminus of Cdc5 and Cwf21 in the chromosomal Sde2–6HA variants shown in Fig 3A. By contrast to the LysSde2‐C, which associated efficiently with Cdc5 and Cwf21, Co‐IP of the precursor AAKGG was strongly diminished (Fig 4B and C). Since Sde2 in the AAKGG strain remains mainly in the precursor form, the residual spliceosomal association of this precursor (Fig 4B and C) might have happened because of the lack of active Sde2‐C. To address this, we performed Cdc5 Co‐IP of a partially processed GAKGG mutant of Sde2, which produces similar levels of precursor and processed proteins; herein, only the processed Sde2‐C associated with Cdc5 (Fig EV4). Thus, spliceosomal association of a small amount of Sde2 precursor is possible, but only in the absence of processed Sde2‐C. Moreover, Sde2UBL appears inhibitory for the precursor's association with the spliceosome, as Cdc5 Co‐IP of free MetSde2‐C, formed either after processing of the GGMGG precursor or from de novo translation, remained largely unaffected (Fig 4D). These results suggested that processing of Sde2UBL facilitates incorporation of LysSde2‐C in the spliceosome, wherein the conserved lysine may not be critical for this association.
Figure EV4. Sde2 UBL is inhibitory for the incorporation of Sde2‐C into the spliceosome.
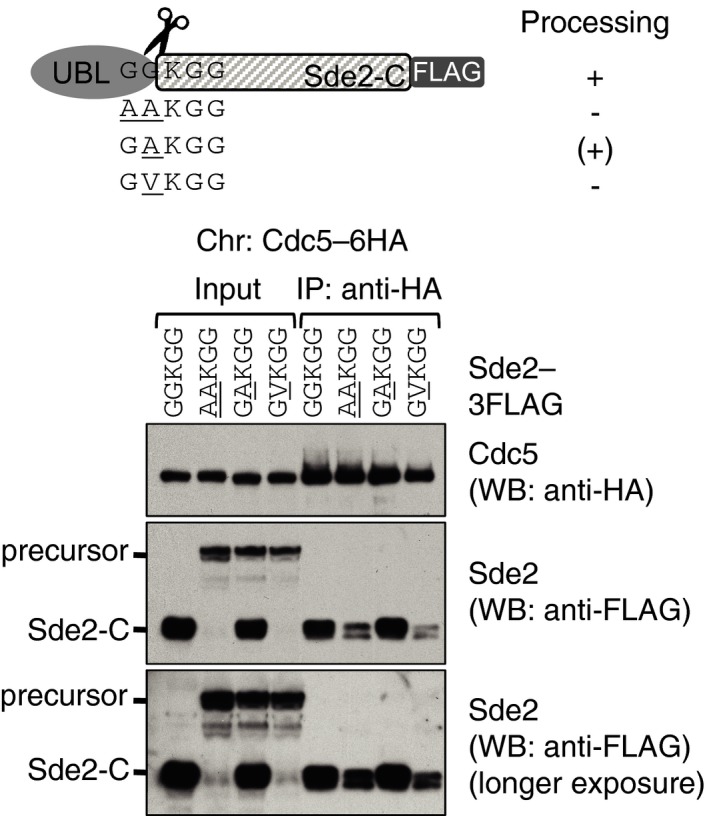
Sde2 variants in the schematics were transformed into Schizosaccharomyces pombe Δsde2 with Cdc5‐6HA chromosomal tag. Anti‐HA IP was carried out to monitor Co‐IP of Sde2‐C and precursor. Only processed Sde2‐C, but not the precursor, associates with Cdc5. Source data are available online for this figure.
Sde2‐C is an intron‐specific pre‐mRNA splicing factor
To elucidate the role of Sde2 in pre‐mRNA splicing, we analysed splicing defects in Δsde2 strain using S. pombe genome‐wide splicing‐sensitive microarrays (Inada & Pleiss, 2010). The arrays have oligonucleotide probes specific for exons, introns and exon–exon junctions for nearly all intron‐containing genes in S. pombe. Since the Δsde2 strain showed temperature‐sensitive growth defects, we analysed cells grown at 30°C or after shifting to 37°C for 15 min to detect early splicing defects. As seen in Fig 5A, splicing of most intron was unaffected in Δsde2 strain under either growth condition, as evidenced by the lack of apparent changes in pre‐mRNA levels. Interestingly, however, splicing defects were observed for nearly 30 genes, where pre‐mRNA levels were increased by at least twofold, and were often accompanied by decrease in the levels of mature mRNA (Fig 5A inset).
Figure 5. Sde2 is required for excision of selected introns from a subset of pre‐mRNAs.
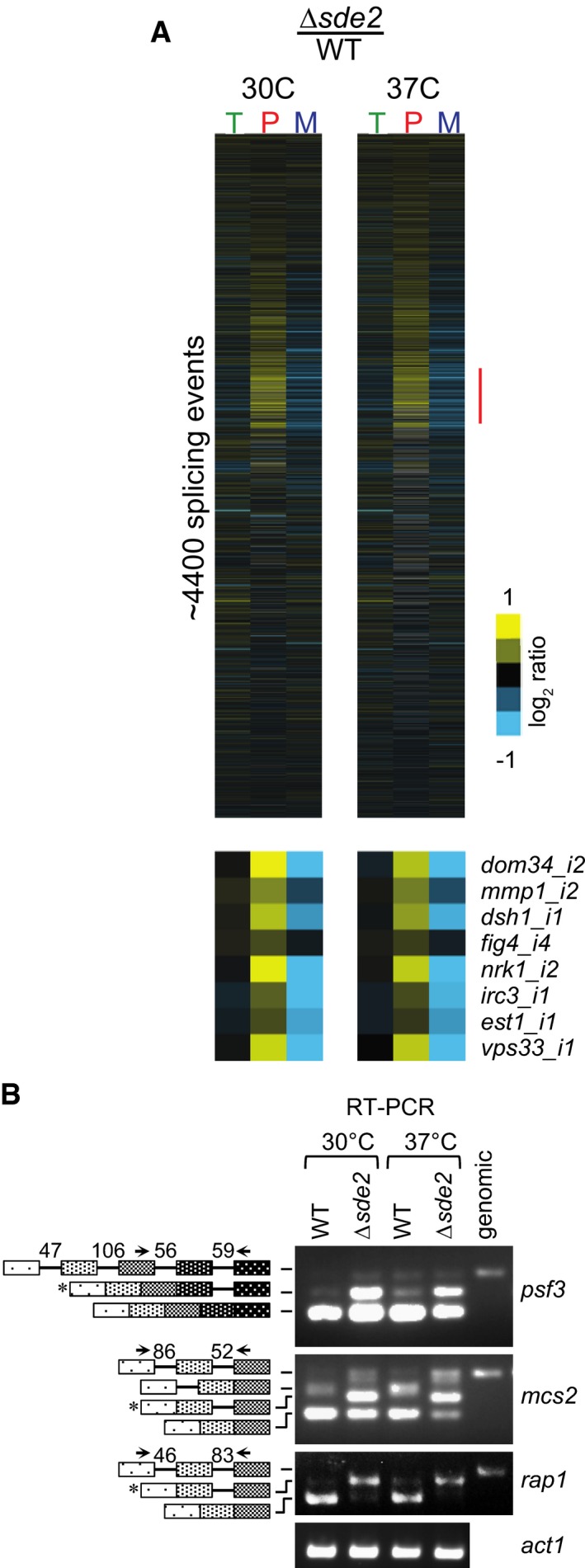
- Analysis of total RNA from Schizosaccharomyces pombe WT and Δsde2 strain using splicing‐sensitive microarray. Microarray heat‐map shows log2 Δsde2/WT ratio of signals for total transcripts (T), intron‐containing transcripts (P) and spliced transcripts (M). 37°C temperature shift was performed for 15 min to monitor early splicing defects. The experiment was repeated with dyes swapped. Yellow colour represents accumulation, black denotes no change, and blue shows reduction of signal in Δsde2.
- Semi‐quantitative RT–PCR shows intron‐containing transcripts for psf3, mcs2 and rap1. cDNA prepared from total RNA isolated from S. pombe WT and Δsde2 mutant at 30°C and 37°C was analysed by PCR. The block diagram (not drawn to the scale) shows exons and introns. Arrows mark primers used for amplification. The numbers above introns (shown as lines in block diagrams) mark their lengths. PCR band from genomic DNA (genomic) corresponds to the pre‐mRNA. RT–PCR of act1 (actin) pre‐mRNA is used as control. Asterisks mark intron‐containing transcripts getting enriched in Δsde2.
Source data are available online for this figure.
We confirmed splicing defects of selected candidates in Δsde2 by RT–PCR assays, followed by agarose gel electrophoresis to visualize intron‐containing transcripts (Fig 5B; Appendix Fig S5A and B). Key targets of Sde2 include multi‐intronic genes rap1 (encodes a telomere binding protein with functions in telomeric silencing), psf3 (encodes a GINS complex subunit with functions in DNA replication), and mcs2 (encodes a TFIIH complex cyclin with functions in transcription). Strikingly, for all these targets one of the RT–PCR bands in Δsde2 sample could not correspond to the products expected from either fully spliced or fully unspliced mRNAs. Consistent with the array analysis for these targets (Table EV1), gel‐purification and sequencing of these bands showed transcripts having retained only one of the intron (for rap1 intron‐2, psf3 intron‐4, and mcs2 intron‐2), accumulated in Δsde2 (Fig 5B; Appendix Fig S5C). The Sde2‐C target introns are enriched with non‐canonical 5′‐ and 3′‐splice sites (Table EV2). Nevertheless, a common feature or sequence element was not obvious in affected pre‐mRNAs or introns. The respective cDNAs of the three targets could not complement growth defects of Δsde2 strain (Appendix Fig S5D); thus, the defects appear to have arisen from cumulative splicing defects of multiple targets.
The hub1‐1 mutant defective in pre‐mRNA splicing (Mishra et al, 2011) showed distinct splicing defects for above transcripts, as different introns of mcs2 and rap1 were retained in this mutant (Fig EV5A). Thus, Sde2 and Hub1 do not seem to have overlapping functions in pre‐mRNA splicing, and the negative genetic interaction between their mutants (Fig EV1A and B) likely resulted from splicing defects of a larger number of transcripts in the double mutant. We reasoned that since LysSde2‐C has rapid turnover rate, overexpression of its stable versions could be toxic and/or dominant‐negative in the cell. Indeed, overexpression of MetSde2‐C or AlaSde2‐C proteins resulted in slow growth phenotypes in hub1‐1 strain (Appendix Fig S6A). This phenotype seems to arise from Δsde2‐like pre‐mRNA splicing defects in hub1‐1 strain (Appendix Fig S6B) indicating that stable forms of Sde2 are dominant‐negative in the cell.
Figure EV5. Sde2 and Hub1 have distinct splicing targets and N‐terminal lysine of Sde2‐C is crucial for interaction with Cactin.
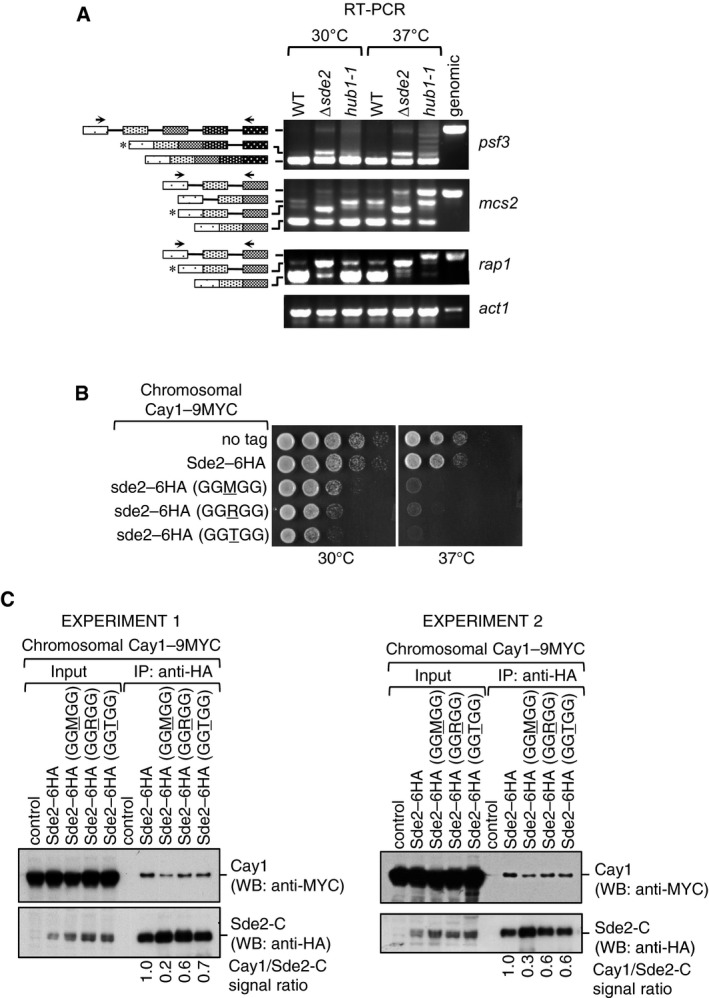
- RT–PCR assays to monitor accumulation of intron‐containing transcripts in Δsde2 and hub1‐1 mutants. Assay is similar to Fig 5B.
- Growth phenotypes of chromosomal Sde2 lysine mutants on rich media at 30°C and 37°C.
- Association of Cactin/Cay1 with lysine mutants of Sde2‐C. Experiments are performed with proteins expressed from chromosomally tagged strains. IP was performed using anti‐HA beads followed by immunoblot assays with anti‐MYC antibody to monitor Co‐IP of Cay1. Anti‐HA antibody was used to monitor IP efficiency of Sde2 mutants. Numbers indicate ratio of Cay1 to Sde2 (MYC/HA) signals obtained from ImageJ quantification of immunoblots. Results from two independent biological replicate experiments are shown.
Source data are available online for this figure.
Sde2 processing is needed for telomeric silencing and genome stability
A functional link between Sde2 and its proteases Ubp5/Ubp15 for intron‐specific pre‐mRNA splicing was confirmed from Δsde2‐like splicing defects observed in the deletion strain of the proteases (Fig 6A). The processing of Sde2 precursor and the lysine at the N‐terminus of Sde2‐C both are essential for the splicing function, as respective mutants also had Δsde2‐like splicing defects (Fig 6B). Sde2‐specific function in telomeric silencing and mini‐chromosome stability was reported previously (Sugioka‐Sugiyama & Sugiyama, 2011). Supporting a role for Sde2 in telomeric silencing, we observed aberrant overexpression of the telomeric DNA helicases tlh1, tlh2 and SPAC212.06c in Δsde2 in our microarrays, which was further confirmed by RT–qPCR assays (Fig 7A). As expected from the small in number yet functionally related targets of Sde2, the defects in telomeric silencing in Sde2 mutants could not be assigned to the splicing defect of a single gene (Appendix Fig S7B and C; Table EV1 and online microarray data). Importantly, the processing‐defective and lysine mutants of Sde2 also showed Δsde2‐like defects in telomeric silencing and mini‐chromosome stability assays (Fig 7B and C), thus revealing a role of ubiquitin‐like processing for intron‐specific pre‐mRNA splicing and associated telomeric silencing and genomic stability.
Figure 6. Sde2 processing in pre‐mRNA splicing.
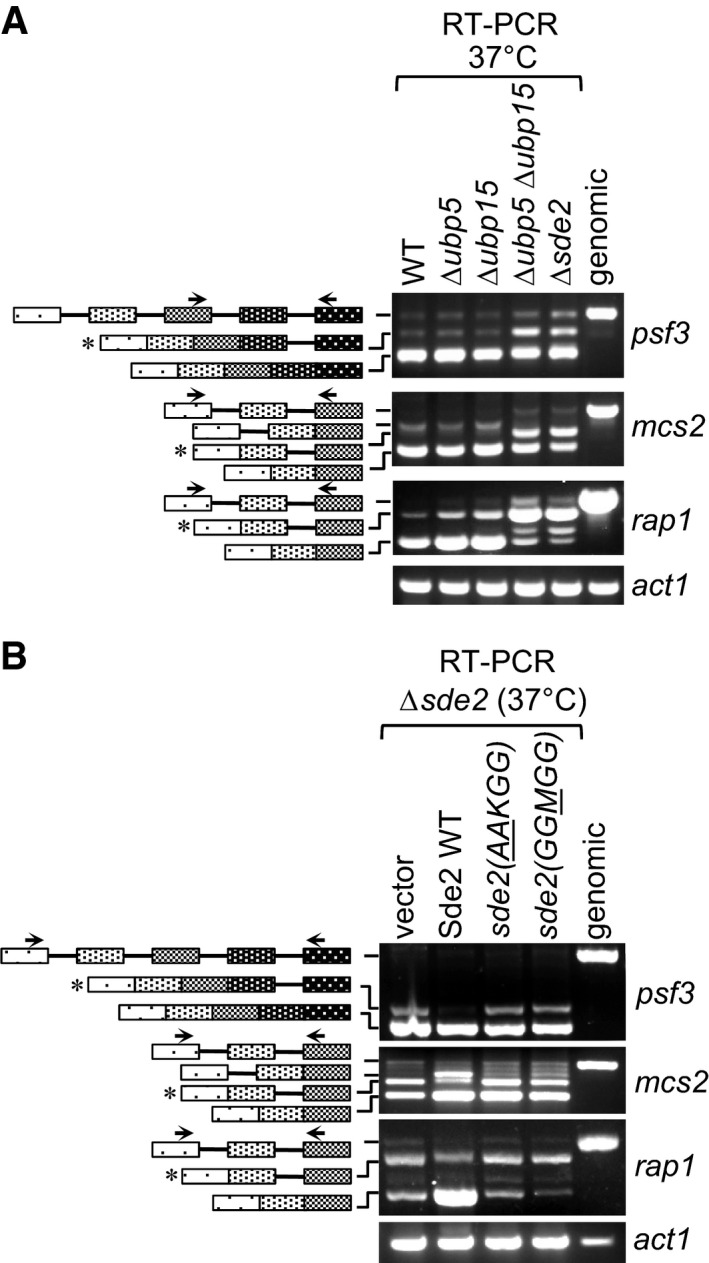
- Δsde2‐like intron‐specific splicing defect in Δubp5 Δubp15 mutants defective in Sde2 processing. The details are as in Fig 5B.
- Intron‐specific splicing defects in processing‐defective and lysine mutants of Sde2. Schizosaccharomyces pombe Δsde2 strain was transformed with vector, Sde2 WT, sde2( AA KGG) and sde2(GG M GG). The details are as in Fig 5B.
Source data are available online for this figure.
Figure 7. Sde2 processing in telomeric silencing and genome stability.
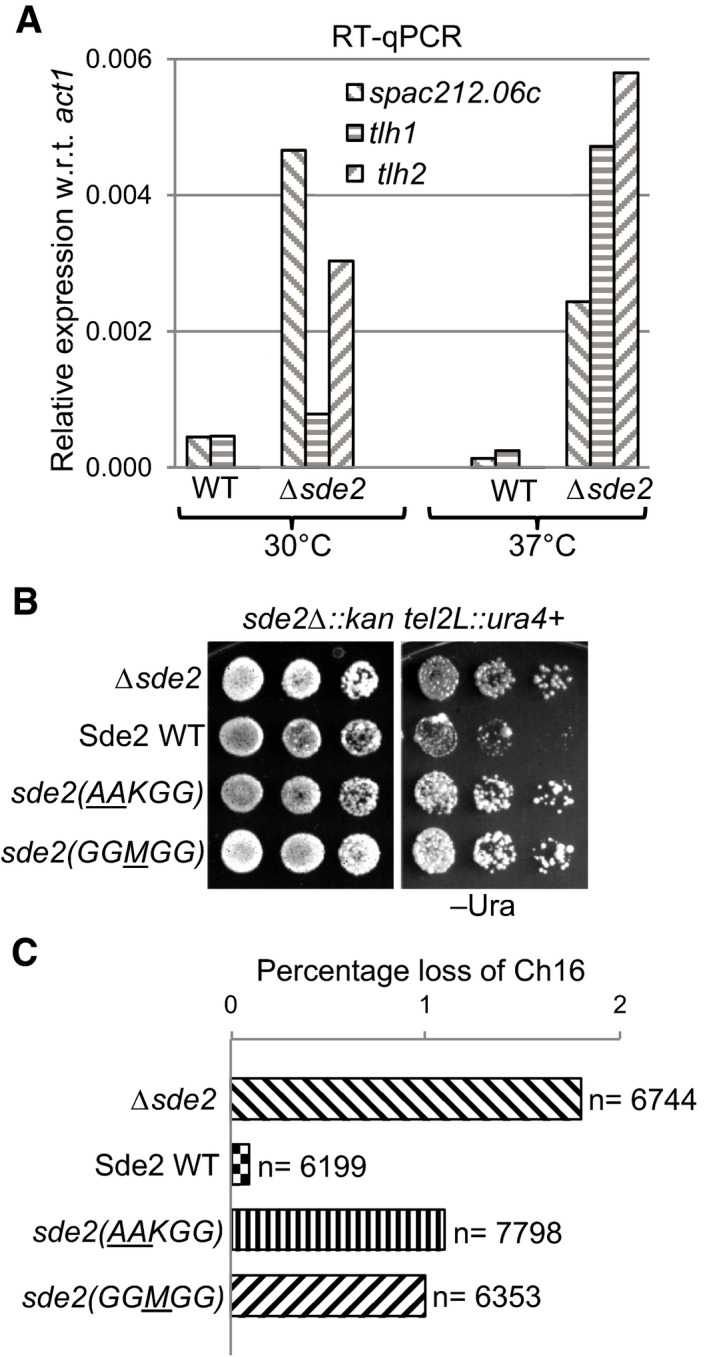
- Aberrant overexpression of telomeric transcripts in Sde2 mutant. Expression levels of telomeric transcripts tlh1, tlh2 and SPAC212.06c were quantitated by RT–qPCR assays in WT, and Δsde2 strain at 30°C and 37°C is normalized with act1.
- Telomeric silencing assay in Sde2 mutants. ura4 + gene inserted at the telomere in Δsde2 strain (Sugioka‐Sugiyama & Sugiyama, 2011) was used as a marker to assay telomeric silencing on indicated agar plates. Suppression of growth on –Ura plate indicates silencing of the reporter at the telomeres.
- Mitotic chromosome stability assay to assess genomic instability in Sde2 mutants. Percentage loss of chromosome 16 in Δsde2 strain (Sugioka‐Sugiyama & Sugiyama, 2011) transformed with Sde2 variants is plotted (n = total number of colonies counted). Processing‐defective and lysine mutants of Sde2 show elevated loss of the chromosome.
Source data are available online for this figure.
Sde2‐C facilitates association of Cay1 with spliceosomes
To understand the mechanism of Sde2 in the spliceosome, we purified and compared spliceosomes from wild‐type and Δsde2 strains harbouring chromosomal Cdc5‐6HA and Prp19‐6HA tags by mass spectrometry. We found that, whereas the composition of spliceosomes in the two strains was almost identical, peptides for Cactin/Cay1 were diminished in spliceosomal purifications from Δsde2 strains (Fig 8A; Table EV3). We confirmed Sde2‐C dependent association of Cay1 with spliceosomes by Co‐IP experiments (Fig 8B). An epistatic relationship between sde2 and cay1 (Fig 8C) and similar pre‐mRNA splicing defects in single and double mutants (Fig 8D) suggest that Cactin also functions as an intron‐specific splicing regulator. A role of S. pombe Cactin in splicing of rap1 pre‐mRNA was reported (Lorenzi et al, 2015), but its spliceosomal association and intron‐specific splicing function was not uncovered.
Figure 8. Sde2‐C facilitates association of Cay1 with spliceosomes.
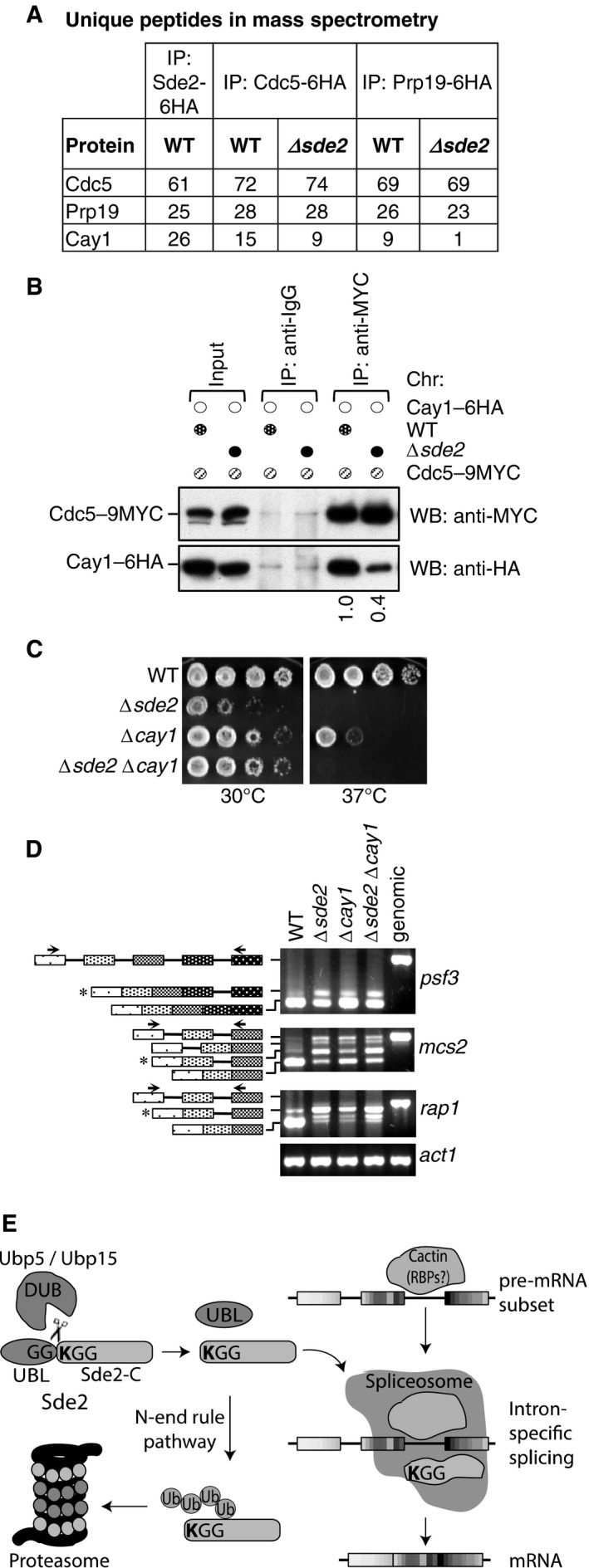
- Sde2 Co‐IPs with Cactin in the spliceosome. Cdc5–6HA and Prp19‐6HA complexes were immunoprecipitated using anti‐HA antibody beads from Schizosaccharomyces pombe lysates wild‐type and Δsde2 strains. Co‐IP proteins were analysed by mass spectrometry. The table shows number of unique peptides obtained for each protein in mass spectrometry.
- Sde2 facilitates recruitment of Cay1 to the spliceosome. Assay is similar to (A). Lysates from S. pombe wild‐type and Δsde2 strain with Cdc5–9MYC and Cay1–6HA tagged were immunoprecipitated using anti‐MYC antibody beads. Co‐IP of Cay1–6HA was analysed by anti‐HA Western blot. Numbers below anti‐HA blot indicate ratio of Cay1 to Cdc5 (HA/MYC) signals obtained from ImageJ quantification of immunoblot signals.
- Sde2 and Cay1 are epistatic. Fivefold serial dilution spotting of indicated mutant strains was done on rich media. Plates were incubated at 30°C and 37°C until growth appeared.
- Cay1 and Sde2 knockout strains have similar pre‐mRNA splicing defects. Assay is similar as in Fig 5B.
- Schematics for Sde2 function and regulation.
Source data are available online for this figure.
Turnover rates of lysine mutants of LysSde2‐C did not correlate well with their ability to complement growth defects in Δsde2 strain (Fig EV3). While the lysine was not critical for the association of Sde2‐C with spliceosomes (Fig 4D), we tested whether the lysine is needed for specific association with Cay1. Indeed, Co‐IP of Cay1 with MetSde‐C was strongly diminished and with ArgSde2‐C and ThrSde2‐C was weaker than the wild‐type LysSde‐C (Fig EV5B and C). Thus, the extent of complementation of Δsde2 by the lysine mutants of Sde2‐C appears to result from two activities, turnover by the N‐end rule pathway and ability to associate with specific pre‐mRNA splicing factors (Fig 8E).
Discussion
The DUBs with dual specificity for Sde2UBL and ubiquitin
We have found that the cell has used ubiquitin‐like processing to obtain an intron‐specific pre‐mRNA splicing regulator, which needs to have a non‐methionine amino acid as its amino end. Multiple lines of evidences confirmed that Sde2 is a bona fide UBL. First, the Sde2UBLGG–Sde2‐C precursor is cleaved at the di‐glycine motif by USPs Ubp5 and Ubp15. Second, Sde2UBLGG–GFP fusions were processed like Sde2 precursor. Third, cleavable ubiquitin and Ned8 could functionally replace Sde2UBL, whereas the processing‐deficient UBL Hub1 could not.
The paralogs Ubp5 and Ubp15 are the two MATH/TRAF domain‐containing ubiquitin proteases in S. pombe, which were reported to process ubiquitin. These proteases display a dynamic localization pattern in the Golgi, cytosol and nucleus (Kouranti et al, 2010). Since Sde2 is processed in the nucleus, the nuclear pool of these proteases would be more relevant for the activation of Sde2. The human ortholog of these DUBs, USP7/HAUSP, was reported to deubiquitinate and stabilize the tumour suppressor p53 (Li et al, 2002; Faesen et al, 2011) and its E3 ubiquitin ligase Mdm2 (Li et al, 2004). Their activities for both ubiquitin and Sde2 clearly show that both DUBs possess dual substrate specificity and process two distant UBLs with sequence identity less than 20%. The complete loss of Sde2 processing in S. pombe deleted of ubp5 and ubp15 highlights specificity of this process, as other nuclear DUBs could not cleave Sde2UBL. The intriguing activity of Ubp5 and Ubp15 to process two distant UBLs requires structural studies of Sde2UBL‐protease complexes to reveal the mechanism of catalysis. In support of conservation of Sde2 processing, Kim and colleagues have recently reported cleavage and degradation of human Sde2 in a PCNA‐dependent manner and its role in replication stress (Jo et al, 2016).
Sde2 precursor is similar to yet distinct from ubiquitin‐ribosomal fusions
Sde2UBL and its processing are important for proper expression of the spliceosomal Sde2‐C. This activity of Sde2UBL in the nucleus resembles the chaperone‐like function of ubiquitin for the expression of ribosomal moieties from ubiquitin–ribosomal fusion proteins in the cytoplasm (Finley et al, 1989; Graciet et al, 2006; Lacombe et al, 2009). As Sde2UBL can be replaced with ubiquitin and Ned8, Sde2UBL function appears to be dedicated to generate the spliceosomal LysSde2‐C. Although a common chaperone‐like activity for all the three UBLs cannot be ruled out, this interpretation needs further investigation. We have not detected any higher molecular weight adducts of S. pombe Sde2UBL , but non‐covalent functions of this UBL may exist in the cell.
The distinguishing feature of Sde2 orthologs is the conserved GGKGG motif, which is required for both processing and function. Processing of Sde2UBL has an activating role, as it is critical for interaction of LysSde2‐C with spliceosomes. Activated LysSde2‐C facilitates association of spliceosomes with Cactin/Cay1, a spliceosomal factor with Sde2‐like function in intron‐specific pre‐mRNA splicing. While the lysine does not appear crucial for the interaction of LysSde2‐C with spliceosomes, the residue is important for interactions with Cactin. The free lysine in LysSde2‐C also plays a regulatory role, as it makes the protein a natural substrate of the N‐end rule pathway of proteasomal degradation. Role of S. pombe Cactin for splicing of rap1 pre‐mRNA was reported previously (Lorenzi et al, 2015). Human Cactin functions in pre‐mRNA splicing through interactions with splicing factors DHX8 and SRRM2 (Zanini et al, 2017). Cactin orthologs were detected in spliceosomal purifications (Bessonov et al, 2008; Doherty et al, 2014), and it also appears to have mRNAs binding ability (Baltz et al, 2012). Cactin possibly recognizes selected pre‐mRNAs and facilitates their splicing with Sde2‐C containing spliceosomes. The lysine has been selected for optimum function and interaction of Sde2‐C in the spliceosome, as proteins neither with stabilizing residues nor with more destabilizing residues performed better than the wild type. To fine balance LysSde2‐C level, the proteasome likely clears off non‐spliceosomal pool of the protein by the N‐end rule pathway.
LysSde2‐C is a unique pre‐mRNA splicing regulator
LysSde2‐C appears to function differently from other pre‐mRNA splicing regulators of the spliceosome. LysSde2‐C promotes efficient excision of selected introns from selected transcripts in S. pombe, but it is not required for general pre‐mRNA splicing. LysSde2‐C thereby becomes a critical control factor for the expression of selected proteins, a majority of which function at the chromatin. Therefore, growth defects or drug sensitivities of Δsde2 strain could not be attributed to splicing defects of individual genes. The lack of any obvious common feature in Sde2 target pre‐mRNAs leads us to postulate that some RNA secondary structures could make their splicing Sde2 dependent. Importantly, Sde2‐specific proteases play a key regulatory role in pre‐mRNA splicing by processing the inactive Sde2 precursor to generate the active spliceosomal LysSde2‐C, as Sde2UBL is inhibitory for its incorporation into the spliceosome. The processing‐deficient and lysine mutants of Sde2 display Δsde2‐like pre‐mRNA splicing defect as well as defects in telomeric silencing and chromosome stability.
Splicing defects of a subset of pre‐mRNAs were reported in the deletion strain of tls1 (Wang et al, 2014). Specific pre‐mRNA splicing defects have also been reported for mutants of U1 snRNA gene (Alvarez et al, 1996), U2AF, prp18 and survival of motor neuron (SMN) gene (Webb et al, 2005; Campion et al, 2010; Vijaykrishna et al, 2016). We have previously reported an intron‐specific pre‐mRNA splicing defect of cdc2 intron‐3 in a hub1 mutant of S. pombe (Mishra et al, 2011). Nevertheless, splicing targets of above factors are distinct, and respective gene deletions in S. pombe are inviable; thus, these factors are likely to be important for general pre‐mRNA splicing. Interestingly, Sde2 orthologs could not be detected in intron‐poor yeasts; therefore, it is tempting to propose that intron‐loss during evolution (Hooks et al, 2014) might have resulted in the loss of Sde2 from these organisms.
In conclusion, the ubiquitin‐like processing of Sde2UBL generates the activated LysSde2‐C protein, which becomes functional in the spliceosome and plays a critical and selective role in pre‐mRNA splicing. This process bears intriguing analogy to the processing of ubiquitin precursors with ribosomal proteins. Thus, the occurrence of precursors with ubiquitin‐folds appears to be a conserved principle for both the ribosome and the spliceosome—the two major RNP complexes known to have certain similarities in their mechanisms of action (Konarska & Query, 2005; Staley & Woolford, 2009). Extending our current findings with Sde2, it is tempting to postulate that ubiquitin processing activates the ribosomal proteins for translation of selective mRNAs.
Materials and Methods
Strains, plasmids and DNA techniques
Schizosaccharomyces pombe strains used in this study are listed in Appendix Table S1, and plasmids are listed in Appendix Table S2. Strains for centromeric and telomeric silencing and mitotic chromosome loss assays in Δsde2 strain were reported previously (Sugioka‐Sugiyama & Sugiyama, 2011) and obtained from NBRP‐yeast, Japan. Preparation of S. pombe competent cells, transformation, chromosomal tagging, gene deletion and isolation of total proteins for Western blot (WB; also referred to as immunoblot) assays by trichloroacetic acid (TCA) precipitation method was done following published protocols for S. cerevisiae (Knop et al, 1999; Janke et al, 2004). For growth and complementation assays, fivefold serial dilutions of cells were spotted on indicated agar plates until growth was seen. Schizosaccharomyces pombe expression constructs under a weak thiamine‐repressible promoter nmt81, with sequences encoding 3MYC epitope tag at the N‐terminus of sde2 gene and 3FLAG epitope tag at its C‐terminus, were used. The expression and processing of the protein were analysed by anti‐MYC and anti‐FLAG immunoblotting. The promoter is induced in the absence of thiamine, and 5 μg/ml thiamine was used to repress the promoter. To shuffle‐out ura4 + plasmids from S. pombe, 5‐fluoroorotic acid (FOA; Zymo Research) (1.0 g/l of media) was used in agar plates. Vectors for complementation assays were under the weak thiamine‐repressible promoter nmt81. Plasmids with point mutations, insertions or deletions were created by QuikChange site‐directed mutagenesis (Agilent) using specific primers. Gene fusions and chimeras were made by overlap extension (SOE) PCR method.
Chromosomal mutagenesis of sde2 gene
A plasmid was made which contained a NotI insert comprising of 500‐bp sde2 promoter, sde2 ORF with the coding sequence of 6HA epitope tag, nat‐NT2 cassette for resistance against Nat antibiotic, and 500 bp of sde2 terminator. The plasmid was mutagenized by site‐directed mutagenesis to obtain AAKGG and GGMGG, GGRGG and GGTGG versions of sde2. To obtain MGG‐sde2‐C, the sequence encoding amino acids 2–85 of sde2 UBL was deleted by SOE PCR. The NotI inserts from the plasmids were purified and transformed in Schizosaccharomyces pombe strains followed by selection on YES‐Nat plates. The chromosomal mutations were screened by growth phenotypes, immunoblotting assays using anti‐HA antibody and/or sequencing of genomic PCR fragments covering the mutated regions.
Protein immunoblot and turnover assays
For immunoblot assays, total protein isolated from logarithmically growing 1.0 OD600 cells by TCA precipitation was separated on SDS–PAGE and transferred on PVDF membrane. To induce protein expression from thiamine‐repressible nmt promoters, cells were first grown as primary cultures in the presence of 1.7 μg/ml thiamine. Protein expression was induced by growing the cultures in the absence of thiamine for 18–20 h at 30°C. For protein turnover assays, logarithmically growing cells at OD600 = 0.7–0.8 were incubated for indicated time points with 1.7 μg/ml thiamine to repress the promoter, and 100 μg/ml of cycloheximide to stop protein translation. Total protein isolated from 1.0 OD600 cells was used to monitor turnover of the protein by immunoblotting.
Antibodies and antibody‐coupled beads
The antibodies anti‐haemagglutinin (HA, Clone HA‐7), anti‐HA (polyclonal), anti‐Myc (polyclonal), anti‐Flag M2 (Clone M2), anti‐mouse‐HRP and anti‐rabbit‐HRP (raised in Goat) were obtained from Sigma‐Aldrich. Antibody‐coupled beads were also from same source. Polyclonal antibody against bacterially purified GST‐tagged Sde2‐C was raised in rabbit (MERCK), and coupled to protein A/G beads (Pierce) for immunoprecipitation experiments.
Screening for Sde2‐specific proteases
Schizosaccharomyces pombe deletion strains of selected proteases from a haploid deletion library (Bioneer) were transformed with pREP81x‐3MYC–sde2–3FLAG plasmid. The transformants were grown for 20 h in the absence of thiamine in EMM‐Leu media. 1.0 OD600 cells were harvested from logarithmically growing cultures and processed for immunoblot analysis.
Processing of Schizosaccharomyces pombe Sde2 in Escherichia coli and Saccharomyces cerevisiae
Escherichia coli BL21(DE3) strain was used to monitor Sde2 processing with Ubp5 and Ubp15. For all co‐transformations, pPROEX‐6HIS–Spsde2 plasmid was used with pCDFduet empty, pCDFduet‐6HIS–Spubp5, pCDFduet‐6HIS–Spubp5(C222S), pCDFduet‐6HIS–Spubp15 and pCDFduet‐6HIS–Spubp15(C239S). Protein expression was induced in logarithmically growing cells using 100 μM IPTG for 16 h at 18°C. 1.0 OD600 cells each were harvested and lysed for 10 min with 240 μl B‐PER reagent (Pierce) containing benzonase (Sigma), 1 mM PMSF (Sigma) and protease inhibitor cocktail (Pierce). Sde2 was detected by anti‐Sde2 immunoblotting. Saccharomyces cerevisiae PJ697a strain was used to monitor processing of S. pombe Sde2. p426ADH‐3MYC–Spsde2–3FLAG plasmid was co‐transformed with p424ADH‐Scubp15, p424ADH‐Spubp5, p424ADH‐Spubp5(C222S), p424ADH‐Spubp15, p424ADH‐Spubp15(C239S) plasmids. Co‐transformants were grown in SC‐Ura‐Trp minimal media at 30°C. 1.0 OD600 cells were harvested from logarithmically growing cells, processed by TCA prep and analysed by anti‐FLAG immunoblotting.
Co‐immunoprecipitation (Co‐IP) assays
The in vivo complex of Sde2 was analysed by immunoprecipitation (IP) of Sde2 complexes followed by mass spectrometry or by Co‐IP assay from chromosomally tagged S. pombe strains. The assay was described previously (Mishra et al, 2011). For IP of Sde2 for mass spectrometry, Δsde2 strain was transformed with pREP81x‐3MYC–sde2–3FLAG. 500 OD600 cells from exponentially growing culture were harvested. Primary cultures were grown in synthetic complete media then transferred to secondary culture in EMM‐Leu media and finally to tertiary culture in EMM‐Leu. 50 OD600 cells were used for Co‐IP assays. Total cell lysates were prepared by mechanical grinding with liquid nitrogen in the presence of cell‐lysis buffer containing 50 mM Tris (pH7.5), 150 mM NaCl, 5 mM MgCl2, 5% glycerol, 1% Triton X‐100, 1 mM PMSF (Sigma), phosphatase inhibitors (Roche) and protease inhibitors (Thermo Scientific). Lysates were cleared from cell debris by centrifugation at 10,000 × g for 10 min. Immunoprecipitation with anti‐FLAG M2 affinity gel (1 μl/OD600 cells) was performed for 3 h at 4°C. Bound proteins were eluted at 65°C in HU buffer and analysed by immunoblot. For Co‐IP assays from chromosomally tagged strains using anti‐MYC beads, total cell lysates was prepared from 100 OD600 grown in YEL media. IP was performed for 3 h using 25 μl of anti‐MYC agarose resin (SIGMA) followed by immunoblot assays to detect Co‐IP proteins.
Pull‐down of ubiquitin conjugates under denaturing conditions
Ubiquitin conjugates were purified following protocols used for the identification of SUMO‐protein conjugates (Sacher et al, 2005). Briefly, S. pombe cells were co‐transformed with pREP1‐6HIS–MYC–Spubi4 plasmid with natMX marker (gift from Li‐Lin Du) and peno1‐3MYC–sde2–3FLAG. Total lysates from 50 OD600 cells prepared under denaturing condition were used to pull‐down HIS–ubiquitin conjugates by Ni‐NTA beads (Qiagen). Western blot with anti‐FLAG antibody was performed to detect ubiquitin conjugates of Sde2.
Splicing‐sensitive microarray
Schizosaccharomyces pombe splicing‐sensitive microarray design, experimental procedure and data analysis were reported earlier (Inada & Pleiss, 2010). Total RNA isolated from logarithmically growing cultures of wild‐type and Δsde2 strains was treated at 30°C and 37°C for 15 min. cDNAs from both strains were labelled with Cy3 and Cy5 dyes. Mixtures of Cy3‐labelled wild‐type sample and Cy5‐labelled Δsde2 sample and from dye‐swapped samples were hybridized to the arrays.
RNA isolation and RT–PCR
RNA isolation and cDNA synthesis were done as described previously (Inada & Pleiss, 2010). Briefly, five OD600 cells in log phase were harvested at 30°C (untreated control) or after 15 min of heat shock at 37°C by filtration. Total RNA was isolated by hot acid phenol method using 15‐ml phase lock gel heavy tubes (5 prime), followed by DNase I (Promega) treatment for 15 min at 30°C. Zymo‐Spin II column (Zymo Research) was used for clean‐up of RNA. cDNA synthesis from 3 μg total RNA was done using reverse transcriptase (RT) and random‐hexamer primers (Invitrogen) at 42°C for 16 h. PCR and the products were analysed by agarose gel electrophoresis. Real‐time quantitative PCR (qPCR) was done using SYBR green dye‐based reagents (Roche). Primers used in RT–PCR assays of splicing targets are listed in Appendix Table S3.
Accession codes
Splicing‐sensitive microarray data were deposited with GEO accession number GSE97097.
Author contributions
PT performed experiments on RNA splicing (Figs 1D, 3F, 4A, 5B, 6B, 7, 8B–D, and EV2B–D and EV5A, Appendix Figs S1A–E, S3C, S4B, S5, S6, S7), PAP. Sde2 processing (Figs 1B and C, 2, 3A–C, 4B–D, 6A, 8E, and EV2A, EV4, EV5B and C, Appendix Figs S1B, S2, S3A, S4B), SD N‐end rule (Figs 3D and E, and EV3, Appendix Fig S3B, D and E), KKK (Fig 8A), SKM and JAP splicing microarrays (Fig 5A), all authors designed the experiments, analysed the data and prepared the manuscript, SKM wrote the article with inputs from JAP.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Source Data for Figure 8
Acknowledgements
We thank S. Jentsch for support to the project, A. Frost and J. Weissman for genetic screens with S. pombe hub1 mutants, T. Ammon for help with experiment in Fig EV1D, A. Das, M. Sanjeev and S. Adusumilli for technical help, NBRP/YGRC and L.L. Du for materials and M. Sharma for comments on the manuscript. S.K.M. is supported by the Ministry of Human Resource and Development (MHRD) and Department of Science and Technology (DST), Government of India, and the Max Planck Society, Germany. P.T. and S.D. are recipients of scholarships from DST, P.A.P. from the University Grant Commission (UGC) and K.K.K from ICMR, Government of India. The authors declare no competing financial interests.
The EMBO Journal (2018) 37: 89–101
References
- Alvarez CJ, Romfo CM, Vanhoy RW, Porter GL, Wise JA (1996) Mutational analysis of U1 function in Schizosaccharomyces pombe: pre‐mRNAs differ in the extent and nature of their requirements for this snRNA in vivo . RNA 2: 404–418 [PMC free article] [PubMed] [Google Scholar]
- Ammon T, Mishra SK, Kowalska K, Popowicz GM, Holak TA, Jentsch S (2014) The conserved ubiquitin‐like protein Hub1 plays a critical role in splicing in human cells. J Mol Cell Biol 6: 312–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ast G (2004) How did alternative splicing evolve? Nat Rev Genet 5: 773–782 [DOI] [PubMed] [Google Scholar]
- Bachmair A, Finley D, Varshavsky A (1986) In vivo half‐life of a protein is a function of its amino‐terminal residue. Science 234: 179–186 [DOI] [PubMed] [Google Scholar]
- Baltz AG, Munschauer M, Schwanhäusser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold‐Brown D, Drew K, Milek M, Wyler E, Bonneau R, Selbach M, Dieterich C, Landthaler M (2012) The mRNA‐bound proteome and its global occupancy profile on protein‐coding transcripts. Mol Cell 46: 674–690 [DOI] [PubMed] [Google Scholar]
- Bayne EH, Portoso M, Kagansky A, Kos‐Braun IC, Urano T, Ekwall K, Alves F, Rappsilber J, Allshire RC (2008) Splicing factors facilitate RNAi‐directed silencing in fission yeast. Science 322: 602–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne EH, Bijos DA, White SA, de Lima Alves F, Rappsilber J, Allshire RC (2014) A systematic genetic screen identifies new factors influencing centromeric heterochromatin integrity in fission yeast. Genome Biol 15: 481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessonov S, Anokhina M, Will CL, Urlaub H, Luhrmann R (2008) Isolation of an active step I spliceosome and composition of its RNP core. Nature 452: 846–850 [DOI] [PubMed] [Google Scholar]
- Campion Y, Neel H, Gostan T, Soret J, Bordonné R (2010) Specific splicing defects in S. pombe carrying a degron allele of the survival of motor neuron gene. EMBO J 29: 1817–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Shulha HP, Ashar‐Patel A, Yan J, Green KM, Query CC, Rhind N, Weng Z, Moore MJ (2014) Endogenous U2·U5·U6 snRNA complexes in S. pombe are intron lariat spliceosomes. RNA 20: 308–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty MF, Adelmant G, Cecchetelli AD, Marto JA, Cram EJ (2014) Proteomic analysis reveals CACN‐1 is a component of the spliceosome in Caenorhabditis elegans . G3: Genes—Genomes—Genetics 4: 1555–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faesen AC, Dirac AMG, Shanmugham A, Ovaa H, Perrakis A, Sixma TK (2011) Mechanism of USP7/HAUSP activation by its C‐terminal ubiquitin‐like domain and allosteric regulation by GMP‐synthetase. Mol Cell 44: 147–159 [DOI] [PubMed] [Google Scholar]
- Finley D, Bartel B, Varshavsky A (1989) The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature 338: 394–401 [DOI] [PubMed] [Google Scholar]
- Fujiwara H, Tanaka N, Yamashita I, Kitamura K (2013) Essential role of Ubr11, but not Ubr1, as an N‐end rule ubiquitin ligase in Schizosaccharomyces pombe . Yeast 30: 1–11 [DOI] [PubMed] [Google Scholar]
- Gordon C, McGurk G, Wallace M, Hastie ND (1996) A Conditional lethal mutant in the fission yeast 26 S protease subunit mts3 is defective in metaphase to anaphase transition. J Biol Chem 271: 5704–5711 [DOI] [PubMed] [Google Scholar]
- Graciet E, Hu RG, Piatkov K, Rhee JH, Schwarz EM, Varshavsky A (2006) Aminoacyl‐transferases and the N‐end rule pathway of prokaryotic/eukaryotic specificity in a human pathogen. Proc Natl Acad Sci USA 103: 3078–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M (2009) Origin and function of ubiquitin‐like proteins. Nature 458: 422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks KB, Delneri D, Griffiths‐Jones S (2014) Intron evolution in Saccharomycetaceae . Genome Biol Evol 6: 2543–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CF, Zhu JK (2014) RNA splicing factors and RNA‐directed DNA methylation. Biology (Basel) 3: 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada M, Pleiss JA (2010) Genome‐wide approaches to monitor pre‐mRNA splicing. Methods Enzymol 470: 51–75 [DOI] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno‐Borchart A, Doenges G, Schwob E, Schiebel E, Knop M (2004) A versatile toolbox for PCR‐based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21: 947–962 [DOI] [PubMed] [Google Scholar]
- Jo U, Cai W, Wang J, Kwon Y, D'Andrea AD, Kim H (2016) PCNA‐dependent cleavage and degradation of SDE2 regulates response to replication stress. PLoS Genet 12: e1006465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallgren SP, Andrews S, Tadeo X, Hou H, Moresco JJ, Tu PG, Yates JR, Nagy PL, Jia S (2014) The proper splicing of RNAi factors is critical for pericentric heterochromatin assembly in fission yeast. PLoS Genet 10: e1004334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy PJ, Vashisht AA, Hoe KL, Kim DU, Park HO, Hayles J, Russell P (2008) A genome‐wide screen of genes involved in cadmium tolerance in Schizosaccharomyces pombe . Toxicol Sci 106: 124–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Magen A, Ast G (2006) Different levels of alternative splicing among eukaryotes. Nucleic Acids Res 35: 125–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, Han S, Jeffery L, Baek ST, Lee H, Shim YS, Lee M, Kim L, Heo KS, Noh EJ, Lee AR et al (2010) Analysis of a genome‐wide set of gene deletions in the fission yeast Schizosaccharomyces pombe . Nat Biotechnol 28: 617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Tanaka K (2010) Regulatory mechanisms involved in the control of ubiquitin homeostasis. J Biochem 147: 793–798 [DOI] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E (1999) Epitope tagging of yeast genes using a PCR‐based strategy: more tags and improved practical routines. Yeast 15: 963–972 [DOI] [PubMed] [Google Scholar]
- Konarska MM, Query CC (2005) Insights into the mechanisms of splicing: more lessons from the ribosome. Genes Dev 19: 2255–2260 [DOI] [PubMed] [Google Scholar]
- Kouranti I, McLean JR, Feoktistova A, Liang P, Johnson AE, Roberts‐Galbraith RH, Gould KL (2010) A global census of fission yeast deubiquitinating enzyme localization and interaction networks reveals distinct compartmentalization profiles and overlapping functions in endocytosis and polarity. PLoS Biol 8: e1000471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe T, García‐Gómez JJ, de la Cruz J, Roser D, Hurt E, Linder P, Kressler D (2009) Linear ubiquitin fusion to Rps31 and its subsequent cleavage are required for the efficient production and functional integrity of 40S ribosomal subunits. Mol Microbiol 72: 69–84 [DOI] [PubMed] [Google Scholar]
- Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W (2002) Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416: 648–653 [DOI] [PubMed] [Google Scholar]
- Li M, Brooks CL, Kon N, Gu W (2004) A dynamic role of HAUSP in the p53‐Mdm2 pathway. Mol Cell 13: 879–886 [DOI] [PubMed] [Google Scholar]
- Lorenzi LE, Bah A, Wischnewski H, Shchepachev V, Soneson C, Santagostino M, Azzalin CM (2015) Fission yeast Cactin restricts telomere transcription and elongation by controlling Rap1 levels. EMBO J 34: 115–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra SK, Ammon T, Popowicz GM, Krajewski M, Nagel RJ, Ares M, Holak TA, Jentsch S (2011) Role of the ubiquitin‐like protein Hub1 in splice‐site usage and alternative splicing. Nature 474: 173–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richert K, Schmidt H, Gross T, Käufer N (2002) The deubiquitinating enzyme Ubp21p of fission yeast stabilizes a mutant form of protein kinase Prp4p. Mol Genet Genomics 267: 88–95 [DOI] [PubMed] [Google Scholar]
- Sacher M, Pfander B, Jentsch S (2005) Identification of SUMO–protein conjugates in ubiquitin and protein degradation. Methods Enzymol 399: 392–404 [DOI] [PubMed] [Google Scholar]
- Sriram SM, Kim BY, Kwon YT (2011) The N‐end rule pathway: emerging functions and molecular principles of substrate recognition. Nat Rev Mol Cell Biol 12: 735–747 [DOI] [PubMed] [Google Scholar]
- Staley JP, Woolford JL (2009) Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines. Curr Opin Cell Biol 21: 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugioka‐Sugiyama R, Sugiyama T (2011) Sde2: a novel nuclear protein essential for telomeric silencing and genomic stability in Schizosaccharomyces pombe . Biochem Biophys Res Commun 406: 444–448 [DOI] [PubMed] [Google Scholar]
- Turcu FER, Ventii KH, Wilkinson KD (2009) Regulation and cellular roles of ubiquitin‐specific deubiquitinating enzymes. Annu Rev Biochem 78: 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen AG, Ploegh HL (2012) Ubiquitin‐like proteins. Annu Rev Biochem 81: 323–357 [DOI] [PubMed] [Google Scholar]
- Vijaykrishna N, Melangath G, Kumar R, Khandelia P, Bawa P, Varadarajan R, Vijayraghavan U (2016) The fission yeast pre‐mRNA processing factor 18 (prp18+) has intron‐specific splicing functions with links to G1‐S cell cycle progression. J Biol Chem 291: 27387–27402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R (2009) The Spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wang J, Tadeo X, Hou H, Andrews S, Moresco JJ, Yates JR, Nagy PL, Jia S (2014) Tls1 regulates splicing of shelterin components to control telomeric heterochromatin assembly and telomere length. Nucleic Acids Res 42: 11419–11432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CJ, Lakhe‐Reddy S, Romfo CM, Wise JA (2005) Analysis of mutant phenotypes and splicing defects demonstrates functional collaboration between the large and small subunits of the essential splicing factor U2AF in vivo . Mol Biol Cell 16: 584–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchman RL, Gordon C, Mayer RJ (2005) Ubiquitin and ubiquitin‐like proteins as multifunctional signals. Nat Rev Mol Cell Biol 6: 599–609 [DOI] [PubMed] [Google Scholar]
- Wilkinson CRM, Dittmar GAG, Ohi MD, Uetz P, Jones N, Finley D (2004) Ubiquitin‐like protein Hub1 is required for pre‐mRNA splicing and localization of an essential splicing factor in fission yeast. Curr Biol 14: 2283–2288 [DOI] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y (2015) The i‐TASSER suite: protein structure and function prediction. Nat Methods 12: 7–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiroda H, Tanaka K (2004) Hub1 is an essential ubiquitin‐like protein without functioning as a typical modifier in fission yeast. Genes Cells 9: 1189–1197 [DOI] [PubMed] [Google Scholar]
- Zanini IMY, Soneson C, Lorenzi LE, Azzalin CM (2017) Human Cactin interacts with DHX8 and SRRM2 to assure efficient pre‐mRNA splicing and sister chromatid cohesion. J Cell Sci 130: 767–778 [DOI] [PubMed] [Google Scholar]
- Zhang L, Ma N, Liu Q, Ma Y (2013) Genome‐wide screening for genes associated with valproic acid sensitivity in fission yeast. PLoS One 8: e68738 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Source Data for Figure 8


